Abstract
A primary challenge to the data-driven analysis is the balance between poor generalizability of population-based research and characterizing more subject-, study- and population-specific variability. We previously introduced a fully automated spatially constrained independent component analysis (ICA) framework called NeuroMark and its functional MRI (fMRI) template. NeuroMark has been successfully applied in numerous studies, identifying brain markers reproducible across datasets and disorders. The first NeuroMark template was constructed based on young adult cohorts. We recently expanded on this initiative by creating a standardized normative multi-spatial-scale functional template using over 100,000 subjects, aiming to improve generalizability and comparability across studies involving diverse cohorts. While a unified template across the lifespan is desirable, a comprehensive investigation of the similarities and differences between components from different age populations might help systematically transform our understanding of the human brain by revealing the most well-replicated and variable network features throughout the lifespan. In this work, we introduced two significant expansions of NeuroMark templates first by generating replicable fMRI templates for infants, adolescents, and aging cohorts, and second by incorporating structural MRI (sMRI) and diffusion MRI (dMRI) modalities. Specifically, we built spatiotemporal fMRI templates based on 6,000 resting-state scans from four datasets. This is the first attempt to create robust ICA templates covering dynamic brain development across the lifespan. For the sMRI and dMRI data, we used two large publicly available datasets including more than 30,000 scans to build reliable templates. We employed a spatial similarity analysis to identify replicable templates and investigate the degree to which unique and similar patterns are reflective in different age populations. Our results suggest remarkably high similarity of the resulting adapted components, even across extreme age differences. With the new templates, the NeuroMark framework allows us to perform age-specific adaptations and to capture features adaptable to each modality, therefore facilitating biomarker identification across brain disorders. In sum, the present work demonstrates the generalizability of NeuroMark templates and suggests the potential of new templates to boost accuracy in mental health research and advance our understanding of lifespan and cross-modal alterations.
Keywords: NeuroMark, hybrid, spatiotemporal functional template, structural template, diffusion template
1. Introduction
Resting-state functional connectivity (FC) (Biswal et al., 1995), which evaluates the statistical relationships between spontaneous BOLD fluctuations from distributed brain regions, has dramatically advanced our understanding of the organization of the functional brain, both in health and psychopathology (Bluhm et al., 2007; Buckner et al., 2013; Honey et al., 2009; Raichle and Snyder, 2007). Unlike voxel-based analysis, such as the first- and second-level analysis using general linear models (Ashburner et al., 2021), the amplitude of low-frequency fluctuation (Zou et al., 2008), and regional homogeneity (Zang et al., 2004), most FC studies require the definition of brain regions to extract regional time courses for measuring their statistical relationships. Atlas-based analysis and decomposition-based analysis (e.g., independent component analysis [ICA]) are the two most used strategies for the segmentation of the brain into regions of interest (ROIs). Atlas-based analysis based on prior knowledge of functional or structural information (Faria et al., 2012; Lord et al., 2016; Rolls et al., 2020), assumes fixed ROIs across subjects and populations (e.g., controls vs. patients) for the investigation of FC. However, subjects and even different scans from the same subject can exhibit distinct spatial coactivation patterns, which implies that fixed brain regions might over-simplify the variation in ROIs across subjects. ICA is an alternative method based on the data-driven decomposition that allows for variations in ROIs across subjects as well as their overlap with one another (Calhoun et al., 2001; Jafri et al., 2008). Combining group ICA with back-reconstruction strategies, one can estimate comparable ROIs across subjects while retaining inter-subject variation in the individual-level ROIs (Beckmann et al., 2009; Du and Fan, 2013).
Benefiting from technological advances, neuroscience studies have successfully characterized individual differences in FC related to the variation in complex neurodevelopmental and psychological phenotypes (Marek et al., 2022). The most challenging topic in recent neuroscience is the reproducibility of population-based research (Aarts et al., 2015; Masouleh et al., 2019; Poldrack et al., 2017). Many neuroimaging consortia have collected much larger samples than before (Barch et al., 2018; Sudlow et al., 2015; Van Essen et al., 2012), providing unprecedented opportunities to perform large-scale analysis across datasets and studies. However, the replicability requirement for biomarker development might be a great challenge to decomposition-based studies because the components identified by data-driven methods can vary across datasets or even across runs. This discrepancy hinders the validation of results and the automation of biomarker development, which might temper the translational values of the findings to clinical usage.
To address this, we developed a fully automated hybrid ICA-based framework called NeuroMark, which can address this issue in previous ICA methods (Du et al., 2020). By incorporating a robust spatial template (NeuroMark_fMRI_1.0) with intra-subject spatially constrained ICA, this framework extracts individual-level functional imaging features comparable across subjects, studies, and datasets. NeuroMark retains the benefits of data-driven strategies by adapting to an individual scan, thus mitigating the pitfalls of fixed ROIs which may not correspond well to the underlying data (e.g., voxels are not coherent), and simplifies the data-driven approach by providing an additional constraint that enables us to fully automate the approach. The independence of individual-level feature extraction in NeuroMark is also useful for classification as it is a single-subject approach, and thus avoids concerns about leakage between training and testing data.
NeuroMark_fMRI_1.0 has been successfully applied to many studies, e.g., (Dhamala et al., 2023; Du et al., 2021; Fu et al., 2021d, 2021a, 2023c; Levey et al., 2022; Salman et al., 2023; Tu et al., 2020; Vaidya et al., 2023; Yan et al., 2023; Zhao et al., 2022), capturing functional network features associated with a wide range of brain disorders. By applying NeuroMark_fMRI_1.0 to subjects with mild cognitive impairment due to Alzheimer’s disease before and after atomoxetine treatment, Levey et al. showed that resting-state FC showed significantly increased within-network FC due to atomoxetine between the insula and the hippocampus (Levey et al., 2022). Vaidya and colleagues implemented NeuroMark_fMRI_1.0 on cross-sessional data including children, adolescents, and young adults, and found negative associations between arsenic exposure and resting-state FC (Vaidya et al., 2023). They further suggested that the within-network FC mediated the alterations in executive function. We subsequently expanded NeuroMark by leveraging a substantial cohort of over 100,000 subjects to create a reproducible and replicable normative template for intrinsic connectivity networks (ICNs) encompassing multiple spatial scales (NeuroMark_fMRI_2.0). The purpose of this template was to augment the generalizability and comparability of functional connectivity studies across diverse cohorts, while also accommodating subject variability. Additionally, we substantiated the existence of the ICN templates in data from individuals who did not meet the quality control (QC) criteria, highlighting the practicality and reliability of our findings. To enhance accessibility and facilitate the dissemination of our research outcomes, we released distinct versions of this multiscale template at https://trendscenter.org/data/. In sum, the NeuroMark framework has proved to be a powerful tool to extract robust FC features for different neuroimaging consortia and datasets.
Neuroimaging studies recruit cohorts in different age groups because brain disorders can have different incidence rates in different age populations. For example, autism and ADHD studies might recruit children or adolescent participants (Durston and Casey, 2006; Konrad and Eickhoff, 2010; Pandolfi et al., 2014; Dardo Tomasi and Volkow, 2012), while neurodegeneration studies might target the aging cohorts (Kitani-Morii et al., 2021; Samra and Ramtahal, 2012; Stern, 2002). Existing literature has suggested the changes in functional networks throughout the lifespan (Cao et al., 2014; Chan et al., 2018; Rieck et al., 2021; Schlee et al., 2012). Although NeuroMark templates have been successfully applied to developmental and aging cohorts (Du et al., 2020, 2021; Zening Fu et al., 2020; Levey et al., 2022; Vaidya et al., 2023) because the adaptive-ICA technique (Du and Fan, 2013) implemented can mitigate the age mismatch between the reference template and the data, there is still an open question of how network templates change throughout the lifespan and whether the NeuroMark templates can capture the reliable network activations if the cohorts show a different age distribution. In addition, different imaging modalities can provide complementary information to each other that might reveal different aspects of disease mechanisms (Gao et al., 2018; Teipel et al., 2015). Previous NeuroMark templates mainly focused on the fMRI data, and it is worth expanding the existing template to different modalities. ICA-based methods have been applied to other modalities, and similarly, because these studies were completely data-driven, the identified components varied across datasets. In contrast, a strength of our guided framework is it can enhance replication, validation, and comparison of the findings across studies. For example, source-based morphometry (SBM) (Xu et al., 2009), which applies ICA to a group of T1 gray matter data to capture “covaried voxels” (spatially distinct regions with common covariation among subjects/sessions), has been successfully utilized in many studies with a variety of components identified (Fornito et al., 2009; Murley et al., 2020; Sui et al., 2012). Developing a gray matter template as a reliable reference for fully automated SBM analyses will help to link the structural features across datasets and studies, which will help advance our understanding of the brain. ICA is also useful for analyzing diffusion MRI (dMRI) data (Jeong et al., 2013; Schouten et al., 2017). Building a reliable dMRI template is another ongoing effect that will boost the imaging analysis across brain disorders.
In this study, we constructed four-dimensional (age × 3D brain) functional templates (NeuroMark_fMRI_3.0) using more than 6000 high-quality fMRI scans from 1800 subjects by four different datasets. The 4D functional templates covered the dynamic brain development in three critical time points, i.e., infant (birth ~ nine months), developmental cohorts (five ~ 21 years), and aged cohorts (36 ~ 100 years). In addition to this, we introduced templates for other modalities constructed based on the T1w scans and diffusion scans. A structural SBM template (NeuroMark_sMRI_3.0) and a diffusion SBM (d-SBM) template (NeuroMark_dMRI_3.0) were built using > 30000 samples from two large neuroimaging consortia (UK biobank and HCP). To show the robustness of the templates, we evaluated the reproducibility of components across data. We also investigated the unique and shared patterns reflected in the functional templates for different age populations. This work represents the first attempt to introduce spatiotemporal functional templates and templates for structural and diffusion data compatible with the adaptive ICA framework, providing a powerful framework for capturing robust imaging makers in brain function and structure.
2. Materials and Methods
2.1. NeuroMark Framework
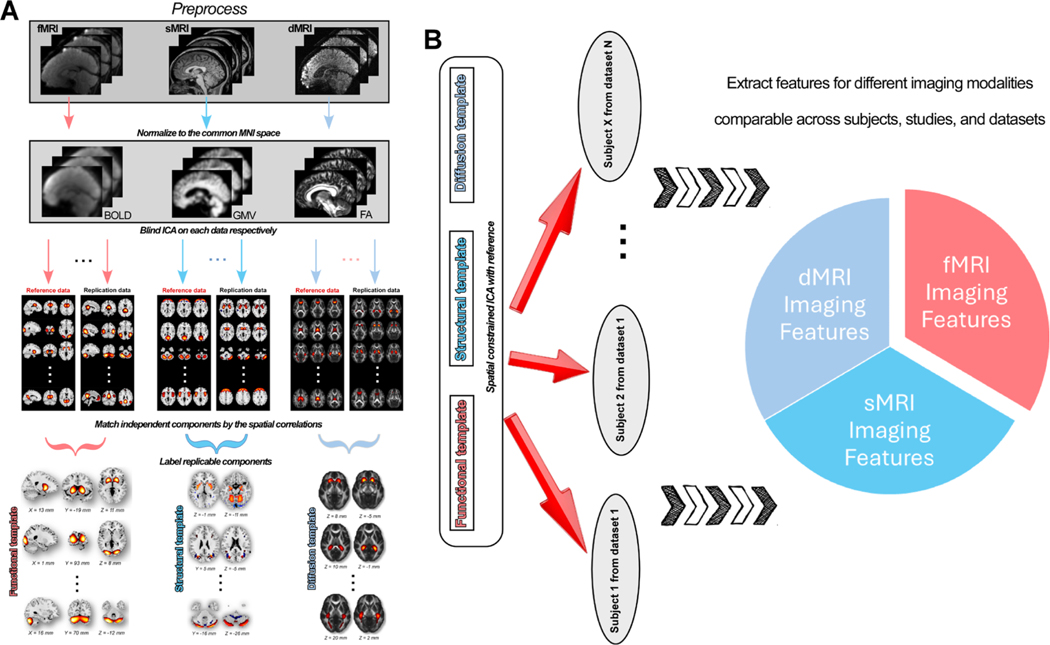
The NeuroMark framework, including the construction of templates and the application to neuroimaging datasets to capture reproducible brain features, is shown in Fig. 1. First, data from different modalities were preprocessed using our well-established preprocessing pipelines. Next, for each modality, ICA was performed on each data. After obtaining independent components (ICs) for each data, we identified the replicable ICs using a greedy spatial correlation analysis. Then the replicated ICs were labeled by evaluating their peak activations and the low-frequency fluctuations. Meaningful ICs, ICN for fMRI, SBM for sMRI, and d-SBM for dMRI, were used to build the template. Finally, the robust templates were used as the reference in an adaptive ICA approach on the target datasets to extract single-subject/scan imaging features. Imaging features from different modalities (e.g., functional network connectivity [FNC], SBM loadings, and d-SBM loadings) can be compared between subjects, datasets, and studies.
Fig. 1.
NeuroMark framework to extract comparable brain features from different image modalities. A) Construction of NeuroMark templates for different imaging modalities. Raw imaging data is preprocessed using standard pipelines and then normalized to the common Montreal Neurological Institute (MNI) space. Group-level independent components (ICs) are estimated from different data. ICs are matched between data for searching replicable components. Replicable ICs are identified as intrinsic connectivity network (ICN) or source-based morphometry (SBM) or diffusion SBM (d-SBM) templates by evaluating the component maps. B) Extracting individual-level imaging features by taking the templates as the reference. Group-information-guided back-reconstruction methods are adopted to single scan to extract imaging features comparable across datasets and studies.
2.2. Building the Developmental Template (5 ~ 21 years)
We adopted the resting-state fMRI dataset from the human connectome project development (HCP-D, https://www.humanconnectome.org/study/hcp-lifespan-development) to build the NeuroMark template for developmental studies (NeuroMark_fMRI_Develomental_3.0). The HCP-D dataset has enrolled 652 healthy children, adolescents, and young adults aged between five years and 21 years old to study brain development in childhood and adolescence. We preprocessed the raw data using a combination of the SPM12 toolbox and the FSL toolbox, where detailed information is provided in the supplementary materials, section “Preprocessing of fMRI Data”.
The HCP-D dataset contains four resting-state sessions collected in the AP or PA direction (Func1_AP, Func2_PA, Func3_AP, and Func4_PA), suitable for searching replicable ICs to build the template. We ran the NeuroMark QC on all the preprocessed functional data to select scans with good imaging quality for the analysis. Specifically, we first excluded scans with relatively large head motions. Scans with head motions larger than 3° rotations or 3 mm translations or with mean framewise displacement (FD) larger than 0.3 mm were excluded. Then we examined the similarity between the single-scan mask and the group mask to check whether they have good normalization to the Echo-planar imaging (EPI) template (Du et al., 2020). Scans were excluded for further analysis if they did not have high enough similarities to the group-averaged mask. Details of QC are described in the supplementary materials, section “Examining Mask of Individual Scan and Average Mask for Subject Selection”. After the QC, we have in total of 2252 scans for the construction of the developmental template, including 562 Func1_AP scans, 569 Func2_PA scans, 557 Func3_AP scans, and 564 Func4_PA scans. The demographic information of each session is provided in Table 1. In this work, we did not balance the number of males and females when constructing the templates because we wanted to include as many subjects as possible to obtain reliable group-level ICs. As an additional analysis, we performed the same procedure using gender-balanced samples, which can be found in the supplementary materials, section “ICA on Gender-balanced Samples shows Highly-similar Network Patterns”. The results demonstrate that all the networks from the NeuroMark 3.0 templates can be reproducible in gender-balanced samples.
Table 1.
Basic Demographics of HCP-Development Data.
| Basic Demographics | Func1_AP | Func2_PA | Func3_AP | Func4_PA |
|---|---|---|---|---|
| Total Subject | 562 | 569 | 557 | 564 |
| Age (month) | 179.34 ± 46.26 | 179.34 ± 45.88 | 179.04 ± 45.95 | 177.63 ± 46.43 |
| Gender (F/M) | 308/254 | 312/257 | 304/253 | 311/253 |
| Race (W/B/A/O) | 361/59/43/99 | 368/57/42/102 | 358/60/43/96 | 366/58/42/98 |
| Mean FD | 0.16 ± 0.05 | 0.14 ± 0.05 | 0.16 ± 0.05 | 0.14 ± 0.05 |
F = female; M = male; W = white; B = black or African American; A = Asian; O = others or unknown; FD = framewise displacement.
