Abstract
The utility of adenovirus (Ad) vectors for gene therapy is restricted by their inability to selectively transduce disease-affected tissues. This limitation may be overcome by the derivation of vectors capable of interacting with receptors specifically expressed in the target tissue. Previous attempts to alter Ad tropism by genetic modification of the Ad fiber have had limited success due to structural conflicts between the fiber and the targeting ligand. Here we present a strategy to derive an Ad vector with enhanced targeting potential by a radical replacement of the fiber protein in the Ad capsid with a chimeric molecule containing a heterologous trimerization motif and a receptor-binding ligand. Our approach, which capitalized upon the overall structural similarity between the human Ad type 5 (Ad5) fiber and bacteriophage T4 fibritin proteins, has resulted in the generation of a genetically modified Ad5 incorporating chimeric fiber-fibritin proteins targeted to artificial receptor molecules. Gene transfer studies employing this novel viral vector have demonstrated its capacity to efficiently deliver a transgene payload to the target cells in a receptor-specific manner.
Human adenoviruses (Ad) of serotypes 2 and 5 (Ad2 and Ad5) have been extensively used for a variety of gene therapy applications. This is largely due to the ability of these vectors to efficiently deliver therapeutic genes to a wide range of different cell types. However, the promiscuous tropism of Ad resulting from the widespread distribution of its primary cellular receptor, the coxsackievirus and Ad receptor (CAR) (1, 28), limits the utility of Ad vectors in those clinical contexts where selective delivery of a therapeutic transgene to diseased tissue is required. Uncontrolled transduction of normal tissues with Ad vectors expressing potentially toxic gene products may lead to a series of side effects, thereby undermining the efficacy of the therapy. Furthermore, cellular targets expressing CAR below certain threshold levels are not susceptible to Ad-based therapies due to their inability to support Ad infection. Therefore, the dependence of the efficiency of the Ad-mediated cell transduction on the levels of CAR expression by the target cell presents a serious challenge for the further development of Ad-based gene therapeutics.
In order to overcome this limitation, the concept of genetic targeting of Ad vectors to specific cell surface receptors has been proposed. Strategies to retarget Ad vectors are based on the currently accepted model of Ad infection (for a review, see reference 16), which postulates that the initial binding of the Ad virion to the cell is mediated by the attachment of the globular knob domain of the Ad fiber protein to CAR. This is then followed by an internalization step triggered by the interaction of the RGD-containing loop of a second Ad capsid protein, the penton base, with cellular integrins. Although recent studies have shown that representatives of different Ad serotypes may utilize cell receptors other than CAR, the two-step mechanism of cell entry established for Ad2 and Ad5 appears to be common to the majority of human Ad serotypes. As the fiber protein is the key mediator of the cell attachment pathway employed by Ad, genetic incorporation of targeting ligands within this viral protein was originally proposed as the strategy to derive targeted, cell type-specific Ad vectors (21).
Each of the rodlike fiber proteins localized at the vertices of icosahedral Ad5 capsid is a homotrimer which consists of three identical copies of a 62-kDa polypeptide (for a review see reference 4). This trimeric molecule has a domain organization, with each domain playing important roles in the structure and function of the virion. Whereas the amino-terminal tail domain is responsible for association of the fiber with the penton base, the carboxy-terminal knob domain, which has a propellerlike structure formed by two β-sheets (34), performs two distinct functions, both vital for virus assembly and propagation. In addition to forming the CAR-binding site, which is formed by residues from the AB loop, β-strand B, and the DE loop (for details, see reference 25), the knob initiates and maintains the trimerization of the entire fiber molecule. This trimerization is critical for the successful formation of the virion, as monomeric fibers cannot associate with the penton base (24). The knob and the tail of the fiber are connected by the β-spiral shaft domain (30), which extends the knob away from the surface of the virion, thereby facilitating interaction with CAR.
Early attempts to generate Ad vectors possessing expanded tropism involved incorporation of short peptide ligands into either the carboxy terminus (32, 33) or the so-called HI loop (7) of the knob of the Ad fiber protein. Although these studies demonstrated the feasibility of genetic targeting of Ad and showed the potential utility of such vectors in the context of several disease models (14, 29), further progress in this direction has been hampered by the structural conflicts often observed as a result of modification of the fiber structure (33). Due to the rather complex structure of the fiber knob domain, even minor modifications to this portion of the molecule may destabilize the fiber, thereby rendering it incapable of trimerization and hence nonfunctional. According to the published data, the upper size limit for a targeting ligand to be incorporated into Ad5 fiber is about 30 amino acid residues (13, 33), which dramatically narrows the repertoire of targeting moieties, thereby restricting the choice of potential receptors and, therefore, cell targets. As a result of this limitation, only a handful of heterologous peptide ligands [oligolysine, FLAG, RGD-4C, RGS(His)6, and HA epitope] have been successfully used in the context of Ad5 fiber modification. The task of Ad targeting is further complicated by the need to ablate the native receptor-binding sites within the fiber of an Ad vector to make it truly targeted.
This study presents an alternative approach to Ad targeting based on replacement of the native fiber in an Ad capsid with a chimeric protein, which results in permanent ablation of native Ad receptor tropism and simultaneously offers flexibility in the generation of novel vector tropism.
MATERIALS AND METHODS
Cell lines.
The 293 human kidney cell line transformed with Ad5 DNA was purchased from Microbix (Toronto, Ontario, Canada). The 211B cell line, a derivative of 293 which constitutively expresses the Ad5 fiber protein (31), was obtained from Dan Von Seggern (The Scripps Research Institute, La Jolla, Calif.). The 293/6H cell line was generated by transfection of 293 cells with the plasmid vector to express an artificial receptor capable of binding proteins containing carboxy-terminal six-histidine tags (8). All cell lines were grown at 37°C in Dulbecco's minimal essential medium (DMEM)-F12 medium supplemented with 10% fetal bovine serum in a humidified atmosphere of 5% CO2.
