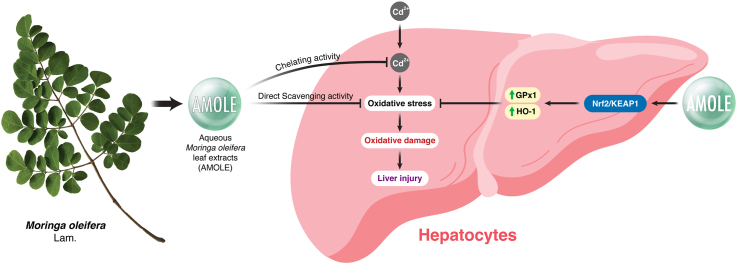
Abstract
Cadmium (Cd) is a highly harmful pollutant that poses a serious threat to human health. The liver is the primary organ for Cd accumulation, and Cd-induced hepatotoxicity has been shown to be strongly correlated with an oxidative imbalance in hepatocytes. Our previous studies in the eukaryotic model organism Saccharomyces cerevisiae revealed that not only co-treatment but also pretreatment with aqueous Moringa oleifera Lam. leaf extract (AMOLE) effectively mitigated Cd toxicity by reducing intracellular Cd accumulation and Cd-mediated oxidative stress. In this study, we therefore investigated the preventive effect of AMOLE against Cd toxicity in human HepG2 hepatocytes. The results showed that, similar to the case of the yeast model, pretreatment with AMOLE prior to Cd exposure also significantly inhibited Cd-induced oxidative stress in HepG2 cells. Untargeted LC-MS/MS-based metabolomic analysis of AMOLE revealed that its major phytochemical constituents were organic acids, particularly phenolic acids and carboxylic acids. Additionally, DPPH-HPTLC fingerprints suggested that quercetin and other flavonoids possibly contribute to the antioxidant activities of AMOLE. Based on our findings, it appears that pretreatment with AMOLE prevented Cd-induced hepatotoxicity via three possible mechanisms: i) direct elimination of free radicals by AMOLE antioxidant compounds; ii) upregulation of antioxidant defensive machinery (GPx1, and HO-1) via Nrf2 signaling cascade to improve cellular antioxidant capacity; and iii) reduction of intracellular Cd accumulation, probably by suppressing Cd uptake. These data strongly suggest the high potential of AMOLE for clinical utility in the prevention of Cd toxicity.
Keywords: Moringa oleifera Lam., Preventive effect, Cadmium, Hepatocyte, Antioxidant machinery, Metal accumulation
Graphical abstract
Highlights
-
•
AMOLE pretreatment reduces Cd toxicity in HepG2 cells.
-
•
AMOLE pretreatment suppresses Cd-induced oxidative stress.
-
•
AMOLE prevents Cd toxicity by promoting the expression of antioxidant enzymes.
-
•
AMOLE pretreatment inhibits Cd accumulation in HepG2 cells.
Abbreviations:
- AMOLE
aqueous Moringa oleifera Lam. leaf extract
- BAL
British Anti-Lewisite
- DCFH-DA
2′,7′-dichlorofluorescein diacetate
- DMPS
2,3-dimercapto-1-propanesulfonic acid
- DMSA
2,3-dimercaptosuccinic acid
- DMSO
dimethyl sulfoxide
- DPPH
2,2-diphenyl-1-picrylhydrazyl
- EDTA
ethylenediaminetetraacetic acid
- FBS
fetal bovine serum
- GPx1
glutathione peroxidase 1
- HPTLC
high-performance thin-layer chromatography
- Keap1
Kelch-like ECH-associated protein 1
- MEM
Minimum Essential Media
- MTT
methylthiazolyldiphenyl-tetrazolium bromide
- Nrf2
nuclear factor erythroid 2-related factor 2
- PBS
phosphate buffer solution
- PEG
polyethylene glycol
- HO-1
heme oxygenase-1
- H2O2
hydrogen peroxide
- Rf
retention factor
- SOD
superoxide dismutase
1. Introduction
Due to rapid urbanization and industrialization, an increase in environmental contamination with heavy metals has caused harmful effects on human health globally. Cadmium (Cd) is a ubiquitous environmental toxicant largely originating from natural occurrences and anthropogenic activities [1]. There are numerous sources of Cd, e.g. tobacco smoking, industrial emissions, and contaminated food and water [2,3]. This harmful heavy metal can enter the human body through ingestion and inhalation. The liver is the major target organ for Cd accumulation and toxicity. Exposure to Cd is associated with a high incidence of serious hepatic injury in humans such as fibrosis, necroinflammation, non-alcoholic fatty liver disease, and non-alcoholic steatohepatitis [4,5]. Chelation therapies, e.g. ethylenediaminetetraacetic acid (EDTA), British Anti-Lewisite (BAL), 2,3-dimercaptosuccinic acid (DMSA), and 2,3-dimercapto-1-propanesulfonic acid (DMPS), are current approaches for the treatment of Cd intoxication [6]. However, the clinical use of these chelating agents has several limitations such as low specificity, poor ability to cross the membrane, and serious adverse effects [7]. Hence, there is a critical need to develop a novel approach with great efficiency and a high safety profile to combat Cd poisoning.
Medicinal plants are the repository of several phytochemicals possessing a broad range of pharmacological activities. In the last decades, there has been a growing interest in the development of new therapeutic strategies to cure several diseases by using botanical sources [8]. Moringa oleifera Lam. (M. oleifera) has been widely used in traditional medicine for centuries with multiple indications, e.g. malnutrition, asthma, diabetes, anemia, cardiovascular diseases, and inflammation [9]. Leaves are the most frequently used part of M. oleifera, which can be freshly eaten, cooked, or used as dried powder. Leaves of M. oleifera contain significant sources of bioactive and phytochemicals, including flavonoids [[10], [11], [12]]. Flavonoids are a class of polyphenolic metabolites with antioxidant activity. The preclinical studies on various models of diseases demonstrated that flavonoids have ability to mitigate Cd poisoning via attenuation of oxidative stress and their metal-chelating properties [[13], [14], [15], [16]]. Moreover, previous studies on rats showed that concomitant administration of ethanolic extracts of M. oleifera leaves with heavy metals can suppress toxicity in male reproductive system [17,18]. Our previous studies on the eukaryotic model organism Saccharomyces cerevisiae revealed that the aqueous M. oleifera leaf extract (AMOLE) can inhibit Cd-induced toxicity through an inhibition of oxidative stress and a reduction of Cd accumulation [10,19]. In addition, we also found that not only the co-treatment but also the pretreatment with AMOLE significantly reduced Cd uptake, leading to an alleviation of endogenous oxidative stress caused by Cd. Therefore, AMOLE is highly promising for the therapeutic prevention of Cd-induced hepatotoxicity.
In this present study, we aimed to explore the preventive potential of AMOLE against Cd-induced hepatotoxicity using human HepG2 hepatocytes as an in vitro model. Our data clearly demonstrated that the pretreatment with AMOLE mitigated the harmful effects of Cd on HepG2 cells. These findings therefore suggest promising applications of AMOLE in the prevention of Cd toxicity and other harmful metals.
2. Materials and methods
2.1. Chemical and reagents
Hydrogen peroxide (H2O2), L-ascorbic acid, methylthiazolyldiphenyl-tetrazolium bromide (MTT), 2′,7′-dichlorofluorescein diacetate (DCFH-DA), rutin, rosmarinic acid, quercetin, 2,2-diphenyl-1-picrylhydrazyl (DPPH), HPTLC silica gel 60 F254, 2-aminoethyl diphenylborinate, dichloromethane, ethyl acetate, polyethylene glycol (PEG), dimethyl sulfoxide (DMSO) and Immobilon Western chemiluminescent HRP substrate were purchased from Sigma-Aldrich (St. Louis, MO, USA).
Cell culture media and supplements (Minimum Essential Media (MEM), fetal bovine serum (FBS) and penicillin/streptomycin were obtained from ThermoFisher Scientific (Waltham, MA, USA). Anti-Nrf2 (rabbit pAb; Cat. No. ab137550; dilution 1:1000), anti-SOD1 (rabbit mAb; Cat. No. ab51254; dilution 1:1000) and anti-GPx1 (rabbit pAb; Cat. No. ab22604; dilution 1:1000) were purchased from Abcam (Cambridge, UK). Anti-HO-1 (rabbit mAb; Cat. No. 43966; dilution 1:1000), anti-Keap1 (rabbit mAb; Cat. No. 8047; dilution 1:1000), anti-SOD2 (rabbit mAb; Cat. No. 13141; dilution 1:1000), anti-GAPDH (rabbit mAb; Cat. No. 5174; dilution 1:1000), and anti-rabbit IgG, HRP-conjugated antibody (Cat. No. 7074; dilution 1:2000) were obtained from Cell Signaling Technology (Danvers, MA, USA).
2.2. Preparation of aqueous M. oleifera leaf extract (AMOLE)
The dried M. oleifera leaf powder was purchased from Khaolaor Laboratories (Samutprakarn, Thailand). The AMOLE was prepared as described previously with some modifications [10,19]. Briefly, 5 g of dried powder leaf was added into 30 mL of water and mixed with vortex for 1 min. The supernatant of extract (AMOLE) was collected by centrifugation at 13,000 ×g for 20 min, followed by filtration with filter paper (Whatman Grade 42). The AMOLE was lyophilized and kept at 4 °C. Before use, the lyophilized AMOLE was redissolved in distilled water and further sterilized with membrane filtration (0.2 μm; Acrodisc Syringe Filters).
2.3. DPPH assay
The radical scavenging activity (RSA) of AMOLE was evaluated with DPPH assay. Serial dilutions of AMOLE (0–6 mg mL−1 in ethanol; 20 μg mL−1) were added into a 96-well microplate and incubated with DPPH solution (150 μM in ethanol; 200 μL) for 30 min. The absorbance of the sample was then determined at 517 nm with CLARIOStar microplate reader (BMG Labtech; Ortenburg; Germany). The positive controls were ascorbic acid and quercetin. The %RSA of each sample was calculated using the following equation.
where ODblank and ODsample are absorbance at 517 nm of 100 % methanol and sample, respectively. The amounts of samples that generated 50 % reduction in RSA were calculated from dose-response plot with GraphPad Prism version 9.4.1 (GraphPad Software Inc., San Diego, CA, USA).
2.4. Preparation of sample and flavonoid standards for HPTLC
Fifty milligrams of AMOLE were mixed with 500 μL of ethanol (v/v) and sonicated for 10 min. Subsequently, the AMOLE solution was centrifuged at 8000 rpm for 5 min and the supernatant was used for HPTLC analysis. The flavonoids standards were dissolved with methanol at the final concentration of 1 mg μL−1 (rutin and rosmarinic acid) or 0.1 mg μL−1 (quercetin).
