Abstract
The envelope protein E of the flavivirus tick-borne encephalitis (TBE) virus promotes cell entry by inducing fusion of the viral membrane with an intracellular membrane after uptake by endocytosis. This protein differs from other well-studied viral and cellular fusion proteins because of its distinct molecular architecture and apparent lack of involvement of coiled coils in the low-pH-induced structural transitions that lead to fusion. A highly conserved loop (the cd loop), which resides at the distal tip of each subunit and is mostly buried in the subunit interface of the native E homodimer at neutral pH, has been hypothesized to function as an internal fusion peptide at low pH, but this has not yet been shown experimentally. It was predicted by examination of the X-ray crystal structure of the TBE virus E protein (F. A. Rey et al., Nature 375:291–298, 1995) that mutations at a specific residue within this loop (Leu 107) would not cause the native structure to be disrupted. We therefore introduced amino acid substitutions at this position and, using recombinant subviral particles, investigated the effects of these changes on fusion and related properties. Replacement of Leu with hydrophilic amino acids strongly impaired (Thr) or abolished (Asp) fusion activity, whereas a Phe mutant still retained a significant degree of fusion activity. Liposome coflotation experiments showed that the fusion-negative Asp mutant did not form a stable interaction with membranes at low pH, although it was still capable of undergoing the structural rearrangements required for fusion. These data support the hypothesis that the cd loop may be directly involved in interactions with target membranes during fusion.
Enveloped viruses enter cells by fusing their membranes with a host cell membrane, either at the cell surface or at an internal site after uptake by endocytosis. This is mediated by metastable surface proteins that undergo a triggered conformational change upon binding to a receptor or exposure to the acidic environment of the endosome, allowing a previously buried portion of the protein, the fusion peptide, to insert into the target membrane (22). Exposure and insertion of the fusion peptide is believed to be the crucial step in the initiation of the fusion process, although further conformational changes may be required for achieving the complete merger of the lipid bilayers.
One structurally related group of well-characterized viral fusion proteins includes the spike proteins of influenza A and C viruses, human immunodeficiency virus and other retroviruses, paramyxoviruses, and filoviruses such as Ebola virus (for reviews, see references 44, 48, and 5). These fusion proteins require proteolytic cleavage for activity (27), and their fusion peptides reside at or near the N-terminal end of the membrane-anchored subunit. When activated by the appropriate trigger, they all adopt a characteristic six-helix rod-like structure with a long coiled coil at the trimer interface. During the formation of the core, the fusion peptide is translocated to the tip of the rod, permitting it to interact with the target membrane and initiate fusion. These observations have led to a general model for virus-induced fusion, which seems to apply also to fusion by some cellular proteins such as SNARES (24, 44, 48).
In contrast, the fusion proteins of several other enveloped viruses (e.g., rhabdoviruses, alphaviruses, and flaviviruses) appear to be different. They are not proteolytically cleaved during maturation, they are not predicted to form coiled coils (43), and they appear to use internal sequences (15, 21, 22, 25) rather than an N-terminal or N-proximal fusion peptide to mediate fusion. The only protein of this class for which a high-resolution structure is currently available is the envelope glycoprotein E of the flavivirus tick-borne encephalitis (TBE) virus (36). The striking lack of structural similarity between the native forms of the fusion proteins of flaviviruses and orthomyxoviruses (41, 49) suggests that they might use fundamentally different mechanisms to carry out essentially identical functions.
The flaviviruses are small enveloped viruses that are responsible for a number of mosquito- and tick-borne diseases such as yellow fever, dengue fever, Japanese encephalitis, West Nile encephalitis, and tick-borne encephalitis (31). The interior of the virion consists of an isometric nucleocapsid containing the unsegmented positive-stranded RNA genome complexed with the capsid protein C. The outer surface contains two membrane-anchored proteins: the envelope glycoprotein (E), which mediates fusion in the endosomal compartment after endocytosis, and the small membrane protein M. The E proteins of all mosquito- and tick-borne flaviviruses have at least 40% amino acid identity and their six intramolecular disulfide bridges are conserved, indicating a common overall structure (36). Flaviviruses are synthesized intracellularly as immature particles containing a larger precursor form of the M protein (prM), which is subsequently cleaved by a cellular protease to yield the mature virion (37).
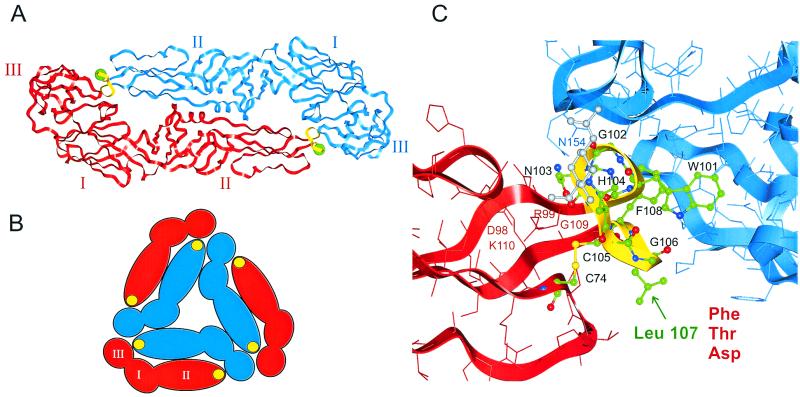
The X-ray crystal structure of the E protein of TBE virus at 2-Å resolution (36) revealed that it is an elongated head-to-tail homodimer that, rather than forming a spike, lies parallel to the surface of the virion, anchored in the membrane at its distal ends (Fig. 1A). The external portion of each subunit consists of three structural domains (I, II, and III), which correspond to previously defined antigenic domains (29). Cryoelectron microscopy studies with recombinant subviral particles (RSPs) have shown that there are specific lateral interactions between neighboring E dimers (13). This results in an icosahedral lattice structure composed of local threefold assemblies of E dimers (Fig. 1B). Exposure to acidic pH induces a conformational change that weakens the subunit interactions within the dimer while strengthening lateral interactions with other E proteins, presumably those in adjacent positions, giving rise to a homotrimeric form of E (2, 46). The exposure of the fusion peptide and binding to the target membrane probably occurs during this reorganization of the viral envelope structure. Fusion then proceeds rapidly without any requirement for specific proteins or lipids in the target membrane (8).
