Abstract
The polyadenylation signal of rice tungro bacilliform virus (RTBV) was characterized by mutational and deletion analysis. The cis-acting signals required to direct polyadenylation conformed to what is known for plant poly(A) signals in general and were very similar to those of the related cauliflower mosaic virus. Processing was directed by a canonical AAUAAA poly(A) signal, an upstream UG-rich region considerably enhanced processing efficiency, and sequences downstream of the cleavage site were not required. When present at the end of a transcription unit, the cis-acting signals for 3′-end processing were highly efficient in both monocot (rice) and dicot (Nicotiana plumbaginifolia) protoplasts. In a promoter-proximal position, as in the viral genome, the signal was also efficiently processed in rice protoplasts, giving rise to an abundant “short-stop” (SS-) RNA. The proportion of SS-RNA was considerably lower in N. plumbaginifolia protoplasts. In infected plants, SS-RNA was hardly detectable, suggesting either that SS-RNA is unstable in infected plants or that read-through of the promoter-proximal poly(A) site is very efficient. SS-RNA is readily detectable in transgenic rice plants (A. Klöti, C. Henrich, S. Bieri, X. He, G. Chen, P. K. Burkhardt, J. Wünn, P. Lucca, T. Hohn, I. Potrylus, and J. Fütterer, 1999. Plant Mol. Biol. 40:249–266), thus the absence of SS-RNA in infected plants can be attributed to poly(A) site bypass in the viral context to ensure production of the full-length pregenomic viral RNA. RTBV poly(A) site suppression thus depends both on context and the expression system; our results suggest that the circular viral minichromosome directs assembly of a transcription-processing complex with specific properties to effect read-through of the promoter-proximal poly(A) signal.
Rice tungro bacilliform virus (RTBV) is a plant pararetrovirus belonging to the Caulimoviridae family (33). RTBV has a circular double-stranded DNA genome (Fig. 1A) replicating via reverse transcription of an RNA intermediate and has many features in common with other plant pararetroviruses and animal retroviruses (reviewed in reference 36) Together with rice tungro spherical virus, RTBV is the causative agent of rice tungro, a devastating disease that affects rice crops in India and Southeast Asia (23). The economic importance of RTBV has prompted much investigation in recent years into the molecular details of various aspects of its biology, in particular its transcriptional and translational regulation (6, 7, 9–11, 19, 25, 45, 46). Like other related viruses, for example, cauliflower mosaic virus (CaMV), RTBV depends on the host transcription machinery. RTBV produces a single, terminally redundant, primary transcript: the pregenomic (pg) RNA. The pgRNA is transcribed by host RNA polymerase II and is polyadenylated at the 3′ end by host 3′-end-processing factors. Thus, the viral poly(A) signal must be recognized as a bona fide plant poly(A) signal. The current model of what constitutes a poly(A) signal in plant systems is based on surprisingly few functional analyses (reviewed in reference 35). Plant poly(A) signals seem to consist of a combination of elements acting in concert to effect 3′-end processing at the poly(A) site or sites: cleavage usually occurs at a YA dinucleotide, under the control of a near upstream element (NUE), which can be AAUAAA or a related A-rich hexamer (37), with the efficiency of processing being greatly enhanced by a more diffuse and ill-defined far upstream element (FUE) (reviewed in references 27 and 35). Computer-aided analysis of several thousand Arabidopsis and rice expressed sequence tags (ESTs) supports this general architecture (15), suggesting that the majority of plant poly(A) signals are likely to fit this model. The poly(A) signals of two dicot-infecting plant pararetroviruses, CaMV (37, 39) and figwort mosaic virus (FMV) (38), have been analyzed so far. The poly(A) signal of RTBV is of interest for two reasons: (i) to increase available data on poly(A) signals functioning in monocot systems and (ii) because of the peculiar requirements for 3′-end-processing regulation that apply to retroelements.
FIG. 1.
(A) Genomic map of RTBV and experimental strategy. The lower part of the figure shows the genome map of RTBV. Viral DNA is represented by a double line, with the box marked R′ indicating the region of the genome that is transcribed twice in the terminally redundant transcript. The thick arrows outside the DNA represent the major viral ORFs (I through IV). Viral transcripts are shown as thin arrows inside the DNA, with the 6.3-kb intron in the spliced transcript encoding ORF IV indicated (dashed line). The basic expression plasmid is represented schematically in the upper part of the panel. The 35S promoter, CAT reporter gene, and RTBV/nos sequences are represented as open boxes. The RTBV, cryptic nos, and nos cleavage sites are indicated with a solid arrow, an open arrowhead, and an open arrow, respectively. The position of the antisense probe transcript for RNase protection analysis is indicated. Homologous probes were used for each construct and were transcribed from linearized plasmid using the T7 promoter present downstream of the nos sequence in the vector. The RTBV sequences inserted in RTPA-L and RTPA-S are indicated with the numbers referring to the transcription start site. Processing efficiencies are given in percent and were roughly the same in N. plumbaginifolia and rice protoplasts, with values from cryptic nos and nos sites being combined as “read through.” Values given are the average of at least three independent transfections. (B) Representative RNase protection assays of RNAs expressed from constructs RTPA-L and RTPA-S. Fragments corresponding to processing at the RTBV, cryptic nos, and nos sites are indicated with a solid arrow, an open arrowhead, and an open arrow, respectively. Signal intensities were always weaker from rice protoplast RNA (RTPA-L not shown). The positions of labeled DNA size markers (pBR322/HpaII) are indicated. The expected sizes of protected fragments at the RTBV, cryptic nos, and nos sites are 515, 818, and 932 nt, or 211, 334, and 448 nt with the RTPA-L and RTBV-S probes, respectively. Signals at the size of the full-length probe in this and other figures are discussed in the text.
As a pararetrovirus, RTBV shares with other retroelements the need for poly(A) site regulation during the production of its terminally redundant RNA. Various mechanisms to achieve poly(A) site bypass have evolved (see Discussion). In RTBV, the 3′-end-processing site first occurs 217 nucleotides (nt) downstream of the transcription start site (Fig. 1A). To produce the pgRNA, the site must be bypassed at this position and used efficiently once the whole circular genome has been transcribed. The poly(A) site of CaMV was reported to be inhibited if in a promoter-proximal position (40), which is how it occurs in the leader sequence of the pregenomic 35S RNA. In this case, poly(A) site bypass is not 100% efficient, and the short-stop (SS-) RNA arising from processing within the leader can be detected in both transfected protoplasts and infected plants (40). An SS-RNA is also seen in plants infected with FMV (38). In this report, we present an analysis of the cis-acting signals of the RTBV poly(A) signal, and show that, in contrast to its behavior at the 3′ end of a transcription unit, processing at a promoter-proximal position depends on both the expression system and the context from which it is expressed. The results suggest that regulation of processing at this site is controlled not only by cis-acting signals but also involves a complex interplay with other transcriptional processes.
MATERIALS AND METHODS
Plasmids.
RTBV sequences were derived from the infectious clone of RTBV previously described by Hay et al. (18). Expression plasmids to test the function of RTBV sequences in 3′-end-processing were based on R-CAT* (37), which expresses the chloramphenicol acetyltransferase (CAT) reporter gene under the control of the CaMV 35S promoter, with the CaMV and nopaline synthase (nos) poly(A) signals in tandem downstream (Fig. 1A). A T7 promoter downstream of the nos sequence allows transcription of homologous antisense probes for RNase protection analyses. RTBV sequences were introduced in place of the CaMV poly(A) signal.
