Abstract
An internal ribosome entry site (IRES) mediates translation initiation of bovine viral diarrhea virus (BVDV) RNA. Studies have suggested that a portion of the Npro open reading frame (ORF) is required, although its exact function has not been defined. Here we show that a subgenomic (sg) BVDV RNA in which the NS3 ORF is preceded only by the 5′ nontranslated region did not replicate to detectable levels following transfection. However, RNA synthesis and cytopathic effects were observed following serial passage in the presence of a noncytopathic helper virus. Five sg clones derived from the passaged virus contained an identical, silent substitution near the beginning of the NS3 coding sequence (G400U), as well as additional mutations. Four of the reconstructed mutant RNAs replicated in transfected cells, and in vitro translation showed increased levels of NS3 for the mutant RNAs compared to that of wild-type (wt) MetNS3. To more precisely dissect the role of these mutations, we constructed two sg derivatives: ad3.10, which contains only the G400U mutation, and ad3.7, with silent substitutions designed to minimize RNA secondary structure downstream of the initiator AUG. Both RNAs replicated and were translated in vitro to similar levels. Moreover, ad3.7 and ad3.10, but not wt MetNS3, formed toeprints downstream of the initiator AUG codon in an assay for detecting the binding of 40S ribosomal subunits and 43S ribosomal complexes to the IRES. These results suggest that a lack of stable RNA secondary structure(s), rather than a specific RNA sequence, immediately downstream of the initiator AUG is important for optimal translation initiation of pestivirus RNAs.
The pestiviruses are animal pathogens causing acute, chronic, and persistent infections that can result in substantial economic losses in the livestock industry (22, 31, 41). Members include bovine viral diarrhea virus (BVDV), classical swine fever virus (CSFV), border disease virus, and the newly proposed member, BVDV-2 (4). The pestiviruses are grouped in the Flaviviridae family along with the hepaciviruses (hepatitis C viruses [HCVs]) and classical flaviviruses; however, similarities in genome organization, translation initiation strategy, and polyprotein processing suggest that the pestiviruses and HCVs are more closely related to each other than to the classical flaviviruses.
The Flaviviridae are small, enveloped viruses containing a single-stranded positive-sense RNA genome (31). The genomes are usually between 9.5 and 12.5 kb in length, with 5′ and 3′ nontranslated regions (NTRs) flanking one long open reading frame (ORF), and lack the 3′ poly(A) tract. Translation of the viral RNA yields a single polyprotein that is co- and posttranslationally processed by host and viral proteases. The classical flaviviruses contain a type 1 cap at the 5′ end that directs translation initiation in a cap-dependent manner. In contrast, the 5′ NTRs of the pestiviruses and HCVs contain structurally and functionally similar internal ribosome entry site (IRES) elements that mediate translation in a cap-independent manner (reviewed in reference 16).
The first protein of the pestivirus ORF, Npro, is a nonstructural autoprotease unique to the pestiviruses, which autocatalytically generates its own C terminus (34). It is followed by the structural proteins (C, Erns, E1, and E2) and the nonstructural proteins (p7, NS2-NS3, NS4A, NS4B, NS5A, and NS5B) (reviewed in reference 22). NS3 harbors serine protease, helicase, and NTPase activities (37, 38, 44–46), and NS5B exhibits RNA-dependent RNA polymerase activity (48). The identification of these activities implies that both NS3 and NS5B are directly involved in viral RNA replication.
Two biotypes can be distinguished following infection of cultured cells. Viruses that do not appear to cause any cell damage are considered noncytopathic (ncp), whereas cytopathic (cp) isolates cause cell shrinkage, membrane blebbing, and cell death (22). For BVDV, only unprocessed NS2-NS3 is observed in ncp isolates. In contrast, production of a discrete NS3 protein in addition to unprocessed NS2-NS3 has been associated with the cp phenotype. Considerable data suggest that cp viruses usually develop from ncp viruses by RNA recombination events resulting in genome duplications, rearrangements, deletions, or cellular insertions. However, one report has demonstrated that single-point mutations in NS2 were responsible for cytopathic effects (CPE) in the BVDV strain Oregon (14).
Characterization of several isolates revealed that cp BVDV and CSFV could be composed of RNAs with large internal deletions and an ncp helper virus (15, 20, 21, 23, 24, 40). In these cases, the 5′ NTR and initiating methionine codon were intact, but in-frame deletions of the structural (C, Erns, E1, and E2) and nonstructural (p7 and NS2) genes were noted. For simplicity and continuity, we will refer to these internally deleted genomes as subgenomic (sg) RNAs (5, 19, 40). The most well-characterized cp sg BVDV RNA, DI9, has a deletion encompassing all of the structural proteins as well as p7 and NS2, leaving the C terminus of Npro fused in frame to NS3 (40). Additional sg BVDV RNAs have been described, in which portions of Npro and E1 are fused to NS3 (DI13) (15) or, in others, a capsid (or NS2)-ubiquitin fusion is inserted between complete or C terminally truncated Npro and NS3 (3). However, in most of the sg CSFV RNAs, the initiating AUG codon is fused in frame to the N terminus of NS3 (21, 24).
Following the identification of the cytopathic sg RNAs, several investigators created cDNA copies to study the molecular requirements of pestivirus replication and cytopathogenicity (5, 20, 23, 25, 39). The first was based on the DI9 genome, and the derived RNA replicated and caused CPE following cotransfection with an ncp helper RNA (20). Subsequent studies showed that the structural proteins, as well as p7 and NS2, were dispensable for viral RNA replication and cytopathogenicity since sg DI9-like RNAs replicated autonomously and caused cell lysis (5, 39). It also appeared that the first 39 nucleotides (nt) downstream of the Npro AUG codon were required for replication of the sg DI9-like RNAs, although the specific Npro amino acid (aa) sequence was not. However, the specific function of these sequences was not examined. In contrast, the CSFV sg RNAs, which lacked Npro, replicated efficiently (23, 25).
To create an experimental system to study pestivirus replication, an sg RNA was constructed based on the sg CSFV genome organization (MetNS3) by using the National Animal Disease Laboratory (NADL) strain of BVDV. We present evidence demonstrating that efficient pestivirus translation initiation correlates with autonomous sg RNA replication. Furthermore, the data suggest that binding of 40S ribosomal subunits and formation of 48S ribosomal initiation complexes require a less-structured or less stable RNA structure downstream of the initiating methionine.
MATERIALS AND METHODS
Cells and viruses.
Madin-Darby bovine kidney (MDBK) cells were generously provided by M. Collett (ViroPharma, Inc.) and were propagated in supplemented Dulbecco's modified minimal essential medium (with sodium pyruvate and 10% horse serum) (19). Cells were maintained at 37°C with 5% CO2.
The BVDV ACNR/NADL and ACNR/cIns−NADL viruses were propagated in MDBK cells as previously described (19).
Plasmids.
Plasmids pACNR/NADL and pACNR/cIns−NADL, representing the cp and ncp infectious cDNAs, respectively, have been described previously (19). The discistronic reporter plasmid, T7DC1-341, was kindly provided by A. Siddiqui (43). It contained a T7 promoter directing transcription of chloramphenicol acetyltransferase (CAT) and an HCV IRES driving expression of luciferase (LUC).
Standard recombinant DNA techniques were used for all plasmid constructions. T4 polymerase (T4) was used to create a blunt end when restriction enzyme digestion created a noncompatible end. cDNA clones were identified by restriction endonuclease digestions, and automated nucleotide sequencing confirmed the sequences of regions amplified by PCR. Brief descriptions of plasmids used in this study are supplied below; specific details are available upon request. When present, numbering refers to NADL, cIns−NADL, or MetNS3 DNA sequences (see below) as indicated.
An XbaI site was created upstream of the T7 promoter in pACNR/NADL (19) as follows. First, a MunI (T4)-XhoI fragment of pACNR/DraIII− was replaced with an XbaI (T4)-XhoI fragment from pTet/BVD5′3′ (46), regenerating the XbaI site in pACNR1180/DraIII−/BVD5′. Second, an HaeII-XhoI fragment from pACNR1180/DraIII−/BVD5′ was subcloned into those sites in pACNR/NADL, creating pxNADL.
The pxcIns−NADL derivative (I. Frolov and C. M. Rice, unpublished data) was created by replacing the NheI-BglII fragment in pxNADL (NADL nt 4930 to 5644) with the corresponding fragment from pACNR/cIns−NADL (nt 4930 to 5374).
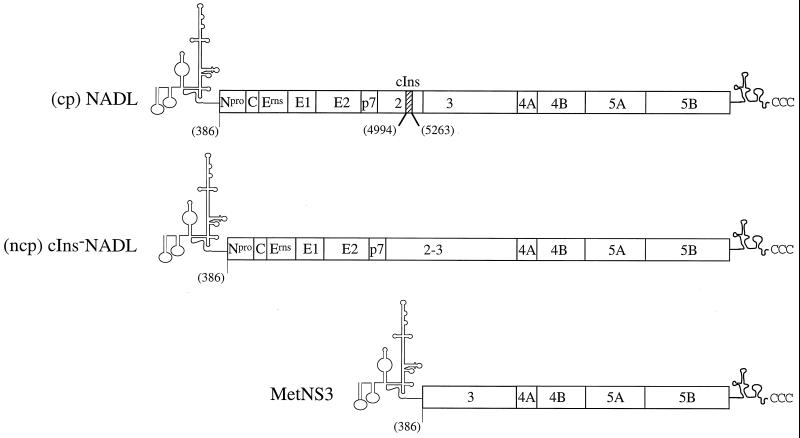
For the sg MetNS3 cDNA that is referred to as the wild type (wt), pMetNS3, the NADL sequence between nt 389 and 5422 was deleted using recombinant PCR. Oligodeoxynucleotides 183 and 184 were used to amplify nt 1 to 388, and the 378 and 344 oligodeoxynucleotides amplified nt 5423 to 5835 (Table 1). Oligodeoxynucleotide primers 378 and 379 contained an ATG codon and a silent nucleotide change (bold), creating an ApaI site (underlined) at the deletion breakpoint (Table 1). The PCR-amplified fragments were digested with ApaI and then ligated, and the product was digested with XhoI-BglII. This fragment was used to replace the corresponding XhoI-BglII fragment (nt 224 to 5644) in pACNR/NADL (Fig. 1).