2.5. High-performance thin-layer chromatography (HPTLC)
Chromatographic separation of AMOLE was conducted on Merck silica gel 60 F254 glass HPTLC plates (20 × 10 cm) with ethyl acetate: toluene: formic acid: water (6:1:1.4:1 (v/v/v/v)) as mobile phase. Briefly, AMOLE (2−6 μL/band) and flavonoid standards (2 μL/band) were applied to the HPTLC plates by Linomat 5 (Camag; Muttenz, Switzerland) with following setting: band length, 8 mm; distance between tracks, 11.4 mm; distance from the lower edge, 8 mm; and distance from the left plate edge, 20 mm. After 70 mm development, the HPTLC plates were visualized at 254 nm using Visualizer 2 (Camag; Muttenz, Switzerland). To observe the presence of flavonoids in the sample, HPTLC plates were further heated at 110 °C for 3 min and subsequently immersed in natural product/polyethylene glycol reagent (NP/PEG). For imaging, the derivatized plates were captured at 366 nm with Visualizer 2.
2.6. HPTLC-DPPH assay
HPTLC-DPPH was conducted to evaluate the antioxidant potential of phytochemicals in ME. After chromatographic development, the HPTLC plate was further derivatized with DPPH solution (0.02 % DPPH in methanol) and incubated in the dark for 2 h. The chromatogram was imaged under white light. Compounds with free radical scavenging properties were detected as yellowish bands against a purple background.
2.7. Sample preparation and liquid chromatography with tandem mass spectrometry (LC-MS/MS) configuration for untargeted metabolomics analysis
The concentration of AMOLE was adjusted to 1 mg mL−1 with deionized water. A total of 15 mL of the extract was subjected to a 3 kDa molecular weight cut-off (GE Healthcare Co., Amersham, UK). The flow-through solution was cleaned up using solid phase extraction as described previously [20]. The cleaned-up extract was dried under a speed vacuum. The experiment was conducted in 3 biological replications (n = 3).
The metabolome profiling was analyzed using Orbitrap HF-X mass spectroscopy with an electrospray ion source operated at 3.5 kV. The dried extract was solubilized in 0.1 % formic acid in a solubilization solution (50 % methanol/50 % Water). Five μL of the samples were loaded on an analytical column (Ascentis® Express C18, 2.7 μm HPLC Column). The column oven temperature was 60 °C. The mobile phase was composed of water with 0.1 % formic acid and acetonitrile with 0.1 % formic acid. The total time of each analysis was 35 min. A blank sample (0.1 % formic acid/water) was administered after every injection. MS spectral data were acquired using a data dependent acquisition dynamical choosing the most abundant precursor ions from the survey scan (55–650 m/z) with charge states (+1 and + 2) for high-energy collision dissociation fragmentation. Dynamic exclusion duration was 10 s. Isolation of precursors was performed with a 1.4 m/z and MS/MS scans were acquired with a starting mass of 100 m/z. Survey scans were acquired at a resolution of 120,000. The resolution for fragmentation spectra was set to 30,000. After integrating the target m/z ions, the chemical formula was predicted by comparing with the online databases, including ChemSpider (http://www.chemspider.com) and mzCloud (https://www.mzcloud.org) to confirm and identify the compounds. The candidate metabolites with the highest MS/MS coverage scores were chosen for annotation. Among these, the metabolites with mzClound best match score >50 and peak area >1 × 109 AU were reported. The functional analysis was analyzed based on the KEGG pathway by MetaboAnalyst 6.0 (accessed on March 21, 2024) [21].
2.8. Cell culture and condition
Human hepatocellular carcinoma HepG2 cells (RRID: CVCL_0027) were purchased from CLS GmbH (Eppelheim, Germany). The authentication of cell lines was conducted by CLS GmbH using short tandem repeat (STR) profiling. The culture medium is MEM supplementing with penicillin (100 U mL−1), streptomycin (100 μg mL−1), and FBS (10 %). HepG2 cells were maintained at 37 °C in a humidified atmosphere with 95 % air and 5 % CO2. Routine monitoring for mycoplasma contamination was performed using the Universal Mycoplasma Detection Kit (ATCC; Manassas, VA, USA).
2.9. Cell viability assay
The survival of cells was evaluated with the MTT viability assay. HepG2 cells were seeded into a 96-well microplate (1 × 104 cells/well) and cultured for 24 h. Following the indicated treatments, the experimental medium was removed; treated cells were gently rinsed twice with phosphate buffer solution (PBS; pH 7.4); MTT solution (0.5 mg mL−1 in serum-free MEM; 100 μL) was subsequently added into each well; and plates were subsequently incubated in dark for 3 h. After incubation, the MTT solution was gently discarded and DMSO (200 μL) was added into each well to solubilize formazan crystals. The optical density of the formazan solution was observed with a CLARIOStar microplate reader at 570 nm.
2.10. Evaluation of intracellular oxidative status
The intracellular oxidative statuses were determined with the DCFH-DA-based approach. Briefly, HepG2 cells were placed into a 96-well, black microplate with clear bottom (1 × 104 cells/well) and cultured for an additional 24 h prior to treatments. Following the indicated treatments, cells were gently rinsed twice with PBS and stained in the dark with DCFH-DA solution (5 μM in serum-free MEM). After 30 min staining, cells were washed with PBS and serum-free medium was added into each well. The intracellular fluorescent signal was recorded with a CLARIOStar microplate reader (excitation wavelength, 485 nm; emission wavelength, 530 nm).
2.11. Western blot analysis
HepG2 cells were seeded into a 6-well plate (3 × 105 cells/well) and cultured for 24 h. The treatment protocols were similar to the cell viability study. Following the indicated treatments, cells were harvested on ice with a cell-scraper and lysed with radio-immunoprecipitation assay (RIPA) buffer (Cat. no. 9806; Cell Signaling Technology) containing Protease/Phosphatase Inhibitor Cocktail (Cat. no. 5872; Cell Signaling Technology). Protein lysates were resolved by SDS-PAGE and transferred onto a polyvinylidene difluoride membrane (Bio-Rad Laboratories, Hercules, CA, USA). The membranes were subsequently blocked with blocking solution (5 % non-fat milk in TBS-T) for 1 h at room temperature and hybridized overnight with primary antibodies at 4 °C. GAPDH was used as a loading control. Following incubation with specific primary antibodies, membranes were washed three times with TBS-T and stained with HRP-conjugated secondary antibody for 1 h at room temperature. The chemiluminescent signal was developed with Immobilon Western chemiluminescent HRP substrate and imaged with an ImageQuant LAS 4000 (GE Healthcare, Chicago, IL, USA). The immunoblot was densitometric analyzed with ImageJ software (version 1.53t; U.S. National Institutes of Health, Bethesda, MD, USA). Full unedited blots are shown in Fig. S1.
2.12. Determination of intracellular Cd contents
The cell culture conditions and treatment protocols were similar to Western blot experiments. After the indicated treatments, cells were detached with trypsin and washed twice with PBS. Cell pellets were then subjected to acid digestion. One mL of each cell pellet was applied to the digestion tube containing 5 mL of 65 % nitric acid (Suprapur®, Supleco). After open reflux digestion at 100 °C in a block digester for 1 h inside a fume hood, 1 mL of H2O2 was added. The samples were heated at 100 °C for another hour. Finally, the digested samples were cooled at room temperature and subsequently filtered with filter paper grade 42. The sample volume was adjusted to 10 mL with pure water. The Cd concentration was measured by flame atomic absorption spectroscopy (FAAS) using a hollow cathode lamp for Cd, with the wavelength 228.8 nm using an air/acetylene flame. The results were expressed as mg L−1. Certified Cd standard solution (MilliporeSigma, Burlington, MA, USA) was used for constructing the calibration curves of the respective analysis. The limit of detection in the Cd measurements was 10 ng L−1.
2.13. Statistical analysis
All statistical analyses were conducted with GraphPad Prism. Statistical difference between treatment groups was calculated with unpaired t-test or one-way ANOVA with Tukey's post hoc analysis. The data was presented as the mean ± standard errors of the mean (SEMs) of at least three independent experiments.
3. Results
3.1. Cd causes toxicity on HepG2 in a dose-dependent fashion
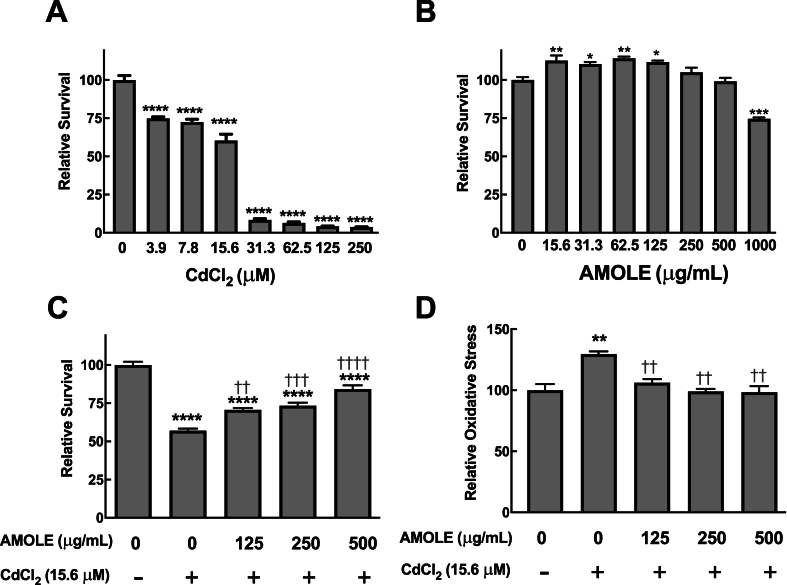
Since Cd primarily accumulates in the liver, it is recognized as the main target organ of Cd toxicity [5,22]. To investigate the hepatotoxicity of Cd, human HepG2 hepatocyte was used as a cell culture model of the human liver. The results of the MTT viability assay demonstrated that exposure to CdCl2 induced hepatotoxicity in a dose-dependent manner, with approximately 40 % of HepG2 cell death observed upon the treatment with 15.6 μM CdCl2 (Fig. 1A). Therefore, this concentration was chosen for further investigation to assess the hepatoprotective effects of AMOLE against CdCl2.
Fig. 1.
Pretreatment with AMOLE suppressed Cd toxicity in HepG2 cells through inhibition of oxidative stress generation. (A) HepG2 cells were treated with CdCl2 (3.9–250 μM; 6 h) or (B) AMOLE (15.6–1000 μg mL−1; 24 h). MTT assay was performed to determine cell survival after treatments. (C, D) HepG2 cells were pre-incubated with AMOLE (125–500 μg mL−1; 24 h), followed by CdCl2 treatment (15.6 μM; 6 h). The cell survival was evaluated with an MTT assay, while intracellular oxidative status was observed with a DCFH-DA-based approach. The cell viability and oxidative status following treatment were calculated relative to the untreated control (n = 3; mean ± SEM; *, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001 v.s. untreated control; ††, p < 0.01; †††, p < 0.001; ††††, p < 0.0001 v.s. CdCl2 group).