(i) RTPA-1.
An SphI-HindIII fragment covering 288 nt of the RTBV promoter and 459 nt downstream of the transcription start site (corresponding to RTBV positions 7117 to 7864) from R · I-CAT (6) was inserted between the PstI and HindIII sites of R-CAT* (using an adapter) to create RTPA-L.
(ii) RTPA-S.
The 256-nt SacI(blunt)-BstBI(blunt) fragment from R·I-CAT (RTBV positions 7428 to 7684) was introduced into R-CAT/SphI(blunt)-HindIII(blunt) to create RTPA-2. The PstI-SacI fragment covering the RTBV sequences in this intermediate was transferred to R-CAT∗/PstI-SacI. Finally, a linker with the sites PstI-SphI-HindIII was added to facilitate subsequent creation of exonuclease III (ExoIII) deletions: oligonucleotides (5′-GGACTGCAAGCTTGCA-3′ and 5′-AGCTTGCATGCCTGCA-3′) were annealed and inserted into the PstI site to produce RTPA-S. The region of the RTBV genome in RTPA-2 and RTPA-S thus differs only by the addition of the upstream linker in RTPA-S. Depending on the experiment, either RTPA-2 or RTPA-S was used as the “wild-type” RTBV poly(A) sequence (as stated in text and figure legends).
(iii) ΔAATAAA.
Deletion of AATAAAG from the RTBV poly(A) signal creates an SphI site at this position. PstI-SphI and SphI-SacI fragments spanning the RTBV region present in RTPA-S were cleaved from PCR products amplified from RTPA-2 using primer pairs (SphI sites are underlined) 5′-AACCCGCATGCTCTTATATTTATCC-3′/5′-CGCAAGACCGGCAACAGG-3′ and 5′-GAATAAGTGATAATAAGCGG-3′/5′-AACCGCATGCAGCGGATAGG-3′ and ligated into R-CAT∗/PstI-SacI to create plasmid ΔAATAAA.
(iv) ΔCS.
The nucleotides ACA at the site of polyadenylation and cleavage were removed by amplifying the RTBV sequences in RTPA-2 with a downstream primer incorporating this 3-nt deletion and a HindIII site (underlined) (5′-CGCAAGCTTTTATCACAAGGGAGGATAAATATAG-3′) and an upstream primer at the 3′ end of the CAT gene (5′-GAATAAGTGATAATAAGCGG-3′). This fragment was digested with PstI and HindIII and cloned into R-CAT∗/PstI-HindIII to create plasmid ΔCS.
(v) ΔDS/+22ΔDS.
Constructs in which RTBV sequences downstream of the ACA at the cleavage site were removed, either completely (ΔDS) or leaving 22 nt of RTBV sequence (+22ΔDS), were made in the same way as ΔCS using downstream primers covering the 3′ end of the required RTBV sequence and including a HindIII site (ΔDS, 5′-GGAAGCTTTGTAGGATAAATATAAG-3′; +22ΔDS, 5′-GGAAGCTTCATGTTTTATCACAAGG-3′).
(vi) ExoIII deletion series.
Truncations at the 5′ end of the RTBV R sequence were generated using ExoIII. RTPA-S was digested with the unique PstI and HindIII sites within the linker at the 5′ end of the RTBV sequence, thus creating an ExoIII-resistant 3′ overhang at the PstI site and an ExoIII-susceptible 5′ overhang at the HindIII site. ExoIII digestion of this linearized template was performed essentially according to the method of Henikoff (20). Following self-ligation and transformation of Escherichia coli, the resulting clones were screened for appropriately sized RTBV fragments.
All mutations were confirmed by DNA sequencing. Plasmids were isolated from E. coli (strain DH5α) using a plasmid purification kit (Qiagen).
The plasmids used to quantify SS and read-through (RT) transcripts in the RTBV leader were R·I-CAT, RΔC183·I-CAT, C·I-CAT, and CΔC183·I-CAT (7), here referred to as RTBV-wt, RTBV-Δ, 35S-wt, and 35S-Δ, respectively.
The internal control plasmid used in some transfections (pDES7) and the plasmid for generation of the corresponding antisense probe (pGS7) were described by Goodall and Filipowiaz (13) and were kindly provided by Hong Xiang Liu, Friedrich Miescher Institute, Basel, Switzerland.
The internal RTBV genome probe (IV-CAT) used in analysis of RNA from infected plants was prepared by in vitro transcription of a ClaI-linearized plasmid containing the EcoRI-PstI fragment of pC4C (10) cloned in pGEM1 (Promega). This fragment covers 135 nt of the RTBV genome at the end of open reading frame (ORF) III and the start of ORF IV as well as 216 nt of the CAT ORF.
C4CΔint was derived from pC4C (10) by deleting RTBV sequences between the BstBI site in the leader (viral position 17) and the ClaI site around 50 nt upstream of the splice acceptor (position 5917).
RNA from RTBV-infected rice plants.
A sample of RNA from rice plants (cultivar TN1) infected with RTBV was kindly provided by Lee Sor-Cheng and Roger Hull, John Innes Centre, Norwich, United Kingdom. Aliquots of 5 μg of total RNA were used in the RNase protection experiments.
Protoplast transfection and RNA analysis.
Preparation and polyethylene glycol (PEG)-mediated transfection of Nicotiana plumbaginifolia protoplasts was performed as described by Goodall et al. (14). Conditions for growth of suspension cultures of the Oryza sativa line Oc and preparation of protoplasts have been described previously (6). Plasmid DNA was introduced into rice protoplasts by PEG-mediated transfection as for N. plumbaginifolia except that PEG 4000 instead of PEG 6000 was used. For both types of protoplasts, 5 μg of test plasmid was routinely used per transfection. Total RNA was isolated from the protoplasts 6 h after transfection and subjected to RNase protection analysis according to published protocols (14). For each mutant tested, a specific, homologous antisense RNA probe was used. Radioactively labeled probes were synthesized by in vitro transcription using T7 RNA polymerase from plasmids linearized either at the PstI site between the CAT and RTBV sequences or at the ScaI site 130 nt upstream within the CAT sequence. In the latter case, only RNase T1 was used in the protection assay to avoid spurious fragments arising from cleavage within AU-rich stretches at the end of the CAT gene. Protected fragments were resolved on 6% polyacrylamide denaturing gels and visualized by autoradiography or by phosphorimaging (Molecular Dynamics). Fragments corresponding to transcripts processed at the RTBV, cryptic nos, and nos sites were identified based on their size. Quantification of the different protected fragments was by phosphorimager analysis. The percentage of transcripts processed at each site for each mutant was calculated taking into account the number of labeled nucleotides in each fragment. Processing efficiencies expressed as percentages represent the mean values from at least three separate transfections, unless otherwise stated. Variations were within 10% of the mean.
RESULTS
All cis-acting signals of the RTBV poly(A) signal reside downstream of the transcription start site.