FIG. 1.
(A) Ribbon diagram of the ectodomain portion of the TBE virus E protein dimer (36) viewed from the top (perpendicular to the virion surface), with the individual subunits colored red and blue. The C-terminal portion of the protein, whose structure is not known, extends downward from domain III at each end of the dimer to anchor the protein in the viral membrane. Domains I, II, and III are labeled by roman numerals, and the cd loop at the tip of domain II is colored yellow. The position of Leu 107 is shown by a green ball. (B) Schematic representation of a threefold assembly of E dimers as determined by cryoelectron microscopy and icosahedral reconstruction of RSPs (13). Domains are indicated by roman numerals, and filled yellow circles represent the cd loop of domain II. (C) Zoom of the top view in panel A, showing details of the cd loop and vicinity. Amino acids 100 to 108 (the cd loop) as well as Cys 74 are shown as ball-and-stick representations and colored by atom (green, C; blue, N; red, O; yellow, S). The first N-acetylglucosamine residue of the N-linked glycan, which covers the cd loop but is attached to Asn 154 of the other subunit, is shown in gray. Other amino acids are represented as sticks.
It was proposed several years ago based on indirect evidence that the sequence element containing amino acids 98 to 110 might serve as an internal fusion peptide (39, 40). This region is highly conserved among flaviviruses, with the exception of position 104, which is occupied by histidine in tick-borne and glycine in mosquito-borne flaviviruses. The X-ray crystal structure (36) showed that this conserved region lies at the distal tip of each subunit (Fig. 1A). Residues 100 to 108 constitute a loop (cd loop) between the antiparallel strands c and d of domain II, which is buried in a hydrophobic pocket formed at the interface between domains I and III of the other monomer. In the native TBE virus E protein dimer, it is also covered by the N-linked oligosaccharide from its partner subunit. This region is highly constrained by multiple interactions, including several internal hydrogen bonds, one salt bridge (Asp 98 and Lys 110), and one disulfide bond (Cys 74 and Cys 105) that connects the cd loop to the bc loop of the same domain (Fig. 1C).
Using the X-ray crystal structure of the TBE virus E protein as a basis for rational mutagenesis, we have attempted to obtain direct experimental evidence for the involvement of this sequence in fusion. As a model system for these studies, we used noninfectious RSPs, which are icosahedrally symmetrical structures composed only of the mature viral surface proteins embedded in a lipid membrane and lacking the nucleocapsid and RNA genome (3, 13, 42). They are assembled intracellularly in cells expressing the prM and E proteins and undergo a maturation process similar to that of the whole virion, including glycosylation, transport through the Golgi complex, furin-mediated cleavage of prM, and secretion (3, 42). At mildly acidic pH, RSPs are capable of inducing cell-cell fusion (42) as well as fusion with artificial membranes (8). In the latter case it was shown that RSPs fuse with the same pH dependence, kinetics, and lipid dependence as whole virions.
Using the known structure of the TBE virus E protein and taking into consideration the structural constraints within the cd loop region, Leu 107 was identified as a target for single amino acid substitutions that could be introduced without interfering with the interactions required for dimer and particle formation. The functional analysis of RSPs containing mutations at position 107 provides evidence that the cd loop might be directly involved in membrane interactions during fusion.
MATERIALS AND METHODS
Construction of mutants.
Recombinant plasmids containing substitutions at codon 107 of the E gene were derived from the wild-type plasmid SV- PEwt (1), a cDNA clone containing the prM and E genes of TBE virus strain Neudoerfl (GenBank accession no. U27495) under the control of the simian virus 40 early promoter, which yields secreted RSPs when expressed in COS cells (3). Mutant plasmids were constructed by replacing a 499-bp AgeI-BpuI fragment (nucleotides 960 to 1459) from SV-PEwt with a 196-bp PCR-generated Sau3AI-BpuI fragment containing the desired mutation and a 303-bp AgeI-Sau3AI fragment from plasmid SV-E07 (1), which is identical to the corresponding region of SV-PEwt except for a silent mutation at nucleotide 1257 that facilitated cloning by eliminating an additional Sau3AI site. The mutated codons at position 107 (TTC for Phe, GAC for Asp, and ACG for Thr) were included in the PCR primers at nucleotides 1291 to 1293. The wild-type and mutant plasmids were propagated in Escherichia coli strain HB101, purified using a Qiagen plasmid mega kit, and sequenced throughout the prM and E coding regions to confirm that only the desired mutations were present.
Production of RSPs.
COS-1 cells were transfected with recombinant plasmids by electroporation as described previously (4). RSPs were harvested by pelleting from cell supernatants 48 h after transfection and purified on sucrose gradients (42). For membrane fusion assays, the particles were metabolically labeled with 1-pyrenehexadecanoic acid (Molecular Probes, Leiden, The Netherlands) as described by Corver et al. (8).
Electron microscopy.
RSPs were placed onto glow-discharged Formvar-carbon-coated cupron grids (Agar Scientific, Stansted Essex, England) and allowed to adsorb for 5 min. Samples were stained for 4 min with 1% uranyl acetate (pH 4.5) and viewed using a Zeiss EM10 electron microscope at a magnification of ×50,000.
Membrane fusion assay.
Fusion of pyrene-labeled RSPs with liposomes was measured by monitoring the decrease in pyrene excimer fluorescence at 480 nm with excitation at 343 nm as described by Corver et al. (8). Briefly, RSPs were mixed with liposomes (total phospholipid, 0.2 mM) consisting of phosphatidylcholine, phosphatidylethanolamine, sphingomyelin, and cholesterol (molar ratio, 1:1:1:1.5) in a continuously stirred fluorimeter cuvette at 37°C (final volume, 0.5 ml). Fluorescence was monitored continuously using a Perkin-Elmer LS-50B9 fluorescence spectrophotometer. The fusion reaction was initiated by the addition of 300 mM 2-(N-morpholino)ethanesulfonic acid (MES) to yield a final pH of 5.5. The degree of fusion was calculated taking the initial excimer fluorescence after mixing to represent 0% fusion and the fluorescence after dispersion of the RSP-liposome mixture with the detergent octa(ethylene glycol)-n-dodecyl monoether (Fluka, Buchs, Switzerland) to represent 100% fusion.