TABLE 1.
Oligodeoxynucleotide primers
| Oligodeoxynucleotide | Sequence | NADL nucleotide positions |
|---|---|---|
| 183 | (+)-TTTTCTAGATAATACGACTCACTATAGTATACGAGAATTAGAAAAGGCACTCGa | 1–27 |
| 344 | (−)-GTCTTTTGGAGGTGAACGACTGCCCC | 5810–5835 |
| 378 | (+)-CATGGCACATGGGGCCCGCCGTGTGTAAGAAGb | 378–388, 5423–5443 |
| 379 | (−)-CACGGCGGGCCCCATGTGCCATGTACAGCAGAGb | 368–388, 5423–5434 |
| 886 | (+)-CCAGAAGGTACCCCATTGTATc | 517–537 |
| 887 | (−)-ACGGCGGGCCCCATATTATCATCGTGTcd | 386–399, 643–655 |
| 888 | (+)-TCTCTGCTGTACATGGCACATGGGACCTGCTGTTTGTAAAAAAATCACAGe | 367–416 |
| 889 | (−)-TCTGCTGTACATGGCACATGGGGCCTGCCGTGTGTAAGe | 369–406 |
Oligodeoxynucleotide primer 183 has been described previously (19).
The ApaI site and silent nucleotide substitution (T394C) are indicated by underlining and bold lettering, respectively.
The numbers for nt 517 to 537 for primer 886 and nt 643 to 655 of primer 887 refer to 5′ BVDV plus EMCVdelB3ABC (9).
The numbers for nt 386 to 399 for primer 887 refer to MetNS3, with 386 being the A of the AUG start codon.
The nucleotide numbers for mutant primers 888 and 889 refer to the MetNS3 sequence, with 386 being the A of the AUG start codon.
FIG. 1.
Schematic representation of BVDV RNAs transcribed from the cDNA templates, cp NADL (top), ncp cIns-NADL (middle), and sg MetNS3 (bottom), used in these studies. The proposed 5′ and 3′ NTR structures are shown, with the polyprotein cleavage products indicated by boxes. The numbers in parentheses refer to the AUG start codon (nt 386) and the boundaries of the cellular insert (cIns, nt 4994 to 5263).
To place the sg MetNS3 sequence in the XbaI-containing vector background (px), an SphI-BglII (nt 247 to 610) fragment with the authentic MetNS3 junction from pMetNS3 was subcloned into those sites in the sg derivative pxNproNS3 (T. Myers et al., unpublished data), creating pxMetNS3. The NADL sequences in pMetNS3 and pxNproNS3 are colinear up to SphI and downstream of BglII.
To create pxMetNS3ad3.7 and pxMetNS3ad3.8, oligodeoxynucleotide primer pairs 888-344 (ad3.7) and 889-344 (ad3.8) were used to amplify the MetNS3 junction (Table 1). For both constructs, the PCR products were digested with BsrGI and BglII and subcloned into those sites in pxMetNS3.
To replace the wt 5′ NTR in MetNS3 with the BVDV-EMCV/delB3ABC chimera 5′ NTR (9) and create chimeric (chm) MetNS3, the MetNS3 junction and most of the BVDV-EMCV/delB3ABC 5′ NTR was amplified by PCR with oligodeoxynucleotides 886 and 887 (Table 1). The PCR product was digested with KpnI and ApaI and combined with the following fragments in a ligation reaction mixture: ApaI-NcoI and NcoI-XbaI from pxMetNS3 and XbaI-KpnI from pBVDV-EMCV/delB3ABC.
To create pxMetNS3ad3.10, containing just the G400U substitution, an XbaI-AseI fragment from pxMetNS3ad3.2 (nt −22 to 434) was subcloned into those sites in wt pxMetNS3.
The initial cDNA copy of adapted MetNS3 RNA was obtained by reverse transcription-PCR (RT-PCR) of MetNS3 RNA isolated from the supernatant of cells infected with passage 4 virus (cIns−NADL and MetNS3). To create pxMetNS3ad3, a portion of the RT-PCR product was digested with SphI and BglII and subcloned into those sites in pxNproNS3, as described above. Five individual cDNA clones (ad3.1, ad3.2, ad3.4, ad3.5, and ad3.6) were sequenced and used for further investigation (see Results).
In vitro transcription of RNA.
Plasmid DNAs containing full-length and sg BVDV NADL sequences were digested with SdaI, and the reporter plasmid, T7DC1-341, was digested with HpaI (LUC expression, transfection studies) or SalI (CAT expression only, in vitro translation). The linearized DNAs were extracted with phenol-chloroform (1:1) and chloroform alone and then were precipitated with ethanol. Pelleted DNAs were resuspended in double-distilled water (ddH2O) and transcribed in vitro using the T7-MEGAscript kit (Ambion, Austin, Tex.) in the presence of 1 μCi of [3H]UTP (Dupont NEN, Boston, Mass.), as recommended by the manufacturer (19). The in vitro-synthesized RNAs were resuspended in ddH2O at approximately 1 μg/μl based on [3H]UTP incorporation. Transcript RNAs and standard RNAs of known concentrations were separated on a 1% agarose gel and stained with ethidium bromide. The final RNA concentrations were determined using a Fluorimager (Molecular Dynamics, Sunnyvale, Calif.) in conjunction with the IPLab Gel program (Scantalytics, Inc., Fairfax, Va.). RNA aliquots were stored at −80°C.
Transfection of cultured cells.
Transfection of MDBK cells by electroporation was performed essentially as described previously (19). Briefly, MDBK cells (approximately 80% confluent) were detached by trypsin treatment; washed three times in sterile, ice-cold phosphate-buffered saline (PBS); and resuspended at between 1.0 × 107 and 2.0 × 107 cells/ml in PBS. Volumes containing 4 to 5 μg of full-length or sg RNA transcripts were combined with 1 μg of LUC reporter RNA (DCI-341), mixed with 0.4 ml of washed MDBK cells, and pulsed immediately (1.5 kV, 25 μF, ∞ resistance, and 2 pulses) using a Bio-Rad Gene Pulser (Hercules, Calif.). The total RNA transfected per reaction mixture was equalized using uninfected MDBK cellular RNA that had been extracted using TRIzol reagent (Gibco-BRL, Grand Island, N.Y.) as described below. The pulsed samples were diluted to 10 ml with Dulbecco's minimal essential medium (supplemented with 10% horse serum and sodium pyruvate), added to either a 100-mm-diameter dish or split equally into four 35-mm-diameter wells, and incubated at 37°C with 5% CO2. The specific infectivity of full-length RNA was determined using an infectious center assay as described previously (19).
Serial infections, or passages, were performed by superinfecting persistently infected MDBK cells (4 × 106) with one-half of the transfected cell supernatant. Persistently infected MDBK cells were established by infecting naïve MDBK cells with ACNR/cIns−NADL (multiplicity of infection [MOI] = ∼2) and splitting the infected cells once prior to superinfection. The cells were incubated at 37°C with 5% CO2 until CPE was observed. CPE was noted usually 24 to 48 h following superinfection, and the passage on persistently infected MDBK cells was repeated. Significant levels of CPE were present by passage 4 in the superinfected cells, at which point the cell supernatants were collected, clarified, and stored at −80°C as a passage 4 virus stock.
Protein analysis.
Transfection efficiency was monitored in the transfected cells by quantitation of LUC activity at 8 h posttransfection (p.t.). Cell monolayers (35-mm-diameter wells) were washed twice with PBS and resuspended in 250 μl of 1% Triton X-100 lysis buffer (25 mM Tris-HCl, pH 7.5; 25 mM dithiothreitol; 2 mM EGTA; 1% [vol/vol] Triton X-100). The LUC activity in one-tenth of the clarified cell extract was measured using a luminometer (EG&G Berthold, Gaithersburg, Md.).
For Western blot analysis, cell lysates from 35-mm-diameter wells were prepared in 300 μl of 0.5% sodium dodecyl sulfate (SDS) lysis buffer (50 mM Tris-HCl, pH 7.5; 150 mM NaCl; 0.5% SDS; 20 μg of phenylmethylsulfonyl fluoride/ml; 1 μg of aprotinin/ml) at the times p.t. indicated in the figure legends. Total protein concentration was determined using the bicinchonic acid protein assay (Pierce, Rockford, Ill.) as recommended by the manufacturer except that all volumes were reduced by one-half. Equivalent amounts of total protein were analyzed by immunoblotting with polyclonal rabbit NS3-specific antisera (G40) following protein separation by SDS–8% polyacrylamide gel electrophoresis (PAGE), as described previously (19).
