Abstract
Avian leukosis virus subgroup J (ALV-J), the most recent member of the avian retroviruses, is predominantly associated with myeloid leukosis in meat-type chickens. We have previously demonstrated that the acutely transforming virus strain 966, isolated from an ALV-J-induced tumor, transformed peripheral blood monocyte and bone marrow cells in vitro and induced rapid-onset tumors, suggesting transduction of oncogenes (L. N. Payne, A. M. Gillespie, and K. Howes, Avian Dis. 37:438–450, 1993). In order to understand the molecular basis for the rapid transformation and tumor induction, we have determined the complete genomic structure of the provirus of the 966 strain. The sequence of the 966 provirus clone revealed that its genome is closely related to that of HPRS-103 but is defective, with the entire pol and parts of the gag and env genes replaced by a 1,491-bp sequence representing exons 2 and 3 of the c-myc gene. LSTC-IAH30, a stable cell line derived from turkey monocyte cultures transformed by the 966 strain of ALV-J, expressed a 72-kDa Gag-Myc fusion protein. The identification of the myc gene in 966 virus as well as in several other ALV-J-induced tumors suggested that the induction of myeloid tumors by this new subgroup of ALV occurs through mechanisms involving the activation of the c-myc oncogene.
Avian leukosis virus subgroup J (ALV-J) is the newest member of the avian oncogenic retroviruses. After the first isolation of the ALV-J prototype virus, HPRS-103, more than 10 years ago in the United Kingdom (21), viruses belonging to this subgroup have spread rapidly to many countries, becoming one of the major pathogens facing the broiler meat industry worldwide (26). The env gene of ALV-J is closely related to that of a novel group of chicken endogenous retroviral elements designated EAV-HP (24), suggesting that ALV-J has emerged by genetic recombination (17). Compared to the pathogenic ALV subgroups, such as A and B, which primarily induce lymphoid leukosis in genetically susceptible birds (18), ALV-J isolates predominantly induce myeloid leukosis (ML), a property thought to be associated with their tropism for the cells of the myeloid lineage (1). Previous studies have shown that HPRS-103 and other ALV-J isolates do not transform chicken bone marrow cell cultures in vitro and that the tumors induced by these viruses occur after long latent periods (19). These observations and the demonstration that the nucleotide sequence of the viral genome does not carry any viral oncogenes (2, 3) suggested that ALV-J-induced oncogenesis occurs by the activation of oncogenes through the mechanism of insertional mutagenesis (13).
Although the tumors induced by HPRS-103 are of late onset, occurring at a median age of 20 weeks (19), we have previously shown that acutely transforming ALVs that induce rapid-onset tumors could be isolated from about 60% of late-onset tumors (20). Many tumors obtained from field cases of ML also contained acutely transforming viruses, suggesting that generation of acutely transforming ALVs is a common feature of ALV-J-induced oncogenesis. Most of these virus isolates were able to transform chicken bone marrow or monocyte cell cultures in vitro and induce rapid-onset tumors when inoculated into susceptible birds, a property attributed to the transduction of oncogenes. The acutely transforming ALV-J strain 966 was recovered from a myeloid tumor induced experimentally by HPRS-103 infection (20). This virus transformed peripheral blood monocyte and bone marrow cells and induced rapid-onset tumors in chickens (20) and turkeys (28). Peripheral blood monocytes and bone marrow cells from different lines of chickens showed variation in the relative susceptibility to transformation by ALV-J strain 966 (1). This variation was correlated with the relative susceptibility to the induction of ML by HPRS-103, suggesting the involvement of common cell-specific viral and/or host factors in oncogenesis induced by these two viruses. In order to identify the viral genes and oncogenes that are involved in the rapid induction of tumors, we have determined the complete sequence of the proviral genome of ALV-J strain 966. In this paper, we demonstrate the genome structure of the provirus of the 966 strain of ALV-J and compare its sequence with that of HPRS-103 and other acutely transforming avian retroviruses. We also present data demonstrating proviral DNA with structures similar to that of 966 virus in different ALV-J-induced tumors and transformed cells. Lastly, we describe the characteristics of a stable cell line, LSTC-IAH30 (14), derived from turkey monocytes transformed by the 966 strain of ALV-J.
MATERIALS AND METHODS
Virus and cell culture.
Stocks of the acutely transforming ALV-J strain 966 (20) were prepared from tissue culture supernatants of transformed line 0 chicken bone marrow cultures. All chickens used were maintained at the specific-pathogen-free Poultry Production Facility at the Institute for Animal Health, Compton, United Kingdom. Bone marrow cells for transformation were prepared from 4 to 6-week-old specific-pathogen-free line 0 chickens and infected with virus as previously described (20). LSTC-IAH30 cells were cultured in equal volumes of Leibovitz L15 medium and McCoy's 5A medium as described previously (14). HPRS-103 virus was grown in chicken embryo fibroblasts (CEF) prepared from 10-day-old line 0 chicken embryos as described previously (21).