HA assay.
Hemagglutination (HA) of goose erythrocytes was carried out at pH 6.4 by the method of Clarke and Casals (7).
Analysis of conformational change.
RSP preparations (E protein concentration, 5 μg/ml) in TAN buffer (0.05 M triethanolamine [pH 8.0], 0.1 M NaCl) plus 0.1% bovine serum albumin were acidified by the addition of a 0.05 M Tris-maleate buffer or 0.05 M MES containing 0.1 M NaCl and 0.1% bovine serum albumin to yield the desired pH. After a 10-min incubation at 37°C, the pH was adjusted to 8.0.
The conformational state of the E protein was assessed in a four-layer enzyme-linked immunosorbent assay (ELISA) using a TBE virus-specific guinea pig antiserum as catching antibody and mouse monoclonal antibodies (MAbs) for detection (20). The pattern of reactivity of the E protein with a panel of 18 conformation-sensitive MAbs was determined as described previously (42).
Sedimentation analysis.
The conversion of E dimers to trimers at low pH was measured by sedimentation analysis as described previously (2, 42). Samples were acidified and back-neutralized as described above and then solubilized for 1 h at room temperature with 0.5% Triton X-100. This material was then applied to a 7 to 20% (wt/wt) sucrose gradient made with TAN buffer and 0.1% Triton X-100 and centrifuged for 20 h at 38,000 rpm in a Beckman SW40 rotor at 15°C. Fractions (0.6 ml) were collected by upward displacement, and E protein was quantitated by four-layer ELISA after a 30-min denaturation with 0.2% sodium dodecyl sulfate at 65°C (19).
Liposome coflotation assay.
RSPs were mixed with liposomes of the same composition used in the fusion assay (final phospholipid concentration, 6 mM) in a buffer consisting of 10 mM triethanolamine (pH 8.0) and 140 mM NaCl. After a 10-min incubation at 37°C, the mixture was acidified by the addition of 150 mM MES to yield a pH of 5.5 and incubated for another 10 min. The mixture was then back-neutralized by the addition of 150 mM triethanolamine to yield a final pH of 7.8 and mixed with sucrose to a final concentration of 20% (wt/wt). The 20% solution containing the sample (0.75 ml) was layered onto a 1-ml cushion of 50% sucrose in TAN buffer, after which two more sucrose layers (15%, 1.25 ml; 5%, 1 ml) were applied. The step gradients were centrifuged for 2 h at 4°C in a Beckman SW55 rotor at 50,000 rpm, and fractions (0.2 ml) were collected by upward displacement. E protein was quantitated by four-layer ELISA after treatment with sodium dodecyl sulfate (19).
RESULTS
Mutagenesis of Leu 107.
To investigate the possible involvement of the cd loop (Fig. 1) in membrane fusion, we made mutant RSPs and investigated their fusion-related properties. Our goal was to introduce amino acid substitutions that would lead to functional changes but would not disrupt dimer interactions or impair particle formation, maturation, and secretion. Close examination, however, revealed that most of the nonglycine residues of the cd loop region are involved in an extensive network of interactions that stabilize either the cd loop structure itself or its interactions with the other subunit of the dimer (36). One exception is Leu 107, whose side chain is on the exterior of the molecule and oriented away from the dimer interface (Fig. 1C), suggesting that amino acid substitutions at this position might be well tolerated.
Leu 107, like most of the cd loop region, is almost completely conserved among flaviviruses, but there are a few exceptions: the tick-borne Powassan virus (30), the mosquito-borne Japanese encephalitis virus strain SA-14-14-2 (32), and dengue virus strain PUO-280 (6) have a phenylalanine at this position. We therefore chose to make this conservative Leu-to-Phe change in addition to substituting a polar (Thr) and a charged (Asp) residue for Leu 107.
Expression and secretion properties of mutants.
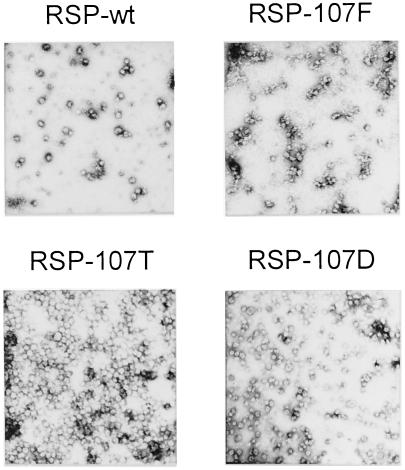
COS-1 cells were transfected with recombinant plasmids encoding the modified E proteins together with the prM protein, which is required for particle assembly and maintenance of the conformation of E during transport and secretion (3). All three of the mutants yielded RSPs with a diameter of approximately 30 nm, which were secreted with essentially the same efficiency as the wild-type control, indicating that the various intermolecular interactions involving prM and E that are necessary for particle assembly and secretion were not affected by the mutations. Analysis of these particles showed that the prM proteins in all cases were properly processed, and no differences in protein composition, degree of maturation, particle density, or sedimentation velocity were observed in comparison with wild-type RSPs (data not shown). It was observed, however, that the Phe mutant (RSP-107F) had a tendency to self-aggregate during the harvesting procedure, resulting in somewhat reduced yields. Negatively stained electron micrographs of purified wild-type and mutant RSPs are shown in Fig. 2.
FIG. 2.
Electron micrographs of wild-type and mutant RSPs stained with uranyl acetate.
Fusion properties of mutant RSPs.
Effects of the mutations on membrane fusion activity were assessed by mixing artificial liposomes with RSPs whose membranes had been fluorescently labeled in vivo with 1-pyrenehexadecanoic acid, acidifying the mixture, and continuously monitoring the decrease in pyrene excimer fluorescence caused by dilution of the probe in the target membrane (8).