Subsaturating concentrations of T7-transcribed sg RNAs (data not shown) were translated in vitro using rabbit reticulocyte lysates (Promega; Madison, Wis.) in the presence of [35S]methionine (Amersham, Piscataway, N.J.). Each translation reaction mixture contained CAT mRNA in addition to the indicated sg RNA for subsequent normalization of the NS3 translation levels. Samples were diluted 10-fold in Laemmli sample buffer and analyzed by SDS–9% PAGE. The radiolabeled bands were detected by autoradiography and quantitated with a Molecular Imager (Bio-Rad Laboratories).
Viral RNA analysis.
Initially, RNA synthesis and accumulation were examined by [3H]uridine (30 μCi/m) (ICN Biomedicals, Inc., Costa Mesa, Calif.) incorporation in the presence of dactinomycin (1 μg/ml) (Fig. 2). Alternatively, 200 μCi of [32P]orthophosphate (H332PO4)/ml was added during the exponential phase of sg RNA synthesis in media containing low levels of phosphate at concentrations that yielded NADL viral titers between 50 and 80% of those obtained in complete media (data not shown). RNAs from the transfected, radiolabeled cells were isolated using TRIzol reagent (Gibco-BRL). To ensure recovery of total RNA, detached cells were collected from the supernatants by low-speed centrifugation and the pelleted cells were combined with the corresponding monolayer lysate. RNAs were isolated according to the manufacturer's protocol, except that carrier tRNA (25 μg) was added to each lysate and an additional chloroform extraction was performed.
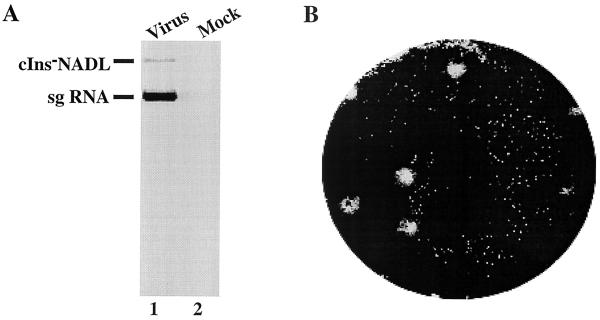
FIG. 2.
Viral RNA accumulation and CPE in cells infected with passage 4 virus. (A) MDBK cells were infected at an MOI of 1 with passage 4 virus (lane 1) or were mock infected (lane 2, media only) and were labeled from 6 to 18 h p.i. with [3H]uridine in the presence of dactinomycin. RNAs were isolated from the cells using TRIzol reagent (Gibco-BRL), denatured with glyoxal, and separated on a 1% agarose gel. The labeled RNAs were visualized by autoradiography. (B) Plaques were detected by crystal violet staining at 3 days p.i. with passage 4 virus as described previously (19).
One-half of the sample was denatured with glyoxal at 51°C and analyzed on a 1% agarose gel in 10 mM sodium phosphate buffer (35). In parallel, twofold dilutions of in vitro-transcribed RNAs were included for a standard RNA concentration curve and as size markers. RNAs were visualized by staining gels in 0.5 μg of ethidium bromide/ml after washing in 50 mM NaOH for 30 min. Nonspecific staining was removed by extensive washing in 10 mM sodium phosphate buffer. The 18S rRNA in each sample was compared with a standard RNA curve using Fluorimager scanning and IP Lab Gel analysis to determine total 18S rRNA concentrations in the cell lysates, as described above. Radiolabeled RNAs were visualized by autoradiography and quantitated with a Molecular Imager as described above.
Assembly and analysis of ribosomal and ribonucleoprotein complexes.
Ribosomal subunits, subunit of eukaryotic initiation factor 2 (eIF2) and eIF3 were purified from rabbit reticulocyte lysates (Green Hectares, Oregon, Wis.) as previously described (28, 29). Ribosomal complexes and complexes between BVDV mRNA and eIF3 were assembled on BVDV mRNA, essentially as previously described (27). BVDV plasmids were linearized with BglII, and mRNAs were transcribed in vitro using T7 RNA polymerase. Aliquots of the mRNAs (0.6 μg) were incubated for 5 min at 30°C in a 40-μl reaction volume that contained 0.4 mM GMP-PNP, Met-tRNAiMet (6 pmol), 40S subunits (6 pmol), eIF2 (3 μg), and eIF3 (7 μg) as indicated in the text. [35S]methionine-labeled Met-tRNAiMet was prepared using rabbit tRNA (Novagen, Madison, Wis.) and aminoacyl-tRNA synthetase purified from Escherichia coli MRE600 (American Type Culture Collection, Manassas, Va.), as described previously (28).
Ribosomal and RNP complexes were analyzed by primer extension (28), using primer 5′-GTAAGCTCGTAGGGAACC-3′ (complementary to nt 5541 to 5558 of the BVDV genomic sequence) and avian myeloblastosis virus reverse transcriptase (Promega) in the presence of [α-32P]dATP (∼6,000 Ci/mmol) as described. The cDNA products were ethanol precipitated, resuspended, and analyzed by electrophoresis through 6% polyacrylamide sequencing gels. The cDNA products were compared with appropriate dideoxynucleotide sequence ladders.
RESULTS
Adaptive mutations are required for replication of sg NADL MetNS3 RNA in cell culture.
To study the genetic requirements of pestivirus replication and cytopathogenicity, we created a classical swine fever-like sg cDNA (MetNS3) derived from an infectious NADL cDNA clone. For MetNS3, the complete NADL 5′ NTR and initiating methionine codon were fused in frame with the first codon of NS3 (14), which was contiguous with the remainder of the NADL sequence (19). MDBK cells were cotransfected with in vitro-synthesized sg MetNS3 and ncp cIns−NADL RNAs (Fig. 1) and observed for the development of CPE (19). Surprisingly, neither CPE nor sg RNA replication was detected in the transfected cells, whereas replication of ncp cIns−NADL virus was readily observed (data not shown). Identical results were obtained after transfection of MetNS3 RNA into persistently infected MDBK cells. However, if the extracellular supernatant was serially passaged on persistently infected MDBK cells, significant CPE was eventually observed (data not shown). Similar to results reported for another virus stock containing both sg and full-length RNA (CP13), the number of PFU was 10- to 100-fold lower than the number of focus-forming units in the passaged virus stocks (15). Thus, the range of MOIs that could be reasonably achieved in subsequent infections was limited.
To confirm that the observed CPE was due to sg MetNS3 RNA replication, naïve MDBK cells were infected with a passage 4 virus stock and the cells were labeled with [3H]uridine from 6 to 18 h postinfection (p.i.) in the presence of dactinomycin. Both 3H-labeled full-length cIns−NADL and sg MetNS3 RNAs were observed in infected cell extracts but not in mock-infected cell extracts (Fig. 2A, lanes 1 and 2, respectively). Moreover, replication of the sg MetNS3 RNA was associated with the appearance of CPE (Fig. 2B). These results suggest that either the sg RNA, the ncp helper RNA, or both accumulate adaptive changes during serial passage. Adaptive mutations in the ncp cIns−NADL RNA were less likely based on our data showing that this virus remains noncytopathic during passage (data not shown). These data also show that the MetNS3 RNA could be packaged in trans by the ncp cIns−NADL helper virus present in the persistently infected MDBK cells.
To determine if the passaged sg MetNS3 RNAs contained adaptive mutations, the RNA population in the passage 4 virus stock was amplified by RT-PCR and sequenced. Mutations specific for the sg RNA population were mapped using primers that amplified the MetNS3 junction. Although nt 1 to 2568, which includes the 5′ NTR (nt 1 to 385), NS3 gene (nt 386 to 2437), and the N-terminal two-thirds of NS4A (nt 2438 to 2629) were sequenced, only a single silent nucleotide substitution (G400U) at the beginning of the NS3 was identified (initiator AUG at nt 386 to 388). To determine if the silent G400U mutation or other less frequent mutations were sufficient to facilitate RNA replication and CPE, we replaced the corresponding wt MetNS3 SphI-BglII fragment (nt 247 to 610) with the mutant RT-PCR product.
Automated sequence analysis of the SphI-BgII fragment of five individual cDNA clones (ad3.1, ad3.2, ad3.4, ad3.5, and ad3.6) revealed at least one additional mutation in each clone (Fig. 3A), with the additional mutation in ad3.2 (A460C) also silent, thereby creating a mutant without any amino acid changes. Clone ad3.6 contained two mutations in common with ad3.5 as well as an additional downstream amino acid substitution, suggesting a common RNA ancestor. The additional mutations not detected by direct sequencing of the RT-PCR products may represent minor variants in the population.
FIG. 3.
RNA and protein accumulation in MDBK cells transfected with the wt and reconstructed MetNS3 RNAs, ad3.1, ad3.2, ad3.4, ad3.5, and ad3.6. (A) Sequence changes identified in five individual cDNA clones after replacing the wt MetNS3 sequence with the corresponding RT-PCR product amplified from passage 4 MetNS3 RNA as described in Materials and Methods. The wt MetNS3 nucleotide sequence is shown at the top with the single-letter amino acid code above. The 1st, 13th, 25th, and 50th amino acids are indicated along with the first nucleotide number of the codons. Hash marks (⫽) represent breaks in the linear sequence. Mutations are indicated in bold, and any resulting amino acid change is above the codon. Dots indicate homology with the wt MetNS3 sequences. (B and C) Analysis of RNA and protein accumulation, respectively, per ∼4 × 106 cells transfected with in vitro-transcribed RNAs. Shown are wt MetNS3 (lanes 2) and the adapted derivatives MetNS3 RNAs (lanes 3 to 7). Mock (—) transfections (lanes 1) lacked RNA. RNA accumulation was detected by radiolabeling the transfected cells with H332PO4 between 16 and 24 h p.t. in the presence of dactinomycin. Total cellular RNA was glyoxylated, separated on a 1% agarose gel, and visualized by autoradiography. The position of the sg RNA is indicated. The radiolabeled bands were quantitated with a Molecular Imager (Bio-Rad Laboratories) and normalized to transfection efficiency and total 18S rRNA as described in Materials and Methods. The percentages of each sg RNA relative to ad3.4 (100%) are indicated. (B). Transfected cell lysates were prepared at 18 h p.t. and separated by SDS–8% PAGE. NS3 levels were detected with polyclonal rabbit NS3-specific antisera by Western blot analysis. Molecular mass markers are indicated on the right and the migration of NS3 is indicated on the left (C).
