Abstract
In this investigation, single nucleotide variants (SNVs) within the chicken interferon-inducible transmembrane protein (chIFITM) genes were explored in Aseel and Kadaknath breeds. Comparative analysis with the GRCg6a reference genome revealed 9 and 16 SNVs in the chIFITM locus for Aseel and Kadaknath breeds, respectively. When referencing the Genome Reference Consortium GRCg7b, Kadaknath exhibited 10 variants, contrasting with none in Aseel. Notably, 17, 8, 2, and 5 SNVs were identified in chIFITM1, chIFITM2, chIFITM3, and chIFITM5 genes, with chIFITM1 showing the highest polymorphism in Kadaknath, featuring 10 intronic variants, including three SNVs (rs16457112, rs16457111, and rs313341707) common to both breeds. Two synonymous exonic variants (g.1817767C > A and g.1819102C > T) were also noted in chIFITM1. Although chIFITM protein sequences were generally conserved, genetic variations clustered predominantly in UTR and intronic regions. Examination of immune response dynamics in live embryos uncovered notable variations in chIFITM gene expression across diverse organs and chicken breeds. Specifically, chIFITM1 mRNA was abundant in cecal tonsils for both breeds and bursa of Aseel (7.61 folds), but it was absent in the heart and lung tissues of both breeds. Conversely, chIFITM3 consistently exhibited heightened expression, particularly in bursa of Aseel (10.23 folds). Whereas mRNA of the chIFITM2 gene was found to be abundant in the heart of Kadaknath (11.03 folds) and lung of both breeds. Furthermore, the expression pattern of chIFITM5 diverged between the two breeds, the heart of Kadaknath chickens showed highest (10.45 folds). The study discovered that breed-specific genetic variants within these genes present a potential pathway for selection and breeding to improve disease resistance in chicken. The observed genetic variation among chicken populations highlights the critical importance of these variants in reinforcing virus resistance, exhibiting applicability across a wide range of breeds.
Keywords: chIFITM, IR-IFITM, IFN γ, Mx, Aseel, Kadaknath, SNVs, Chicken
Highlights
-
•
The chIFITM1 gene was found to be highly polymorphic in Kadaknath chicken breed.
-
•
The robust and consistent expression of the chIFITM3 gene across various tissues substantiates its potent antiviral efficacy against viral infections.
-
•
The coding sequences of all chIFITM genes demonstrate a high level of conservation.
1. Introduction
Interferon-induced transmembrane protein (IFITM) gene family was first discovered as an IFN-stimulable response element (ISRE) in human neuroblastoma cells [1]. They were primarily named as 9–27 (IFITM1), 1-8D (IFITM2), and 1–8U (IFITM3) [2] and found that it has antiviral activity towards various viruses affecting humans, such as vesicular stomatitis virus (Alber and Staeheli 1996), highly pathogenic Influenza A virus, West Nile Virus, Dengue Virus [3], Zika Virus [4]. Besides, it was also found that they had a strong inhibitory effect on the replication by restricting the viral attachment with the cell membrane of animal and avian virus [5]. Recent evolutionary study reports showed that IFITM genes were positively selected as potent innate genes [[6], [7], [8]] and showed resistance against various livestock diseases such as African swine fever virus (ASFV) [9], Avian and swine influenza viruses [[10], [11], [12], [13], [14], [15]], Lyssaviruses [11], foot and mouth disease virus [16], Avian reovirus [17], Infectious bronchitis virus [18,19] and Newcastle disease virus [20].
A decade ago, human IFITM genes were characterized and found five IFITM homologs (IFITM1, IFITM2, IFITM3, IFITM5, and IFITM10) in the human IFITM gene locus [21]. Further, three more—IFITM4, IFITM6, and IFITM9 have been discovered in zebra fish [6]. In addition to huIFITMs, IFITMs from mice, Swine [15,22,23], Ducks [24], Goose [25] and Canine [26], Chicken [27] have also been studied by various scientists. Recently, re-sequencing was utilized to characterize the chIFITM area, and it found that this locus does not appear to have a lot of repetitive sequences or a very high GC content [10].