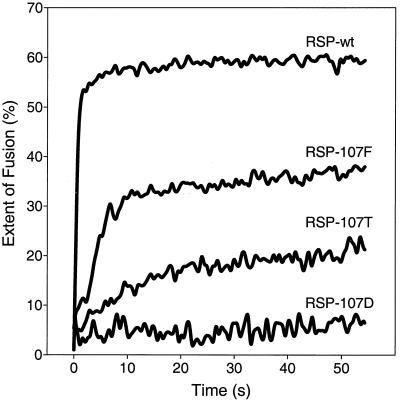
Consistent with earlier results (8), RSP-wt, which contained the wild-type Leu at position 107, fused rapidly within the first seconds after acidification (Fig. 3). RSP-107F, with Phe at this position, also fused with liposomes at low pH, but both the initial rate and final extent of fusion were lower than with RSP-wt. The Thr mutant, RSP-107T, retained only a low level of fusion activity, and no fusion at all was observed with the Asp mutant, RSP-107D.
FIG. 3.
Low-pH-induced fusion of pyrene-labeled RSPs with liposomes. Fusion was measured at 37°C by continuous monitoring of pyrene excimer fluorescence (8), and the extent of fusion was calculated as described in Materials and Methods. At least three replicates were performed, and representative curves are shown. The time zero represents the time of acidification.
The pattern shown in the fusion curves was also partly reflected in differences in the ability of the mutants to induce HA of goose erythrocytes at pH 6.4, a property of flaviviruses that requires low pH (7) and therefore is probably due to interactions of the fusogenic form of E with erythrocyte membranes rather than to receptor binding, as is the case with influenza virus. The specific HA titer of RSP-107F was the same as that of RSP-wt (titer of 128 at a final E protein concentration of 1 μg/ml), but no HA activity could be detected for RSP-107T or RSP-107D in any of five separate preparations. The fusion and HA data together indicate that specific mutations at position 107 can impair the ability of RSPs to interact with membranes at low pH. Since the most dramatic effect was observed with the Asp mutant, this was chosen for more detailed analysis.
Conformational changes at low pH.
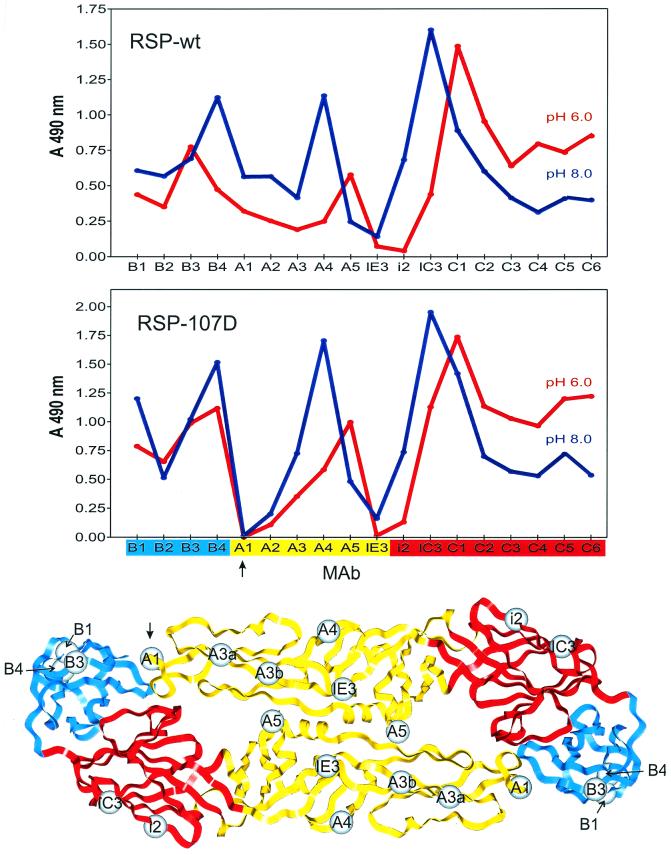
To compare low-pH-induced conformational changes in the E proteins of the wild-type RSP and the nonfusing RSP-107D, we measured the binding activities of a panel of E-specific MAbs whose epitopes in domains I, II, and III have been mapped previously (36). As shown in the top panel of Fig. 4, these conformation-dependent epitopes are altered by low-pH treatment, resulting in a characteristic change in the MAb reactivity pattern of the wild-type E protein (42). With a few significant exceptions (see below), the reactivity profiles with RSP-107D before and after low-pH treatment (Fig. 4, middle) were very similar to those of the wild type, indicating that the Leu-to-Asp mutation did not fundamentally impair the switching mechanism or prevent the E protein from attaining its final low-pH conformation.
FIG. 4.
Antibody binding profiles before and after low-pH treatment. The binding activities of 18 E-protein-specific MAbs with low-pH-treated and untreated RSP-wt (upper panel) and RSP-107D (middle panel) were compared by four-layer ELISA. The blue patterns were obtained with untreated samples, and the red patterns were obtained with samples that had been preincubated at pH 6.0 and back-neutralized. The colors on the x axis of the middle panel represent the structural domains (depicted in the lower panel) to which the antibodies bind (red, domain I; yellow, domain II; blue, domain III). The spheres show the positions of mutations defining individual MAb binding sites that have been mapped by selection of neutralization escape variants of TBE virus (29) or mutations in RSPs that abolish binding of an individual MAb (A1 [this study] or B3 [unpublished data]). The epitopes for the nonneutralizing MAbs B2 and C1 to C6 have not been precisely mapped, but the antigenic domains to which they belong were established previously (29). Note that binding to MAb A1 (arrows) is abolished by the Leu 107-to-Asp mutation.
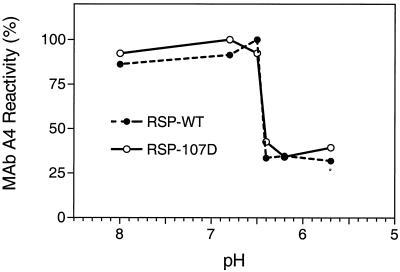
To investigate the pH dependence of this change, we compared the abilities of RSP-107D and RSP-wt that had been pretreated at different pHs to bind the conformation-sensitive MAb A4. As shown in Fig. 5, the mutation clearly did not cause a shift in the pH threshold of the conformational change, which was about 6.5 in each case.
FIG. 5.
pH dependence of the conformational change in the E protein. RSP-wt and RSP-107D were pretreated at different pHs, and binding activity of MAb A4 with these preparations was measured by four-layer ELISA.