Adapted ad3 RNA replicate autonomously.
Recent reports demonstrated that an sg BVDV DI9 RNA could replicate autonomously in cultured cells and was able to induce CPE in the absence of an ncp helper virus (5, 39). The sg DI9 or DI9-like RNAs do not encode structural proteins. Thus, viral packaging, release, and spread were absent, and RNA replication and cytopathogenicity were examined as first-cycle events in transfected cells. To determine if the reconstructed sg MetNS3 RNAs were capable of autonomous replication, MDBK cells were transfected with in vitro-synthesized RNA and a LUC reporter RNA as a transfection control (43). Transfected cells were split into 35-mm-diameter wells to examine viral RNA accumulation, protein expression, CPE, and LUC activity.
Cells transfected with the mutant sg RNAs ad3.1, ad3.2, ad3.4, and ad3.5 developed CPE that included vacuole formation, cell shrinkage, and detachment and were distinguishable from that induced by dactinomycin treatment alone (data not shown). Accumulation of 32P-labeled sg RNA was observed for ad3.1, ad3.2, ad3.4, and ad3.5 (Fig. 3B, lanes 3, 4, 5, and 6), whereas neither in vivo-labeled ad3.6 RNA nor wt MetNS3 RNA was detectable under these conditions. RNA accumulation in ad3.4-transfected cells was reproducibly higher than it was in the others and was set at 100% for quantitation comparisons with other samples (see Materials and Methods). The levels of accumulated ad3.1 and ad3.2 RNAs were somewhat reduced (83 and 51% of that of ad3.4, respectively; Fig. 3B, lanes 3 and 4), whereas the level of ad3.5 was significantly lower (29%; Fig. 3B, lane 6). As expected, the mock- and wt MetNS3-transfected cells did not show detectable accumulation of discrete 32P-labeled RNA products (Fig. 3A, lanes 1 and 2). In vivo-labeled 32P-labeled RNAs comigrated with in vitro-synthesized MetNS3 sg RNA transcripts (data not shown). These data demonstrate that two silent mutations in the ad3.2 RNA are sufficient for autonomous replication and CPE (Fig. 3B, lane 4), and it is likely that the prevalent G400U substitution is important for replication. Furthermore, the additional mutation in ad3.6 was probably detrimental to NS3 activity.
To examine polyprotein synthesis and processing, transfected cell extracts were analyzed for NS3 protein levels by Western blotting (7, 19). All replication-competent sg RNAs expressed NS3 (Fig. 3C, lanes 3, 4, 5, and 6) and the level of NS3 expression correlated with the level of RNA accumulation. Furthermore, NS3 expressed from the replicons comigrated with NS3 produced in NADL virus-infected cells (data not shown). In the absence of detectable replication, primary translation of the input transcripts did not produce detectable levels of NS3 (Fig. 3C, lanes 2 and 7). Moreover, a lack of unprocessed polyprotein precursor containing NS3 indicated that the NS3-NS4A site was efficiently processed in the replicons and demonstrated the presence of an active NS3 protease (Fig. 3C, lanes 3, 4, 5, and 6).
Based on these data, we hypothesized that the silent mutations in the protein coding sequence could be affecting a cis element required for RNA replication or perhaps a step prior to replication, such as translation. To address these possibilities, we first examined primary translation in a cell-free system independent of replication.
Translation of wt MetNS3 RNA in vitro is reduced compared to adapted MetNS3 RNAs.
To analyze the effect of the wt and mutant NS3 sequences on NADL IRES function, the sg MetNS3 RNAs were translated in vitro in the presence of [35S]methionine to detect the proteins. The level of NS3 translated from each RNA was normalized by comparing the amount of NS3 to the level of CAT expressed from a separate transcript (see Materials and Methods). As shown in Fig. 4, the level of 35S-labeled wt NS3 was between 8- to 10-fold less than the levels of adapted NS3, suggesting that there was a defect in the translation initiation of wt MetNS3 (Fig. 4, compare lane 3 with lanes 4, 5, 6, 7, and 8). In contrast, translation of ad3.6 NS3 appeared similar to that of NS3 expressed from the replicating sg RNAs, suggesting that the inability of ad3.6 to replicate may be due to an impairment in NS3 function and not to reduced translation initiation (Fig. 4, compare lane 8 to lanes 4, 5, 6, and 7). As controls, we confirmed that the in vitro-synthesized NS3 proteins comigrated with authentic NS3 from NADL-infected cells (data not shown) and that no 35S-labeled protein products were observed in the mock reaction mixture (Fig. 4, lane 1).
FIG. 4.
In vitro translation of wt and adapted NS3 proteins. The wt and reconstructed MetNS3 RNAs were cotranslated with a capped monocistronic CAT RNA in the presence of [35S]methionine as described in Materials and Methods. Reaction products were diluted 10-fold in Laemmli sample buffer and separated by SDS–9% PAGE, and the labeled proteins were visualized by autoradiography. The mock reaction mixture (m; lane 1) contained ddH2O only, whereas mock plus CAT (m+c), wt MetNS3, ad3.1, ad3.2, ad3.4, ad3.5, and ad3.6 RNAs each contained CAT RNA (lanes 2, 3, 4, 5, 6, 7, and 8, respectively). The radiolabeled bands were quantitated with a Molecular Imager (Bio-Rad Laboratories). The data are representative of two experiments.
Type 2 IRES-mediated translation initiation allows replication of wt MetNS3 RNA.
If the lack of detectable wt MetNS3 RNA replication was due to reduced translation, increasing translation efficiency should enhance replication. On the other hand, if wt MetNS3 lacks a cis-acting replication signal or inhibits RNA synthesis directly, increasing the level of translation should not significantly influence sg RNA replication. To distinguish between these possibilities, the wt MetNS3 5′ NTR was replaced with a chm BVDV-EMCV 5′ NTR that contained the three 5′ terminal BVDV hairpins required for BVDV replication (Fig. 5A) fused to the canonical EMCV IRES element (9). Since the EMCV IRES functions independently of sequences downstream of the initiator methionine codon (12), chm MetNS3 RNA would allow us to determine the effect of the wt MetNS3 coding sequences on RNA replication independently of translation.
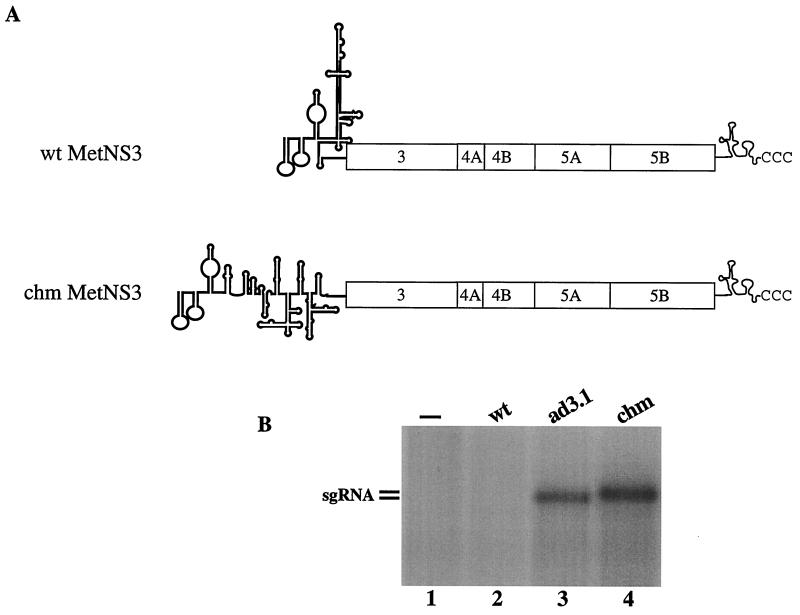
FIG. 5.
Replication of wt MetNS3 RNA containing a type 2 IRES. (A) Schematic representation of the wt BVDV (type 3 IRES) and chm MetNS3 (type 2 IRES) RNAs as described for Fig. 1. (B) MDBK cells were transfected with no RNA (lane 1), wt MetNS3 RNA (lane 2), ad3.1 RNA (lane 3), and chm MetNS3 RNA (lane 4), and RNA synthesis and accumulation were assessed by H332PO4 incorporation as described for Fig. 3. An autoradiogram of the dried gel is pictured, and the results are representative of two experiments.