Extraction of DNA from transformed cells, Southern blotting, and hybridization.
DNA was extracted from ALV-J-induced tumor tissues or transformed bone marrow cells after digestion with proteinase K, phenol-chloroform extraction, and ethanol precipitation (22). Samples of DNA (10 μg) digested with different restriction endonucleases were separated by agarose gel electrophoresis and transferred to nylon membranes, fixed by baking them at 80°C, and prehybridized in 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) containing 0.25% low-fat dry milk BLOTTO (12) for 2 h at 68°C. Hybridization was carried out overnight under optimum conditions (melting temperature [Tm], −25°C) with C-myc or HPRS-103 gag probes labeled with [α-32P]dCTP to a specific activity of approximately 1 × 108 to 2 × 108 dpm/μg. The c-myc probe was derived from a gel-purified EcoRI-HindIII fragment from the pBR322 plasmid containing intron 2 and exon 3 of chicken c-myc (kindly provided by Don Ewert, Wistar Institute, Philadelphia, Pa.). The gag probe consisted of a 351-bp PCR product derived from the full-length HPRS-103 clone using primers 5′-TCGGGGGAGTTAAAAACCTG-3′ and 5′-CCAGGGAAGGATACAAACCA-3′ (2). The membranes were washed under conditions of high stringency (Tm, −10°C) and exposed to autoradiography film (Hyperfilm MP; Amersham International) at −70°C overnight or longer if required.
Identification of 966 provirus from transformed bone marrow cell genomic library.
Genomic DNA was extracted from bone marrow cells transformed with 966 virus by standard methods (22). A genomic DNA library was constructed using partially digested and size-fractionated MboI DNA fragments in λGEM-12 vector (Promega) following the manufacturer's instructions. The library was screened with c-myc and gag probes as described above. One of the DNA clones that hybridized with both probes was isolated and subcloned into pGEM-3Z (Promega) and sequenced in both orientations using dye terminator cycle sequencing in an Applied Biosystems 377 DNA automatic sequencing system with primers derived from the published sequence of HPRS-103 (2). Database searches and sequence analyses were carried out with the Genetics Computer Group (Madison, Wisconsin) version 10 software.
PCR and sequencing.
To examine whether the transduction of myc as a gag-myc fusion gene is a feature associated with ALV-J-induced tumors, we have determined the sequence of the gag-myc regions from the DNA extracted from four HPRS-103-induced myelocytomas as well as from chicken blastoderm cells transformed with an acutely transforming variant (AVO4-1B) of ALV-J (29). A nested-PCR method described earlier for determining proviral c-myc integration sites during various stages of tumor induction (8) was used for PCR. The sequences of the primers L1 and L2 from the long terminal repeat (LTR) region and M1 and M2 from c-myc exon 2 have been described (8). Proviral sequences were amplified using the Expand long-template PCR amplification system (Roche Molecular Biochemicals) with 1 μM (each) L1 and M1 primers in a total volume of 50 μl following the protocol provided by the manufacturer. The reaction mixture containing the primers, deoxynucleoside triphosphate, and the DNA template in a 25-μl volume was denatured at 98°C for 10 min and held at 90°C until the 25-μl reaction mixture containing the buffer and the enzyme (Roche Molecular Biochemicals) was added. Thermal cycling was carried out for two cycles at 96°C for 2 min and 61.5°C for 5 min and then for 23 cycles at 95°C for 5 min and 61.5°C for 5 min, followed by one cycle at 68°C for 10 min. A second-round PCR was carried out under the same conditions with 2 μM (each) L2 and M2 primers (8) using 10 μl of a 1:500 dilution of the first-round PCR products. PCR products obtained from four individual tumors together with three distinct PCR products from AVO4-1B-transformed cells were agarose gel purified, cloned into the pGEM-T vector (Promega), and sequenced using vector-specific primers. The sequences were analyzed using Genetics Computer Group version 10 software.
IF test and Western blotting.
The immunofluorescence (IF) test was performed on LSTC-IAH30 cells using Gag- or Myc-specific antibodies on either unfixed or acetone-fixed cytospin preparations. The cells were stained initially with either HPRS-103-specific chicken serum (27) or v-Myc-specific polyclonal rabbit serum (kindly provided by Markus Hartl, University of Innsbruck, Innsbruck, Austria). After the primary antibodies were washed away in phosphate-buffered saline (PBS), the cells were stained with fluorescein isothiocyanate-labeled anti-chicken and anti-rabbit immunoglobulins (Sigma, Poole, United Kingdom). For Western blotting analysis, approximately 107 HPRS-103 virus-infected and uninfected CEF or LSTC-IAH30 cells were lysed in ice-cold PBS containing 0.1% sodium dodecyl sulfate, 0.1% NP-40, 0.5% deoxycholate, and 170 μg of phenylmethylsulfonyl fluoride per ml. The cell lysates were separated on sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis gels and transferred by electroblotting to a nitrocellulose membrane. The Western blots were blocked with PBS containing 0.05% Tween 20 and 5% milk protein before being incubated with rabbit anti-Gag (21) or anti-Myc (9) serum at 4°C overnight. After being washed with PBS containing 0.1% Tween 20, the blot was incubated with peroxidase-conjugated goat anti-rabbit immunoglobulin G (Sigma), and positive reactions were detected by chemiluminescence.