There was a clear difference, however, in the abilities of RSP-wt and RSP-107D at both pHs to bind MAb A1, whose reactivity in three separate experiments appeared to be completely abolished by the Leu 107-to-Asp mutation, and to a lesser extent MAb A2, whose reactivity was severely reduced (Fig. 4). Similar results were also obtained with RSP-107T and RSP-107F, with MAb A1 binding completely abolished by these mutations as well (data not shown). Earlier binding competition experiments (17) showed that the epitopes for both of these nonneutralizing MAbs lie close to that of the neutralizing MAb A3 (Fig. 4, bottom), whose binding site has been mapped by two separate mutations in domain II (amino acids 67 and 71) that allow the virus to escape from neutralization (29). MAb A1 is broadly cross-reactive even with distantly related flaviviruses (17), indicating that its epitope is highly conserved. It is noteworthy that Powassan virus, one of the few flaviviruses with a Phe at position 107 instead of Leu, does not bind MAb A1 (30). It is therefore likely that the highly conserved cd loop is a structural component of the MAb A1 binding site. Since the reactivities of MAb A3 and other domain-II-specific neutralizing MAbs were not affected by the Leu 107-to-Asp mutation (Fig. 4), it appears that any structural perturbations in domain II that might have been caused by this substitution were minor and restricted to the tip of the domain. However, slight differences could be observed in the distal face of domain III (B MAbs), which, due to the geometry of the icosahedral lattice, would be in close proximity to one of the domain II cd loops of a neighboring dimer (Fig. 1B).
Quaternary structure changes at low pH.
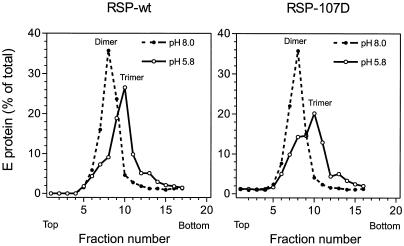
Earlier studies with virions and RSPs have shown that the E protein is irreversibly converted from a homodimer to a homotrimer upon exposure to low pH, and these forms can be distinguished by sedimentation analysis after solubilization with detergent (2, 42). To examine the effect of the Leu 107-to-Asp mutation on this structural transition, RSP-wt and RSP-107D were preincubated at pH 5.8 or 8.0, back-neutralized, solubilized with 0.5% Triton X-100, and analyzed by sedimentation at pH 8.0 on sucrose gradients. As shown in Fig. 6, the pH 8.0 forms of the wild-type and mutant E proteins exhibited identical sedimentation behavior, confirming that the mutant protein is initially homodimeric. After preexposure to low pH, a faster-sedimenting form of E, shown in earlier studies to be a homotrimer (2, 42), was observed with both RSP-wt and RSP-107D. In the case of RSP-107D, the majority of the E protein had been converted to the trimeric form, but in three separate experiments the trimerization was less complete than it was with RSP-wt, suggesting that the 107D mutation, while still allowing trimers to form, caused a decrease in trimerization efficiency. A similar reduction was also observed with RSP-107T and, to a lesser extent, with RSP-107F.
FIG. 6.
Conversion of E homodimers to homotrimers at low pH. RSPs that had been pretreated at the indicated pHs were solubilized with detergent, and sedimentation analysis on continuous sucrose gradients was used to assess the oligomeric state of the E protein. The positions of the E dimer and trimer peaks are indicated.
Low-pH-induced association with lipid membranes.
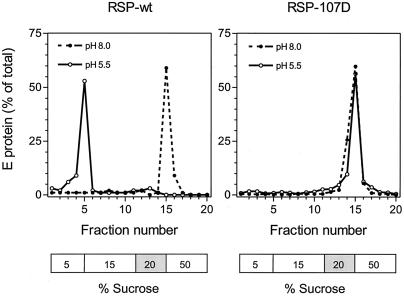
To investigate the initial low-pH-triggered interaction of the E protein with the target membrane, we used a liposome coflotation assay in which RSPs were mixed with liposomes at either pH 8.0 or 5.5 and then analyzed by centrifugation in a sucrose step gradient (see Materials and Methods). At pH 8.0, all of the E protein from both RSP-wt and RSP-107D remained in the 20% sucrose layer, where it had been initially applied (Fig. 7). After treatment at pH 5.5, however, most of the wild-type E protein migrated upward with the liposomes to the 5% sucrose layer, whereas essentially all of the protein carrying the 107D mutation remained in the 20% sucrose layer (Fig. 7). This shows that RSP-107D does not form a stable association with liposomes after low-pH treatment. In several experiments it was found that the Phe and Thr mutants, unlike RSP-107D, were able to interact with liposomes at low pH but to a lesser extent than the wild type. Incomplete binding was always observed with RSP-107F, whereas RSP-107T gave an irregular distribution of protein in the sucrose gradient, suggesting that complexes of the 107T mutant with liposomes were unstable and dissociated during centrifugation (data not shown). This finding is consistent with the fusion data in Fig. 3 and further suggests that substitutions at position 107 might directly interfere with early interactions between the E protein and the target membrane.
FIG. 7.
Liposome coflotation assay. RSPs were incubated with liposomes at the indicated pHs and analyzed by sucrose step gradient centrifugation. The positions of the sucrose layers are indicated below each graph, and the 20% sucrose layer is shaded to indicate the position where the samples were initially applied before centrifugation.
DISCUSSION
The data presented here provide functional evidence that a highly conserved loop at the tip of each subunit of the flavivirus E protein is important for fusion activity and may be directly involved in interactions with target membranes during the initial stages of membrane fusion. The notion that this portion of the protein serves as an internal fusion peptide is consistent with earlier observations that antipeptide antibodies recognizing the region from amino acids 98 to 110 are capable of blocking low-pH-induced fusion of TBE virus with artificial membranes (47) and react more strongly with the low-pH form of the dengue 2 virus E protein than with the native neutral form (40). We have recently observed that the flavivirus cross-reactive MAb A1, whose binding was shown in the present study to be abolished by amino acid substitutions at Leu 107, is also capable of inhibiting virus-liposome fusion and binding of isolated E proteins to artificial membranes (K. Stiasny et al., unpublished data). While all of these data suggest that the cd loop itself might insert into the target membrane during fusion, the actual contact region will ultimately have to be identified by more direct methods such as photolabeling with radioactive lipids (12, 18).