In vitro, chm MetNS3 was translated efficiently and at levels twofold higher than those of the adapted sg RNAs (data not shown). Replication of chm MetNS3 RNA was assayed following transfection of MDBK cells, and similar levels of 32P-labeled sg RNAs were detected for chm MetNS3 and ad3.1 (Fig. 5B, compare lane 4 with lane 3). Furthermore, replication of the sg RNAs was associated with cytopathology in the transfected cell monolayers (data not shown). These results demonstrate that wt MetNS3 does not have a cis-acting RNA replication defect, suggesting that inefficient translation may be limiting wt MetNS3 replication.
MetNS3 RNAs with less-stable RNA structures downstream of the AUG codon replicate efficiently.
It was possible that the nucleotide sequence or RNA structure downstream of the AUG codon in wt MetNS3 inhibited translation initiation, similar to that reported for HCV IRES-driven translation (10, 11). The 5′ NS3 sequence in wt MetNS3 is GC-rich (Fig. 3A) and might predispose this region to form a stable RNA structure. In keeping with this hypothesis, the NS3 sequence downstream of the initiator methionine in the replication-competent sg CSFV MetNS3 RNAs is AU-rich and the nucleotide at position 400 is a U (21).
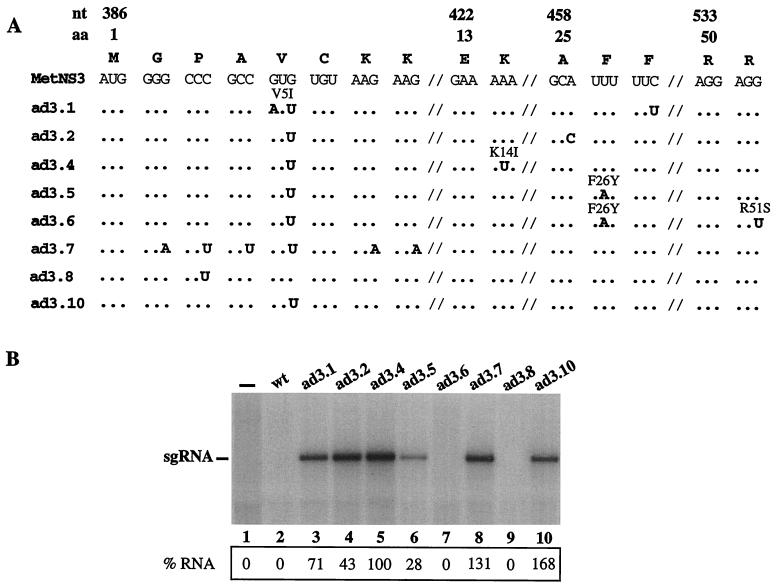
To address this possibility, we created a MetNS3 RNA with a predicted less-stable or unstructured RNA element downstream of the AUG codon having silent A or U substitutions at the third base position of the second, third, fourth, fifth, seventh, and eighth NS3 codons (Fig. 6A; ad3.7). Two additional MetNS3 cDNAs were also constructed: ad3.8 (Fig. 6A, C394U [ApaI site]), to determine if the silent U394C substitution created during the engineering of wt MetNS3 was responsible for the replication-defective phenotype, and ad3.10 (G400U), to test if the prevalent G400U mutation alone was sufficient for translation and autonomous replication (Fig. 6A).
FIG. 6.
Replication of MetNS3 RNAs with less-stable RNA structures downstream of the initiator AUG codon. (A) Diagram of the sequences for the MetNS3 RNAs as described for Fig. 3. ad3.7, ad3.8, and ad3.10 were engineered to contain the indicated mutations. (B) Autoradiogram of RNA accumulation in transfected MDBK cells as described for Fig. 3. The percentages of radiolabeled sg RNAs relative to that of ad3.4 (100%) are indicated. Values are normalized to transfection efficiency and total 18S rRNA.
Next, we examined (indirectly) the RNA secondary structure(s) surrounding the initiating AUG codon (nt 374 to 430) by using the mfold algorithm (18, 49). Two to five stem-loop structures encompassing the AUG codon were formed for the wt, ad3.7, and ad3.10 sg RNAs (data not shown). Replication-incompetent wt MetNS3 formed two similar stem-loop structures, with ΔG of −11.5 and −11.6 kcal/mol at 37°C, and three stem-loops with similar thermodynamic stability, with ΔG from −11.0 to −11.6 kcal/mol, were formed for ad3.8 (predicted stem-loop shown in Fig. 9). In contrast, ad3.7 and ad3.10 formed four and five stem-loop structures, respectively, with significantly less-stable structures (ΔG = −3.7 to −4.6 kcal/mol and −4.9 to −5.6 kcal/mol at 37°C, respectively). Due to the region folded, the remaining replicons and ad3.6 had the same structures and stability as ad3.10. Consistently, the G400U substitution in the replicons and ad3.10 disrupted base pairing in the stem, which was associated with decreased thermodynamic stability of the stem-loop structure. For ad3.7, the multiple substitutions disrupted the basic structure of the stem-loop, with the U at position 400 being paired in some cases and unpaired in others.
FIG. 9.
Model for the mechanism of 48S ribosomal preinitiation complex formation on type 3 IRES elements. (A) A schematic representation of type 3 IRES structures with either less-structured RNA (left) or stable stem-loop RNA structures (right) near the initiator AUG codon. (B) In the absence of stable RNA secondary structures, a single 40S ribosomal subunit binds multiple noncontiguous sites on wt pestivirus and HCV RNAs, including near the initiator AUG codon (11, 13, 27, 29, 30, 32). (C) An eIF2-GTP-tRNA ternary complex binds to a binary (40S RNA) complex forming a 48S complex at the initiation codon on wt RNAs but not on RNAs with stable RNA secondary structure(s) near the initiating AUG codon.
Structure-probing experiments of the RNA region surrounding the initiator AUG in wt MetNS3, ad3.7, and ad3.10 were performed by mapping the RNase T1- and RNase V1-sensitive bases. Although our results were not conclusive, the data did suggest enhanced RNase T1 sensitivity at nt 402 in the ad3.7 and ad3.10 RNAs, consistent with the hypothesis that these RNAs may contain less structure downstream of the initiating methionine (data not shown).
To determine if the RNAs with predicted less-stable secondary structures would replicate autonomously, the sg MetNS3 RNAs were synthesized in vitro from the engineered cDNA templates and transfected into MDBK cells, and RNA replication was measured by incorporation of 32P as described previously. Both the AU-rich RNA (ad3.7) and the RNA containing the single adaptive G400U (ad3.10) substitution replicated efficiently, as demonstrated by the accumulation of distinct 32P-labeled RNAs (Fig. 6B, lanes 8 and 10). In contrast, the single U394C change was not responsible for repressed replication since ad3.8 did not accumulate detectable levels of 32P-labeled RNA (Fig. 6B, lane 9). These results show that sg RNAs with predicted less-stable RNA secondary structures (ad3.7 and ad3.10) downstream of the AUG replicate autonomously and that the G400U substitution alone is sufficient.
As expected, ad3.7 and ad3.10 showed similar levels of NS3 protein synthesis in vitro (55 and 65% of the level of ad3.4 NS3, respectively), whereas ad3.8 NS3 levels were consistently less than 30% of that of ad3.4 (data not shown). These data show that sg RNAs that translate efficiently in vitro replicate efficiently and that reduced translation in vitro correlates with undetectable levels of RNA replication in transfected MDBK cells.
Adapted MetNS3, but not wt MetNS3, binds 40S ribosomes and forms 48S ribosomal initiation complexes.
Translation initiation of BVDV, CSFV, and the related HCV has been evaluated in vitro using purified translation components and in vitro-transcribed RNAs (27, 29). Therefore, to analyze the efficiency of IRES-mediated translation initiation in the wt MetNS3, ad3.7 (AU-rich), and ad3.10 (G400U) RNAs, we reconstituted this process in vitro to the stage of 48S initiation complex formation. The ribosomal complexes were analyzed by toeprinting, which involves primer extension by reverse transcriptase on a template RNA to which a protein or ribosomal complex is bound. Synthesis of the cDNA is arrested at the leading edge of the complex, yielding toeprints that can be accurately mapped on sequencing gels.
The BVDV IRES is highly structured (see Fig. 8), and as a result, reverse transcriptase arrest occurs at several sites even in the absence of translation components. Three of these sites occur on wt genomic BVDV mRNA (27) and also on wt MetNS3 and adapted variant RNAs (at A320, G327, and U352; Fig. 7, lanes 1, 5, and 9). Prominent reverse transcriptase stops were also detected at GCCG (nt 395 to 398) on wt MetNS3 and ad3.10 RNAs (Fig. 7, lanes 1 and 9), indicative of a stable secondary structure at or immediately upstream of these residues. Corresponding stops on ad3.7 RNA were less intense, indicating that the ad3.7 mRNA is less structured downstream of the initiation codon (Fig. 7, lane 5).
FIG. 8.
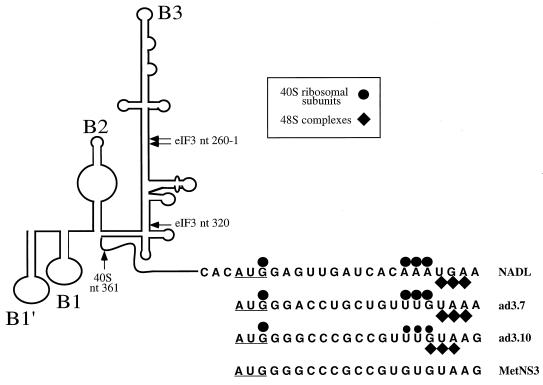
Schematic representation of model secondary and tertiary RNA structures of BVDV as described previously by Pestova et al. (27). The NADL nucleotide sequences are of nt 383 to 406 of the BVDV 5′ NTR-Npro sequence (the initiation AUG codon is underlined), and nt 386 to 406 are shown for MetNS3, ad3.7, and ad3.10. Primer extension toeprints of the MetNS3 RNAs caused by binding of 40S ribosomes and 48S preinitiation complexes are shown. Smaller symbols indicate less intense toeprints observed for the RNA.
