Abstract
Human immunodeficiency virus (HIV)-specific helper T lymphocytes (HTL) play a key role in the immune control of HIV type 1 (HIV-1) infection, and as such are an important target of potential HIV-1 vaccines. In order to identify HTL epitopes in HIV-1 that might serve as vaccine targets, conserved HIV-1-derived peptides bearing an HLA-DR binding supermotif were tested for binding to a panel of the most representative HLA-DR molecules. Eleven highly cross-reactive binding peptides were identified: three in Gag and eight in Pol. Lymphoproliferative responses to this panel of peptides, as well as to the HIV-1 p24 and p66 proteins, were evaluated with a cohort of 31 HIV-1-infected patients. All 11 peptides were recognized by peripheral blood mononuclear cells from multiple HIV-infected donors. Many of the responsive HIV-infected subjects showed recognition of multiple peptides, indicating that HIV-1-specific T-helper responses may be broadly directed in certain individuals. A strong association existed between recognition of the parental recombinant HIV-1 protein and the corresponding HTL peptides, suggesting that these peptides represent epitopes that are processed and presented during the course of HIV-1 infection. Lastly, responses to the supermotif peptides were mediated by CD4+ T cells and were restricted by major histocompatibility complex class II molecules. The epitopes described herein are potentially important components of HIV-1 therapeutic and prophylactic vaccines.
Virus-specific helper T lymphocytes (HTL) have been shown to play an important role in maintaining effective cytotoxic T-lymphocyte (CTL) function and in controlling viremia during several chronic viral infections (24). Human immunodeficiency virus type 1 (HIV-1) infection is marked by a gradual loss of CD4+ T lymphocytes in general and a specific loss or failure to develop functional HIV-1-specific HTL in the majority of chronically infected individuals (47, 52). Several investigators have observed defects in the ability of HTL from most HIV-1-infected individuals to respond by proliferation or cytokine production to HIV-1 peptide antigens and other “recall” antigens (35, 47). This HTL dysfunction is likely to be important in the immunopathogenesis of HIV-1 infection in that dysfunctional HTL are unable to appropriately assist in expansion, differentiation, and maintenance of HIV-1-specific CTL, which are thought to be crucial for effective control of HIV-1 replication.
There is also mounting evidence that HTL that are reactive against HIV-1 antigens may play an important role in delaying disease progression in some circumstances. Vigorous HIV-1 p24-specific HTL proliferative responses were more frequently found in individuals with long-term nonprogressive HIV-1 infection than in those with more standard disease progression, and these responses were found to correlate inversely with viral load in chronically infected individuals not receiving antiretroviral therapy (37). Similarly, Pitcher et al., using a novel method based on cytokine secretion, found there to be a higher frequency of Gag-specific HTL in the peripheral blood of HIV-1-infected patients with nonprogressive disease (33). Furthermore, both HIV-1-specific HTL and CTL responses have been identified in HIV-1-exposed but persistently seronegative individuals (39, 48), including exposed health care workers (11, 32), children born to infected mothers (12, 40), female sex workers (16, 38), and partners of HIV-1-infected individuals (9, 17, 30) suggesting that HIV-specific cellular immune responses may contribute to the control or prevention of HIV-1 infection in these settings.
These studies suggest that a broad and coordinated HIV-1-specific cellular immune response, including both HTL and CTL responses, may correlate with control of HIV-1 infection in exposed individuals. Taken together, these data support the concept that induction of HIV-1-specific HTL responses might be important for both treatment and prevention of HIV-1 disease. The development of vaccines to induce protective or therapeutic cellular immune responses to HIV-1 is complicated by the presence of numerous viral variants or quasispecies (13). Epitope-based vaccines offer the advantage of focusing immune responses on multiple conserved epitopes. An additional attractive feature of multiepitope vaccines is the potential for eliciting a broad-based response directed against both dominant and subdominant epitopes. This is of relevance because weak and narrowly directed HIV-1-specific immune responses have been associated with a more rapid disease progression (22).
One potential obstacle to the development of epitope-based vaccines has been the large degree of polymorphism of HLA molecules, which complicates the identification of epitopes that are suitable for use in a patient population. Previous studies have demonstrated that the majority of HLA class I and class II molecules can be grouped in broad supertypes with overlapping peptide binding specificity (44). In the case of the HLA-DR molecules, a single superfamily encompassing DRB1 alleles expressed in the majority of humans has been defined (49).
Based on these data, we are currently investigating an HIV-1 vaccine design which includes multiple, conserved CTL and HTL epitopes. A number of broadly cross-reactive minimal CTL epitopes in conserved regions of HIV-1 proteins have been identified (26). By contrast, relatively few HIV-1-specific HTL epitopes have been identified. To address this issue, we sought to identify conserved, major histocompatibility complex (MHC) class II-restricted epitopes in HIV-1 that would be recognized by a large fraction of the global population.
MATERIALS AND METHODS
Sequence analysis.
Using an algorithm analysis program, eight HIV-1 antigens (Gag, Pol, Nef, Rev, Tat, Vif, Vpr, and Vpu) were scanned for the presence of the HLA-DR supermotif. The sequences of these antigens were scanned for the presence of 15 amino acid sequences containing the previously described HLA-DR supermotif (31, 49). The analysis included complete sequences from 62 HIV-1 isolates; 3 A, 18 B, 8 C, 4 D, 2 F, 3 G, 3 H, 2 J, 1 N, 2 O, and 16 recombinant (AC, ADI, AE, AG, AGI, AGJ, and BF) isolates. The three Gag peptide sequences (Gag 171, 294, and 298) are contained within the HIV-1 p24 protein. The peptides Pol 303, 335, 596, 711, and 712 are contained in HIV-1 reverse transcriptase, and Pol peptides 758, 915, and 956 are contained within HIV-1 integrase.
Peptide synthesis.
Peptides were synthesized at Epimmune on an Applied Biosystems (Foster City, Calif.) 430A peptide synthesizer using 9-fluorenylmethoxy carbonyl chemistry. After the synthesis was completed, the peptide was cleaved from the resin, the protecting groups were removed, and the peptides were then purified by reversed-phase high-performance liquid chromatography. The purity of the peptides was confirmed by amino acid sequence and/or composition analysis to be routinely greater than 95%.
Cell lines.
The following Epstein-Barr virus-transformed homozygous cell lines were used as sources of human HLA class II molecules: LG2 (allele DRB1*0101 [antigen DR1]); GM3107 (DRB5*0101 [DR2w2a]); MAT (DRB1*0301 [DR3]); PREISS (DRB1*0401 [DR4w4]); SWEIG (DRB1*1101 [DR5w11]); PITOUT (DRB1*0701 [DR7]); KT3 (DRB1*0405 [DR4w15]); Herluf (DRB1*1201 [DR5w12]); HO301 (DRB1*1302 [DR6w19]); OLL (DRB1*0802 [DR8w2]); and HID (DRB1*0901 [DR9], supplied as a kind gift by Paul Harris, Columbia University). In one instance, transfected fibroblasts were used: L466.1 (DRB1*1501 [DR2w2b]) (2, 50). Cells were maintained in vitro by culturing in RPMI 1640 medium supplemented with 2 mM l-glutamine (GIBCO, Grand Island, N.Y.), 50 μM 2-mercaptoethanol, and 10% heat-inactivated fetal calf serum (Irvine Scientific, Santa Ana, Calif.). Cells were also supplemented with 100 μg of streptomycin/ml and 100 U of penicillin (Irvine Scientific)/ml. Large quantities of cells were grown in spinner cultures. Cells were lysed for 30 min at 4°C with a lysis buffer of 50 mM Tris-HCl, pH 8.5, 1% NP-40 (Fluka Biochemika, Buchs, Switzerland), 150 mM NaCl, and 2 mM phenylmethylsulfonyl fluoride (CalBioChem, La Jolla, Calif.). Lysates were cleared of debris and nuclei by centrifugation at 15,000 × g for 30 min.
Affinity purification of HLA-DR molecules.
Class II molecules were purified by affinity chromatography as previously described (18, 43) using the monoclonal antibody (MAb) LB3.1 coupled to Sepharose CL-4B beads. Lysates were filtered twice through two precolumns of inactivated Sepharose CL4-B and protein A-Sepharose and then passed over the anti-DR column. The anti-DR column was then washed with 10 column volumes of 10 mM Tris-HCl, pH 8.0, in a solution containing 1% NP-40, phosphate-buffered saline (PBS), 2 column volumes of PBS, and 2 column volumes of PBS containing 0.4% n-octylglucoside. Finally, DR molecules were eluted with 50 mM diethylamine in 0.15 M NaCl containing 0.4% n-octylglucoside, pH 11.5. A 1/25 volume of 2.0 M Tris, pH 6.8, was added to the eluate to reduce the pH to ∼8.0. The eluate was then concentrated by centrifugation in Centriprep 30 concentrators at 2,000 rpm (Amicon, Beverly, Mass.).
HLA-DR peptide-binding assays.
A panel of 12 different specific HLA-DR peptide assays was utilized in the present study. These assays were chosen to be representative of the most common HLA-DR alleles. In brief, purified human HLA-DR molecules (5 to 500 nM) were incubated with various unlabeled HIV-1 peptides and 1 to 10 nM 125I-radiolabeled probe peptides for 48 h. Assays were performed at pH 7.0 with the exception of that for DRB1*0301, which was performed at pH 4.5 (45). HLA-DR peptide complexes were separated from free peptide by gel filtration on TSK200 columns (TosoHaas, Montgomeryville, Pa.), and the fraction of bound peptide was calculated as described previously (43).
The radiolabeled probes used were HA Y307–319 (for DRB1*0101), TT 830–843 (for DRB5*0101, DRB1*1101, DRB1*0701, DRB1*0802, and DRB1*0901), MBP Y85–100 (for DRB1*1501), MT 65 kD Y3–13 with Y7 replaced with F (for DRB1*0301), a nonnatural peptide with the sequence YARFQSQTTLKQKT (for DRB1*0401 and DRB1*0405) (2, 50), a naturally processed peptide (sequence EALIHQLKINPYVLS) (15) of unknown origin eluted from a DRB1*1201+ C1R cell line, and an analog of TT 830–843 (sequence QYIKANAKFIGITE) (for DRB1*1302) (6).