Genetic engineering.
All recombinant DNA molecules generated in this study were designed by standard methods of genetic engineering (described in reference 26). Restriction endonucleases, T4 DNA ligase, and Klenow enzyme were from New England Biolabs (Beverly, Mass.). PCR was performed with Pfu DNA polymerase purchased from Stratagene (La Jolla, Calif.). In order to PCR amplify segments of the fibritin gene, bacteriophage T4 DNA obtained from Sigma (St. Louis, Mo.) was used as a template. Recombinant Ad genomes were derived by homologous DNA recombination in Escherichia coli BJ5183 essentially as described previously (3). Details of all DNA manipulation experiments are available upon request.
Bacterial expression of recombinant fibritin proteins.
Recombinant forms of the fibritin-derived proteins bearing six-His tags were expressed in E. coli M15(pREP4) cells by using vectors of the pQE series (Qiagen, Valencia, Calif.). Induction of protein expression and subsequent purification by immobilized metal ion affinity chromatography of recombinant products on Ni-nitrilotriacetic acid (NTA)-agarose were done according to the manufacturer's recommendations.
Ad vectors.
Firefly luciferase-expressing Ad utilized in this study, Ad5Luc1 and Ad5LucFF/6H, are first-generation vectors with E1 deleted based on human Ad serotype 5. The viruses were rescued by transfection of either 293 (12) or 211B (31) cells with recombinant Ad genomes generated in E. coli. The particle titer of CsCl-purified viruses amplified in 293 or 211B cells was determined as described by Mittereder et al. (23), whereas the infectious titer of the vectors was obtained in a spot assay as developed by Bewig and Schmidt (2).
ELISA.
Two-hundred-nanogram aliquots of soluble CAR protein (sCAR) (6) were adsorbed to the wells of an enzyme-linked immunosorbent assay (ELISA) plate, unbound sCAR was aspirated, and the wells were blocked with blocking buffer (0.05% Tween 20, 2% bovine serum albumin in phosphate-buffered saline [PBS]). Purified viruses diluted in blocking buffer to concentrations ranging from 0.12 to 10 μg/ml were added to the wells in 100-μl aliquots and allowed to bind with the receptor for 1 h. The plate was washed with washing buffer (0.05% Tween 20, 0.5% bovine serum albumin in PBS), and the bound virus was probed with the rabbit anti-Ad2 serum (American Type Culture Collection, Manassas, Va.). Following incubation for another hour, the wells were washed, incubated with goat anti-rabbit immunoglobulin G conjugated to horseradish peroxidase (DAKO, Carpinteria, Calif.), washed again, and developed with o-phenylenediamine (Sigma) as recommended by the manufacturer. The absorbance of samples at 490 nm was then determined with a microtiter plate reader.
Gene transfer experiments.
Cells grown in the wells of 24-well plates to 90 to 100% confluence were washed with DMEM-F12–2% fetal calf serum and then infected for 30 min with an Ad vector diluted in 0.4 ml of the same medium. The multiplicities of infection (MOIs) were 40, 400, and 4,000 virus particles per cell. The infected monolayers were then incubated at 37°C in an atmosphere of 5% CO2. Twenty hours postinfection, the virus-containing medium was aspirated and the cells were washed with PBS and lysed in 0.25 ml of Luciferase Reporter Lysis buffer (Promega, Madison, Wis.). The luciferase activities in the cell lysates were then measured according to the manufacturer's protocol. Each data point was set in triplicate and calculated as the mean of three determinations.
Inhibition of Ad5LucFF/6H-mediated gene transfer to 293/6H cells.
The inhibition experiment was done essentially as described above for gene transfer to 293 and 293/6H cells. Prior to infection with Ad5LucFF/6H (MOI = 40 virions per cell), the cells were incubated for 10 min at room temperature with 0.2 ml of serial 10-fold dilutions of either recombinant fibritin or Ad5 fiber knob (17) in PBS. Then, a 0.2-ml aliquot of vector was added to each well, and the incubation was continued for another 30 min. The medium containing the virus and the inhibiting protein was removed, and the cell monolayers were washed with DMEM-F12–2% fetal calf serum and overlaid with fresh medium. The levels of luciferase activity detected in the cell lysates 20 h postinfection were normalized to those registered in mock-infected controls.
RESULTS
Design, expression, and characterization of a recombinant fiber-fibritin-ligand chimera.
This work was driven by the hypothesis that genetic targeting of Ad could best be achieved by “splitting” the functions normally performed by the knob domain of the Ad5 fiber between two different protein moieties which would substitute for the knob. Specifically, we chose to replace the knob of the fiber with a heterologous trimerization motif to maintain trimerization of the knobless fiber and to simultaneously introduce a ligand capable of targeting the virion to a novel receptor. Therefore, in marked contrast to the previous attempts to fit a desired ligand into the highly complex framework of the fiber knob domain, we employed a radical replacement of the fiber with a protein chimera, rationally designed to carry out the fiber's functions.
