Abstract
Ovine pulmonary carcinoma (OPC) is a contagious neoplasm of alveolar epithelial type II (ATII) or Clara cells caused by a type D/B chimeric retrovirus, jaagsiekte sheep retrovirus (JSRV). Here we report the isolation, sequencing, pathogenicity, and integration site of a JSRV provirus isolated from a sheep lung tumor cell line (JS7). The sequence of the virus was 93 to 99% identical to other JSRV isolates and contained all of the expected open reading frames. To produce virions and test its infectivity, the JS7 provirus (JSRVJS7) was cloned into a plasmid containing a cytomegalovirus promoter and transfected into 293T cells. After intratracheal inoculation with virions from concentrated supernatant fluid, JSRV-associated OPC lesions were found in one of four lambs, confirming that JSRVJS7 is pathogenic. In JS7-cell DNA, the viral genome was inserted in the protein-coding region for the surfactant protein A (SP-A) gene, which is highly expressed in ATII cells, in an orientation opposite to the direction of transcription of the SP-A gene. No significant transcription was detected from either the viral or the SP-A gene promoter in the JS7 cell line at passage level 170. The oncogenic significance of the JSRV proviral insertion involving the SP-A locus in the JS7 tumor cell line is unknown.
Ovine pulmonary carcinoma (OPC), also known as sheep pulmonary adenomatosis and jaagsiekte, is a contagious neoplasm of sheep that shares several features with human bronchioloalveolar carcinoma (BAC) (9, 23). Both OPC and BAC are tumors of alveolar epithelial type II (ATII) cells or nonciliated bronchiolar (Clara) cells, and both exhibit multifocal growth in the periphery of the lung in adults (4, 20, 27). A common feature of OPC and some cases of human BAC is production of copious quantities of lung fluid, an apparent secretory product of the tumor cells. BAC is only weakly associated with smoking and accounts for about 24% of human lung cancer cases (3, 38). The etiology of BAC is unknown, whereas OPC is caused by jaagsiekte sheep retrovirus (JSRV) (25, 39). The lung fluid produced in OPC cases contains JSRV, and the disease can be experimentally induced by intratracheal inoculation of lung fluid or cell-free tumor filtrate into lambs (8, 34). The time required to experimentally induce lung tumors is inversely proportional to the amount of reverse transcriptase (RT) activity in the inoculum (37). Recently it was shown that JSRV derived from a molecular proviral clone was able to cause OPC when inoculated intratracheally into newborn lambs (25).
The genome of JSRV was originally cloned as cDNA produced from virus particles in lung fluid from South African (39) and Peruvian (12) OPC cases. Analysis of the nucleotide sequence of JSRV shows a simple retroviral gag-pol-env organization with an additional alternate open reading frame, designated orfX, contained in the pol gene (1, 39). JSRV is very closely related to the enzootic nasal tumor virus which is associated with transmissible intranasal tumors of sheep and goats (6). JSRV is also closely related to a family of endogenous sheep retroviruses in the sheep genome (1, 2, 12, 13, 24). Using exogenous JSRV-specific U3 and TM probes, Southern blot hybridization revealed a single proviral integration site in the JS7 lung tumor cell line (1), suggesting this cell line as a good candidate for isolation of a JSRV provirus and characterization of its integration site.
JSRV proviral DNA, mRNA, and capsid protein are consistently detected in OPC tumor cells, but virus, whether purified from lung fluid or produced from an infectious clone, replicates only at low levels in sheep cell lines (26). The oncogenic role of JSRV in OPC is not well defined, and much of the virology, molecular biology, and pathogenesis of the JSRV-OPC system remain to be elucidated. The prolonged incubation period and slow, progressive clinical course of naturally occurring OPC suggest the possibility that virus-induced genetic changes in tumor cells may be important in the pathogenesis of the disease. The long-term goal of our work is to determine whether insertional mutagenesis by JSRV plays a role in the pathogenesis of naturally occurring OPC by determining whether shared integration sites exist, and if so, whether host genes are activated, rearranged, or deleted in tumor cells. An additional objective of the present study was to obtain a full-length JSRV proviral clone for subsequent analysis and pathogenicity studies. We report here the cloning of an infectious and pathogenic JSRV provirus from the JS7 cell line derived from an OPC case. This cell line contained a single proviral insertion in the gene encoding pulmonary surfactant protein A (SP-A). The SP-A gene is highly expressed in ATII cells and Clara cells, and its protein is a major component of pulmonary surfactant, which is produced in abundance in OPC-affected sheep.
MATERIALS AND METHODS
Cell culture.
The JS7 line was derived from OPC tumor tissue obtained from an adult sheep with naturally occurring disease using a three-step procedure described previously (16). The cell line 293T has also been described (26). Cells were cultured in a humidified 5% CO2 incubator in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum (Sigma), 100 U of penicillin per ml, and 100 μg of streptomycin per ml (DMEM).
Immunoblotting.
Protein immunoblot analysis of ovine lung fluid and supernatant from OPC cell lines followed the procedure of Sharp and Herring (35) using goat antiserum to Mason-Pfizer monkey virus p27 and iodinated rabbit anti-sheep F(ab)2 to detect the 26-kDa JSRV capsid antigen (CA). Immunoblot analysis of concentrated JSRVJS7 virus from transfected 293T cells was performed using rabbit antiserum to JSRV CA as previously described (22).
Electron microscopy.
Cell monolayers between passages 6 and 37 were fixed in situ for 10 min with 1% glutaraldehyde in 0.1 M phosphate buffer (pH 7.2) before physical detachment and centrifugation to pellet the cells. The cell pellet was fixed for a further 2 h in 3% glutaraldehyde and postfixed in 1% osmium tetroxide. Ultrathin sections were cut and stained with uranyl acetate and lead citrate.
Library construction.
Genomic DNA (100 μg) from JS7 cells (passage 170) was partially digested with 0.85 × 10−3 U of restriction endonuclease Sau3A per μg for 30 min at 37°C to generate fragments between 12 and 23 kb; these were ligated to BamHI-digested arms of the lambda BlueStar vector (Novagen). The library was generated as recommended by the manufacturer. The final titer of the JS7 library before amplification was 1.35 × 106 PFU/μg. After amplification the titer was calculated to be 4.7 × 109 PFU/μg.