Nucleotide sequence accession numbers.
The accession numbers for the sequences in this study are as follows: ALV-J strain 966, AF265231; ALV-J strain HPRS-103, Z46390; MC29, V01174; CMII, M15241; MH2, K02082; OK10, M11352; FH3, M74581; and chicken c-myc, J00889.
RESULTS
DrdI fragments of 966 proviral DNA clone are smaller than those of HPRS-103.
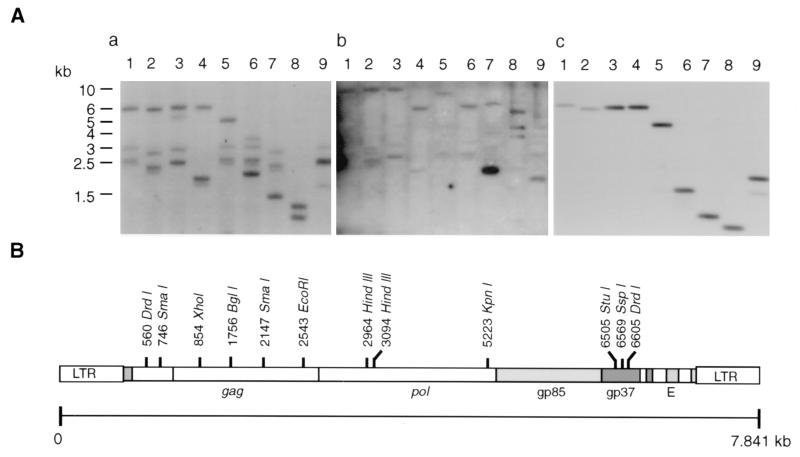
The restriction endonuclease DrdI cleaves twice in the HPRS-103 genome sequence close to the termini and so separates the integrated viral DNA and the host DNA (Fig. 1B). Hybridization of the DrdI-digested DNA from 966 virus-transformed cells detected three species of DrdI DNA fragments with sizes between 2.5 and 3 kb hybridizing with both the gag and c-myc probes (Fig. 1A, blots a and b, lanes 1). These bands were substantially smaller than the 6-kb band of the nondefective helper HPRS-103 virus in the same lane (Fig. 1A, blot a, lane 1) or the DrdI-digested HPRS-103 full-length plasmid DNA (Fig. 1A, blot c, lane 1). The same three DNA bands were also seen after double digestion with DrdI and either KpnI (lane 5) or EcoRI (lane 6), indicating deletion of the last two enzyme sites in these DNA species. After double digestion with DrdI and either XhoI (lane 2) or SmaI (lane 7), all three bands were present but were reduced in size to approximately the same extent, indicating the presence of XhoI and SmaI sites in similar locations. DrdI- and StuI-digested DNA (lane 3) gave two bands comigrating with the 2.5- and 3-kb bands in lane 1, indicating the absence of a StuI site in those DNA species. An approximately 5-kb band observed in lane 3 is not seen in lanes 1 (DrdI digested) or lane 2 (DrdI and XhoI digested), indicating that it is approximately the same size as HPRS-103 but has an extra StuI site. Neither this band nor that from the HPRS-103 helper virus hybridized with the c-myc probe (Fig. 1A, blot b, lane 3). Hybridization of the three smaller bands to myc (Fig. 1A, blot b, lanes 2, 5, and 6) suggested that the genomic DNA from 966 virus-transformed cells contained a variant of HPRS-103, with substantial deletions, that contained the myc gene.
FIG. 1.
Southern blot hybridization of 966 virus-infected genomic DNA and HPRS-103 proviral DNA clone with gag and myc probes. (A) Southern blots of restriction endonuclease-digested 966 virus-infected genomic DNA (10 μg/lane) were probed with either gag (blot a) or c-myc exon 3 (blot b). Blot c is a Southern blot of HPRS-103 full-length plasmid DNA probed with gag. DNA samples were either digested with DrdI alone (lane 1) or doubly digested with DrdI and XhoI (lane 2), StuI (lane 3), SspI (lane 4), KpnI (lane 5), EcoRI (lane 6), SmaI (lane 7), BglI (lane 8), or HindIII (lane 9). (B) Schematic diagram showing the restriction map of HPRS-103 full-length clone.
966 virus proviral genome carries a gag-myc fusion gene.