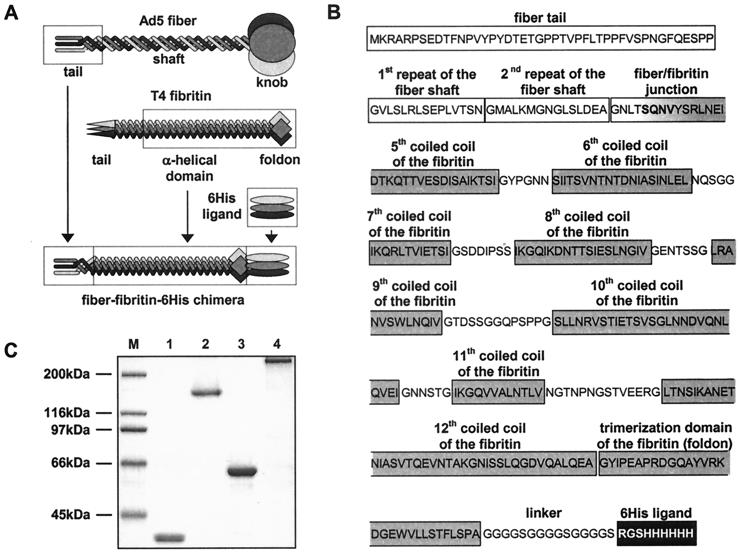
The fiber-replacing molecule engineered in this study incorporated the tail and two amino-terminal repeats of the shaft domain of the Ad5 fiber protein genetically fused with a truncated form of the bacteriophage T4 fibritin protein, which was employed as the heterologous trimerizing moiety in order to compensate for the knob deletion (Fig. 1A and B). The choice of the T4 fibritin as the component of the fiber chimera was dictated by a number of its structural features. The fibritin protein is a product of the wac gene, which forms the “collar” and the “whiskers” of the T4 capsid, where it mediates assembly of the long tail fibers and their subsequent attachment to the tail baseplate. Trimerization of this rodlike, 486-amino-acid-long protein is initiated and maintained by the short (30-amino-acid-long) carboxy-terminal domain, or “foldon,” which is stabilized by a number of hydrophobic interactions and hydrogen bonds (27). The central α-helical domain of fibritin, which consists of 12 segments of parallel triple coiled coils separated by flexible loop structures, passively follows the trimerization initiated at the carboxy terminus of the molecule. The trimeric structure of fibritin is extremely stable and is not compromised by either extensive amino-terminal deletions (up to 92% of the molecule) (19) or carboxy-terminal insertions up to at least 163 amino acids long (V. V. Mesyanzhinov, personal communication). For the purposes of this study, it is important to mention that no receptor-binding function has been shown for fibritin.
FIG. 1.
Generation of Ad5 fiber-T4 fibritin chimera containing targeting ligand. (A) Schema showing key components of the fiber-fibritin-ligand chimera and their sources. The tail of the fiber anchors the fiber–fibritin–six-His chimera in the Ad virion; a fragment of the fibritin protein provides trimerization of the molecule, while the six-His ligand mediates binding to an AR. (B) Amino acid sequence and domain structure of the FF/6H chimera. FF/6H protein is a 373-amino-acid-long molecule which consists of the amino-terminal segment of Ad5 fiber sequence genetically fused with the carboxy-terminal portion of the T4 fibritin protein, followed by the linker and the six-His-containing ligand. The beginning of the third pseudorepeat of the fiber shaft domain (GNTLSQNV) is joined to the fibritin sequence starting with the fragment of the insertion loop (SQN) preceding the fifth coiled-coil segment of the α-helical central domain of the fibritin (VYSRLNEIDTKQTTVESDISAIKTSI). The sequence SQNV present in the native structures of both fusion partners was chosen as the hinge between the two molecules in order to minimize potential structural conflicts between the β-spiral configuration of the fiber shaft and the triple α-helix of the central domain of the fibritin. The segments of the fibritin sequence localized between every two adjacent coiled coils are the insertion loops which provide some degree of flexibility needed for optimal ligand presentation. A peptide linker is incorporated between the carboxy-terminal trimerization domain (foldon) of the fibritin and the six-histidine-containing ligand to extend the ligand away from the carrier protein in order to facilitate binding to the target receptor. The domain organization of the fiber and the fibritin proteins is as published previously by Van Raaij et al. (30) and Tao et al. (27), respectively. (C) SDS-PAGE analysis of E. coli-expressed, immobilized metal ion chromatography-purified FF/6H chimeric protein. Lane M, molecular mass protein ladder; lanes 1 and 2, FF/6H protein; lanes 3 and 4, wild-type Ad5 fiber. The samples in lanes 1 and 3 were denatured by being boiled, which resulted in degradation of trimeric proteins to monomers, while lanes 2 and 4 contain proteins in their native trimeric configuration.
In order to provide a receptor-binding ligand, we chose to incorporate into the design of this fiber-fibritin chimera a carboxy-terminal six-His sequence connected to the fibritin protein via a short peptide linker. The purpose of this maneuver was to demonstrate the feasibility of targeting fibritin-containing Ad vectors to alternative cell surface receptors by directing the modified vector to an artificial receptor (AR), which is expressed on the surface of 293/6H cells previously derived in our laboratory. The extracellular domain of this AR is an anti-five-His single-chain antibody, which is genetically fused with the transmembrane domain of the platelet-derived growth factor receptor (8). In addition to receptor binding, this six-His sequence was employed to facilitate the detection and purification of the fiber–fibritin–six-His chimera, FF/6H, and Ad virions incorporating this protein.
For the purposes of preliminary characterization, the FF/6H chimeric protein was initially expressed in E. coli and purified on an Ni-NTA-agarose column. Subsequent sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis of the purified chimeric protein proved that it is trimeric and that the FF/6H trimers are as stable in an SDS-containing gel as the trimers of the wild-type Ad5 fiber (Fig. 1C). Efficient binding of the FF/6H protein to a Ni-NTA-containing matrix proved that the six-His ligand was available for binding in the context of this trimeric molecule. According to this analysis, truncated T4 fibritin incorporated into the FF/6H protein was able to direct trimerization of the chimera and also successfully served the purposes of ligand presentation, thereby satisfying the two key functional criteria of an ideal fiber-replacing molecule.
Fiber-fibritin-ligand chimera efficiently incorporates into mature Ad5 virions.
