Abstract
Interaction between the human immunodeficiency virus type 1 (HIV-1) envelope and the relevant chemokine receptors is crucial for subsequent membrane fusion and viral entry. Although the V3 region of gp120 is known to determine the cell tropism as well as the coreceptor usage, the significance of the binding of the V3 region to the chemokine receptor has not been fully understood. To address this issue, we adopted the pseudotyped virus infection assay in which the V3 region of the T-cell line-tropic (T-tropic) NL4-3 envelope was replaced with a portion of stromal cell-derived factor 1 (SDF-1), the ligand of CXCR4. The V3 region of the NL4-3 envelope expression vector was replaced with three different stretches of SDF-1 cDNA. Expression of each chimeric envelope protein was confirmed by immunoprecipitation and Western blotting. Luciferase reporter viruses were prepared by cotransfection of the pNL4-3.Luc.E−R− vector and each chimeric envelope expression vector, and the infection assay was then carried out. We showed that pseudotyped viruses with one of the chimeric envelopes, NL4-3/SDF1-51, could infect U87.CD4.CXCR4 but not U87.CD4 or U87.CXCR4 cells and that this infection was inhibited by the ligand of CXCR4, SDF-1β, by anti-human SDF-1 antibody, or by an anti-CD4 antibody, Leu3a, in a dose-dependent manner. Furthermore, chimeric NL4-3/SDF1-51 gp120 significantly inhibited binding of labeled SDF-1 to CXCR4. It was suggested that replacement of the V3 region of the NL4-3 envelope with SDF-1 preserved the CD4-dependent infectivity of T-tropic HIV-1. These results indicate that binding between the V3 region and the relevant coreceptor is important for viral entry, whether its amino acid sequence is indigenous to the virus or not.
Human immunodeficiency virus type 1 (HIV-1) enters target cells through the interaction between the viral envelope glycoproteins and the cellular receptors, CD4 and one of the coreceptors that are members of the seven-transmembrane domain, G-protein-coupled receptor superfamily (1, 3, 10, 17, 18, 20, 22, 23, 30). The cell tropism of HIV-1 is thought to be determined at the level of viral entry by the interaction of the viral envelope with certain types of coreceptors that support either T-cell line-tropic (T-tropic) or macrophage-tropic (M-tropic) HIV-1 infection (13). It has been widely accepted that CXCR4 and CCR5 are the major coreceptors for T- and M-tropic HIV-1 strains, respectively.
The viral envelope can be envisioned as a fusogenic apparatus, catalyzing pH-independent fusion between the viral membrane displaying the gp120-gp41 complex and the target cell membrane displaying CD4 plus one of the coreceptors (2, 46). Numerous studies have demonstrated that CD4 binding induces a conformational change in gp120 that exposes, creates, or stabilizes the coreceptor-binding determinants (42, 47, 54). The gp120 molecule contains five variable loops designated V1 to V5 interspersed with five relatively conserved regions designated C1 to C5 (54). It has been speculated that subsequent to its binding to the CD4 molecule, gp120 changes its conformation and exposes the cryptic V3 loop together with the V1/V2 loop embedded in the gp120 molecule (39, 49, 53). Evidence has indicated that the V3 region contains a critical determinant of envelope fusogenicity and tropism (2, 11, 45). Although it is known that the V3 region functions not alone but rather in concert with other gp120 regions, including V1, V2, and C4, binding experiments using mutant gp120 molecules with a deletion at the V3 region or anti-V3 loop neutralizing antibodies have strongly suggested that the V3 region is crucial for interaction with the relevant coreceptor (52). In accordance with this, we previously reported that T-tropic HIV-1-derived V3 loop peptides directly bind to CXCR4 and inhibit T-tropic HIV-1 infection (40). These results imply that the binding capability of the V3 region to the relevant coreceptor rather than its indigenous amino acid sequence is important for entry of HIV-1 into target cells. Thus, it remains to be tested whether a nonviral amino acid sequence with an appropriate binding affinity to the relevant coreceptor can replace the V3 region without losing viral infectivity.
In the present study, to evaluate the significance of binding between the V3 region and the relevant coreceptor in viral entry, we prepared pseudotyped virus whose V3 region of the T-tropic NL4-3 envelope was replaced with three different fragments of stromal cell-derived factor 1 (SDF-1), the ligand of CXCR4, and carried out the pseudotyped virus infection assay. Here, we show that pseudotyped HIV-1 with one of these chimeric envelopes can infect CD4+ CXCR4+ target cells.
MATERIALS AND METHODS
Cells and culture conditions.
HEK293T, originally referred to as 293tsA1609neo, is a human embryonic kidney cell line and was cultured in Dulbecco's modified Eagle's medium (DMEM) (Life Technologies, Inc., Rockville, Md.) supplemented with 10% fetal calf serum (FCS) (Life Technologies, Inc.) and penicillin-streptomycin-glutamine (Life Technologies, Inc.) (24, 36). U87MG is a human astroglial cell line (38). Three stable transfectants of U87MG, U87.CD4 expressing CD4, U87.CD4.CXCR4 expressing CD4 as well as CXCR4, and U87.CD4.CCR5 expressing CD4 as well as CCR5, were obtained from the NIH AIDS Research and Reference Reagent Program (Rockville, Md.) (4) and cultured in DMEM with 15% FCS. U87.CXCR4 and U87.CCR5 cells were generated by transfection of CXCR4 and CCR5 expression vectors into U87MG cells in our laboratory.
Vectors.
The luciferase reporter HIV-1 clone pNL4-3.Luc.E−R− (8, 12, 25) provided by the NIH AIDS Research and Reference Reagent Program was used for the reporter gene assay of pseudotyped virus infection. pNL4-3.Luc.E−R− consists of pNL4-3 with the gene for the luciferase reporter fused in frame to NotI and XhoI sites at the 5′ end of the nef coding region. In addition, a frameshift was introduced near the 5′ end of env to block production of gp160 and to limit the virus to a single round of replication. We also used the enhanced green fluorescence protein (EGFP) as a reporter gene (14, 35). The coding region of EGFP cDNA was amplified by PCR using primers containing a 5′ NotI and a 3′ XhoI site from the pEGFP-C3 vector (Clontech, Palo Alto, Calif.). The PCR product was then cleaved with NotI and XhoI and inserted into pNL4-3.Luc.E−R− at these sites. This resulted in a replacement of the luciferase reporter gene with EGFP (pNL4-3.EGFP.E−R−).