The 8-kbp RTBV genome (Fig. 1A) directs transcription of the terminally redundant pgRNA (∼8.2 kb). The 5′ and 3′ ends of this RNA have been mapped to positions 7405 and 7621, respectively, on the genomic DNA (3, 34, 44). We first wanted to ascertain the extent of the region of the RTBV genome required to signal 3′-end processing. For the other two plant pararetroviral poly(A) signals analyzed to date (CaMV [37, 39] and FMV [38]), all cis-acting elements of the poly(A) signal are present within the terminal redundancy; that is, no specific sequences upstream of the transcription start site or downstream of the site of poly(A) addition are required.
The experimental system used to test cis-acting elements of the RTBV poly(A) signal is shown in Fig. 1A. RTBV sequences were cloned between the CAT reporter gene and a second, downstream, poly(A) signal—nos. After transient expression of such a construct, RNase A/T1 mapping analysis can distinguish and quantify transcripts correctly processed at the RTBV site and those which read through to be processed at the nos poly(A) sites. The latter can direct processing in one of two regions: the wild-type nos poly(A) site, which is a collection of closely spaced sites near the 3′ end of the sequence, or a cryptic site just downstream of an AATAAA motif, which can become activated if the FUE of a heterologous poly(A) signal is present upstream (37, 39).
Initially, RTBV sequences corresponding to genome positions 7117 to 7864 (i.e., from 288 nt upstream of the transcription start site to 459 nt downstream) were inserted (RTPA-L; Fig. 1B). RTPA-L was transiently expressed in protoplasts of N. plumbaginifolia and rice. In both types of protoplasts, processing at the RTBV site was almost 100%. Truncating the RTBV sequences at the 5′ end to +24 relative to the start site, and at the 3′ end to 63 nt downstream of the poly(A) site made no difference (RTPA-S; Fig. 1A and B). Thus, sequences upstream of the transcription start site do not contribute to processing efficiency, and all necessary signals reside within this fragment.
In addition, in both protoplast types, a single protected fragment corresponding to processing at the 3′ end mapped in viral RNA from infected rice plants (34) was observed, indicating that the RTBV poly(A) signal functions correctly in our test constructs, even in a heterologous dicot system.
Fragments shorter than the one that corresponds to processing at the RTBV site were sometimes observed with RTPA-L, but these fragments were not always present; if present, they varied in intensity in different experiments; and they were not observed with RTPA-S. We conclude that the longer labeled transcript probe for RTPA-L was more susceptible to nonspecific degradation during the experiments, and that the bands that were sometimes observed might represent transcripts protected by pieces of truncated probe.
In this and other experiments, we often observed protected fragments that corresponded to the full length of the probe. These fragments could represent protection of either residual DNA in the sample or transcription events that read through all of the available 3′-end-processing signals on the construct and continue into the vector. Since all samples were extensively DNase treated, we consider the latter explanation more likely. Similar observations have been confirmed experimentally by others (26) as being due to transcription that continues right around the circular plasmid template. These bands were not included in calculations of processing efficiency in our experiments.
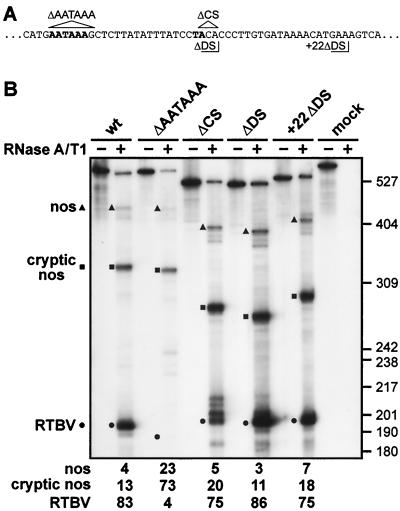
AATAAA is an essential part of the poly(A) signal, but the cleavage site and specific sequences downstream are not required.
The AATAAA motif present 18 nt upstream of the poly(A) site was deleted in the context of RTPA-2 (RTBV sequences are as in RTPA-S but lack the short linker at the 5′ end, see Materials and Methods). Processing at the RTBV site was almost abolished, with the vast majority of transcripts reading through to be processed at the cryptic nos site (Fig. 2B). Thus, AATAAA is an essential part of the RTBV poly(A) signal and corresponds to the NUE (see the introduction).
FIG. 2.
AATAAA is required for 3′-end processing; the cleavage site and specific downstream sequences are not. (A) Sequences surrounding the AATAAA and cleavage sites (both in bold type): nucleotides deleted in ΔAATAAA and ΔCS are indicated by triangles; the end points of RTBV sequences in ΔDS and +22ΔDS are delimited by right-angled lines. (B) RNase protection assay to examine processing events at the RTBV poly(A) site in a wild-type construct (RTPA-2), and constructs carrying deletions of either AATAAA (ΔAATAAA), the cleavage site (ΔCS), or sequences directly downstream of the cleavage site (ΔDS) or from 22 nt downstream of the cleavage site (+22ΔDS) as shown in panel A. Protected fragments corresponding to transcripts processed at the RTBV, cryptic nos, and nos sites are indicated, with the positions of size markers (pBR322/HpaII) shown on the right. Processing efficiencies (in percent) are given underneath. The gel shown is from analysis of RNA from N. plumbaginifolia protoplasts.
The site of cleavage and poly(A) addition maps to genomic position 7621. The contribution of RTBV sequences downstream of this position was tested by deleting nucleotides 3′ of the processing site only leaving either 2 or 22 nucleotides (Fig. 2A [ΔDS or +22ΔDS, respectively]) fused to the polylinker sequences preceding the nos poly(A) signal. Neither of these modifications had a significant effect on the efficiency or accuracy of processing at the RTBV site (Fig. 2B), indicating that specific sequences downstream do not form part of the signal. To test the cleavage site itself, the nucleotides ACA (7621 to 7623) were removed (Fig. 2A [ΔCS]), this time with 14 nt of RTBV sequence left downstream. These three nucleotides were deleted to ensure that there was no remaining YA dinucleotide at the position of the wild-type cleavage site. Although the overall processing efficiency at, or near, the RTBV site was similar to the wild-type construct, accuracy was affected. The majority of transcripts (40% of the total at the RTBV site) were processed at a CC dinucleotide at the position of the wild-type A at position 7621. Other transcripts were processed ∼4, ∼8, and ∼12 nucleotides downstream of this position (27%, 17%, and 16% of transcripts, respectively). These results demonstrate that although cleavage can occur at a site other than YA if a YA dinucleotide is not available, positioning of processing is not as tightly controlled and alternative sites are cleaved.
The results shown in Fig. 2B were obtained with RNA from N. plumbaginifolia protoplasts. Essentially the same results were obtained in rice protoplasts, but the amounts of RNA recovered were substantially lower (data not shown). In this series of experiments, the level of readthrough of the poly(A) signal was a little higher than in the initial analysis [cf. RTPA-S in Fig. 1 with wt (RTPA-2) in Fig. 2] for unknown reasons. This was the case in both types of protoplasts.