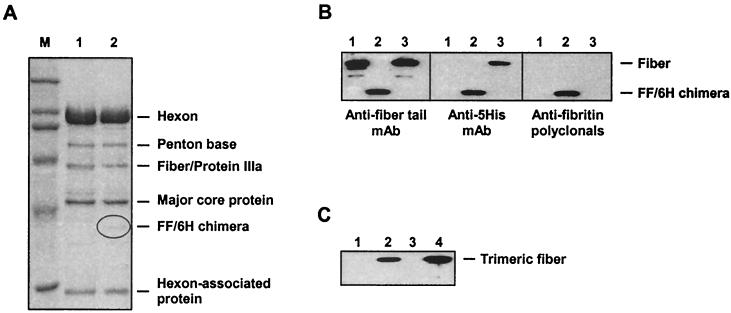
In order to evaluate the functional utility of the FF/6H chimeras incorporated into a mature Ad particle, we employed homologous DNA recombination in E. coli (15) to insert the FF/6H-encoding gene into the genome of the firefly luciferase-expressing Ad5 with E1 deleted, in place of the wild-type fiber gene. The virus of interest, Ad5LucFF/6H, was then rescued by transfection of 211B cells with the resultant Ad genome. 211B cells, a derivative of 293 cells which constitutively express the wild-type Ad5 fiber protein (31), were chosen for this transfection experiment in order to guarantee the success of the viral rescue. Ad5LucFF/6H was further expanded on 211B cells and purified by double banding in a CsCl gradient. At this point, the virus stock contained mosaic virions bearing a mixture of the wild-type fibers and FF/6H chimeras (data not shown). In order to obtain a homogenous population of Ad5LucFF/6H virions lacking the wild-type fibers, but exclusively incorporating FF/6H proteins, the original virus stock was then used to infect 293/6H cells at an MOI of 1,000 virus particles per cell. CsCl gradient purification of Ad5LucFF/6H virions isolated from the lysates of infected 293/6H cells 72 h postinfection (at which point a complete cytopathic effect was observed) resulted in a yield of 3 × 104 virus particles per cell, which is well within the range of yields characteristic for Ad5 vectors with E1 deleted.
Our next goal was to demonstrate that the FF/6H chimeras had been incorporated into the Ad5LucFF/6H capsids. Since fiberless Ad5 virions have been successfully purified on CsCl gradients by others (18, 31), it was possible that the putative Ad5LucFF/6H virions isolated in our study lacked FF/6H proteins. This was ruled out by SDS-PAGE of purified Ad5LucFF/6H virions and a Western blot analysis utilizing antisera specific to all three major components of the FF/6H chimera, the fiber tail, the fibritin, and the six-His ligand (Fig. 2A and B). These assays showed that the capsid of Ad5LucFF/6H virions consists of completely matured Ad proteins and incorporates full-size FF/6H chimeras. As expected, no wild-type fibers were found in this preparation of Ad5LucFF/6H. The lack of wild-type Ad5 fibers in this stock of the vector was further confirmed by a Western blot of Ad5LucFF/6H using the anti-Ad5 fiber knob-specific monoclonal antibody 1D6.14 (9) (Fig. 2C).
FIG. 2.
Analysis of Ad5LucFF/6H capsid composition. (A) SDS-PAGE of CsCl-purified Ad5LucFF/6H virions. Samples containing 4 × 1010 particles of either the wild-type Ad5 (lane 1) or Ad5LucFF/6H (lane 2) were boiled in Laemmli sample buffer and resolved on an SDS–10% PAGE gel. Of note, the resolution of this minigel is not sufficient for separation of the fiber and protein IIIa, which comigrate in lane 1. (B) Western blot analysis of FF/6H chimeras incorporated into Ad5LucFF/6H virions. Proteins of Ad5LucFF/6H virions denatured by being boiled in sample buffer (lanes 2) were separated on an SDS–10% PAGE gel and then probed with the anti-Ad fiber tail MAb 4D2, the anti-five-His MAb Penta-His, and anti-fibritin mouse polyclonal antibodies. Wild-type Ad5 (lanes 1) and Ad5LucFc6H, a virus containing wild-type fibers with carboxy-terminal six-His tags (lanes 3), were used as controls. (C) Western blot of Ad5Luc1 and Ad5LucFF/6H virions with anti-fiber knob antibody. Virions denatured by incubation in a sample buffer containing SDS were resolved on an SDS–10% PAGE gel and incubated with the anti-Ad5 fiber knob MAb 1D6.14 (9), which recognize the knob trimer. Lanes 1 and 3, Ad5LucFF/6H (3 × 109 and 6 × 109 virus particles per lane, respectively); lanes 2 and 4, Ad5Luc1 (same amounts of the virus as in lanes 1 and 3). Detection of viral proteins was done with the ECL Plus kit from Amersham Pharmacia Biotech (Piscataway, N.J.).
These findings were further corroborated in an experiment involving binding of purified Ad5LucFF/6H virions to Ni-NTA-resin; in contrast to the Ad vector containing wild-type fibers, which did not bind to the matrix, Ad5LucFF/6H clearly demonstrated six-His-mediated binding to the resin (Fig. 3). Therefore, in addition to its ability to assume a trimeric configuration and bind to a receptor-mimicking molecule, the FF/6H chimera also retained the capacity to be incorporated into mature Ad capsids.
FIG. 3.
Binding of Ad5LucFF/6H virions to Ni-NTA-agarose. Wild-type Ad5 or Ad5LucFF/6H were incubated with an aliquot of Ni-NTA-resin for 1 h. The matrix was pelleted by centrifugation, and the supernatant was removed and then incubated with a second aliquot of Ni-NTA-agarose. Aliquots of material subsequently eluted from the resin, as well as an aliquot of the material present in the supernatant after two sequential incubations with the resin, were separated on an SDS–10% PAGE gel (wild-type Ad5 in lanes 1 through 4 and Ad5LucFF/6H in lanes 5 through 8) and then stained (A) or probed with either the anti-fiber tail MAb 4D2 (B) or the anti-five-His MAb Penta-His (C). Lane M, molecular mass marker; lanes 1 and 5, aliquot of the virus prior to incubation with Ni-NTA-agarose; lanes 2 and 6, material bound to the first aliquot of the resin; lanes 3 and 7, material bound to the second aliquot of the resin; lanes 4 and 8, material remaining in the supernatant after two sequential bindings to the resin. Incomplete binding of Ad5LucFF/6H virions to Ni-NTA-agarose is most likely due to the small size of the pores in the Sepharose CL-6B used as the matrix for manufacturing Ni-NTA-agarose. According to the manufacturer's specifications, the size of those pores does not allow protein molecules with a molecular mass larger that 4 MDa to enter the pores. Thus, the Ni-NTA groups which are localized on the surfaces of the Sepharose particles (a relatively small percentage) are accessible to the six-His-tagged virions, whereas those hidden inside the pores (the majority) are not.