In preliminary experiments, the titers of the HLA-DR preparation were determined in the presence of fixed amounts of radiolabeled peptides to determine the concentration of HLA-DR molecules necessary to bind 10 to 20% of the total radioactivity. All subsequent inhibition and direct binding assays were then performed using these HLA-DR concentrations.
Peptide inhibitors were typically tested at concentrations ranging from 120 μg/ml to 1.2 ng/ml. In appropriate stoichiometric conditions, the 50% inhibitory concentration (IC50) of an unlabeled test peptide for the purified HLA-DR is a reasonable approximation of the affinity of interaction (Kd). Peptides were tested in two to four completely independent experiments.
Population coverage analysis.
To obtain population coverage estimates for each peptide, cumulative gene frequencies of DR antigens that bound peptide were calculated. The impact of the two DR5 subtypes, DR11 and DR12, was considered separately since they are known to have different binding specificities. The contribution of B3, B4, and B5 gene products was also considered, based on known linkage disequilibrium frequencies. The redundancy of coverage by the panel of epitopes is defined as the total number of different DR-peptide combinations potentially presented in a given individual. The number of DR epitopes presented by each individual in the model population was determined by tabulating the number of DR-peptide combinations associated with binding with an IC50 of ≤1,000 nM.
Study population.
HIV-1-infected study subjects were selected from a cohort of chronically HIV-1-infected individuals followed in the Adult Infectious Diseases Group Practice at the University of Colorado Health Sciences Center. HIV-1 RNA levels in plasma were measured using the Roche HIV-1 Monitor kit, with analytical sensitivities of 200 and 20 copies/ml for the quantitative and ultraquantitative assays, respectively. HIV-1-negative subjects were normal healthy adult volunteers. All study subjects participated voluntarily, and the study was approved by the University of Colorado Health Sciences Center Institutional Review Board.
Lymphocyte proliferation assay.
Peripheral blood was collected in heparinized cell preparation tubes (Vacutainer Systems; Becton Dickinson, Franklin Lakes, N.J.), and peripheral blood mononuclear cells (PBMCs) were isolated by density gradient centrifugation. Cells were resuspended at a concentration of 106 cells/ml in RPMI with 10% human AB serum (Sigma, St. Louis, Mo.), and 100 μl was added to plates containing 100 μl of HIV-1 p24 and p66 baculovirus-expressed recombinant proteins (NY5 and IIIB strains, respectively; final concentration, 1 μg/ml; Protein Sciences Corporation, Meriden, Conn.), baculovirus control protein (final concentration, 0.05 μg/ml), and supermotif HTL peptides (final concentration, 2.5 μg/ml). Phytohemagglutinin (PHA) (5 μg/ml; Sigma) and whole Candida protein (10 μg/ml; Greer, Lenoir, N.C.) were used as positive controls in each assay. In some experiments, HLA-DR was blocked by the addition of 10 μg of affinity-purified anti-HLA-DR monoclonal antibody (hybridoma L243; American Type Tissue Collection, Manassas, Va.)/ml at the beginning of culture. Cells were incubated at 37°C in a humidified 5% CO2 atmosphere for 6 days. Plates were pulsed with tritiated thymidine for 6 h and harvested, and radioactivity was measured on a beta-counter (Packard). The stimulation index (S.I.) was calculated by dividing the thymidine incorporation in the presence of antigen by the incorporation in the absence of antigen (containing media alone for peptides or baculovirus control protein for recombinant HIV-1 proteins). Background responses (to media alone or to baculovirus control protein) were lower than 1,000 cpm in all assays, and mean background counts did not differ significantly between HIV− and HIV+ donor groups. PBMCs from all donors tested responded significantly to phytohemagglutinin and Candida protein.
Flow-cytometric detection of antigen-induced intracellular cytokines.
The frequency of antigen-specific CD4+ T cells in PBMCs secreting gamma interferon (IFN-γ) was determined using a previously reported method (33), with minor modifications. Briefly, PBMCs (1 × 106 to 2 × 106) were placed in 12-by-75-mm culture tubes containing 3 μg of anti-CD28 and -CD49d antibodies (Becton Dickinson, San Diego, Calif.)/ml in RPMI 10% human serum (Sigma) and one of the following stimulation conditions: HIV-1 p24 or p66 antigens (5 μg/ml; Protein Sciences), HTL epitope peptides (10 μg/ml), staphylococcal enterotoxin B (Sigma), and baculovirus control protein or medium alone. These cultures were incubated at a 5° slant at 37°C in a humidified 5% CO2 atmosphere for 14 h with Brefeldin A added after the initial 4 h. Cells were surface stained with anti-CD4 monoclonal antibody (Caltag, Burlingame, Calif.) for 30 min at 4°C. Cells were washed twice with PBS containing 1% bovine serum albumin and fixed for 15 min at room temperature, made permeable, and stained with anti-IFN-γ and -CD69 MAbs (Caltag) for 30 min at 4°C. Permeablized cells were washed and resuspended in 1% formaldehyde. Samples were analyzed on a FACScan flow cytometer. Generally 200,000 to 400,000 events in the lymphocyte gate were collected and analyzed using Cell Quest software.
Expansion of antigen-specific CD4+ T cells.
PBMCs (106 PBMCs/ml) were cultured with HIV-1 p24 antigen (5 μg/ml) for 1 week in RPMI 10% human serum at 37°C in a humidified 5% CO2 atmosphere. The frequency of cultured CD4+ T cells secreting IFN-γ in response to antigenic stimulation was determined as described above, with the exception that autologous antigen-presenting cells (CD3+ T-cell-depleted autologous PBMCs) were added 1:1 to cultured cells during the 14-h incubation with HIV-1 protein and peptides.
HLA typing.
Serological and molecular HLA-DR typing was carried out by the University of Colorado Health Sciences Center Histocompatibility Laboratory. Serologic HLA-DRB1 and HLA-DRB3/4/5 typing was performed on all HIV-1-infected donor specimens using standard microcomplement-dependent cytotoxicity. High-resolution HLA-DRB1 typing was selectively performed using the sequence-based typing method with commercial kits from Perkin-Elmer.
RESULTS
Selection of a set of the most prevalent human HLA-DR molecules.
The goal of the present study was to identify a number of conserved, HIV-1-derived, HLA-DR-restricted epitopes. We were particularly interested in epitopes with the potential of being recognized by the majority of humans, irrespective of ethnicity. Correspondingly, a set of HLA-DR types representing an overwhelming majority of the world population was selected for use in the epitope screening process. The specific set of HLA-DR antigens utilized in this study is shown in Table 1. The only major HLA-DR antigen not included in the analysis was HLA-DR10, since this antigen is expressed at a low frequency. As a rule, the most common allelic subtype of each HLA-DR antigen was used for the purpose of measuring peptide-binding affinities, the one exception being HLA-DR4, for which both the DRB1*0401 and DRB1*0405 subtypes were utilized, since differences in the peptide binding specificities of the products of these particular alleles are known (25, 29). In addition to the B1 gene, humans also express the B3, B4, and B5 genes, the products of which, when complexed with the invariant HLA-DR alpha chain, generate the serologic HLA-DR51, -52, and -53 antigens. Due to linkage disequilibrium, specific B1 allele products are generally found associated with the expression of particular B3, B4, and B5 alleles. Accordingly, representative allelic products of these antigens were also included in the analysis.
TABLE 1.
Frequencies of HLA-DR antigens in common ethnicities and the patient populationa
| Antigen | Representative allele(s) | Phenotypic frequency (%) of antigen in group
|
nc | |||||
|---|---|---|---|---|---|---|---|---|
| Asian | Black | Euro. Cauc. | N. Amer. Cauc. | Avg | Patient populationb | |||
| DR1 | DRB1*0101 | 6.0 | 13.1 | 19.3 | 22.5 | 15.2 | 12.9 | 4 |
| DR2 | DRB1*1501 | 34.7 | 29.2 | 27.6 | 27.3 | 29.7 | 32.2 | 10 |
| DR3 | DRB1*0301 | 5.2 | 22.4 | 24.7 | 21.0 | 18.3 | 6.4 | 2 |
| DR4 | DRB1*0401, DRB1*0405 | 35.4 | 8.9 | 29.3 | 31.1 | 26.2 | 45.2 | 14 |
| DR6 | DRB1*1302 | 27.2 | 43.8 | 26.5 | 28.0 | 31.4 | 41.9 | 13 |
| DR7 | DRB1*0701 | 4.2 | 14.8 | 25.5 | 23.4 | 17.0 | 29.0 | 9 |
| DR8 | DRB1*0802 | 18.6 | 10.7 | 5.4 | 6.7 | 10.4 | 9.7 | 3 |
| DR9 | DRB1*0901 | 23.5 | 3.9 | 2.0 | 2.0 | 7.8 | 0.0 | 0 |
| DR11 | DRB1*1101 | 7.7 | 23.1 | 18.2 | 18.5 | 16.9 | 19.4 | 6 |
| DR12 | DRB1*1201 | 15.3 | 9.6 | 3.4 | 2.2 | 7.6 | 3.2 | 1 |
| DR51 | DRB5*0101 | 22.0 | 16.5 | 32.2 | 23.9 | 23.9 | 31.2d | 10 |
| DR52 | DRB3*0101 | 54.2 | 55.2 | 39.7 | 53.2 | 51.4 | 70.9 | 22 |
| DR53 | DRB4*0101 | 48.9 | 21.1 | 59.6 | 50.9 | 45.8 | 77.4 | 24 |
Phenotypic frequencies of DR antigens were derived from gene frequencies reported previously (17a). Euro. Cauc., European Caucasian; N. Amer. Cauc., North American Caucasian.
The patient population consists of 31 DRB1-typed patients.
The number of patients expressing the indicated DR antigen.
Phenotypic frequencies of DR51, DR52, and DR53 are based on predicted linkage disequilibrium.
The estimated phenotypic frequencies of the selected HLA-DR antigens in different ethnic groups (23) are also presented in Table 1. Each DR antigen would be projected to be expressed in 8 to 31% of an average population for the B1 antigens and up to 50% in the case of DRB3, -B4, and -B5 antigens. The patient population used in this study consisted of 31 HIV-1-infected patients from a restricted geographical area. On an individual HLA-DR antigen basis, the observed phenotypic frequencies in the patient cohort are similar to the estimated frequencies for an average population. The only HLA-DR antigen not represented in our patient population was HLA-DR9, which is likely a result of the relatively small group of patients studied and the fact that HLA-DR9 is on average expressed at a lower frequency. When considered from the standpoint of HLA-DR expression, these results indicate that the cohort utilized generally reflects the HLA-DR frequency expected in a global population.