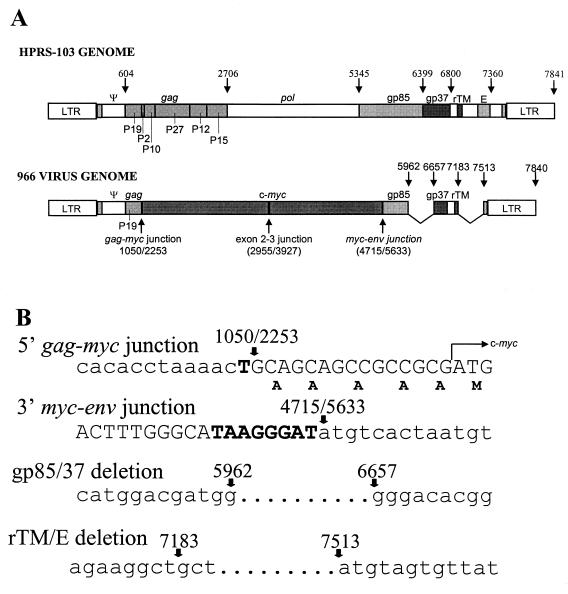
A λ-GEM12 genomic DNA library constructed from 966 virus-transformed bone marrow cells was screened with gag and c-myc probes, and one clone that hybridized with both probes was subcloned into the pGEM3Z vector. The sequence of this clone showed that it was derived from HPRS-103, with the 5′ LTR and the region encoding the 149 residues of the Gag protein showing more than 98% identity to HPRS-103. However, the nucleotide sequence representing positions 1051 to 5632 of HPRS-103 had been replaced by a 1,491-bp v-myc sequence in frame with the gag sequence. The amino acid sequence of the myc gene in the 966 provirus genomic clone was identical to that of the c-myc sequence, indicating that the transduced myc gene has not undergone any mutations, unlike what is observed in the sequences of many other acutely transforming viruses. The insertion of the c-myc sequence results in the deletion of the region of gag encoding part of p19 and the entire region of other Gag subunits, together with the entire pol and part of the env regions (Fig. 2A).
FIG. 2.
Schematic diagram of HPRS-103 and 966 viral genomes showing recombination and deletion junctions. (A) HPRS-103 structure showing the nucleotide positions of the genes and regions in the genome (top) and 966 virus genome structure showing the positions of insertion of c-myc and other deletions (bottom). (B) Nucleotide sequences at the junctions of recombination of myc and deletions in the env and E (XSR) element. The five A residues inserted before the translational start site (M) of myc are shown.
There was a single-base-pair (T) substitution at the 5′ HPRS-103–v-myc junction that was followed by the myc sequence. At the 3′ junction, HPRS-103 and v-myc shared an 8-base-pair sequence, TAAGGGAT (Fig 2B), that could probably be the site for homologous recombination. The recombination event between gag and myc in the 966 virus occurred within the p19 coding region of gag using a splice acceptor site at the 5′ end of chicken c-myc exon 2 (Fig. 2B). This would insert five alanine residues between the end of the Gag sequence and methionine, the major translation start site for c-myc. The coding capacity of the 966 virus gag-myc sequence is 571 amino acids (comprising the first 149 residues of Gag, five alanine residues, and 417 residues of the Myc protein) with a predicted size of approximately 60 kDa. The env gene of 966 virus also showed another deletion (between positions 5962 and 6657) that would remove a region of the genome that included the junction of the SU and TM domains of the envelope glycoprotein. A second deletion (between positions 7183 and 7513) removed the E (XSR) element from the genome (Fig. 2B).
Other acutely transforming strains of ALV-J also show gag-myc genome structure.
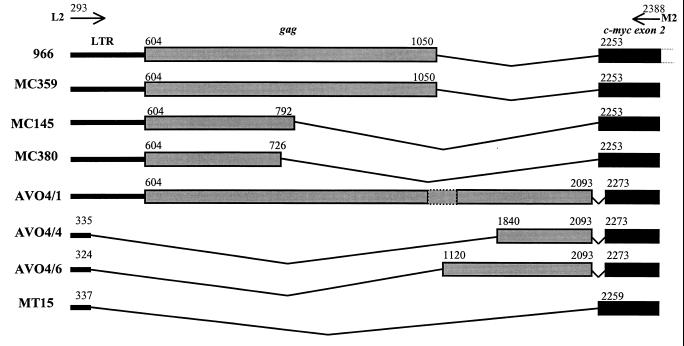
Sequence data on the clones of L2-M2 PCR products obtained from the four myeloid tumors (MC359, MC145, MC380, and MT15) and the three distinct PCR products (AVO4/1, AVO4/4, and AVO4/6) obtained from the AVO4-1B-transformed cells showed a genome structure closely related to that of the 966 provirus. The genome structures of the proviruses from these tumors and transformed cells are presented diagrammatically together with that of 966 virus (Fig. 3). In the clones representing three of the tumors (MC359, MC145, and MC380) and AVO4/1, although there was variation in the lengths of the gag sequences, the 5′ ends were conserved, including the translational start codon at nucleotide 604. The other products contained either no gag (MT15) or gag lacking 5′ sequences (AVO4/4 and AVO4/6). The junction of myc was identical to that of the 966 virus in three of the myeloid tumors, whereas the clones from AVO4-1B-transformed cells and the tumor MT15 showed deletions of small regions of the myc sequence (Fig. 3).