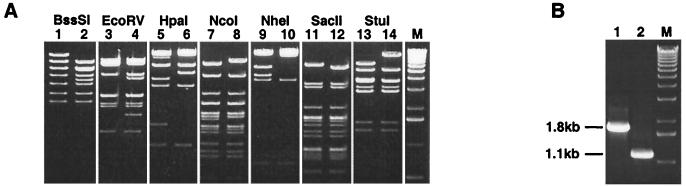
Restriction enzyme analysis of the Ad5LucFF/6H genome, diagnostic PCR utilizing a pair of primers flanking the fiber gene in the Ad5 genome (Fig. 4), and partial sequencing of Ad5LucFF/6H DNA all demonstrated that the viral genome was stable and that the only fiber-encoding gene present was the FF/6H gene. This set of experiments completed the molecular characterization of Ad5LucFF/6H by confirming both the identity and the integrity of the virus capsid and its genome.
FIG. 4.
Analysis of Ad5LucFF/6H genome structure. (A) DNA isolated from purified Ad5LucFF/6H virions was subjected to restriction enzyme analysis using a number of restriction endonucleases which cleave either the wild-type fiber or the FF/6H gene sequence. Odd-numbered lanes, control Ad5Luc1 DNA; even-numbered lanes, Ad5LucFF/6H DNA. (B) Diagnostic PCR utilizing a pair of primers flanking the fiber gene in the Ad5 genome was employed to show the absence of the wild-type fiber gene sequence in the Ad5LucFF/6H genome. The primers used in this test were designed to amplify both the wild-type fiber gene (1.8 kb) and the FF/6H gene (1.1 kb). Lane 1, PCR product amplified from wild-type Ad5 DNA; lane 2, PCR product amplified from Ad5LucFF/6H DNA; lane M, 1-kb ladder.
Ad5LucFF/6H vector containing FF/6H protein chimeras lacks tropism to CAR and is capable of efficient and target receptor-specific gene delivery.
As Ad5LucFF/6H was designed for CAR-independent delivery of transgenes to AR-expressing cells, we wished to prove that this vector is not capable of binding to CAR and that its cell attachment occurs solely via interaction with AR.
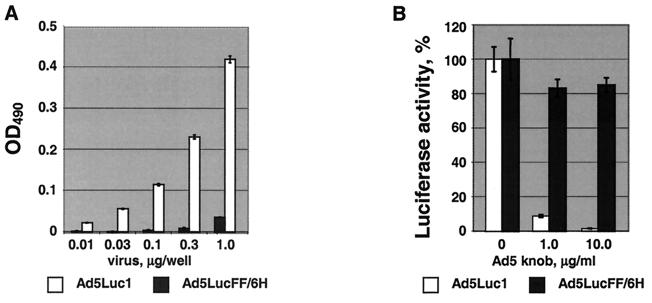
This vector's lack of CAR-binding capacity was first confirmed by an ELISA utilizing a soluble form of the human CAR protein, sCAR (6), and purified Ad virions. As expected, this assay showed that in contrast to the Ad vector incorporating wild-type Ad5 fibers (Ad5Luc1, which binds to sCAR in an efficient, dose-dependent manner), Ad5LucFF/6H clearly fails to bind CAR (Fig. 5A). This lack of Ad5LucFF/6H tropism to CAR was further confirmed in a gene transfer experiment employing E. coli-expressed Ad5 fiber knob protein (17) as a CAR-blocking competitor. By binding to CAR present on 293/6H cells, the knob protein blocked 91 and 98% of Ad5Luc1-mediated gene delivery at concentrations of 1 and 10 μg/ml, respectively, whereas no significant inhibition was seen for the Ad5LucFF/6H vector (Fig. 5B). In the aggregate, these data proved the inability of Ad5LucFF/6H to bind to cognate Ad5 receptor and, therefore, to use the native mechanism of cell attachment.
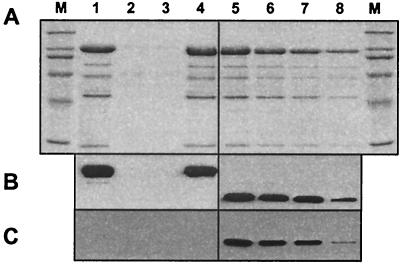
FIG. 5.
Analyses of Ad5LucFF/6H binding to CAR. (A) ELISA employing recombinant human sCAR protein. Baculovirus-expressed sCAR protein adsorbed on an ELISA plate was incubated with various amounts of purified Ad virions. Virions bound to sCAR were then detected with anti-Ad2 polyclonal antibody. Each point represents a mean of three readings obtained in one experiment, with the error bars showing standard deviations. The background, which was equal to 0.06, has been subtracted from all data points. (B) Ad5 fiber knob-mediated inhibition of 293/6H cell transduction. 293/6H cells expressing AR and grown in the wells of a 24-well plate were preincubated with PBS or E. coli-expressed recombinant Ad5 fiber knob protein at concentrations of 1 and 10 μg/ml prior to infection with the Ad vectors. Luciferase activities detected in the lysates of cells infected in the presence of the knob are shown as percentages of the activity registered in cells in which infection was blocked with the inhibitor. Each data point corresponds to an average of three measurements less the background. The error bars show the standard deviations.