For screening, phage (9.9 × 104 PFU) were adsorbed to ER1647 host cells and then plated. Once plaques reached 1 to 2 mm, they were transferred to duplicate nitrocellulose membranes, denatured (1.5 M NaCl, 0.5 M NaOH), neutralized (1.5 M NaCl, 0.5 M Tris-HCl [pH 8.0]), and dried in an 80°C vacuum oven for 15 min. Membranes were prehybridized in a solution containing 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 5× Denhardt's reagent, 0.5% sodium dodecyl sulfate (SDS), 100 μg of fragmented salmon sperm DNA per ml, and 50% deionized formamide at 42°C overnight. [32P]αdCTP-labeled probes were produced using PCR (33). Primers for the amplification of U3 were JB24 (AAGAATTTTTAAAAGCTCTTAAGG) and JB25R (ACAATGCTATATTTATAAAGTACA). Primers for the amplification of TM were JB26 (TGGAAAACCCTGATCGGTCTAGGA) and JB27R (TAGTTCTATATTTCATATGTAGCA). PCR conditions for generation of probes were the following: 1 ng of template, 1× PCR buffer, 200 μM dGTA mix, 300 nmol of each primer, 2.5 U of Taq polymerase (Sigma), and 50 μCi of [α-32P]dCTP (6,000 Ci/mmol; Amersham). Amplification cycles consisted of 94°C for 1 min (1 cycle); 94°C for 30 s, 50°C for 30 s, 72°C for 30 s (30 cycles); and 72°C for 5 min (1 cycle). Probes were purified through an Elutip (Schleicher and Schuell), boiled, and added to the membranes. Membranes were washed twice for a total of 30 min at low stringency (2× SSC, 0.1% SDS at 25°C), once at medium stringency (1× SSC, 0.1% SDS at 25°C) for 15 min, and once at medium stringency at 37°C for 30 min. The membranes were exposed to Kodak X-Omat film overnight at −70°C. Positive plaques were picked and rescreened, and the phage DNA was isolated (Qiagen).
Sequence generation and analysis.
The lambda JSRVJS7 clone was sequenced in the forward and reverse directions using an ABI 377 automated sequencer. Sequence data was assembled and analyzed using Lasergene (DNASTAR, Inc.). Other JSRV and endogenous sheep retrovirus sequences used in comparative analysis were obtained from GenBank.
Plasmids and virus production.
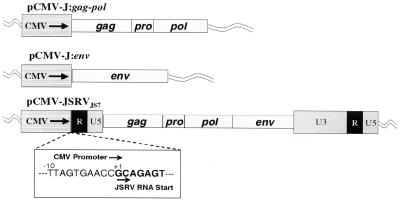
All plasmid propagation, DNA ligation, and PCR amplification was performed as previously described (33). PCR amplification cycles consisted of 94°C for 2 min (1 cycle); 94°C for 30 s, 55°C for 30 s, 72°C for 45 s (30 cycles); and 72°C for 5 min (1 cycle). The full-length JSRVJS7 proviral genome was cloned into the vector pCMV-Script (Stratagene) by replacing the upstream U3 region with the human cytomegalovirus (CMV) immediate-early promoter contained within the pCMV-Script vector while maintaining the native start site of transcription for the virus. This was accomplished by PCR amplification of region 232 to 588 of pCMV-Script with primers TA-1F (AGTGTATCATATGCCAAGTAC) and TA-2R (CTCTTCGTGCGGTTCACTAAACCAGCTCTG), creating a PCR product containing a 5′ NdeI site and a 3′ Eam1104I site. Region 3820 to 4185 in JSRVJS7 was amplified with primers TA-3F (CTCTTCAGCAGAGTATCAGCCATTTTGGTC) and TA-4R (GCGGCCGCAAGAAAATTAATTAATTTGGG), which created a PCR product with a 5′ Eam1104I site and a 3′-internal PacI site. A DNA restriction fragment spanning region 4171 to 11392 of JSRVJS7 was created by digesting the provirus clone with the restriction enzymes PacI and NotI. Following digestion and purification of each PCR product and of the DNA restriction fragment, a three-way ligation was performed to create the full-length CMV-driven provirus, pCMV-JSRVJS7. Plasmid pCMV-J:gag-pol was created by amplification of the region 4084 to 111285 using primers TA-9F (TCTGAGCTCATGGGACAAACGCATAGTCGT) and TA-10R (CTGCGGCCGCGGTATAATGCGTCCGAATTT), creating a PCR product with a 5′ SacI site and a 3′-internal PacI site. A DNA restriction fragment spanning region 4171 to 9090 of JSRVJS7 was created by digesting the provirus clone with the restriction enzymes PacI and BamHI. The 3′ end of the gag-pol gene was then created by PCR amplification of the region 9070 to 9268 in the provirus with primer TA-11F (GATGGAAGGATCCATTTACGA) and TA-12R (TGTGGTACCTCACTCGTGGGCTCGCTCAGC), creating a PCR product with a 5′ BamHI site and a 3′ KpnI site. A three-way ligation into the corresponding sites in the pCMV-Script vector was then performed. Plasmid pCMV-J:env was produced by PCR amplification of the region 9165 to 11012 using primers TA-13F (TGTGAGCTCATGCCGAAGCGCCGCGCTGGA) and TA-14R (TGTGGTACCTCACGGGTCGTCCCCCGCATC), creating a PCR product with a 5′ SacI site and a 3′ KpnI site, which was also ligated into the pCMV-Script vector.
Transfection of pCMV-JSRVJS7, pCMV-J:gag-pol, and pCMV-J:env was performed using LipofectAmine (Life Technologies) as described by the manufacturer. Briefly, 293T cells were plated in 60-mm tissue culture plates and then incubated for ∼15 h or until the cells reached 70% confluence. Two micrograms of each plasmid was then mixed with 18 μl of LipofectAmine in 400 μl of serum-free DMEM (SFM) and then subjected to a 40-min incubation at room temperature; 1.6 ml of SFM was then added to the transfection mixture. Cells were washed with SFM twice to remove any traces of serum; this was followed by addition of transfection mixture to the cells. Cells were incubated for 5 h; this was followed by addition of 2 ml of DMEM containing 20% fetal bovine serum. Cells were then incubated for 15 h, and then the transfection mixture was removed and replaced with DMEM. Virus was harvested at 24, 48, and 72 h posttransfection. JSRVJS7 virus was concentrated ∼20-fold using the Minitan tangential-flow ultrafiltration system (Millipore) with a 300-kDa molecular size exclusion filter as described by the manufacturer.