The tip of domain II is buried in the subunit interface of the neutral-pH form of the E homodimer (Fig. 1) and is therefore inaccessible for membrane interactions in its native state. However, the quantitative conversion of E homodimers to homotrimers that occurs at the pH of fusion would require at least a partial dissociation of the native dimer, which could then lead to the exposure of the cd loop. We have shown previously that truncated E dimers that lack the stem-anchor region and therefore cannot trimerize nevertheless dissociate at mildly acidic pH (46). It is likely that ectodomain dissociation in the native virion is responsible not only for initiating the reorganization of the viral envelope (dimer-trimer transition) but also for a transient exposure of the fusion peptide in a configuration that facilitates interaction with the target membrane. Consistent with this idea, we have recently obtained evidence that dimer dissociation is required for binding of E ectodomain fragments to liposomes (Stiasny et al., unpublished). Subsequent steps in the fusion reaction are likely to require more extensive conformational changes. However, in contrast to the prevailing model based on the influenza virus hemagglutinin and structurally related fusion proteins (24, 44, 48), it is difficult to envision the region containing the TBE virus fusion peptide being converted to an extended α-helical structure at low pH, given the constraints placed upon it by the three disulfide bridges in domain II, one of which (Cys 74-Cys 105) involves the fusion peptide itself. Moreover, the extraordinarily rapid rate of fusion by TBE virus (8) essentially rules out the involvement of any steps preceding the lipid-mixing phase that are not extremely favorable kinetically, such as the reduction of disulfide bonds.
The notion that an internal fusion peptide can reside in a disulfide-stabilized loop structure has already been supported by studies with avian sarcoma-leukosis virus. The fusion protein of this retrovirus contains a probable fusion peptide at an internal position starting about 21 amino acids from the N terminus (9, 23), and it was shown by mutagenesis that cysteine residues flanking this region are important for fusion activity (10).
Although the fusion proteins of influenza virus and flaviviruses have very dissimilar structures, it is intriguing that their fusion peptides both contain the tetrapeptide sequence GLFG. Mutagenesis studies with influenza virus hemagglutinin have demonstrated that at least the first glycine residue of this tetrapeptide is critically important for fusion (16, 35, 45). It is therefore likely that this motif has properties that are conducive to membrane interactions, a notion that is also supported by experiments with synthetic peptides (11, 33). A similar tetrapeptide sequence, GFLG, is found in the N-terminal fusion peptides of several retroviruses (11, 33) and hepatitis B virus (38). In the case of gp41 of human immunodeficiency virus, the conserved Phe appears to be important for maintaining the functionally active conformation of the fusion peptide (34). The sequence GFFG occurs naturally in a wild-type strain of the flavivirus Powassan virus at the position corresponding to GLFG in TBE virus (30), and we show here that TBE virus RSPs carrying this sequence (RSP-107F) are still capable of undergoing fusion with membranes, albeit less efficiently.
A number of viral fusion proteins appear to belong to a structural superfamily whose members have in common the capability of adopting a characteristic core structure involving a trimeric coiled coil of α helices. These proteins all have either N-terminal or N-proximal fusion peptides that reside at or near the tip of the coiled coil in the fusion-active form. On the other hand, fusion proteins not belonging to this structural class, i.e., those that are not cleaved and are not predicted to form coiled coils, appear to have fusion peptides within internal loop structures, distant from the N terminus, as has been shown by mutagenesis (26, 28) in the case of the alphaviruses and by both mutagenesis (14, 50) and direct labeling experiments (12) in the case of rhabdoviruses. The TBE virus E protein is currently the only viral fusion protein of the non-coiled-coil type for which a high-resolution structure is available and thus provides the first example of the native structure of a probable internal fusion peptide. Comparison of the properties of these functionally analogous but structurally and mechanistically distinct protein classes should facilitate the identification of essential features shared by both and thereby help to reveal some of the fundamental principles common to protein-mediated fusion of membranes.
ACKNOWLEDGMENTS
We thank Melby Wilfinger, Angela Dohnal, Walter Holzer, and Silvia Röhnke for excellent technical assistance.
Part of this work was supported by a grant from the International Human Frontier Science Program.
REFERENCES
- 1.Allison S L, Mandl C W, Kunz C, Heinz F X. Expression of cloned envelope protein genes from the flavivirus tick-borne encephalitis virus in mammalian cells and random mutagenesis by PCR. Virus Genes. 1994;8:187–198. doi: 10.1007/BF01703077. [DOI] [PubMed] [Google Scholar]
- 2.Allison S L, Schalich J, Stiasny K, Mandl C W, Kunz C, Heinz F X. Oligomeric rearrangement of tick-borne encephalitis virus envelope proteins induced by an acidic pH. J Virol. 1995;69:695–700. doi: 10.1128/jvi.69.2.695-700.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allison S L, Stadler K, Mandl C W, Kunz C, Heinz F X. Synthesis and secretion of recombinant tick-borne encephalitis virus protein E in soluble and particulate form. J Virol. 1995;69:5816–5820. doi: 10.1128/jvi.69.9.5816-5820.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Allison S L, Stiasny K, Stadler K, Mandl C W, Heinz F X. Mapping of functional elements in the stem-anchor region of tick-borne encephalitis virus envelope protein E. J Virol. 1999;73:5605–5612. doi: 10.1128/jvi.73.7.5605-5612.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker K A, Dutch R E, Lamb R A, Jardetzky T S. Structural basis for paramyxovirus-mediated membrane fusion. Mol Cell. 1999;3:309–319. doi: 10.1016/s1097-2765(00)80458-x. [DOI] [PubMed] [Google Scholar]