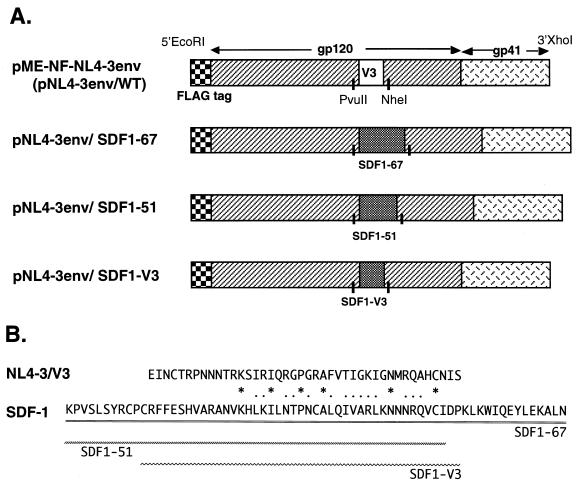
We first constructed a T-tropic HIV-1 envelope expression vector, pME-NF-NL4-3env (pNL4-3env) in which the NL4-3 envelope gene with an amino-terminal FLAG tag was inserted into pME18S. We also constructed pME-NF-YU2env (pYU2env) using the YU2 envelope gene taken from the pYU2 vector, an M-tropic HIV-1 infectious molecular clone (28, 29). The fragment encoding the V3 region, 42 amino acids (EINCTRPNNNTRKSIRIQRGPGRAFVTIGKIGNMRQAHCNIS) in the PvuII-NheI fragment of the NL4-3 envelope, was replaced with a fragment of SDF-1 cDNA encoding the first 51 amino acid residues, the full-length protein (67 amino acids), or a portion weakly homologous to the V3 region of HIV-1 (amino acids 11 to 53) (Fig. 1). SDF-1 cDNA fragments were amplified by PCR from HUT102 cDNA with a PvuII-tagged forward primer containing the 5′ flanking sequence of the V3 region and an NheI-tagged reverse primer containing the 3′ flanking sequence of the V3 region. These PCR products were cloned into pNL4-3env by using PvuII and NheI sites. The chimeric envelopes used in this report are depicted in Fig. 1. We also replaced the V3 region with macrophage inflammatory protein-1α (MIP-1α) cDNA encoding the full-length protein (70 amino acids) in the same way.
FIG. 1.
(A) Constructs of NL4-3 chimeric envelope expression vectors. As a WT envelope expression vector, we used pME-NF-NL4-3env with a FLAG tag at the 5′ terminal end of the NL4-3 envelope gene. The V3 region of the NL4-3 envelope was replaced with a fragment of SDF-1 by using PvuII and NheI restriction sites. (B) Amino acid sequences of the V3 region of the NL4-3 envelope (NL4-3/V3) and human SDF-1. Homology between NL4-3/V3 and SDF-1 is indicated as follows: identical, asterisks; related, dots. Two different overlapping portions of SDF-1 used for chimeric envelope expression vectors are shown with three separate lines.
Immunoprecipitation and Western blotting.
The chimeric envelope vectors were transfected into HEK293T cells with Lipofectamine (Life Technologies, Inc.). After 48 h, cells were washed once with phosphate-buffered saline (PBS) and lysed in lysis buffer (1% Triton X-100, 5 mM EDTA, 150 mM NaCl, 10 mM Tris-HCl, pH 7.6) at 4°C for 30 min with gentle mixing. Each cell lysate of HEK293T cells was subjected to immunoprecipitation by anti-FLAG monoclonal antibody (MAb) (M2) (Sigma Chemical Co., St. Louis, Mo.). The immunoprecipitates were fractionated on 7.5% polyacrylamide gels. The samples were transferred to a 0.45-μm-pore-size polyvinylidene difluoride (PVDF) membrane and immunoblotted with anti-SDF-1 polyclonal antibody (Ab) (R & D Systems, Minneapolis, Minn.), anti-gp120 (V3) MAb (MAb 902) (NIH AIDS Research and Reference Reagent Program) (9, 37), or anti-FLAG MAb (M2). Then the membrane was washed and incubated with horseradish peroxidase-conjugated secondary antibodies and developed with the enhanced chemiluminescence system according to the supplier's instructions (Amersham Pharmacia Biotech, Buckinghamshire, United Kingdom).
Purification of HIV-1 virions by sucrose density equilibrium gradients and analysis of the virion-associated proteins.
To analyze the incorporation of HIV-1 envelope protein in the virion, HIV-1 pseudotyped virions were purified by sucrose density equilibrium gradients. Briefly, virions were pelleted by centrifugation, resuspended in 1 ml of PBS, layered on top of the sucrose gradient, prepared in PBS ranging from 20 to 60%, and centrifuged for 13 h at 30,000 rpm in an SW-41Ti rotor (Beckman, Palo Alto, Calif.). Gradient fractions were collected from the top of the gradient and pelleted by centrifugation. These samples were used for analyzing protein profiles of the virion by Western blotting. Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to 0.45-μm-pore-size PVDF, and immunoblotted with anti-gp120 (conserved region) MAb (ABI, Columbia, Md.) or anti-HIV-1 p24 MAb (ZeptoMetrix, Buffalo, N.Y.).
Pseudotyped virus and luciferase assay.
Pseudotyped luciferase reporter viruses were prepared in HEK293T cells by the contransfection of pNL4-3.Luc.E−R− vector and the envelope expression vectors using the calcium phosphate method. Briefly, HEK293T cells were seeded in 100-mm-diameter culture dishes at a density of 106 cells per dish the day before the transfection. The complexes of plasmid DNA and calcium chloride were added to the cells. After changing to fresh medium the next day, pseudotyped viruses in the supernatants were harvested after 48 h of transfection. Viral supernatants were filtered with a 0.45-μm-pore-size filter unit (Millipore, Bedford, Mass.). The titers of pseudotyped viruses were measured by an enzyme-linked immunosorbent assay kit for the p24 antigen (RETRO-TEK; ZeptoMetrix). U87.CD4, U87.CXCR4, U87.CD4.CXCR4, and U87.CD4.CCR5 cells were seeded at 8 × 104 cells/well in a six-well plate 1 day before infection. An adjusted amount of viruses was added to each well. On day 3 postinfection, the cells were washed with PBS and lysed in 500 μl of passive lysis buffer (Promega, Madison, Wis.). The luciferase activity in 20 μl of the lysate was measured with a luminometer (EG & G Berthold, Bad Wildbad, Germany) using the commercially available substrate, Luciferase Assay Reagent (Promega).
Infection assay using EGFP reporter viruses.
The pseudotyped EGFP reporter viruses were prepared in HEK293T cells by the cotransfection of pNL4-3.EGFP.E−R− vector and the envelope expression vectors using the calcium phosphate method as described above. As with the luciferase reporter viruses, an equal amount of viruses was added to cells. On day 3 postinfection, the cells were harvested, fixed with PBS containing 0.2% bovine serum albumin, 0.2 mM EDTA, and 2% paraformaldehyde, and analyzed by flow cytometry using a FACScan (Becton Dickinson, San Jose, Calif.).
Binding assays.
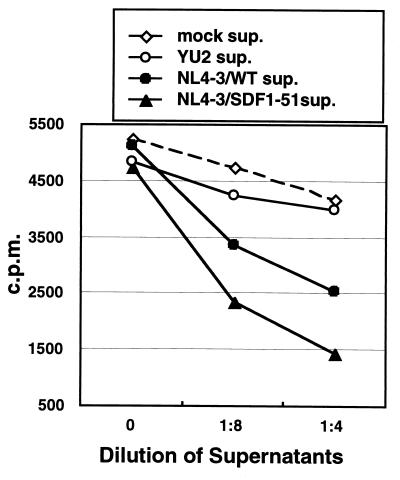
We prepared supernatants of HEK293T cells which were transiently transfected with the envelope expression vectors (pNL4-3env, pNL4-3env/SDF1-51, pYU2env, and mock control of pME18S). Soluble gp120 proteins in the supernatants were concentrated 50-fold by the Ultrafree-15 centrifugal filter device (Millipore) and were subjected to Western blot analysis to measure and compare the amount of gp120 proteins. U87.CD4.CXCR4 cells were incubated with the supernatants containing equally adjusted amounts of gp120 proteins at 4°C for 1 h. Subsequently, 125I–SDF-1α (NEN Life Science Products, Boston, Mass.) in binding buffer (DMEM containing 25 mM HEPES, 0.3% bovine serum albumin, and 0.05% NaN3) was added to the cells. After incubation at 4°C for 1 h, cells were washed twice with binding buffer and the cell-bound radioactivities were counted in a gamma counter (Abbott Laboratories, Abbott Park, Ill.).