RT assay.
RT activity was quantitated by measuring the amount of bromodeoxyuridine (BrdU) incorporation into an immobilized RNA template as described by the manufacturer (Lenti-RT; Cavidi Tech AB). BrdU incorporation was quantified by the binding of a BrdU-specific antibody conjugated to alkaline phosphatase (AP) followed by colorimetric measurement of AP activity. In this assay, AP activity is proportional to the RT activity in the sample.
In vivo infections.
Four specific-pathogen-free newborn lambs, as described previously (25), were each inoculated intratracheally with 5 ml of concentrated supernatant collected from 293T cells transiently transfected with pCMV-JSRVJS7. Two lambs were inoculated with phosphate-buffered saline alone and were used as negative controls, whereas two lambs inoculated with concentrated lung fluid from an OPC case were used as positive controls. The lambs were killed between 18 and 20 weeks after inoculation, depending on development of clinical evidence of disease, and the lungs were examined at necropsy.
Histological examination and immunohistochemistry.
Four sections (4 to 6 μm thick) of each lung were stained with hematoxylin and eosin and examined by light microscopy. Sections were also examined for the presence of JSRV CA protein by immunohistochemistry as described previously (22), except that an antigen retrieval step (microwave treatment at 800 W twice for 7 min) was included. OPC tumor tissue was used as a positive control.
Nucleotide sequence accession numbers.
The nucleotide sequence of JSRVJS7 and the proviral integration site flanking sequences containing the SP-A gene has been deposited in GenBank under accession number AF357971.
RESULTS
Derivation of the JS7 cell line.
As described in Materials and Methods, the JS7 cell line was derived by culture of tumor cells from a naturally occurring case of OPC in Scotland. JS7 cells had epithelial morphology (Fig. 1A) and features of ATII cells, including surface microvilli, tonofilaments, desmosomes, intracytoplasmic glycogen, and lamellar body-like structures (Fig. 1B); with maintenance in culture, some features of ATII differentiation were lost. JSRV CA was detected by immunoblotting in culture supernatants of JS7 cells during early passages (Fig. 1C) but could not be demonstrated after passage 11 (data not shown). Replication of JSRV could not be induced in high-passage JS7 cells, but intratracheal inoculation of the cells at passage 137 into lambs induced JSRV-associated OPC of recipient karyotype (15).
FIG. 1.
Features of the JS7 cell line. (A) Confluent monolayer showing the polygonal, epithelial appearance of JS7 cells and presence of perinuclear granules (arrows) (bar = 20 μm). (B) Transmission electron micrograph showing ultrastructural features of JS7 cells including surface microvilli (mv), desmosomes (arrows), and intracytoplasmic lamellar body-like structures (arrowheads) (bar = 10 μm). (C) Protein immunoblot analysis using goat antiserum to Mason-Pfizer monkey virus p27 and iodinated rabbit anti-sheep F(ab)2 to detect the 26-kDa JSRV CA (arrowhead) in lung fluid from a natural case of OPC (lane a) and in supernatants from a JS7 cell culture, passage 8 (lane b). Immunoglobulin light chain (faint fuzzy band above the JSRV CA band in lung fluid [lane a]) is absent in preparations from the JS7 culture supernatant.
Cloning of the JSRVJS7 provirus.
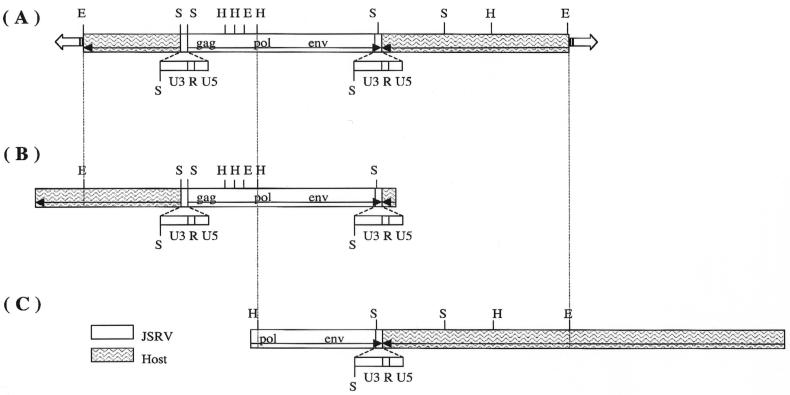
Since the JS7 cell line had a single integration site, it was used as source material for cloning the JSRV provirus (1). A genomic library was constructed from JS7 cell DNA in a bacteriophage lambda vector. This library was screened by plaque hybridization with JSRV-specific U3 and TM probes (2). Two lambda clones that hybridized to the probes were identified and purified. One of these, clone 2-1 (Fig. 2B), contained a complete JSRV provirus, and the other, clone 5-1 (Fig. 2C), contained only the 3′ half of the provirus. Southern blot analysis demonstrated that they are independent clones of the same JSRV provirus locus (see Fig. 2A). The provirus is referred to as JSRVJS7.
FIG. 2.
Alignment of JSRVJS7 provirus restriction maps. (A) Results of Southern blot analysis of JS7 genomic DNA digested with restriction endonucleases EcoRI (E), HindIII (H), and SacI (S). The membrane was hybridized with probes specific to the U3 and TM regions of JSRV (1). (B) Full-length proviral lambda clone 2-1. (C) Partial-length proviral lambda clone 5-1.