FIG. 3.
Schematic diagram comparing the sequences of the PCR products amplified from DNA extracted from HPRS-103-induced tumors (MC359, MC145, MC380, and MT15) and AVO4-1B-transformed cells (AVO4/1, -4, and -6) with that of 966 virus. The positions of the L2 and M2 primers and the coordinates of the viral and c-myc sequences are shown.
LSTC-IAH30 cell line expresses 72-kDa Gag-Myc fusion protein.
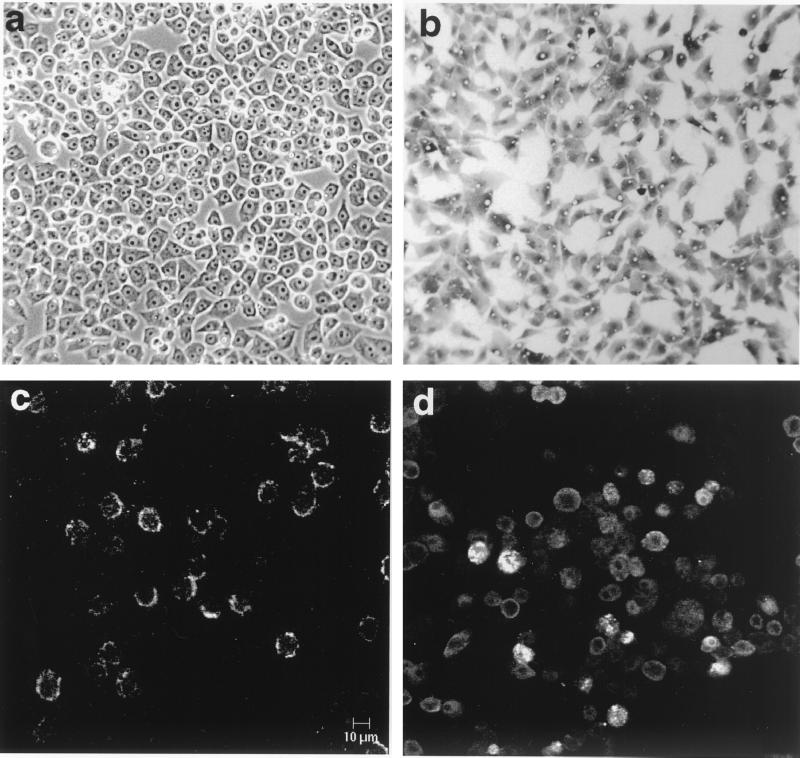
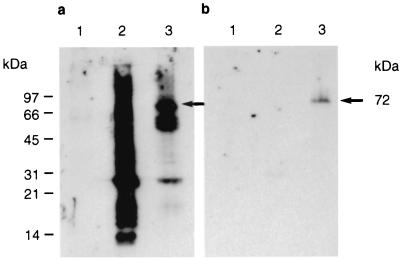
LSTC-IAH30 is a stable cell line derived from turkey monocytes transformed with ALV-J strain 966. LSTC-IAH30 cells, grown for more than 100 passages in the laboratory, are adherent cells having an epithelioid appearance and ovoid nuclei showing an intensely basophilic cytoplasm with lipid vacuoles characteristic of macrophages (Fig. 4a and b). IF tests on unfixed LSTC-IAH30 cells using HPRS-103-specific chicken serum showed cell surface fluorescent staining (Fig. 4c), indicating the expression of ALV-J-specific proteins in these cells. On acetone-fixed cytospin preparations, v-Myc protein could be detected by IF tests on a high proportion of LSTC-IAH30 cells, although the levels of expression varied among individual cells (Fig. 4d). In order to determine the size of the Gag-Myc fusion protein, lysates from LSTC-IAH30 cells, together with HPRS-103-infected and uninfected CEF, were tested by Western blotting using rabbit anti-Gag or anti-Myc serum. The anti-Gag serum produced a smear with the HPRS-103-infected cell lysates, which included the strong p27 band representing the viral capsid (CA) protein. LSTC-IAH30 cell lysates also reacted with p27, although two additional bands of approximately 72 and 62 kDa were seen reacting to anti-Gag serum (Fig. 5a). Of these two bands, the 72-kDa protein also reacted with the anti-Myc serum, indicating that it represented the Gag-Myc fusion protein (Fig. 5b).
FIG. 4.