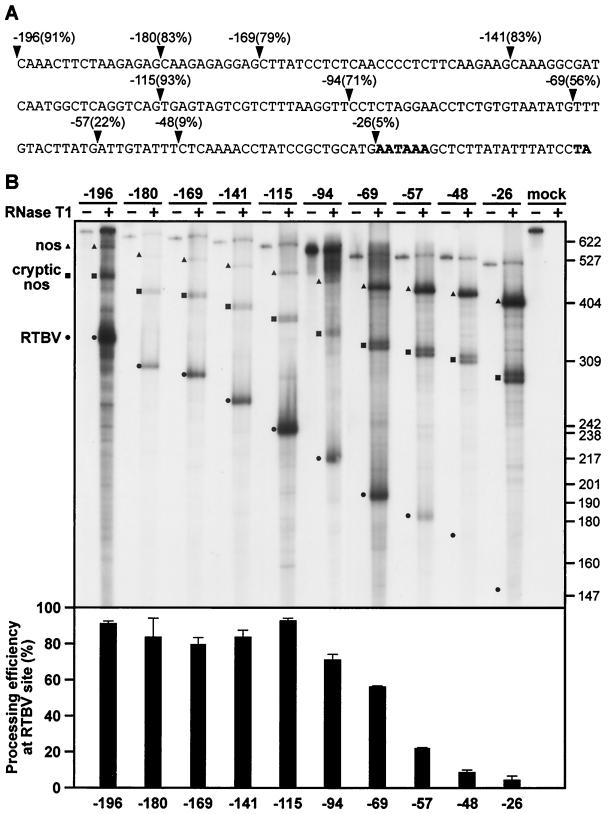
TG-rich upstream sequences enhance efficiency of processing.
The region upstream of the AATAAA poly(A) signal is relatively T rich. Removal of the first 100 nt from the 5′ end of the RTBV sequences downstream of the CAT ORF in RTPA-S did not significantly lower processing efficiency (Fig. 3). Further deletion of sequences more proximal to the AATAAA was extremely detrimental. In particular, T- and TG-rich stretches lying between −69 and −48 nt upstream of the processing site make a substantial contribution to the efficiency of processing at the RTBV site. Accuracy was unaffected by these deletions, and where the RTBV signal was disabled by removal of upstream sequences, transcripts were processed at one of the downstream nos sites. Thus, the AATAAA signal alone is insufficient to direct processing at the RTBV site. Again, results were similar in rice and N. plumbaginifolia protoplasts, but RNA yields were higher from the latter (rice data not shown). The region identified as the FUE contains several copies of TTTGTA-like motifs. Such elements have been shown to be functionally important in the FUEs of other plant poly(A) signals, in the yeast Ty element, and in the animal viruses simian virus 40 and ground squirrel hepatitis virus (see reference 35, and references therein). A recent “in silico” survey of Arabidopsis and rice ESTs has revealed a statistically significant increased occurrence of TTGTAT and TTGTAA (or similar motifs) in the last 100 nt of plant mRNAs (15), suggesting a role for such sequences in signaling 3′-end formation, probably as sites of recognition for processing factors (see Discussion).
FIG. 3.
Sequences upstream of AATAAA enhance processing efficiency at the RTBV site. (A) The RTBV sequence in RTPA-S up to the site of poly(A) addition is shown. Arrowheads mark the endpoints of the ExoIII deletion series, with the number of nucleotides remaining upstream of the cleavage site and the corresponding processing efficiency indicated. (B) Representative RNase T1 protection assay of the deletion series shown in panel A. Protected fragments corresponding to transcripts processed at the RTBV, cryptic nos, and nos sites are indicated, with the positions of size markers (pBR322/HpaII) shown on the right. Processing efficiencies at the RTBV site are shown in the chart underneath (error bars represent standard deviations). The gel shown is from analysis of RNA from N. plumbaginifolia protoplasts.
Recognition of the RTBV poly(A) signal is not inhibited by a promoter-proximal position in rice protoplasts.
The pregenomic RNA of RTBV is terminally redundant (Fig. 1A), requiring that the poly(A) signal must be bypassed when first encountered by the transcription machinery. The poly(A) site of CaMV was reported to be inhibited if in a promoter-proximal position, which is how it occurs in the leader sequence of the pregenomic 35S RNA (40). Since the cis-acting sequences of the RTBV poly(A) site are so similar to those of CaMV and given the relatedness of the two viruses, we investigated whether the RTBV poly(A) signal might also be regulated in this manner.
Rice protoplasts were transfected with CAT reporter constructs under the control of either the CaMV 35S promoter or the RTBV promoter fused to the RTBV pregenomic leader sequence (Fig. 4A). Transcripts were mapped using a probe transcribed from RTPA-L (see Materials and Methods). This probe allowed visualization of transcripts which have been processed at the poly(A) site within the leader (SS), as well as those where transcription has continued (RT).
FIG. 4.
Processing at the RTBV poly(A) site in a promoter-proximal position gives rise to SS-RNA in transfected protoplasts. (A) Constructs consisting of the RTBV (to −681) or CaMV 35S (to −343) upstream promoter sequences, the RTBV leader sequence (either complete [wt] or deleted between +8 and +83 [Δ]) and the CAT reporter gene fused to RTBV ORF I are shown. All constructs are terminated by the CaMV polyadenylation signal. The location of the homologous region of the antisense probe used for RNase A/T1 mapping, and the extent of protected fragments corresponding to SS and RT transcripts, are indicated. (B and C) RNase protection analysis. Total RNA isolated from transfected rice (B) or N. plumbaginifolia (C) protoplasts was subjected to RNase A/T1 protection analysis with an antisense probe transcribed from RTPA-L. Protected fragments corresponding to RTBV SS and RT transcripts for the four constructs used are indicated, with the percentage of SS shown below the gels. ∗, Values with the RTBV-Δ construct in N. plumbaginifolia were too low to quantify reliably. Other values given are the average from two independent experiments (variation was within 10% of the mean). i.c., internal control; pDES7 (see Materials and Methods).
Unexpectedly, in all constructs tested, the vast majority of transcripts were in fact processed at the poly(A) site within the leader (Fig. 4B). Bringing the poly(A) signal even closer to the promoter by deletion of leader sequences +8 to +83 did not significantly alter the SS:RT transcript ratio. The choice of promoter had no influence; the proportion of SS transcripts was similar, although the expression level was substantially higher with the 35S promoter. (Note that, in the context of the RTBV promoter, transcript levels are further reduced in the presence of the leader deletion, since this region contains an enhancer element for the RTBV promoter [7].)
The same constructs were also expressed in N. plumbaginifolia protoplasts. During the analysis of the cis-acting signals documented above, that is, with the RTBV poly(A) signal at the 3′ end of the reporter construct, results in both types of protoplast were essentially the same. However, the leader-containing constructs behaved differently in N. plumbaginifolia protoplasts. As shown in Fig. 4C, the proportion of SS transcripts in these cells was very much lower than in rice, with the majority of transcripts corresponding to RT. The RTBV promoter was expressed very poorly in these cells, especially in the presence of the leader deletion, and the level of SS-RNA was too low to quantify reliably, yet it was clear that in the case of RTBV-wt, much more RT-RNA than SS-RNA was present (Fig. 4C).
These results indicate that there is a difference in the efficiency of RTBV poly(A) signal recognition in this promoter-proximal position in the two species tested. Alternatively, the stability of the SS-RNA varies in different species.
SS-RNA is not detectable in RTBV-infected rice plants.