RESULTS
Expression of NL4-3/SDF-1 chimeric envelopes.
We constructed chimeric envelope expression vectors based on T-tropic NL4-3 in which the V3 region of gp120 was replaced with portions of SDF-1, a ligand of CXCR4. We designed three kinds of cDNA fragments of SDF-1 to replace the V3 loop, namely the first 51 amino acids of SDF-1 (SDF1-51), the full-length protein (SDF1-67), and 43 amino acids (amino acids 11 to 53) which are weakly homologous to the V3 loop of NL4-3 (HIV-IIIB) (SDF1-V3) (Fig. 1).
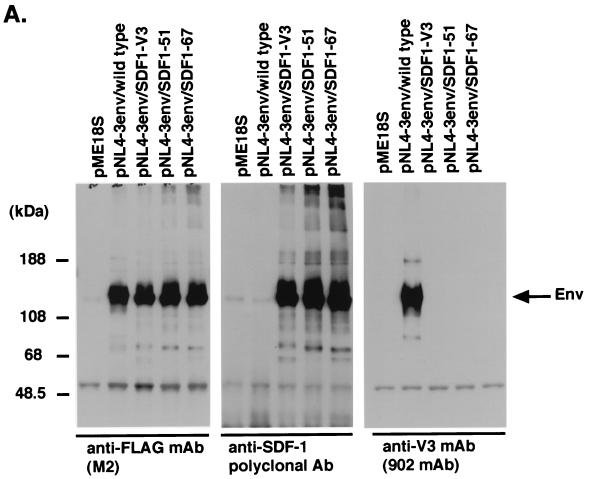
We examined whether each chimeric envelope protein was expressed by immunoprecipitation and Western blotting. Four envelope expression vectors, namely pNL4-3env/SDF1-67, pNL4-3env/SDF1-51, pNL4-3env/SDF1-V3, and pNL4-3env/wild type (WT), and pME18S as a negative control were transfected into HEK293T cells by Lipofectamine transfection. Cell lysates of transiently transfected HEK293T cells were subjected to immunoprecipitation and immunoblotting. All envelope proteins were detected by anti-FLAG MAb (M2) except for the mock control pME18S vector. Anti-SDF-1 polyclonal Ab interacted with the NL4-3/SDF-1 chimeric envelopes but not with the WT, while MAb 902 reacted with the WT but not with the NL4-3/SDF-1 chimeric envelopes (Fig. 2A).
FIG. 2.
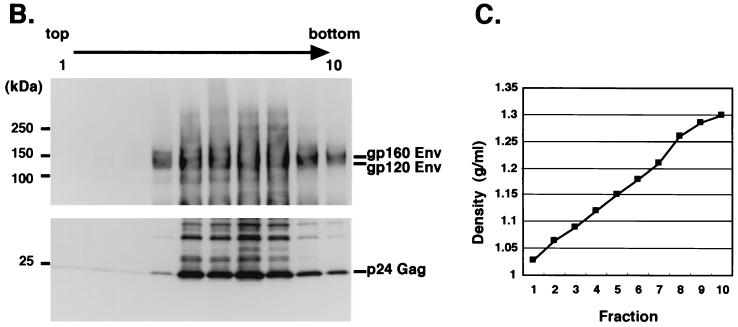
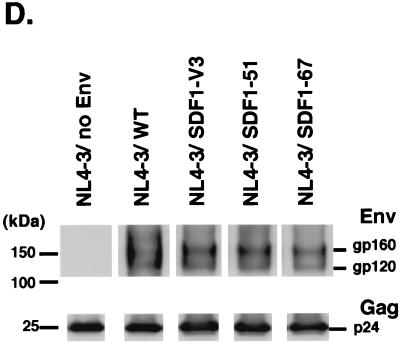
(A) Immunoprecipitation and Western blotting of NL4-3 chimeric envelope proteins. The cell lysate of HEK293T cells transfected with each envelope expression vector was subjected to immunoprecipitation by anti-FLAG MAb (M2). The immunoprecipitates were fractionated on 7.5% polyacrylamide gels. The samples were transferred to a 0.45-μm-pore-size PVDF membrane and immunoblotted with anti-SDF-1 polyclonal Ab, anti-V3 MAb (902 mAb), or anti-FLAG MAb (M2). (B) Sucrose density equilibrium gradient analysis of viral supernatants. Fractions from a sucrose density equilibrium gradient containing pseudotyped viruses (NL4-3/WT virus) were analyzed by SDS-PAGE and Western blotting using anti-gp120 (conserved region) MAb (top) and anti-HIV-1 p24 MAb (bottom). (C) Densities of each fraction are indicated. (D) Western blotting of the sixth fraction from the top of the gradient containing each pseudotyped virus.
Incorporation of the chimeric envelope proteins in the virion.
Next, we analyzed whether the chimeric envelope proteins were incorporated into pseudotyped virions. We purified each pseudotyped virion by density equilibrium gradient analysis, and the fractions from the top of the gradient to the bottom were subjected to SDS-PAGE followed by Western blotting. As shown in Fig. 2B and C, the envelope proteins of NL4-3/WT were detected as a doublet of gp120 and gp160 and colocalized with HIV-1 Gag (p24) proteins. Similarly, the other chimeric envelope proteins were also demonstrated to be incorporated in the virions (Fig. 2D). It appeared that the incorporations of three NL4-3/SDF1 chimeric envelope proteins were comparable to that of NL4-3/WT except that the gp120/gp160 ratio was slightly lower than that of NL4-3/WT.
Infectivity of pseudotyped viruses with the NL4-3/SDF-1 chimeric envelope.
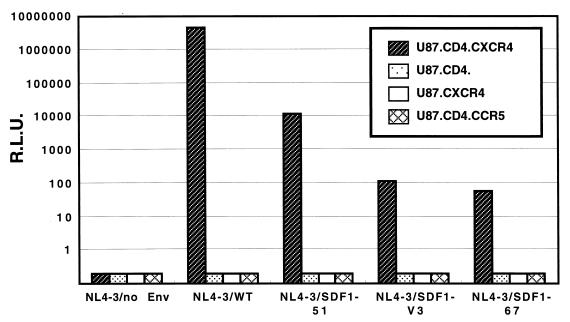
Luciferase reporter virus with each chimeric envelope was prepared in HEK293T cells by the cotransfection of pNL4-3.Luc.E−R− vector and one of the envelope expression vectors. The titration of pseudotyped viruses was performed with an enzyme-linked immunosorbent assay of the p24 antigen. We used U87.CD4.CXCR4, U87.CD4, U87.CD4.CCR5, and U87.CXCR4 cells as target cells to which each pseudotyped virus, equalized by the amount of p24 antigen, was added. Measurement of luciferase activities of the cell lysates showed that pseudotyped viruses with NL4-3/SDF1-51 as well as NL4-3/WT could infect U87.CD4.CXCR4 but not U87.CD4, U87.CXCR4, or U87.CD4.CCR5 cells (Fig. 3). Although the efficiency of infection of the former was relatively low, significant levels of infection were reproducibly observed. The pseudotyped viruses with the NL4-3/SDF1-V3 and NL4-3/SDF1-67 chimeric envelopes could infect U87.CD4.CXCR4 cells with somewhat lower efficiency than NL4-3/SDF1-51. Four independent experiments gave similar results.