Group ICA was performed on each session data respectively, resulting in four groups of components for building the template. For each session, principal component analysis (PCA) was performed on each scan to reduce fMRI data to 120 principal components (PCs), which preserved more than 95% variance of the original data. Individual-level PCs were concatenated across subjects, and then a second-level PCA was performed to reduce the group data to 100 PCs. The Infomax algorithm (Bell and Sejnowski, 1995) was implemented on the 100 PCs to estimate 100 ICs, where this procedure was repeated 20 times based on the ICASSO method (Himberg and Hyvärinen, 2003). Finally, the best ICA run was used to generate 100 reliable group ICs for each session. After having the four groups of ICs, we examined the replicability of ICs for the template construction. We chose Func1_AP as the reference data and the other three sessions as the replication data. We also used the other sessions as the reference data for validation, where the results were highly consistent. For each IC from the reference data, we calculated the absolute value of the correlation coefficient between its spatial map and the spatial maps of the ICs from the other sessions. Based on the spatial correlation, we identified the matched IC from each replication data that showed the maximum correlation value. Therefore, for each IC from the reference data, there are three matched ICs from the three replication sessions. After identifying the matched ICs, we considered the reference IC replicable if it has a average spatial correlation with the matched ICs larger than 0.4, a stricter threshold than previous work (Smith et al., 2009). The replicable ICs were further labeled as meaningful ICNs or noise components by evaluating their spatial activations. The ICNs were finally clustered into different functional domains according to prior functional and anatomical knowledge (Allen et al., 2014) and were taken to build the template for the developmental cohorts. The ICNs captured from the Func1_AP session were used to build the template, but it should be noted that ICNs from other sessions show similar spatial patterns that can be used as the template as well. Hereinafter, we use Nd to denote the number of ICNs in the template.
2.3. Building the Aging Template (36 ~ 100+ years)
The NeuroMark template for aging cohorts (Neuro-Mark_fMRI_Aging_3.0) was built using the resting-state fMRI dataset from the lifespan human connectome project aging (HCP-A, https://www.humanconnectome.org/study/hcp-lifespan-aging). The HCP-A dataset recruited 725 healthy subjects aged between 36~100+ years old to investigate the healthy aging of brain function. We preprocessed the HCP-A data using the same pipelines for the HCP-D data.
The same QC criteria were applied to the preprocessed data to select scans for the construction of the aging template. We had in total of 2395 scans for building the aging template, including 589 Func1_AP scans, 626 Func2_PA scans, 573 Func3_AP scans, and 607 Func4_PA scans. The demographic information for each session is provided in Table 2. Group ICA was performed on each session to estimate the ICs respectively. Four groups of ICs were obtained, each with 100 ICs showing the coactivation patterns across subjects. The Func1_AP session was the reference data, and the other three sessions were the replication data. We performed the spatial correlation analysis introduced above and then inspected the replicable ICs as the ICNs, which were used to build the NeuroMark aging template. The ICNs captured from the Func1_AP were used as the network template. We use Na to denote the number of ICNs in the aging template. Results were highly consistent when we used the other session data as the reference. We also split the HCP aging samples into middle-aged adults (36 ~ 64 years) and older adults (65 ~ 100+ years) and then built the template for each age group. The results were summarized in the supplementary materials, section “Aging Templates for Middle-aged Adults (36 ~ 64 years) and Older Adults (65 ~ 100+ years)”.
Table 2.
Basic Demographics of HCP-Aging Data.
| Basic Demographics | Func1_AP | Func2_PA | Func3_AP | Func4_PA |
|---|---|---|---|---|
| Total Subject | 589 | 626 | 573 | 607 |
| Age (month) | 712.77 ± 186.96 | 720.41 ± 186.04 | 718.47 ± 187.35 | 720.15 ± 184.88 |
| Gender (F/M) | 336/253 | 361/265 | 325/248 | 355/252 |
| Race (W/B/A/O) | 424/80/45/40 | 461/80/45/40 | 416/74/44/39 | 450/75/43/39 |
| Mean FD | 0.18 ± 0.05 | 0.17 ± 0.05 | 0.19 ± 0.05 | 0.17 ± 0.06 |
F = female; M = male; W = white; B = black or African American; A = Asian; O = others or unknown; FD = framewise displacement.
2.4. Building the Infant Template (newborn ~ nine months)
To build the infant template (NeuroMark_fMRI_Infant_3.0), we used our infant datasets, namely the Neuroimaging of Infants at High- and Low-Risk for ASD (NI-HLA) and the ACE Center 2017: Project 3 – Neuroimaging (ACE), collected at the Center for Systems Imaging at Emory University School of Medicine using a 32-channel head coil (Feinberg et al., 2010; Moeller et al., 2010). Infants were scanned during natural sleep at multiple time points between birth and nine months of age. The preprocessing of infant datasets has a slight difference, which is mainly in the normalization of the images. The other steps are the same as the preprocessing of HCP datasets. Details of the preprocessing are provided in the supplementary materials, section “Preprocessing of fMRI Data”.
We performed similar QC on the preprocessed infant data to select good scans for building the infant template. We excluded scans with head motions larger than 3° rotations or 3 mm translations and scans with bad normalization to the MNI space. After the QC, we had 155 scans from 45 subjects for the NI-HLA dataset and 175 scans from 60 subjects for the ACE dataset. The demographic information for each dataset is provided in Table 3. Group ICA was first performed on each dataset. The replicability of the group ICs between datasets was examined using a similar spatial correlation analysis. Here, we used the NI-HLA dataset as the reference data and the ACE dataset as the replication data. We searched for the matched ICs from the replication data based on the maximum correlation value. ICs were reproducible if their matched ICs showed spatial correlations larger than 0.4. The replicable ICs were further inspected and labeled as ICNs if their peak activations fell into the well-known gray matter regions (Allen et al., 2014). There are ICNs captured from both datasets, but in the present work, we used the ICNs from the NI-HLA dataset to build the infant template. We use Ni to denote the number of ICNs in the infant template.
Table 3.
Basic Demographics of Infant Data.
| Basic Demographics | NI-HLA Dataset | ACP Dataset |
|---|---|---|
| Scans | 155 | 175 |
| Subjects | 73 | 70 |
| Age (month) [scans] | 3.78 ± 1.81 | 3.66 ± 1.72 |
| Gender (F/M) [scans] | 66/89 | 82/93 |
| Mean FD [scans] | 0.17 ± 0.08 | 0.23 ± 0.07 |
F = female; M = male; FD = framewise displacement. Age, gender, and mean FD information is measured across scans.
2.5. Similarity across Functional Templates from Different Age Populations
After having the developmental, aging, and infant functional templates, we explored how and what similar and different patterns are shared across them. Specifically, we used the developmental template as the reference and measured its spatial similarities with the other two templates. For each ICN from the developmental template, we calculated its correlation coefficients with ICNs from the aging and infant templates. We identified the matched ICN from the aging and infant templates, which showed the maximum correlation values with the ICN from the developmental template. If the matched ICN showed a correlation higher than 0.4, we consider this developmental ICN reproducible in either the aging or infant template, or both templates. We repeated this analysis by setting the aging template and infant template as the reference, respectively. This procedure allows us to comprehensively investigate the shared and unique patterns across NeuroMark templates for different age groups.
2.6. Structural Template for Adult Cohorts
The structural template (NeuroMark_sMRI_3.0) was built using two large datasets, namely the human connectome project 1200 subject release (HCP, https://www.humanconnectome.org/study/hcp-young-adult/document/1200-subjects-data-release) and the UK-biobank project (UKB, https://www.ukbiobank.ac.uk/). The HCP 1200 subject release includes 3T MRI data from healthy young adult participants aged 22 ~ 35 years old collected from 2012 to 2015. We downloaded the raw T1 images from the HCP official website (https://www.humanconnectome.org/). The UKB data includes a population-based cohort of over 500,000 individuals aged 39 ~ 73 years old from 22 centers across the United Kingdom between 2006 and 2010. For building the structural template, we focused on a subset of healthy participants who have completed T1 MRI data. UKB subjects having any ICD-10 coded neurological/psychiatric diseases or congenital neurological disorders, reporting themselves that they were told to have a specific neurological/psychiatric disorder (which may or may not have been ICD-coded), and with incomplete MRI data were excluded. Our study is under Application ID 34175. We downloaded the raw T1 data from the UKB access management system and preprocessed the data using the SPM12 toolbox. Detailed preprocessing steps are provided in the supplementary materials, section “Preprocessing of sMRI Data and dMRI Data”.
We ran QC on the preprocessed structural data to select scans with good imaging quality for the analysis. After QC, we have 34822 T1 scans for the UKB data and 1110 scans for the HCP data. The demographic information of each data is provided in Table 4. For each dataset, PCA was performed to reduce the data into 120 principal components (PCs) which preserve more than 95% variance of the original data. The infomax ICA algorithm was conducted to decompose the 120 PCs into 100 ICs and such procedure was repeated 20 times in ICASSO, in which the best run was selected to ensure the estimation stability. ICs from the two datasets were matched by comparing their corresponding spatial maps. We used the UKB data as the reference and the HCP data as the replication. For each IC from the UKB data, we first identified the best-matched IC from the HCP which shows the highest spatial similarity to the reference. This was achieved by using a greedy elimination approach to match the ICs starting with the highest correlation values in the pair-wise correlation matrix of the ICs from UKB and HCP. If the spatial correlation of the matched IC from HCP is larger than 0.4, we consider this UKB IC replicable. We then labeled those replicable ICs as SBM networks if they exhibit peak activations in gray matter and have low spatial overlap with known vascular, ventricular, motion, and susceptibility artifacts. Finally, the SBMs from the UKB dataset were used to construct the structural template.
Table 4.
Basic Demographics of Structural Data.
| Basic Demographics | UKB Dataset | HCP Dataset |
|---|---|---|
| Subjects | 34882 | 1110 |
| Age (year) | 60.64 ± 15.23 | 28.79 ± 3.69 |
| Gender (F/M) | 19095/15727 | 603/507 |
| Race (W/B/O) | 32162/198/2462 | 830/165/115 |
F = female; M = male; W = white; B = black or African American; O = others or unknown.
2.7. Diffusion Template for Adult Cohorts
The diffusion template (NeuroMark_dMRI_3.0) was constructed using the same datasets, i.e., the HCP1200 release and the UKB project, described in ‘Structural Template for Adult Cohorts’. Note that, for building the diffusion template, we only selected a subset of 1135 healthy subjects from the UKB data, as it is computationally infeasible to run group ICA on such big diffusion data. The preprocessing of dMRI data involved a series of steps using FSL (www.fmrib.ox.ac.uk/fsl) and ANTs (Avants et al., 2022), which are detailed in the supplementary materials, section “Preprocessing of sMRI Data and dMRI Data”.
After the inspection of the preprocessed fractional anisotropy (FA) maps, we have 1000 dMRI scans for the UKB data and 1006 scans for the HCP data retained for building the diffusion template. The demographic information of each data is provided in Table 5. For each dataset, PCA was performed to reduce the data to 100 PCs which preserved more than 95% variance of the original data. The infomax ICA algorithm was conducted to decompose PCs to 100 ICs and such procedure was repeated 20 times in ICASSO, in which the best run was selected to ensure the estimation stability (Luo et al., 2020). ICs from the two datasets were matched by comparing their corresponding spatial maps. We used the UKB data as the reference and the HCP data as the replication. For each IC from the UKB data, we first identified the best-matched IC from the HCP which shows the highest spatial similarity to the reference IC. This was achieved by using a greedy elimination approach to match the ICs starting with the highest correlation values in the pair-wise correlation matrix of the ICs from UKB and HCP. If the spatial correlation between the UKB IC and its matched IC from HCP is larger than 0.4, we consider this IC replicable. We characterized those replicable ICs as ‘diffusion source-based morphometry’(d-SBM) networks if they exhibit peak covariations in white matter and have low spatial overlap with known vascular, ventricular, motion, and susceptibility artifacts. Finally, the d-SBMs estimated from the UKB dataset were chosen to construct the FA template.
Table 5.
Basic Demographics of Diffusion Data.
| Basic Demographics | UKB Dataset | HCP Dataset |
|---|---|---|
| Subjects | 1000 | 1006 |
| Age (year) | 56.42±7.29 | 26.89±3.44 |
| Gender (F/M) | 512/488 | 540/466 |
| Race (W/B/O) | 975/3/22 | 764/133/109 |
F = female; M = male; W = white; B = black or African American; O = others or unknown.
2.8. Applying NeuroMark Templates to Extract Single-subject Brain Imaging Features
With the network templates, the hybrid NeuroMark framework can extract individual-level imaging features from the MRI data. Individual-level imaging feature extraction is performed independently for each scan, which can avoid information leakage between training and testing data that might bias the analysis, such as the classification and prediction. Multiple methods incorporated in our group ICA of fMRI toolbox (GIFT) (http://trendscenter.org/software/) can estimate individual-level ICs using the NeuroMark templates as guidance. Semi-blind spatial constrained ICA was the first algorithm available in the toolbox utilizing the prior spatial information to perform ICA (Lin et al., 2010). This method is within the framework of constrained ICA with fixed-point learning and enables robust estimation of consistently identified spatial networks. This method can more accurately detect some brain networks compared to conventional ICA approaches. The most commonly used approach in NeuroMark studies is the group-information-guided ICA (GIG-ICA) (Du and Fan, 2013). GIG-ICA consists of two objective functions, one is to optimize the independence of single-subject ICNs, and the other is to optimize the correspondence between single-subject ICNs and the template prior. This method improves accuracy and intra-class coefficients (ICCs) in single-subject IC estimation compared to other network back-reconstruction methods (Du et al., 2017, 2016). GIG-ICA can capture more single-subject variation in network features that might be beneficial for classifying patients from healthy cohorts (Salman et al., 2019). Two newer methods are available in the toolbox for estimating single-subject components, namely constrained entropy bound minimization (cEBM) and constrained independent vector analysis (cIVA). cEBM adds increased flexibility by making use of the available prior knowledge in the spatial maps while allowing flexible density modeling of the sources without an orthogonality requirement for the demixing matrix (Yang et al., 2023). cIVA is an effective two-stage method to extract spatial and temporal features using IVA, mitigating the problems with big data while preserving the variability across subjects and time (Bhinge et al., 2019). It can use the NeuroMark template in a parameter-tuned constrained IVA to estimate time-varying representations of these signals while preserving the variability by tuning the constraint parameter.
Implementing NeuroMark with the new templates, brain features from different modalities can be calculated from multiple studies and datasets. Researchers only need to choose the appropriate template that best fits their datasets and study designs. For example, if a study focuses on ASD-related functional brain alterations in adolescents, the developmental template might be appropriate. NeuroMark can capture network features, such as spatial networks, functional network connectivity (FNC) between networks, frequency information of network fluctuations, etc., from both static and dynamic perspectives. Static FNC can be measured by computing the Pearson correlations between time courses (TCs) of ICNs, representing inter-relationships between different ICNs. Dynamic FNC is estimated via a sliding window approach (Allen et al., 2014; Fu et al., 2019), in which a tapered window obtained by convolving a rectangle with a Gaussian is used to segment the entire TCs into multiple short periods. If a study is interested in the white matter injury in dementia datasets, it can use the NeuroMark framework with the diffusion template. Reliable and comparable FA features can be extracted for the exploration of abnormal diffusion in radiographically normal-appearing regions across disorders.