The nucleotide sequence of the cloned DNA insert of the lambda phage clone 2-1, containing the complete JSRVJS7 provirus, was determined. The insert contained 3,549 bp of sheep genomic DNA downstream from the proviral long terminal repeat (LTR) and 399 bp upstream to the other proviral LTR (Fig. 2B). (Upstream and downstream refer to the orientation relative to the SP-A promoter. Thus, the upstream LTR is the 3′ LTR of the provirus and the downstream LTR is the 5′ LTR of the provirus.) The JSRVJS7 provirus is 7,841 bp in length, and the gag, pro, pol, orfX, and env reading frames are all open. The JSRVJS7 isolate is 99.2% identical to another isolate of Scottish origin, JSRV21 (25), but is only 93% identical to the South African isolate of JSRV (39). Amino acid sequence identity among the JSRV isolates in the gag, pro, and pol regions ranges between 95 and 99%. These regions are generally the most conserved in retroviruses. The predicted amino acid sequences of orfX in the two Scottish JSRV isolates are 98% identical but are only 90 to 91% identical to the African JSRV isolates (1). JSRV21 has a 5-bp deletion in U3 of the LTR relative to JSRVJS7, but no known retroviral transcription factor binding site is lost or gained due to this deletion.
Pathogenicity studies of JSRVJS7.
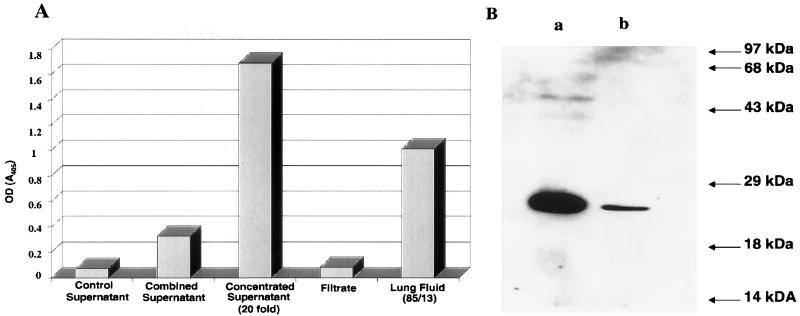
The determination of whether JSRVJS7 is capable of inducing OPC in lambs was crucial to our future studies using this clone. To investigate the pathogenicity of the JSRVJS7 proviral clone, it was necessary to produce infectious virus. Cotransfection of pCMV-JSRVJS7, pCMV-J:gag-pol, and pCMV-J:env (Fig. 3) into 293T cells resulted in transient production of JSRVJS7 virus as demonstrated by RT activity in tissue culture supernatants 24 h after transfection (data not shown). The production of viral particles was confirmed in cell culture supernatants concentrated by using a 300-kDa membrane filter. As seen in Fig. 4A, RT activity was found only in the concentrated supernatant; the filtrate contained no detectable RT, demonstrating that RT activity was associated with a particle size larger than the molecular mass of RT. Additional evidence for packaged virus included ultracentrifugation concentration of viral particles (data not shown) and Western immunoblotting analysis of concentrated JSRVJS7 showing a band of approximately 27 kDa when blots were probed with a rabbit anti-capsid polyclonal antibody (Fig. 4B).
FIG. 3.
CMV-based JSRVJS7 plasmid constructs. Schematic representation of the full-length JSRVJS7 provirus clone in which the 5′ U3 region was replaced by the CMV promoter inserted 5′ to the JSRVJS7 RNA start site. The insert shows the context of the region joining the CMV promoter with the 5′ R region of JSRVJS7. Also shown are pCMV-J:gag-pol and pCMV-J:env. Both of these constructs were designed to overexpress viral proteins by using the CMV promoter to drive transcription. Standard retrovirus notation is used. These constructs were transfected into 293T cells for production of JSRV virions.
FIG. 4.
Production of JSRV in 293T cells. (A) RT activity in supernatants of 293T cells or in lung fluid of an OPC-affected lamb detected by measuring the amount of BrdU incorporation into an immobilized RNA template (Lenti-RT; Cavidi Tech AB). Colorimetric quantitation of BrdU incorporation was achieved by the binding of a BrdU-specific antibody conjugated with alkaline phosphatase. Supernatant was prepared from 293T cells transiently transfected with the JSRV plasmids described in Fig. 3 and pooled. Combined supernatant was then concentrated ∼20-fold by ultrafiltration; the ultrafiltrate was also examined for RT activity. RT activity in lung fluid from an experimental case of OPC (85/13) was used as an indicator for acceptable levels of JSRVJS7 concentrated virus for lamb inoculations. (B) Western immunoblot analysis of concentrated JSRVJS7 virus from transfected 293T cells using rabbit antiserum to JSRV CA. Lane a represents 30 μl of the lung fluid (85/13) described above. Lane b represents 30 μl of concentrated supernatant from 293T cells. OD, optical density.
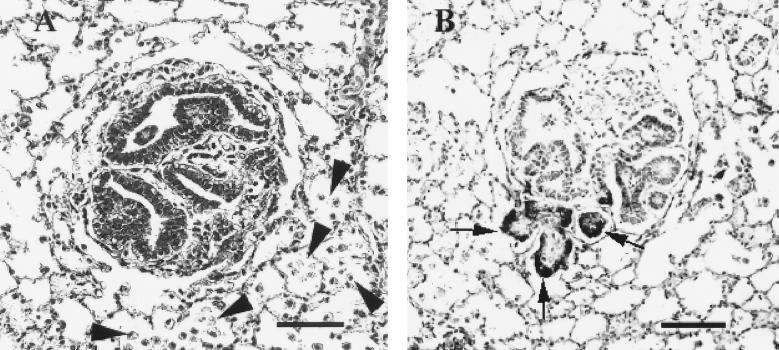
Four lambs were inoculated with concentrated stocks of JSRVJS7 produced after transfection of 293T cells. When lambs were killed at 18 to 20 weeks postinoculation and examined histologically, the lungs of one lamb had several papillary foci within alveoli and bronchioles accompanied by increased numbers of vacuolated cells resembling alveolar macrophages within adjacent alveoli, hallmark features of OPC (Fig. 5A). As a positive control, two lambs were inoculated with lung fluid from a field case of OPC, and both developed gross and histological lesions diagnostic for OPC (data not shown). Neither of the two medium-inoculated lambs developed OPC lesions. To confirm the presence of JSRV in the lesions found in the lamb inoculated with JSRVJS7, immunohistochemistry was performed using an antibody against the capsid protein of JSRV. Positive staining indicated that not only was the virus present, but transcription and translation of the viral genome were taking place at the site of the lesion (Fig. 5B).