Selection of highly cross-reactive HLA-DR binding peptides.
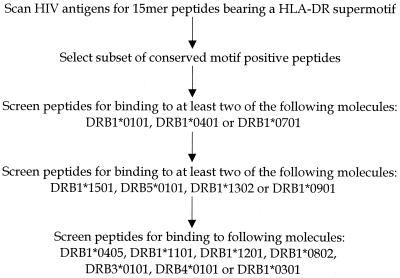
To identify peptides with highly cross-reactive HLA-DR binding capacity, the amino acid sequences of Gag, Pol, Nef, Rev, Tat, Vif, Vpr, and Vpu were scanned for the presence of the HLA-DR supermotif described by O'Sullivan et al. (31) and Southwood et al. (49) (Fig. 1). Specifically, 15-amino-acid peptides containing a 9-residue core region comprised of a DR supermotif, and 3 N- and C-terminal flanking amino acids, were selected. Although most of the energy of HLA-DR–peptide interactions appears to be contributed by the nine-residue core region, the N- and C-terminal flanking amino acids were included in the selection process since these residues are clearly necessary in most instances for T-cell recognition.
FIG. 1.
The amino acid sequences of Gag, Pol, Nef, Rev, Tat, Vif, Vpr, and Vpu were scanned for the presence of 15-amino-acid peptides containing the HLA-DR supermotif. Conserved peptides were tested for HLA-DR binding affinity in a sequential panel of HLA-DR binding assays.
A subset of the motif-bearing peptides was selected for further analysis on the basis of conservancy (Fig. 1). Peptides were tested for binding to the various HLA-DR antigens only if the contiguous nine-amino-acid core region was conserved in >50% of the clade B isolates analyzed. While selecting peptides based on conservation may preclude the identification of variable epitopes, this approach should result in the selection of epitopes that will be antigenic in a greater percentage of the HIV-1-infected population. Correspondingly, these epitopes may be more suitable for use as vaccine candidates.
Previous studies (49) have suggested that an IC50 of 1,000 nM represents an affinity threshold associated with immunogenicity in the context of DR molecules. Accordingly, this threshold was utilized as a cutoff value for epitope prediction. Conserved, motif-bearing peptides were initially screened for binding to the DRB1*0101, DRB1*0401, and DRB1*0701 subtypes (Fig. 1). Peptides binding at least two of these three DR molecules with an affinity of ≥1,000 nM were screened for binding to the DRB1*1501, DRB1*1302, DRB1*0901, and DRB5*0101 subtypes. Finally, peptides binding at least two of the four secondary panel DR molecules were screened for binding to the DRB1*0405, DRB*1101, DRB*1201, DRB1*0802, DRB3*0101, DRB4*0101, and DRB*0301 subtypes.
As a result of this analysis, 11 conserved highly cross-reactive HLA-DR binding peptides were identified. The binding affinities of each of these peptides to the various HLA-DR allele products are shown in Table 2. It should be noted that all of the peptides presented in Table 2 bound seven or more of the HLA-DR molecules examined at an IC50 of 1,000 nM or lower. In conclusion, the selection process described resulted in the identification of 11 highly cross-reactive HLA-DR binding peptides that would be predicted to be antigenic in HIV-1-infected patients.
TABLE 2.
HLA-DR binding peptides.
| Peptide | Binding capacity (IC50 [nM]) for:
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DR1 (DRB1*0101)a | DR2 (DRB1*1501) | DR3 (DRB1*0301) | DR4 (DRB1*0401) | DR4 (DRB1*0405) | DR5 (DRB1*1101) | DR5 (DRB1*1201) | DR6 (DRB1*1302) | DR7 (DRB1*0701) | DR8 (DRB1*0802) | DR9 (DRB1*0901) | DR51 (DRB5*0101) | DR52 (DRB3*0101) | DR53 (DRB4*0101) | |
| Gag 171 | 72 | 65 | 17647 | 60 | 400 | —b | — | 412 | 455 | 7313 | 117 | 13 | — | 135 |
| Gag 294 | 82 | 138 | — | 1,667 | 380 | 213 | 1656 | 98 | 192 | 63 | 536 | 225 | 1205 | 161 |
| Gag 298 | 4.2 | 5.1 | 188 | 633 | 404 | 54 | 124 | 0.36 | 379 | 49 | 58 | 24 | 12,051 | 121 |
| Pol 303 | 185 | 70 | — | 294 | 136 | 1,818 | — | — | 30 | 803 | 39 | 4,167 | 18,076 | — |
| Pol 335 | 357 | 217 | — | 3,571 | 109 | 741 | — | 13 | 68 | 3,267 | 33 | 667 | 887 | 14,500 |
| Pol 596 | 7.2 | 222 | 13,636 | 28 | 20 | 317 | 1,355 | 90 | 15 | 350 | 39 | 2.1 | 8,545 | 527 |
| Pol 711 | 3.6 | 21 | 3,226 | 9.3 | 27 | 37 | 6,478 | 3,500 | 18 | 31 | 144 | 4.9 | — | 14 |
| Pol 712 | 8.3 | 25 | — | 156 | 165 | 71 | 12,598 | 2,500 | 179 | 196 | 250 | 24 | — | 290 |
| Pol 758 | 38 | — | — | 11 | 15 | 95 | — | 4,375 | 472 | 1,960 | 872 | 125 | — | 951 |
| Pol 915 | 161 | 650 | — | 909 | 452 | 182 | 18,625 | 125 | 1,786 | 1,441 | 2,586 | 690 | 14,688 | 1,000 |
| Pol 956 | 2.9 | 3.4 | — | 357 | 49 | 53 | 124 | 25 | 25 | 75 | 577 | 80 | — | 611 |
Representative allele used to test binding to the corresponding antigen.
— indicates an IC50 of >20 μM.
Conservation and expected population coverage of HLA-DR binding peptides.
As shown in Table 3, the HLA-DR highly cross-reactive binding peptides are well conserved in a variety of HIV-1 isolates. While the selection process excluded peptides that are less than 50% conserved in clade B isolates, in fact, the overall conservation of these peptides is in general much higher. On average, this set of peptides is 82% conserved in clade B isolates. As expected, the conservation of these peptides is lower when other clades are included in this analysis. Nevertheless, these peptides are conserved on average in 57% of all the HIV-1 isolates examined.
TABLE 3.
Conservation and expected population coverage of HLA-DR binding peptides
| Peptide | Sequence | Conservation (%)a
|
No. of DR antigens bound | Projected population coverage (%)b | |
|---|---|---|---|---|---|
| Total | Clade B | ||||
| Gag 171–185 | QGQMVHQAISPRTLN | 41 | 52 | 9 | 77.0 |
| Gag 294–308 | GEIYKRWIILGLNKI | 58 | 95 | 10 | 89.7 |
| Gag 298–312 | KRWIILGLNKIVRMY | 85 | 94 | 13 | 95.4 |
| Pol 303–317 | FRKYTAFTIPSINNE | 59 | 68 | 7 | 77.0 |
| Pol 335–349 | SPAIFQSSMTKILEP | 52 | 79 | 9 | 88.6 |
| Pol 596–610 | WEFVNTPPLVKLWYQ | 79 | 84 | 11 | 89.7 |
| Pol 711–725 | EKVYLAWVPAHKGIG | 32 | 94 | 10 | 84.1 |
| Pol 712–726 | KVYLAWVPAHKGIGG | 32 | 89 | 10 | 84.1 |
| Pol 758–772 | HSNWRAMASDFNLPP | 48 | 68 | 8 | 77.2 |
| Pol 915–929 | KTAVQMAVFIHNFKR | 87 | 94 | 8 | 84.1 |
| Pol 956–970 | QKQITKIQNFRVYYR | 56 | 95 | 12 | 92.5 |
Conservation of the contiguous sequence based on the 1999 Los Alamos HIV molecular immunology database.
Projected population coverage was calculated considering the phenotypic frequencies of the antigens which bind the corresponding peptide.
Also shown in Table 3 is the projected population coverage of each peptide. Calculations of population coverage were based on the phenotypic frequencies of the HLA-DR antigens and assumed that the HLA-DR molecules are representative of all subtypes of the same antigen. All of the peptides would be predicted to be potentially recognized in at least 77% and up to 95% of an average population.
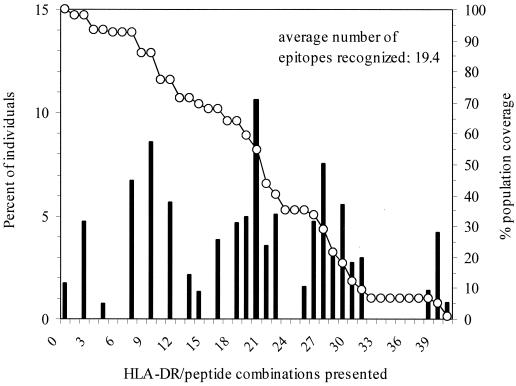
We also calculated the projected population coverage, defined as the total number of different DR-peptide combinations potentially presented in a given individual by the panel of 11 epitopes. Since 11 peptides were identified and up to four different DR molecules can be expressed in each individual, the theoretical maximum number of different DR-peptide combinations presented is 44. The percentage of individuals yielding any given number of DR-peptide combinations is shown in Fig. 2. Less than 2% of members of an average population are not predicted to show any peptide binding. The average number of DR-peptide combinations presented is 19.3. In conclusion, this set of conserved peptides would be projected to bind to HLA-DR antigens expressed in a large fraction of the human population.
FIG. 2.
Percentages of individuals projected to present the indicated number of HLA-DR–epitope combinations in a composite population, derived from gene frequencies in Asian, black, European Caucasian, and North American Caucasian populations (black bars). Also shown on the right axis is the cumulative plot of percent projected population coverage (open circles).
Antigenicity of the highly cross-reactive HLA-DR binding peptides in HIV-1-infected patients.