The chicken IFITM gene locus is surrounded by the genes for the centromeric Acid Trehalase-Like 1 (ATHL1) gene and the telomeric β −1,4-N Acetyl-Galactosaminyl Transferase 4 (B4GALNT4) in the q arm of Chromosome 5 of its genome [6,11,27,28]. These two genes are highly conserved among species, from mammals to amphibians [11]. Relatively high levels of sequence homology (41.9–67.2 per cent) were found between the chicken and human IFITM genes [10]. The chicken IFITM family comprises five members of the gene, and they were grouped as immunity-related IFITM (IFITM1, IFITM2, and IFITM3) and IFITM5 and IFITM10 were exerts indistinct immune function [29,30]. The vertebrate IFITM genes are typically found in two loci, one containing the IFITM10 gene and the other having IFITM5 and IR-IFITM (IFITM 1, 2, 3) genes [6,27,28]. The IFITM5 gene encodes a membrane protein that is well-known to play a role in the formation, osteoblast maturation and mineralization of bones [5,22] and it emerged from the bony fish species. The goal of IFITM10 is still not understood and it was revealed that it may be restricted to the tetrapod species [5,6,11,29]. Furthermore, it was discovered that the chIFITM2 and chIFITM1 loci are oriented oppositely compared to their human counterparts. Also, the transcriptional units of the chIFITM1,2, 3, and 5 genes have a consistent orientation [27]. It has been found that the IR-chIFITM genes located at chromosome 5q are in the following order: centromeric - B4GALNT4 - chIFITM3 - chIFITM2 - chIFITM1 -chIFITM5 - ATHL1 – telomeric [28]. The chicken IFITM locus was previously described as IFITM1 and has been annotated as ‘chIFITM3’ and IFITM3 was called ‘chIFITM1’ [11]. All the chIFITM genes continue to have their characteristic genetic structure, with two exons separated by a single intron [11,27].
The evolution of IFITM genes in Avian species had shown patterns of gene duplication and positive selection [28]. Particularly in the IR-IFITM sub-family, which had shown many duplications specific to certain species and lineages. Considering the long interaction of vertebrates with viral infections and positive selection, suggests that the IR-IFITM gene variations could be linked to defense mechanisms [6]. Complementing this perspective, more recent studies have indicated that the SNVs rs12252 and rs6598045 within the huIFITM3 gene are significantly associated with the intensity of IAV infections and the death rates from COVID-19 [[31], [32], [33], [34], [35], [36]]. Further, subtle changes in the IFITM3 gene correlate with higher susceptibility to ulcerative colitis [37] and also a raised risk of developing hemorrhagic fever with renal syndrome [38,39]. Concurrently, specific SNPs in the chIFITM1 and 3 genes are closely related to the susceptibility of chickens to the H7N9 virus [32]. Consistent with other ISGs, it is suggested that genetic variations might be associated with infection susceptibility or severity both within and between species. While variations in IFITM have been correlated with differential susceptibility to infections in various bird species [40], a thorough investigation into the IFITM locus genetic variations, especially in Indian chicken breeds like Aseel and Kadaknath, has yet to be carried out. These chicken breeds are reared in the backyard, which have adapted to their native environment, including weather, endemic diseases, and feeding preferences. As a result, they may have distinct and continuous positive selection for their immunity, illness resistance, and tolerance to tropical environments [41]. Immunological responses of Aseel, Kadaknath, and White Leghorn chickens to sheep RBS cells and NDV demonstrated that Aseel and Kadaknath chickens had greater immunological reactivity than White Leghorn chicks [20,42]. However, there is no research on the IFITM gene in India's native chickens like Aseel and Kadaknath, hence this study has been made for analyzing the SNV variations of the IR-chIFITM genes in Aseel and Kadaknath chicken breeds. It is essential to uncover genetic alterations that might be highlighted significant differences between these two breeds.
2. Materials and methods
2.1. Experimental birds and sample collection
A total of 72 blood samples (36 Aseel and 36 Kadaknath) were collected from different poultry flocks viz., Poultry Farm, Veterinary College and Research Institute, Namakkal; Regional Research and Education Centre, Pudukkottai, and local poultry farmer's flock (Dindigul, Karur and Mohanur) of Tamil Nadu, South India. The genomic DNA from the blood samples of Aseel and Kadaknath was extracted using DNeasy Blood and Tissue Kit (Qiagen, Germany Cat. No. 69504) as per the manufacturer's protocol. The genomic DNA samples of both breeds were pooled into two pools per breed (18 samples per pool) [43].