3. Results
3.1. Functional Template for Children and Adolescents
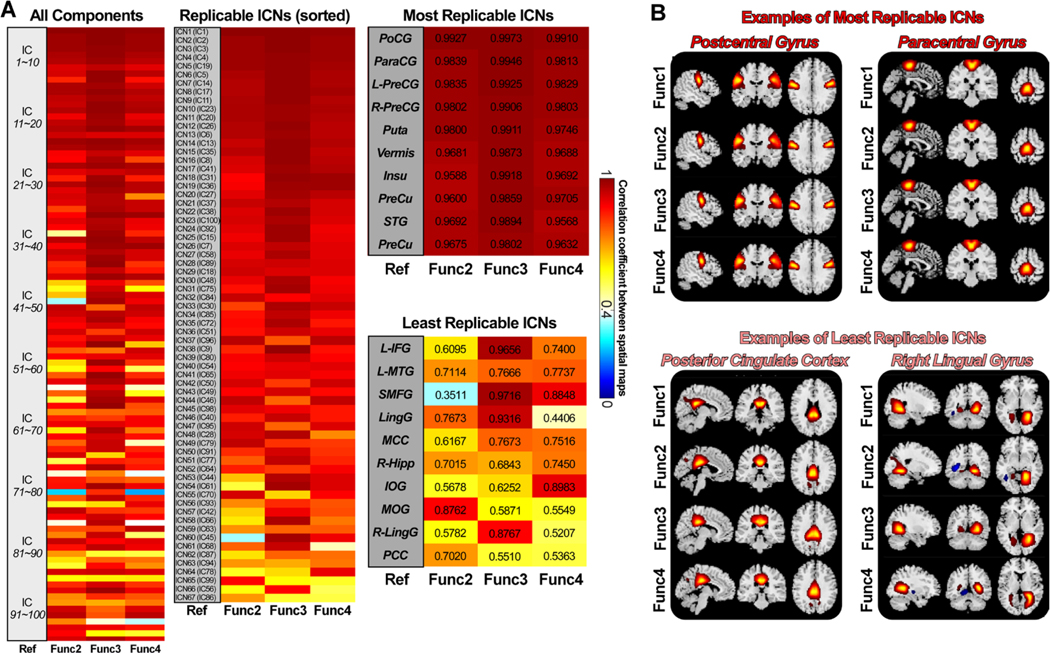
Fig. 2 displays the matching results of ICs of different session data from the HCP-D cohorts. Fig. 2A shows the spatial correlation between the matched ICs, where the Func1_AP session is the reference data and the other sessions are the replication data. The first subplot displays the correlations between all 100 ICs and their best-matched ICs from each replication data. Most ICs are highly replicable across sessions with spatial similarity > 0.4. Specifically, all 100 ICs have replicable ICs in the other sessions with mean correlations larger than r = 0.4. 94 ICs have replicable ICs with mean correlations larger than r = 0.6, and 76 ICs have replicable ICs with mean correlations larger than r = 0.8. The spatial correlation reflects the similarity between matched components obtained by using the brain voxels within the brain mask.
Fig. 2.
Spatial matching results of the ICs of different sessions from the HCP-D data. A) Spatial correlations between matched ICs from the reference data (Func1_AP) and the replication data. Correlations between meaningful ICNs are also displayed (average r > 0.4 for the replication data), sorted by the average correlation. ICNs are used for the construction of the developmental template. B) Examples of the most and the least replicable ICNs between reference and replication data. ICNs show robust activation patterns across sessions.
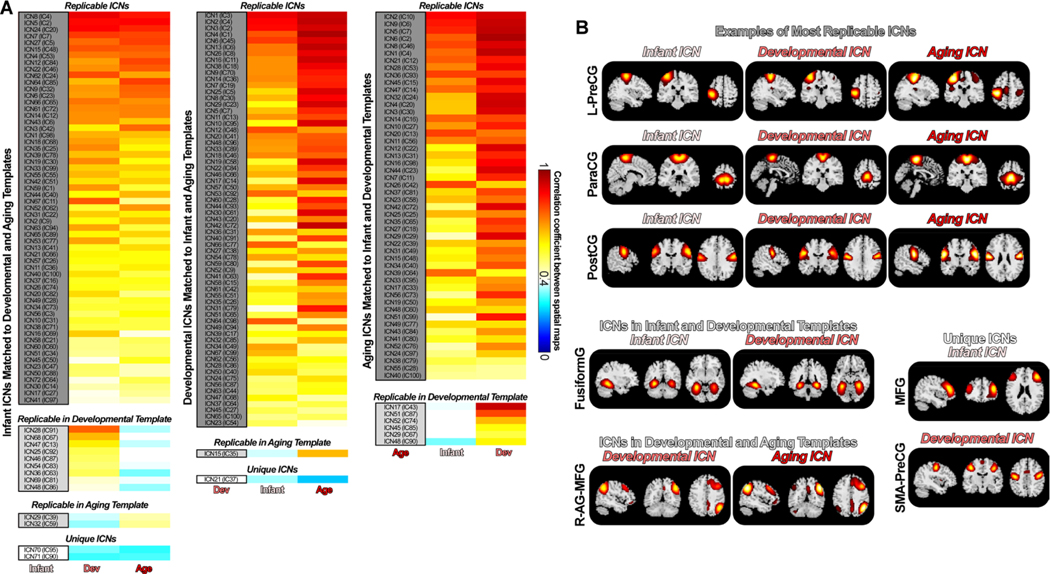
Among the replicated ICs, 67 ICs were characterized as meaningful ICNs by examining their spatial activation and the low-frequency fluctuations in the TCs. These results are highly consistent if we used other session data as the reference. The second subplot in Fig. 2A displays the correlations for the 67 ICNs, sorting from highest replicability to lowest replicability (maximum average correlation is 0.9937, and the minimum average correlation is 0.5964). We labeled these networks from ICN1 to ICN67, which were used to construct the developmental template. In the third subplot of Fig. 2A, we provided the labels of the ten most replicable ICNs and ten least replicable ICNs, along with their spatial correlations. Postcentral gyrus (PoCG), paracentral gyrus (ParaCG), precentral gyrus (PreCG), putamen (Puta), vermis8_9 (Vermis), insula (Insu), precuneus (PreCu) and superior temporal gyrus (STG) are the most replicable ICNs in the developmental template. In contrast, the hippocampus (Hipp), inferior frontal gyrus (IFG), middle temporal gyrus (MTG), vermis 4_5 (Vermis), middle cingulate cortex (MCC), middle occipital gyrus (MOG), fusiform gyrus (Fusi) and post cingulate cortex (PCC) are the least replicable ICNs. Fig. 2B provides several examples of the most and least replicable ICNs across reference and replication data. The overall results suggest that ICNs from the developmental template are highly replicable, showing similar spatial activation patterns across sessions.
3.2. Functional Template for Aging Cohorts
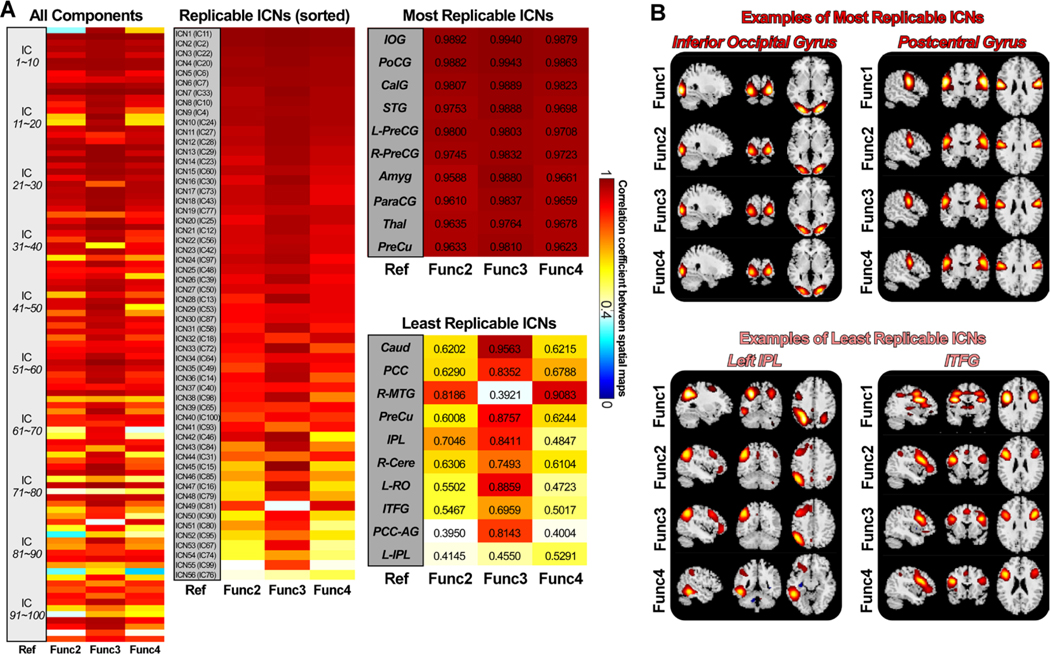
Fig. 3 shows the matching results of the ICs of different sessions from the HCP-A data. Fig. 3A provides the spatial correlations of the matching ICs between the reference and the replication data. Again, most ICs are highly replicable, showing consistent spatial activations across sessions. 99 ICs from the reference data (Func1_AP) are replicable in the other three sessions, with the most matching ICs showing mean spatial correlation > 0.4. 93 ICs have replicable ICs with mean correlations larger than r = 0.6, and 73 ICs have replicable ICs with mean correlations larger than r = 0.8. 56 of the 99 replicable ICs were identified as ICNs, which were used to construct the NeuroMark aging template. These ICNs show a maximum correlation of 0.9904 and a minimum correlation of 0.4662 (averaged across replication data). These results are highly consistent if we use other session data as the reference data. We observed that ICs from the data collected in the same phase-encoding direction show higher similarity than those from the data collected in different directions. In the aging template, inferior occipital gyrus (IOG), PoCG, Calcarine gyrus (CalG), STG, amygdala (Amyg), ParaCG, thalamus (Thal), and Precu are the most replicable ICNs. Anterior cingulate cortex (ACC), MCC, Caudate (Caud), PCC, PreCu+MOG, inferior parietal lobule (IPL), cerebellum, Rolandic operculum (RO), and inferior opercular frontal gyrus (IOFG) are the least replicable ICNs. Although ICNs might show variations across sessions, they are highly similar across the reference and the replication data.
Fig. 3.
Spatial matching results of the ICs of different sessions from the HCP-A data. A) Spatial correlations between matched ICs from the reference data (Func1_AP) and the replication data. Correlations between meaningful ICNs are also displayed (average r > 0.4 for the replication data), sorted by the average correlation. ICNs are used for the construction of the aging template. B) Examples of the most and the least replicable ICNs between reference and replication data. ICNs show consistent activation patterns across sessions.
3.3. Functional Template for Infants
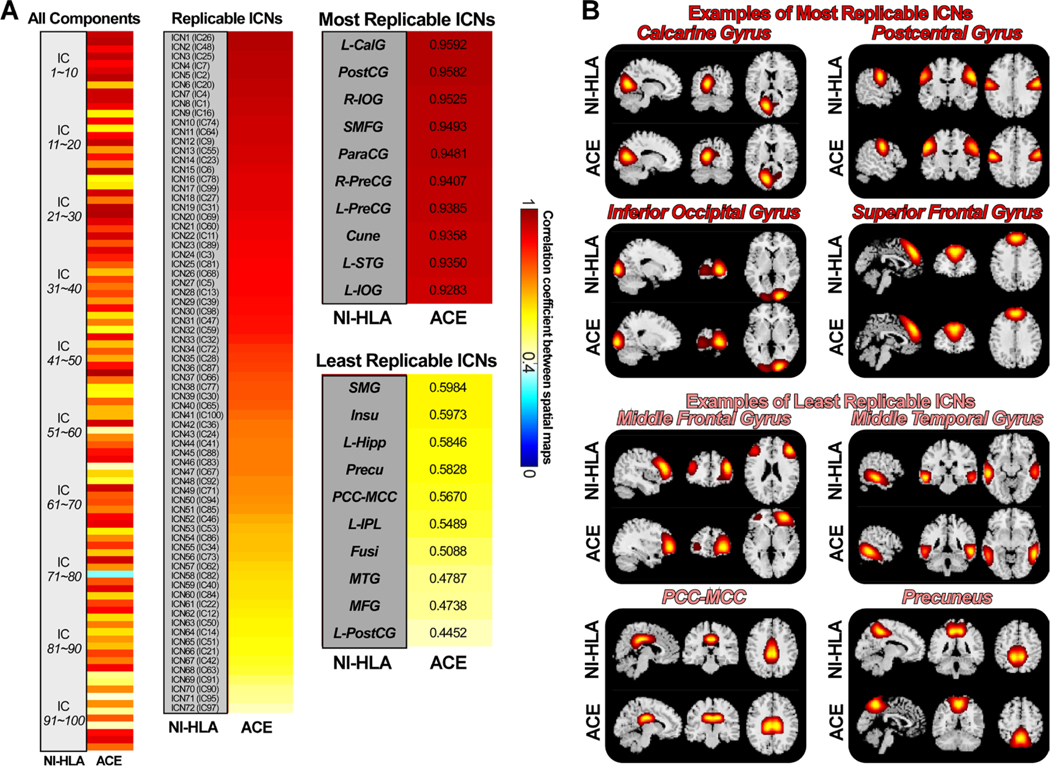
Fig. 4 displays the spatial similarities of ICs estimated from two infant datasets. Unlike the developmental and aging templates built on the data from different sessions, the infant template was built using different datasets. Therefore, relatively smaller correlations were identified between these two groups of ICs. However, most ICs are still highly replicable. 99 of the 100 ICs from the NI-HLA data have the best-matched ICs from the ACE data showing a spatial correlation larger than 0.4. 86 ICs have replicable ICs with correlations larger than r = 0.6, and 43 ICs have replicable ICs with correlations larger than r = 0.8. Different from the developmental and aging cohorts with aggregated networks, infant components showed more widely distributed patterns. We labeled 72 reproducible ICs as meaningful ICNs, sorted in descending order in Fig. 4B. The most replicable ICN is the left CalG, which shows spatial similarity as 0.9592, while the least replicable ICN is the left PostCG, which shows spatial similarity as 0.4452. PostCG, IFG, superior medial frontal gyrus (SMFG), ParaCG, right PostCG, left PreCG, Cuneus (Cune), STG, and IOG are the most replicable ICNs. Supramarginal gyrus (SMG), Insu, Hipp, Precu, PCC, IPL, Fusi, MTG, and middle frontal gyrus (MFG) are the least replicable ICNs in the infant template.
Fig. 4.
Spatial matching results of the ICs from two infant datasets. A) Spatial correlations between matched ICs from the reference data (NI-HLA) and the replication data (ACE). Correlations between meaningful ICNs are also displayed (r > 0.4 for the replication data), sorted by the correlation value. ICNs are used for the construction of the infant template. B) Examples of the most and the least replicable ICNs between the reference and replication data. ICNs show similar activation patterns between datasets.
3.4. Comparison of Functional Templates
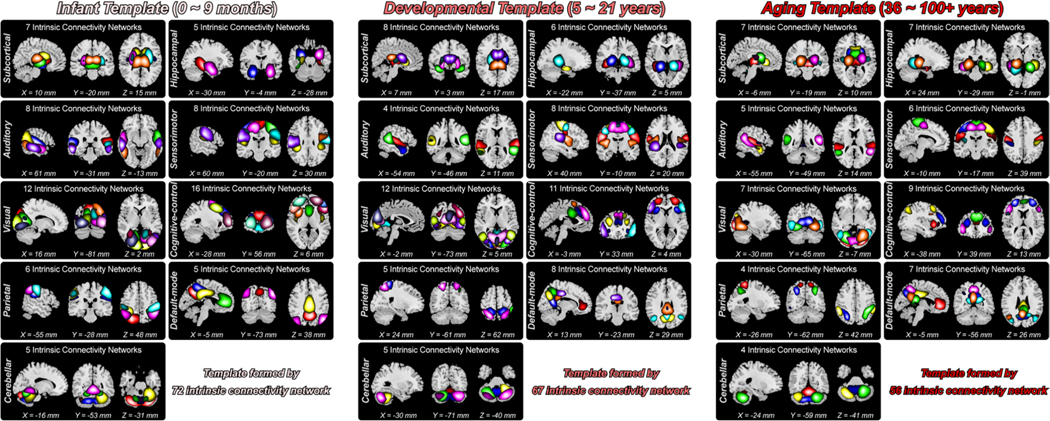
Fig. 5 displays the composite spatial maps of 72 ICNs from the infant template, 67 ICNs from the developmental template, and 56 ICNs from the aging template. In each template, the ICNs were labeled and arranged into different domains according to the prior functional and anatomical information (Allen et al., 2014; Du et al., 2020). In line with our previous work, we first defined the seven most commonly used domains as the subcortical domain (SC), auditory domain (AUD), sensorimotor domain (SM), visual domain (VS), cognitive-control domain (CC), default-mode domain (DM), and cerebellar domain (CB). In addition to these domains, we introduced two domains, the hippocampal domain (HP) and the parietal domain (PA), which have attracted increasing attention in current neuroscience. Therefore, ICNs of each template were finally assigned to nine domains for the visualization of their spatial activations. The activation map of each ICN represented the voxels that mostly contributed to the ICN because the skewness of each ICN was changed to be positive. Specifically, the infant template includes seven ICNs in SC, five ICNs in HP, eight ICNs in AUD, eight ICNs in SM, 12 ICNs in VS, 16 ICNs in CC, six ICNs in PA, five ICNs in DM, and five ICNs in CB. The developmental template includes eight ICNs in SC, six ICNs in HP, four ICNs in AUD, eight ICNs in SM,12 ICNs in VS, 11 ICNs in CC, five ICNs in PA, eight ICNs in DM, and five ICNs in CB. The aging template includes seven ICNs in SC, seven ICNs in HP, five ICNs in AUD, six ICNs in SM, seven ICNs in VS, nine ICNs in CC, four ICNs in PA, seven ICNs in DM, and four ICNs in CB. The detailed label, peak coordinate, and spatial map of each ICN are provided in the supplementary materials, sections “Template and Intrinsic Connectivity Networks”.