FIG. 5.
Experimental induction of OPC by JSRVJS7. Lung tissue from a JSRVJS7-inoculated lamb was fixed in neutral formalin, sectioned at 4 to 6 μm, and stained. (A) Microscopic tumor nodule detected in lamb number 1615 (hematoxylin and eosin stain; bar = 40 μm). Note the increased numbers of alveolar macrophages in alveoli surrounding the tumor nodule (arrows). (B) JSRV antigen presence in the same tumor nodule detected by immunohistochemistry using rabbit antiserum to JSRV capsid developed using Vector Red and using Carazzi's hematoxylin as a counterstain (bar = 40 μm).
The JSRVJS7 integration site.
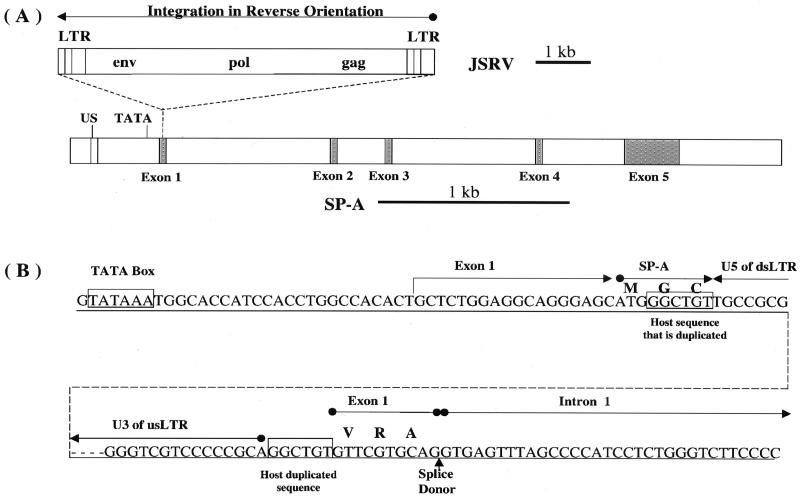
As part of an investigation of insertional mutagenesis by JSRV provirus, the JSRV integration site in the JS7 cell line was studied. A comparison of the sequences flanking the provirus with the GenBank database indicated that the virus had inserted into the sheep gene that encodes SP-A. The JSRVJS7 provirus was inserted into the promoter proximal region of the SP-A gene in reverse orientation with respect to the SP-A gene (Fig. 6). The insertion had all of the hallmarks of a typical retrovirus insertion, including a TG at the downstream end and a CA at the upstream end of the provirus, and a duplication of 6 bp of host DNA at the site of insertion (5). The 399 bp of sheep genomic sequence just upstream of the LTR showed similarity to the promoter regions of the human (17) and baboon (19) SP-A genes. There was a 26-bp region, centered about 170 bp upstream of the TATA box of the sheep SP-A promoter, which was 96% (25 of 26) identical to a 26-bp sequence in human and baboon promoters, centered about 180 bp upstream of their TATA boxes. The significance of this sequence is unknown, but its location suggests that it may be a transcription factor-binding site. The upstream LTR of the provirus was about 50 bp downstream of the putative SP-A TATA box, and transcription from the 5′ LTR of the provirus was predicted to be in the opposite direction of the transcription of the SP-A gene. Consistent with this interpretation, there were regions of significant similarity to the SP-A genes of several mammalian species within the 3,549 bases of genomic sequence just upstream of the 5′ LTR of the provirus in clone 2-1 (Fig. 2B). The SP-A genes of human, baboon, rabbit, rat, and mouse have five exons. The regions of high sequence similarity corresponded to the exons of the mammalian genes, and the regions of little or no similarity corresponded to the introns of the mammalian genes. This suggests that the exon-intron organization of the sheep SP-A gene conforms to that of other mammals.
FIG. 6.
SP-A is the site of JSRVJS7 proviral integration. (A) JSRVJS7-provirus is integrated in a reverse orientation to the 5′ untranslated region of SP-A exon 1. Both JSRVJS7 and SP-A sequence fragments are shown to scale. Ovine SP-A exons are shaded and introns are white. US is a 25-nucleotide upstream “enhancer” region which shares high homology to sequences in human and baboon. (B) The boundaries of the integration site are shown in detail. Exon 1 is predicted to begin approximately 25 nucleotides downstream of the TATA box. Integration of JSRVJS7 interrupts the proposed six-amino-acid exon 1 SP-A translation product. The dotted line represents the JSRVJS7 provirus. GGCTGT is the retroviral target sequence duplicated by the host upon integration. Amino acids are indicated above some of the codons in the nucleotide sequences as follows: M, Met; G, Gly; C, Cys; V, Val; R, Arg; A, Ala. us, upstream; ds, downstream.
The amino acid sequence of the sheep SP-A was predicted by using the sequences of cDNAs from other species (dog, GenBank accesssion no. M11769; guinea pig, U40869; human, M68519; rat, X13176; rabbit, L19387; pig, L41350; and mouse, S48768) to guide the prediction of splice sites. If translation of the sheep SP-A message begins at the first AUG codon in the second exon, a protein showing high sequence similarity to the proteins predicted from the cDNAs of other mammals would result, and it would be identical to an SP-A cDNA sequence recently reported for the sheep (28; GenBank accession No. AF076633). Although the 5′ end of the sheep SP-A gene transcript has not been definitively determined, we assume that it is approximately 25 bases downstream of the TATA box as indicated in Fig. 6B. The first AUG codon is about 23 bases downstream of the predicted 5′ end of the message and is part of the sequence AGCAUGG, which is in good agreement with the optimal consensus translation initiation sequence PuCCAUGG (29). The 6-amino-acid peptide encoded by the first exon could be joined to the rest of the SP-A to form a different N terminus if the AGGT indicated in Fig. 6B is used as the splice donor during removal of the first intron. The rat SP-A gene also has an ATG in the first exon, resulting in two translation initiation sites for SP-A. Furthermore, two isoforms of the rat SP-A arise as a consequence of differential use of alternative signal peptidase cleavage sites, which is in turn influenced by which ATG is used for translation initiation (7).