Sixty (30 Aseel and 30 Kadaknath) SPF chicken embryos, 19 days old, were acquired from the Department of Poultry Science at VCRI in Namakkal and utilized in the viral challenge study. With the exception of a control group of 15 Aseel and 15 Kadaknath, the remaining 15 Aseel and 15 Kadaknath embryos were exposed to a velogenic strain of Newcastle disease virus (NDV) at a dosage of 50 percent Embryo Infective Dose (105 EID50). Subsequently, these embryos were maintained in an egg incubator at a temperature of 38 °C and a relative humidity range of 65–75 percent until tissue harvesting at various hours after infection.
2.2. Primer designing
Primers for chIFITM genes were designed for PCR amplification of chIFITM1, 2, 3 and 5 genes, and also for real time qPCR amplification of targeted genes (Table 1). Primers were designed based on the Chicken (taxid:9031) sequences (NC_052536.1) available in the NCBI by using the online primer designing tool Primer-NCBI BLAST/Primer 3 software (https://www.ncbi.nlm.nih.gov/tools/primer-blast/). A total of 10 g of pooled genomic DNA per sample was utilized for PCR to amplify the complete chIFITM1, 2, 3 and 5 genes, which ranged between 1.8 and 2.1 kb in size.
Table 1.
Primers designed for amplification of chIFITM genes.
| Gene Name | Forward/Reverse | Sequence (5′-3′) | Primer length (bp) | Amplicon Size (bp) |
|---|---|---|---|---|
| chIFITM1 | Forward | TATCCTTTACACCACAGCTGGC | 22 | 1799 |
| Reverse | TTGTCCAATCACCTTGTCCTAGC | 23 | ||
| chIFITM2 | Forward | CAGCTAGGACAAGGTGATTGGAC | 23 | 2173 |
| Reverse | ATGGCTAGAGAAGCATGGAACTT | 23 | ||
| chIFITM3 | Forward | AACATCCAGGCTCACCGCTA | 20 | 1764 |
| Reverse | TCACCCATCCGGGAGCTTAG | 20 | ||
| chIFITM5 | Forward | ACTGCCTTACCAGACATCTTCAG | 23 | 1928 |
| Reverse | ACACATTTGGCTGGACTAGATGA | 23 |
2.3. Sequencing and SNV prediction
The Illumina deep sequencing method was used to sequence the samples. The samples have been sequenced with 15× coverage with an average read length of 150bp. Sequence reads were analyzed for quality by FastaQC and low-quality reads were trimmed using Trimmomatic [45]. The mean phred score value for the sequence quality across each base position (150bp) in the read ranges from 36.29 to 33.31. After trimming, the sequences were stored in fastaQ format for further analysis [46]. All sequences were submitted to the Sequence Read Archive (SRA) of NCBI in FASTQ format, and they were assigned with the following accession numbers i.e., SRR24893278 and SRR24793329 for the Aseel female and male samples, while SRR24881115 and SRR24881091 for the Kadaknath female and male samples. The trimmed sequences were mapped with the reference genome [45,46] The reference genome Red Jungle fowl (GRCg6a) [47] and Cross of Broiler mother x White Leghorn layer father (GRCg7b) [48] were used to assess and categorize the location and type of consequence predicted for each SNV. The results were visualized using the Integrated Genome Browser [49]. SAMtools 0.1.19 and BCFtools [50,51] managed the final variant calling, producing a combined VCF for all samples [45,52,53]. All SNVs were screened in the VCF file (Variant call file) and the SNVs were characterized based on location (Intronic, Exon, upstream, downstream, splice site, 5′ UTR, or 3’ UTR).
2.4. Validation of chIFITM genes with gene-specific primers
The chIFITM genes were amplified to validate the gene using specifically designed primers in a Thermal Cycler (Bio-Rad). A total reaction volume of PCR mixture was 25 μL, with 1 μL DNA template, 1 μL of each primer (10 pM), 12.5 μL Ampliqon Taq 2× PCR Master mix (comprising 1.5 mM MgCl2, Taq DNA polymerase, and 100 μM dNTPs), and 9.5 μL nuclease-free water. The amplification condition was set with initial denaturation at 95 °C for 3 min; 35 cycles of denaturation at 95 °C for 30 s, annealing at 57.5 °C/56.5 °C/63.6 °C/62.5 °C for 30 s (based on the specific primer set in use), and extension at 72 °C for 1.5 min and the final extension at 72 °C for 5 min. The PCR products were evaluated using 2 % agarose gel electrophoresis. The complete gene size of DNA fragments of chIFITM1, 2, 3 and 5 genes were amplified, and the corresponding PCR product sizes were 1799, 2173, 1764 and 1928 bp respectively.