FIG. 3.
Infectivity of pseudotyped virus with NL4-3/SDF1 chimeric envelope. Pseudotyped viruses were added to U87.CD4.CXCR4, U87.CD4, U87.CXCR4, and U87.CD4.CCR5 target cells. After 3 days, the luciferase activities were measured. R.L.U., relative luciferase units.
Infectivity of pseudotyped viruses with the NL4-3/SDF1-51 chimeric envelope using EGFP reporter virus.
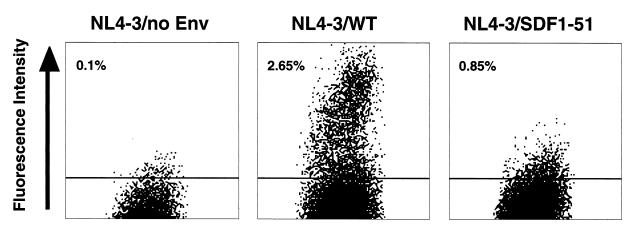
To confirm the infectivity of HIV-1 with an SDF-1 chimeric envelope, we also examined the infection assay using EGFP reporter viruses and U87.CD4.CXCR4 cells. On day 3 postinfection, we analyzed the infectivity by flow cytometry. As shown in Fig. 4, many bright EGFP+ cells were detected after infection of the EGFP reporter pseudotyped viruses with the NL4-3/WT envelope. Infection of the pseudotyped virus with the NL4-3/SDF1-51 chimeric envelope generated a significant number of dull EGFP+ cells that were not present in infection with the virus with NL4-3/no Env. These results provide more supportive evidence that HIV-1 with the NL4-3/SDF1-51 chimeric envelope can infect CD4+ CXCR4+ target cells.
FIG. 4.
Infectivity of EGFP reporter pseudotyped viruses measured by flow cytometric analysis of the target cells. EGFP reporter viruses with NL4-3/WT envelope, NL4-3/SDF1-51, or NL4-3/no Env were added to U87.CD4.CXCR4 cells. After 3 days, the cells were analyzed by FACScan. Numbers represent percentages of GFP-positive cells (upper gated events).
Effects of SDF-1β and anti-SDF-1 Ab on the infection of the pseudotyped virus with the NL4-3/SDF1-51 chimeric envelope.
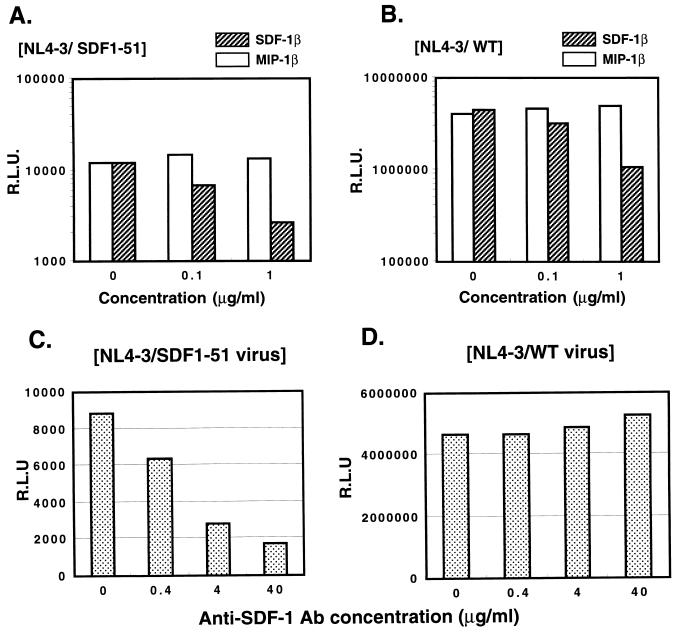
Next, we examined whether the ligand of CXCR4, SDF-1β, could inhibit the infectivity of the pseudotyped virus with the NL4-3/SDF1-51 chimeric envelope as well as the NL4-3/WT virus. Before the addition of the pseudotyped virus, cells were incubated with recombinant human SDF-1β (rhSDF-1β) (R & D Systems) or recombinant human MIP-1β (rhMIP-1β) (R & D Systems) at 37°C for 30 min. As shown in Fig. 5A and B, pretreatment with rhSDF-1β resulted in inhibition of the infectivity of the pseudotyped virus with the NL4-3/SDF1-51 chimeric envelope as well as with the NL4-3/WT virus in a dose-dependent manner, while pretreatment with rhMIP-1β did not.
FIG. 5.
(A and B) Effects of rhSDF-1β or MIP-1β on infectivity of pseudotyped virus with NL4-3/SDF1-51 chimeric envelope. U87.CD4.CXCR4 cells were preincubated with various concentrations of rhSDF-1β or rhMIP-1β at 37°C for 30 min. Then pseudotyped viruses with the NL4-3/SDF1-51 chimeric envelope (NL4-3/SDF1-51) (A) or NL4-3 WT viruses (NL4-3/WT) (B) were added to the cells. (C and D) Effects of anti-human SDF-1 Ab on the infectivity of pseudotyped virus with NL4-3/SDF1-51 chimeric envelope and NL4-3/WT envelope. Pseudotyped viruses NL4-3/SDF1-51 (C) and NL4-3/WT (D) preincubated with anti-human SDF-1 Ab were added to U87.CD4.CXCR4 cells. R.L.U., relative luciferase units.
Then we examined the effect of anti-SDF-1 Ab on the infection of the NL4-3/SDF1-51 virus. Pseudotyped viruses NL4-3/SDF1-51 and NL4-3/WT preincubated with anti-human SDF-1 Ab (R & D Systems) or healthy goat immunoglobulin G (IgG) (R & D Systems) were added to U87.CD4.CXCR4 cells. As shown in Fig. 5C and D, the infectivity of the NL4-3/SDF1-51 virus was inhibited by anti-SDF-1 Ab, while that of the NL4-3/WT virus was not. Healthy goat IgG did not affect the infectivities of either of them (data not shown). Therefore, it was suggested that the pseudotyped virus with the NL4-3/SDF1-51 chimeric envelope could infect U87.CD4.CXCR4 cells through interaction between the V3 region of the chimeric envelope and CXCR4.