The 6-bp host sequence that was duplicated in the JSRV integration reaction begins 21 bases downstream of the predicted 5′ end of the SP-A transcript and 3 bases downstream from the first AUG in the putative mRNA. Thus, the insertion interrupts the exon 1 open reading frame of SP-A. This finding raises the question of what effect the JSRVJS7 provirus may have on the expression of the SP-A gene, and conversely what effect insertion in an antisense orientation with respect to the SP-A promoter may have on the production of viral transcripts. To address this question, total RNA was isolated from JS7 cells at passage 171 and analyzed for the presence of transcripts from the SP-A promoter and from the JSRV 5′ LTR promoter by Northern blot hybridization. JSRV DNA was used as a probe, since it should detect proviral transcripts from both viral and SP-A gene promoters. No bands were seen with the JSRV probe, although an actin mRNA band was detected when an actin probe was used (data not shown). When the SP-A gene was used as a probe, no bands were detected on the Northern blot (data not shown). More sensitive RT-PCR assays were used in an attempt to detect low levels of transcription in both the sense and antisense directions through the JSRV provirus. Weak signals were detected for both sense and antisense JSRV RNAs (data not shown). JS7 cells also were analyzed for the presence of SP-A protein by Western blotting using cross-reactive antibodies against rat SP-A (18). Although the antibodies against rat SP-A recognize the sheep antigen, sheep SP-A was not found in JS7 cells (data not shown). These results suggest that neither the SP-A promoter nor the JSRV LTR promoter is active in JS7 cells at the present passage level.
DISCUSSION
In this work, a full-length provirus, JSRVJS7, was isolated from JS7 cells, an OPC tumor cell line that had been maintained in culture for over 170 passages. Derived from a naturally occurring OPC tumor in Scotland, JSRVJS7 is an example of a type 2 JSRV geographic variant found in Europe and North America, distinguished from the type 1 JSRV genotype found in Africa (1, 2). The nucleotide sequence of JSRVJS7 is more than 99% identical to JSRV21, another Scottish isolate of recent origin (25), but is only 93% identical to the more distantly related type 1 prototype JSRV from South Africa (39). The virus has all of the expected open reading frames and replicated in JS7 cells through passage 8 but, interestingly, not after passage 11. The presence of open reading frames in the JSRVJS7 sequence, therefore, indicates that the lack of virus production at higher passages of the JS7 cell line is not due to nonsense mutations in the provirus. This finding is also consistent with the recovery of virus from tumors induced in lambs inoculated with high-passage JS7 cells (15).
Upon intratracheal inoculation of newborn lambs, the JSRVJS7 provirus clone was shown to be infectious and capable of inducing pulmonary papillary nodules, characteristic of OPC. JSRV presence in the tumor cells was confirmed by demonstrating intracellular JSRV capsid protein. The lesions were similar to those induced by clone JSRV21, a provirus which had been isolated directly from tumor DNA rather than from a cell line (25). Because both of these pathogenicity studies were performed only with animals that are known to contain at least 15 copies of endogenous sheep retroviruses closely related to JSRV (13) and because lesions were detected after a 4- to 5-month incubation period, the possibility that endogenous sheep retroviruses may contribute to JSRV pathogenicity through recombination cannot be excluded. Thorough sequence analysis of JSRV isolates recovered from animals inoculated with JSRV molecular clones will be required to investigate this possibility.
The relatively rapid onset of disease induced by JSRVJS7 is similar to the incubation period after intratracheal inoculation of newborn lambs with extracts from OPC-affected lungs (8, 31) and contrasts with naturally occurring OPC, which occurs most frequently in adult sheep (8, 32). This may reflect different oncogenic mechanisms in early and late stages of disease or the effect of different ATII cell replication rates in young lambs and adults, with consequent effects on viral replication, integration, and mutagenesis. The experimentally induced disease in young lambs occurs within weeks of exposure to JSRV, and the lesions usually consist of small adenomatous foci disseminated thoughout the pulmonary parenchyma, suggesting possible polyclonal origin and an epigenetic basis of proliferation. On the other hand, the lesions of naturally occurring OPC in adult sheep most often consist of large, occasionally fibrotic, tumor masses involving one or more lung lobes and a 10% rate of metastasis to regional lymph nodes, features more consistent with oligoclonal or monoclonal origin and more likely to be related to an insertional mutagenesis origin. We believe that different stages of OPC are related to different pathogenetic mechanisms and that OPC is a model of multistep carcinogenesis, as has been suggested for the murine leukemia system (10).
JSRV causes a pulmonary tumor consisting of ATII cells or Clara cells or their precursors. ATII cells produce pulmonary surfactant, a complex of lipids and proteins that form a film at the air-liquid interface of alveoli (11). The chief components of pulmonary surfactant are the lipids dipalmitoylphosphatidylcholine and phosphatidylglycerol and the surfactant proteins SP-A, SP-B, SP-C, and SP-D (36). A morphological hallmark of ATII cells is the lamellar body, an intracellular storage form of surfactant; lamellar bodies are frequently found in OPC tumor cells and were present in JS7 cells, particularly at low passage levels. Although leukocytes, and perhaps other cell types, are infected by JSRV (14), the virus is apparently only oncogenic in these pulmonary epithelial cell types. This may reflect the observation that JSRV gene expression, under the control of LTR promoters or enhancers, is upregulated in pulmonary epithelial cells (21). Receptor-ligand interactions, perhaps involving a recently described JSRV receptor (30; A. D. Miller, University of Washington, personal communication), or genetic events associated with JSRV integration also may be related to ATII proliferation.
The finding of JSRV integration in a gene that is highly expressed in the OPC target cell, the ATII cell, is of potential interest, but its relevance to the pathogenesis of the disease is unclear. In a Southern blot study of tumor DNA of eight sheep naturally affected by OPC using clone 2-1 (Fig. 2B) as a probe, no rearrangements were detected in the SP-A gene (data not shown); however, dilution of tumor cell DNA in the sample by leukocyte and stromal cell DNA may have reduced sensitivity of detection. Unfortunately, DNA from the original tumor source of the JS7 cell line was unavailable to determine whether the tumor was clonal with respect to the SP-A integration site. JSRV insertion into the SP-A gene also could be a fortuitous retroviral insertion into the relatively open chromatin structure of an actively expressed gene in ATII cells.