2.5. In ovo gene expression profiling
Various organ tissues such as the Lung, Heart, Liver, Intestine and Bursa of Fabricius were collected from NDV infected and control Aseel and Kadaknath chicken embryos at 3, 6, 12, 24 and 48 hpi. Tissues were immediately stored in RNAlater® (ThermoFisher) at −80 °C until RNA isolation. The total RNA was extracted by Trizole method using RNAiso Plus, M/s Takara, (Cat. No: 9109). The relative expression of specific mRNA was quantified [54] by a real-time thermal cycler (Roche LightCycler® 96). Gene expression studies, include all the four targeted genes (chIFITM1, 2, 3 and 5) alone with two positive immune-related genes (IFN- γ and Mx) were assessed and β-actin was the gene used as the reference gene. The Mx gene is included as positive immune gene against viruses and also it was an Interferon Stimulating Gene family. The quantitative Real-time PCR (qRT-PCR) was performed to quantify the targeted genes using the designed primers (Table 2) and Bio-Rad Univer SYBR green Master mix (Cat.No: 1725271). The real-time PCR cycling conditions were, initial denaturation at 95 °C for 10 min followed by 40 cycles of denaturation at 95 °C for 10 min, followed by 40 cycles of 95 °C for 10s and 60 °C–62 °C (β Actin, IFN γ, Mx, chIFITM (1, 2, 3 and 5) for 45s and 72 °C for 15s. For each, the sample dissociation curve (melt curve) was generated after the completion of amplification. A negative control containing all ingredients except the cDNA template (non-template control; NTC) was set up invariably for each master mix made for conducting the reactions. Each sample was run in duplicate. Transcript levels were normalized to those for the chicken housekeeping gene β-actin. The alteration in mRNA levels of each gene in NDV-infected embryos was quantified as a fold change using the 2−ΔΔCt method [55], in comparison to the non-infected mock control.
Table 2.
List of primer sequences for qRT-PCR.
| Gene | Primer | Sequence (5'--3′) | Reference |
|---|---|---|---|
| chIFITM1 | Forward | GCAGGATGTGACCACCACTA | NM_001350059.2 |
| Reverse | CTTCGCTGTCCTCCCATAGC | ||
| chIFITM2 | Forward | AACAGGCGGAGGTGAGCAT | NM_001350058.2 |
| Reverse | AAGATGAGCGAGGGGAAGCA | ||
| chIFITM3 | Forward | CGTGAAGTCCAGGGATCGCA | NM_001350061.2 |
| Reverse | GCAACCAGGGCGATGATGAG | ||
| chIFITM5 | Forward | CCAACCCCACTTCTGGACGA | NM_001199498.1 |
| Reverse | ATCACTCCGAAGGGCACGAC | ||
| chMx | Forward | GTCCAAGAGGCTGAATAACAGAG | NM_204609 |
| Reverse | GTCGGATCTTTCTGTCATATTGG | ||
| chIFN-γ | Forward | TGAGCCAGATTGTTTCGATG | [44] |
| Reverse | CTTGGCCAGGTCCATGATA | ||
| chβ-Actin | Forward | TATGTGCAAGGCCGGTTTC | |
| Reverse | TGTCTTTCTGGCCCATACCAA |
2.6. Prediction of chIFITM proteins and comparison
The potential open reading frames (ORFs) of the respective chIFITM genes were predicted by ORF finder in NCBI database (https://www.ncbi.nlm.nih.gov/gorf/gorf.html). Based on this, the corresponding coding sequences for Aseel and Kadaknath IFITM genes were taken. The DNA sequences of these breeds were subsequently translated into their peptide sequences using the EMBOSS Transeq tool available online (https://www.ebi.ac.uk/Tools/st/emboss_transeq/). A comprehensive alignment of these translated amino acid sequences, benchmarked against the reference chicken IFITM protein sequence sourced from the Uniprot database, was executed using Kalign ClustalW [12].