The high level of SS-RNA in transfected rice protoplasts was surprising, since it would seem to be disadvantageous to the virus to terminate the majority of transcription events prematurely. To determine whether the results described above reflect the situation in planta, we attempted to map SS transcripts in RNA from RTBV-infected rice plants. The probe used was derived from RTPA-L, thus allowing simultaneous detection of all transcripts covering this region of the pgRNA (Fig. 5A). Against a background smear of protected fragments, transcripts corresponding to the expected sizes of the 5′ and 3′ ends of the pgRNA were identified (Fig. 5B). In addition, a signal at a higher molecular weight indicating a fragment of longer than genome length (RT2, Fig. 5B) was observed (although a fragment of this size would also be observed if a single probe molecule simultaneously hybridized with the 5′ end and the region of the 3′ end just preceding the terminal redundancy). Two additional fragments (corresponding to around 265 and 190 nt) could be discerned against the background smear, but it is unclear what these fragments represent. There was no clear signal above background at the position expected for SS-RNA (217 nt). Since this probe was able to detect SS-RNA in other experiments (see Fig. 4) and can also detect the 3′ end of the viral RNA that contains the same sequence as the SS-RNA, SS-RNA would be expected to be revealed if present. Thus, we conclude that SS-RNA does not accumulate to detectable levels in RTBV-infected plants.
FIG. 5.
SS-RNA is not detected in RTBV-infected rice plants. (A) The RTBV pgRNA is represented twice, by solid and dashed curved lines, with the SS-RNA as a straight line colinear with the 3′ end of the pgRNA. The antisense probes transcribed from RTPA-L or RTPA-2 are represented above the upper and lower diagrams, respectively, and thick lines indicate the position and expected sizes (in nucleotides) of protected RNA fragments corresponding to the 5′ and 3′ ends of the pgRNA and the SS-RNA. The diagram is not to scale, and the nos sequences on the probes are not indicated. (B) RNase protection analysis of total RNA (5-μg aliquots) from RTBV-infected rice plants using probes covering the terminal redundancy (RTPA-L), the 3′ end of the terminal redundancy (RTPA-2), or a 135-nt fragment of the RTBV pgRNA spanning the end of ORF III and the beginning of ORF IV (IV-CAT [10] protects a 135-nt internal fragment on the pgRNA). Fragments obtained with probe RTPA-L corresponding to the 5′ (RT) and 3′ ends of the RTBV pgRNA are indicated on the left, as well as the expected position of SS-RNA. A longer fragment (RT2) is also indicated (see text for possible explanations). Fragments protected by the RTPA-2 and IV-CAT probes are labeled on the right. Note that the RTPA-2 probe does not distinguish between SS-RNA and the 3′ end of pgRNA.
As a control, RNA from infected rice was mapped with two additional probes: RTPA-2 and a probe covering an internal region of the RTBV genome (IV-CAT; see Fig. 6A). RTPA-2 again allowed identification of the 5′ end of the pgRNA but does not distinguish between the 3′ end of pgRNA and SS-RNA. This probe was used to check that the probe sequence covering this region of viral RNA would indeed lead to a protected fragment of the predicted size under the conditions of the experiment, that is, there was no significant internal digestion by RNase A in A+U-rich regions of the hybrid. The IV-CAT probe confirmed the integrity of the viral RNA in the sample (Fig. 5B).
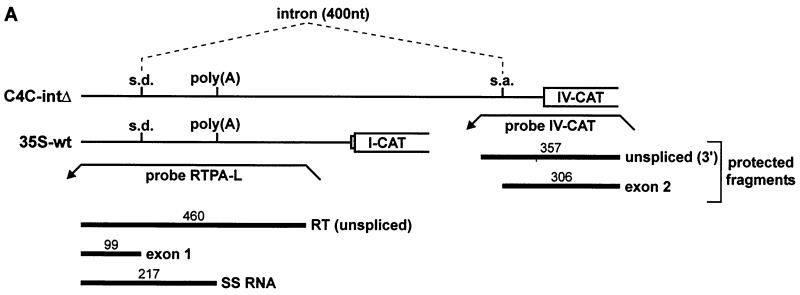
FIG. 6.
SS-RNA is unaffected by splicing of the viral intron. (A) Constructs C4C-intΔ and 35S-wt are depicted schematically, with the positions of the splice donor (s.d.), splice acceptor (s.a.), and poly(A) site indicated. Splicing of the 400-nt intron is indicated by dashed lines. Probes RTPA-L and IV-CAT are shown as leftward pointing arrows. The position and size (in nucleotides) of protected fragments are shown underneath. (B) Representative RNase protection analyses of the constructs shown in panel A, expressed in either rice (left panel) or N. plumbaginifolia (right panel) protoplasts. Probes RTPA-L and IV-CAT were used together in each sample. Protected fragments are labeled and named as in panel A. In rice protoplasts, some splicing events occurred on the 35S-wt transcript, presumably using cryptic acceptor sites within the CAT ORF or in the vector sequences. (C) The amount of SS-RNA in percent (calculated as SS/[SS+RT+exon 1]).
This experiment measures the steady-state levels of viral RNA isolated from the infected plants. With the RTPA-L probe, the protected RT fragment (corresponding to the 5′ end of the pgRNA) was much more abundant than the 3′-end protected fragment. One would expect a 1:1 ratio, and thus the longer 3′ fragments should appear more intense than the RT fragment (since the probe is uniformly labeled). Although we do not have a satisfactory explanation for this observation, it would appear to be an artifact specific to this probe, since the RTPA-2 probe gives a more proportional distribution of 5′ to 3′ fragments.
Very little spliced viral RNA was observed with any of the probes used (fragments in the 70- to 100-nt range; area of gel not shown), in keeping with previous observations that the splicing of the pgRNA occurs with low efficiency (10).
SS-RNA levels are unaffected by the presence of the viral intron.
In the viral pgRNA, the poly(A) site is located within an intron splicing and 3′-end processing are not independent processes in vivo, and there are well-documented examples where splice sites influence poly(A) site use (see Discussion). In the transient expression experiments described above, only the splice donor was present in the constructs tested. To examine the possibility that the splicing process affects recognition of 3′-end-processing signals and would thus influence the production of SS-RNA, a construct (C4C-intΔ) in which the poly(A) site is present within an intron was tested in protoplasts. C4C-intΔ carries the poly(A) site within an internally deleted version of the viral intron (see Materials and Methods), and can be compared to 35S-wt, which contains only the splice donor (Fig. 6A).
C4C-intΔ and 35S-wt (Fig. 6A) were expressed in rice and N. plumbaginifolia protoplasts to assess the effect of the intron on SS-RNA production. A typical RNase A/T1 protection analysis is shown in Fig. 6B. Splicing with C4C-intΔ occurred with an efficiency of ∼50% in both systems. The amount of SS-RNA was measured in percent relative to the sum of SS+RT+exon 1 for each case (Fig. 6C). In rice protoplasts, the level of SS-RNA was completely unaffected by the presence of an intact intron surrounding the poly(A) site. In N. plumbaginifolia protoplasts, only a very minor effect was observed (SS-RNA levels were slightly reduced in the presence of the intron). Thus, the presence of a functional intron surrounding the poly(A) site does not influence the relative production of SS-RNA in protoplasts.
DISCUSSION
The transcription termination-polyadenylation signals of the plant pararetroviruses are of interest not only as models of these processes in plant systems, but also because their location within the transcribed region just downstream of the promoter suggests some regulation of their usage during virus infection. To date, there is no evidence that virus-encoded factors play a role in this regulation (see below), thus the virus exploits cellular RNA processing mechanisms to effect differential poly(A) site recognition.