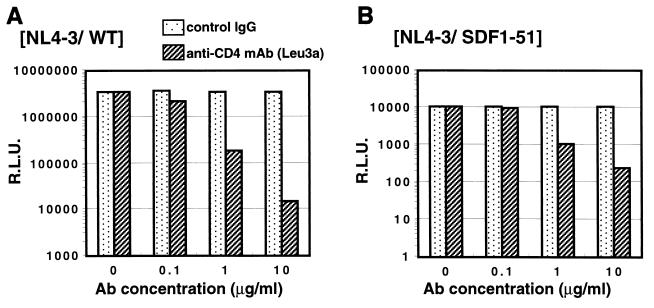
Effect of anti-CD4 MAb Leu3a on the infectivity of the pseudotyped virus with the NL4-3/SDF1-51 chimeric envelope.
It is known that the infectivity of most strains of HIV-1, including NL4-3, is CD4 dependent and can be inhibited by the anti-CD4 MAb Leu3a (16, 41). So we next examined whether anti-CD4 MAb could inhibit the infectivity of the pseudotyped virus with the NL4-3/SDF1-51 chimeric envelope. U87.CD4.CXCR4 cells were pretreated with an anti-CD4 MAb, Leu3a (Becton Dickinson). After 1 h, the pseudotyped viruses were added to the cells. As shown in Fig. 6, the preincubation with Leu3a strongly inhibited the infectivity of pseudotyped viruses with the NL4-3/SDF1-51 chimeric envelope as well as the NL4-3/WT virus. Together with the difference in infectivity for U87.CD4.CXCR4 and U87.CXCR4 cells, this indicated that the infectivity of the pseudotyped viruses with the NL4-3/SDF1-51 chimeric envelope was CD4 dependent.
FIG. 6.
Effect of anti-CD4 MAb Leu3a on infectivity of pseudotyped virus with NL4-3/SDF1-51 chimeric envelope. U87.CD4.CXCR4 cells were preincubated with the indicated concentrations of anti-CD4 MAb Leu3a or mouse IgG control antibody. After 1 h, NL4-3/WT viruses (A) or NL4-3/SDF1-51 viruses (B) were added to the cells. R.L.U., relative luciferase units.
Effects of soluble gp120 on the binding of SDF-1α to CXCR4.
We showed that the NL4-3/SDF1-51 virus could infect U87.CD4.CXCR4 cells but that its efficiency was much less than that of the NL4-3/WT virus. To confirm the binding capability of the chimeric envelope proteins to CXCR4, we examined whether these proteins could inhibit binding of labeled SDF-1α to CXCR4. The gp120 proteins were prepared as the supernatants of HEK293T cells transiently transfected with the envelope vectors (pNL4-3env, pNL4-3env/SDF1-51, pYU2env, and the mock control, pME18S). After concentration, the gp120 proteins were visualized by Western blotting, and adjusted amounts were used (data not shown). As shown in Fig. 7, NL4-3/SDF1-51 gp120 could significantly inhibit binding of SDF-1α in a dose-dependent manner which was comparable to that of NL4-3/WT gp120, while M-tropic YU2 gp120 as well as mock supernatants showed no effects. These results strongly suggested that this chimeric envelope could specifically bind to CXCR4.
FIG. 7.
Effects of soluble gp120 on the binding of 125I–SDF-1α to CXCR4. Soluble gp120 proteins were prepared in supernatants of HEK293T cells transiently transfected with the envelope expression vectors (pNL4-3env, pNL4-3env/SDF1-51, YU2env, and mock control of pME18S). The indicated dilutions of each supernatant were incubated with U87.CD4.CXCR4 cells at 4°C for 1 h. Subsequently, 125I–SDF-1α in binding buffer was added to cells. After incubation at 4°C for 1 h, cells were washed twice and the cell-bound radioactivities were counted in a gamma counter.
Infectivity of pseudotyped viruses with the NL4-3/MIP-1α chimeric envelope.
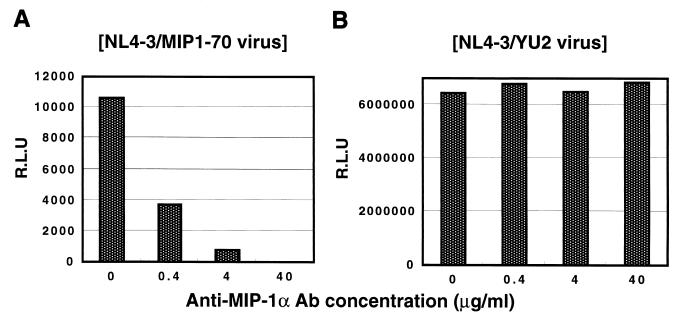
We subsequently examined whether the replacement of the V3 region with MIP-1α, a ligand for CCR5, resulted in acquisition of infectivity of CCR5+ cells. We constructed a chimeric envelope expression vector based on T-tropic NL4-3 in which the V3 region of gp120 was replaced with the full-length, 70-amino-acid MIP-1α protein (NL4-3/MIP1-70). As shown in Fig. 8, the pseudotyped virus with the NL4-3/MIP1-70 chimeric envelope could infect U87.CD4.CCR5 cells. Although the efficiency was much lower than that of pseudotyped viruses with the YU2 WT envelope the infectivity of the NL4-3/MIP1-70 virus seems to be specific and CCR5 mediated, since it was inhibited by neutralizing anti-human MIP-1α MAb (R & D Systems) in a dose-dependent manner, while the infectivity of the YU2 envelope virus (NL4-3/YU2) was not affected. This virus did not infect U87.CD4 or U87.CCR5 cells but might have slight infectivity for U87.CD4.CXCR4 cells (less than 10% of that for U87.CD4.CCR5 cells; data not shown), which might be due to the backbone structure of T-tropic NL4-3 gp120.
FIG. 8.
Effects of anti-human MIP-1α Ab on infectivity of pseudotyped virus with NL4-3/MIP1-70 chimeric envelope (A) and NL4-3/YU2 envelope (B). Pseudotyped viruses NL4-3/MIP1-70 and NL4-3/YU2 preincubated with anti-human MIP1α MAb were added to U87.CD4.CCR5 cells. R.L.U., relative luciferase units.
DISCUSSION
Entry of HIV-1 into the host cell begins with the binding of the gp120 envelope glycoprotein to CD4, which serves as the primary receptor. CD4 binding induces conformational changes in the gp120 glycoprotein, leading to the exposure and/or formation of a binding site for the relevant chemokine receptors (32, 42). These chemokine receptors, mainly CCR5 and CXCR4 for HIV-1, serve as second receptors for virus entry. Among the defined regions of gp120, the V3 loop region is the principal determinant of chemokine receptor specificity and has a close association with the cell tropism of HIV-1 (11, 45).