Morphological features and antigen expression of LSTC-IAH30 cells. (a) Unstained live adherent cells with an epithelioid appearance and ovoid nuclei; (b) May-Grunwald-Giemsa-stained cells showing basophilic cytoplasm and lipid vacuoles; (c) surface fluorescence on unfixed cells stained with ALV-J-specific chicken serum; (d) acetone-fixed cells stained with v-Myc-specific polyclonal serum.
FIG. 5.
Western blotting analysis of cell lysates from uninfected CEF (lane 1), HPRS-103-infected CEF (lane 2), and LSTC-IAH30 cells (lane 3) using polyclonal rabbit anti-Gag (a) and anti-v-Myc (b) sera.
DISCUSSION
Among proto-oncogenes, c-myc appears to be a preferred target for transduction by several ALV strains, and so far five myc-containing avian retroviruses—MC29, MH2, OK10, CMII, and FH3—have been characterized. Here we describe the structure of strain 966, another acutely transforming myc-containing ALV derived from an ALV-J-induced myelocytoma. The characteristics of the v-myc protein encoded by each of these viruses were different. In MC29, FH3, and CMII, myc is expressed as Gag-Myc fusion proteins of 110, 149, and 90 kDa, respectively (4–6), while OK10 encoded two proteins of 200 and 60 kDa (10). In the present study, we demonstrate that ALV-J strain 966 has a genome structure similar to that of other acutely transforming viruses and encoded a 72-kDa Gag-Myc fusion protein.
Since most of the myc-containing avian retroviruses carried more or less identical regions of the myc gene, the differences between the sizes of Gag-Myc fusion proteins in these viruses are largely due to the differences in the length of the Gag part of the fusion protein. For example, both MC29 and 966 viruses share the same 422-amino-acid v-myc region. However, while MC29 carries about 450 amino acids derived from the N terminus of gag, the 966 sequence contained only the first 149 residues. These differences in the lengths of the gag sequences do not appear to be significant for transformation, since HBI virus, a variant of MC29 with deletion of the entire gag region, was shown to be transformation competent (23). However, studies of FH3 virus indicated that gag sequences might have a modulatory role in the transformation process, particularly in a cell-type-specific manner. FH3 virus fails to transform fibroblasts in vitro, although it induces rapid transformation of macrophages (6). Subsequent studies have shown that the fibroblast transformation incapability of FH3 virus is directly correlated with a 212-amino-acid region in the gag gene product (25). However, as this region is deleted from 966 virus, one could conclude that it might not be the sole determinant in the inhibition of fibroblast transformation and that the importance of the C-terminal gag region in FH3 might not be universal for myc-containing viruses.
Comparison of the sequences of v-Myc from several viruses indicated that cell type restriction of transformation could possibly be overcome by mutations in the v-Myc protein (7). The number of such substitutions varied among isolates (one in CMII and FH3, two in OK10, five in MC29, and 27 in MH2), and most of these mutations mapped to the transformation-associated domains of Myc. Some of the mutations, such as those at the threonine 61 residue seen in the OK10, MH2, and MC29 viruses, are thought to stabilize Myc against rapid degradation, thereby enhancing its transformation potential (15). The Myc protein of 966 virus does not show any substitution mutations compared to c-myc, and it would seem likely that the inability of 966 virus to transform fibroblasts could be related to the absence of vital point mutations seen in fibroblast transformation-competent viruses. These data support the hypothesis that Myc-induced transformation is a tissue-specific phenomenon requiring both point mutations and overexpression for the transformation of fibroblasts but only the ALV LTR-driven overexpression for macrophage transformation (25).
Transduction of oncogenes by retroviruses and generation of acutely transforming viruses is common in many retroviral infections. The frequency of generating acutely transforming viruses is highly variable and is dependent on factors such as the location and orientation of the retroviral insertion, recombination junctions, presence or absence of packaging signals, and primer binding sites (13). It has been reported that under optimum conditions, such as those occurring in c-erbB activation by ALV, the frequency can be as high as 50% (16). In ALV-J-induced tumors, we have previously shown that acutely transforming viruses could be isolated from more than 60% of tumors (20). To determine the frequency of transduction of the myc gene in ALV-J-induced tumors, we have examined the genome structures of acutely transforming viruses from several independent myeloid tumors and transformed cells, using PCR with primers specifically designed to amplify 3′ LTR-host junction fragments (8). PCR products representing the gag-myc region were obtained from six of these tumors and transformed cells, while one of the products represented the LTR-myc region. The sequence of these PCR products showed a gag-myc region very closely resembling that of 966 virus, although the recombination junction and the length of the gag or the myc region varied among the individual PCR products (Fig. 3). The transduction of oncogenes occurs when the readthrough transcripts containing the oncogene sequences transcribed by the integrated proviruses are copackaged with the viral RNA followed by recombination. Since our study demonstrated that many of the ALV-J-induced tumors and transformed cells contained acutely transforming viruses with transduced myc sequences, we conclude that the induction of myeloid tumors by nonacute ALV-J strains occurs by insertional mutagenesis involving the c-myc oncogene. Very limited data are available on the frequency of generation of acutely transforming viruses in infections with other ALV subgroups (16). However, the high frequency of generation of acutely transforming viruses that we demonstrated earlier (20) and the detection of PCR products with gag-myc structure in many ALV-J-induced tumors and transformed cells observed in this study suggest that transduction of the myc oncogene is a characteristic of ALV subgroup J.