The cis-acting components of the RTBV poly(A) signal revealed by this study concur with the current model of a plant poly(A) signal: cleavage at UA, a perfect (and essential) AAUAAA (NUE), and an FUE consisting of U- and UG-rich sequences. In common with other plant poly(A) signals, specific sequences downstream of the cleavage site are not required.
Although RTBV and CaMV infect monocots and dicots, respectively, the cis-acting components of their poly(A) signals are almost identical (Fig. 7), and we have shown here that the RTBV poly(A) signal behaves similarly in both systems when present at the 3′ end of a transcription unit.
FIG. 7.
Comparison of the poly(A) signals of RTBV and CaMV. The sequences of the terminal redundancies of RTBV and CaMV are shown. The region is delimited by the transcription start site (black dot) and the poly(A) site (bold type UA). NUEs are set in bold type, and FUEs are underlined, with a dashed underline indicating sequences with only a slight effect on processing efficiency. Perfect and one-base mismatches of UUUGUA are indicated with black and white arrows, respectively. (Adapted from reference 35.)
Although 3′-end-processing factors in plants remain wholly uncharacterized, the conservation of some cis-acting elements and the presence in plant databases of probable homologues of 3′-end-processing factors from other eukaryotes suggest that the basic mechanism is likely to be universal. A great deal is known about the biochemistry of cleavage and polyadenylation in mammalian and yeast systems (reviewed in references 43 and 47), allowing us to speculate on the details of these processes in plants.
In general, plant polyadenylation signals have more in common with those of Saccharomyces cerevisae, where positioning elements (PE) and efficiency elements (EE) upstream of the cleavage site direct processing via their interaction with a battery of protein factors (47). The equivalence in position, function, and even sequence characteristics of the yeast PE and EE to the NUEs and FUEs of plant poly(A) signals suggests that the processing complexes are likely to be yeast-like. On the other hand, plant protein sequences in the databases with homology to 3′-end-processing factors are more related to metazoan than to yeast-processing factors (e.g., plant ESTs with homology to subunits of mammalian cleavage and specificity factor [CPSF] and cleavage stimulation factor [CstF] can be found [22; H. M. Rothnie, unpublished observations]). Thus, by analogy with mammalian systems, a CPSF-like factor might interact with the AAUAAA of the RTBV signal. In mammals, sequences downstream of the cleavage site are contacted by CstF, and processing machinery involving at least two other cleavage factors and poly(A) polymerase assembles around the CPSF–pre-mRNA–CstF complex (43). Some mammalian poly(A) signals also have a requirement for upstream sequence elements (USEs), the characterization of which suggests possible roles for plant FUEs. The function of the FUE might be to stabilize a CPSF-NUE interaction, either directly as in the case human immunodeficiency virus type 1 (HIV-1), equine infectious anemia virus, and the human lamin B2 poly(A) signals (5, 12, 16) or via the interaction of the U1A protein of U1 snRNP with both the USE and CPSF, as in the case of the simian virus 40 late poly(A) signal (28, 29). It is interesting that this latter USE bears a striking resemblance to the FUEs of plant poly(A) signals, with several repeats of UUUGUA or related sequences (35). Alternatively, or in addition, the FUE may be functionally analogous to the mammalian downstream element and bind cleavage factors. In mammals, binding of a cleavage factor to an upstream sequence has been reported in the case of the human C2 complement poly(A) signal, which is unusual in that it lacks a recognizable downstream component. Instead, a U-rich USE enhances 3′-end formation, in part by facilitating CPSF-dependent binding of CstF (31). The involvement of CstF in processing of a pre-mRNA that lacks a downstream element suggests a possible mechanism for processing of plant pre-mRNAs without the need to invoke novel, plant-specific processing factors.
The RTBV poly(A) signal functioned efficiently in a promoter-distal location in both protoplast systems tested. In a promoter-proximal situation, as in the RTBV genome, the signal was efficiently recognized in rice protoplasts, but it was recognized considerably less well in N. plumbaginifolia protoplasts. While both systems are apparently capable of supplying all necessary trans-acting factors for efficient 3′-end processing at a distal signal, they display differences at a proximal signal. This in turn indicates that the interaction of processing factors with the pre-mRNA depends on more than just the primary RNA sequence. The efficiency of processing at a promoter-proximal signal in rice protoplasts was surprising, since in a virus infection this would reduce the production of full-length viral pgRNA considerably. However, our analysis of RNAs in infected rice plants revealed that no or very little SS-RNA can be detected under these conditions. In contrast, the efficiency of poly(A) site bypass in the case of CaMV is not 100%, and SS-RNA is readily detectable in transfected protoplasts (65% of transcripts in N. plumbaginifolia protoplasts) and to a lesser extent in infected plants (40). SS-RNA is also seen in plants infected with FMV (38). In CaMV-infected plants, the proportion of SS-RNA varies in resistant and susceptible host plant species (41). A higher SS-35S RNA ratio was a feature of host plants showing only mild or undetectable symptoms. The significance of this observation and the function, if any, of SS-RNA in the life cycle of CaMV remain unclear. Similar analyses of RTBV in different hosts or at different stages of viral infection have not been performed, thus it remains theoretically possible that SS-RNA is also produced by RTBV under certain conditions. However, the situation observed here, that is, the absence of SS-RNA, would appear to be the most advantageous for viral replication; bypass of the 5′-proximal poly(A) site means that the majority of transcription initiation events result in full-length pgRNA.
Since SS-RNA is stable in transgenic rice plants (25), its absence in infected plants must be due to lack of synthesis. Thus, the virus must differentiate between promoter-proximal and distal poly(A) signals to suppress processing at the former site. Mechanisms of retroelement poly(A) site regulation have been studied in other systems (reviewed in reference 36). Various scenarios include (i) inherent inefficiency of the poly(A) signal, (ii) an incomplete poly(A) signal at the 5′ end of the transcript, (iii) a requirement for enhancing sequences upstream of the transcription start site, (iv) occlusion by proximity of the promoter or cap-site, (v) a role for a virally encoded factor, (vi) an influence of RNA structure, or (vii) the relative juxtaposition of other processing signals on the RNA. In the case of RTBV, the first three scenarios can be ruled out, since the signal is efficient and wholly contained within the terminal redundancy. Proximity to the promoter is also probably not the explanation, as evidenced by the huge amounts of SS-RNA observed in transfected rice protoplasts, even when the poly(A) site is moved even closer to the start site (Fig. 4).
So far, we have been unsuccessful in our attempts to show an influence of a virally encoded factor on production or processing of mRNAs containing the RTBV leader and poly(A) site. (H. M. Rothnie, Orlene Guerra Peraza, and Saule Zhanybekova, unpublished data). In addition, the differential effectiveness of the promoter-proximal signal in rice and N. plumbaginifolia protoplasts shows that suppression of the signal can at least partially occur in the absence of RTBV-derived factors.
It is unlikely that RNA structure plays a role in regulation of RTBV poly(A) signal recognition, as has been suggested for HIV-1 (8, 24). The main structural element of the RTBV pgRNA 5′ end is an extended hairpin, which is conserved among all plant pararetroviruses and which includes the poly(A) signal in a double-stranded region in both RTBV and CaMV (32). However, this structure should be the same in the RTBV pgRNA and in the RNAs used for the analysis in protoplasts, since both contain the relevant region completely.