3.2. AMOLE pretreatment inhibits toxic effects of Cd on HepG2 cells
Prior to examining the protective activities of AMOLE, the safety concentration of AMOLE on HepG2 cells had to be first evaluated. Twenty-four-hour treatment with AMOLE at 15.6 μg mL−1 to 500 μg mL−1 did not cause harmful effects on HepG2 cells (Fig. 1B). The cytotoxicity of AMOLE was observed at 1000 μg mL−1. Hence, AMOLE at 125–500 μg mL−1 was used for further investigation due to their safety profiles.
To investigate the beneficial effects of AMOLE against CdCl2 toxicity, HepG2 cells were pre-treated with AMOLE for 24 h prior to CdCl2 exposure. Pretreatment with AMOLE significantly inhibited CdCl2-induced injury in HepG2 cells (Fig. 1C). These results suggest the potential benefits of AMOLE as a preventive approach against the hepatotoxicity of Cd. Nevertheless, it should be noted that the protective effects of AMOLE were not observed when co-treated with CdCl2 (Fig. S2).
3.3. AMOLE pretreatment suppresses Cd-induced oxidative stress on HepG2 cells
Previous reports showed that liver toxicity of Cd is strongly correlated with oxidative stress [23,24]. To observe cellular oxidative status, DCFH-DA-based assay was performed after CdCl2 treatment. As expected, exposure to CdCl2 caused an induction of oxidative stress in HepG2 cells by approximately 1.3 times. On the other hand, the redox status of AMOLE-treated HepG2 cells was maintained at basal level even after CdCl2 challenges (Fig. 1D). This data suggest that the beneficial activity of AMOLE against CdCl2-induced hepatotoxicity could be due to an inhibition of oxidative stress.
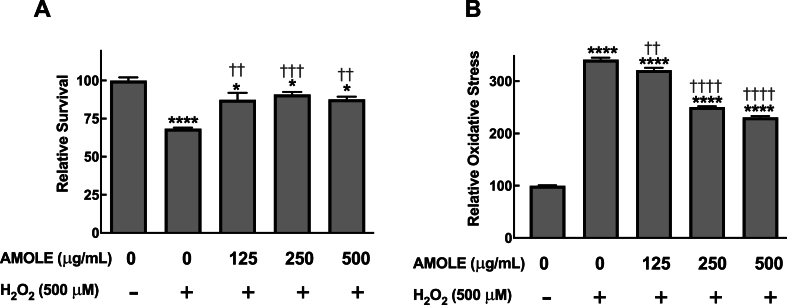
3.4. AMOLE pretreatment inhibits H2O2-induced toxicity on HepG2 cells
Hydrogen peroxide (H2O2) is a widely used agent for inducing oxidative stress in vitro [[25], [26], [27]]. In this study, we used H2O2 as an additional model of oxidative stress inducer to validate the protective activities of AMOLE against oxidative injury. Treatment with H2O2 reduced cell viability of HepG2 by approximately 30 % (Fig. 2A). In addition, the oxidative stress levels of HepG2 cells were strikingly increased by 3.4 times after H2O2 challenge (Fig. 2B). Similar to the case of CdCl2 hepatotoxicity, pretreatment with AMOLE significantly inhibited the H2O2 cytotoxicity on HepG2 cells (Fig. 2A). Moreover, results from the DCFH-DA assay showed that the AMOLE pretreatment attenuated stimulation of oxidative stress in H2O2-challenged HepG2 cells (Fig. 2B). Hence, these results strongly confirm that the inhibitory activities of AMOLE against oxidative imbalance could be a potential mechanism for hepatopreventive effects of AMOLE.
Fig. 2.
Pretreatment with AMOLE prevented H2O2-induced toxicity in HepG2 cells through suppression of oxidative stress. HepG2 cells were pretreated with AMOLE (125–500 μg mL−1; 24 h), then exposed to H2O2 (500 μM; 1 h). (A) The cell viability following treatment was observed with MTT assay (B) The intracellular oxidative status was evaluated by DCFH-DA assay. The cell viability and oxidative status following treatment were calculated relative to the untreated control (n = 3; mean ± SEM; *, p < 0.05; ****, p < 0.0001 v.s. untreated control; ††, p < 0.01; †††, p < 0.001; ††††, p < 0.0001 v.s. H2O2 group).
3.5. AMOLE possesses strong antioxidant activities
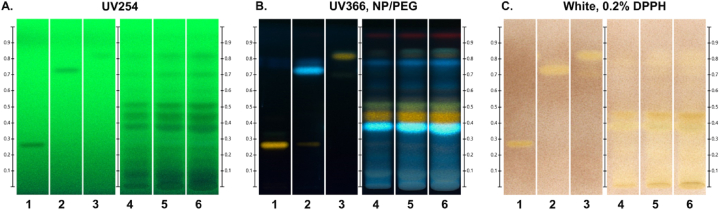
Our above-mentioned results revealed that the hepatopreventive effects of AMOLE are likely related to its ability to inhibit endogenous oxidative stress (Fig. 1, Fig. 2), suggesting that AMOLE may possess antioxidant properties capable of neutralizing harmful free radicals induced by CdCl2. To evaluate the free radical scavenging capacities of AMOLE, the traditional DPPH assay was conducted. Results from the DPPH assay showed that AMOLE has a good antioxidant profile with IC50 value for DPPH scavenging activity of AMOLE at 312.2 ± 19.9 μg mL−1 (IC50 of ascorbic acid = 4.9 ± 0.1 μg mL−1; quercetin = 5.2 ± 0.1 μg mL−1).
Rutin, rosmarinic acid, and quercetin are flavonoids with antioxidant capacity that have been previously reported in M. oleifera with different extraction systems [[28], [29], [30]]. The presence of flavonoids in a sample can be detected and characterized with HPTLC-based approach. The flavonoids were visualized on HPTLC plates as dark bands under UV light at 254 nm (Fig. 3A). Following derivatization with NP/PEG reagent, flavonoids were presented on HPTLC plate as bright blue, red-orange and yellow-green fluorescence chromatograms under UV light at 365 nm (Fig. 3B). The HPTLC fingerprints showed that only the band of quercetin (Rf = 0.82), but not rutin or rosmarinic acid, was detected in AMOLE (Fig. 3A and B). Notably, in addition to quercetin, HPTLC phytochemical profiling demonstrated that other flavonoids were contained in AMOLE (Fig. 3A and B). To investigate the antioxidant capacity of active components in AMOLE, the HPTLC plate was further sprayed with DPPH reagent. DPPH-HPTLC chromatograms demonstrated that AMOLE has potent antioxidant activity, as determined by an increase in the intensity of the yellow zone (Fig. 3C; Lane 4–6). Moreover, the DPPH-HPTLC fingerprints suggested that quercetin and other flavonoids possibly contribute to the antioxidant activities of AMOLE (Fig. 3).
Fig. 3.
The antioxidant activity of AMOLE is possibly due to quercetin and other flavonoids. HPTLC chromatogram of flavonoid standards (1, rutin (Rf, 0.27); 2, rosmarinic acid (Rf, 0.72); 3, quercetin (Rf, 0.82)), and AMOLE (4, AMOLE 200 ng; 5, AMOLE 400 ng; 6, AMOLE 600 ng). (A) The HPTLC plate was visualized with UV light at 254 nm. (B) The HPTLC plate was visualized under UV light at 366 nm after derivatization with NP/PEG reagent. (C) The HPTLC plate was imaged under white light after spraying with 0.2 % DPPH reagent.
3.6. Untargeted mass spectrometry-based metabolome profiling of AMOLE
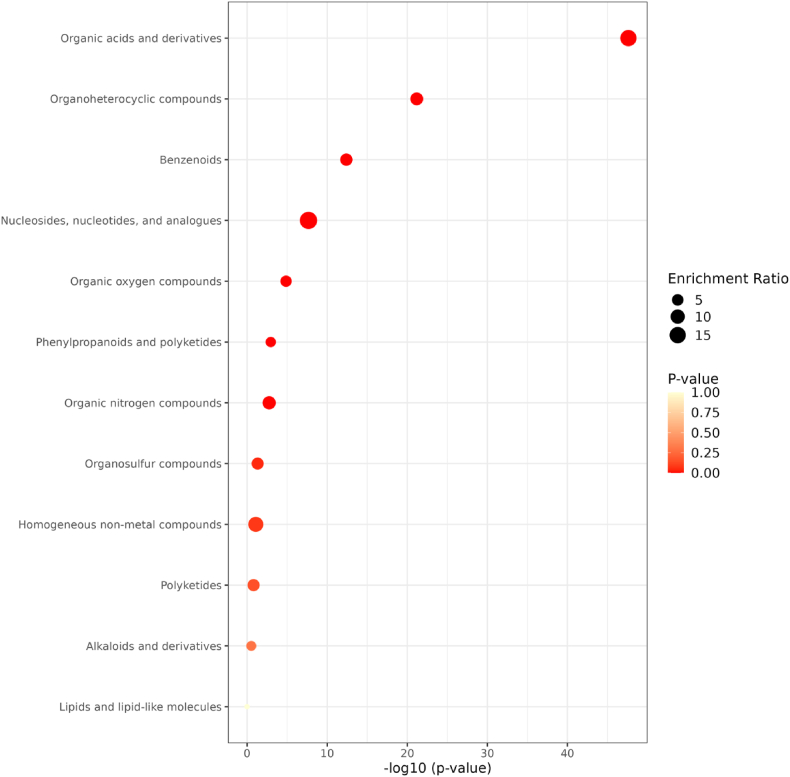
In our previous report, partial phenolic and flavonoid constituents of AMOLE were qualitatively and quantitatively analyzed using the high-performance liquid chromatography (HPLC) approach [10]. In this study, to further identify the comprehensive phytochemical composition of AMOLE, the untargeted metabolomics profiling analysis of AMOLE was performed using LC-MS/MS. Metabolite identification was successfully completed, with a total of 849 compounds identified. To prioritize the identification of metabolites with high confidence levels from the extract, specific criteria were employed. Particularly, metabolites were deemed significant if they exhibited a similarity matching score of over 60 in the MS2 spectrum and a retention time (RT) deviation of less than 0.2 min between two biological replications. Applying these criteria, a total of 302 metabolites were fully annotated and identified, subsequently utilized for further analysis. Table 1 lists the most abundant metabolites associated with the KEGG pathway (Fig. 4). All identified metabolites were classified and grouped into organic acid and their derivative with the highest rank (p-value 2.45e−48 and FDR 8.57e−47) (Fig. 4).
Table 1.