Fig. 5. Composite maps of the identified ICNs for the infant, developmental and aging templates.
ICNs covered almost the whole brain are clustered into nine functional domains, including 1) subcortical domain, 2) hippocampal domain, 3) auditory domain, 4) sensorimotor domain, 5) visual domain, 6) cognitive-control domain, 7) parietal domain, 8) default-mode domain, and 9) cerebellar domain. One color in the composite maps corresponds to an ICN.
We observed that ICNs show highly similar patterns across templates, although the younger template tends to be more segregated while the older template tends to be more aggregated. We investigated the shared and unique aspects of the lifespan functional templates by examining their spatial similarities. Fig. 6 displays the comparison results across templates. We first used the infant template as the reference and found most infant ICNs replicated in the developmental and aging templates. 81.9% of ICNs (59 out of 72) from the infant template are observed in the developmental and aging templates (r > 0.4). Nine infant ICNs can only be replicated in the developmental template, while two infant ICNs can only be replicated in the aging template. Two ICNs, namely high MFG (ICN 70, averaged r with older templates = 0.2989) and low MTG (ICN 71, averaged r with older templates = 0.3042), cannot be identified in the older templates. 97% of ICNs (65 out of 67) from the developmental template are replicable in the infant and aging templates (r > 0.4), except for two ICNs. ICN 15 from the developmental template, labeled as lingual gyrus (LingG), is not identified in the infant template. ICN 21, which is more likely an aggregated network combining PreCG with the supplementary motor area (SMA), is not captured in the infant and aging templates. 100% of ICNs from the aging template have replicated ICs from the developmental template. Six aging ICNs are not identified in the infant template, most of which are aggregated networks containing both frontal and parietal regions. For example, ICN 17 and ICN 51 from the aging template, which covers the posterior brain including the right IPL and angular gyrus (AG), and the anterior brain including the frontal gyrus, cannot be observed in the infant template. Fig. 6B shows some examples of ICNs across the NeuroMark templates along the lifespan. While most ICNs are highly replicable across the templates, there are some ICNs with specific activations to given age populations, which further proves the importance of introducing NeuroMark templates for different age groups for better capturing the brain functional features across the lifespan.
Fig. 6.
Similarities and differences across lifespan NeuroMark templates. ICNs show both similar and unique patterns across templates. A) Spatial correlations between ICNs from the reference template and the other templates. Most of the ICNs are replicable across age groups. B) Examples of shared and unique ICNs across the infant, developmental and aging templates. Infant ICNs tend to be distributed across the whole brain, while aging ICNs tend to be aggregated, covering multiple distanced brain regions.
3.5. NeuroMark Structural and Diffusion Templates
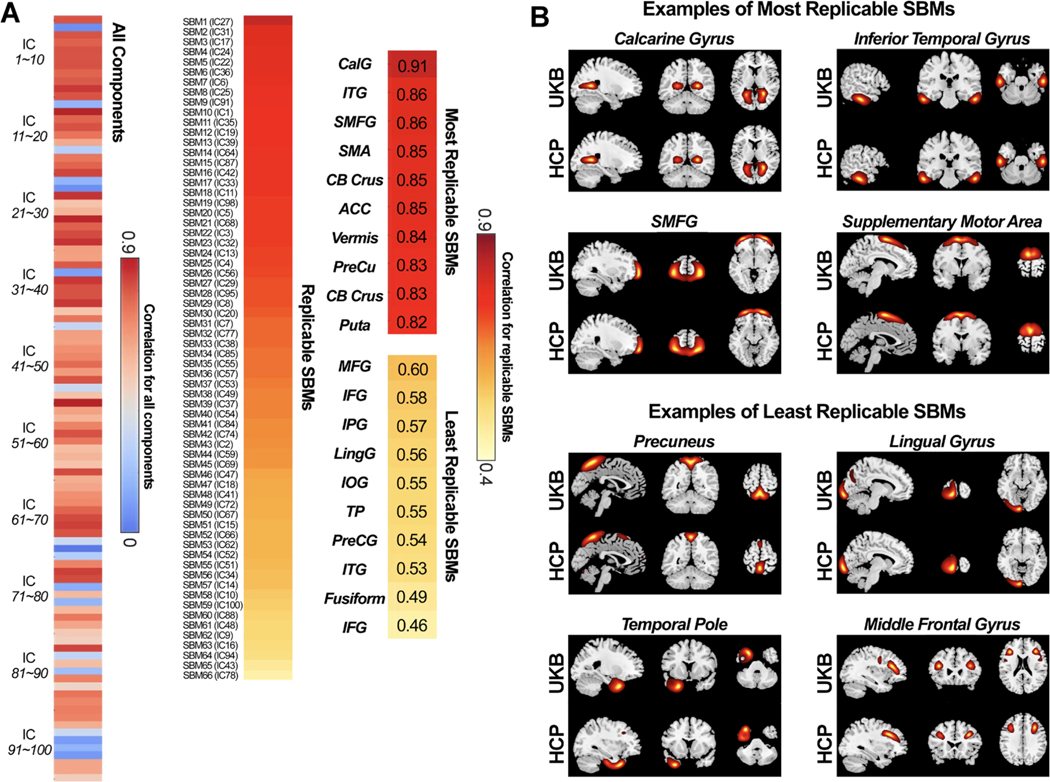
Fig. 7 shows the spatial correlations between the best-matched ICs from the UKB and HCP datasets. Out of the 100 ICs identified from the UKB dataset, 66 components were replicable in the HCP dataset, having the best-matched ICs with spatial correlation larger than 0.4. All 66 replicable ICs show covariations in the meaningful gray matter areas and were labeled as SBM networks for building the structural template. The most replicable IC was the CalG (r = 0.91), which is also one of the most replicable ICs in the functional templates. Other top replicable ICs include inferior temporal gyrus (ITG), SMFG, and SMA. The least replicable SBM networks were Precuneus, IOG, and Fusi. As seen in Fig. 7B, the replicable SBMs show highly similar covariations despite being captured from two independent datasets.
Fig. 7.
Spatial matching results of the ICs from two structural datasets. A) Spatial correlations between matched ICs from the reference data (UKB) and the replication data (HCP). Correlations between meaningful SBM networks are also displayed (r > 0.4 for the replication data), sorted by the correlation value. SBM networks are used for the construction of the NeuroMark structural template. B) Examples of the most and the least replicable SBM networks between the reference and replication data. SBM networks show similar covariation patterns of gray matter between datasets.
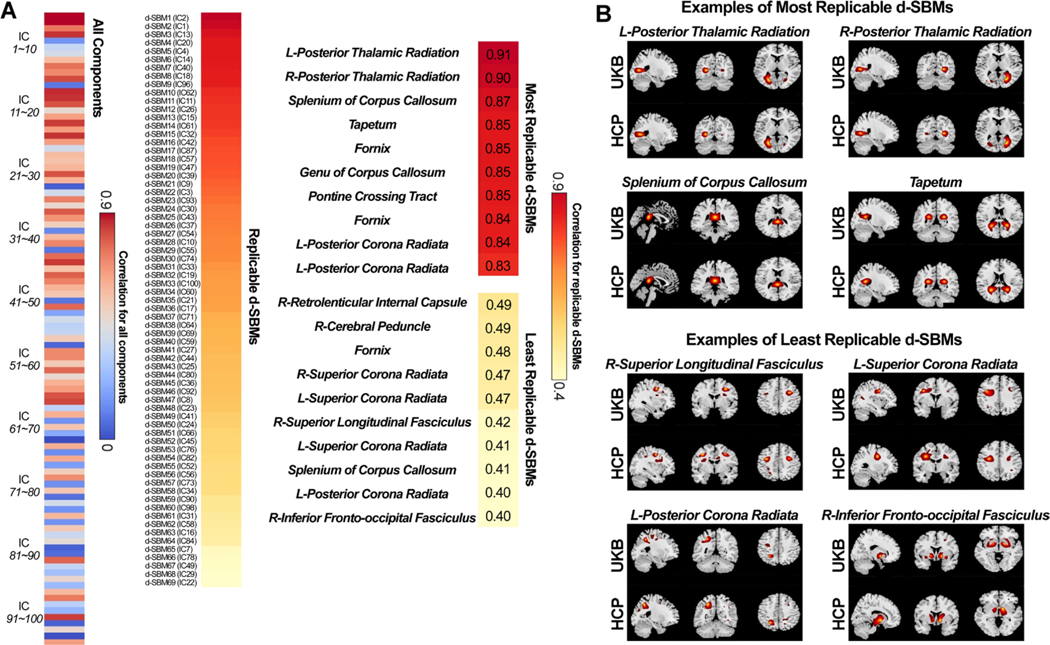
Fig. 8 shows the spatial correlations between the best-matched diffusion ICs from the UKB and HCP datasets. Out of the 100 ICs identified from the UKB dataset, 69 components were replicable in the HCP dataset, having the best-matched ICs with a spatial correlation larger than 0.4. All the 69 replicable ICs show covariations in the meaningful white matter areas and therefore were labeled as d-SBM networks for building the diffusion template. Two white matter atlases (Laboratory of Brain Anatomical MRI, Johns Hopkins University, FSL6.0) were used for labeling. As the d-SBM networks were derived from ICA in FA maps, the labeling in Fig. 8 referred to the 50-region ICBM-DTI-81 white-matter labels atlas, which were originally generated from diffusion tensor maps as well. In addition, to seek better grouping, we used a 20-tract JHU white-matter tractography atlas to separate all 69 d-SBM networks into more detailed categories (commissure, association, and project) based on the highest overlapping and tract orientations. More details on the labeling and grouping are provided in the supplementary materials, section “Diffusion Template and Source-based Morphometry Networks”. The most replicable ICs were the left and right posterior thalamic radiation (including optic radiation) (r = 0.91, and r = 0.90), which are, intriguingly, directly connected to the (or one of) most replicable ICs, i.e., CalG, in both the functional and structural templates. Other top replicable ICs include tapetum, genu of corpus callosum, and pontine crossing tract. The least replicable among the d-SBM networks were inferior frontal-occipital fasciculus, superior corona radiata, and superior longitudinal fasciculus. As seen in Fig. 8B, the replicable d-SBM networks show highly similar covariations between two independent datasets.
Fig. 8.
Spatial matching results of the ICs from two diffusion datasets. A) Spatial correlations between matched ICs from the reference data (UKB) and the replication data (HCP). Correlations between meaningful d-SBM networks are also displayed (r > 0.4 for the replication data), sorted by the correlation value. d-SBM networks are used for the construction of the NeuroMark diffusion template. B) Examples of the most and the least replicable d-SBM networks between the reference and replication data. d-SBM networks show similar covariation patterns of white matter between datasets.
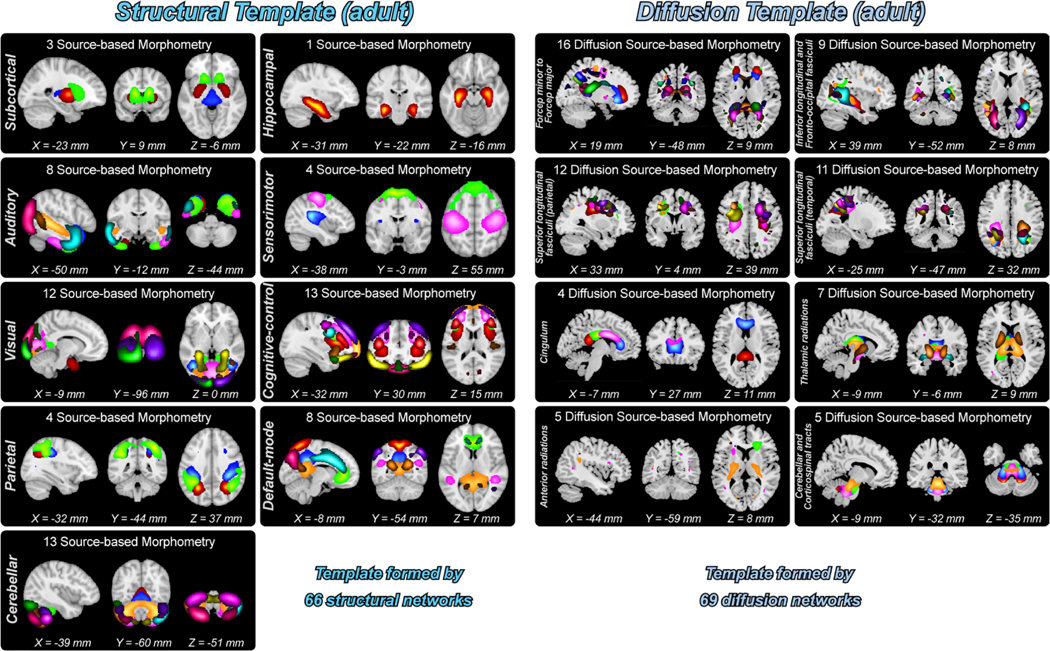
Fig. 9 shows the composite maps of 66 SBMs from the structural template and 69 d-SBMs from the diffusion template. Similar to the functional templates, the SBMs from the structural template were grouped into nine domains. Specifically, the structural template includes three ICNs in SC, one ICN in HP, eight ICNs in AUD, four ICNs in SM, 12 ICNs in VS, 13 ICNs in CC, four ICNs in PA, eight ICNs in DM, and 13 ICNs in CB. On the other hand, the d-SBMs from the diffusion template were grouped into eight categories, which are, Commissure-prefrontal (forceps minor) to occipital (forceps major) tracts including 16 d-SBMs, Association-inferior longitudinal and inferior frontal-occipital fasciculi including nine d-SBMs, Association-superior longitudinal fasciculi (parietal) including 12 d-SBMs, Association-superior longitudinal fasciculi (temporal) including 11 d-SBMs, Association-cingulum including four d-SBMs, Projection-thalamic radiations including seven d-SBMs, Projection-anterior radiations including five d-SBMs, and Projection-cerebellar and corticospinal tracts including five d-SBMs.
Fig. 9.
Composite maps of SBMs and d-SBMs for the structural and diffusion templates. SBMs and d-SBMs are clustered into different categories. One color in the composite maps corresponds to an SBM or d-SBM.
4. Discussion
4.1. Automating and Hybrid NeuroMark Approach
In this study, we expanded the NeuroMark framework by introducing the first 4D functional templates (NeuroMark_fMRI_3.0), as well as the first structural (NeuroMark_sMRI_3.0) and diffusion templates (NeuroMark_dMRI_3.0). With these templates, NeuroMark can capture features according to the age of the populations and across modalities, which might boost accuracy in mental health research and advance our understanding of lifespan and cross-modalities alterations.
ICA is a promising data-driven approach widely used in neuroscience to extract meaningful imaging features across imaging modalities (Calhoun et al., 2009; Liu et al., 2009; Sui et al., 2011). However, there is a potential limitation in the conventional ICA approach that the components estimated by the data-driven method might change across runs, making the results difficult to compare and validate across studies. With the development of neuroscience, there is an increasing need for comparing the results between brain disorders and validating the results across studies and datasets (Marek et al., 2022; Weller and Kinder-Kurlanda, 2016). Precise characterization of reliable neuroimaging features holds great promise in predicting brain disorders and advancing our understanding of complex human behavior (Aarts et al., 2015). In addition, there is a consensus that many brain disorders show clinically overlapping and unique symptoms, reflected by the similarity and distinction in their neuroimaging data. For example, autism spectrum disorder (ASD) and Schizophrenia (SZ) have overlapping clinical features (Konstantareas and Hewitt, 2001) and similar brain alterations (Fu et al., 2020; Mastrovito et al., 2018; Yoshihara et al., 2018), which were previously regarded as the same brain disorders in different stages (Eisenberg and Kanner, 1956). SZ also shares overlapping symptoms with bipolar disorder (BP), where 60% of bipolar patients have psychotic features (Goes et al., 2007) and similar neurocognitive deficits (Glahn et al., 2004). SZ and BD show similar brain alterations in motor and parietal regions but were separated by functional differences in the medial frontal and visual cortex (Sui et al., 2011). Alzheimer’s disease (AD) and vascular cognitive impairment and dementia (VCID) are the two most common causes of dementia in elderly individuals, which share similar brain lesions, symptoms, and progressive course that confound the clinical distinction (Fu et al., 2020; Rosenberg et al., 2016). From a clinical perspective, it would be greatly beneficial to investigate and compare brain changes among brain disorders for increased sensitivity and specificity and further advancing diagnosis and accurate treatments. Neuroimaging consortia have collected many large sample data, such as UK Biobank (Douaud et al., 2021), ABCD (Casey et al., 2018), HCP (Van Essen et al., 2012), ADNI (Jack et al., 2008), OASIS (O’Connor and Davitt, 2012; Tullai-McGuinness et al., 2009), ABIDE (Di Martino, 2012), BSNIP (Mokhtari et al., 2016), and ADHD200 (Milham et al., 2012), providing an unprecedented opportunity for the investigation of comparability and reproducibility of population-based research. The development of current neuroscience magnifies the potential problem of conventional ICA approaches, which might hinder the replication and comparison of imaging features.