An inhibitory influence by splice sites on 3′-end processing has been documented in several cases, and at least two mechanisms have been implicated, both involving interactions of components of the U1 snRNP with the cleavage-polyadenylation complex (1, 2, 17, 42). In RTBV, the splice donor is around 100 nt upstream of the NUE and, although the presence of the donor alone did not preclude SS-RNA production in protoplasts, we considered the possibility that the presence of the poly(A) site within a functional intron could effect its suppression. However, our results showed this not to be the case.
In transgenic rice plants, the production of SS-RNA depends largely on the promoter used to drive otherwise identical transcription units (25). This suggests that the nature of the transcription complex is directly involved in determining downstream processing events. In recent years, the many steps involved in transcription, processing, and export of mRNAs have ceased to be viewed in isolation. Indeed, research into the individual events has converged to reveal a complex, integrated process in which all processes are closely coordinated. Specifically, several key factors involved in 3′-end processing associate with the transcription complex already at the promoter (reviewed in references 4, 21, and 30). A differential association of processing factors with this complex in the different assay systems could offer an explanation for all our findings. This would suggest that only the circular viral minichromosome possesses the proper structure and control signals to assemble a complex in rice vascular cells that allows efficient read-through of the promoter-proximal processing signal.
ACKNOWLEDGMENTS
We are very grateful to Lee Sor-Cheng and Roger Hull (John Innes Centre, Norwich, United Kingdom) for providing us with RNA from RTBV-infected rice plants. We highly acknowledge the expertise of Matthias Müller in maintenance of plant cultures and preparation of protoplasts. We thank Etienne Herzog and Karin Wiebauer for commenting on the manuscript. Thanks also to Mike Rothnie for help with preparation of figures.
This work was supported by the Novartis Research Foundation.
REFERENCES
- 1.Ashe M P, Furger A, Proudfoot N J. Stem-loop 1 of the U1 snRNP plays a critical role in the suppression of HIV-1 polyadenylation. RNA. 2000;6:170–177. doi: 10.1017/s1355838200991957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashe M P, Pearson L H, Proudfoot N J. The HIV-1 5′ poly(A) site is inactivated by U1 snRNP interaction with the downstream major splice donor site. EMBO J. 1997;16:5752–5763. doi: 10.1093/emboj/16.18.5752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bao Y, Hull R. Mapping the 5′-terminus of rice tungro bacilliform viral genomic RNA. Virology. 1993;197:445–448. doi: 10.1006/viro.1993.1609. [DOI] [PubMed] [Google Scholar]
- 4.Bentley D. Coupling RNA polymerase II transcription with pre-mRNA processing. Curr Opin Cell Biol. 1999;11:347–351. doi: 10.1016/S0955-0674(99)80048-9. [DOI] [PubMed] [Google Scholar]
- 5.Brackenridge S, Proudfoot N J. Recruitment of a basal polyadenylation factor by the upstream sequence element of the human lamin B2 polyadenylation signal. Mol Cell Biol. 2000;20:2660–2669. doi: 10.1128/mcb.20.8.2660-2669.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen G, Müller M, Potrykus I, Hohn T, Fütterer J. Rice tungro bacilliform virus: transcription and translation in protoplasts. Virology. 1994;204:91–100. doi: 10.1006/viro.1994.1513. [DOI] [PubMed] [Google Scholar]
- 7.Chen G, Rothnie H M, He X, Hohn T, Fütterer J. Efficient transcription from the rice tungro bacilliform virus promoter requires elements downstream of the transcription start site. J Virol. 1996;70:8411–8421. doi: 10.1128/jvi.70.12.8411-8421.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Das A T, Klaver B, Berkhout B. A hairpin structure in the R region of the human immunodeficiency virus type 1 RNA genome is instrumental in polyadenylation site selection. J Virol. 1999;73:81–91. doi: 10.1128/jvi.73.1.81-91.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fütterer J, Potrykus I, Bao Y, Li L, Burns T M, Hull R, Hohn T. Position-dependent ATT initiation during plant pararetrovirus rice tungro bacilliform virus translation. J Virol. 1996;70:2999–3010. doi: 10.1128/jvi.70.5.2999-3010.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fütterer J, Potrykus I, Valles-Brau M P, Dasgupta I, Hull R, Hohn T. Splicing in a plant pararetrovirus. Virology. 1994;198:663–670. doi: 10.1006/viro.1994.1078. [DOI] [PubMed] [Google Scholar]
- 11.Fütterer J, Rothnie H M, Hohn T, Potrykus I. Rice tungro bacilliform virus open reading frames II and III are translated from polycistronic pregenomic RNA by leaky scanning. J Virol. 1997;71:7984–7989. doi: 10.1128/jvi.71.10.7984-7989.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gilmartin G M, Fleming E S, Oetjen J, Graveley B R. CPSF recognition of an HIV-1 mRNA 3′-processing enhancer: multiple sequence contacts involved in poly(A) site definition. Genes Dev. 1995;9:72–83. doi: 10.1101/gad.9.1.72. [DOI] [PubMed] [Google Scholar]
- 13.Goodall G J, Filipowicz W. The AU-rich sequences present in the introns of plant nuclear pre-mRNAs are required for splicing. Cell. 1989;58:473–483. doi: 10.1016/0092-8674(89)90428-5. [DOI] [PubMed] [Google Scholar]
- 14.Goodall G J, Wiebauer K, Filipowicz W. Analysis of pre-mRNA processing in transfected plant protoplasts. Methods Enzymol. 1990;181:148–161. doi: 10.1016/0076-6879(90)81117-d. [DOI] [PubMed] [Google Scholar]
- 15.Graber J H, Cantor C R, Mohr S C, Smith T F. In silico detection of control signals: mRNA 3′-end-processing sequences in diverse species. Proc Natl Acad Sci USA. 1999;96:14055–14060. doi: 10.1073/pnas.96.24.14055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graveley B R, Gilmartin G M. A common mechanism for the enhancement of mRNA 3′ processing by U3 sequences in two distantly related lentiviruses. J Virol. 1996;70:1612–1617. doi: 10.1128/jvi.70.3.1612-1617.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gunderson S I, Polycarpou-Schwarz M, Mattaj I W. U1 snRNP inhibits pre-mRNA polyadenylation through a direct interaction between U1 70K and poly(A) polymerase. Mol Cell. 1998;1:255–264. doi: 10.1016/s1097-2765(00)80026-x. [DOI] [PubMed] [Google Scholar]
- 18.Hay J M, Jones M C, Blakebrough M L, Dasgupta I, Davies J W, Hull R. An analysis of the sequence of an infectious clone of rice tungro bacilliform virus, a plant pararetrovirus. Nucleic Acids Res. 1991;19:2615–2621. doi: 10.1093/nar/19.10.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He X, Hohn T, Fütterer J. Transcriptional activation of the rice tungro bacilliform virus gene is critically dependent on an activator element located immediately upstream of the TATA box. J Biol Chem. 2000;275:11799–11808. doi: 10.1074/jbc.275.16.11799. [DOI] [PubMed] [Google Scholar]