The most abundant metabolites detected from AMOLE.
| Compounds | Formula | Mass (Da) | RT (min) | Area (AU) |
|---|---|---|---|---|
| Cyclohexenone | C6H8O | 96.06 | 7.44 | 8.50E+10 |
| HO-dPEG8-OH | C16H34O9 | 370.22 | 7.12 | 1.67E+10 |
| Benzaldehyde | C7H6O | 106.04 | 5.60 | 1.66E+10 |
| Choline | C5H13NO | 103.10 | 4.77 | 6.28E+09 |
| Cinnamic acid | C9H8O2 | 148.05 | 6.99 | 6.02E+09 |
| L-Norleucine | C6H13NO2 | 131.09 | 6.41 | 4.99E+09 |
| Heptaethylene Glycol | C14H30O8 | 326.19 | 7.09 | 4.91E+09 |
| D-(+)-Proline | C5H9NO2 | 115.06 | 4.93 | 4.59E+09 |
| D-(+)-Pyroglutamic Acid | C5H7NO3 | 129.04 | 4.82 | 4.29E+09 |
| 1-[(3-Carboxypropyl)amino]-1-deoxy-beta-D-fructofuranose | C10H19NO7 | 265.12 | 4.80 | 3.82E+09 |
| Furfural | C5H4O2 | 96.02 | 4.90 | 3.79E+09 |
| L-Isoleucine | C6H13NO2 | 131.09 | 6.13 | 3.06E+09 |
| (2S)-3-Methyl-2-({[(3S,4S,5R)-2,3,4-trihydroxy-5-(hydroxymethyl)tetrahydro-2-furanyl]methylamino)butanoic acid | C11H21NO7 | 279.13 | 5.19 | 2.94E+09 |
| Valine | C5H11NO2 | 117.08 | 5.16 | 2.84E+09 |
| (2S)-4-Methyl-2-({[(3S,4S,5R)-2,3,4-trihydroxy-5-(hydroxymethyl)tetrahydro-2-furanyl]methyl}amino)pentanoic acid | C12H23NO7 | 293.15 | 6.39 | 2.73E+09 |
| Adenosine | C10H13N5O4 | 267.10 | 5.78 | 2.71E+09 |
| Adenine | C5H5N5 | 135.05 | 4.99 | 2.62E+09 |
| DL-Serine | C3H7NO3 | 105.04 | 4.82 | 2.21E+09 |
| 13-(dodecan-2-yl)-6-(1-hydroxyethyl)-3-(hydroxymethyl)-12-methyl-9-(propan-2-yl)-1-oxa-4,7,10-triazacyclotridecane-2,5,8,11-tetrone | C28H51N3O7 | 563.35 | 7.28 | 2.15E+09 |
| 3-(3,4-dihydroxyphenyl)propanoic acid | C9H10O4 | 164.05 | 5.78 | 2.08E+09 |
| trans-3-Indoleacrylic acid | C11H9NO2 | 187.06 | 7.05 | 1.83E+09 |
| Benzaldehyde | C7H6O | 106.04 | 7.02 | 1.69E+09 |
| 2-Amino-9-(2-deoxypentofuranosyl)-3,9-dihydro-6H-purin-6-one | C10H13N5O4 | 267.10 | 4.83 | 1.69E+09 |
| Glu-Glu | C10H16N2O7 | 276.10 | 4.83 | 1.61E+09 |
| DL-Arginine | C6H14N4O2 | 174.11 | 4.64 | 1.56E+09 |
| 8-hydroxy-deoxyguanosine | C10H13N5O5 | 283.09 | 5.92 | 1.51E+09 |
| Crotonic acid | C4H6O2 | 86.04 | 4.79 | 1.50E+09 |
| O-ureido-D-serine | C4H9N3O4 | 163.06 | 4.53 | 1.44E+09 |
| (2S)-3-Phenyl-2-({[(3S,4S,5R)-2,3,4-trihydroxy-5-(hydroxymethyl)tetrahydro-2-furanyl]methyl}amino)propanoic acid | C15H21NO7 | 327.13 | 6.97 | 1.34E+09 |
| Lotaustralin | C11H19NO6 | 261.12 | 5.19 | 1.32E+09 |
| N-Methylpyrrolidone | C5H9NO | 99.07 | 7.17 | 1.31E+09 |
| Maleamic acid | C4H5NO3 | 115.03 | 4.84 | 1.16E+09 |
| Phosphoric acid | H3PO4 | 97.98 | 4.89 | 1.09E+09 |
| Methyl 3,4,5-trimethoxycinnamate | C13H16O5 | 252.10 | 5.60 | 1.03E+09 |
| NOP | C13H12O3 | 216.08 | 5.60 | 1.01E+09 |
| Glutarylcarnitine | C12H21NO6 | 275.14 | 6.39 | 1.00E+09 |
Fig. 4.
Functional enrichment of AMOLE metabolomicprofile by dot plot analysis. The plot illustrates the functional annotation of the identified metabolites. The horizontal axis represents the various classifications of metabolite entries, while the vertical axis indicates the -log (p-value), color-coded from red to yellow (ranging from 2.45e-48 to 0.297). The size of the dots corresponds to the enrichment ratio, which is calculated by dividing the number of input metabolites assigned to a specific pathway by the total number of metabolites annotated to that pathway. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
3.7. AMOLE prevents Cd toxicity in HepG2 cells by enhancing the expression of antioxidant enzymes
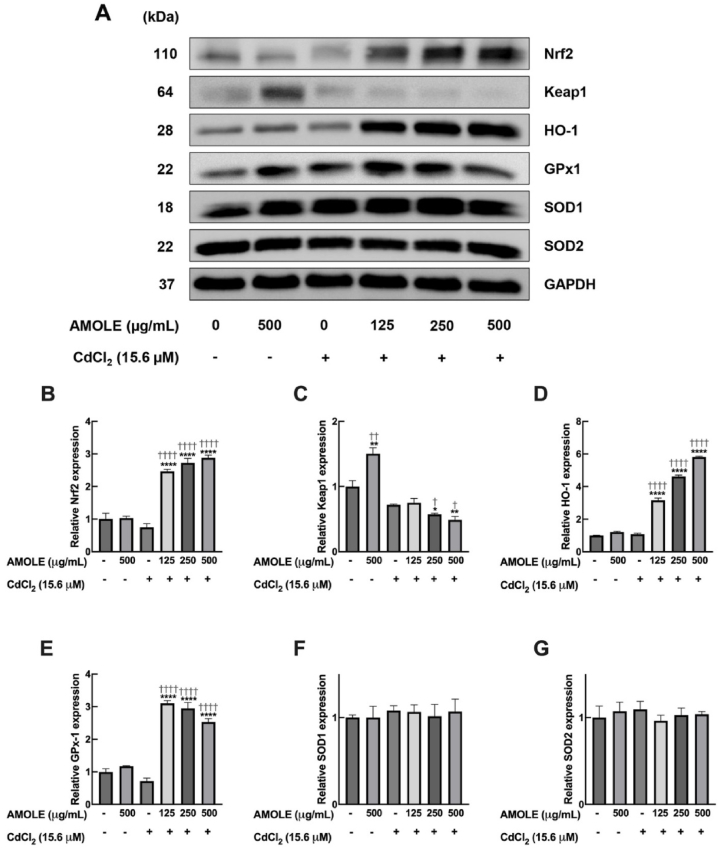
Another potential mechanism for the preventive activity of AMOLE against Cd could be an upregulation of cellular antioxidant enzymes in hepatocytes. The nuclear factor erythroid 2-related factor 2 (Nrf2) is a master regulator of redox equilibrium. Stimulation of the Nrf2 pathway leads to an upregulation of downstream antioxidant machinery, e.g. heme oxygenase-1 (HO-1), glutathione peroxidase 1 (GPx1), and superoxide dismutases (SODs) [25,31,32]. Moreover, previous studies on various cell types have demonstrated that the activation of the Nrf2 cascade is a crucial mechanism for the adaptive response that protects cells from Cd-induced oxidative damage [[33], [34], [35], [36]]. Hence, the stimulation of the Nrf2 system could be a potential mechanism underlying the protective activities of AMOLE against Cd toxicity. Data from Western blot analysis demonstrated that the pretreatment with AMOLE promoted the total levels of Nrf2 protein in HepG2 cells upon CdCl2 exposure (Fig. 5A and B). This data suggests that the activation of Nrf2 cascade could be associated with the beneficial activities of AMOLE against Cd toxicity. Kelch-like ECH-associated protein 1 (Keap1) is an essential negative regulator of Nrf2. Under physiological conditions, Keap1 targets Nrf2 for ubiquitin-proteasomal degradation. Hence, the total level of Nrf2 at homeostatic conditions is maintained at a relatively low level. In response to oxidative insults, Nrf2 dissociates from Keap1 and subsequently enters into the nucleus to stimulate the transcription of several antioxidant genes [37]. Parallel to total Nrf2 protein levels, supplementation with AMOLE repressed Keap1 expression upon CdCl2 exposure (Fig. 5A and C). These results support the contribution of the Nrf2 cascade in the preventive mechanism of AMOLE against CdCl2 toxicity. Of note, the effects of AMOLE on Keap1 expression are associated with CdCl2 exposure. In the absence of CdCl2, AMOLE strongly upregulated Keap1 protein levels. Whereas, in the presence of CdCl2 stimulation, AMOLE downregulated Keap1 expression (Fig. 5A and C).
Fig. 5.
Pretreatment with AMOLE upregulated the antioxidant machinery of HepG2 cells in response to CdCl2. HepG2 cells were pre-treated with AMOLE (125–500 μg mL−1; 24 h), then incubated with CdCl2 (15.6 μM; 6 h). After treatment, the expressions of indicated proteins were detected with Western blot analysis. (A) The expression of indicated proteins after the treatment was detected with Western blot analysis. (B-G) The band densities of indicated proteins from Western blot experiments were estimated and normalized against internal control GAPDH. The expressions of certain proteins in treatment groups were calculated relative to the untreated control (n = 3; mean ± SEM; *, p < 0.05; **, p < 0.01; ****, p < 0.0001 v.s. untreated control; †, p < 0.05; ††, p < 0.01; ††††p < 0.0001 v.s. CdCl2 group).