- 6.Blok J, Samuel S, Gibbs A J, Vitarana U T. Variation of the nucleotide and encoded amino acid sequences of the envelope gene from eight dengue-2 viruses. Arch Virol. 1989;105:39–53. doi: 10.1007/BF01311115. [DOI] [PubMed] [Google Scholar]
- 7.Clarke D H, Casals J. Techniques for hemagglutination and hemagglutination inhibition with arthropod-borne viruses. Am J Trop Med Hyg. 1958;7:561–573. doi: 10.4269/ajtmh.1958.7.561. [DOI] [PubMed] [Google Scholar]
- 8.Corver J, Ortiz A, Allison S L, Schalich J, Heinz F X, Wilschut J. Membrane fusion activity of tick-borne encephalitis virus and recombinant subviral particles in a liposomal model system. Virology. 2000;269:37–46. doi: 10.1006/viro.1999.0172. [DOI] [PubMed] [Google Scholar]
- 9.Delos S E, Gilbert J M, White J M. The central proline of an internal viral fusion peptide serves two important roles. J Virol. 2000;74:1686–1693. doi: 10.1128/jvi.74.4.1686-1693.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Delos S E, White J M. Critical role for the cysteines flanking the internal fusion peptide of avian sarcoma/leukosis virus envelope glycoprotein. J Virol. 2000;74:9738–9741. doi: 10.1128/jvi.74.20.9738-9741.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Durell S R, Martin I, Ruysschaert J M, Shai Y, Blumenthal R. What studies of fusion peptides tell us about viral envelope glycoprotein-mediated membrane fusion. Mol Membr Biol. 1997;14:97–112. doi: 10.3109/09687689709048170. . (Review.) [DOI] [PubMed] [Google Scholar]
- 12.Durrer P, Gaudin Y, Ruigrok R W, Graf R, Brunner J. Photolabeling identifies a putative fusion domain in the envelope glycoprotein of rabies and vesicular stomatitis viruses. J Biol Chem. 1995;270:17575–17581. doi: 10.1074/jbc.270.29.17575. [DOI] [PubMed] [Google Scholar]
- 13.Ferlenghi, I., M. Clarke, T. Rutten, S. L. Allison, J. Schalich, F. X. Heinz, S. C. Harrison, F. A. Rey, and S. D. Fuller. Molecular organization of a recombinant subviral particle from tick-borne encephalitis virus. Mol. Cell, in press. [DOI] [PubMed]
- 14.Fredericksen B L, Whitt M A. Vesicular stomatitis virus glycoprotein mutations that affect membrane fusion activity and abolish virus infectivity. J Virol. 1995;69:1435–1443. doi: 10.1128/jvi.69.3.1435-1443.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaudin Y, Tuffereau C, Durrer P, Brunner J, Flamand A, Ruigrok R. Rabies virus-induced membrane fusion. Mol Membr Biol. 1999;16:21–31. doi: 10.1080/096876899294724. [DOI] [PubMed] [Google Scholar]
- 16.Gething M J, Doms R W, York D, White J. Studies on the mechanism of membrane fusion: site-specific mutagenesis of the hemagglutinin of influenza virus. J Cell Biol. 1986;102:11–23. doi: 10.1083/jcb.102.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guirakhoo F, Heinz F X, Kunz C. Epitope model of tick-borne encephalitis virus envelope glycoprotein E: analysis of structural properties, role of carbohydrate side chain, and conformational changes occurring at acidic pH. Virology. 1989;169:90–99. doi: 10.1016/0042-6822(89)90044-5. [DOI] [PubMed] [Google Scholar]
- 18.Harter C, James P, Bachi T, Semenza G, Brunner J. Hydrophobic binding of the ectodomain of influenza hemagglutinin to membranes occurs through the “fusion peptide.”. J Biol Chem. 1989;264:6459–6464. [PubMed] [Google Scholar]
- 19.Heinz F X, Stiasny K, Puschner A G, Holzmann H, Allison S L, Mandl C W, Kunz C. Structural changes and functional control of the tick-borne encephalitis virus glycoprotein E by the heterodimeric association with protein prM. Virology. 1994;198:109–117. doi: 10.1006/viro.1994.1013. [DOI] [PubMed] [Google Scholar]
- 20.Heinz F X, Tuma W, Guirakhoo F, Kunz C. A model study of the use of monoclonal antibodies in capture enzyme immunoassays for antigen quantification exploiting the epitope map of tick-borne encephalitis virus. J Biol Stand. 1986;14:133–141. doi: 10.1016/0092-1157(86)90032-6. [DOI] [PubMed] [Google Scholar]
- 21.Helenius A. Alphavirus and flavivirus glycoproteins: structures and functions. Cell. 1995;81:651–653. doi: 10.1016/0092-8674(95)90523-5. [DOI] [PubMed] [Google Scholar]
- 22.Hernandez L D, Hoffman L R, Wolfsberg T G, White J M. Virus-cell and cell-cell fusion. Annu Rev Cell Dev Biol. 1996;12:627–661. doi: 10.1146/annurev.cellbio.12.1.627. [DOI] [PubMed] [Google Scholar]
- 23.Hernandez L D, White J M. Mutational analysis of the candidate internal fusion peptide of the avian leukosis and sarcoma virus subgroup A envelope glycoprotein. J Virol. 1998;72:3259–3267. doi: 10.1128/jvi.72.4.3259-3267.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hughson F M. Enveloped viruses: a common mode of membrane fusion? Curr Biol. 1997;7:R565–R569. doi: 10.1016/s0960-9822(06)00283-1. [DOI] [PubMed] [Google Scholar]
- 25.Kielian M. Membrane fusion and the alphavirus life cycle. Adv Virus Res. 1995;45:113–151. doi: 10.1016/s0065-3527(08)60059-7. [DOI] [PubMed] [Google Scholar]
- 26.Kielian M, Klimjack M R, Ghosh S, Duffus W A. Mechanisms of mutations inhibiting fusion and infection by Semliki Forest virus. J Cell Biol. 1996;134:863–872. doi: 10.1083/jcb.134.4.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klenk H D, Garten W. Activation cleavage of viral spike proteins. In: Wimmer E, editor. Cellular receptors for animal viruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1994. pp. 241–280. [Google Scholar]