In order to determine whether these highly cross-reactive HLA-DR binding peptides are recognized during the course of HIV-1 infection, the antigenicity of the 11 supermotif peptides (three in Gag p24, five in Pol p66, and three in Pol integrase) was assessed using recall lymphocyte stimulation assays in an initial panel of 22 HIV-1-infected donors and 13 uninfected donors. The clinical characteristics of the HIV-1-infected subjects are shown in Table 4. The 22 HIV-1-infected donors initially tested were receiving combination antiretroviral therapy (three or more agents), with peripheral CD4 counts ranging from 43 to 856 cells/mm3 (mean, 482 cells/mm3) and plasma HIV-1 RNA viral loads ranging from <20 to 89,520 viral copies/ml of plasma (73% with viral loads of <400 copies/ml of plasma HIV-1 RNA).
TABLE 4.
Clinical characteristics and HLA-DR serotypes of HIV-1-infected study subjects
| Subject | Count (cells/mm3)
|
Plasma HIV RNA (no. of copies/ml)c | Current antiretroviral regimend | Duration of current regimen (months) | Length of time HIV-1+ (yr)f | HLA-DRB1 typingg | HTL peptide response | |
|---|---|---|---|---|---|---|---|---|
| CD4a | CD4 nadirb | |||||||
| UH1 | 43 | 27 | 89,520 | ABC/D4T/EFV/SQV/RTV | 6e | 6 | 6, 7 | − |
| UH2 | 139 | 8 | <20 | D4T/3TC/EFV | 4e | 4 | 4, 8 | − |
| UH3 | 188 | 50 | 2,821 | ABC/D4T/EFV/NFV | 12e | 5 | 1, 6 | + |
| UH4 | 255 | 111 | <400 | D4T/3TC/IND | 12e | 8 | 4, 13 | − |
| UH5 | 259 | 161 | 1,526 | EFV/SQV/RTV | 3e | 4 | 13, 15 | + |
| UH6 | 384 | 116 | <20 | D4T/3TC/IND | 38e | 7 | 7, 15 | + |
| UH7 | 387 | 190 | <400 | D4T/3TC/NFV | 10e | 9 | 4, 11 | − |
| UH8 | 403 | 19 | <20 | D4T/3TC/NFV | 23e | 11 | 4, 14 | + |
| UH9 | 436 | 180 | <20 | ABC/DDI/D4T/EFV/NFV | 9e | 9 | 3, 13 | − |
| UH10 | 458 | 50 | 1,903 | ABC/D4T/EFV | 22e | 4 | 4, 15 | + |
| UH11 | 487 | 456 | <200 | AZT/3TC/IND | 12 | 1h | 1, 15 | + |
| UH12 | 504 | 17 | <20 | D4T/DDI/RTV | 40e | 4 | 14, 15 | − |
| UH13 | 524 | 245 | <200 | D4T/3TC/EFV/NFV | 31e | 4 | 16, 17 | + |
| UH14 | 548 | 165 | 170 | AZT/3TC/IND | 20 | 6 | 8, 15 | − |
| UH15 | 577 | 273 | <20 | D4T/3TC/IND | 26e | 13 | 7, 11 | − |
| UH16 | 619 | 247 | 72,600 | AZT/3TC/SQV/RTV | 19e | 8 | 7, 15 | − |
| UH17 | 735 | 351 | 62 | AZT/3TC/APV | 29e | 7 | 4, 7 | + |
| UH18 | 738 | 317 | <20 | D4T/3TC/NFV | 24e | 7 | 4, 13 | + |
| UH19 | 786 | 411 | 529 | D4T/3TC/NVP | 1 | <1 | 4, 12 | + |
| UH20 | 803 | 581 | <400 | AZT/3TC/APV | 27e | 7 | 11 | + |
| UH21 | 856 | 490 | <20 | AZT/3TC/NFV | 25e | 10 | 13, 15 | + |
| UH22 | 466 | 0 | <20 | D4T/3TC/RTV | 44 | 7 | 8, 13 | + |
Peripheral CD4+ T-lymphocyte count at the time of study.
Lowest documented peripheral CD4+ T-lymphocyte count.
Copies of HIV-1 RNA/ml of plasma at the time of study.
Antiretroviral drug abbreviations: ABC, abacavir; APV, amprenavir; AZT, zidovudine; D4T, stavudine; EFV, efavirenz; IND, indinavir; NFV, nelfinavir; NVP, nevirapine; RTV, ritonavir; SQV, saquinavir; 3TC, lamivudine.
This subject received prior antiretroviral therapy.
Time elapsed since subject first was documented to be HIV-1 positive by serologic testing.
As determined by serologic HLA typing.
Documented acute HIV-1 infection 1 year prior to study.
Recall HTL responses were determined by stimulating PBMCs from HIV-1-infected or uninfected donors with each peptide individually and measuring tritiated thymidine incorporation after 6 days. Antigen-specific T-cell proliferation was calculated as an S.I., defined as the ratio of thymidine incorporation in the presence of antigen divided by the incorporation in the absence of antigen. In order to accurately determine the significance of peptide-specific responses in HIV-1+ donors, a comparison was made to responses to each peptide obtained using a normal donor cohort. In addition to establishing true background proliferation for each HTL peptide, rather than assigning arbitrary criteria for significance, this normal donor analysis assured us that the responses seen in the HIV-1+ donor group were not the result of in vitro priming of HTL peptide-specific T cells. For the purpose of data analysis, a significant proliferative response to the supermotif peptides, based on peptide responses of HIV− control donors (mean S.I. plus 2 standard deviations [SD]), was determined to be an S.I. of ≥2 for all peptides, with the following exceptions: Gag 171 (S.I. ≥ 2.3), Pol 335 (S.I. ≥ 2.8), and Pol 915 (S.I. ≥ 2.8).
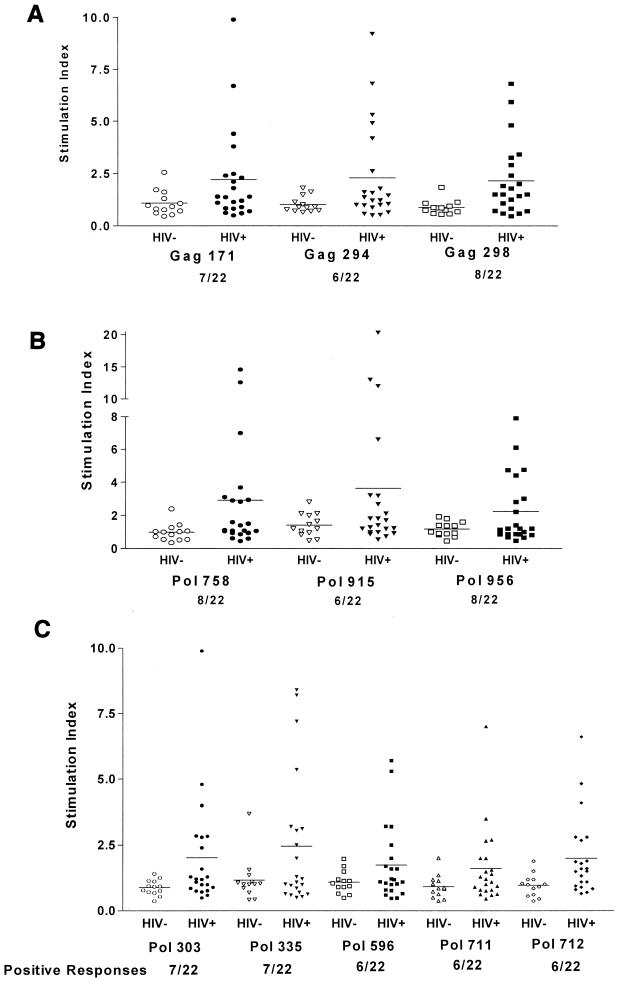
Proliferative responses to each of the 11 highly cross-reactive HLA-DR binding peptides by PBMCs from 22 HIV-1-infected and 13 seronegative donors are depicted in Fig. 3. Using the criteria described above for a significant peptide response, all 11 peptides were recognized in recall proliferative responses by PBMCs from at least six HIV-1-infected patients. Three peptides, Gag 298, Pol 758, and Pol 956, elicited responses in PBMCs of 8 of the 22 subjects. Even when applying more conservative criteria for establishing the significance of a peptide-specific response by using a cutoff S.I. of greater than or equal to 5, a value often used to establish significant proliferative responses to whole-protein antigens, each HTL peptide was still recognized by PBMCs from one or more HIV+ donors. In addition, PBMCs from donors with disparate MHC class II antigens recognized the same peptide, suggesting that multiple HLA-DR molecules can present a given peptide (Tables 4 and 5). The responses of greatest magnitude were seen against the Pol 758 and Pol 915 peptides, both sequences in the HIV-1 integrase protein.
FIG. 3.
Proliferative responses to supermotif HIV-1 HTL epitope peptides. Individual proliferative responses of 22 HIV-1-infected donors and 13 uninfected donors to 11 highly cross-reactive HLA-DR binding HIV-1 peptides, 3 derived from HIV-1 Gag sequences (A), 3 from HIV-1 Pol integrase sequences (B), and 5 from Pol p66 sequences (C), in a 6-day proliferation assay are shown. Proliferative responses to each of the 11 HIV-1 supermotif peptides are depicted as separate points in terms of S.I., with the mean for each group depicted as a solid bar. The number of HIV+ donors that significantly responded to each peptide is listed below the figure. A significant proliferative response to the supermotif peptides was defined based on responses of HIV− control donors, with an S.I. of ≥2 considered significant for all peptides except Gag 171 (S.I. ≥ 2.3) and Pol peptides 335 and 915 (S.I. ≥ 2.8).
TABLE 5.