We then further investigated the effects of AMOLE on the protein expression of Nrf2's downstream targets, including HO-1, GPx1, SOD1 and SOD2. HO-1 is a stress-responsive cytoprotective enzyme that plays a role in the maintenance of cellular redox status [38]. Results from Western blot analysis demonstrated that the HO-1 protein levels of HepG2 cells were strongly upregulated by AMOLE in a concentration-dependent fashion upon CdCl2 treatment (Fig. 5A and D). These data suggested a contribution of HO-1 in the preventive activities of AMOLE. GPx1 is an antioxidant enzyme that contributes to the detoxification of H2O2 and lipid hydroperoxide [39]. Supplementation with AMOLE significantly promoted the GPx1 expression of HepG2 cells approximately 2.5–3 times after CdCl2 exposure (Fig. 5A and E). This information suggested that the upregulation of GPx1 could be involved in beneficial mechanisms of AMOLE against the deleterious effects of Cd. We also observed the effects of AMOLE on the expression of two major isoforms of human SODs, i.e. SOD1 and SOD2. These two isoforms have distinct characteristics in catalytic metal ions (i.e., Cu/Zn-SOD1 and Mn-SOD2) and localization (SOD1, cytoplasm, peroxisome and mitochondrial membrane space; SOD2, mitochondrial matrix) [40,41]. The SODs family is antioxidant machinery that plays a vital role in the elimination of superoxide free radicals. Unlike HO-1 and GPx1, pretreatment with AMOLE did not alter the levels of protein expression of both SOD1 and SOD2 after CdCl2 challenge (Fig. 5A, F, and G). These results can imply that the induction of SODs may not be involved in the hepatopreventive effects of AMOLE against Cd. Altogether, results from Western blot analysis demonstrated that the supplementation with AMOLE could upregulate Nrf2's downstream targets, HO-1 and GPx1, in response to Cd exposure. The increase in the expression of these antioxidant enzymes could enhance the capacity of hepatic cells to combat Cd-induced oxidative stress.
3.8. AMOLE pretreatment inhibits Cd accumulation in HepG2 cells
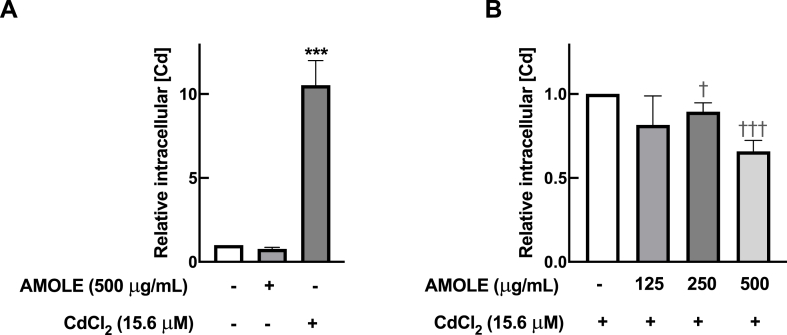
In addition to the direct scavenging activity and the upregulation of antioxidant machinery, it is possible that AMOLE exerts their hepatopreventive activities against Cd by suppressing Cd accumulation. To evaluate the levels of intracellular Cd after the treatments, FAAS analysis was conducted. The levels of intracellular concentration of Cd in HepG2 cells were dramatically increased by 10.5 times after exposure to CdCl2 (Fig. 6A). The pre-incubation with AMOLE significantly suppressed the accumulation of Cd in hepatocytes; specifically, 250 μg mL−1 and 500 μg mL−1 of AMOLE reduced the intracellular concentration of Cd by 11 % and 35 %, respectively, after CdCl2 challenge (Fig. 6B). Hence, these findings suggested that another potential mechanism by which AMOLE prevents hepatocytes from Cd toxicity could be the prevention of Cd accumulation.
Fig. 6.
Pretreatment with AMOLE suppressed intracellular Cd accumulation in HepG2 cells. HepG2 cells were pre-treated with AMOLE (125–500 μg mL−1; 24 h), then exposed to CdCl2 (15.6 μM; 6 h). Following treatments, the intracellular concentrations of Cd of hepatocytes were measured with FAAS method. The intracellular concentrations of Cd were calculated relatively to (A) untreated control (n = 5; mean ± SEM; ***, p < 0.001 v.s. untreated control); and (B) Cd-treated cells (n = 5; mean ± SEM; †, p < 0.05; †††p < 0.001 v.s. Cd-treated group).
4. Discussion
This research study offers novel and invaluable knowledge into the protective benefits of AMOLE against Cd poisoning on hepatocytes. Results from MTT viability assay demonstrated the therapeutic potentials of AMOLE in prevention of Cd-induced-hepatotoxicity (Fig. 1C). The major mechanism for liver injuries by Cd is an induction of cellular oxidative stress [[42], [43], [44], [45]]. Consistent with these previous reports, we showed that intracellular oxidative stress was induced in HepG2 cells after Cd exposure, as determined by an increase in oxidation of DCFH fluorescent dye. The pretreatment with AMOLE inhibited the generation of oxidative imbalance caused by Cd (Fig. 1D). These results suggested that the preventive effects of AMOLE against Cd-induced liver injuries could be due to an attenuation of oxidative stress. The contribution of AMOLE to maintaining cellular redox balance was validated using a strong oxidative inducer, H2O2. Parallel to Cd experiments (Fig. 1), the pretreatment with AMOLE on HepG2 cells significantly inhibited the generation of oxidative stress and oxidative damage upon H2O2 exposure (Fig. 2). These data strengthen our hypothesis that the inhibition of oxidative imbalance is an essential factor for the hepatopreventive roles of AMOLE. Nevertheless, since it has been reported that Cd can trigger the intracellular formation of H2O2 [[46], [47], [48]], the elevated oxidative stress upon Cd exposure and/or H2O2 treatment may be partially due to an increase in intracellular levels of H2O2. Although DCFH-DA, a fluorescent dye frequently used for assessing oxidative stress, can be used for detecting intracellular H2O2, its deesterified form DCFH does not directly react with H2O2 and can be oxidized by other oxidizing species [49]. Therefore, it is not a reliable probe for measuring intracellular H2O2. To better understand the relationship between intracellular H2O2 production and Cd toxicity, future studies should incorporate the H2O2 removal enzyme catalase into the DCFH-DA-based assay [33,50,51]. Moreover, previous studies have demonstrated that Cd-induced oxidative stress can lead to hepatotoxicity through multiple pathways, such as apoptosis [52,53], ferroptosis [54], and autophagy [55,56]. Consequently, future research focusing on the specific characteristics of cell death is essential to better elucidate the beneficial effects of AMOLE against Cd poisoning.
In this study, we proposed three possible mechanisms for the hepatopreventive benefits of AMOLE against Cd toxicity. First, AMOLE could directly neutralize free radicals that are generated during oxidative stress, possibly through its capability to scavenge free radicals. According to DPPH-HPTLC fingerprint, we have identified that quercetin is one of the essential flavonoids in AMOLE that could contribute to the free-radical-scavenging properties (Fig. 3). In addition to flavonoids, phenolic acids are also well-known as major antioxidant compounds in plants, which are capable of directly neutralizing free radicals. In this study, the results of metabolomics analysis revealed that the primary constituents of AMOLE were organic acid compounds including phenolic acids such as cinnamic acid (Fig. 4). In this study, since the hepatocytes were pre-incubated with AMOLE for 24 h and washed with PBS prior to Cd challenge, substantial quantities of phenolic acids and/or flavonoids in AMOLE must be accumulated and presented intracellularly during oxidative stress to directly eradicate free radical species. Further analyses on intracellular concentrations of phenolic acids and flavonoids in hepatocytes are required to support this proposed hypothesis.
Second, AMOLE enhances the capacity of hepatocytes to eradicate oxidative insults by upregulation of cellular antioxidant machinery. HO-1 is an inducible enzyme that plays crucial roles in the anti-inflammatory and antioxidant activities of hepatocytes in response to toxic xenobiotics [57,58]. Under physiological conditions, expression of HO-1 is maintained at low levels. The levels of HO-1 can be profoundly upregulated by several inducers, e.g. heavy metals, reactive oxygen species, and inflammatory cytokines. Enhanced expression of HO-1 is an adaptive response for maintaining the redox status of cells [59]. GPx1 is the most abundant isoform of the GPx family. GPx1 plays a pivotal role in the elimination of hydrogen peroxide, hydroperoxides, and soluble lipid hydroperoxides [39]. Our Western blot data clearly demonstrated that AMOLE supplementation promotes the expression of HO-1 and GPx1 of hepatocytes in response to Cd exposure (Fig. 5A, D, and E). Increased levels of HO-1 and GPx1 by AMOLE could contribute to an enhanced resistance of hepatocytes against oxidative insults.
The upregulation of HO-1 and GPx1 by AMOLE could be closely regulated by the Nrf2 system. Our results demonstrated that AMOLE supplementation significantly enhanced the expression of Nrf2 and suppressed the expression of Keap1 in Cd-treated hepatocytes (Fig. 5A−C). The data from our present study is consistent with previous observations on M. oleifera leaf extract with different preclinical models. The ethanolic M. oleifera leaf extract improved oxidative metabolism of skeletal muscle cells by the activation of Nrf2 network and the enhancement of activities of antioxidant machinery (SOD, CAT, GPx, glutathione transferase) [60]. AMOLE exerted its antioxidant and anti-inflammatory activities on rats with titanium-dioxide-induced nephrotoxicity via upregulation of Nrf2/HO-1 cascade [61]. In an alcoholic liver disease model, supplementation with AMOLE ameliorated liver inflammation and liver injury in vivo through regulation of Nrf2 pathway [62]. Moreover, HPTLC-DPPH chromatogram suggested that quercetin could be one of the principal phytochemicals of AMOLE (Fig. 3). Numerous studies on preclinical models of neurodegenerative diseases [63,64], cardiomyopathy [65], glaucoma [66], idiopathic pulmonary fibrosis [67], and liver injury [[68], [69], [70]] demonstrated that quercetin indirectly counteract oxidative stress by modulating Nrf2 pathway. In addition to quercetin, HPTLC-DPPH fingerprint also demonstrated that AMOLE contains several bioflavonoids with promising antioxidant activities (Fig. 3). A number of flavonoids have been reported as Nrf2 activators. The stimulation of Nrf2 cascade by these flavonoids has been shown to be associated with their protective activities against oxidative damage [71,72]. In addition to flavonoids, it has been reported that pretreatment with certain phenolic acids could reduce Cd toxicity. For instance, the pretreatment with ferulic acid, a natural polyphenol derived from cinnamic acid, could protect hepatocytes from damage caused by Cd through its role in the activation of the Keap1/Nrf2 signaling pathway [73]. Hence, some flavonoids and/or organic acids in AMOLE could contribute to the activation of the Nrf2 system in response to Cd toxicity. Altogether, these previous reports supported our hypothesis that the beneficial effects of AMOLE against Cd are tightly regulated by the Nrf2 signaling network. Our current study investigated the impact of AMOLE on the Nrf2 cascade by assessing changes in protein expression levels of key modulators and effectors after the treatments. To gain a deeper understanding of the roles of this redox modulator in the hepatopreventive effects of AMOLE, the nuclear translocation of Nrf2 and its binding to antioxidant response element (ARE) in the promoter regions of certain antioxidant genes should be explored. Additionally, other transcriptional regulators such as AP-1 [74] and NF-κB [75], as well as other antioxidant enzymes such as glutathione-S-transferase [76,77] have been previously reported for their synergistic roles with Nrf2 activation in combating against oxidative insults. Therefore, further investigations into these pathways are required to fully elucidate the protective mechanism of AMOLE against Cd-induced oxidative stress.