The V3 region of any strain of HIV-1 has a simple loop structure with one disulfide bond. It was previously reported that the T-tropic HIV-1-derived synthetic cyclized V3 peptides but not the linear or M-tropic V3 peptides could directly bind to CXCR4 and inhibit T-tropic HIV-1 infection (40). In addition to this, Murakami et al. reported the inhibition of T-tropic HIV-1 infection by a small-molecule CXCR4 antagonist, T22, which also has a simple loop structure with two disulfide bonds, but not by a linear control peptide, 4-Ala-T-I (33, 34). Thus, it appears that the loop structure is important for the binding of the peptides to CXCR4, resulting in the inhibition of HIV-1 infection. To test whether the entry and the infection of HIV-1 depends on the loop structure and the binding between the V3 region and the relevant coreceptor rather than on its indigenous amino acid sequence, we examined the effect of the replacement of the V3 region of gp120 with exogenous nonviral amino acid sequences on viral infectivity. We prepared chimeric envelope vectors in which the V3 region of the T-tropic NL4-3 envelope was replaced with stretches of SDF-1, a ligand for CXCR4. SDF-1 was originally described as a chemokine produced by a bone marrow stromal cell line composed of two alternative splicing forms, SDF-1α (68 amino acid residues) and SDF-1β (72 amino acid residues) (44, 48). After processing at the C terminus, a form that is called SDF-1 (67 amino acid residues) is generated from stromal cells (5). SDF-1 adopts a chemokine-like fold consisting of three antiparallel β-strands and an overlying α-helix (15). SDF-1 has two N-terminal sites for binding and activating of its receptor, CXCR4. Receptor activation requires Lys-1 and Pro-2 within the N-terminal region. Amino acid residues 12 to 17 of the loop region, the RFFESH site, are important for optimal binding. The SDF-1 RFFESH loop binds to the N-terminal segment of CXCR4, but inhibition of HIV-1 entry by SDF-1 does not require Lys-1 and Pro-2, which can activate CXCR4, but does require the RFFESH loop. We prepared three chimeric envelope vectors, with the first 51 amino acids of SDF-1 (SDF1-51), the full-length protein (SDF1-67), and 43 amino acids (amino acids 11 to 53) which are weakly homologous to the V3 loop of NL4-3 (HIV-IIIB) (SDF1-V3), and carried out the pseudotyped virus infection assay.
Among the pseudotyped viruses with these chimeric envelopes, the NL4-3/SDF1-51 virus showed considerable and reproducible infectivity for U87.CD4.CXCR4 cells. The other chimeric envelope viruses, NL4-3/SDF1-V3 and NL4-3/SDF1-67, could also infect the cells, but with somewhat lower efficiency. Since the incorporation of the envelope proteins in the NL4-3/SDF1-V3 and NL4-3/SDF1-67 virions was comparable to that in the NL4-3/SDF1-51 virus, the lower infectivity of the former two viruses may be due to a subtle structural difference. The infectivity of the NL4-3/SDF1-51 virus could be specifically inhibited by rhSDF-1β as well as anti-human SDF-1 Ab. This suggests that the interaction between a portion of SDF-1 of gp120 and the coreceptor CXCR4 is essential for the infectivity of the NL4-3/SDF1-51 virus. Similarly, it was shown that pretreatment with anti-CD4 MAb Leu3a inhibited the infectivity of the NL4-3/SDF1-51 virus, which together with the difference in infectivities for U87.CD4.CXCR4 and U87.CXCR4 cells indicated that infection of this virus was CD4 dependent. Based on these findings, we presume the following steps: first, the NL4-3/SDF1-51 virus binds to CD4 on the target cell; second, the replaced portion of SDF1-51 in the V3 region of the envelope becomes exposed by a conformational change of the NL4-3/SDF1-51 envelope; and third, binding between the portion of SDF1-51 and CXCR4 leads to infection with the virus.
To summarize, it is suggested that replacement of the V3 region of the NL4-3 envelope with SDF-1 preserves the infectivity of T-tropic HIV-1. We think that our data argue for the above-mentioned hypothesis that the binding between the V3 region and the corresponding coreceptor is important for viral entry, regardless of whether its amino acid sequence is virus derived or exogenous.
It was noted, however, that the infectivity of the pseudotyped viruses with the SDF1-51 chimeric envelope was much less than that of the WT virus. The reason for this may be a low binding affinity between the V3 region of the NL4-3/SDF1-51 chimeric envelope and CXCR4 compared with that of the NL4-3/WT envelope. With regard to this point, we could not measure the direct binding of chimeric envelope gp120 to CXCR4 but showed that chimeric NL4-3/SDF1-51 gp120 as well as NL4-3/WT gp120 could significantly inhibit binding of SDF-1α to CXCR4. Since the regions of CXCR4 interacting with the HIV envelope are not the same as the SDF-1-binding epitopes (21, 50), we could not compare precisely the binding affinity of envelope proteins in this assay. Nonetheless, it was suggested that this chimeric envelope protein specifically bound to CXCR4 with such a level of affinity that it could inhibit the ligand binding.
Accordingly, the following two possibilities should be taken into consideration as to the low infectivity of the NL4-3/SDF1-51 virus. One is somewhat poorer processing of the SDF-1 chimeric envelope in the virion than in the WT. It was noticed that the gp120/gp160 ratio of envelope in the NL4-3/SDF1-51 virus was slightly lower than that of the NL4-3/WT virus. In accordance with this, the amount of soluble gp120 of NL4-3/SDF1-51 in the supernatants was less than that of NL4-3/WT (data not shown). The other possibility is that a subtle structural difference is induced by the insertion of the portion of SDF-1. Kwong et al. and Wyatt et al. reported the crystal structure of the HIV gp120 envelope in complex with CD4 and a neutralizing antibody (27, 55). Based on this analysis, Rizzuto et al. suggested that a conserved gp120 structure adjacent to the V3 loop containing neutralizing epitopes induced by CD4 binding (CD4i epitope) is important for chemokine receptor binding (39). CD4 binding induces a conformational change in gp120 and results in movement of the V2 loop, which probably partially occludes the V3 loop and CD4i epitopes. Although the interaction between the V3 region and the other region of gp120 has not been fully understood, we speculate that the configuration of the portion of SDF-1 as well as the whole structure of the envelope might be changed by the V3 replacement, which could affect Env-coreceptor binding. Additionally, chemokine receptor binding may trigger a further conformational change in the envelope glycoprotein complex that ultimately leads to the fusion of the viral and target cell membrane. It is believed that some of these changes include exposure of the ectodomain of the gp41 transmembrane envelope glycoprotein (7, 19, 51). It is possible that the replacement of the V3 region with a certain exogenous peptide may affect this additional conformational change and also the noncovalent association of gp120 and gp41 subunits in the trimeric complex.
The systemic delivery of genes will open new applications for gene therapy (43). The development of retrovirus vectors is being considered for this purpose. Mammalian retrovirus vectors commonly used for gene transfer are classified on the basis of their host range as either ecotropic, which only infect murine cells, or amphotropic, which infect both murine and nonmurine cells. The host range of retrovirus vectors has become broad by using vesicular stomatitis virus glycoprotein as an envelope protein (6, 56). On the other hand, the modification of the retroviral envelope gene is becoming a promising strategy for targeting retroviral vectors (43). In the previous studies, the envelope genes of Moloney murine leukemic virus, spleen necrosis virus, and avian leukemia virus A were replaced with some fusion gene such as a single chain antibody and a ligand against a target receptor. For example, Kasahara et al. reported a tissue-specific targeting strategy by introducing the polypeptide hormone erythropoietin into the ecotropic Moloney murine leukemic virus envelope (26). There has been a report on retroviral targeting using a WT envelope of HIV (31) but not using the HIV chimeric envelope by replacing the V3 region with a certain exogenous peptide segment which binds to corresponding coreceptors. In the present study, we engineered pseudotyped viruses with chimeric envelopes in which the V3 region was replaced with a ligand for CXCR4, SDF-1, and demonstrated the infectivity of these viruses for CD4+ and CXCR4+ cells.