In this context, we developed the NeuroMark framework (Du et al., 2020), which makes use of the pre-defined reference and the data-driven method to extract reliable imaging features that can be linked to brain disorders and complex human behavior. Unlike conventional ICA-based approaches, NeuroMark relies on pre-defined templates to extract imaging features for each scan. The use of the pre-defined template not only guarantees the correspondence of imaging features across measurements but also increases the likelihood of detecting meaningful biomarkers by reducing the search space. The use of data-driven approaches in feature extraction allows the NeuroMark to retain more subject-specific variations benefiting the characterization of inter-subject variability (Du and Fan, 2013). The NeuroMark framework, therefore, absorbs the strengths of the ICA-based and atlas-based approaches and has shown promising abilities to capture functional biomarkers in many recent applications. It has successfully extracted a wide range of imaging features, including spatial activation of networks (Du et al., 2020; Hajjar et al., 2022), time-courses of the networks (Hajjar et al., 2022; Zhao et al., 2022), static FNC (Du et al., 2021; Levey et al., 2022; Rahaman et al., 2023; Vaidya et al., 2023), and dynamic FNC (Dini et al., 2021; Fu et al., 2021d, 2021c, 2021b; Tu et al., 2020, 2019), across healthy populations (Abrol et al., 2023; Lewis et al., 2023; López-Vicente et al., 2021) and many brain disorders, such as mild cognitive impairment (MCI) (Levey et al., 2022; Li et al., 2021), SZ (Du et al., 2021; Fu et al., 2020; Fu et al., 2021b; M.S.E. Sendi et al., 2021a), ASD (Du et al., 2021; Fu et al., 2020), COVID-19 (Fu et al., 2021d), AD (Hajjar et al., 2022; M.S.E. Sendi et al., 2021b), and major depressive disorder (MDD) (Fu et al., 2023a, 2023b, 2021c; Mohammad S.E. Sendi et al., 2021), etc. The imaging features captured by the NeuroMark framework have been combined with many advanced analytic approaches to quantify the compatibility, reproducibility, and reliability of brain makers across studies and datasets.
4.2. 4D Functional Templates, from Infant to Aging Cohorts
Although NeuroMark brings a lot of conveniences to standardize biomarker development, there are still concerns about the template used in previous studies. We have listed all the available NeuroMark templates and their brief information in Table 6. The previous NeuroMark_fMRI_1.0 is constructed using two large-sample healthy populations from the HCP and GSP datasets, where the subjects’ average age is 28.79 years old and 21.54 years old. NeuroMark_fMRI_1.0 mainly reflects the network structure for young adults at 20–30 years old. However, this template has been applied to many datasets where the participants are in significantly different age groups. For example, this template has been used in AD and MCI studies where the participants are most likely aging cohorts. Recent work has adopted this template on the MCI participants aged 50–90 years old for characterizing atomoxetine-associated functional brain connectivity changes (Levey et al., 2022). NeuroMark has also been used to extract ASD-related imaging features, in which subjects are typically children or adolescents (Fu et al., 2020). More recently, a study focusing on Arsenic exposure has implemented this template to different age populations for the exploration of brain abnormalities among children, adolescents, and young adults in India (Vaidya et al., 2023). Normal aging in functional brain networks has long been appreciated in literature (Chan et al., 2018; Schlee et al., 2012), where age-related dedifferentiation can manifest as neural representations showing reductions in the specialization and specificity of networks (Goh, 2011). Functional network differences between children and adults have also been reported in many studies (Jolles et al., 2011; Liu et al., 2018; Sato et al., 2021). There is an open question of whether a common template in NeuroMark can capture reliable functional activations reflected in different age populations.
Table 6.
List of NeuroMark Templates.
| Version | Template Name | Brief Information | Reference |
|---|---|---|---|
| NeuroMark 1 | NeuroMark_fMRI_1.0 | Functional, young adult | (Du et al., 2020) |
| NeuroMark 2 | NeuroMark_fMRI_2.0 | Functional, multi-model-order, lifespan | (Iraji et al., 2023) |
| NeuroMark_fMRI_2.1 | Functional, multi-model-order, lifespan | ||
| NeuroMark 3 | NeuroMark_fMRI_Infant_3.0 | Functional, infant | Current |
| NeuroMark_fMRI_Developmental_3.0 | Functional, youth | ||
| NeuroMark_fMRI_Aging_3.0 | Functional, aging | ||
| NeuroMark_sMRI_3.0 | Structural, adult | ||
| NeuroMark_dMRI_3.0 | Diffusion, adult |
Therefore, in this work, we introduced age-specific templates adaptable to the NeuroMark framework and examined their similarities and differences. The new functional templates were built using 5000 scans from subjects aged from infancy to aging. To the best of our knowledge, this is the first study that introduced network templates for different age populations that provide hypothesis-driven references to the hybrid data-driven approach. We applied a common normalization strategy to wrap the data into a standard MNI space that can make the resulting templates comparable across age groups. Meanwhile, we replicated the components across data, which therefore generated more reliable ICN templates for future neuroimaging studies. As shown in the comparison of NeuroMark templates for different age groups, the infant, developmental, and aging templates encompass highly similar ICNs. More than 80% of the ICNs are replicable across templates built for different age populations. NeuroMark has an objective function that optimizes feature extraction based on single-subject data properties (Du and Fan, 2013). This helps to reduce differences (such as age, conditions, etc.) between the target data and the template. As a result, we believe that NeuroMark templates are suitable for use with various clinical datasets to capture reliable neuroimaging features. Future studies using datasets in a wide age range can also concentrate on those shared ICNs that reliably exist along the lifespan. NeuroMark_fMRI_3.0 also shows age-specific ICNs, which might provide important network information at an appropriate age or for a specific population. Age-specific templates combined with the NeuroMark framework might precisely capture the functional network features in various applications.
An interesting finding in this work is that the infant template is more likely to split the widespread regions into different networks, which implies that the infant brain might show more functional segregation. On the contrary, the aging template contains networks with coactivated voxels located at distributed brain regions. This result might explain the phenomenon of more ICNs captured in the younger template and fewer ICNs captured in the older template (because an ICN in the aging template might split into multiple networks in the infant template). This finding also provides evidence supporting dedifferentiation of the aging brain (Koen and Rugg, 2019), where the functional dedifferentiation theory suggests that functional networks in the elderly are characterized by increased coupling between remote regions from different networks (Chan et al., 2014; Geerligs et al., 2015; Grady et al., 2016; D. Tomasi and Volkow, 2012), such as the default-mode, salience, and frontoparietal networks.
4.3. Structural and Diffusion Templates for Capturing Brain Markers from Other Modalities
In addition to the 4D functional templates, we introduced network templates for other MRI modalities. Multimodal neuroimaging techniques might provide complementary information, aiding in the detection of brain changes related to mental health and complex human behavior. Recent neuroscience has been paying increasing attention to cross-modality analysis and believes that different modalities represent different perspectives of the functional, structural, or anatomical properties, potentially revealing the latent biomarkers that might be missed from unimodal analysis (Calhoun and Sui, 2016). The investigation of brain imaging across modalities can inform us how brain structure shapes brain function or the reverse and advance our understanding of how functional or structural aspects of physiology could drive human behavior and cognition.
Similar to functional analysis, one can combine information across voxels of structural or diffusion data to identify networks and perform testing on the subject covariation of these networks rather than testing each voxel separately. As a multivariate extension to voxel-based morphometry (VBM) (Ashburner and Friston, 2000), SBM (Xu et al., 2009) is a data-driven approach that captures the covariations of brain voxels across scans. SBM has been successfully applied to many studies, identifying imaging biomarkers that are linked to a wide range of brain disorders (Duan et al., 2021; Gupta et al., 2015; Park et al., 2022; Steenwijk et al., 2016; Sui et al., 2012). However, as an ICA-based method, SBM shares the same weakness in that the identified components vary across datasets and studies, significantly influencing the replication and validation of the results. Therefore, we built robust sources to guide the individual source computation in sMRI and dMRI. Two large datasets, UKB and HCP were used for constructing the network templates. NeuroMark_sMRI_3.0 and NeuroMark_dMRI_3.0 allow for the extraction of covaried voxels from the gray matter volume (GMV) data and the FA data, where the resulting network features are comparable across subjects, studies, and datasets. With the structural and diffusion templates, the NeuroMark framework can automate the searching of structural and diffusion biomarkers within and across clinical conditions. In addition, studies can also combine ICA-based features from different modalities with advanced analytic approaches, such as machine learning methods, which can distill a rich set of imaging features into a single index for accurately predicting human behavior (Sui et al., 2020). Machine learning and multivariate methods have been successfully applied to neuroimaging studies in recent years, identifying robust biomarkers for complex human behavior (Chen et al., 2022; Chopra et al., 2022; Jiang et al., 2022b, 2022a; Kucyi et al., 2021; Lee et al., 2021; Woo et al., 2017b, 2017a). Multivariate associations between brain features and behavioral variables are robustly related to univariate effect sizes (r = 0.79, p < 1.0 × 10−3) (Marek et al., 2022), suggesting the potential to integrate univariate and multivariate analyses to reveal mechanistic linkages between brain imaging and behavior.
4.4. Limitations and Future Directions
NeuroMark_fMRI_3.0 introduces functional templates for different age populations. We note that temporal information should also be considered for other modalities. Future NeuroMark will develop structural and diffusion templates for different age groups and comprehensively investigate the similarities and differences across ages. NeuroMark can also focus on building templates for different clinical cohorts, which might capture disorder-specific network covariation patterns. Although previous applications of NeuroMark in many brain disorders have shown that the data-driven approach in NeuroMark can adapt to populational differences, it would be of great interest to allow NeuroMark to adapt to disorder-specific networks. Literature has demonstrated evidence of different network covariations, underlying distinct disorders, and disease mechanisms (Nomi and Uddin, 2015; Rubinov and Bullmore, 2013; Skidmore et al., 2011; Tijms et al., 2013). However, it is also useful to preserve the correspondence to control groups, to do this we can potentially adjust the framework to allow for this additional aspect.
In our future studies, we plan to extend our UKB application to gain full access to the dMRI data. The process of developing cross-modality templates can be divided into two parts. Firstly, the templates can be constructed using the same subjects while only considering the unimodal imaging variation. Then, the templates will be matched and compared to investigate the overlapping and unique patterns across modalities. Secondly, we can develop multimodal fusion templates that can be placed on a spectrum that ranges from analyzing each modality separately in the context of a similar study to the symmetric analysis of multimodalities to extract the joint information that is often hidden.
There are currently numerous approaches to multimodal fusion, and many are now being applied to systematically evaluate neuroimaging biomarkers of brain disorders, such as joint ICA (Calhoun et al., 2006), parallel ICA (Liu et al., 2009), and multiset canonical correlation analysis (mCCA) (Correa et al., 2010). However, data-driven approaches can suffer from variability across runs and studies, which can hinder the development and replication of fusion studies.
More recently, we introduced a hybrid multimodal fusion method, namely mCCA with reference plus joint ICA (mCCAR+jICA) (Calhoun, 2018), which can jointly analyze the shared components while linking the patterns to the behavioral or symptom variable. This idea can be easily extended to the hybrid NeuroMark fusion framework by considering the covariations across fMRI, sMRI, and dMRI.
Different from the post-analysis on the NeuroMark features that are extracted independently from each modality, the benefit of the hybrid NeuroMark fusion framework in this context is the ability to decompose components considering the intermodal relationship and the hypothesis-driven reference, in which the covarying networks are comparable and replicable while still benefiting from data-driven methods. Therefore, constructing priori fusion-based multimodal templates to guide the individual joint sources computation will be an ongoing effort.
5. Conclusion
The mixture of big data, algorithmic advances, and neuroinformatics solutions has propelled the neuroimaging field forward rapidly. Undoubtedly, employment of neuroimaging biomarkers will move from single data types to incorporating all relevant available data, in essence developing multimodal fingerprints. While we have not advanced as quickly as we might have hoped in explicating the mysteries of neurological and mental disorders, there is considerable reason to be optimistic about the not-too-distant future. The brain is extremely complex, which makes focusing on brain disorders even more challenging. NeuroMark, combining a priori neuroimaging and data-driven methods, is a promising tool that provides a way forward here, continuously advancing the field. The NeuroMark 3.0 templates, which include spatiotemporal functional templates and templates from other modalities, will have plenty of applications in future neuroimaging studies. For example, the 4D NeuroMark_fMRI_3.0 template will be very important for investigating lifespan patterns of the brain that might advance our understanding of brain aging in healthy and disease cohorts. Secondly, the NeuroMark_sMRI_3.0 and NeuroMark_dMRI_3.0 templates can be applied to MRI data from different modalities to generalize and standardize the calculation of network features that leverage the benefits of a data-driven approach and provide comparability across multiple analyses. Taken together, we believe that NeuroMark 3.0 templates can complement and exploit the richness of neuroimaging data, which will shed light on the biological mechanisms of the linkages between brain image and human behavior.
Supplementary Material
Funding and Disclosure
This work was supported by National Institutes of Health (No. R01MH118695, R01EB020407, R01MH117107 and R01MH119251), the National Science Foundation (2112455), the National Institute of Mental Health Intramural Research Program, and the China Natural Science Foundation (No. 82022035).
Footnotes
Ethics Statement
The infant datasets used in this work in under the study protocol approved by the Emory University Institutional Review Board (IRB). Legal guardians provided written consent for their infant’s participation at time of study enrollment. The UKB and HCP datasets are open-source de-identified human data. Therefore, no further ethical review was required.
CRediT authorship contribution statement
Zening Fu: Writing – review & editing, Writing – original draft, Visualization, Validation, Software, Resources, Methodology, Investigation, Formal analysis, Conceptualization. IshaanBatta: Visualization, Methodology, Investigation, Formal analysis. Lei Wu: Writing – review & editing, Investigation, Formal analysis. Anees Abrol: Writing – original draft, Validation, Software. Oktay Agcaoglu: Writing – review & editing, Validation. Mustafa S Salman:. Yuhui Du: Writing – review & editing, Supervision. Armin Iraji: Writing – original draft, Software, Methodology, Conceptualization. Sarah Shultz: Writing – review & editing, Resources, Funding acquisition. Jing Sui: Validation, Methodology, Funding acquisition. Vince D. Calhoun: Writing – review & editing, Validation, Supervision, Software, Resources, Investigation, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Supplementary materials
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.neuroimage.2024.120617.
Data and Code Availability Statement
The HCP developmental and aging data used in the present study can be accessed upon application from NDA (https://nda.nih.gov/) with the approval of the ABCD consortium. The HCP 1200 release data can be accessed upon application from Connectome Coordination Facility (https://www.humanconnectome.org/). The UK biobank data can be accessed upon application from UK biobank biomedical database (https://www.ukbiobank.ac.uk/).
NeuroMark 3.0 templates are accessible on our official website (https://trendscenter.org/data/) and GitHub (https://github.com/trendscenter/gift/tree/master/GroupICAT/icatb/icatb_templates). The codes of the NeuroMark framework have been integrated into the group ICA Toolbox (GIFT 4.0c, https://trendscenter.org/software/gift/). Other MATLAB codes of this study can be obtained from the corresponding author.
Data availability
Data will be made available on request.