- 28.Levy-Mintz P, Kielian M. Mutagenesis of the putative fusion domain of the Semliki Forest virus spike protein. J Virol. 1991;65:4292–4300. doi: 10.1128/jvi.65.8.4292-4300.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mandl C W, Guirakhoo F, Holzmann H, Heinz F X, Kunz C. Antigenic structure of the flavivirus envelope protein E at the molecular level, using tick-borne encephalitis virus as a model. J Virol. 1989;63:564–571. doi: 10.1128/jvi.63.2.564-571.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mandl C W, Holzmann H, Kunz C, Heinz F X. Complete genomic sequence of Powassan virus: evaluation of genetic elements in tick-borne versus mosquito-borne flaviviruses. Virology. 1993;194:173–184. doi: 10.1006/viro.1993.1247. [DOI] [PubMed] [Google Scholar]
- 31.Monath T P, Heinz F X. Flaviviruses. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Straus S E, editors. Virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 961–1034. [Google Scholar]
- 32.Nitayaphan S, Grant J A, Chang G J, Trent D W. Nucleotide sequence of the virulent SA-14 strain of Japanese encephalitis virus and its attenuated vaccine derivative, SA-14-14-2. Virology. 1990;177:541–552. doi: 10.1016/0042-6822(90)90519-w. [DOI] [PubMed] [Google Scholar]
- 33.Pécheur E I, SainteMarie J, Bienvenue A, Hoekstra D. Peptides and membrane fusion: towards an understanding of the molecular mechanism of protein-induced fusion. J Membr Biol. 1999;167:1–17. doi: 10.1007/s002329900466. [DOI] [PubMed] [Google Scholar]
- 34.Pritsker M, Rucker J, Hoffman T L, Doms R W, Shai Y. Effect of nonpolar substitutions of the conserved Phe(11) in the fusion peptide of HIV-1 gp41 on its function, structure, and organization in membranes. Biochemistry. 1999;38:11359–11371. doi: 10.1021/bi990232e. [DOI] [PubMed] [Google Scholar]
- 35.Qiao H, Armstrong R T, Melikyan G B, Cohen F S, White J M. A specific point mutant at position 1 of the influenza hemagglutinin fusion peptide displays a hemifusion phenotype. Mol Biol Cell. 1999;10:2759–2769. doi: 10.1091/mbc.10.8.2759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rey F A, Heinz F X, Mandl C, Kunz C, Harrison S C. The envelope glycoprotein from tick-borne encephalitis virus at 2 Å resolution. Nature. 1995;375:291–298. doi: 10.1038/375291a0. [DOI] [PubMed] [Google Scholar]
- 37.Rice C M. Flaviviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, Chanock R M, Melnick J L, Monath T P, Roizman B, Straus S E, editors. Virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 931–595. [Google Scholar]
- 38.Rodriguez-Crespo I, Nunez E, Gomez G J, Yelamos B, Albar J P, Peterson D L, Gavilanes F. Phospholipid interactions of the putative fusion peptide of hepatitis B virus surface antigen S protein. J Gen Virol. 1995;76:301–308. doi: 10.1099/0022-1317-76-2-301. [DOI] [PubMed] [Google Scholar]
- 39.Roehrig J T, Hunt A R, Johnson A J, Hawkes R A. Synthetic peptides derived from the deduced amino acid sequence of the E-glycoprotein of Murray Valley encephalitis virus elicit antiviral antibody. Virology. 1989;171:49–60. doi: 10.1016/0042-6822(89)90509-6. [DOI] [PubMed] [Google Scholar]
- 40.Roehrig J T, Johnson A J, Hunt A R, Bolin R A, Chu M C. Antibodies to dengue 2 virus E-glycoprotein synthetic peptides identify antigenic conformation. Virology. 1990;177:668–675. doi: 10.1016/0042-6822(90)90532-v. [DOI] [PubMed] [Google Scholar]
- 41.Rosenthal P B, Zhang X, Formanowski F, Fitz W, Wong C H, Meier E H, Skehel J J, Wiley D C. Structure of the haemagglutinin-esterase-fusion glycoprotein of influenza C virus. Nature. 1998;396:92–96. doi: 10.1038/23974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schalich J, Allison S L, Stiasny K, Mandl C W, Kunz C, Heinz F X. Recombinant subviral particles from tick-borne encephalitis virus are fusogenic and provide a model system for studying flavivirus envelope glycoprotein functions. J Virol. 1996;70:4549–4557. doi: 10.1128/jvi.70.7.4549-4557.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Singh M, Berger B, Kim P S. LearnCoil-VMF: computational evidence for coiled-coil-like motifs in many viral membrane-fusion proteins. J Mol Biol. 1999;290:1031–1041. doi: 10.1006/jmbi.1999.2796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Skehel J J, Wiley D C. Coiled coils in both intracellular vesicle and viral membrane fusion. Cell. 1998;95:871–874. doi: 10.1016/s0092-8674(00)81710-9. [DOI] [PubMed] [Google Scholar]
- 45.Steinhauer D A, Wharton S A, Skehel J J, Wiley D C. Studies of the membrane fusion activities of fusion peptide mutants of influenza virus hemagglutinin. J Virol. 1995;69:6643–6651. doi: 10.1128/jvi.69.11.6643-6651.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stiasny K, Allison S L, Marchler-Bauer A, Kunz C, Heinz F X. Structural requirements for low-pH-induced rearrangements in the envelope glycoprotein of tick-borne encephalitis virus. J Virol. 1996;70:8142–8147. doi: 10.1128/jvi.70.11.8142-8147.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Volkova T D, Vorovitch M F, Ivanov V T, Timofeev A V, Volpina O M. A monoclonal antibody that recognizes the predicted tick-borne encephalitis virus E protein fusion sequence blocks fusion. Arch Virol. 1999;144:1035–1039. doi: 10.1007/s007050050566. [DOI] [PubMed] [Google Scholar]
- 48.Weissenhorn W, Dessen A, Calder L J, Harrison S C, Skehel J J, Wiley D C. Structural basis for membrane fusion by enveloped viruses. Mol Membr Biol. 1999;16:3–9. doi: 10.1080/096876899294706. [DOI] [PubMed] [Google Scholar]
- 49.Wilson I A, Skehel J J, Wiley D C. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 Å resolution. Nature. 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 50.Zhang L, Ghosh H P. Characterization of the putative fusogenic domain in vesicular stomatitis virus glycoprotein G. J Virol. 1994;68:2186–2193. doi: 10.1128/jvi.68.4.2186-2193.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]