2.7. Statistical analysis
Gene expression was assessed in five distinct tissues at five distinct time points in each of the two breeds. The time points in each breed's tissue were 3, 6, 12, 24, and 48 h following infection. ANOVA was used to analyze the significance of the mRNA expression levels within breeds and confirm any variation in time intervals within breeds. The significance of the gene expression level at various times in each tissue between breeds was later analyzed using the t-test because only two breeds were compared. All analyses were performed using the R program (version 4.2), with a p-value <0.05 considered statistically significant. Graphic illustrations were also created using R.
3. Results
The sequences were evaluated after being deep sequenced using Illumina-based NGS technology platforms. The preliminary analysis targeted the chIFITM locus, identifying the presence of variants and their distribution in each chIFITM genes of the respective chicken breeds relative to the reference chicken genome (Table 3). The results showed the presence of nine and sixteen SNVs across the chIFITM locus in Aseel and Kadaknath breeds, respectively with respective to the reference genome Red Jungle fowl (GRCg6a) across coding and non-coding regions of the locus. In contrast, using the Cross of Broiler mother x White Leghorn layer father (GRCg7b) as the reference genome, the Kadaknath chicken breed exhibited ten variants in the chIFITM locus and for the Aseel breed, no SNVs were identified.
Table 3.
Number of SNVs in chIFITMs gene as per the reference genomes.
| Gene Symbol |
GRCg6a |
GRCg7b |
||
|---|---|---|---|---|
| Aseel | Kadaknath | Aseel | Kadaknath | |
| chIFITM 1 | 3 | 14 | – | 3 |
| chIFITM 2 | 4 | – | – | 4 |
| chIFITM 3 | 1 | 1 | – | – |
| chIFITM 5 | 1 | 1 | – | 3 |
| Total number of SNVs | 9 | 16 | – | 10 |
3.1. IR-IFITM gene variants
Based on the reference sequences from the Genome Reference Consortium (GRCg6a and GRCg7b) accessible in the NCBI, the coding and non-coding regions of the three antiviral IR-chIFITM and chIFITM5 (two exons and one intron per IFITM) genes were investigated for nucleotide variation.
The chIFITM1 gene was located in the locus LOC422993 of chromosome number 5:g.1817672-1819250bp position and found 14 SNVs in the region of the chIFITM1 gene with respect to the Genome Reference Consortium GRCg6a (Table 4). This comprised 10 SNVs in the intronic region, three of which were present in both Aseel and Kadaknath breeds, whereas the remaining seven were only discovered in the Kadaknath chIFITM1 gene. In the exonic region of the chIFITM1 gene in Kadaknath, single nucleotide variations were discovered, i.e., g.1817767C > A and g.1819102C > T in Exon 1 and 2, respectively, whereas g.1819135A > G and g.1819157C > T were present in three prime untranslated region (3’ UTR) region of Exon 2. These SNVs were confirmed to be available in the Ensembl genome database.
Table 4.
SNVs of chIFITM1 as per the Red Jungle fowl reference genome (GRCg6a).
| Position as per GRCg6a | Reference Allele |
Alternate Allele |
Variation Type | Consequence | Location | Feature Sequence | Existing Variation |
Breed |
|---|---|---|---|---|---|---|---|---|
| 1817767 | C | T | SNP | Synonymous variant | EXON 1 | NM_001350059.1 | rs317414539 | Kadaknath |
| 1819102 | C | A | EXON 2 | NM_001350059.1 | rs316752027 | Kadaknath | ||
| 1819135 | A | G | 3′ UTR variant | NM_001350059.1 | rs16457103 | Kadaknath | ||
| 1819157 | C | T | NM_001350059.1 | rs317795576 | Kadaknath | |||
| 1817999 | G | C | Intronic variant | INTRON 1 | ERR7564039.23466 | rs16457112 | Aseel/Kadaknath | |
| 1818006 | A | G | ERR7564039.23466 | rs16457111 | Aseel/Kadaknath | |||
| 1818213 | T | C | ERR7564039.23466 | rs313341707 | Aseel/Kadaknath | |||
| 1818260 | C | T | ERR7564039.23466 | rs313434559 | Kadaknath | |||
| 1818278 | A | G | ERR7564039.23466 | rs313015881 | Kadaknath | |||
| 1818455 | T | C | ERR7564039.23466 | rs16457108 | Kadaknath | |||
| 1818462 | G | A | ERR7564039.23466 | rs314739981 | Kadaknath | |||
| 1818473 | A | G | ERR7564039.23466 | rs16457107 | Kadaknath | |||
| 1818835 | T | C | ERR7564023.10248 | rs13649904 | Kadaknath | |||
| 1818841 | G | A | ERR7564023.10248 | rs16457105 | Kadaknath |
While the chIFITM2 gene was positioned next to the chIFITM1 gene at the locus LOC107053353 in the position g.1819414 - 1821290 of chicken chromosomal number 5, in chIFITM2 of the Aseel chicken, four distinct nucleotide variances were found in relation to the reference genome GRCg6a (Table 5). A complex nucleotide variant was noticed as CCAC > TCAT at five prime untranslated region (5′UTR) of Exon1, which was the combination of two SNPs viz., rs16457102 and rs16457101. Detection of a downstream gene variant at exon 1 of the chIFITM2 gene revealed multi-nucleotide polymorphism (MNP) as g.1820823CC > TG. This demonstrated the merging of two existing SNPs viz., rs735111122 and rs739838225, in SNPdb. Two A > G transition mutations were identified at the 3′UTR and 5′UTR of exon 1 and 2, respectively, of chIFITM2.