FIG. 7.
Toeprint analysis of 40S ribosomal and 43S preinitiation complex binding to the BVDV IRES. Ribosomal complexes were assembled under standard reaction conditions on BVDV mRNAs and in the presence of translation components as indicated and were analyzed by primer extension. Full-length cDNA (E) and other cDNA products terminated at sites indicated on the right. The sequence GCCG395-8 refers to nt 395 to 398 in MetNS3 mRNA and to equivalent residues in ad3.7 and ad3.10 mRNA.
The BVDV IRES is able to bind 40S ribosomal subunits in the absence of initiation factors to form stable binary complexes that result in the appearance of strong toeprints at U361 in the pseudoknot, at G388, and at A400, A401, and A402, at positions +15 to 17 relative to the initiation codon. These noncontiguous stops indicate that ribosomal binding to the BVDV IRES is stabilized by multiple interactions, and the appearance of toeprints downstream of the initiation codon indicates that the initiation codon and flanking residues are placed in the mRNA-binding cleft of the 40S subunit. A comparable stop at U361 appeared on wt MetNS3, ad3.7, and ad3.10 RNAs in the presence of 40S subunits, but stops at G388 appeared only on ad3.7 and ad3.10 RNAs, with strong +15 to 17 stops appearing only on ad3.7 RNA (Fig. 7, lanes 2, 6, and 10). Significantly weaker stops appeared at the +15 to 17 position on ad3.10 RNA and were not detected on MetNS3 RNA. These data show that the wt MetNS3 RNA has defects that render it unable to bind to the 40S subunit in the same manner as wt BVDV RNA and that the weaker toeprints for ad3.10 indicate that the 40S subunit binds it in the vicinity of the initiation codon with less affinity than ad3.7.
Assembly of 48S ribosomal initiation complexes on the BVDV IRES requires binding of 43S preinitiation complexes to the mRNA. For BVDV, a 43S complex composed of only 40S subunits, eIF2, GMP-PNP, or GTP and Met-tRNAiMet is sufficient for this interaction. GMP-PNP is a nonhydrolyzable GTP analog that causes 48S complexes to accumulate. Addition of eIF2–GMP-PNP–tRNA to assembly reaction mixtures containing BVDV mRNA and 40S subunits resulted in a reduction in the intensity of toeprints at positions 400 to 402 (+15 to 17) and the appearance of prominent new toeprints, UGA, at positions 403 to 405 (+18 to 20) (27). These coordinated changes in the pattern of toeprints at the leading edge of the 40S subunit are likely to be due to codon-anticodon base pairing, accompanied by a small conformational change in the BVDV mRNA at and downstream of the initiation codon. An identical pattern of change was detected when eIF2–GMP-PNP–tRNA was included with 40S subunits in assembly reaction mixtures on ad3.7. Toeprints caused by bound 43S complexes were weakly detectable on ad3.10 RNA, albeit at +17 to 19 positions, but were not apparent on wt MetNS3 RNA (Fig. 7, lanes 3, 7, and 11). These observations suggest that the defect in the ability of ad3.10 RNA, and particularly that of wt MetNS3 RNA, to bind to 40S subunits impairs or completely abrogates the ability to form 48S complexes.
The presence of eIF3 in reconstituted assembly reaction mixtures resulted in the appearance of toeprints at A260C261 and A320 on wt BVDV RNA caused by direct and specific binding of this factor to the apical half of IRES domain III (27). An identical pattern of toeprints was detected when wt MetNS3, ad3.7, and ad3.10 RNAs were incubated with eIF3 (Fig. 7, lanes 4, 8, and 12). These results show that the defect in wt MetNS3 that impairs its activity in mediating translation in rabbit reticulocyte lysates (Fig. 4), binding 40S subunits, and assembling of 48S complexes in vitro (Fig. 7, lanes 2 and 3) did not significantly affect IRES structure, as evidenced by normal eIF3 binding. A summary of the toeprinting results is shown schematically in Fig. 8. Taken together, these results indicate that the reduced translation activity of wt MetNS3 can be attributed to impaired 40S binding, such that the initiation codon and flanking residues are not placed in the 40S mRNA-binding cleft.
DISCUSSION
In this study, we created an sg NADL RNA (MetNS3) based on the genome structure of the cp CSFV isolates, in which the 5′ NTR and initiating AUG codon were fused to the N terminus of NS3 (Fig. 1; 21, 23). In contrast to earlier reports in the literature for sg CSFV RNA of similar structure, neither replication nor induction of CPE was observed for sg NADL MetNS3. However, following serial passage in the presence of ncp helper virus, detectable sg RNA replication and CPE were detected (Fig. 2). These results suggested that perhaps the Npro sequence was essential, similar to reports for the BVDV DI9-like RNAs (5, 39). However, efficient replication of the reconstructed RNAs, ad3.1, ad3.2, ad3.4, and ad3.5 (Fig. 3A and B, lanes 3 to 6), showed that a specific Npro nucleotide or amino acid sequence was not required. Furthermore, comparison of the Npro and NS3 sequences over the first 114 nt (38 aa) revealed very little homology at the nucleotide (29 to 31% identity) or amino acid (8 to 10.5% identity) level.
The combined results of reduced wt MetNS3 translation in vitro (Fig. 4, lane 3) and autonomous replication of the 5′ NTR chimera RNA, chm MetNS3 (Fig. 5B, lane 4), suggested that the sequences downstream of the initiating AUG codon were directly influencing BVDV IRES-directed translation initiation and not RNA synthesis. This conclusion is not unprecedented since translation initiation directed by the BVDV or closely related HCV IRES elements has been reported to require downstream sequences within the 5′ portion of the ORFs (6, 11, 17, 30). In contrast, others have reported efficient HCV IRES-directed translation in the absence of HCV coding sequences (32, 42, 43). Honda et al. (10) suggested that the conflicting reports for HCV IRES-mediated translation were due to a stable RNA secondary structure near the initiating AUG codon, because mutations which increased the stability of the stem-loop surrounding the HCV initiator AUG codon were inversely correlated with translation efficiency.
In support of the latter ideas, we observed that sg RNAs (ad3.1, ad3.2, ad3.4, ad3.5, ad3.7, and ad3.10) that were predicted to have less-stable RNA secondary structures near the AUG codon were able to translate (Fig. 4, lanes 4 to 7, and data not shown) and replicate efficiently (Fig. 3, lanes 3 to 6; Fig. 6, lanes 3 to 6, 8, and 10). In contrast, sg RNAs (wt MetNS3 and ad3.8) with secondary structures predicted to be more stable were translated poorly (Fig. 4, lane 3, and data not shown) and did not replicate to detectable levels (Fig. 3B, lane 2; Fig. 6B, lanes 2 and 9). Furthermore, enhanced RNase T1 cleavage at nt 402 was observed in the replication-competent sg RNAs of ad3.7 and ad3.10, with the prevalent G400U substitution, but not in wt MetNS3 RNA. These data suggest that the presence of a U at nt 400 may promote formation of an RNase T1-accessible, single-stranded RNA in this region. The inability of ad3.6 to replicate to detectable levels, even though it was translated efficiently (Fig. 4, lane 8), may have been due to the influence of the additional amino acid mutation on NS3 function (R51S; Fig. 3A).
Our data do not preclude the possibility that additional RNA-protein interactions modulate BVDV-IRES-mediated translation initiation in vivo. However, the inability of the 40S ribosomal subunits and 43S complexes to bind wt MetNS3 RNA in vitro provides strong evidence that the wt MetNS3 RNA inhibits translation initiation through an intrinsic property of the RNA (Fig. 7, lanes 2 and 3). Furthermore, base substitutions which are predicted to decrease the thermodynamic stability of RNA structures within this region allowed 40S binding and subsequent 48S complex formation, albeit at different levels for ad3.7 and ad3.10 (Fig. 7, compare lanes 6 and 7 with 10 and 11). Although the ad3.10 RNA was significantly less efficient at binding 40S ribosomal subunits and 43S ribosomal preinitiation complexes in vitro than ad3.7, the levels of sg RNA replication in cultured cells were similar (Fig. 6, lanes 8 and 10).
The binding of 40S ribosomal subunits to pestivirus and HCV IRES elements closely resembles the mechanism of 30S ribosomal subunits binding to prokaryotic mRNAs (13, 27, 29). In prokaryotes, the 30S ribosome binding site is extremely sensitive to the presence of stable RNA secondary structures, such that small increases in stability (2.4 kcal/mol) can decrease binding by an order of magnitude (reviewed in reference 8). Hence, it appears that the stability of the RNA secondary structure at the 30S ribosome binding site is quite close to the threshold for being inhibitory. Our results are consistent with the observation that increased RNA secondary structure can significantly inhibit ribosome binding and that a single G-to-U substitution predicted to decrease the stability of the RNA secondary structure permitted 40S ribosome binding.