References
- Aarts AA, Anderson JE, Anderson CJ, Attridge PR, Attwood A, Axt J, Babel M, Bahník Š,Baranski E, Barnett-Cowan M, Bartmess E, Beer J, Bell R, Bentley H, Beyan L, Binion G, Borsboom D, Bosch A, Bosco FA, Bowman SD, Brandt MJ, Braswell E, Brohmer H, Brown BT, Brown K, Brüning J, Calhoun-Sauls A, Callahan SP, Chagnon E, Chandler J, Chartier CR, Cheung F, Christopherson CD, Cillessen L, Clay R, Cleary H, Cloud MD, Conn M, Cohoon J, Columbus S, Cordes A, Costantini G, Alvarez LDC, Cremata E, Crusius J, DeCoster J, DeGaetano MA, Penna ND, Den Bezemer B, Deserno MK, Devitt O, Dewitte L, Dobolyi DG, Dodson GT, Donnellan MB, Donohue R, Dore RA, Dorrough A, Dreber A, Dugas M, Dunn EW, Easey K, Eboigbe S, Eggleston C, Embley J, Epskamp S, Errington TM, Estel V, Farach FJ, Feather J, Fedor A, Fernández-Castilla B, Fiedler S, Field JG, Fitneva SA, Flagan T, Forest AL, Forsell E, Foster JD, Frank MC, Frazier RS, Fuchs H, Gable P, Galak J, Galliani EM, Gampa A, Garcia S, Gazarian D, Gilbert E, Giner-Sorolla R, Glöckner A, Goellner L, Goh JX, Goldberg R, Goodbourn PT, Gordon-McKeon S, Gorges B, Gorges J, Goss J, Graham J, Grange JA, Gray J, Hartgerink C, Hartshorne J, Hasselman F, Hayes T, Heikensten E, Henninger F, Hodsoll J, Holubar T, Hoogendoorn G, Humphries DJ, Hung COY, Immelman N, Irsik VC, Jahn G, Jäkel F, Jekel M, Johannesson M, Johnson LG, Johnson DJ, Johnson KM, Johnston WJ, Jonas K, Joy-Gaba JA, Kappes HB, Kelso K, Kidwell MC, Kim SK, Kirkhart M, Kleinberg B, Knežević G, Kolorz FM, Kossakowski JJ, Krause RW, Krijnen J, Kuhlmann T, Kunkels YK, Kyc MM, Lai CK, Laique A, Lakens D, Lane KA, Lassetter B, Lazarević LB, Le Bel EP, Lee KJ, Lee M, Lemm K, Levitan CA, Lewis M, Lin L, Lin S, Lippold M, Loureiro D, Luteijn I, MacKinnon S, Mainard HN, Marigold DC, Martin DP, Martinez T, Masicampo EJ, Matacotta J, Mathur M, May M, Mechin N, Mehta P, Meixner J, Melinger A, Miller JK, Miller M, Moore K, Möschl M, Motyl M, Müller SM, Munafo M, Neijenhuijs KI, Nervi T, Nicolas G, Nilsonne G, Nosek BA, Nuijten MB, Olsson C, Osborne C, Ostkamp L, Pavel M, Penton-Voak IS, Perna O, Pernet C, Perugini M, Pipitone RN, Pitts M, Plessow F, Prenoveau JM, Rahal RM, Ratliff KA, Reinhard D, Renkewitz F, Ricker AA, Rigney A, Rivers AM, Roebke M, Rutchick AM, Ryan RS, Sahin O, Saide A, Sandstrom GM, Santos D, Saxe R, Schlegelmilch R, Schmidt K, Scholz S, Seibel L, Selterman DF, Shaki S, Simpson WB, Sinclair HC, Skorinko JLM, Slowik A, Snyder JS, Soderberg C, Sonnleitner C, Spencer N, Spies JR, Steegen S, Stieger S, Strohminger N, Sullivan GB, Talhelm T, Tapia M, Te Dorsthorst A, Thomae M, Thomas SL, Tio P, Traets F, Tsang S, Tuerlinckx F, Turchan P, Valášek M, Van’t Veer AE, Van Aert R, Van Assen M, Van Bork R, Van De Ven M, Van Den Bergh D, Van Der Hulst M, Van Dooren R, Van Doorn J, Van Renswoude DR, Van Rijn H, Vanpaemel W, Echeverría AV, Vazquez M, Velez N, Vermue M, Verschoor M, Vianello M, Voracek M, Vuu G, Wagenmakers EJ, Weerdmeester J, Welsh A, Westgate EC, Wissink J, Wood M, Woods A, Wright E, Wu S, Zeelenberg M, Zuni K, 2015. Estimating the reproducibility of psychological science. Science (80-.) 349. 10.1126/science.aac4716. [DOI] [PubMed] [Google Scholar]
- Abrol A, Fu Z, Du Y, Wilson TW, Wang YP, Stephen JM, Calhoun VD, 2023. Developmental and aging resting functional magnetic resonance imaging brain state adaptations in adolescents and adults: A large N (>47K) study. Hum. Brain Mapp 44, 2158–2175. 10.1002/hbm.26200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen EA, Damaraju E, Plis SM, Erhardt EB, Eichele T, Calhoun VD, 2014. Tracking whole-brain connectivity dynamics in the resting state. Cereb. Cortex 24, 663–676. 10.1093/cercor/bhs352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner J, Barnes G, Chen C-C, Daunizeau J, Flandin G, Friston K, Gitelman D, Glauche V, Henson R, Hutton C, Jafarian A, Kiebel S, Kilner J, Litvak V, Mattout J, Moran R, Penny W, Phillips C, Razi A, Stephan K, Tak S, Tyrer A, Zeidman P, 2021. SPM12 Manual. Funct. Imaging Lab. Wellcome Cent. Hum. Neuroimaging [Google Scholar]
- Ashburner J, Friston KJ, 2000. Voxel-based morphometry - The methods. Neuroimage 11, 805–821. 10.1006/nimg.2000.0582. [DOI] [PubMed] [Google Scholar]
- Avants B, Tustison NJ, Song G, 2022. Advanced Normalization Tools: V1.0. Insight J. 10.54294/uvnhin. [DOI] [Google Scholar]
- Barch DM, Albaugh MD, Avenevoli S, Chang L, Clark DB, Glantz MD, Hudziak JJ, Jernigan TL, Tapert SF, Yurgelun-Todd D, Alia-Klein N, Potter AS, Paulus MP, Prouty D, Zucker RA, Sher KJ, 2018. Demographic, physical and mental health assessments in the adolescent brain and cognitive development study: Rationale and description. Dev. Cogn. Neurosci 32, 55–66. 10.1016/j.dcn.2017.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann C, Mackay C, Filippini N, Smith S, 2009. Group comparison of resting-state FMRI data using multi-subject ICA and dual regression. Neuroimage 47, S148. 10.1016/s1053-8119(09)71511-3. [DOI] [Google Scholar]
- Bell AJ, Sejnowski TJ, 1995. An information-maximization approach to blind separation and blind deconvolution. Neural Comput 7, 1129–1159. 10.1162/neco.1995.7.6.1129. [DOI] [PubMed] [Google Scholar]
- Bhinge S, Mowakeaa R, Calhoun VD, Adali T, 2019. Extraction of Time-Varying Spatiotemporal Networks Using Parameter-Tuned Constrained IVA. IEEE Trans. Med. Imaging 38, 1715–1725. 10.1109/TMI.2019.2893651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal B, Zerrin Yetkin F, Haughton VM, Hyde JS, 1995. Functional connectivity in the motor cortex of resting human brain using echo-planar mri. Magn. Reson. Med 34, 537–541. 10.1002/mrm.1910340409. [DOI] [PubMed] [Google Scholar]
- Bluhm RL, Miller J, Lanius RA, Osuch EA, Boksman K, Neufeld RWJ, Thebérge J, Schaefer B, Williamson P, 2007. Spontaneous low-frequency fluctuations in the BOLD signal in schizophrenic patients: Anomalies in the default network. Schizophr. Bull 33, 1004–1012. 10.1093/schbul/sbm052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner RL, Krienen FM, Yeo BTT, 2013. Opportunities and limitations of intrinsic functional connectivity MRI. Nat. Neurosci 16, 832–837. 10.1038/nn.3423. [DOI] [PubMed] [Google Scholar]
- Calhoun V, 2018. Data-driven approaches for identifying links between brain structure and function in health and disease. Dialogues Clin. Neurosci 20, 87–100. 10.31887/dcns.2018.20.2/vcalhoun. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VD, Adali T, Giuliani NR, Pekar JJ, Kiehl KA, Pearlson GD, 2006. Method for multimodal analysis of independent source differences in schizophrenia: Combining gray matter structural and auditory oddball functional data. Hum. Brain Mapp 27, 47–62. 10.1002/hbm.20166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VD, Adali T, Pearlson GD, Pekar JJ, 2001. A method for making group inferences from functional MRI data using independent component analysis. Hum. Brain Mapp 14, 140–151. 10.1002/hbm.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VD, Liu J, Adali T, 2009. A review of group ICA for fMRI data and ICA for joint inference of imaging, genetic, and ERP data. Neuroimage 45. 10.1016/j.neuroimage.2008.10.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calhoun VD, Sui J, 2016. Multimodal Fusion of Brain Imaging Data: A Key to Finding the Missing Link(s) in Complex Mental Illness. Biol. Psychiatry Cogn. Neurosci. Neuroimaging 1, 230–244. 10.1016/j.bpsc.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao M, Wang JH, Dai ZJ, Cao XY, Jiang LL, Fan FM, Song XW, Xia MR, Shu N, Dong Q, Milham MP, Castellanos FX, Zuo XN, He Y, 2014. Topological organization of the human brain functional connectome across the lifespan. Dev. Cogn. Neurosci 7, 76–93. 10.1016/j.dcn.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casey BJ, Cannonier T, Conley MI, Cohen AO, Barch DM, Heitzeg MM, Soules ME, Teslovich T, Dellarco DV, Garavan H, Orr CA, Wager TD, Banich MT, Speer NK, Sutherland MT, Riedel MC, Dick AS, Bjork JM, Thomas KM, Chaarani B, Mejia MH, Hagler DJ, Daniela Cornejo M, Sicat CS, Harms MP, Dosenbach NUF, Rosenberg M, Earl E, Bartsch H, Watts R, Polimeni JR, Kuperman JM, Fair DA, Dale AM, 2018. The Adolescent Brain Cognitive Development (ABCD) study: Imaging acquisition across 21 sites. Dev. Cogn. Neurosci 32, 43–54. 10.1016/j.dcn.2018.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan MY, Na J, Agres PF, Savalia NK, Park DC, Wig GS, 2018. Socioeconomic status moderates age-related differences in the brain’s functional network organization and anatomy across the adult lifespan. Proc. Natl. Acad. Sci. U. S. A 115, E5144–E5153. 10.1073/pnas.1714021115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan MY, Park DC, Savalia NK, Petersen SE, Wig GS, 2014. Decreased segregation of brain systems across the healthy adult lifespan. Proc. Natl. Acad. Sci. U. S. A 111, E4997–E5006. 10.1073/pnas.1415122111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Tam A, Kebets V, Orban C, Ooi LQR, Asplund CL, Marek S, Dosenbach NUF, Eickhoff SB, Bzdok D, Holmes AJ, Yeo BTT, 2022. Shared and unique brain network features predict cognitive, personality, and mental health scores in the ABCD study. Nat. Commun 13 10.1038/s41467-022-29766-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra S, Dhamala E, Lawhead C, Ricard JA, Orchard ER, An L, Chen P, Wulan N, Kumar P, Rubenstein A, Moses J, Chen L, Levi P, Holmes A, Aquino K, Fornito A, Harpaz-Rotem I, Germine LT, Baker JT, Yeo BT, Holmes Avram J., 2022. Reliable and generalizable brain-based predictions of cognitive functioning across common psychiatric illness. bioRxiv. 10.1101/2022.12.08.22283232, 2022.12.08.22283232. [DOI] [Google Scholar]
- Correa NM, Eichele T, Adali T, Li YO, Calhoun VD, 2010. Multi-set canonical correlation analysis for the fusion of concurrent single trial ERP and functional MRI. Neuroimage 50, 1438–1445. 10.1016/j.neuroimage.2010.01.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhamala E, Yeo BTT, Holmes AJ, 2023. One Size Does Not Fit All: Methodological Considerations for Brain-Based Predictive Modeling in Psychiatry. Biol. Psychiatry 93, 717–728. 10.1016/j.biopsych.2022.09.024. [DOI] [PubMed] [Google Scholar]
- Di Martino A, 2012. The autism brain imaging data exchange: Towards largescale evaluation of the intrinsic brain in autism. Neuropsychopharmacology 38, S303–S304. [Google Scholar]
- Dini H, Sendi MSE, Sui J, Fu Z, Espinoza R, Narr KL, Qi S, Abbott CC, van Rooij SJH, Riva-Posse P, Bruni LE, Mayberg HS, Calhoun VD, 2021. Dynamic Functional Connectivity Predicts Treatment Response to Electroconvulsive Therapy in Major Depressive Disorder. Front. Hum. Neurosci 15 10.3389/fnhum.2021.689488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douaud G, Lee S, Alfaro-Almagro F, Arthofer C, Wang C, Lange F, Andersson JLR, Griffanti L, Duff E, Jbabdi S, Taschler B, Winkler A, Nichols TE, Collins R, Matthews PM, Allen N, Miller KL, Smith SM, 2021. Brain imaging before and after COVID-19 in UK Biobank. medRxiv. 10.1101/2021.06.11.21258690, 2021.06.11.21258690. [DOI] [Google Scholar]
- Du Y, Allen EA, He H, Sui J, Wu L, Calhoun VD, 2016. Artifact removal in the context of group ICA: A comparison of single-subject and group approaches. Hum. Brain Mapp 37, 1005–1025. 10.1002/hbm.23086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Fan Y, 2013. Group information guided ICA for fMRI data analysis. Neuroimage 69, 157–197. 10.1016/j.neuroimage.2012.11.008. [DOI] [PubMed] [Google Scholar]
- Du Y, Fu Z, Sui J, Gao S, Xing Y, Lin D, Salman M, Abrol A, Rahaman MA, Chen J, Hong LE, Kochunov P, Osuch EA, Calhoun VD, 2020. NeuroMark: An automated and adaptive ICA based pipeline to identify reproducible fMRI markers of brain disorders. NeuroImage Clin 28. 10.1016/j.nicl.2020.102375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Fu Z, Xing Y, Lin D, Pearlson G, Kochunov P, Hong LE, Qi S, Salman M, Abrol A, Calhoun VD, 2021. Evidence of shared and distinct functional and structural brain signatures in schizophrenia and autism spectrum disorder. Commun. Biol 4, 1–16. 10.1038/s42003-021-02592-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, Lin D, Yu Q, Sui J, Chen J, Rachakonda S, Adali T, Calhoun VD, 2017. Comparison of IVA and GIG-ICA in brain functional network estimation using fMRI data. Front. Neurosci 11 10.3389/fnins.2017.00267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan K, Premi E, Pilotto A, Cristillo V, Benussi A, Libri I, Giunta M, Bockholt HJ, Liu J, Campora R, Pezzini A, Gasparotti R, Magoni M, Padovani A, Calhoun V, 2021. Alterations of frontal-temporal gray matter volume associate with clinical measures of older adults with COVID-19. Neurobiol. Stress 100326. 10.1016/j.ynstr.2021.100326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durston S, Casey BJ, 2006. What have we learned about cognitive development from neuroimaging? Neuropsychologia 44, 2149–2157. 10.1016/j.neuropsychologia.2005.10.010. [DOI] [PubMed] [Google Scholar]
- Eisenberg L, Kanner L, 1956. Childhood Schizophrenia Symposium, 1955. 6. Early Infantile Autism, 1943–55. Am. J. Orthopsychiatry 26, 556–566. 10.1111/j.1939-0025.1956.tb06202.x. [DOI] [PubMed] [Google Scholar]
- Faria AV, Joel SE, Zhang Y, Oishi K, van Zjil PCM, Miller MI, Pekar JJ, Mori S, 2012. Atlas-based analysis of resting-state functional connectivity: Evaluation for reproducibility and multi-modal anatomy-function correlation studies. Neuroimage 61, 613–621. 10.1016/j.neuroimage.2012.03.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg DA, Moeller S, Smith SM, Auerbach E, Ramanna S, Glasser MF, Miller KL, Ugurbil K, Yacoub E, 2010. Multiplexed echo planar imaging for sub-second whole brain fmri and fast diffusion imaging. PLoS One 5, e15710. 10.1371/journal.pone.0015710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornito A, Yücel M, Patti J, Wood SJ, Pantelis C, 2009. Mapping grey matter reductions in schizophrenia: An anatomical likelihood estimation analysis of voxel-based morphometry studies. Schizophr. Res 108, 104–113. 10.1016/j.schres.2008.12.011. [DOI] [PubMed] [Google Scholar]