Third, AMOLE could alleviate Cd toxicity in hepatocytes via a reduction in Cd accumulation. Supplementation with AMOLE significantly suppressed intracellular accumulation of Cd on HepG2 cells after CdCl2 treatment (Fig. 6). This finding on hepatocytes is supported by our previous work using the lower eukaryotic model, S. cerevisiae. We demonstrated that AMOLE supplementation could alleviate toxicities from heavy metals and metalloids, including arsenic (As), nickel (Ni), lead (Pb), and Cd. These protective activities of AMOLE on yeast cells are strongly associated with the inhibition of intracellular accumulation of these harmful metal(loid)s [10,19]. Particularly, gallic acid, one of the important phenolic constituents of AMOLE, has been shown to effectively protect yeast cells against arsenite (As(III)) toxicity by reducing As(III) accumulation and As(III)-induced ROS production [10]. Additionally, it has been reported that the exogenous application of organic acids (such as citric acid, malic acid, jasmonic acid, and salicylic acid) can mitigate the toxicities of various heavy metals, including Cd, in plants through their roles in reducing metal accumulation, probably due to their metal-chelating properties [78]. Nevertheless, effective chelation occurs only when the metal chelator is present with the target metal. In this study, since the hepatocytes were pretreated with AMOLE before being transferred to fresh media containing CdCl2, it is unlikely that the reduced Cd accumulation by the AMOLE pretreatment was due to the metal-chelating properties of AMOLE. A potential mechanism by which the AMOLE pretreatment suppressed Cd accumulation may be an alteration in the expression of some metal transporters, such as ZIP14, DMT1, and ATP7A, which are involved in Cd transport in mammalian cells [[79], [80], [81]]. Supporting this idea, it has been reported that these transporters are expressed in human hepatocytes [[82], [83], [84]]. Previously, it has been demonstrated that pre-soaking of sunflower (Helianthus annuus) seeds with gallic acid significantly reduced Cd uptake and enhanced the activities of antioxidant enzymes including catalase, ascorbate peroxidase, and glutathione reductase [85]. Hence, we postulated that AMOLE could disturb Cd trafficking into hepatocytes via modulation in the expression of these metal transporters and probably improve cellular antioxidant capacity by promoting the antioxidant enzyme activities. Further investigations are necessitated to explore the activities of AMOLE on the expression of these metal transporters and/or antioxidant enzymes of hepatocytes in response to Cd exposure.
5. Conclusion
The pretreatment with AMOLE exerts beneficial effects against Cd-induced cytotoxicity in HepG2 cells by preventing the generation of oxidative stress. Our results suggest that the hepatopreventive action of AMOLE, which is transported and accumulated in hepatocytes during the pretreatment, may be attributed to three possible mechanisms: i) direct scavenging of endogenous free radicals through its antioxidant bioactive compounds, such as flavonoids and phenolic acids; ii) enhanced upregulation of the antioxidant defensive machinery (HO-1 and GPx1) of hepatocytes via activation of the Nrf2 signaling system; and iii) inhibition of intracellular Cd accumulation, possibly by reducing Cd uptake via certain Cd transporters. Our findings strongly indicate the high potential of AMOLE for clinical application in preventive medical approaches against adverse effects of Cd poisoning.
Data availability
Data will be made available on request.
CRediT authorship contribution statement
Visarut Buranasudja: Writing – review & editing, Writing – original draft, Visualization, Resources, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Kittipong Sanookpan: Methodology, Investigation, Formal analysis. Sornkanok Vimolmangkang: Methodology, Investigation, Formal analysis. Asma Binalee: Methodology, Investigation, Formal analysis. Kamil Mika: Investigation, Formal analysis. Sucheewin Krobthong: Writing – original draft, Visualization, Methodology, Investigation, Formal analysis, Data curation. Kittikhun Kerdsomboon: Writing – original draft, Methodology, Formal analysis, Data curation. Supeecha Kumkate: Writing – review & editing, Supervision, Methodology, Funding acquisition, Formal analysis, Conceptualization. Toemthip Poolpak: Methodology, Investigation, Funding acquisition, Formal analysis, Data curation. Siraprapa Kidhakarn: Methodology, Investigation, Formal analysis. Kwang Mo Yang: Methodology, Investigation, Formal analysis. Tossapol Limcharoensuk: Methodology, Investigation, Formal analysis. Choowong Auesukaree: Writing – review & editing, Supervision, Resources, Project administration, Methodology, Funding acquisition, Formal analysis, Data curation, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This research study was supported by Mahidol University (Basic Research Fund: fiscal year 2022 (BRF1-042/2565)). We thank the Research Instrument Center of the Faculty of Pharmaceutical Sciences, Chulalongkorn University, the Center of Excellence on Environmental Health and Toxicology (EHT), and Nabsolute Co., Ltd. for providing research facilities. We also thank Mr. Chayapol Tungphatthong for his assistance in the illustration of the graphical abstract.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e37424.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Chunhabundit R. Cadmium exposure and potential health risk from foods in contaminated area, Thailand. Toxicol. Res. 2016;32(1):65–72. doi: 10.5487/TR.2016.32.1.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ganguly K., Levanen B., Palmberg L., Akesson A., Linden A. Cadmium in tobacco smokers: a neglected link to lung disease? Eur. Respir. Rev. 2018;27(147) doi: 10.1183/16000617.0122-2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kubier A., Wilkin R.T., Pichler T. Cadmium in soils and groundwater: a review. Appl. Geochem. 2019;108:1–16. doi: 10.1016/j.apgeochem.2019.104388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han S., Sung G.H., Lee S., Han K.J., Han H.J. Serum cadmium is associated with hepatic steatosis and fibrosis: Korean national health and nutrition examination survey data IV-VII. Medicine (Baltim.) 2022;101(4) doi: 10.1097/MD.0000000000028559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hyder O., Chung M., Cosgrove D., Herman J.M., Li Z., Firoozmand A., Gurakar A., Koteish A., Pawlik T.M. Cadmium exposure and liver disease among US adults. J. Gastrointest. Surg. 2013;17(7):1265–1273. doi: 10.1007/s11605-013-2210-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernhoft R.A. Cadmium toxicity and treatment. Sci. World J. 2013;2013 doi: 10.1155/2013/394652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flora S.J., Pachauri V. Chelation in metal intoxication. Int. J. Environ. Res. Publ. Health. 2010;7(7):2745–2788. doi: 10.3390/ijerph7072745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pan S.Y., Zhou S.F., Gao S.H., Yu Z.L., Zhang S.F., Tang M.K., Sun J.N., Ma D.L., Han Y.F., Fong W.F., Ko K.M. New perspectives on how to discover drugs from herbal medicines: CAM's outstanding contribution to modern therapeutics. Evid. Based Complement. Alternat. Med. 2013;2013 doi: 10.1155/2013/627375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Meireles D., Gomes J., Lopes L., Hinzmann M., Machado J. A review of properties, nutritional and pharmaceutical applications of Moringa oleifera: integrative approach on conventional and traditional Asian medicine. Adv. Tradit. Med. 2020;20(4):495–515. doi: 10.1016/j.toxlet.2013.01.010. [DOI] [Google Scholar]
- 10.Kerdsomboon K., Chumsawat W., Auesukaree C. Effects of Moringa oleifera leaf extracts and its bioactive compound gallic acid on reducing toxicities of heavy metals and metalloid in Saccharomyces cerevisiae. Chemosphere. 2021;270 doi: 10.1016/j.chemosphere.2020.128659. [DOI] [PubMed] [Google Scholar]
- 11.Siddhuraju P., Becker K. Antioxidant properties of various solvent extracts of total phenolic constituents from three different agroclimatic origins of drumstick tree (Moringa oleifera Lam.) leaves. J. Agric. Food Chem. 2003;51(8):2144–2155. doi: 10.1021/jf020444+. [DOI] [PubMed] [Google Scholar]
- 12.Wang Y., Gao Y., Ding H., Liu S., Han X., Gui J., Liu D. Subcritical ethanol extraction of flavonoids from Moringa oleifera leaf and evaluation of antioxidant activity. Food Chem. 2017;218:152–158. doi: 10.1016/j.foodchem.2016.09.058. [DOI] [PubMed] [Google Scholar]
- 13.Abib R.T., Peres K.C., Barbosa A.M., Peres T.V., Bernardes A., Zimmermann L.M., Quincozes-Santos A., Fiedler H.D., Leal R.B., Farina M., Gottfried C. Epigallocatechin-3-gallate protects rat brain mitochondria against cadmium-induced damage. Food Chem. Toxicol. 2011;49(10):2618–2623. doi: 10.1016/j.fct.2011.07.006. [DOI] [PubMed] [Google Scholar]
- 14.An Z., Qi Y., Huang D., Gu X., Tian Y., Li P., Li H., Zhang Y. EGCG inhibits Cd2+-induced apoptosis through scavenging ROS rather than chelating Cd2+ in HL-7702 cells. Toxicol. Mech. Methods. 2014;24(4):259–267. doi: 10.3109/15376516.2013.879975. [DOI] [PubMed] [Google Scholar]
- 15.Chen J., Du L., Li J., Song H. Epigallocatechin-3-gallate attenuates cadmium-induced chronic renal injury and fibrosis. Food Chem. Toxicol. 2016;96:70–78. doi: 10.1016/j.fct.2016.07.030. [DOI] [PubMed] [Google Scholar]
- 16.Ghosh J., Das J., Manna P., Sil P.C. Protective effect of the fruits of Terminalia arjuna against cadmium-induced oxidant stress and hepatic cell injury via MAPK activation and mitochondria dependent pathway. Food Chem. 2010;123(4):1062–1075. doi: 10.1016/j.foodchem.2010.05.062. [DOI] [Google Scholar]