In addition to the deletion of the gag sequences, the 966 viral clone also showed a deletion in the env gene resulting in the loss of the junction between the two envelope subunits. Because of this deletion and the loss of the region containing the splice acceptor site of the envelope (Fig. 2), the 966 virus derived from this clone will be defective and would be expected to rely entirely on helper virus for envelope functions. Another deletion further downstream would result in the loss of the E (XSR) element (2) from the 966 virus clone. The effect of this deletion on 966 virus is not clear, as ALV-Js with this region deleted have been reported to have been isolated from the field.
The data presented here represent the first detailed molecular characterization of an acutely transforming variant of the most recently identified subgroup J ALV. Members of this new subgroup are unique in that they induce myeloid tumors in meat-type chickens. Activation of c-myc by ALV subgroups A and B has been well documented in ALV-induced lymphoid tumors (11). The involvement of the myc gene in the induction of myeloid tumors by ALV-J confirms that irrespective of the cell targets for transformation, ALVs make use of the common myc-mediated pathway for the induction of tumors. If myc is involved in the induction of tumors in both these cell types, then the determinants for cell tropism could be dependent on the interaction of cell-specific factors and unique viral sequences.
ACKNOWLEDGMENTS
This work was partly funded by the Ministry of Agriculture, Fisheries and Food, United Kingdom, and Ross Breeders Ltd., Newbridge, Scotland. We thank Don Ewert, Wistar Institute, Philadelphia, for providing the c-myc plasmid; Markus Hartl, University of Innsbruck, Innsbruck, Austria, for providing the v-Myc-specific antiserum; and Bernard Clark for photographic assistance.
REFERENCES
- 1.Arshad S S, Howes K, Barron G S, Smith L M, Russell P H, Payne L N. Tissue tropism of the HPRS-103 strain of J subgroup avian leukosis virus and of a derivative acutely transforming virus. Vet Pathol. 1997;34:127–137. doi: 10.1177/030098589703400205. [DOI] [PubMed] [Google Scholar]
- 2.Bai J, Payne L N, Skinner M A. HPRS-103 (exogenous avian leukosis virus, subgroup J) has an env gene related to those of endogenous elements EAV-0 and E51 and an E element found previously only in sarcoma viruses. J Virol. 1995;69:779–784. doi: 10.1128/jvi.69.2.779-784.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benson S J, Ruis B L, Garbers A L, Fadly A M, Conklin K F. Independent isolates of the emerging subgroup J avian leukosis virus derive from a common ancestor. J Virol. 1998;72:10301–10304. doi: 10.1128/jvi.72.12.10301-10304.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bister K, Hayman M J, Vogt P K. Defectiveness of avian myelocytomatosis virus MC29: isolation of long-term nonproducer cultures and analysis of virus-specific polypeptide synthesis. Virology. 1977;82:801–822. doi: 10.1016/0042-6822(77)90017-4. [DOI] [PubMed] [Google Scholar]
- 5.Bister K, Loliger H-C, Duesberg P H. Oligoribonucleotide map and protein of CMII: detection of conserved and nonconserved genetic elements in avian acute leukemia viruses CMII, MC29, and MH2. J Virol. 1979;32:208–219. doi: 10.1128/jvi.32.1.208-219.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen C, Biegalke B J, Eisenman R N, Linial M L. FH3, a v-myc avian retrovirus with limited transforming ability. J Virol. 1989;63:5092–5100. doi: 10.1128/jvi.63.12.5092-5100.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farina S F, Huff J L, Parsons T. Mutations within the 5′ half of the avian retrovirus MC29 v-myc gene alter or abolish transformation of chicken embryo fibroblasts and macrophages. J Virol. 1992;66:2698–2708. doi: 10.1128/jvi.66.5.2698-2708.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gong M, Semu H L, Bird K J, Stramer B J, Ruddell A. Differential selection of cells with proviral c-myc and c-erbB integrations after avian leukosis virus infection. J Virol. 1998;72:5517–5525. doi: 10.1128/jvi.72.7.5517-5525.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hartl M, Bister K. Structure and transcriptional regulation of BKJ, a novel AP-1 target gene activated during jun- or fos-induced fibroblast transformation. Oncogene. 1998;17:2901–2913. doi: 10.1038/sj.onc.1202219. [DOI] [PubMed] [Google Scholar]