The mechanisms by which JSRV induces neoplasia of the secretory epithelium of the lower respiratory tract may involve viral protein expression or genetic changes in the principal target cells for transformation, the ATII cells. The availability of pathogenic molecular clones of JSRV will be crucial in studies to determine whether viral proteins are capable of directly transforming cells. In addition, further studies to determine whether the SP-A gene is frequently rearranged in natural tumors will be necessary to evaluate the overall significance of the SP-A integration site. Aside from the potential significance of the SP-A integration site in oncogenesis, JSRV-induced changes in the ATII cell cycle, differentiation state, or alterations of SP-A production may lead to new information about the biology of this important pneumocyte.
ACKNOWLEDGMENTS
This work was supported by grant CA 59116 from the National Cancer Institute, National Institutes of Health. Some of the work in Scotland was supported by the Scottish Executive, Rural Affairs Department. J. C. DeMartini is a member of the University of Colorado Cancer Center.
We thank Lorenzo Gonzalez for performing necropsy examinations and histopathology and Patricia Dewar for technical assistance.
REFERENCES
- 1.Bai J, Bishop J V, Carlson J O, DeMartini J C. Sequence comparison of JSRV with endogenous proviruses: envelope genotypes and a novel ORF with similarity to a G-protein coupled receptor. Virology. 1999;258:333–343. doi: 10.1006/viro.1999.9728. [DOI] [PubMed] [Google Scholar]
- 2.Bai J, Zhu R-Y, Stedman K, Cousens C, Carlson J, Sharp J M, DeMartini J C. Unique long terminal repeat U3 sequences distinguish exogenous jaagsiekte sheep retroviruses associated with ovine pulmonary carcinoma from endogenous loci in the sheep genome. J Virol. 1996;70:3159–3168. doi: 10.1128/jvi.70.5.3159-3168.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barsky S H, Cameron R, Osann K E, Tomita D, Holmes E C. Rising incidence of bronchioloalveolar lung carcinoma and its unique clinicopathologic features. Cancer. 1994;73:1163–1170. doi: 10.1002/1097-0142(19940215)73:4<1163::aid-cncr2820730407>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 4.Bonne C. Morphological resemblance of pulmonary adenomatosis (Jaagsiekte) in sheep and certain cases of cancer of the lung in man. Am J Cancer. 1939;35:491. [Google Scholar]
- 5.Coffin J M. Retroviridae and their replication. In: Fields B N, editor. Virology. New York, N.Y: Raven Press, Ltd.; 1990. pp. 1437–1499. [Google Scholar]
- 6.Cousens C, Minguijon E, Dalziel R G, Ortin A, Garcia M, Park J, Gonzalez L, Sharp J M, de las Heras M. Complete sequence of enzootic nasal tumor virus, a retrovirus associated with transmissible intranasal tumors of sheep. J Virol. 1999;73:3986–3993. doi: 10.1128/jvi.73.5.3986-3993.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Damodarasamy M, Zhang M, Dienger K, McCormack F X. Two rat surfactant protein A isoforms arise by a novel mechanism that includes alternative translation initiation. Biochemistry. 2000;39:10189–10195. doi: 10.1021/bi000448c. [DOI] [PubMed] [Google Scholar]
- 8.DeMartini J C, Rosadio R H, Sharp J M, Russell H I, Lairmore M D. Experimental coinduction of type D retrovirus-associated pulmonary carcinoma and lentivirus-associated lymphoid interstitial pneumonia in lambs. JNCI. 1987;79:167–177. [PubMed] [Google Scholar]
- 9.DeMartini J C, York D F. Retrovirus-associated neoplasms of the respiratory system of sheep and goats. Ovine pulmonary carcinoma and enzootic nasal tumor. Vet Clin N Am Food Anim Pract. 1997;13:55–70. doi: 10.1016/s0749-0720(15)30364-9. [DOI] [PubMed] [Google Scholar]
- 10.Fan H. Leukemogenesis by Moloney murine leukemia virus: a multistep process. Trends Microbiol. 1997;5:74–82. doi: 10.1016/S0966-842X(96)10076-7. [DOI] [PubMed] [Google Scholar]
- 11.Goerke J. Pulmonary surfactant: functions and molecular composition. Biochim Biophys Acta. 1998;1408:79–89. doi: 10.1016/s0925-4439(98)00060-x. [DOI] [PubMed] [Google Scholar]
- 12.Hecht S J, Carlson J O, DeMartini J C. Analysis of a type D retroviral capsid gene expressed in ovine pulmonary carcinoma and present in both affected and unaffected sheep genomes. Virology. 1994;202:480–484. doi: 10.1006/viro.1994.1366. [DOI] [PubMed] [Google Scholar]
- 13.Hecht S J, Stedman K, Carlson J O, DeMartini J C. Distribution of endogenous type B and D retroviral sequences in ungulates and other mammals. Proc Natl Acad Sci USA. 1996;93:3297–3302. doi: 10.1073/pnas.93.8.3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holland M J, Palmarini M, Garcia-Goti M, Gonzalez L, McKendrick I, de las Heras M, Sharp J M. Jaagsiekte retrovirus is widely distributed both in T and B lymphocytes and in mononuclear phagocytes of sheep with naturally and experimentally acquired pulmonary adenomatosis. J Virol. 1999;73:4004–4008. doi: 10.1128/jvi.73.5.4004-4008.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jassim F A. Identification and characterization of transformed cells in jaagsiekte, a contagious lung tumor of sheep. Ph.D. dissertation. Edinburgh, Scotland: University of Edinburgh; 1988. [Google Scholar]
- 16.Jassim F A, Sharp J M, Marinello P D. Three-step procedure for isolation of epithelial cells from the lungs of sheep with jaagsiekte. Res Vet Sci. 1987;43:407–409. [PubMed] [Google Scholar]