- 17.Elblehi S.S., El Euony O.I., El-Nahas A.F. Partial ameliorative effect of Moringa leaf ethanolic extract on the reproductive toxicity and the expression of steroidogenic genes induced by subchronic cadmium in male rats. Environ. Sci. Pollut. Res. Int. 2019;26(23):23306–23318. doi: 10.1007/s11356-019-05607-y. [DOI] [PubMed] [Google Scholar]
- 18.Sadek K.M. Chemotherapeutic efficacy of an ethanolic Moringa oleifera leaf extract against chromium-induced testicular toxicity in rats. Andrologia. 2014;46(9):1047–1054. doi: 10.1111/and.12196. [DOI] [PubMed] [Google Scholar]
- 19.Kerdsomboon K., Tatip S., Kosasih S., Auesukaree C. Soluble Moringa oleifera leaf extract reduces intracellular cadmium accumulation and oxidative stress in Saccharomyces cerevisiae. J. Biosci. Bioeng. 2016;121(5):543–549. doi: 10.1016/j.jbiosc.2015.09.013. [DOI] [PubMed] [Google Scholar]
- 20.Krobthong S., Yingchutrakul Y., Sittisaree W., Tulyananda T., Samutrtai P., Choowongkomon K., Lao-On U. Evaluation of potential anti-metastatic and antioxidative abilities of natural peptides derived from Tecoma stans (L.) Juss. ex Kunth in A549 cells. PeerJ. 2022;10 doi: 10.7717/peerj.13693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Xia J., Psychogios N., Young N., Wishart D.S. MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic Acids Res. 2009;37(Web Server issue):W652–W660. doi: 10.1093/nar/gkp356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim D.W., Ock J., Moon K.W., Park C.H. Association between Pb, Cd, and Hg exposure and liver injury among Korean adults. Int. J. Environ. Res. Publ. Health. 2021;18(13) doi: 10.3390/ijerph18136783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Renugadevi J., Prabu S.M. Naringenin protects against cadmium-induced oxidative renal dysfunction in rats. Toxicology. 2009;256(1–2):128–134. doi: 10.1016/j.tox.2008.11.012. [DOI] [PubMed] [Google Scholar]
- 24.Shaikh Z.A., Vu T.T., Zaman K. Oxidative stress as a mechanism of chronic cadmium-induced hepatotoxicity and renal toxicity and protection by antioxidants. Toxicol. Appl. Pharmacol. 1999;154(3):256–263. doi: 10.1006/taap.1998.8586. [DOI] [PubMed] [Google Scholar]
- 25.Buranasudja V., Muangnoi C., Sanookpan K., Halim H., Sritularak B., Rojsitthisak P. Eriodictyol attenuates H2O2-induced oxidative damage in human dermal fibroblasts through enhanced capacity of antioxidant machinery. Nutrients. 2022;14(12) doi: 10.3390/nu14122553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Buranasudja V., Rani D., Malla A., Kobtrakul K., Vimolmangkang S. Insights into antioxidant activities and anti-skin-aging potential of callus extract from Centella asiatica (L.) Sci. Rep. 2021;11(1) doi: 10.1038/s41598-021-92958-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang X., Wang L., Lu H., Zong Z., Chen Z., Li Y., Luo X., Li Y. Preservation of hydrogen peroxide-induced oxidative damage in HepG-2 cells by rice protein hydrolysates pretreated with electron beams. Sci. Rep. 2020;10(1):8415. doi: 10.1038/s41598-020-64814-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chin C.Y., Jalil J., Ng P.Y., Ng S.F. Development and formulation of Moringa oleifera standardised leaf extract film dressing for wound healing application. J. Ethnopharmacol. 2018;212:188–199. doi: 10.1016/j.jep.2017.10.016. [DOI] [PubMed] [Google Scholar]
- 29.El-Seadawy I.E., Kotp M.S., El-Maaty A.M.A., Fadl A.M., El-Sherbiny H.R., Abdelnaby E.A. The impact of varying doses of moringa leaf methanolic extract supplementation in the cryopreservation media on sperm quality, oxidants, and antioxidant capacity of frozen-thawed ram sperm. Trop. Anim. Health Prod. 2022;54(6):344. doi: 10.1007/s11250-022-03344-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodríguez-Pérez C., Quirantes-Piné R., Fernández-Gutiérrez A., Segura-Carretero A. Optimization of extraction method to obtain a phenolic compounds-rich extract from Moringa oleifera Lam. leaves. Ind. Crops Prod. 2015;66:246–254. doi: 10.1016/j.indcrop.2015.01.002. [DOI] [Google Scholar]
- 31.Bajpai V.K., Alam M.B., Quan K.T., Kwon K.R., Ju M.K., Choi H.J., Lee J.S., Yoon J.I., Majumder R., Rather I.A., Kim K., Lee S.H., Na M. Antioxidant efficacy and the upregulation of Nrf2-mediated HO-1 expression by (+)-lariciresinol, a lignan isolated from Rubia philippinensis, through the activation of p38. Sci. Rep. 2017;7 doi: 10.1038/srep46035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Velagapudi R., El-Bakoush A., Olajide O.A. Activation of Nrf2 pathway contributes to neuroprotection by the dietary flavonoid tiliroside. Mol. Neurobiol. 2018;55(10):8103–8123. doi: 10.1007/s12035-018-0975-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bartolini D., Arato I., Mancuso F., Giustarini D., Bellucci C., Vacca C., Aglietti M.C., Stabile A.M., Rossi R., Cruciani G., Rende M., Calafiore R., Luca G., Galli F. Melatonin modulates Nrf2 activity to protect porcine pre-pubertal Sertoli cells from the abnormal H2O2 generation and reductive stress effects of cadmium. J. Pineal Res. 2022;73(1) doi: 10.1111/jpi.12806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He X., Chen M.G., Ma Q. Activation of Nrf2 in defense against cadmium-induced oxidative stress. Chem. Res. Toxicol. 2008;21(7):1375–1383. doi: 10.1021/tx800019a. [DOI] [PubMed] [Google Scholar]
- 35.Lawal A.O., Ellis E.M. Nrf2-mediated adaptive response to cadmium-induced toxicity involves protein kinase C delta in human 1321N1 astrocytoma cells. Environ. Toxicol. Pharmacol. 2011;32(1):54–62. doi: 10.1016/j.etap.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 36.Migni A., Mancuso F., Baroni T., Di Sante G., Rende M., Galli F., Bartolini D. Melatonin as a repairing agent in cadmium- and free fatty acid-induced lipotoxicity. Biomolecules. 2023;13(12):1758. doi: 10.3390/biom13121758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bellezza I., Giambanco I., Minelli A., Donato R. Nrf2-Keap1 signaling in oxidative and reductive stress. Biochim. Biophys. Acta Mol. Cell Res. 2018;1865(5):721–733. doi: 10.1016/j.bbamcr.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 38.Gozzelino R., Jeney V., Soares M.P. Mechanisms of cell protection by heme oxygenase-1. Annu. Rev. Pharmacol. Toxicol. 2010;50:323–354. doi: 10.1146/annurev.pharmtox.010909.105600. [DOI] [PubMed] [Google Scholar]
- 39.Lubos E., Loscalzo J., Handy D.E. Glutathione peroxidase-1 in health and disease: from molecular mechanisms to therapeutic opportunities. Antioxidants Redox Signal. 2011;15(7):1957–1997. doi: 10.1089/ars.2010.3586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eleutherio E.C.A., Silva Magalhaes R.S., de Araujo Brasil A., Monteiro Neto J.R., de Holanda Paranhos L. SOD1, more than just an antioxidant. Arch. Biochem. Biophys. 2021;697 doi: 10.1016/j.abb.2020.108701. [DOI] [PubMed] [Google Scholar]
- 41.Wang Y., Branicky R., Noe A., Hekimi S. Superoxide dismutases: dual roles in controlling ROS damage and regulating ROS signaling. J. Cell Biol. 2018;217(6):1915–1928. doi: 10.1083/jcb.201708007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ahamed M., Akhtar M.J., Khan M.A.M., Alhadlaq H.A. Reduced graphene oxide mitigates cadmium-induced cytotoxicity and oxidative stress in HepG2 cells. Food Chem. Toxicol. 2020;143 doi: 10.1016/j.fct.2020.111515. [DOI] [PubMed] [Google Scholar]
- 43.Athmouni K., Belhaj D., Mkadmini Hammi K., El Feki A., Ayadi H. Phenolic compounds analysis, antioxidant, and hepatoprotective effects of Periploca angustifolia extract on cadmium-induced oxidative damage in HepG2 cell line and rats. Arch. Physiol. Biochem. 2018;124(3):261–274. doi: 10.1080/13813455.2017.1395890. [DOI] [PubMed] [Google Scholar]
- 44.Nguyen K.C., Willmore W.G., Tayabali A.F. Cadmium telluride quantum dots cause oxidative stress leading to extrinsic and intrinsic apoptosis in hepatocellular carcinoma HepG2 cells. Toxicology. 2013;306:114–123. doi: 10.1016/j.tox.2013.02.010. [DOI] [PubMed] [Google Scholar]
- 45.Renugadevi J., Prabu S.M. Cadmium-induced hepatotoxicity in rats and the protective effect of naringenin. Exp. Toxicol. Pathol. 2010;62(2):171–181. doi: 10.1016/j.etp.2009.03.010. [DOI] [PubMed] [Google Scholar]
- 46.Lee W.K., Probst S., Scharner B., Deba T., Dahdouh F., Thevenod F. Distinct concentration-dependent oxidative stress profiles by cadmium in a rat kidney proximal tubule cell line. Arch. Toxicol. 2024;98(4):1043–1059. doi: 10.1007/s00204-023-03677-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Son Y.O., Lee J.C., Hitron J.A., Pan J., Zhang Z., Shi X. Cadmium induces intracellular Ca2+- and H2O2-dependent apoptosis through JNK- and p53-mediated pathways in skin epidermal cell line. Toxicol. Sci. 2010;113(1):127–137. doi: 10.1093/toxsci/kfp259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang J., Wang Q., Li J., Shen Q., Wang F., Wang L. Cadmium induces hydrogen peroxide production and initiates hydrogen peroxide-dependent apoptosis in the gill of freshwater crab, Sinopotamon henanense. Comp. Biochem. Physiol. C Toxicol. Pharmacol. 2012;156(3−4):195–201. doi: 10.1016/j.cbpc.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 49.Dikalov S.I., Harrison D.G. Methods for detection of mitochondrial and cellular reactive oxygen species. Antioxidants Redox Signal. 2014;20(2):372–382. doi: 10.1089/ars.2012.4886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bartolini D., Commodi J., Piroddi M., Incipini L., Sancineto L., Santi C., Galli F. Glutathione S-transferase pi expression regulates the Nrf2-dependent response to hormetic diselenides. Free Radic. Biol. Med. 2015;88(Pt B):466–480. doi: 10.1016/j.freeradbiomed.2015.06.039. [DOI] [PubMed] [Google Scholar]