- 10.Hayflick J, Seeburg P H, Ohlsson R, Pfeifer-Ohlsson S, Watson D, Papas T, Duesberg P H. Nucleotide sequence of two overlapping myc-related genes in avian carcinoma virus OK10 and their relation to the myc genes of other viruses and the cell. Proc Natl Acad Sci USA. 1985;82:2718–2722. doi: 10.1073/pnas.82.9.2718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hayward W S, Neel B C, Astrin S M. Activation of a cellular onc gene by promoter insertion in ALV-induced lymphoid leukosis. Nature (London) 1981;290:475–480. doi: 10.1038/290475a0. [DOI] [PubMed] [Google Scholar]
- 12.Johnson D A, Gautsch J W, Sportsman J R, Elder J H. Improved technique utilizing non-fat dry milk for analysis of proteins and nucleic acids transferred to nitrocellulose. Gene Anal Tech. 1984;1:3–8. [Google Scholar]
- 13.Kung H-J, Liu J-L. Retroviral oncogenesis. In: Nathanson N, editor. Viral pathogenesis. Philadelphia, Pa: Lippincott-Raven Publishers; 1997. pp. 235–266. [Google Scholar]
- 14.Lawson S, Rothwell L, Lambrecht B, Howes K, Venugopal K, Kaiser P. Turkey and chicken interferon-γ, which share high sequence identity, are biologically cross-reactive. Dev Comp Immunol. 2001;25:69–82. doi: 10.1016/s0145-305x(00)00044-6. [DOI] [PubMed] [Google Scholar]
- 15.Lee C M, Reddy E P. The v-myc oncogene. Oncogene. 1999;18:2997–3003. doi: 10.1038/sj.onc.1202786. [DOI] [PubMed] [Google Scholar]
- 16.Miles B D, Robinson H L. High-frequency transduction of c-erbB in avian leukosis virus-induced erythroblastosis. J Virol. 1985;54:295–303. doi: 10.1128/jvi.54.2.295-303.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Payne L N. HPRS-103: a retrovirus strikes back. The emergence of subgroup J avian leukosis virus. Avian Pathol. 1998;27:S36–S45. [Google Scholar]
- 18.Payne L N. Biology of avian retroviruses. In: Levy J A, editor. The Retroviridae. I. New York, N.Y: Plenum Press; 1992. pp. 299–404. [Google Scholar]
- 19.Payne L N, Gillespie A M, Howes K. Myeloid leukaemogenicity and transmission of the HPRS-103 strain of avian leukosis virus. Leukemia. 1992;6:1167–1176. [PubMed] [Google Scholar]
- 20.Payne L N, Gillespie A M, Howes K. Recovery of acutely transforming viruses from myeloid leukosis induced by the HPRS-103 strain of avian leukosis virus. Avian Dis. 1993;37:438–450. [PubMed] [Google Scholar]
- 21.Payne L N, Brown S R, Bumstead N, Howes K, Frazier J A, Thouless M E. A novel subgroup of exogenous avian leukosis virus in chickens. J Gen Virol. 1991;72:801–807. doi: 10.1099/0022-1317-72-4-801. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 23.Shaw J, Hayman M J, Enrietto P J. Analysis of a deleted MC29 provirus: gag sequences are not required for fibroblast transformation. J Virol. 1985;56:943–950. doi: 10.1128/jvi.56.3.943-950.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Smith L M, Toye A A, Howes K, Bumstead N, Payne L N, Venugopal K. Novel endogenous retroviral sequences in the chicken genome closely related to HPRS-103 (subgroup J) avian leukosis virus. J Gen Virol. 1999;80:261–268. doi: 10.1099/0022-1317-80-1-261. [DOI] [PubMed] [Google Scholar]
- 25.Tikhonenko A T, Linial M L. Gag as well as myc sequences contribute to the transforming phenotype of the avian retrovirus FH3. J Virol. 1992;66:946–955. doi: 10.1128/jvi.66.2.946-955.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Venugopal K. Avian leukosis virus subgroup J: a rapidly evolving group of oncogenic retroviruses. Res Vet Sci. 1999;67:113–119. doi: 10.1053/rvsc.1998.0283. [DOI] [PubMed] [Google Scholar]
- 27.Venugopal K, Howes K, Barron G S, Payne L N. Recombinant env-gp85 of HPRS-103 (subgroup J) avian leukosis virus: antigenic characteristics and usefulness as a diagnostic reagent. Avian Dis. 1995;41:283–288. [PubMed] [Google Scholar]
- 28.Venugopal K, Howes K, Flannery D M J, Payne L N. Subgroup J avian leukosis virus infection in turkeys: induction of rapid onset tumors by acutely transforming virus strain 966. Avian Pathol. 2000;29:319–325. doi: 10.1080/03079450050118449. [DOI] [PubMed] [Google Scholar]
- 29.Venugopal K, Howes K, Flannery D M J, Payne L N. Isolation of acutely transforming subgroup J avian leukosis viruses that induce erythroblastosis and myelocytomatosis. Avian Pathol. 2000;29:497–504. doi: 10.1080/030794500750047252. [DOI] [PubMed] [Google Scholar]