- 17.Kouretas D, Karinch A M, Rishi A, Melchers K, Floros J. Conservation analysis of rat and human SP-A gene identifies 5′ flanking sequences of rat SP-A that bind rat lung nuclear proteins. Exp Lung Res. 1993;19:485–503. doi: 10.3109/01902149309064359. [DOI] [PubMed] [Google Scholar]
- 18.Kuroki Y, Mason R J, Voelker D R. Pulmonary surfactant apoprotein A structure and modulation of surfactant secretion by rat alveolar type II cells. J Biol Chem. 1988;263:3388–3394. [PubMed] [Google Scholar]
- 19.Li J, Gao E, Seidner S R, Mendelson C R. Differential regulation of baboon SP-A1 and SP-A2 genes: structural and functional analysis of 5′-flanking DNA. Am J Physiol. 1998;275:L1078–L1088. doi: 10.1152/ajplung.1998.275.6.L1078. [DOI] [PubMed] [Google Scholar]
- 20.Liebow A A. Bronchiolo-alveolar carcinoma. Adv Intern Med. 1960;10:329–358. [PubMed] [Google Scholar]
- 21.Palmarini M, Datta S, Omid R, Murgia C, Fan H. The long terminal repeat of jaagsiekte sheep retrovirus is preferentially active in differentiated epithelial cells of the lungs. J Virol. 2000;74:5776–5787. doi: 10.1128/jvi.74.13.5776-5787.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palmarini M, Dewar P, de las Heras M, Inglis N F, Dalziel R G, Sharp J M. Epithelial tumour cells in the lungs of sheep with pulmonary adenomatosis are major sites of replication for jaagsiekte retrovirus. J Gen Virol. 1995;76:2731–2737. doi: 10.1099/0022-1317-76-11-2731. [DOI] [PubMed] [Google Scholar]
- 23.Palmarini M, Fan H, Sharp J M. Sheep pulmonary adenomatosis: a unique model of retrovirus-associated lung cancer. Trends Microbiol. 1997;5:478–483. doi: 10.1016/S0966-842X(97)01162-1. [DOI] [PubMed] [Google Scholar]
- 24.Palmarini M, Hallwirth C, York D, Murgia C, de Oliveira T, Spencer T, Fan H. Molecular cloning and functional analysis of three type D endogenous retroviruses of sheep reveal a different cell tropism from that of the highly related exogenous jaagsiekte sheep retrovirus. J Virol. 2000;74:8065–8076. doi: 10.1128/jvi.74.17.8065-8076.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Palmarini M, Sharp J M, de las Heras M, Fan H. Jaagsiekte sheep retrovirus is necessary and sufficient to induce a contagious lung cancer in sheep. J Virol. 1999;73:6964–6972. doi: 10.1128/jvi.73.8.6964-6972.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Palmarini M, Sharp J M, Lee C, Fan H. In vitro infection of ovine cell lines by Jaagsiekte sheep retrovirus. J Virol. 1999;73:10070–10078. doi: 10.1128/jvi.73.12.10070-10078.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Perk K, Hod I. Sheep lung carcinoma: an epidemic analogue of a human neoplasm. JNCI. 1982;69:747–750. [PubMed] [Google Scholar]
- 28.Pietschmann S M, Pison U. cDNA cloning of ovine pulmonary SP-A, SP-B, and SP-C: isolation of two different sequences for SP-B. Am J Physiol. 2000;278:L765–L778. doi: 10.1152/ajplung.2000.278.4.L765. [DOI] [PubMed] [Google Scholar]
- 29.Poirier Y, Kozak C, Jolicoeur P. Identification of a common helper provirus integration site in Abelson murine leukemia virus-induced lymphoma DNA. J Virol. 1988;62:3985–3992. doi: 10.1128/jvi.62.11.3985-3992.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rai S K, DeMartini J C, Miller A D. Retrovirus vectors bearing jaagsiekte sheep retrovirus Env transduce human cells by using a new receptor localized to chromosome 3p21.3. J Virol. 2000;74:4698–4704. doi: 10.1128/jvi.74.10.4698-4704.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosadio R H, Lairmore M D, Russell H I, DeMartini J C. Retrovirus-associated ovine pulmonary carcinoma (sheep pulmonary adenomatosis) and lymphoid interstitial pneumonia. I. Lesion development and age susceptibility. Vet Pathol. 1988;25:475–483. doi: 10.1177/030098588802500611. [DOI] [PubMed] [Google Scholar]
- 32.Rosadio R H, Sharp J M, Lairmore M D, Dahlberg J E, DeMartini J C. Lesions and retroviruses associated with naturally occurring ovine pulmonary carcinoma (sheep pulmonary adenomatosis) Vet Pathol. 1987;25:58–66. doi: 10.1177/030098588802500108. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 34.Sharp J M, Angus K, Gray E, Scott F. Rapid transmission of sheep pulmonary adenomatosis (jaagsiekte) in young lambs. Arch Virol. 1983;78:89–95. doi: 10.1007/BF01310861. [DOI] [PubMed] [Google Scholar]
- 35.Sharp J M, Herring A J. Sheep pulmonary adenomatosis: demonstration of a protein which cross reacts with the major core proteins of Mason-Pfizer monkey virus and mouse mammary tumor virus. J Gen Virol. 1983;64:2223–2227. doi: 10.1099/0022-1317-64-10-2323. [DOI] [PubMed] [Google Scholar]
- 36.Veldhuizen R, Nag K, Orgeig S, Possmayer F. The role of lipids in pulmonary surfactant. Biochim Biophys Acta. 1998;1408:90–108. doi: 10.1016/s0925-4439(98)00061-1. [DOI] [PubMed] [Google Scholar]
- 37.Verwoerd D W, Tustin R C, Payne A L. Jaagsiekte: an infectious pulmonary adenomatosis of sheep. In: Olsen R G, Krakowka S, Blackslee J R, editors. Comparative pathobiology of viral diseases. Boca Raton, Fla: CRC Press, Inc.; 1985. pp. 53–76. [Google Scholar]
- 38.Wu A H, Yu M C, Thomas D C, Pike M C, Henderson B E. Personal and family history of lung disease as risk factors for adenocarcinoma of the lung. Cancer Res. 1988;48:7279–7284. [PubMed] [Google Scholar]
- 39.York D F, Vigne R, Verwoerd D W, Querat G. Nucleotide sequence of the Jaagsiekte retrovirus, an exogenous and endogenous type D and B retrovirus of sheep and goats. J Virol. 1992;66:4930–4939. doi: 10.1128/jvi.66.8.4930-4939.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]