- Fu Z, Abbott CC, Miller J, Deng Z.De, McClintock SM, Sendi MSE, Sui J, Calhoun VD, 2023a. Cerebro-cerebellar functional neuroplasticity mediates the effect of electric field on electroconvulsive therapy outcomes. Transl. Psychiatry 13. 10.1038/s41398-023-02312-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z, Abbott CC, Sui J, Calhoun VD, 2023b. Predictive signature of static and dynamic functional connectivity for ECT clinical outcomes. Front. Pharmacol 14 10.3389/fphar.2023.1102413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z, Iraji A, Caprihan A, Adair JC, Sui J, Rosenberg GA, Calhoun VD, 2020a. In search of multimodal brain alterations in Alzheimer’s and Binswanger’s disease. NeuroImage Clin 26. 10.1016/j.nicl.2019.101937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z, Iraji A, Sui J, Calhoun VD, 2021a. Whole-Brain Functional Network Connectivity Abnormalities in Affective and Non-Affective Early Phase Psychosis. Front. Neurosci 15 10.3389/fnins.2021.682110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z, Iraji A, Turner JA, Sui J, Miller R, Pearlson GD, Calhoun VD, 2021b. Dynamic state with covarying brain activity-connectivity: On the pathophysiology of schizophrenia. Neuroimage 224. 10.1016/j.neuroimage.2020.117385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z, Liu J, Salman MS, Sui J, Calhoun VD, 2023c. Functional connectivity uniqueness and variability? Linkages with cognitive and psychiatric problems in children. Nat. Ment. Heal 10.1038/s44220-023-00151-8. [DOI] [Google Scholar]
- Fu Z, Sui J, Espinoza R, Narr K, Qi S, Sendi MSE, Abbot CC, Calhoun VD, 2021c. Whole-brain Functional Connectivity Dynamics associated with Electroconvulsive Therapy Treatment Response. Biol. Psychiatry Cogn. Neurosci. Neuroimaging 10.1016/j.bpsc.2021.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Zening, Sui J, Turner JA, Du Y, Assaf M, Pearlson GD, Calhoun VD, 2020. Dynamic functional network reconfiguration underlying the pathophysiology of schizophrenia and autism spectrum disorder. Hum. Brain Mapp 10.1002/hbm.25205 hbm.25205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z, Tu Y, Calhoun VD, Zhang Y, Zhao Q, Chen J, Meng Q, Lu Z, Hu L, 2021d. Dynamic functional network connectivity associated with post-traumatic stress symptoms in COVID-19 survivors. Neurobiol. Stress 15. 10.1016/j.ynstr.2021.100377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Z, Tu Y, Di X, Du Y, Sui J, Biswal BB, Zhang Z, de Lacy N, Calhoun VD, 2019. Transient increased thalamic-sensory connectivity and decreased whole-brain dynamism in autism. Neuroimage. 10.1016/j.neuroimage.2018.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao ZB, Wang W, Zhao XL, Chen T, Fu LP, Xu BX, Wang ZF, 2018. Multi-modality molecular imaging characteristics of dementia with Lewy bodies. J. Int. Med. Res 46, 2317–2326. 10.1177/0300060518764749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geerligs L, Renken RJ, Saliasi E, Maurits NM, Lorist MM, 2015. A Brain-Wide Study of Age-Related Changes in Functional Connectivity. Cereb. Cortex 25, 1987–1999. 10.1093/cercor/bhu012. [DOI] [PubMed] [Google Scholar]
- Glahn DC, Bearden CE, Niendam TA, Escamilla MA, 2004. The feasibility of neuropsychological endophenotypes in the search for genes associated with bipolar affective disorder. Bipolar Disord 6, 171–182. 10.1111/j.13995618.2004.00113.x. [DOI] [PubMed] [Google Scholar]
- Goes FS, Zandi PP, Miao K, McMahon FJ, Steele J, Willour VL, MacKinnon DF, Mondimore FM, Schweizer B, Nurnberger JI, Rice JP, Scheftner W, Coryell W, Berrettini WH, Kelsoe JR, Byerley W, Murphy DL, Gershon ES, DePaulo JR, McInnis MG, Potash JB, 2007. Mood-incongruent psychotic features in bipolar disorder: Familial aggregation and suggestive linkage to 2p11-q14 and 13q21–33. Am. J. Psychiatry 164, 236–247. 10.1176/ajp.2007.164.2.236. [DOI] [PubMed] [Google Scholar]
- Goh JOS, 2011. Functional dedifferentiation and altered connectivity in older adults: Neural accounts of cognitive aging. Aging Dis 2, 30–48. [PMC free article] [PubMed] [Google Scholar]
- Grady C, Sarraf S, Saverino C, Campbell K, 2016. Age differences in the functional interactions among the default, frontoparietal control, and dorsal attention networks. Neurobiol. Aging 41, 159–172. 10.1016/j.neurobiolaging.2016.02.020. [DOI] [PubMed] [Google Scholar]
- Gupta CN, Calhoun VD, Rachakonda S, Chen J, Patel V, Liu J, Segall J, Franke B, Zwiers MP, Arias-Vasquez A, Buitelaar J, Fisher SE, Fernandez G, Van Erp TGM, Potkin S, Ford J, Mathalon D, McEwen S, Lee HJ, Mueller BA, Greve DN, Andreassen O, Agartz I, Gollub RL, Sponheim SR, Ehrlich S, Wang L, Pearlson G, Glahn DC, Sprooten E, Mayer AR, Stephen J, Jung RE, Canive J, Bustillo J, Turner JA, 2015. Patterns of gray matter abnormalities in schizophrenia based on an international mega-analysis. Schizophr. Bull 41, 1133–1142. 10.1093/schbul/sbu177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajjar I, Okafor M, Wan L, Yang Z, Nye JA, Bohsali A, Shaw LM, Levey AI, Lah JJ, Calhoun VD, Moore RH, Goldstein FC, 2022. Safety and biomarker effects of candesartan in non-hypertensive adults with prodromal Alzheimer’s disease. Brain Commun 4. 10.1093/braincomms/fcac270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himberg J, Hyvärinen A, 2003. ICASSO: Software for investigating the reliability of ICA estimates by clustering and visualization. Neural Networks Signal Process. In: - Proc. IEEE Work. 2003-Janua, pp. 259–268. 10.1109/NNSP.2003.1318025. [DOI] [Google Scholar]
- Honey CJ, Sporns O, Cammoun L, Gigandet X, Thiran JP, Meuli R, Hagmann P, 2009. Predicting human resting-state functional connectivity from structural connectivity. Proc. Natl. Acad. Sci. U. S. A 106, 2035–2040. 10.1073/pnas.0811168106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iraji A, Fu Z, Faghiri A, Duda M, Chen J, Rachakonda S, DeRamus T, Kochunov P, Adhikari BM, Belger A, Ford JM, Mathalon DH, Pearlson GD, Potkin SG, Preda A, Turner JA, van Erp TGM, Bustillo JR, Yang K, Ishizuka K, Faria A, Sawa A, Hutchison K, Osuch EA, Theberge J, Abbott C, Mueller BA, Zhi D, Zhuo C, Liu S, Xu Y, Salman M, Liu J, Du Y, Sui J, Adali T, Calhoun VD, 2023. Identifying canonical and replicable multi-scale intrinsic connectivity networks in 100k+ resting-state fMRI datasets. Hum. Brain Mapp 44, 5729–5748. 10.1002/hbm.26472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack CR, Bernstein MA, Fox NC, Thompson P, Alexander G, Harvey D, Borowski B, Britson PJ, Whitwell JL, Ward C, Dale AM, Felmlee JP, Gunter JL, Hill DLG, Killiany R, Schuff N, Fox-Bosetti S, Lin C, Studholme C, DeCarli CS, Krueger G, Ward HA, Metzger GJ, Scott KT, Mallozzi R, Blezek D, Levy J, Debbins JP, Fleisher AS, Albert M, Green R, Bartzokis G, Glover G, Mugler J, Weiner MW, 2008. The Alzheimer’s Disease Neuroimaging Initiative (ADNI): MRI methods. J. Magn. Reson. Imaging 27, 685–691. 10.1002/jmri.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jafri MJ, Pearlson GD, Stevens M, Calhoun VD, 2008. A method for functional network connectivity among spatially independent resting-state components in schizophrenia. Neuroimage 39, 1666–1681. 10.1016/j.neuroimage.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeong JW, Asano E, Yeh FC, Chugani DC, Chugani HT, 2013. Independent component analysis tractography combined with a ball-stick model to isolate intravoxel crossing fibers of the corticospinal tracts in clinical diffusion MRI. Magn. Reson. Med 70, 441–453. 10.1002/mrm.24487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R, Calhoun VD, Noble S, Sui J, Liang Q, Qi S, Scheinost D, 2022a. A functional connectome signature of blood pressure in >30 000 participants from the UK biobank. Cardiovasc. Res 10.1093/cvr/cvac116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang R, Scheinost D, Zuo N, Wu J, Qi S, Liang Q, Zhi D, Luo N, Chung YC, Liu S, Xu Y, Sui J, Calhoun V, 2022b. A Neuroimaging Signature of Cognitive Aging from Whole-Brain Functional Connectivity. Adv. Sci 9 10.1002/advs.202201621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolles DD, Van Buchem MA, Crone EA, Rombouts SARB, 2011. A comprehensive study of whole-brain functional connectivity in children and young adults. Cereb. Cortex 21, 385–391. 10.1093/cercor/bhq104. [DOI] [PubMed] [Google Scholar]
- Kitani-Morii F, Kasai T, Horiguchi G, Teramukai S, Ohmichi T, Shinomoto M, Fujino Y, Mizuno T, 2021. Risk factors for neuropsychiatric symptoms in patients with Parkinson’s disease during COVID-19 pandemic in Japan. PLoS One 16, e0245864. 10.1371/journal.pone.0245864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koen JD, Rugg MD, 2019. Neural Dedifferentiation in the Aging Brain. Trends Cogn. Sci 23, 547–559. 10.1016/j.tics.2019.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konrad K, Eickhoff SB, 2010. Is the ADHD brain wired differently? A review on structural and functional connectivity in attention deficit hyperactivity disorder. Hum. Brain Mapp 31, 904–916. 10.1002/hbm.21058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konstantareas MM, Hewitt T, 2001. Autistic Disorder and Schizophrenia: Diagnostic Overlaps. J. Autism Dev. Disord 31, 19–28. 10.1023/A:1005605528309. [DOI] [PubMed] [Google Scholar]
- Kucyi A, Esterman M, Capella J, Green A, Uchida M, Biederman J, Gabrieli JDE, Valera EM, Whitfield-Gabrieli S, 2021. Prediction of stimulus-independent and task-unrelated thought from functional brain networks. Nat. Commun 12 10.1038/s41467-021-22027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JJ, Kim HJ, Čeko M, Park B.yong, Lee SA, Park H, Roy M, Kim SG, Wager TD, Woo CW, 2021. A neuroimaging biomarker for sustained experimental and clinical pain. Nat. Med 27, 174–182. 10.1038/s41591-020-1142-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levey AI, Qiu D, Zhao L, Hu WT, Duong DM, Higginbotham L, Dammer EB, Seyfried NT, Wingo TS, Hales CM, Tansey MG, Goldstein DS, Abrol A, Calhoun VD, Goldstein FC, Hajjar I, Fagan AM, Galasko D, Edland SD, Hanfelt J, Lah JJ, Weinshenker D, 2022. A phase II study repurposing atomoxetine for neuroprotection in mild cognitive impairment. Brain 145, 1924–1938. 10.1093/brain/awab452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis N, Miller R, Gazula H, Calhoun V, 2023. Fine temporal brain network structure modularizes and localizes differently in men and women: insights from a novel explainability framework. Cereb. Cortex 10.1093/cercor/bhac462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li K, Fu Z, Qi S, Luo X, Zeng Q, Xu X, Huang P, Zhang M, Calhoun VD, 2021. Polygenic Hazard Score Associated Multimodal Brain Networks Along the Alzheimer’s Disease Continuum. Front. Aging Neurosci 13 10.3389/fnagi.2021.725246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin QH, Liu J, Zheng YR, Liang H, Calhoun VD, 2010. Semiblind spatial ICA of fMRI using spatial constraints. Hum. Brain Mapp 31, 1076–1088. 10.1002/hbm.20919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Pearlson G, Windemuth A, Ruano G, Perrone-Bizzozero NI, Calhoun V, 2009. Combining fMRI and SNP data to investigate connections between brain function and genetics using parallel ICA. Hum. Brain Mapp 30, 241–255. 10.1002/hbm.20508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Gao Y, Di Q, Hu J, Lu C, Nan Y, Booth JR, Liu L, 2018. Differences between child and adult large-scale functional brain networks for reading tasks. Hum. Brain Mapp 39, 662–679. 10.1002/hbm.23871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Vicente M, Agcaoglu O, Pérez-Crespo L, Estévez-López F, Heredia-Genestar JM, Mulder RH, Flournoy JC, van Duijvenvoorde ACK, Güroğlu B, White T, Calhoun V, Tiemeier H, Muetzel RL, 2021. Developmental Changes in Dynamic Functional Connectivity From Childhood Into Adolescence. Front. Syst. Neurosci 15 10.3389/fnsys.2021.724805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lord A, Ehrlich S, Borchardt V, Geisler D, Seidel M, Huber S, Murr J, Walter M, 2016. Brain parcellation choice affects disease-related topology differences increasingly from global to local network levels. Psychiatry Res. - Neuroimaging 249, 12–19. 10.1016/j.pscychresns.2016.02.001. [DOI] [PubMed] [Google Scholar]
- Luo N, Sui J, Abrol A, Chen J, Turner JA, Damaraju E, Fu Z, Fan L, Lin D, Zhuo C, Xu Y, Glahn DC, Rodrigue AL, Banich MT, Pearlson GD, Calhoun VD, 2020. Structural Brain Architectures Match Intrinsic Functional Networks and Vary across Domains: A Study from 15 000+ Individuals. Cereb. Cortex 10.1093/cercor/bhaa127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marek S, Tervo-Clemmens B, Calabro F, Nature DM-U, 2022, 2022. Reproducible brain-wide association studies require thousands of individuals _ Enhanced Reader.pdf. nature.com. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masouleh SK, Eickhoff SB, Hoffstaedter F, Genon S, 2019. Empirical examination of the replicability of associations between brain structure and psychological variables. Elife 8. 10.7554/eLife.43464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastrovito D, Hanson C, Hanson SJ, 2018. Differences in atypical resting-state effective connectivity distinguish autism from schizophrenia. NeuroImage Clin 18, 367–376. 10.1016/j.nicl.2018.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milham PM, Damien F, Maarten M, Stewart HM, 2012. The ADHD-200 Consortium: A model to advance the translational potential of neuroimaging in clinical neuroscience. Front. Syst. Neurosci 1–5. 10.3389/fnsys.2012.00062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moeller S, Yacoub E, Olman CA, Auerbach E, Strupp J, Harel N, Uğurbil K, 2010. Multiband multislice GE-EPI at 7 tesla, with 16-fold acceleration using partial parallel imaging with application to high spatial and temporal whole-brain fMRI. Magn. Reson. Med 63, 1144–1153. 10.1002/mrm.22361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mokhtari M, Narayanan B, Hamm JP, Soh P, Calhoun VD, Ruaño G, Kocherla M, Windemuth A, Clementz BA, Tamminga CA, Sweeney JA, Keshavan MS, Pearlson GD, 2016. Multivariate genetic correlates of the auditory paired stimuli-based p2 event-related potential in the psychosis dimension from the BSNIP study. Schizophr. Bull 42, 851–862. 10.1093/schbul/sbv147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murley AG, Coyle-Gilchrist I, Rouse MA, Simon Jones P, Li W, Wiggins J, Lansdall C, Rodríguez PV, Wilcox A, Tsvetanov KA, Patterson K, Lambon Ralph MA, Rowe JB, 2020. Redefining the multidimensional clinical phenotypes of frontotemporal lobar degeneration syndromes. Brain 143, 1555–1571. 10.1093/brain/awaa097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nomi JS, Uddin LQ, 2015. Face processing in autism spectrum disorders: From brain regions to brain networks. Neuropsychologia 71, 201–216. 10.1016/j.neuropsychologia.2015.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connor M, Davitt JK, 2012. The Outcome and Assessment Information Set (OASIS): A Review of Validity and Reliability. Home Health Care Serv. Q 31, 267–301. 10.1080/01621424.2012.703908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandolfi V, Magyar CI, Norris M, 2014. Validity Study of the CBCL 6–18 for the Assessment of Emotional Problems in Youth With ASD. J. Ment. Health Res. Intellect. Disabil 7, 306–322. 10.1080/19315864.2014.930547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HRP, Quidé Y, Schofield PR, Williams LM, Gatt JM, 2022. Grey matter covariation and the role of emotion reappraisal in mental wellbeing and resilience after early life stress exposure. Transl. Psychiatry 12, 1–10. 10.1038/s41398-022-01849-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poldrack RA, Baker CI, Durnez J, Gorgolewski KJ, Matthews PM, Munafò MR, Nichols TE, Poline JB, Vul E, Yarkoni T, 2017. Scanning the horizon: Towards transparent and reproducible neuroimaging research. Nat. Rev. Neurosci 18, 115–126. 10.1038/nrn.2016.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahaman MA, Chen J, Fu Z, Lewis N, Iraji A, van Erp TGM, Calhoun VD, 2023. Deep multimodal predictome for studying mental disorders. Hum. Brain Mapp 44, 509–522. 10.1002/hbm.26077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raichle ME, Snyder AZ, 2007. A default mode of brain function: A brief history of an evolving idea. 10.1016/j.neuroimage.2007.02.041. [DOI] [PubMed] [Google Scholar]