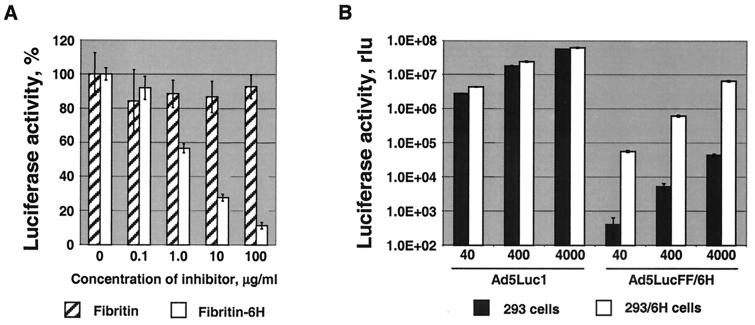
We next sought to test whether the six-His tags of the fiber-fibritin proteins incorporated into Ad5LucFF/6H virions are capable of functioning as receptor-binding ligands and mediating binding to AR expressed by 293/6H cells (8). This was addressed by another gene transfer experiment employing two different forms of recombinant fibritin proteins as blocking agents, only one of which, fibritin-6H, contained a carboxy-terminal six-His tag (Fig. 6A). The purpose of using fibritin, which lacks the carboxy-terminal tag, was to provide additional evidence that the backbone of the fibritin molecule does not contribute to binding to the AR or any other cell surface receptor. Dose-dependent inhibition of Ad5LucFF/6H infection of 293/6H cells with fibritin-6H, but not with the fibritin lacking the six-His tag, proved that this ligand is the component of the virion solely responsible for binding of the virus to target cells.
FIG. 6.
Evaluation of the efficiency and receptor specificity of Ad5LucFF/6H-mediated gene transfer. (A) Specificity of Ad5LucFF/6H binding to the AR. 293/6H cells grown in monolayer culture were preincubated with various concentrations of either the truncated form of fibritin or fibritin carrying a carboxy-terminal six-His tag, fibritin-6H, prior to infection with Ad5LucFF/6H. Luciferase activities detected in the lysates of infected cels 20 h postinfection are given as percentages of the activity in the absence of blocking protein. (B) Gene transfer to 293 and 293/6H cells. Cells seeded in 24-well plates were infected with various doses of Ad5Luc1 or Ad5LucFF/6H. The MOI was equal to 40, 400, or 4,000 virus particles per cell. Twenty hours postinfection, the cells were collected and lysed, and the luciferase activities of the lysates were measured in relative light units (rlu). On both graphs, each data point was set in triplicate and calculated as the mean of three determinations. The error bars show standard deviations.
We then wished to compare the efficiency of the cell entry pathway used by Ad5LucFF/6H with the natural, CAR-mediated mechanism of cell attachment utilized by unmodified Ad vectors. Based on the significant difference in the dissociation constants (kd) previously determined for the Ad5 fiber-CAR interaction, 4 × 10−9 M (5), and for the five-His–anti-five-His monoclonal antibody (MAb) 3D5 interaction, 4.75 × 10−7 M (20), we anticipated lower efficiency of the AR-mediated cell entry pathway compared to the native route of Ad infection. With these considerations in mind, in the next gene transfer experiment we used both vectors at MOIs ranging from 40 to 4,000 virus particles per cell in order to compare the results obtained for the two vectors at various virus doses. The doses of both viruses were normalized by the particle titers of the virus preparations. Although this experiment showed that Ad5LucFF/6H was capable of efficient transgene delivery to the target cells, it also revealed that at equal MOIs the levels of transgene expression in Ad5Luc1-infected 293/6H cells were 9.5- to 77-fold higher than those registered in 293/6H cells infected with Ad5LucFF/6H. Importantly, there was an increase of 2 orders of magnitude in Ad5LucFF/6H-expressed luciferase activities detected in 293/6H cells expressing AR compared to that in parental 293 cells infected with the same vector. Transduction of 293 cells revealed a 1,300- to 6,900-fold difference in luciferase expression in the Ad5Luc1- and Ad5LucFF/6H-infected cells. The poor transduction of 293 cells by Ad5LucFF/6H further proved the inability of this vector with the fiber deleted to use CAR, which is abundantly expressed by these cells (7), for cell attachment.
In order to find out whether the replication of Ad5Luc1 and Ad5LucFF/6H in 293/6H cells had any confounding effect on the relative efficiencies of their gene transfer capabilities, we compared the infectivities of these vectors on 293/6H cells by using the spot assay developed by Bewig and Schmidt (2). This technique excludes any potential effects of viral replication on the ratio of the vectors' transduction capacities, thereby providing a direct measure of Ad infectivity. According to this assay, the differential between the capacities of these vectors to infect 293/6H cells was equal to 51, which is similar to that observed earlier in our gene transfer experiments.
Taken together, these data strongly suggest that as a result of the replacement of the fiber in the Ad5LucFF/6H capsid with the FF/6H chimera, this vector possesses the capacity to achieve cell infection in a CAR-independent, receptor-specific manner via interaction of the virus with the AR.
DISCUSSION
In this study, we have developed a novel approach to the modification of Ad vector tropism by replacing the receptor-binding fiber protein in the Ad capsid with an artificial protein chimera. The rational design of this chimera, based on the general structural similarity of the Ad5 fiber and bacteriophage T4 fibritin, has resulted in the derivation of a novel ligand-presenting molecule. The most important difference between this protein and the wild-type fiber is the disengagement of the trimerization and the receptor-binding functions normally performed by the fiber knob domain. As a result of this distribution of functions, the receptor specificity of the reengineered Ad5 vector may now be defined by a domain of the chimera which plays no role in the trimerization of the molecule and may therefore be manipulated without the risk of destabilizing the ligand-presenting protein and the virion. Previous successful reports of the use of T4 fibritin for ligand display (10, 22) suggest that a wide variety of heterologous targeting ligands, including large polypeptide molecules, could be employed in the context of the fiber-fibritin chimera described here.