In addition to the replacement with SDF-1, we investigated whether the replacement of the V3 region with another ligand besides CXCR4 endowed the virus with infectivity for its cognate receptor-expressing cells. We showed that the pseudotyped virus with a chimeric T-tropic HIV-1 envelope in which the V3 region was replaced with MIP-1α could infect CCR5+ cells. Further studies are required to generalize the concept that replacing the V3 region with a certain exogenous peptide segment which can bind to the cognate receptor can change and select the target cells. Our new strategy of replacement of the V3 region with other ligand molecules may enable development of tissue- or cell-specific targeting of HIV-based vectors.
ACKNOWLEDGMENTS
This study was supported by grants-in-aid from the Ministry of Education, Science, Sports, and Culture of Japan.
The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: U87MG from Bruce Chesebro; U87.CD4, U87.CD4.CXCR4, and U87.CD4.CCR5 from HongKui Deng and Dan R. Littman; pNL4-3.Luc.E−R− from Nathaniel Landau; pYU2 from Beatrice and George Shaw; and hybridoma 902 (anti-gp120) from Bruce Chesebro. We thank K. Fukunaga for excellent technical assistance.
REFERENCES
- 1.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC-CKR5: a RANTES, MIP-1α, MIP-1β receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 2.Berger E A, Murphy P M, Farber J M. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu Rev Immunol. 1999;17:657–700. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- 3.Berson J F, Long D, Doranz B J, Rucker J, Jirik F R, Doms R W. A seven-transmembrane domain receptor involved in fusion and entry of T-cell-tropic human immunodeficiency virus type 1 strains. J Virol. 1996;70:6288–6295. doi: 10.1128/jvi.70.9.6288-6295.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Björndal Å, Deng H, Jansson M, Fiore J R, Colognesi C, Karlsson A, Albert J, Scarlatti G, Littman D R, Fenyö E M. Coreceptor usage of primary human immunodeficiency virus type 1 isolates varies according to biological phenotype. J Virol. 1997;71:7478–7487. doi: 10.1128/jvi.71.10.7478-7487.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bleul C C, Fuhlbrigge R C, Casasnovas J M, Aiuti A, Springer T A. A highly efficacious lymphocyte chemoattractant, stromal cell-derived factor 1 (SDF-1) J Exp Med. 1996;184:1101–1109. doi: 10.1084/jem.184.3.1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burns J C, Friedmann T, Driever W, Burrascano M, Yee J-K. Vesicular stomatitis virus G glycoprotein pseudotyped retroviral vectors: concentration to very high titer and efficient gene transfer into mammalian and nonmammalian cells. Proc Natl Acad Sci USA. 1993;90:8033–8037. doi: 10.1073/pnas.90.17.8033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan D C, Fass D, Berger J M, Kim P S. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1998;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 8.Chen B K, Saksela K, Andino R, Baltimore D. Distinct modes of human immunodeficiency virus type 1 proviral latency revealed by superinfection of nonproductively infected cell lines with recombinant luciferase-encoding viruses. J Virol. 1994;68:654–660. doi: 10.1128/jvi.68.2.654-660.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chesabro B, Wehrly K. Development of a sensitive quantitative focal assay for human immunodeficiency virus infectivity. J Virol. 1988;62:3779–3788. doi: 10.1128/jvi.62.10.3779-3788.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. The β-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 11.Cocchi F, DeVico A L, Garzino-Demo A, Cara A, Gallo R C, Lusso P. The V3 domain of the HIV-1 gp120 envelope glycoprotein is critical for chemokine-mediated blockade of infection. Nat Med. 1996;2:1244–1247. doi: 10.1038/nm1196-1244. [DOI] [PubMed] [Google Scholar]
- 12.Connor R I, Chen B K, Choe S, Landau N R. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 13.Connor R I, Sheridan K E, Ceradini D, Choe S, Landau N R. Change in coreceptor use correlates with disease progression in HIV-1-infected individuals. J Exp Med. 1997;185:621–628. doi: 10.1084/jem.185.4.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cormack B P, Valdivia R H, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 15.Crump M P, Gong J-H, Loetscher P, Rajarathnam K, Amara A, Arenzana-Seisdedos F, Virelizier J-L, Baggiolini M, Sykes B D, Clark-Lewis I. Solution structure and basis for functional activity of stromal cell-derived factor-1; dissociation of CXCR4 activation from binding and inhibition of HIV-1. EMBO J. 1997;16:6996–7007. doi: 10.1093/emboj/16.23.6996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dalgleish A G, Beverley P C L, Clapham P R, Crawford D H, Greaves M F, Weiss R A. The CD4(T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature. 1984;312:763–767. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- 17.Deng H K, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, DiMarzio P, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 18.Deng H K, Unutmaz D, KewalRamani V N, Littman D R. Expression cloning of new receptors used by simian and human immunodeficiency viruses. Nature. 1997;388:296–300. doi: 10.1038/40894. [DOI] [PubMed] [Google Scholar]
- 19.Doms R W, Moore J P. HIV-1 membrane fusion: target of opportunity. J Cell Biol. 2000;151:F9–F13. doi: 10.1083/jcb.151.2.f9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. A dual-tropic primary HIV-1 isolate that uses fusin and the β-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 21.Doranz B J, Orsini M J, Turner J D, Hoffman T L, Berson J F, Hoxie J A, Peiper S C, Brass L F, Doms R W. Identification of CXCR4 domains that support coreceptor and chemokine receptor functions. J Virol. 1999;73:2752–2761. doi: 10.1128/jvi.73.4.2752-2761.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC–CKR-5. Nature. 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 23.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of seven transmembrane, G protein-coupled receptor. Science. 1996;272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 24.Graham F L, Smiley J, Russell W C, Nairn R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J Gen Virol. 1977;36:59–74. doi: 10.1099/0022-1317-36-1-59. [DOI] [PubMed] [Google Scholar]
- 25.He J, Choe S, Walker R, Di Marzio P, Morgan D O, Landau N R. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J Virol. 1995;69:6705–6711. doi: 10.1128/jvi.69.11.6705-6711.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kasahara N, Dozy A M, Kan Y W. Tissue-specific targeting of retroviral vectors through ligand-receptor interactions. Science. 1994;266:1373–1376. doi: 10.1126/science.7973726. [DOI] [PubMed] [Google Scholar]