- Rieck JR, Baracchini G, Nichol D, Abdi H, Grady CL, 2021. Reconfiguration and dedifferentiation of functional networks during cognitive control across the adult lifespan. Neurobiol. Aging 106, 80–94. 10.1016/j.neurobiolaging.2021.03.019. [DOI] [PubMed] [Google Scholar]
- Rolls ET, Huang CC, Lin CP, Feng J, Joliot M, 2020. Automated anatomical labelling atlas 3. Neuroimage 206. 10.1016/j.neuroimage.2019.116189. [DOI] [PubMed] [Google Scholar]
- Rosenberg GA, Wallin A, Wardlaw JM, Markus HS, Montaner J, Wolfson L, Iadecola C, Zlokovic BV, Joutel A, Dichgans M, Duering M, Schmidt R, Korczyn AD, Grinberg LT, Chui HC, Hachinski V, 2016. Consensus statement for diagnosis of subcortical small vessel disease. J. Cereb. Blood Flow Metab 36, 6–25. 10.1038/jcbfm.2015.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubinov M, Bullmore E, 2013. Schizophrenia and abnormal brain network hubs. Dialogues Clin. Neurosci 15, 339–349. 10.31887/dcns.2013.15.3/mrubinov. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salman MS, Du Y, Lin D, Fu Z, Fedorov A, Damaraju E, Sui J, Chen J, Mayer AR, Posse S, Mathalon DH, Ford JM, Van Erp T, Calhoun VD, 2019. Group ICA for identifying biomarkers in schizophrenia: ‘Adaptive’ networks via spatially constrained ICA show more sensitivity to group differences than spatiotemporal regression. NeuroImage Clin 22, 101747. 10.1016/j.nicl.2019.101747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salman MS, Verner E, Bockholt HJ, Fu Z, Misiura M, Baker BT, Osuch E, Sui J, Calhoun VD, 2023. Multi-study evaluation of neuroimaging-based prediction of medication class in mood disorders. Psychiatry Res. - Neuroimaging 333. 10.1016/j.pscychresns.2023.111655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samra A, Ramtahal J, 2012. Recurrent subacute visual loss presenting in a 52-year- … Recurrent subacute visual loss presenting in a 52-year- … Brain 139, 16–17. 10.1093/brain. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato JR, Biazoli CE, Zugman A, Pan PM, Bueno APA, Moura LM, Gadelha A, Picon FA, Amaro E, Salum GA, Miguel EC, Rohde LA, Bressan RA, Jackowski AP, 2021. Long-term stability of the cortical volumetric profile and the functional human connectome throughout childhood and adolescence. Eur. J. Neurosci 54, 6187–6201. 10.1111/ejn.15435. [DOI] [PubMed] [Google Scholar]
- Schlee W, Leirer V, Kolassa S, Thurm F, Elbert T, Kolassa IT, 2012. Development of large-scale functional networks over the lifespan. Neurobiol. Aging 33, 2411–2421. 10.1016/j.neurobiolaging.2011.11.031. [DOI] [PubMed] [Google Scholar]
- Schouten TM, Koini M, Vos F.de, Seiler S, Rooij M.de, Lechner A, Schmidt R, Heuvel M.van den, Grond, van der J, Rombouts SARB, 2017. Individual classification of Alzheimer’s disease with diffusion magnetic resonance imaging. Neuroimage 152, 476–481. 10.1016/j.neuroimage.2017.03.025. [DOI] [PubMed] [Google Scholar]
- Sendi MSE, Zendehrouh E, Ellis CA, Liang Z, Fu Z, Mathalon DH, Ford JM, Preda A, van Erp TGM, Miller RL, Pearlson GD, Turner JA, Calhoun VD, 2021a. Aberrant dynamic functional connectivity of default mode network in schizophrenia and links to symptom severity. bioRxiv. 10.1101/2021.01.03.425152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sendi MSE, Zendehrouh E, Miller RL, Fu Z, Du Y, Liu J, Mormino EC, Salat DH, Calhoun VD, 2021b. Alzheimer’s Disease Projection From Normal to Mild Dementia Reflected in Functional Network Connectivity: A Longitudinal Study. Front. Neural Circuits 14. 10.3389/fncir.2020.593263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sendi Mohammad S.E., Zendehrouh E, Sui J, Fu Z, Zhi D, Lv L, Ma X, Ke Q, Li X, Wang C, Abbott CC, Turner JA, Miller RL, Calhoun VD, 2021. Abnormal Dynamic Functional Network Connectivity Estimated from Default Mode Network Predicts Symptom Severity in Major Depressive Disorder. Brain Connect 11, 838–849. 10.1089/brain.2020.0748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skidmore F, Korenkevych D, Liu Y, He G, Bullmore E, Pardalos PM, 2011. Connectivity brain networks based on wavelet correlation analysis in Parkinson fMRI data. Neurosci. Lett 499, 47–51. 10.1016/j.neulet.2011.05.030. [DOI] [PubMed] [Google Scholar]
- Smith SM, Fox PT, Miller KL, Glahn DC, Fox PM, Mackay CE, Filippini N, Watkins KE, Toro R, Laird AR, Beckmann CF, 2009. Correspondence of the brain’s functional architecture during activation and rest. Proc. Natl. Acad. Sci. U. S. A 106, 13040–13045. 10.1073/pnas.0905267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenwijk MD, Geurts JJG, Daams M, Tijms BM, Wink AM, Balk LJ, Tewarie PK, Uitdehaag BMJ, Barkhof F, Vrenken H, Pouwels PJW, 2016. Cortical atrophy patterns in multiple sclerosis are non-random and clinically relevant. Brain 139, 115–126. 10.1093/brain/awv337. [DOI] [PubMed] [Google Scholar]
- Stern Y, 2002. What is cognitive reserve? Theory and research application of the reserve concept. J. Int. Neuropsychol. Soc 8, 448–460. 10.1017/S1355617702813248. [DOI] [PubMed] [Google Scholar]
- Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, Downey P, Elliott P, Green J, Landray M, Liu B, Matthews P, Ong G, Pell J, Silman A, Young A, Sprosen T, Peakman T, Collins R, 2015. UK Biobank: An Open Access Resource for Identifying the Causes of a Wide Range of Complex Diseases of Middle and Old Age. PLoS Med 12. 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui J, Adali T, Yu Q, Chen J, Calhoun VD, 2012. A review of multivariate methods for multimodal fusion of brain imaging data. J. Neurosci. Methods 204, 68–81. 10.1016/j.jneumeth.2011.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui J, Jiang R, Bustillo J, Calhoun V, 2020. Neuroimaging-based Individualized Prediction of Cognition and Behavior for Mental Disorders and Health: Methods and Promises. Biol. Psychiatry 88, 818–828. 10.1016/j.biopsych.2020.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui J, Pearlson G, Caprihan A, Adali T, Kiehl KA, Liu J, Yamamoto J, Calhoun VD, 2011. Discriminating schizophrenia and bipolar disorder by fusing fMRI and DTI in a multimodal CCA+ joint ICA model. Neuroimage 57, 839–855. 10.1016/j.neuroimage.2011.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teipel S, Drzezga A, Grothe MJ, Barthel H, Chételat G, Schuff N, Skudlarski P, Cavedo E, Frisoni GB, Hoffmann W, Thyrian JR, Fox C, Minoshima S, Sabri O, Fellgiebel A, 2015. Multimodal imaging in Alzheimer’s disease: Validity and usefulness for early detection. Lancet Neurol 14, 1037–1053. 10.1016/S1474-4422(15)00093-9. [DOI] [PubMed] [Google Scholar]
- Tijms BM, Wink AM, de Haan W, van der Flier WM, Stam CJ, Scheltens P, Barkhof F, 2013. Alzheimer’s disease: connecting findings from graph theoretical studies of brain networks. Neurobiol. Aging 34, 2023–2036. 10.1016/j.neurobiolaging.2013.02.020. [DOI] [PubMed] [Google Scholar]
- Tomasi Dardo, Volkow ND, 2012. Abnormal functional connectivity in children with attention-deficit/hyperactivity disorder. Biol. Psychiatry 71, 443–450. 10.1016/j.biopsych.2011.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomasi D, Volkow ND, 2012. Aging and functional brain networks. Mol. Psychiatry 17, 549–558. 10.1038/mp.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Y, Fu Z, Mao C, Falahpour M, Gollub RL, Park J, Wilson G, Napadow V, Gerber J, Chan ST, Edwards RR, Kaptchuk TJ, Liu T, Calhoun V, Rosen B, Kong J, 2020. Distinct thalamocortical network dynamics are associated with the pathophysiology of chronic low back pain. Nat. Commun 11, 1–12. 10.1038/s41467-020-17788-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu Y, Fu Z, Zeng F, Maleki N, Lan L, Li Z, Park J, Wilson G, Gao Y, Liu M, Calhoun V, Liang F, Kong J, 2019. Abnormal thalamocortical network dynamics in migraine. Neurology 92, e2706–e2716. 10.1212/WNL.0000000000007607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tullai-McGuinness S, Madigan EA, Fortinsky RH, 2009. Validity testing the Outcomes and Assessment Information Set (OASIS). Home Health Care Serv. Q 28, 45–57. 10.1080/01621420802716206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidya N, Holla B, Heron J, Sharma E, Zhang Y, Fernandes G, Iyengar U, Spiers A, Yadav A, Das Surajit, Roy S, Ahuja CK, Barker GJ, Basu D, Bharath RD, Hickman M, Jain S, Kalyanram K, Kartik K, Krishna M, Krishnaveni G, Kumaran K, Kuriyan R, Murthy P, Orfanos DP, Purushottam M, Kurpad SS, Singh L, Singh R, Subodh BN, Toledano M, Walter H, Desrivières S, Chakrabarti A, Benegal V, Schumann G, (cVEDA), C. on V. to E.D. and A, Varghese M, Thennarasu K, Metha U, Girimaji S, Jacob P, Jayarajan D, Kumar K, Narayanan G, Khullar M, Khandelwal N, Ghosh Abhishek, Joshi N, Chanu ND, M.C. F, Ph. V, Phurailatpam C, Bhattacharya D, Haque B, Nagraj A, Ghosh Arpita, Basu A, Pandit M, Das Subhadip, Maurya P, Gourisankar A, T GR, B S, Rangaswamy M, Fall C, KN K, MC R, Urs C, N S, R S, K D, Rao A, R P, Tripathy S, Parashar N, B NK, Seshadri A, Kumar S, Baligar S, Arumugam T, Safai A, Cyril A, Roy A, D D, D S, R BB, 2023. Neurocognitive Analysis of Low-level Arsenic Exposure and Executive Function Mediated by Brain Anomalies Among Children, Adolescents, and Young Adults in India. JAMA Netw. Open 6. 10.1001/JAMANETWORKOPEN.2023.12810 e2312810–e2312810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Essen DC, Ugurbil K, Auerbach E, Barch D, Behrens TEJ, Bucholz R, Chang A, Chen L, Corbetta M, Curtiss SW, Della Penna S, Feinberg D, Glasser MF, Harel N, Heath AC, Larson-Prior L, Marcus D, Michalareas G, Moeller S, Oostenveld R, Petersen SE, Prior F, Schlaggar BL, Smith SM, Snyder AZ, Xu J, Yacoub E, 2012. The Human Connectome Project: A data acquisition perspective. Neuroimage 62, 2222–2231. 10.1016/j.neuroimage.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller K, Kinder-Kurlanda KE, 2016. A manifesto for data sharing in social media research. In: WebSci 2016 - Proc. 2016 ACM Web Sci. Conf., pp. 166–172. 10.1145/2908131.2908172. [DOI] [Google Scholar]
- Woo CW, Chang LJ, Lindquist MA, Wager TD, 2017a. Building better biomarkers: Brain models in translational neuroimaging. Nat. Neurosci 20, 365–377. 10.1038/nn.4478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo CW, Schmidt L, Krishnan A, Jepma M, Roy M, Lindquist MA, Atlas LY, Wager TD, 2017b. Quantifying cerebral contributions to pain beyond nociception. Nat. Commun 8 10.1038/ncomms14211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Groth KM, Pearlson G, Schretlen DJ, Calhoun VD, 2009. Source-based morphometry: The use of independent component analysis to identify gray matter differences with application to schizophrenia. Hum. Brain Mapp 30, 711–724. 10.1002/hbm.20540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W, Pearlson GD, Fu Z, Li X, Iraji A, Chen J, Sui J, Volkow ND, Calhoun VD, 2023. A brain-wide risk score for psychiatric disorder evaluated in a large adolescent population reveals increased divergence among higher-risk groups relative to controls. Biol. Psychiatry 10.1016/j.biopsych.2023.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Ghayem F, Gabrielson B, Akhonda MABS, Calhoun VD, Adali T, 2023. Constrained Independent Component Analysis Based on Entropy Bound Minimization for Subgroup Identification from Multi-subject fMRI Data. ICASSP 2023. Rhodes Island, Greece ieeexplore.ieee.org. [Google Scholar]
- Yoshihara Y, Lisi G, Yahata N, Fujino J, Matsumoto Y, Miyata J, Sugihara G, Urayama S, Kubota M, Yamashita M, Hashimoto R, Ichikawa N, Cahn W, Haren NEM, Mori S, Okamoto Y, Kasai K, Kato N, Imamizu H, Kahn RS, Sawa A, Kawato M, Murai T, Morimoto J, Takahashi H, 2018. Overlapping but asymmetrical relationships between schizophrenia and autism revealed by brain connectivity. bioRxiv 403212. 10.1101/403212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zang Y, Jiang T, Lu Y, He Y, Tian L, 2004. Regional homogeneity approach to fMRI data analysis. Neuroimage 22, 394–400. 10.1016/j.neuroimage.2003.12.030. [DOI] [PubMed] [Google Scholar]
- Zhao M, Yan W, Luo N, Zhi D, Fu Z, Du Y, Yu S, Jiang T, Calhoun VD, Sui J, 2022. An attention-based hybrid deep learning framework integrating brain connectivity and activity of resting-state functional MRI data. Med. Image Anal 78 10.1016/j.media.2022.102413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou QH, Zhu CZ, Yang Y, Zuo XN, Long XY, Cao QJ, Wang YF, Zang YF, 2008. An improved approach to detection of amplitude of low-frequency fluctuation (ALFF) for resting-state fMRI: Fractional ALFF. J. Neurosci. Methods 172, 137–141. 10.1016/j.jneumeth.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The HCP developmental and aging data used in the present study can be accessed upon application from NDA (https://nda.nih.gov/) with the approval of the ABCD consortium. The HCP 1200 release data can be accessed upon application from Connectome Coordination Facility (https://www.humanconnectome.org/). The UK biobank data can be accessed upon application from UK biobank biomedical database (https://www.ukbiobank.ac.uk/).
NeuroMark 3.0 templates are accessible on our official website (https://trendscenter.org/data/) and GitHub (https://github.com/trendscenter/gift/tree/master/GroupICAT/icatb/icatb_templates). The codes of the NeuroMark framework have been integrated into the group ICA Toolbox (GIFT 4.0c, https://trendscenter.org/software/gift/). Other MATLAB codes of this study can be obtained from the corresponding author.
Data will be made available on request.