Fibritin chimeras analogous to the one described in this work may be viewed as versatile ligand-displaying molecules suitable for the genetic modification of virtually any human or animal Ad vector. The problem of the elimination of undesirable natural tropism of native fibers contained in the Ad virion may thus be solved by replacement of native fibers with such fibritin chimeras. This approach has a significant advantage over maneuvers involving the identification and subsequent mutagenesis of the native receptor-binding sites within the fibers of numerous Ad species, some of which are able to bind to different types of primary receptors. In addition, the strategy of fiber replacement eliminates the risk of reversion of the mutated fiber gene to the wild type during multiple rounds of propagation, which, if it happened, would compromise the efficiency of any vector targeting schema.
An additional advantage offered by Ad vectors incorporating the fibritin-based chimeras for the purposes of human gene therapy becomes apparent in light of recently published data on interference of anti-fiber antibodies present in the sera of some gene therapy patients with the Ad vectors used in clinical protocols (11). Importantly, these antibodies have been shown to have a synergistic effect on Ad vector neutralization when present together with anti-penton base antibodies. Thus, deletion of most of the fiber sequence in the fibritin-bearing Ad vectors would make them refractory to this type of immune response and therefore more efficient as therapeutic agents.
ACKNOWLEDGMENTS
We thank V. Mesyanzhinov for providing anti-fibritin antibody and for sharing with us unpublished data on carboxy-terminal modifications of the fibritin protein. D. Von Seggern is thanked for generously providing 211B cells. We are grateful to I. Dmitriev for making recombinant Ad5 fiber and sCAR proteins available to us. We are indebted to J. T. Douglas for stimulating discussions, as well as for providing 1D6.14 for our studies.
This work was supported by the following grants: NCI N01 CO-97110, NIH R01 CA74242, NIH R01 HL50255, and NIH R01 CA83821.
REFERENCES
- 1.Bergelson J M, Cunningham J A, Droguett G, Kurt-Jones E A, Krithivas A, Hong J S, Horwitz M S, Crowell R L, Finberg R W. Isolation of a common receptor for coxsackie B viruses and adenoviruses 2 and 5. Science. 1997;275:1320–1323. doi: 10.1126/science.275.5304.1320. [DOI] [PubMed] [Google Scholar]
- 2.Bewig B, Schmidt W E. Accelerated titering of adenoviruses. BioTechniques. 2000;28:870–873. doi: 10.2144/00285bm08. [DOI] [PubMed] [Google Scholar]
- 3.Chartier C, Degryse E, Gantzer M, Dieterle A, Pavirani A, Mehtali M. Efficient generation of recombinant adenovirus vectors by homologous recombination in Escherichia coli. J Virol. 1996;70:4805–4810. doi: 10.1128/jvi.70.7.4805-4810.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chroboczek J, Ruigrok R W, Cusack S. Adenovirus fiber. Curr Top Microbiol Immunol. 1995;199:163–200. doi: 10.1007/978-3-642-79496-4_10. [DOI] [PubMed] [Google Scholar]
- 5.Davison E, Kirby I, Elliott T, Santis G. The human HLA-A*0201 allele, expressed in hamster cells, is not a high-affinity receptor for adenovirus type 5 fiber. J Virol. 1999;73:4513–4517. doi: 10.1128/jvi.73.5.4513-4517.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dmitriev I, Kashentseva E, Rogers B E, Krasnykh V, Curiel D T. Ectodomain of coxsackievirus and adenovirus receptor genetically fused to epidermal growth factor mediates adenovirus targeting to epidermal growth factor receptor-positive cells. J Virol. 2000;74:6875–6884. doi: 10.1128/jvi.74.15.6875-6884.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dmitriev I, Krasnykh V, Miller C R, Wang M, Kashentseva E, Mikheeva G, Belousova N, Curiel D T. An adenovirus vector with genetically modified fibers demonstrates expanded tropism via utilization of a coxsackievirus and adenovirus receptor-independent cell entry mechanism. J Virol. 1998;72:9706–9713. doi: 10.1128/jvi.72.12.9706-9713.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Douglas J T, Miller C R, Kim M, Dmitriev I, Mikheeva G, Krasnykh V, Curiel D T. A system for the propagation of adenoviral vectors with genetically modified receptor specificities. Nat Biotechnol. 1999;17:470–475. doi: 10.1038/8647. [DOI] [PubMed] [Google Scholar]
- 9.Douglas J T, Rogers B E, Rosenfeld M E, Michael S I, Feng M, Curiel D T. Targeted gene delivery by tropism-modified adenoviral vectors. Nat Biotechnol. 1996;14:1574–1578. doi: 10.1038/nbt1196-1574. [DOI] [PubMed] [Google Scholar]
- 10.Efimov V P, Nepluev I V, Mesyanzhinov V V. Bacteriophage T4 as a surface display vector. Virus Genes. 1995;10:173–177. doi: 10.1007/BF01702598. [DOI] [PubMed] [Google Scholar]
- 11.Gahery-Segard H, Farace F, Godfrin D, Gaston J, Lengagne R, Tursz T, Boulanger P, Guillet J G. Immune response to recombinant capsid proteins of adenovirus in humans: antifiber and anti-penton base antibodies have a synergistic effect on neutralizing activity. J Virol. 1998;72:2388–2397. doi: 10.1128/jvi.72.3.2388-2397.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graham F L, Smiley J, Russell W C, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 13.Hong J S, Engler J A. Domains required for assembly of adenovirus type 2 fiber trimers. J Virol. 1996;70:7071–7078. doi: 10.1128/jvi.70.10.7071-7078.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kasono K, Blackwell J L, Douglas J T, Dmitriev I, Strong T V, Reynolds P, Kropf D A, Carroll W R, Peters G E, Bucy R T, Curiel D T, Krasnykh V. Selective gene delivery to head and neck cancer cells via an integrin targeted adenovirus vector. Clin Cancer Res. 1999;5:2571–2579. [PubMed] [Google Scholar]