- 27.Kwong P D, Wyatt R, Robinson J, Sweet R W, Sodroski J, Hendrickson W A. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y, Kappes J C, Conway J A, Price R W, Shaw G M, Hahn B H. Molecular characterization of human immunodeficiency virus type 1 cloned directly from uncultured human brain tissue: identification of replication-competent and -defective viral genomes. J Virol. 1991;65:3973–3985. doi: 10.1128/jvi.65.8.3973-3985.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y, Hui H, Burgess C J, Price R W, Sharp P M, Hahn B H, Shaw G M. Complete nucleotide sequence, genome organization, and biological properties of human immunodeficiency virus type 1 in vivo: evidence for limited defectiveness and complementation. J Virol. 1992;66:6587–6600. doi: 10.1128/jvi.66.11.6587-6600.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liao F, Alkhatib G, Peden K W C, Sharma G, Berger E A, Farber J M. STRL33, a novel chemokine receptor-like protein, functions as a fusion cofactor for both macrophage-tropic and T cell line-tropic HIV-1. J Exp Med. 1997;185:2015–2023. doi: 10.1084/jem.185.11.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mammano F, Salvatori F, Indraccolo S, Rossi A D, Chieco-Bianchi L, Gottlinger H G. Truncation of the human immunodeficiency virus type 1 envelope glycoprotein allows efficient pseudotyping of Moloney murine leukemia virus particles and gene transfer into CD4+ cells. J Virol. 1997;71:3341–3345. doi: 10.1128/jvi.71.4.3341-3345.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moore J P, McKeating J A, Weiss R A, Sattentau Q J. Dissociation of gp120 from HIV-1 virions induced by soluble CD4. Science. 1990;250:1139–1142. doi: 10.1126/science.2251501. [DOI] [PubMed] [Google Scholar]
- 33.Murakami T, Nakajima T, Koyanagi Y, Tachibana K, Fujii N, Tamamura H, Yoshida N, Waki M, Matsumoto A, Yoshie O, Kishimoto T, Yamamoto N, Nagasawa T. A small molecule CXCR4 inhibitor that blocks T cell line-tropic HIV-1 infection. J Exp Med. 1997;186:1389–1393. doi: 10.1084/jem.186.8.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murakami T, Zhang T-Y, Koyanagi Y, Tanaka Y, Kim J, Suzuki Y, Minoguchi S, Tamamura H, Waki M, Matsumoto A, Fujii N, Shida H, Hoxie J A, Peiper S C, Yamamoto N. Inhibitory mechanism of the CXCR4 antagonist T22 against human immunodeficiency virus type 1 infection. J Virol. 1999;73:7489–7496. doi: 10.1128/jvi.73.9.7489-7496.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Page K A, Liegler T, Feinberg M B. Use of a green fluorescent protein as a marker for human immunodeficiency virus type 1 infection. AIDS Res Hum Retrovir. 1997;13:1077–1081. doi: 10.1089/aid.1997.13.1077. [DOI] [PubMed] [Google Scholar]
- 36.Pear W S, Nolan G P, Scott M L, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci USA. 1993;90:8392–8396. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pincus S, Wehrly K, Chesabro B. Treatment of HIV tissue culture infection with monoclonal antibody-ricin A chain conjugates. J Immunol. 1989;142:3070–3075. [PubMed] [Google Scholar]
- 38.Ponten J, Maclntyre E H. Long-term culture of normal and neoplastic human glia. Acta Pathol Microbiol. 1968;74:465–486. doi: 10.1111/j.1699-0463.1968.tb03502.x. [DOI] [PubMed] [Google Scholar]
- 39.Rizzuto C D, Wyatt R, Hernandez-Ramos N, Sun Y, Kwong P D, Hendrickson W A, Sodroski J. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science. 1998;280:1949–1953. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- 40.Sakaida H, Hori T, Yonezawa A, Sato A, Isaka Y, Yoshie O, Hattori T, Uchiyama T. T-tropic human immunodeficiency virus type 1 (HIV-1)-derived V3 loop peptides directly bind to CXCR-4 and inhibit T-tropic HIV-1 infection. J Virol. 1998;72:9763–9770. doi: 10.1128/jvi.72.12.9763-9770.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sattentau Q J, Dalgleish A J, Weiss R A, Beverley P C L. Epitopes of the CD4 antigen and HIV infection. Science. 1986;234:1120–1123. doi: 10.1126/science.2430333. [DOI] [PubMed] [Google Scholar]
- 42.Sattentau Q J, Moore J P. Conformational changes induced in the human immunodeficiency virus envelope glycoprotein by soluble CD4 binding. J Exp Med. 1991;174:407–415. doi: 10.1084/jem.174.2.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schnierle B S, Groner B. Retroviral targeted delivery. Gene Ther. 1996;3:1069–1073. [PubMed] [Google Scholar]
- 44.Shirozu M, Nakano T, Inazawa J, Tashiro K, Tada H, Shinohara T, Honjo T. Structure and chromosomal localization of the human stromal cell-derived factor 1 (SDF1) gene. Genomics. 1995;28:495–500. doi: 10.1006/geno.1995.1180. [DOI] [PubMed] [Google Scholar]
- 45.Speck R F, Wehrly K, Platt E J, Atchison R E, Charo I F, Kabat D, Chesebro B, Goldsmith M A. Selective employment of chemokine receptors as human immunodeficiency virus type 1 coreceptors determined by individual amino acids within the envelope V3 loop. J Virol. 1997;71:7136–7139. doi: 10.1128/jvi.71.9.7136-7139.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stein B S, Gowda S D, Lifson J D, Penhallow R C, Beusch K G, Engleman E G. pH independent HIV entry into CD4-positive T cells via virus envelope fusion to the plasma membrane. Cell. 1987;49:659–668. doi: 10.1016/0092-8674(87)90542-3. [DOI] [PubMed] [Google Scholar]
- 47.Sullivan N, Sun Y, Sattentau Q, Thali M, Wu D, Denisova G, Gershoni J, Robinson J, Moore J, Sodroski J. CD4-induced conformational changes in the human immunodeficiency virus type 1 gp120 glycoprotein: consequences for virus entry and neutralization. J Virol. 1998;72:4694–4703. doi: 10.1128/jvi.72.6.4694-4703.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tashiro K, Tada H, Heilker R, Shirozu M, Nakano T, Honjo T. Signal sequence trap: a cloning strategy for secreted proteins and type I membrane protein. Science. 1993;261:600–603. doi: 10.1126/science.8342023. [DOI] [PubMed] [Google Scholar]
- 49.Thali M, Moore J P, Furman C, Charles M, Ho D D, Robinson J, Sodroski J. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J Virol. 1993;67:3978–3988. doi: 10.1128/jvi.67.7.3978-3988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Trkola A, Paxton W A, Monard S P, Hoxie J A, Siani M A, Thompson D A, Wu L, Mackay C R, Horuk R, Moore J P. Genetic subtype-independent inhibition of human immunodeficiency virus type 1 replication by CC and CXC chemokines. J Virol. 1998;72:396–404. doi: 10.1128/jvi.72.1.396-404.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Weissenhorn W, Dessen A, Harrison S C, Skehel J J, Wiley D C. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 52.Wu L, Gerard N P, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso A A, Desjardin E, Newman W, Gerard C, Sodroski J. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 53.Wyatt R, Moore J, Accola M, Desjardin E, Robinson J, Sodroski J. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J Virol. 1995;69:5723–5733. doi: 10.1128/jvi.69.9.5723-5733.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 55.Wyatt R, Kwong P D, Desjardins E, Sweet R W, Robinson J, Hendrickson W A, Sodroski J G. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;393:705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- 56.Yee J-K, Miyanohara A, LaPorte P, Bouic K, Burns J C, Friedmann T. A general method for the generation of high-titer, pantropic retroviral vectors: highly efficient infection of primary hepatocytes. Proc Natl Acad Sci USA. 1994;91:9564–9568. doi: 10.1073/pnas.91.20.9564. [DOI] [PMC free article] [PubMed] [Google Scholar]