Proliferative responses of PBMCs from peptide-responsive HIV-infected donors to HIV-1 proteins and HIV-1 HTL peptides
| HIV+ donora | Response of PBMCs tob:
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HIV-1 proteins
|
HIV-1 supermotif peptides
|
||||||||||||
| P24 | P66 | Gag 171 | Gag 294 | Gag 298 | Pol 303 | Pol 335 | Pol 596 | Pol 711 | Pol 712 | Pol 758 | Pol 915 | Pol 956 | |
| UH3 | 2.6 | 3.8 | — | — | — | — | 8.2 | — | — | — | — | — | — |
| UH5 | 1.8 | 16.6 | 2.3 | 2.6 | 2.4 | — | — | — | 7 | — | — | — | 4.4 |
| UH6 | 2.9 | 6.4 | 2.4 | 5.3 | — | 2.4 | — | — | 2.7 | 2.8 | 2.9 | — | 2.8 |
| UH8 | 32.2 | 22.1 | — | — | — | 2.9 | 3.1 | — | — | — | 2.8 | 3.2 | — |
| UH10 | 14 | 86.9 | 2.5 | — | 3.3 | — | — | — | — | — | — | — | — |
| UH11 | 2.5 | 6.5 | — | — | 4.8 | 4 | 3.2 | 2.5 | 2 | 4.1 | 3 | — | — |
| UH13 | 3.8 | 6 | — | — | — | — | — | — | — | — | — | — | 4.7 |
| UH17 | 5.5 | 4.2 | — | — | — | — | — | 5.7 | — | — | 12.6 | 12 | 7.9 |
| UH18 | 4 | 3.8 | 6.7 | 9.2 | 3.4 | 9.9 | 7.2 | 5.3 | 3.5 | 6.6 | 14.6 | 6.6 | 3 |
| UH19 | 5.2 | 318.3 | 3.8 | — | 5.9 | 4.8 | 5.4 | 3.2 | 2.7 | 4.8 | 3.7 | 20.3 | 6.1 |
| UH20 | 62.2 | 22.1 | 9.9 | 4.2 | 6.8 | 2.8 | 3.1 | 3.2 | — | 2.7 | 2.9 | 13 | 4.7 |
| UH21 | 95 | 8.2 | 4.4 | 4.9 | 2.9 | 2.8 | 8.4 | 2 | 2 | 2.8 | 7 | 3.2 | 2.2 |
| UH22 | 69.6 | 32 | — | 6.8 | 2 | — | — | — | — | — | — | — | — |
The following HIV+ donors failed to respond to any of the HIV HTL epitope peptides: UH1, UH2, UH4, UH7, UH9, UH12, UH14, UH15, and UH16.
Values denote S.I. Peptide responses that failed to reach significance are denoted by a dash (—). A significant S.I. based on HIV− control responses ≥2 for all peptides with the following exceptions: Gag 171 (S.I. > 2.3), Pol 335 (S.I. > 2.8), and Pol 915 (S.I. > 2.8).
Overall, PBMCs from 13 of the initial 22 HIV+ donors tested responded to one or more of the supermotif peptides (Table 5). Of these peptide responders, several recognized multiple peptides. There was a trend toward broader peptide recognition and a greater magnitude of peptide-specific responses in cells from donors with higher CD4 counts (Tables 4 and 5). On average, the HIV-1-infected donors whose cells responded to one or more of the peptides responded to an average of six of the HLA-DR highly cross-reactive binding peptides. Notably, PBMCs from four patients, UH18, -19, -20, and -21, responded to 10 or more of the HLA-DR supertype peptides tested.
Analysis of HLA-DR restriction and functional frequency of HIV-1 HTL supertype peptide-specific CD4+ T cells.
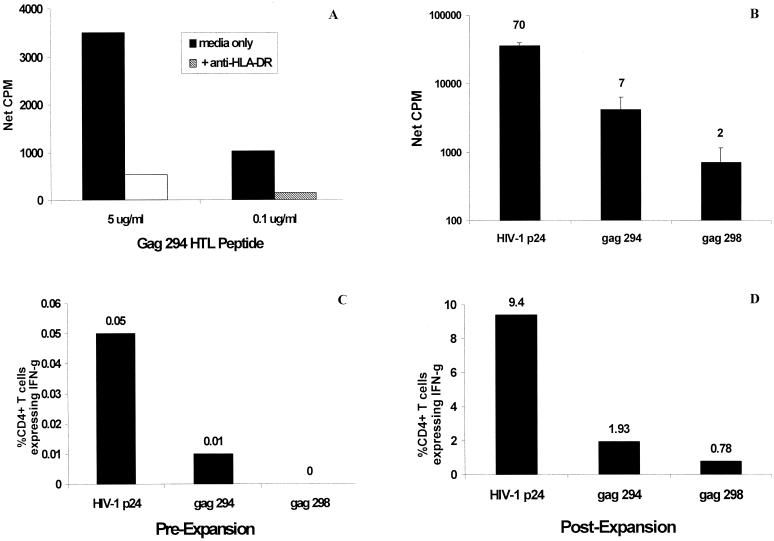
The MHC class II-restricted nature of the peptide-specific responses was confirmed by specifically blocking proliferation with an anti-HLA-DR MAb, a representative example of which was used for Fig. 4A. The presence of an HLA-DR antibody resulted in an 80% reduction in proliferation to the peptide examined. The frequency of IFN-γ-producing CD4+ T cells responding to HIV-1 p24 and p66 proteins and to the corresponding supermotif peptides was determined for selected HIV+ donors by antigen-induced intracellular cytokine analysis. In the donor cohort tested, frequencies of HIV p24- and p66-specific, IFN-γ-producing CD4+ T cells among fresh PBMCs ranged from 0.02 to 0.35% of CD4+ T cells. Of nine donors evaluated for responses to supermotif peptides, the frequency among PBMCs of CD4+ T cells producing IFN-γ was either low, not exceeding 0.06% of CD4+ T cells, or below the level of detection of the assay. However, supermotif peptide-specific CD4+ T-cell responses could be easily measured in cultured PBMCs from the same donors following a week of in vitro stimulation with the parental HIV-1 protein. A representative example is shown in Fig. 4. Figure 4B depicts the baseline lymphoproliferative responses of PBMCs from donor UH22 to recombinant p24 protein and the Gag 294 and 298 supermotif peptides. The ability of CD4+ cells from this donor to secrete IFN-γ in response to the p24 protein and the Gag peptides was determined with fresh PBMCs (Fig. 4C) and following a stimulation period of PBMCs with p24 antigen (Fig. 4D). Among PBMCs, a low but detectable frequency of p24-specific, IFN-γ-secreting cells was noted (1 in 2,000 CD4+ T cells), whereas the response to Gag 294 was measurable (1 in 10,000 CD4+ T cells) but not reliably within the detection limits of the assay. An IFN-γ response to Gag 298, if present, was below the limits of detection of the assay. However, following culturing with p24 antigen, the CD4 responses to both p24 and the Gag peptides comprised a significantly greater fraction of the total CD4+ T cells, well within the detectable range of the assay. Taken together, these results suggest that the observed lymphoproliferative responses to the HIV-1 supermotif peptides are mediated, at least in part, by functional CD4+ T cells capable of proliferation and the secretion of IFN-γ, a TH1-type cytokine. The fact that these HIV-1 peptide- and protein-specific T-helper cells exist in the peripheral blood at a low frequency in the treated HIV+ patient cohort that we studied is consistent with the findings of others and provides a rationale for targeting this donor population with therapeutic vaccination strategies.
FIG. 4.
Analysis of HLA-DR restriction and functional frequencies of HIV-1 HTL supertype peptide-specific CD4+ T cells. (A) Blocking of UH22 PBMC proliferation to the peptides using specific anti-HLA-DR MAb. (B) Lymphoproliferative responses of PBMCs from HIV+ donor UH22 to HIV-1 p24 protein and two Gag supertype epitope peptides contained within the p24 sequence (Gag 294 and 298). The corresponding S.I. are indicated above each bar. The frequencies of CD4+ lymphocytes from donor UH22 producing IFN-γ in response to p24 protein or the Gag peptides both in fresh PBMCs (C) and following in vitro expansion of PBMCs with p24 antigen (D) are depicted.
Antigenicity of HLA-DR binding peptides correlates with recognition of naturally processed antigen.
Also shown in Table 5 are the proliferative responses observed to stimulation with recombinant p24 and p66 antigen. A positive association was noted between responses to the parental protein and responses to the corresponding peptides contained within that protein. Eight of the nine subjects (89%) that recognized at least one of the Gag peptides contained within the p24 antigen (Gag 171, 294, and 298) also recognized the HIV-1 p24 protein with an S.I. of ≥2.5. Of the subjects that recognized peptides contained within the p66 protein (Pol 303, 335, 596, 711, and 712), 100% recognized the corresponding whole antigen. The finding that the peptide-specific responses were not seen in uninfected controls and that they correlated with responses to intact antigen implies that these peptides are processed and presented during the course of HIV-1 infection. In summary, the panel of highly cross-reactive HLA-DR binding peptides represents a set of class II-restricted epitopes targeted in a number of HIV-1-infected individuals.
Lack of peptide-specific proliferative responses is correlated with a defect in recall responses to HIV-1 antigens.
Collectively, one or more of the HLA-DR supertype peptides was antigenic in 59% of the HIV-1-infected patients evaluated. Since the set of peptides studied has the potential of being recognized in 70 to 95% of these individuals, less-frequent antigenicity might be a reflection of either different epitopes being recognized (immunodominance) or fewer epitopes being recognized (immunodeficiency). This prompted further analysis to compare the various parameters associated with the 13 peptide responders to the 9 peptide nonresponders. Differences in the peripheral CD4 counts and HIV-1 RNA levels between the responder group and the nonresponder group did not reach statistical significance (two-tailed Mann-Whitney test; 95% confidence). Although viral loads were not significantly different between the two groups due to the large number of “suppressed” individuals in each group, it is notable that cells from the two subjects with the highest viral burdens (donors UH1 and UH16) failed to respond to the HIV-1 HTL peptides (Table 4). While these data are limited, this observation supports other reports that suggest an adverse effect of HIV-1 replication on the generation or maintenance of HIV-1-specific HTL responses.