- 15.Krasnykh V, Dmitriev I, Mikheeva G, Miller C R, Belousova N, Curiel D T. Characterization of an adenovirus vector containing a heterologous peptide epitope in the HI loop of the fiber knob. J Virol. 1998;72:1844–1852. doi: 10.1128/jvi.72.3.1844-1852.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krasnykh V N, Douglas J T, van Beusechem V W. Genetic targeting of adenoviral vectors. Mol Ther. 2000;1:391–405. doi: 10.1006/mthe.2000.0062. [DOI] [PubMed] [Google Scholar]
- 17.Krasnykh V N, Mikheeva G V, Douglas J T, Curiel D T. Generation of recombinant adenovirus vectors with modified fibers for altering viral tropism. J Virol. 1996;70:6839–6846. doi: 10.1128/jvi.70.10.6839-6846.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Legrand V, Spehner D, Schlesinger Y, Settelen N, Pavirani A, Mehtali M. Fiberless recombinant adenoviruses: virus maturation and infectivity in the absence of fiber. J Virol. 1999;73:907–919. doi: 10.1128/jvi.73.2.907-919.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Letarov A V, Londer Y Y, Boudko S P, Mesyanzhinov V V. The carboxy-terminal domain initiates trimerization of bacteriophage T4 fibritin. Biochemistry. 1999;64:817–823. [PubMed] [Google Scholar]
- 20.Lindner P, Bauer K, Krebber A, Nieba L, Kremmer E, Krebber C, Honegger A, Klinger B, Mocikat R, Pluckthun A. Specific detection of His-tagged proteins with recombinant anti-His tag scFv-phosphatase or scFv-phage fusions. BioTechniques. 1997;22:140–149. doi: 10.2144/97221rr01. [DOI] [PubMed] [Google Scholar]
- 21.Michael S I, Hong J S, Curiel D T, Engler J A. Addition of a short peptide ligand to the adenovirus fiber protein. Gene Ther. 1995;2:660–668. [PubMed] [Google Scholar]
- 22.Miroshnikov K A, Marusich E I, Cerritelli M E, Cheng N, Hyde C C, Steven A C, Mesyanzhinov V V. Engineering trimeric fibrous proteins based on bacteriophage T4 adhesins. Protein Eng. 1998;11:329–332. doi: 10.1093/protein/11.4.329. [DOI] [PubMed] [Google Scholar]
- 23.Mittereder N, March K L, Trapnell B C. Evaluation of the concentration and bioactivity of adenovirus vectors for gene therapy. J Virol. 1996;70:7498–7509. doi: 10.1128/jvi.70.11.7498-7509.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Novelli A, Boulanger P A. Assembly of adenovirus type 2 fiber synthesized in cell-free translation system. J Biol Chem. 1991;266:9299–9303. [PubMed] [Google Scholar]
- 25.Roelvink P W, Mi Lee G, Einfeld D A, Kovesdi I, Wickham T J. Identification of a conserved receptor-binding site on the fiber proteins of CAR-recognizing adenoviridae. Science. 1999;286:1568–1571. doi: 10.1126/science.286.5444.1568. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 27.Tao Y, Strelkov S V, Mesyanzhinov V V, Rossmann M G. Structure of bacteriophage T4 fibritin: a segmented coiled coil and the role of the C-terminal domain. Structure. 1997;5:789–798. doi: 10.1016/s0969-2126(97)00233-5. [DOI] [PubMed] [Google Scholar]
- 28.Tomko R P, Xu R, Philipson L. HCAR and MCAR: the human and mouse cellular receptors for subgroup C adenoviruses and group B coxsackieviruses. Proc Natl Acad Sci USA. 1997;94:3352–3356. doi: 10.1073/pnas.94.7.3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vanderkwaak T J, Wang M, Gomez-Navarro J, Rancourt C, Dmitriev I, Krasnykh V, Barnes M, Siegal G P, Alvarez R, Curiel D T. An advanced generation of adenoviral vectors selectively enhances gene transfer for ovarian cancer gene therapy approaches. Gynecol Oncol. 1999;74:227–234. doi: 10.1006/gyno.1999.5432. [DOI] [PubMed] [Google Scholar]
- 30.van Raaij M J, Mitraki A, Lavigne G, Cusack S. A triple beta-spiral in the adenovirus fibre shaft reveals a new structural motif for a fibrous protein. Nature. 1999;401:935–938. doi: 10.1038/44880. [DOI] [PubMed] [Google Scholar]
- 31.Von Seggern D J, Kehler J, Endo R I, Nemerow G R. Complementation of a fibre mutant adenovirus by packaging cell lines stably expressing the adenovirus type 5 fibre protein. J Gen Virol. 1998;79:1461–1468. doi: 10.1099/0022-1317-79-6-1461. [DOI] [PubMed] [Google Scholar]
- 32.Wickham T J, Roelvink P W, Brough D E, Kovesdi I. Adenovirus targeted to heparan-containing receptors increases its gene delivery efficiency to multiple cell types. Nat Biotechnol. 1996;14:1570–1573. doi: 10.1038/nbt1196-1570. [DOI] [PubMed] [Google Scholar]
- 33.Wickham T J, Tzeng E, Shears L L, Roelvink P W, Li Y, Lee G M, Brough D E, Lizonova A, Kovesdi I. Increased in vitro and in vivo gene transfer by adenovirus vectors containing chimeric fiber proteins. J Virol. 1997;71:8221–8229. doi: 10.1128/jvi.71.11.8221-8229.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xia D, Henry L J, Gerard R D, Deisenhofer J. Crystal structure of the receptor-binding domain of adenovirus type 5 fiber protein at 1.7 Å resolution. Structure. 1994;2:1259–1270. doi: 10.1016/s0969-2126(94)00126-x. [DOI] [PubMed] [Google Scholar]