It is also possible that either an A or C at position 400 would have allowed translation and sg RNA replication. The translation and autonomous replication of ad3.7 suggest that the identity of the primary sequence, at least at the third base position, is not as important as the resulting effects on secondary structure. This hypothesis is compatible with the observation that changes in RNA secondary structure have a far greater impact on 30S ribosome binding than the identity of the specific nucleotide substitution (8). The predicted stability of RNAs with either an A or C at nt 400 (ΔG between −6.5 and −8.0 kcal/mol) was increased relative to that of ad3.10 and ad3.7 but less than that of wt MetNS3 or ad3.8. Since our results suggested that the thermodynamic threshold (ΔG) for 40S binding to the MetNS3 RNA is between −5.6 and −11.0 kcal/mol, it will interesting to determine the effect of the A and C substitutions at position 400.
Another explanation is that within the cellular milieu, additional host factors influence translation initiation. Several studies have demonstrated interactions between La protein and polypyrimidine tract binding protein (PTB) with the related HCV IRES element (reviewed in reference 16). La protein specifically interacts with the HCV RNA in the context of the initiator methionine codon and may contribute to efficient translation initiation (1, 2). La protein is an RNA binding protein with helicase activity and, hence, may unwind double-stranded RNA in preparation for translation initiation. In addition, La has been shown to associate with small ribosomal subunits in the cytoplasm (26). The possible role(s) of PTB in HCV IRES-mediated translation initiation is somewhat less definitive but may involve PTB acting as an RNA chaperone or perhaps through additional protein-protein interactions (16). For BVDV, recent results have shown that although PTB weakly binds BVDV IRES RNA, it does not influence BVDV IRES-mediated translation in vitro (36). Therefore, it is possible that La or other, as-yet-unidentified cellular proteins influence BVDV IRES-directed translation initiation.
The importance of maintaining wt BVDV 5′ Npro sequence for efficient replication was supported by the observation that at least 28 aa of Npro were found in 11 sg RNAs isolated from four different animals (3). Our data are also consistent with the results obtained with the D19-like replicons, for which a portion of Npro was necessary for sg RNA replication (5, 39). Efficient translation initiation, including 40S and 43S binding, was associated with sg RNA replication, and since translation was not examined in the nonreplicating DI9-like RNAs, it is entirely possible that they were defective in protein synthesis.
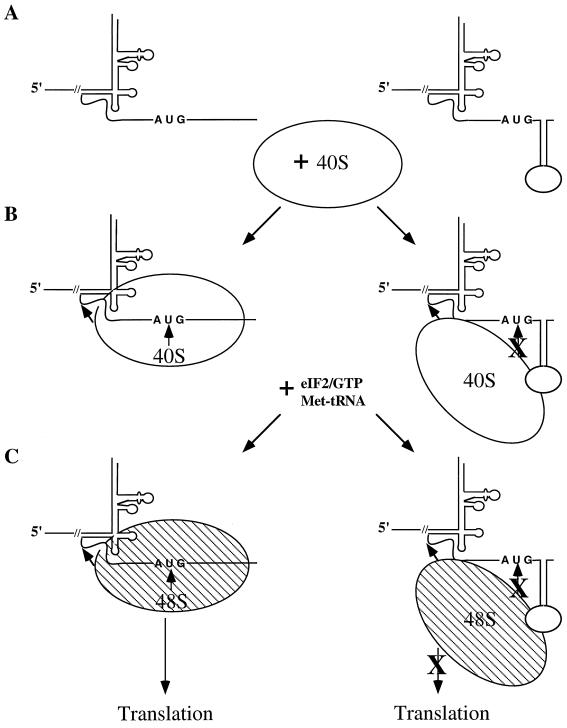
These data and those of others (10, 11, 13, 13a, 27, 29, 33) led us to propose a model for type 3 IRES-mediated translation initiation (Fig. 9). Effective binding of the 40S ribosomal subunit to the initiator AUG codon requires the surrounding RNA to be relatively unstructured (Fig. 9A, left) since stable RNA secondary structure(s) immediately downstream of the canonical IRES (Fig. 9A, stem-loop, right) appear to inhibit 40S binding (Fig. 9B). In the absence of 40S binding, 48S initiation complex formation and subsequent translation initiation are blocked (Fig. 9C). The ability to influence type 3 IRES-driven translation initiation by increasing RNA secondary structure downstream of the initiator AUG codon may have evolved as a mechanism for controlling viral gene expression and subsequent RNA replication in virus-infected cells. In addition, translation initiation could be modulated by binding of viral or host proteins or of both, such that the stability of the RNA secondary structure is either increased or decreased.
Taken together, these results also suggest that pestivirus translation and RNA replication are coupled processes in vivo. Similarly, Yu et al. (47) reported that the 5′ terminal stem loop of the BVDV IRES contains a cis-acting element involved in regulation of both translation and replication. The inapparent replication of wt MetNS3 RNA in the presence of the ncp helper virus also suggests that complementation of RNA replication does not occur in trans, at least to detectable levels. It is likely that the regulation of translation and replication in vivo is tightly regulated and that BVDV modulates various host responses by controlling viral protein and RNA levels.
In summary, using engineered sg BVDV replicons, we have shown that efficient translation initiation likely requires a less-structured RNA surrounding the AUG initiation codon. Moreover, it appears that certain levels of viral protein expression are necessary to support a complete RNA replication cycle. We have demonstrated interactions of the sg RNAs with the 40S and 43S ribosomal complexes and that the formation of the ribosomal preinitiation complexes associates with biological function in vitro.
ACKNOWLEDGMENTS
We thank Rebecca Moran for expert technical assistance and Tatyana Pestova for advice and reagents. We are also grateful to many colleagues for helpful discussions during the course of this work and to Beate Kümmerer, Keril Blight, Arash Grakoui, Holly Hanson, Jane McKeating, and Sondra Schlesinger for careful reading of the manuscript. We are indebted to Bill Goldman for valuable advice and for providing access to his Fluorimager.
T.M.M. was supported by a fellowship from the U.S. Department of Agriculture (9802328). This work was supported in part by grants from the Public Health Service (CA57973 and AI44118).
REFERENCES
- 1.Ali N, Pruijn G J, Kenan D J, Keene J D, Siddiqui A. Human La antigen is required for the hepatitis C virus internal ribosome entry site (IRES)-mediated translation. J Biol Chem. 2000;275:27531–27540. doi: 10.1074/jbc.M001487200. [DOI] [PubMed] [Google Scholar]
- 2.Ali N, Siddiqui A. The La antigen binds 5′ noncoding region of the hepatitis C virus RNA in the context of the initiator AUG codon and stimulates internal ribosome entry site-mediated translation. Proc Natl Acad Sci USA. 1997;94:2249–2254. doi: 10.1073/pnas.94.6.2249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becher P, Orlich M, Konig M, Thiel H J. Nonhomologous RNA recombination in bovine viral diarrhea virus: molecular characterization of a variety of subgenomic RNAs isolated during an outbreak of fatal mucosal disease. J Virol. 1999;73:5646–5653. doi: 10.1128/jvi.73.7.5646-5653.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Becher P, Orlich M, Kosmidou A, Konig M, Baroth M, Thiel H J. Genetic diversity of pestiviruses: identification of novel groups and implications for classification. Virology. 1999;262:64–71. doi: 10.1006/viro.1999.9872. [DOI] [PubMed] [Google Scholar]
- 5.Behrens S-E, Grassmann C W, Thiel H-J, Meyers G, Tautz N. Characterization of an autonomous subgenomic pestivirus RNA replicon. J Virol. 1998;72:2364–2372. doi: 10.1128/jvi.72.3.2364-2372.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chon S K, Perez D R, Donis R O. Genetic analysis of the internal ribosome entry segment of bovine viral diarrhea virus. Virology. 1998;251:370–382. doi: 10.1006/viro.1998.9425. [DOI] [PubMed] [Google Scholar]
- 7.Collett M S, Larson R, Belzer S K, Retzel E. Proteins encoded by bovine viral diarrhea virus: the genomic organization of a pestivirus. Virology. 1988;165:200–208. doi: 10.1016/0042-6822(88)90673-3. [DOI] [PubMed] [Google Scholar]
- 8.de Smit M H, van Duin J. Control of prokaryotic translational initiation by mRNA secondary structure. Prog Nucleic Acid Res Mol Biol. 1990;38:1–35. doi: 10.1016/s0079-6603(08)60707-2. [DOI] [PubMed] [Google Scholar]
- 9.Frolov I, McBride M S, Rice C M. cis-acting RNA elements required for replication of bovine viral diarrhea virus-hepatitis C virus 5′ nontranslated region chimeras. RNA. 1998;4:1418–1435. doi: 10.1017/s1355838298981031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Honda M, Brown E A, Lemon S M. Stability of a stem-loop involving the initiator AUG controls the efficiency of internal initiation of translation on hepatitis C virus RNA. RNA. 1996;2:955–968. [PMC free article] [PubMed] [Google Scholar]
- 11.Honda M, Ping L H, Rijnbrand R C, Amphlett E, Clarke B, Rowlands D, Lemon S M. Structural requirements for initiation of translation by internal ribosome entry within genome-length hepatitis C virus RNA. Virology. 1996;222:31–42. doi: 10.1006/viro.1996.0395. [DOI] [PubMed] [Google Scholar]
- 12.Hunt S L, Kaminski A, Jackson R J. The influence of viral coding sequences on the efficiency of internal initiation of translation of cardiovirus RNAs. Virology. 1993;197:801–807. doi: 10.1006/viro.1993.1661. [DOI] [PubMed] [Google Scholar]