Table 5.
SNVs of chIFITM2 as per the Red Jungle fowl reference genome (GRCg6a).
| Position as per GRCg6a | Reference Allele |
Alternate Allele |
Variation Type | Consequence | Location | Feature Sequence | Existing Variation |
Breed |
|---|---|---|---|---|---|---|---|---|
| 1819423 -1819426 | CCAC | TCAT | Complex | 5′ UTR variant | EXON 1 | NM_001350058.1 | rs16457102/rs16457101 | Aseel |
| 1820823 -1820824 | CC | TG | MNP | 3′ UTR variant | EXON 1 | ERR7564035.14760 | rs735111122/rs739838225 | Aseel |
| 1820837 | A | G | SNP | 3′ UTR variant | EXON 1 | ERR7564035.14760 | rs732321039 | Aseel |
| 1821251 | A | G | 5′ UTR variant | EXON 2 | ERR2368565.61130 | rs14508756 | Aseel |
As per the Genome Reference Consortium GRCg6a, the chIFITM3 gene was located at the LOC770612 locus of chromosome 5g.1814234-1815550, which was flanked at the 5′prime by the B4GALNT4 genes and the 3′prime by the chIFITM1 genes. A transition SNP at g.1814938G > A in the 3′UTR of Exon 1 in and a transversion SNP at g.1815386A > C of Exon 2 were found, respectively (Table 6) in Aseel and Kadaknath breeds.
Table 6.
SNVs of chIFITM3 as per the Red Jungle fowl reference genome (GRCg6a).
| Position as per GRCg6a | Reference Allele | Alternate Allele | Variation Type | Consequence | Location | Feature Sequence | Existing Variation | Breed |
|---|---|---|---|---|---|---|---|---|
| 1814938 | G | A | SNP | 3′ UTR variant | EXON 1 | ERR7564031.18410 | rs16457122 | Aseel |
| 1815386 | A | C | SNP | 5′ UTR variant | EXON 2 | ERR7564047.38625 | rs10725914 | Kadaknath |
In comparison to the reference genome of GRCg7b, the chIFITM1, 2, and 3 were positioned at chromosome 5:g.1776991-1778570, 1778734-1780610 and 1773554 – 1774870 respectively. Three new transition mutations were found in chIFITM1 exon1. Similarly, four novel nucleotide polymorphisms were observed at LOC107053353 of Kadaknath chicken breed, including two transitions, one multi-nucleotide Variation (MNV) and one INDEL (insertion) (Table 7).
Table 7.
SNVs in Kadaknath as per the reference genome Cross of Broiler mother x White Leghorn layer father (GRCg7b).