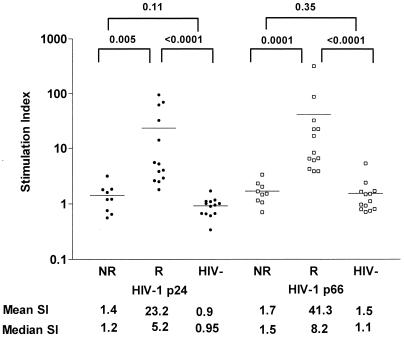
Statistically significant differences between the peptide responders and nonresponders were noted when individuals in each group were assessed for recall responses to recombinant HIV-1 antigens (Fig. 5). HIV-1-infected subjects that responded to the HLA-DR binding peptides generally had strong responses to whole HIV-1 proteins, with responses against HIV-1 p24 and p66 in the responder group being significantly stronger than responses elicited in the nonresponder group (P = 0.005 and 0.0001, respectively). In contrast, those HIV-1-infected donors whose PBMCs failed to respond to HLA-DR binding peptides likewise had poor responses to whole HIV-1 proteins, with responses to p24 and p66 not significantly different from responses of uninfected donors. These data indicate that the subjects whose PBMCs fail to recognize the HTL epitope peptides also have a reduced capacity to recognize whole HIV-1 proteins, suggesting that these patients possess a general defect in their CD4+ T-cell-mediated HIV-1-specific immune responses.
FIG. 5.
Proliferative responses of PBMCs of HIV-1+ HTL epitope peptide responders (R), HIV-1+ HTL epitope peptide nonresponders (NR), and uninfected donors (HIV-) to recombinant HIV-1 p24 and p66 protein antigens. PBMCs from 22 HIV-1-infected donors (13 of whom had significant proliferative responses to 1 or more of the 11 supermotif HIV-1 HTL peptides and 9 of whom failed to respond to any of the peptides tested) and 13 uninfected donors were tested for their ability to proliferate in response to recombinant HIV-1 p24 and p66 proteins. The mean S.I. for each group is depicted in the figure as a solid line, and the mean S.I. and median S.I. for each group are shown numerically below the figure. P values for differences between each group (Mann-Whitney test) are shown at the top of the figure.
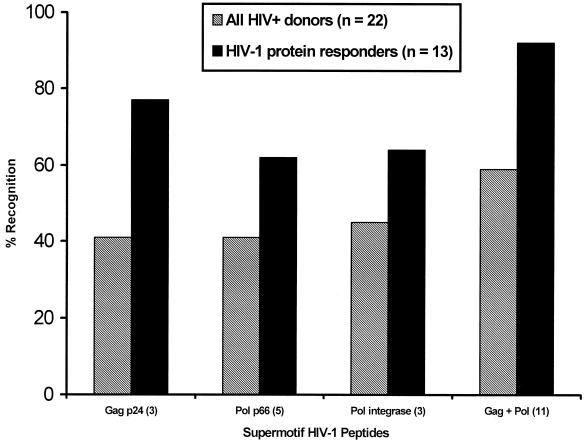
Given the association between significant responses to whole proteins and supermotif peptides, and the greater likelihood that significant epitope-specific responses could be detected in subjects with HIV-specific T-helper responses of greater magnitude, responses to the supertype peptides were evaluated in a select cohort of HIV+ donors who displayed strong HIV-specific T-helper responses. Out of more than 50 HIV-infected donors screened, 13 displayed strong lymphoproliferative responses to both the HIV-1 p24 and the HIV-1 p66 proteins (S.I. > 5; net cpm > 1,000). Collectively, cells from 92% of those HIV+ donors with the strongest responses to HIV-1 p24 and p66 recognized one or more of the 11 supertype peptides, compared to 59% for the HIV+ donors not selected based on the presence of HIV-specific T-helper-cell responses (Fig. 6). Likewise, recognition of the Gag or Pol peptides by the p24 or p66 protein responders, respectively, was 30 to 50% greater than the recognition noted in the unselected donor cohort. Additionally, in this group of subjects with intact HIV-specific T-helper responses, responses for a given donor were quite broad. On average, two of three Gag, three of five Pol p66, and two of three Pol integrase peptides were recognized by cells from each donor in this cohort. These results confirm the antigenicity of these supermotif peptides in HIV-infected individuals and underscore the high frequency with which they may be targeted in HIV-infected persons capable of mounting an HIV-specific HTL response.
FIG. 6.
Recognition of HIV-1 HTL supertype epitope peptides by HIV-infected donors selected for responses to both recombinant HIV-1 p24 and p66 proteins. Recognition of one or more supertype peptides, based on lymphoproliferation, is compared between an unselected group of 22 HIV+ donors and a group of 13 HIV+ donors selected by the presence of significant lymphoproliferative responses to the two parental proteins, HIV-1 p24 and p66, using the following criteria: S.I. > 5; net cpm > 1,000. Responses are compared for groups of peptides contained within a given HIV-1 protein and for the set of 11 peptides as a whole.
DISCUSSION
We report in the present study the identification of a panel of highly conserved HLA-DR-restricted epitopes derived from HIV-1 antigens. These epitopes were initially identified by screening HIV-1 antigens for peptides that contained the HLA-DR-supertype binding motif, and more than 8,000 peptides that might bind one or more HLA-DR molecules were found. Peptides that were conserved in the majority of isolates were tested for binding using panels of purified HLA-DR antigens representative of the worldwide population. Through this process, 11 peptides capable of binding a minimum of seven HLA-DR types were defined.
All of the HLA-DR highly cross-reactive binding peptides were recognized in recall HTL responses from HIV-1-infected patients. The fact that each of the peptides was antigenic is most likely a result of focusing screening exclusively on conserved peptides capable of binding multiple HLA-DR alleles. Similar correlations between cross-reactive binding capacity and antigenicity have been observed for MHC class I binding epitopes (46). One consequence of focusing on conserved and highly cross-reactive HLA-DR binding peptides is that some epitopes that are poorly conserved between viral isolates or that have narrow HLA-DR binding affinities have not been studied. Given the moderate responses to whole HIV-1 protein antigens in many cases, we expected that responses directed against individual HTL epitopes would be of low frequency. The finding that peptide-specific responses were in general weaker than responses to whole antigen may be evidence that epitopes other than those examined in this study are also recognized.
Historically, measuring CD4+ T-cell responses to individual HTL epitopes in the setting of HIV-1 infection, a disease marked by depletion of peripheral CD4+ lymphocytes and T-helper-cell dysfunction, has been difficult. Although viral suppression induced by highly active antiretroviral therapy (HAART) has been shown to increase the peripheral CD4+ T-cell count and lead to significant immune reconstitution (4), HIV-1-specific T-helper-cell responses are generally not restored following the institution of HAART (28, 34, 36) and may actually decline with prolonged viral suppression (33). However, significant proliferative responses to the HIV-1 p24 and p66 proteins were noted with a considerable number of our HIV-1+ donors receiving combination antiretroviral therapy, suggesting that this would be a reasonable population in which to study responses to individual HTL epitopes. The high percentage of HIV-infected individuals in this study that were responsive to HIV-1 p24 and p66 antigens might be explained by the heterogeneity of the donor cohort itself. Although all were receiving antiretroviral therapy, with the majority achieving viral suppression at the time of the study, this donor cohort includes individuals with a varied range of CD4 counts, duration of HIV-1 infection, and duration of antiretroviral therapy (Table 4). In this complex outpatient population, we would postulate that such factors as host-dependent variations in disease progression, periods of unstructured treatment interruption (i.e., missed doses), and the initiation of therapy in less-advanced stages of HIV-1 disease might contribute to the high percentage of individuals with significant HIV-specific T-helper-cell responses detected in our patient cohort (3, 21, 37). It has been suggested that HIV-1-infected individuals achieving near or complete viral suppression on HAART might potentially benefit from therapeutic vaccination approaches designed to boost HIV-1-specific cellular immunity. Although the primary goal of this study was to evaluate the antigenicity of the HIV-1 HTL supermotif peptides, a secondary but important goal was to determine the breadth of preexisting HTL responses in a donor population likely to benefit from such a therapeutic vaccine approach.
The observation that these peptides are capable of stimulating recall lymphoproliferative responses implies that they are processed and presented during the course of HIV-1 infection. The fact that PBMCs from many HIV-1-infected donors recognized multiple peptides illustrates that the naturally occurring HTL response to HIV-1 antigens may be broadly directed in some individuals, extending beyond a limited number of immunodominant epitopes. Failure to respond to the individual epitopes was correlated with the failure to recognize whole HIV-1 antigens. This is consistent with the findings of other investigators that have shown that HIV-1 infection leads to a progressive loss of CD4+ T-lymphocyte function even in the absence of clinical symptoms (35) and that virus-specific responses are generally affected before responses to other recall antigens (10, 27). The panel of epitopes discussed in this study may serve as a tool to characterize in more detail the deficiencies in HIV-1-specific immune responses.
Although cumulatively more research has focused on the identification of HIV-1-derived CTL epitopes, a number of previous studies have defined HTL epitopes. Hale and colleagues identified a series of overlapping epitopes that were immunogenic in a mouse model (20). Subsequent studies illustrated that these peptides were recognized in humans (5). Similar approaches have been used to identify HTL epitopes from other HIV-1 antigens (8, 14, 41). In fact, one of the epitopes examined herein, Pol 711, has been demonstrated to be immunogenic in mice (19). While these studies have illustrated that there is an overlap between mice and humans in the recognition of class II-restricted epitopes, it is likely that many human epitopes would be undetected with this approach.
Another approach for the identification of HTL epitopes has involved screening HIV-1-seropositive individuals for proliferative responses to peptides spanning selected viral antigens (1, 42). In fact, two of the peptides characterized in this study, Gag 171 and Gag 298, are nested within epitopes reported by Rosenberg and colleagues and found to be associated with the control of viremia (37). Although this approach has resulted in the identification of relevant human epitopes in the setting of HIV-1 disease, the immunological impact of HIV-1 infection and active viral replication on CD4+ T lymphocytes has made it technically difficult to measure responses to multiple peptides, identify MHC class II restricting elements, or define the minimal epitopes recognized. The present report extends these prior studies, making use of refined HLA-DR motif algorithms and binding assays to guide the epitope identification process, resulting in the definition of a number of previously unknown epitopes with well-characterized HLA-DR binding affinities.
One of the primary goals of this study was to identify HTL epitopes that could be utilized in broadly applicable vaccines. The epitope identification process we describe incorporates two features particularly well suited to meet these ends, namely conservation and population coverage. HIV-1, like other retroviruses, rapidly mutates, resulting in viral strains that can escape antiviral therapy and immune recognition (7). To be truly effective, vaccines must induce responses that recognize immunologically conserved regions of the virus. Identification of nonvariable epitopes permits the design of a peptide-based vaccine that would focus the immune response on highly conserved viral sequences, decreasing the likelihood that the virus will be capable of mutating to escape immune recognition. Furthermore, utilizing only conserved epitopes allows the development of a vaccine approach that would be effective against numerous viral variants and, as such, have a more global application.