- 13.Kolupaeva V G, Pestova T V, Hellen C U. An enzymatic footprinting analysis of the interaction of the 40S ribosomal subunits with the internal ribosomal entry site of hepatitis C virus. J Virol. 2000;74:6242–6250. doi: 10.1128/jvi.74.14.6242-6250.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13a.Kolupaeva V G, Pestova T V, Hellen C U T. Ribosome binding to the internal ribosome entry site of classical swine fever virus. RNA. 2000;6:1791–1807. doi: 10.1017/s1355838200000662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kümmerer B M, Stoll D, Meyers G. Bovine viral diarrhea virus strain Oregon: a novel mechanism for processing of NS2-3 based on point mutations. J Virol. 1998;72:4127–4138. doi: 10.1128/jvi.72.5.4127-4138.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kupfermann H, Thiel H-J, Dubovi E J, Meyers G. Bovine viral diarrhea virus: characterization of a cytopathogenic defective interfering particle with two internal deletions. J Virol. 1996;70:8175–8181. doi: 10.1128/jvi.70.11.8175-8181.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lemon S H, Honda M. Internal ribosome entry sites within the RNA genomes of hepatitis C virus and other flaviviruses. Semin Virol. 1997;8:274–288. [Google Scholar]
- 17.Lu H H, Wimmer E. Poliovirus chimeras replicating under the translational control of genetic elements of hepatitis C virus reveal unusual properties of the internal ribosomal entry site of hepatitis C virus. Proc Natl Acad Sci USA. 1996;93:1412–1417. doi: 10.1073/pnas.93.4.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mathews D H, Sabina J, Zuker M, Turner D H. Expanded sequence dependence of thermodynamic paramenters improves prediction of RNA secondary structure. J Mol Biol. 1999;288:911–940. doi: 10.1006/jmbi.1999.2700. [DOI] [PubMed] [Google Scholar]
- 19.Mendez E, Ruggli N, Collett M S, Rice C M. Infectious bovine viral diarrhea virus (strain NADL) RNA from stable cDNA clones: a cellular insert determines NS3 production and viral cytopathogenicity. J Virol. 1998;72:4737–4745. doi: 10.1128/jvi.72.6.4737-4745.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Meyers G, Tautz N, Becher P, Thiel H-J, Kümmerer B M. Recovery of cytopathogenic and noncytopathogenic bovine viral diarrhea viruses from cDNA constructs. J Virol. 1996;70:8606–8613. doi: 10.1128/jvi.70.12.8606-8613.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meyers G, Thiel H-J. Cytopathogenicity of classical swine fever virus caused by defective interfering particles. J Virol. 1995;69:3683–3689. doi: 10.1128/jvi.69.6.3683-3689.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meyers G, Thiel H-J. Molecular characterization of pestiviruses. Adv Virus Res. 1996;47:53–118. doi: 10.1016/s0065-3527(08)60734-4. [DOI] [PubMed] [Google Scholar]
- 23.Meyers G, Thiel H-J, Rumenapf T. Classical swine fever virus: recovery of infectious viruses from cDNA constructs and generation of recombinant cytopathogenic defective interfering particles. J Virol. 1996;70:1588–1595. doi: 10.1128/jvi.70.3.1588-1595.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mittelholzer C, Moser C, Tratschin J-D, Hoffman M A. Generation of cytopathogenic subgenomic RNA of classical swine fever virus in persistently infected porcine cell culture. Virus Res. 1997;51:125–137. doi: 10.1016/S0168-1702(97)00081-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moser C, Stettler P, Tratschin J D, Hofmann M A. Cytopathogenic and noncytopathogenic RNA replicons of classical swine fever virus. J Virol. 1999;73:7787–7794. doi: 10.1128/jvi.73.9.7787-7794.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Peek R, Pruijn G J, Van Venrooij W J. Interaction of the La (SS-B) autoantigen with small ribosomal subunits. Eur J Biochem. 1996;236:649–655. doi: 10.1111/j.1432-1033.1996.0649d.x. [DOI] [PubMed] [Google Scholar]
- 27.Pestova T V, Hellen C U. Internal initiation of translation of bovine viral diarrhea virus RNA. Virology. 1999;258:249–256. doi: 10.1006/viro.1999.9741. [DOI] [PubMed] [Google Scholar]
- 28.Pestova T V, Hellen C U, Shatsky I N. Canonical eukaryotic initiation factors determine initiation of translation by internal ribosomal entry. Mol Cell Biol. 1996;16:6859–6869. doi: 10.1128/mcb.16.12.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pestova T V, Shatsky I N, Fletcher S P, Jackson R J, Hellen C U. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reynolds J E, Kaminski A, Kettinen H J, Grace K, Clarke B E, Carroll A R, Rowlands D J, Jackson R J. Unique features of internal initiation of hepatitis C virus RNA translation. EMBO J. 1995;14:6010–6020. doi: 10.1002/j.1460-2075.1995.tb00289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rice C M. Flaviviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 1. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 931–960. [Google Scholar]
- 32.Rijnbrand R, Bredenbeek P J, van der Straaten T, Whetter L, Inchauspe G, Lemon S, Spaan W. Almost the entire 5′ non-translated region of hepatitis C virus is required for cap-independent translation. FEBS Lett. 1995;365:115–119. doi: 10.1016/0014-5793(95)00458-l. [DOI] [PubMed] [Google Scholar]
- 33.Rijnbrand R, van der Straaten T, van Rijn P A, Spaan W J, Bredenbeek P J. Internal entry of ribosomes is directed by the 5′ noncoding region of classical swine fever virus and is dependent on the presence of an RNA pseudoknot upstream of the initiation codon. J Virol. 1997;71:451–457. doi: 10.1128/jvi.71.1.451-457.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rümenapf T, Stark R, Heimann M, Thiel H-J. N-terminal protease of pestiviruses: identification of putative catalytic residues by site-directed mutagenesis. J Virol. 1998;72:2544–2547. doi: 10.1128/jvi.72.3.2544-2547.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 36.Sanderbrand S A, Tautz N, Thiel H-J, Ochs K, Beck E, Niepmann M. Translation from the internal ribosome entry site of bovine viral dirrhea virus is independent of the interaction with polypyrimidine tract-binding protein. Vet Microbiol. 2000;77:215–227. doi: 10.1016/s0378-1135(00)00278-9. [DOI] [PubMed] [Google Scholar]
- 37.Tamura J K, Warrener P, Collett M S. RNA-stimulated NTPase activity associated with the p80 protein of the pestivirus bovine viral diarrhea virus. Virology. 1993;193:1–10. doi: 10.1006/viro.1993.1097. [DOI] [PubMed] [Google Scholar]
- 38.Tautz N, Elbers K, Stoll D, Meyers G, Thiel H-J. Serine protease of pestiviruses: determination of cleavage sites. J Virol. 1997;71:5415–5422. doi: 10.1128/jvi.71.7.5415-5422.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tautz N, Harada T, Kaiser A, Rinck G, Behrens S, Thiel H J. Establishment and characterization of cytopathogenic and noncytopathogenic pestivirus replicons. J Virol. 1999;73:9422–9432. doi: 10.1128/jvi.73.11.9422-9432.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tautz N, Thiel H-J, Dubovi E J, Meyers G. Pathogenesis of mucosal disease: a cytopathogenic pestivirus generated by an internal deletion. J Virol. 1994;68:3289–3297. doi: 10.1128/jvi.68.5.3289-3297.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thiel H-J, Plagemann P G W, Moennig V. Pestiviruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 1. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1059–1073. [Google Scholar]
- 42.Tsukiyama-Kohara K, Iizuka N, Kohara M, Nomoto A. Internal ribosome entry site within hepatitis C virus RNA. J Virol. 1992;66:1476–1483. doi: 10.1128/jvi.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang C, Sarnow P, Siddiqui A. Translation of human hepatitis C virus RNA in cultured cells is mediated by an internal ribosome-binding mechanism. J Virol. 1993;67:3338–3344. doi: 10.1128/jvi.67.6.3338-3344.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Warrener P, Tamura J K, Collett M S. An RNA-stimulated NTPase activity associated with yellow fever virus NS3 protein expressed in bacteria. J Virol. 1993;67:989–996. doi: 10.1128/jvi.67.2.989-996.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wiskerchen M, Collett M S. Pestivirus gene expression: protein p80 of bovine viral diarrhea virus is a proteinase involved in polyprotein processing. Virology. 1991;184:341–350. doi: 10.1016/0042-6822(91)90850-b. [DOI] [PubMed] [Google Scholar]
- 46.Xu J, Mendez E, Caron P R, Lin C, Murcko M A, Collett M S, Rice C M. Bovine viral diarrhea virus NS3 serine proteinase: polyprotein cleavage sites, cofactor requirements, and molecular model of an enzyme essential for pestivirus replication. J Virol. 1997;71:5312–5322. doi: 10.1128/jvi.71.7.5312-5322.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yu H, Grassmann C W, Behrens S E. A stem-loop motif formed by the immediate 5′ terminus of the bovine viral diarrhea virus genome modulates translation as well as replication of the viral genome. J Virol. 2000;74:5825–5835. doi: 10.1128/jvi.74.13.5825-5835.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhong W, Gutshall L L, Del Vecchio A M. Identification and characterization of an RNA-dependent RNA polymerase activity within the nonstructural protein 5B region of bovine viral diarrhea virus. J Virol. 1998;72:9365–9369. doi: 10.1128/jvi.72.11.9365-9369.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zuker M, Mathews D H, Turner D H. Algorithms and thermodynamics for RNA secondary structure prediction: a practical guide. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1999. [Google Scholar]