| Position as per (GRCg7b) | Reference Allele |
Alternate Allele |
Variation Type | Consequence | Gene Symbol |
Location | Feature Sequence |
|---|---|---|---|---|---|---|---|
| 1777551 | G | A | SNP | 3′ UTR variant | chIFITM1 | EXON 1 | ERR7564039.23466 |
| 1777587 | A | G | EXON 1 | ERR7564039.23466 | |||
| 1777972 | A | G | 5′ UTR variant | EXON 1 | ERR7564023.10394 | ||
| 1778830 | A | G | chIFITM2 | EXON 1 | NM_001350058.1 | ||
| 1779054 | NNNNNNN | TTTTTTT | MNP | EXON 1 | NM_001350058.1 | ||
| 1779409 | A | AC | INS | Intron variant | INTRON 1 | NM_001350058.1 | |
| 1780160 | A | G | SNP | Intron variant | INTRON 1 | ERR7564035.14760 | |
| 1781267 | C | T | SNP | 5′ UTR variant | chIFITM5 | EXON 1 | NM_001199498.1 |
| 1781282 | G | A | EXON 1 | NM_001199498.1 | |||
| 1781411 | A | G | EXON 1 | NM_001199498.1 |
3.2. chIFITM5 gene variants
According to the reference sequence GRCg6a, the chIFITM5 gene was placed after chIFITM2 and before ATHL1 at the location of chromosome 5:1821594-1823163 (+). The SNVs were identified at the 5′ UTR variant in Exon 1 of Kadaknath (g. 1821812C > T) and Exon 2 of Aseel (g. 1822662C > G), and upon verification in the Ensemble database, it has been confirmed that two variations were present, each with its own unique reference, viz., SNP cluster IDs of rs3386028259 and rs74110888 (Table 8). As per the reference genome of GRCg7b, the chIFITM5 gene was located at chromosome 5:g.1780914-1782483. Three new transition mutations at 5′UTR of exon 1 were noticed while compared with the reference genome, GRCg7b.
Table 8.
SNVs of chIFITM5 as per the Red Jungle fowl genome (GRCg6a).
| Position as per GRCg6a | Reference Allele |
Alternate Allele |
Variation Type | Consequence | Location | Feature Sequence | Existing Variation |
Breed |
|---|---|---|---|---|---|---|---|---|
| 1821812 | C | T | SNP | 5′ UTR variant | EXON 1 | NM_001199498.1 | rs3386028259 | Kadaknath |
| 1822662 | C | G | SNP | 5′ UTR variant | EXON 2 | SRR13267647.1959 | rs741108889 | Aseel |
3.3. Prediction of IR-chIFITM protein and comparison
The chIFITM genes of Kadaknath and Aseel chickens were analyzed through sequencing, and the results revealed a total of 27 SNVs in IR-chIFITM genes, with 17 SNVs in chIFITM1, 8 SNVs in chIFITM2, and 2 SNVs in chIFITM3 genes. Six SNVs were found in the chIFITM5 gene. Most of the SNVs were found in the UTR regions and introns of chIFITM genes in both breeds. Two SNVs were specifically located in the open reading frame (ORF) of the Kadaknath chIFITM1 gene, resulting in a change of codons. Further analysis (Table 9) indicated that these exonic variations led to synonymous mutations.
Table 9.
Predicted consequences of SNVs in Kadaknath as per the GRCg6a.
| Symbol | Genomic position | cDNA position | CDS position | Protein position | Amino acids | Codons Ref/Alt |
Existing variation |
|---|---|---|---|---|---|---|---|
| chIFITM1 | 1817767 | 96 | 42 | 14 | S | tcC/tcT | rs317414539 |
| 1819102 | 388 | 334 | 112 | R | Cgg/Agg | rs316752027 |
3.4. In ovo chIFITM gene expression against NDV
We conducted an in-ovo ND viral challenging investigation as a continuation of the in-vitro viral challenging study to gain a deeper understanding of the tissue-specific chIFITM gene expression patterns. There was a significant difference in immune gene expression (Table 10) between breeds (p < 0.05), genes (p < 0.01), and tissues (p < 0.0001). On the other hand, the interactions among various factors profoundly influenced immune gene expression (p < 0.01).
Table 10.
Analysis of Variance (ANOVA) of immune gene expression in chicken embryo against NDV.