- 20.Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984;28:351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- 21.Hirose Y, Manley J L. RNA polymerase II and the integration of nuclear events. Genes Dev. 2000;14:1415–1429. [PubMed] [Google Scholar]
- 22.Jenny A, Keller W. Cloning of cDNAs encoding the 160kDa subunit of the bovine cleavage and polyadenylation specificity factor. Nucleic Acids Res. 1995;23:2629–2635. doi: 10.1093/nar/23.14.2629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones M C, Gough K, Dasgupta I, Subba Rao B L, Cliffe J, Qu R, Shen P, Kaniewska M, Blakebrough M, Davies J W, Beachy R N, Hull R. Rice tungro disease is caused by an RNA and a DNA virus. J Gen Virol. 1991;72:757–761. doi: 10.1099/0022-1317-72-4-757. [DOI] [PubMed] [Google Scholar]
- 24.Klasens B I F, Thiesen M, Virtanen A, Berkhout B. The ability of the HIV-1 AAUAAA signal to bind polyadenylation factors is controlled by local RNA structure. Nucleic Acids Res. 1999;27:446–454. doi: 10.1093/nar/27.2.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klöti A, Henrich C, Bieri S, He X, Chen G, Burkhardt P K, Wünn J, Lucca P, Hohn T, Potrykus I, Fütterer J. Upstream and downstream elements determine the specificity of the rice tungro bacilliform virus promoter and influence RNA production after transcription initiation. Plant Mol Biol. 1999;40:249–266. doi: 10.1023/a:1006119517262. [DOI] [PubMed] [Google Scholar]
- 26.Lambermon M H L, Simpson G G, Wieczorek Kirk D A, Hemmings-Mieszczak M, Klahre U, Filipowicz W. UBP1, a novel hnRNP-like protein that functions at multiple steps of higher plant nuclear pre-mRNA maturation. EMBO J. 2000;19:1638–1649. doi: 10.1093/emboj/19.7.1638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li Q, Hunt A G. The polyadenylation of RNA in plants. Plant Physiol. 1997;115:321–325. doi: 10.1104/pp.115.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lutz C S, Alwine J C. Direct interaction of the U1snRNP-A protein with the upstream efficiency element of the SV40 late polyadenylation signal. Genes Dev. 1994;8:576–586. doi: 10.1101/gad.8.5.576. [DOI] [PubMed] [Google Scholar]
- 29.Lutz C S, Murthy K G K, Schek N, O'Conner P, Manley J L, Alwine J C. Interaction between the U1 snRNP-A protein and the 160-kD subunit of cleavage-polyadenylation specificity factor increases polyadenylation efficiency in vitro. Genes Dev. 1996;10:325–337. doi: 10.1101/gad.10.3.325. [DOI] [PubMed] [Google Scholar]
- 30.Minvielle-Sebastia L, Keller W. mRNA polyadenylation and its coupling to other RNA processing reactions and to transcription. Curr Opin Cell Biol. 1999;11:352–357. doi: 10.1016/S0955-0674(99)80049-0. [DOI] [PubMed] [Google Scholar]
- 31.Moreira A, Takagaki Y, Brackenridge S, Wollerton M, Manley J L, Proudfoot N J. The upstream sequence element of the C2 complement poly(A) signal activates mRNA 3′ end formation by two distinct mechanisms. Genes Dev. 1998;12:2522–2534. doi: 10.1101/gad.12.16.2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pooggin M M, Fütterer J, Skryabin K G, Hohn T. A short open reading frame terminating in front of a stable hairpin is the conserved feature in pregenomic RNA leaders of plant pararetroviruses. J Gen Virol. 1999;80:2217–2228. doi: 10.1099/0022-1317-80-8-2217. [DOI] [PubMed] [Google Scholar]
- 33.Pringle C R. Virus taxonomy—1999. Arch Virol. 2000;144:421–429. doi: 10.1007/s007050050515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qu R, Bhattacharyya M, Laco G S, de Kochko A, Rao B L S, Kaniewska M B, Elmer J S, Rochester D E, Smith C E, Beachy R N. Characterization of the genome of rice tungro bacilliform virus: comparison with Commelina yellow mottle virus and caulimoviruses. Virology. 1991;185:354–364. doi: 10.1016/0042-6822(91)90783-8. [DOI] [PubMed] [Google Scholar]
- 35.Rothnie H M. Plant mRNA 3′-end formation. Plant Mol Biol. 1996;32:43–61. doi: 10.1007/BF00039376. [DOI] [PubMed] [Google Scholar]
- 36.Rothnie H M, Chapdelaine Y, Hohn T. Pararetroviruses and retroviruses: a comparative review of viral structure and gene expression strategies. Adv Virus Res. 1994;44:1–67. doi: 10.1016/s0065-3527(08)60327-9. [DOI] [PubMed] [Google Scholar]
- 37.Rothnie H M, Reid J, Hohn T. The contribution of AAUAAA and the upstream element UUUGUA to the efficiency of mRNA 3′-end formation in plants. EMBO J. 1994;13:2200–2210. doi: 10.1002/j.1460-2075.1994.tb06497.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanfaçon H. Analysis of figwort mosaic virus (plant pararetrovirus) polyadenylation signal. Virology. 1994;198:39–49. doi: 10.1006/viro.1994.1006. [DOI] [PubMed] [Google Scholar]
- 39.Sanfaçon H, Brodmann P, Hohn T. A dissection of the cauliflower mosaic virus polyadenylation signal. Genes Dev. 1991;5:141–149. doi: 10.1101/gad.5.1.141. [DOI] [PubMed] [Google Scholar]
- 40.Sanfaçon H, Hohn T. Proximity to the promoter inhibits recognition of cauliflower mosaic virus polyadenylation signal. Nature. 1990;346:81–84. doi: 10.1038/346081a0. [DOI] [PubMed] [Google Scholar]
- 41.Sanfaçon H, Wieczorek A. Analysis of cauliflower mosaic virus RNAs in Brassica species showing a range of susceptibility to infection. Virology. 1992;190:30–39. doi: 10.1016/0042-6822(92)91189-2. [DOI] [PubMed] [Google Scholar]
- 42.Vagner S, Rüegsegger U, Gunderson S I, Keller W, Mattaj I W. Position-dependent inhibition of the cleavage step of pre-mRNA 3′-end processing by U1snRNP. RNA. 2000;6:178–188. doi: 10.1017/s1355838200991854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wahle E, Rüegsegger U. 3′-End processing of pre-mRNA in eukaryotes. FEMS Microbiol Rev. 1999;23:277–295. doi: 10.1111/j.1574-6976.1999.tb00400.x. [DOI] [PubMed] [Google Scholar]
- 44.Yin Y, Beachy R N. The regulatory regions of the rice tungro bacilliform virus promoter and interacting nuclear factors in rice (Oryza sativa L.) Plant J. 1995;7:969–980. doi: 10.1046/j.1365-313x.1995.07060969.x. [DOI] [PubMed] [Google Scholar]
- 45.Yin Y, Chen L, Beachy R. Promoter elements required for phloem-specific gene expression from the RTBV promoter in rice. Plant J. 1997;12:1179–1188. doi: 10.1046/j.1365-313x.1997.12051179.x. [DOI] [PubMed] [Google Scholar]
- 46.Yin Y, Zhu Q, Dai S, Lamb C, Beachy R N. RF2a, a bZIP transcriptional activator of the phloem-specific rice tungro bacilliform virus promoter, functions in vascular development. EMBO J. 1997;16:5247–5259. doi: 10.1093/emboj/16.17.5247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhao J, Hyman L, Moore C. Formation of mRNA 3′-ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]