- 51.Bartolini D., Piroddi M., Tidei C., Giovagnoli S., Pietrella D., Manevich Y., Tew K.D., Giustarini D., Rossi R., Townsend D.M., Santi C., Galli F. Reaction kinetics and targeting to cellular glutathione S-transferase of the glutathione peroxidase mimetic PhSeZnCl and its D,L-polylactide microparticle formulation. Free Radic. Biol. Med. 2015;78:56–65. doi: 10.1016/j.freeradbiomed.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lawal A.O., Marnewick J.L., Ellis E.M. Heme oxygenase-1 attenuates cadmium-induced mitochondrial-caspase 3- dependent apoptosis in human hepatoma cell line. BMC Pharmacol. Toxicol. 2015;16:41. doi: 10.1186/s40360-015-0040-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhou X.L., Wan X.M., Fu X.X., Xie C.G. Puerarin prevents cadmium-induced hepatic cell damage by suppressing apoptosis and restoring autophagic flux. Biomed. Pharmacother. 2019;115 doi: 10.1016/j.biopha.2019.108929. [DOI] [PubMed] [Google Scholar]
- 54.He Z., Shen P., Feng L., Hao H., He Y., Fan G., Liu Z., Zhu K., Wang Y., Zhang N., Hu X., Fu Y., Wu J. Cadmium induces liver dysfunction and ferroptosis through the endoplasmic stress-ferritinophagy axis. Ecotoxicol. Environ. Saf. 2022;245 doi: 10.1016/j.ecoenv.2022.114123. [DOI] [PubMed] [Google Scholar]
- 55.Niture S., Lin M., Qi Q., Moore J.T., Levine K.E., Fernando R.A., Kumar D. Role of autophagy in cadmium-induced hepatotoxicity and liver diseases. J. Toxicol. 2021;2021 doi: 10.1155/2021/9564297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zou H., Sun J., Wu B., Yuan Y., Gu J., Bian J., Liu X., Liu Z. Effects of cadmium and/or lead on autophagy and liver injury in rats. Biol. Trace Elem. Res. 2020;198(1):206–215. doi: 10.1007/s12011-020-02045-7. [DOI] [PubMed] [Google Scholar]
- 57.Sasson A., Kristoferson E., Batista R., McClung J.A., Abraham N.G., Peterson S.J. The pivotal role of heme oxygenase-1 in reversing the pathophysiology and systemic complications of NAFLD. Arch. Biochem. Biophys. 2021;697 doi: 10.1016/j.abb.2020.108679. [DOI] [PubMed] [Google Scholar]
- 58.Yao P., Hao L., Nussler N., Lehmann A., Song F., Zhao J., Neuhaus P., Liu L., Nussler A. The protective role of HO-1 and its generated products (CO, bilirubin, and Fe) in ethanol-induced human hepatocyte damage. Am. J. Physiol. Gastrointest. Liver Physiol. 2009;296(6):G1318–G1323. doi: 10.1152/ajpgi.00555.2007. [DOI] [PubMed] [Google Scholar]
- 59.Ryter S.W., Alam J., Choi A.M. Heme oxygenase-1/carbon monoxide: from basic science to therapeutic applications. Physiol. Rev. 2006;86(2):583–650. doi: 10.1152/physrev.00011.2005. [DOI] [PubMed] [Google Scholar]
- 60.Duranti G., Maldini M., Crognale D., Horner K., Dimauro I., Sabatini S., Ceci R. Moringa oleifera leaf extract upregulates Nrf2/HO-1 expression and ameliorates redox status in C2C12 Skeletal Muscle Cells. Molecules. 2021;26(16) doi: 10.3390/molecules26165041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Abdou K.H., Moselhy W.A., Mohamed H.M., El-Nahass E.S., Khalifa A.G. Moringa oleifera leaves extract protects titanium dioxide nanoparticles-induced nephrotoxicity via Nrf2/HO-1 signaling and amelioration of oxidative stress. Biol. Trace Elem. Res. 2019;187(1):181–191. doi: 10.1007/s12011-018-1366-2. [DOI] [PubMed] [Google Scholar]
- 62.Kim C.G., Chang S.N., Park S.M., Hwang B.S., Kang S.A., Kim K.S., Park J.G. Moringa oleifera mitigates ethanol-induced oxidative stress, fatty degeneration and hepatic steatosis by promoting Nrf2 in mice. Phytomedicine. 2022;100 doi: 10.1016/j.phymed.2022.154037. [DOI] [PubMed] [Google Scholar]
- 63.Li X., Wang H., Gao Y., Li L., Tang C., Wen G., Zhou Y., Zhou M., Mao L., Fan Y. Protective effects of quercetin on mitochondrial biogenesis in experimental traumatic brain injury via the Nrf2 signaling pathway. PLoS One. 2016;11(10) doi: 10.1371/journal.pone.0164237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sun G.Y., Chen Z., Jasmer K.J., Chuang D.Y., Gu Z., Hannink M., Simonyi A. Quercetin attenuates inflammatory responses in BV-2 microglial cells: role of MAPKs on the Nrf2 pathway and induction of heme oxygenase-1. PLoS One. 2015;10(10) doi: 10.1371/journal.pone.0141509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sharma A., Parikh M., Shah H., Gandhi T. Modulation of Nrf2 by quercetin in doxorubicin-treated rats. Heliyon. 2020;6(4) doi: 10.1016/j.heliyon.2020.e03803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Miyamoto N., Izumi H., Miyamoto R., Kondo H., Tawara A., Sasaguri Y., Kohno K. Quercetin induces the expression of peroxiredoxins 3 and 5 via the Nrf2/NRF1 transcription pathway. Invest. Ophthalmol. Vis. Sci. 2011;52(2):1055–1063. doi: 10.1167/iovs.10-5777. [DOI] [PubMed] [Google Scholar]
- 67.Boots A.W., Veith C., Albrecht C., Bartholome R., Drittij M.J., Claessen S.M.H., Bast A., Rosenbruch M., Jonkers L., van Schooten F.J., Schins R.P.F. The dietary antioxidant quercetin reduces hallmarks of bleomycin-induced lung fibrogenesis in mice. BMC Pulm. Med. 2020;20(1):112. doi: 10.1186/s12890-020-1142-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sanjay S., Girish C., Toi P.C., Bobby Z. Quercetin modulates NRF2 and NF-kB/TLR-4 pathways to protect against isoniazid- and rifampicin-induced hepatotoxicity in vivo. Can. J. Physiol. Pharmacol. 2021;99(9):952–963. doi: 10.1139/cjpp-2021-0008. [DOI] [PubMed] [Google Scholar]
- 69.Wang J., Wang K., Ding L., Zhao P., Zhang C., Wang H., Yang Z., Liu Z. Alleviating effect of quercetin on cadmium-induced oxidative damage and apoptosis by activating the Nrf2-keap1 pathway in BRL-3A cells. Front. Pharmacol. 2022;13 doi: 10.3389/fphar.2022.969892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhu M., Zhou X., Zhao J. Quercetin prevents alcohol-induced liver injury through targeting of PI3K/Akt/nuclear factor-kB and STAT3 signaling pathway. Exp. Ther. Med. 2017;14(6):6169–6175. doi: 10.3892/etm.2017.5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Khan H., Tundis R., Ullah H., Aschner M., Belwal T., Mirzaei H., Akkol E.K. Flavonoids targeting NRF2 in neurodegenerative disorders. Food Chem. Toxicol. 2020;146 doi: 10.1016/j.fct.2020.111817. [DOI] [PubMed] [Google Scholar]
- 72.Li Y.R., Li G.H., Zhou M.X., Xiang L., Ren D.M., Lou H.X., Wang X.N., Shen T. Discovery of natural flavonoids as activators of Nrf2-mediated defense system: structure-activity relationship and inhibition of intracellular oxidative insults. Bioorg. Med. Chem. 2018;26(18):5140–5150. doi: 10.1016/j.bmc.2018.09.010. [DOI] [PubMed] [Google Scholar]
- 73.Shi Y., Shi L., Liu Q., Wang W., Liu Y. Molecular mechanism and research progress on pharmacology of ferulic acid in liver diseases. Front. Pharmacol. 2023;14 doi: 10.3389/fphar.2023.1207999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ishii T., Itoh K., Takahashi S., Sato H., Yanagawa T., Katoh Y., Bannai S., Yamamoto M. Transcription factor Nrf2 coordinately regulates a group of oxidative stress-inducible genes in macrophages. J. Biol. Chem. 2000;275(21):16023–16029. doi: 10.1074/jbc.275.21.16023. [DOI] [PubMed] [Google Scholar]
- 75.Gao W., Guo L., Yang Y., Wang Y., Xia S., Gong H., Zhang B.K., Yan M. Dissecting the crosstalk between Nrf2 and NF-kappaB response pathways in drug-induced toxicity. Front. Cell Dev. Biol. 2021;9 doi: 10.3389/fcell.2021.809952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bartolini D., Giustarini D., Pietrella D., Rossi R., Galli F. Glutathione S-transferase P influences the Nrf2-dependent response of cellular thiols to seleno-compounds. Cell Biol. Toxicol. 2020;36(4):379–386. doi: 10.1007/s10565-020-09517-5. [DOI] [PubMed] [Google Scholar]
- 77.Bartolini D., Torquato P., Piroddi M., Galli F. Targeting glutathione S-transferase P and its interactome with selenium compounds in cancer therapy. Biochim. Biophys. Acta. Gen. Subj. 2019;1863(1):130–143. doi: 10.1016/j.bbagen.2018.09.023. [DOI] [PubMed] [Google Scholar]
- 78.Vega A., Delgado N., Handford M. Increasing heavy metal tolerance by the exogenous application of organic acids. Int. J. Mol. Sci. 2022;23(10):5438. doi: 10.3390/ijms23105438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bannon D.I., Abounader R., Lees P.S., Bressler J.P. Effect of DMT1 knockdown on iron, cadmium, and lead uptake in Caco-2 cells. Am. J. Physiol. Cell Physiol. 2003;284(1):C44–C50. doi: 10.1152/ajpcell.00184.2002. [DOI] [PubMed] [Google Scholar]
- 80.Fujishiro H., Yano Y., Takada Y., Tanihara M., Himeno S. Roles of ZIP8, ZIP14, and DMT1 in transport of cadmium and manganese in mouse kidney proximal tubule cells. Metallomics. 2012;4(7):700–708. doi: 10.1039/c2mt20024d. [DOI] [PubMed] [Google Scholar]
- 81.Ohta H., Ohba K. Involvement of metal transporters in the intestinal uptake of cadmium. J. Toxicol. Sci. 2020;45(9):539–548. doi: 10.2131/jts.45.539. [DOI] [PubMed] [Google Scholar]
- 82.Aydemir T.B., Troche C., Kim M.H., Cousins R.J. Hepatic ZIP14-mediated zinc transport contributes to endosomal insulin receptor trafficking and glucose metabolism. J. Biol. Chem. 2016;291(46):23939–23951. doi: 10.1074/jbc.M116.748632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li Z.P., Kwok M.L., Law T.Y.S., Cheung K.C.P., Chan K.M. Functional analyses of copper transporter genes in the human liver cell line HepG2. Toxicol. Vitro. 2020;66 doi: 10.1016/j.tiv.2020.104856. [DOI] [PubMed] [Google Scholar]
- 84.Min K.S., Takano M., Amako K., Ueda H., Tanaka K. Involvement of the essential metal transporter Zip14 in hepatic Cd accumulation during inflammation. Toxicol. Lett. 2013;218(1):91–96. doi: 10.1016/j.toxlet.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 85.Saidi I., Guesmi F., Kharbech O., Hfaiedh N., Djebali W. Gallic acid improves the antioxidant ability against cadmium toxicity: impact on leaf lipid composition of sunflower (Helianthus annuus) seedlings. Ecotoxicol. Environ. Saf. 2021;210 doi: 10.1016/j.ecoenv.2021.111906. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.