Secondly, the epitope screening process used in this study is based on the ability of peptides to bind multiple HLA-DR alleles. van der Burg and colleagues recently reported the identification of a single HIV-1-derived class II epitope that they estimate would be recognized by 50 to 60% of the population (51). On average, any of the peptides described here would be expected to bind in at least 77% and up to 95% of the human population. From the standpoint of vaccine development, this panel of peptides has the potential to induce HTL responses on average to more than 19 epitopes. Consequently the population coverage afforded by this panel of peptides is very high. This prediction is further supported by the observation that within the HIV-1-infected patient cohort studied here, patients with disparate HLA-DR types were found to respond to the same peptide. The set of HIV-1 epitopes defined in this study, incorporated into an appropriate vaccine format, should allow redundant coverage of a significant fraction of the global population.
Despite the initial success of HAART in controlling HIV-1 infection, issues relating to cost, toxicity, and viral escape limit the usefulness of this treatment. As such, the development of effective prophylactic and therapeutic vaccines remains crucial. The use of multivalent epitope vaccines is a promising approach to fighting infectious disease, since it allows for directing the immune response to epitopes that may have a greater impact on the disease outcome. The identification of highly conserved, widely recognized epitopes represents one of the critical steps in developing such vaccines. The 11 class II-restricted epitopes disclosed in this report meet these criteria and may make such an approach feasible.
ACKNOWLEDGMENTS
We thank the participants of this study for their cooperation. We thank Steven Johnson, Wheaton Williams, John Gerber, and Michael Grodesky for assistance with subject recruitment. We also thank Robert Schooley, Jerry Bill, and Mark Newman for their input and support.
This work was supported in part by NIH grants AI01459 and AI43664 (to C.C.W.).
REFERENCES
- 1.Adams S L, Biti R A, Stewart G J. T-cell response to HIV in natural infection: optimized culture conditions for detecting responses to gag peptides. J Acquir Immune Defic Syndr Hum Retrovirol. 1997;15:257–263. doi: 10.1097/00042560-199708010-00002. [DOI] [PubMed] [Google Scholar]
- 2.Alexander J, Sidney J, Southwood S, Ruppert J, Oseroff C, Maewal A, Snoke K, Serra H M, Kubo R T, Sette A. Development of high potency universal DR-restricted helper epitopes by modification of high affinity DR-blocking peptides. Immunity. 1994;1:751–761. doi: 10.1016/s1074-7613(94)80017-0. [DOI] [PubMed] [Google Scholar]
- 3.Al Harthi L, Siegel J, Spritzler J, Pottage J, Agnoli M, Landay A. Maximum suppression of HIV replication leads to the restoration of HIV-specific responses in early HIV disease. AIDS. 2000;14:761–770. doi: 10.1097/00002030-200005050-00001. [DOI] [PubMed] [Google Scholar]
- 4.Autran B, Carcelain G, Li T S, Mathez D, Tubiana R, Katlama C, Debre P, Leibowitch J. Positive effects of combined antiretroviral therapy on CD4+ T cell homeostasis and function in advanced HIV disease. Science. 1997;277:112–116. doi: 10.1126/science.277.5322.112. [DOI] [PubMed] [Google Scholar]
- 5.Berzofsky J A, Pendleton C D, Clerici M, Ahlers J, Lucey D R, Putney S D, Shearer G M. Construction of peptides encompassing multideterminant clusters of human immunodeficiency virus envelope to induce in vitro T cell responses in mice and humans of multiple MHC types. J Clin Investig. 1991;88:876–884. doi: 10.1172/JCI115389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boitel B, Blank U, Mege D, Corradin G, Sidney J, Sette A, Acuto O. Strong similarities in antigen fine specificity among DRB1* 1302-restricted tetanus toxin tt830–843-specific TCRs in spite of highly heterogeneous CDR3. J Immunol. 1995;154:3245–3255. [PubMed] [Google Scholar]
- 7.Borrow P, Lewicki H, Wei X, Horwitz M S, Peffer N, Meyers H, Nelson J A, Gairin J E, Hahn B H, Oldstone M B, Shaw G M. Antiviral pressure exerted by HIV-1-specific cytotoxic T lymphocytes (CTLs) during primary infection demonstrated by rapid selection of CTL escape virus. Nat Med. 1997;3:205–211. doi: 10.1038/nm0297-205. [DOI] [PubMed] [Google Scholar]
- 8.Cease K B, Margalit H, Cornette J L, Putney S D, Robey W G, Ouyang C, Streicher H Z, Fischinger P J, Gallo R C, DeLisi C. Helper T-cell antigenic site identification in the acquired immunodeficiency syndrome virus gp120 envelope protein and induction of immunity in mice to the native protein using a 16-residue synthetic peptide. Proc Natl Acad Sci USA. 1987;84:4249–4253. doi: 10.1073/pnas.84.12.4249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clerici M, Giorgi J V, Chou C C, Gudeman V K, Zack J A, Gupta P, Ho H N, Nishanian P G, Berzofsky J A, Shearer G M. Cell-mediated immune response to human immunodeficiency virus (HIV) type 1 in seronegative homosexual men with recent sexual exposure to HIV-1. J Infect Dis. 1992;165:1012–1019. doi: 10.1093/infdis/165.6.1012. [DOI] [PubMed] [Google Scholar]
- 10.Clerici M, Hakim F T, Venzon D J, Blatt S, Hendrix C W, Wynn T A, Shearer G M. Changes in interleukin-2 and interleukin-4 production in asymptomatic, human immunodeficiency virus-seropositive individuals. J Clin Investig. 1993;91:759–765. doi: 10.1172/JCI116294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clerici M, Levin J M, Kessler H A, Harris A, Berzofsky J A, Landay A L, Shearer G M. HIV-specific T-helper activity in seronegative health care workers exposed to contaminated blood. JAMA. 1994;271:42–46. [PubMed] [Google Scholar]
- 12.Clerici M, Sison A V, Berzofsky J A, Rakusan T A, Brandt C D, Ellaurie M, Villa M, Colie C, Venzon D J, Sever J L. Cellular immune factors associated with mother-to-infant transmission of HIV. AIDS. 1993;7:1427–1433. doi: 10.1097/00002030-199311000-00004. [DOI] [PubMed] [Google Scholar]
- 13.Coffin J M. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 14.De Groot A S, Clerici M, Hosmalin A, Hughes S H, Barnd D, Hendrix C W, Houghten R, Shearer G M, Berzofsky J A. Human immunodeficiency virus reverse transcriptase T helper epitopes identified in mice and humans: correlation with a cytotoxic T cell epitope. J Infect Dis. 1991;164:1058–1065. doi: 10.1093/infdis/164.6.1058. [DOI] [PubMed] [Google Scholar]
- 15.Falk K, Rotzschke O, Stevanovic S, Jung G, Rammensee H G. Pool sequencing of natural HLA-DR, DQ, and DP ligands reveals detailed peptide motifs, constraints of processing, and general rules. Immunogenetics. 1994;39:230–242. doi: 10.1007/BF00188785. [DOI] [PubMed] [Google Scholar]
- 16.Fowke K R, Nagelkerke N J, Kimani J, Simonsen J N, Anzala A O, Bwayo J J, MacDonald K S, Ngugi E N, Plummer F A. Resistance to HIV-1 infection among persistently seronegative prostitutes in Nairobi, Kenya. Lancet. 1996;348:1347–1351. doi: 10.1016/S0140-6736(95)12269-2. [DOI] [PubMed] [Google Scholar]
- 17.Furci L, Scarlatti G, Burastero S, Tambussi G, Colognesi C, Quillent C, Longhi R, Loverro P, Borgonovo B, Gaffi D, Carrow E, Malnati M, Lusso P, Siccardi A G, Lazzarin A, Beretta A. Antigen-driven C-C chemokine-mediated HIV-1 suppression by CD4+ T cells from exposed uninfected individuals expressing the wild-type CCR-5 allele. J Exp Med. 1997;186:455–460. doi: 10.1084/jem.186.3.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17a.Gjertson D W, Terasaki P I, editors. HLA, 1998. Lenexa, Kans: American Society for Histocompatibility and Immunogenetics; 1998. [Google Scholar]
- 18.Gorga J C, Horejsi V, Johnson D R, Raghupathy R, Strominger J L. Purification and characterization of class II histocompatibility antigens from a homozygous human B cell line. J Biol Chem. 1987;262:16087–16094. [PubMed] [Google Scholar]
- 19.Haas G, David R, Frank R, Gausepohl H, Devaux C, Claverie J M, Pierres M. Identification of a major human immunodeficiency virus-1 reverse transcriptase epitope recognized by mouse CD4+ T lymphocytes. Eur J Immunol. 1991;21:1371–1377. doi: 10.1002/eji.1830210607. [DOI] [PubMed] [Google Scholar]
- 20.Hale P M, Cease K B, Houghten R A, Ouyang C, Putney S, Javaherian K, Margalit H, Cornette J L, Spouge J L, DeLisi C. T cell multideterminant regions in the human immunodeficiency virus envelope: toward overcoming the problem of major histocompatibility complex restriction. Int Immunol. 1989;1:409–415. doi: 10.1093/intimm/1.4.409. [DOI] [PubMed] [Google Scholar]
- 21.Haslett P A, Nixon D F, Shen Z, Larsson M, Cox W I, Manandhar R, Donahoe S M, Kaplan G. Strong human immunodeficiency virus (HIV)-specific CD4+ T cell responses in a cohort of chronically infected patients are associated with interruptions in anti-HIV chemotherapy. J Infect Dis. 2000;181:1264–1272. doi: 10.1086/315381. [DOI] [PubMed] [Google Scholar]
- 22.Hay C M, Ruhl D J, Basgoz N O, Wilson C C, Billingsley J M, DePasquale M P, D'Aquila R T, Wolinsky S M, Crawford J M, Montefiori D C, Walker B D. Lack of viral escape and defective in vivo activation of human immunodeficiency virus type 1-specific cytotoxic T lymphocytes in rapidly progressive infection. J Virol. 1999;73:5509–5519. doi: 10.1128/jvi.73.7.5509-5519.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Imanishi T, Akaza T, Kimura A, Tokunga K, Gojobori T. Allele and haplotype frequencies for HLA and complement loci in various ethnic groups. In: Tsuki K, Aizaqa M, Sasazuki T, editors. HLA proceedings of the Eleventh International Histocompatibility Workshop and Conference. Tokyo, Japan: Oxford University Press; 1991. p. 1065. [Google Scholar]
- 24.Kalams S A, Walker B D. The critical need for CD4 help in maintaining effective cytotoxic T lymphocyte responses. J Exp Med. 1998;188:2199–2204. doi: 10.1084/jem.188.12.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kinouchi R, Kobayasi H, Sato K, Kimura S, Katagiri M. Peptide motifs of HLA-DR4/DR53 (DRB1*0405/DRB4*0101) molecules. Immunogenetics. 1994;40:376–378. doi: 10.1007/BF01246679. [DOI] [PubMed] [Google Scholar]
- 26.Korber B, Moore J, Brander C, Koup R, Haynes B, Walker B. Theoretical biology and biophysics. Los Alamos, N. Mex: Los Alamos National Laboratory; 1998. HIV molecular immunology database 1998. [Google Scholar]
- 27.Laurence J, Friedman S M, Chartash E K, Crow M K, Posnett D N. Human immunodeficiency virus infection of helper T cell clones. Early proliferative defects despite intact antigen-specific recognition and interleukin 4 secretion. J Clin Investig. 1989;83:1843–1848. doi: 10.1172/JCI114090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lederman M M, Connick E, Landay A, Kuritzkes D R, Spritzeler J, St. Clair M, Kotzin B L, Fox L, Chiozzi M H, Leonard J M, Rousseau F, Wade M, Roe J D, Martinez A, Kessler H. Immunologic responses associated with 12 weeks of combination antiretroviral therapy consisting of zidovudine, lamivudine, and ritonavir: results of AIDS Clinical Trials Group Protocol 315. J Infect Dis. 1998;178:70–79. doi: 10.1086/515591. [DOI] [PubMed] [Google Scholar]
- 29.Marshall K W, Liu A F, Canales J, Perahia B, Jorgensen B, Gantzos R D, Aguilar B, Devaux B, Rothbard J B. Role of the polymorphic residues in HLA-DR molecules in allele-specific binding of peptide ligands. J Immunol. 1994;152:4946–4957. [PubMed] [Google Scholar]
- 30.Mazzoli S, Trabattoni D, Lo C S, Piconi S, Ble C, Meacci F, Ruzzante S, Salvi A, Semplici F, Longhi R, Fusi M L, Tofani N, Biasin M, Villa M L, Mazzotta F, Clerici M. HIV-specific mucosal and cellular immunity in HIV-seronegative partners of HIV-seropositive individuals. Nat Med. 1997;3:1250–1257. doi: 10.1038/nm1197-1250. [DOI] [PubMed] [Google Scholar]
- 31.O'Sullivan D, Arrhenius T, Sidney J, del Guercio M F, Albertson M, Wall M, Oseroff C, Southwood S, Colon S M, Gaeta F C. On the interaction of promiscuous antigenic peptides with different DR alleles. Identification of common structural motifs. J Immunol. 1991;147:2663–2669. [PubMed] [Google Scholar]
- 32.Pinto L A, Sullivan J, Berzofsky J A, Clerici M, Kessler H A, Landay A L, Shearer G M. ENV-specific cytotoxic T lymphocyte responses in HIV seronegative health care workers occupationally exposed to HIV-contaminated body fluids. J Clin Investig. 1995;96:867–876. doi: 10.1172/JCI118133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pitcher C J, Quittner C, Peterson D M, Connors M, Koup R A, Maino V C, Picker L J. HIV-1-specific CD4+ T cells are detectable in most individuals with active HIV-1 infection, but decline with prolonged viral suppression. Nat Med. 1999;5:518–525. doi: 10.1038/8400. [DOI] [PubMed] [Google Scholar]
- 34.Plana M, Garcia F, Gallart T, Miro J M, Gatell J M. Lack of T-cell proliferative response to HIV-1 antigens after 1 year of highly active antiretroviral treatment in early HIV-1 disease. Immunology Study Group of Spanish EARTH-1 Study. Lancet. 1998;352:1194–1195. doi: 10.1016/s0140-6736(05)60532-6. [DOI] [PubMed] [Google Scholar]
- 35.Pontesilli O, Carlesimo M, Varani A R, Ferrara R, Guerra E C, Bernardi M L, Ricci G, Mazzone A M, D'Offizi G, Aiuti F. HIV-specific lymphoproliferative responses in asymptomatic HIV-infected individuals. Clin Exp Immunol. 1995;100:419–424. doi: 10.1111/j.1365-2249.1995.tb03716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rinaldo C R, Liebmann J, Huang X-L, Fan Z, Al-Shboul Q, McMahon D K, Day R D, Riddler S A, Mellors J W. Prolonged suppression of human immunodeficiency virus type 1 (HIV-1) viremia in persons with advanced disease results in enhancement of CD4 T cell reactivity to microbial antigens but not to HIV-1 antigens. J Infect Dis. 1999;179:329–336. doi: 10.1086/314599. [DOI] [PubMed] [Google Scholar]
- 37.Rosenberg E S, Billingsley J M, Caliendo A M, Boswell S L, Sax P E, Kalams S A, Walker B D. Vigorous HIV-1-specific CD4+ T cell responses associated with control of viremia. Science. 1997;278:1447–1450. doi: 10.1126/science.278.5342.1447. [DOI] [PubMed] [Google Scholar]
- 38.Rowland-Jones S, Sutton J, Ariyoshi K, Dong T, Gotch F, McAdam S, Whitby D, Sabally S, Gallimore A, Corrah T. HIV-specific cytotoxic T-cells in HIV-exposed but uninfected Gambian women. Nat Med. 1995;1:59–64. doi: 10.1038/nm0195-59. [DOI] [PubMed] [Google Scholar]
- 39.Rowland-Jones S L, McMichael A. Immune responses in HIV-exposed seronegatives: have they repelled the virus? Curr Opin Immunol. 1995;7:448–455. doi: 10.1016/0952-7915(95)80087-5. [DOI] [PubMed] [Google Scholar]
- 40.Rowland-Jones S L, Nixon D F, Aldhous M C, Gotch F, Ariyoshi K, Hallam N, Kroll J S, Froebel K, McMichael A. HIV-specific cytotoxic T-cell activity in an HIV-exposed but uninfected infant. Lancet. 1993;341:860–861. doi: 10.1016/0140-6736(93)93063-7. [DOI] [PubMed] [Google Scholar]
- 41.Sastry K J, Arlinghaus R B. Identification of T-cell epitopes without B-cell activity in the first and second conserved regions of the HIV Env protein. AIDS. 1991;5:699–707. doi: 10.1097/00002030-199106000-00009. [DOI] [PubMed] [Google Scholar]
- 42.Schrier R D, Gnann J W, Jr, Landes R, Lockshin C, Richman D, McCutchan A, Kennedy C, Oldstone M B, Nelson J A. T cell recognition of HIV synthetic peptides in a natural infection. J Immunol. 1989;142:1166–1176. [PubMed] [Google Scholar]
- 43.Sette A, Buus S, Colon S, Miles C, Grey H M. Structural analysis of peptides capable of binding to more than one Ia antigen. J Immunol. 1989;142:35–40. [PubMed] [Google Scholar]
- 44.Sette A, Sidney J. HLA supertypes and supermotifs: a functional perspective on HLA polymorphism. Curr Opin Immunol. 1998;10:478–482. doi: 10.1016/s0952-7915(98)80124-6. [DOI] [PubMed] [Google Scholar]
- 45.Sette A, Southwood S, O'Sullivan D, Gaeta F C, Sidney J, Grey H M. Effect of pH on MHC class II-peptide interactions. J Immunol. 1992;148:844–851. [PubMed] [Google Scholar]
- 46.Sette A, Vitiello A, Reherman B, Fowler P, Nayersina R, Kast W M, Melief C J, Oseroff C, Yuan L, Ruppert J. The relationship between class I binding affinity and immunogenicity of potential cytotoxic T cell epitopes. J Immunol. 1994;153:5586–5592. [PubMed] [Google Scholar]
- 47.Shearer G M, Clerici M. Early T-helper cell defects in HIV infection. AIDS. 1991;5:245–253. doi: 10.1097/00002030-199103000-00001. [DOI] [PubMed] [Google Scholar]
- 48.Shearer G M, Clerici M. Protective immunity against HIV infection: has nature done the experiment for us? Immunol Today. 1996;17:21–24. doi: 10.1016/0167-5699(96)80564-0. [DOI] [PubMed] [Google Scholar]
- 49.Southwood S, Sidney J, Kondo A, del Guercio M F, Appella E, Hoffman S, Kubo R T, Chesnut R W, Grey H M, Sette A. Several common HLA-DR types share largely overlapping peptide binding repertoires. J Immunol. 1998;160:3363–3373. [PubMed] [Google Scholar]
- 50.Valli A, Sette A, Kappos L, Oseroff C, Sidney J, Miescher G, Hochberger M, Albert E D, Adorini L. Binding of myelin basic protein peptides to human histocompatibility leukocyte antigen class II molecules and their recognition by T cells from multiple sclerosis patients. J Clin Investig. 1993;91:616–628. doi: 10.1172/JCI116242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van der Burg S H, Kwappenberg K M, Geluk A, van der Kruk M, Pontesilli O, Hovenkamp E, Franken K L, van Meijgaarden K E, Drijfhout J W, Ottenhoff T H, Melief C J, Offringa R. Identification of a conserved universal Th epitope in HIV-1 reverse transcriptase that is processed and presented to HIV-specific CD4+ T cells by at least four unrelated HLA-DR molecules. J Immunol. 1999;162:152–160. [PubMed] [Google Scholar]
- 52.Wahren B, Morfeldt-Mansson L, Biberfeld G, Moberg L, Ljungman P, Nordlund S, Bredberg-Raden U, Werner A, Lower J, Kurth R. Impaired specific cellular response to HTLV-III before other immune defects in patients with HTLV-III infection. N Engl J Med. 1986;315:393–394. [PubMed] [Google Scholar]