| Source of variance | Df | SS | MS | F value | Pr(>F) | Significance |
|---|---|---|---|---|---|---|
| Breed | 1 | 5.9 | 5.85 | 6.356 | 0.01193 | * |
| Gene | 5 | 105.5 | 21.1 | 22.916 | <2e-16 | *** |
| Tissue | 4 | 206.3 | 51.58 | 56.013 | <2e-16 | *** |
| Time point: Breed | 5 | 14.5 | 2.9 | 3.145 | 0.00815 | ** |
| Time point: Gene | 25 | 176.7 | 7.07 | 7.676 | <2e-16 | *** |
| Breed: Gene | 5 | 634.3 | 126.87 | 137.761 | <2e-16 | *** |
| Time point: Tissue | 20 | 177.2 | 8.86 | 9.62 | <2e-16 | *** |
| Breed: Tissue | 4 | 618.4 | 154.6 | 167.879 | <2e-16 | *** |
| Gene: Tissue | 18 | 1742 | 96.78 | 105.09 | <2e-16 | *** |
| Time point: Breed: Gene | 25 | 320.5 | 12.82 | 13.921 | <2e-16 | *** |
| Time point: Breed: Tissue | 20 | 252.5 | 12.62 | 13.707 | <2e-16 | *** |
| Time point: Gene: Tissue | 90 | 1014 | 11.27 | 12.234 | <2e-16 | *** |
| Breed: Gene: Tissue | 18 | 831.1 | 46.17 | 50.139 | <2e-16 | *** |
| Time point: Breed: Gene: Tissue | 90 | 929.8 | 10.33 | 11.219 | <2e-16 | *** |
| Residuals | 672 | 618.9 | 0.92 |
*p < 0.05; **p < 0.01; ***p < 0.001.
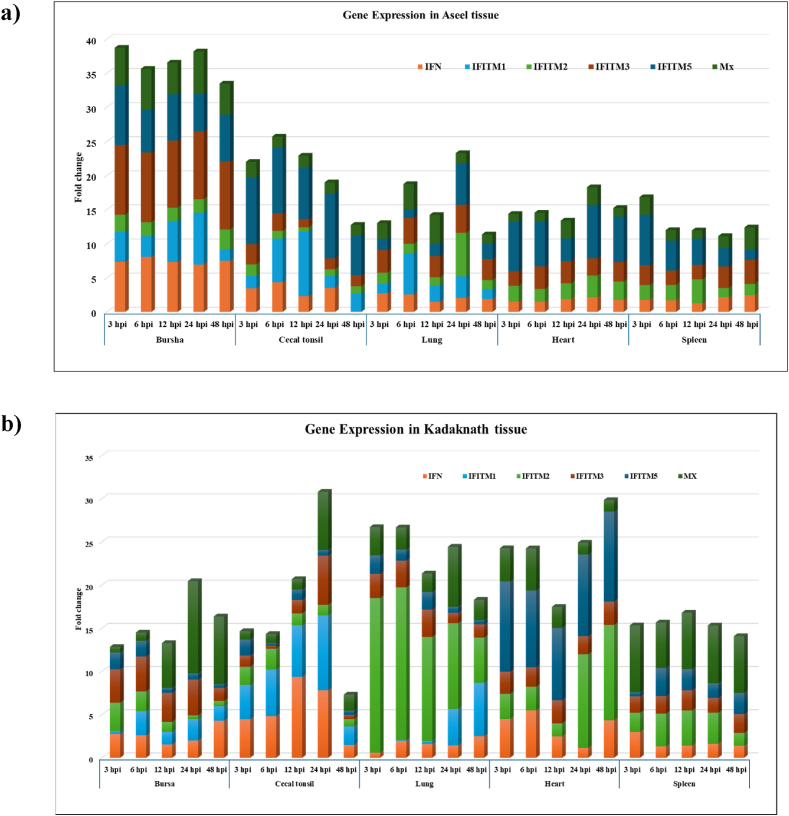
The average fold change of IR-chIFITM gene expression in response to NDV across different tissues, including the bursa, cecal tonsils, heart, lung, and spleen was calculated. There were significant differences noted in the expression of immune genes within breeds at various post-infection time points across all tissues in Aseel and Kadaknath. Similarly, disparities among breeds in terms of transcript levels of immune genes at different post-infection stages were noticed.
3.4.1. IR-chIFITM gene expression
The study involved an analysis of the relative quantification of mRNA levels for three IR-chIFITM genes, namely chIFITM1, chIFITM2, and chIFITM3, in various tissues of infected chicken embryos from two different breeds, Aseel and Kadaknath, at different time points post-infection (hpi). In the bursal tissue, all three IR-chIFITM genes were significantly up-regulated in Aseel, with the most pronounced expression observed for chIFITM1 and chIFITM3 up to 24 and 6 hpi and showed highest fold changes as of 7.61 and 10.23, respectively. Meanwhile, chIFITM2 exhibited a moderate up-regulation, with the highest fold change of 2.92 at 48 hpi. Significant upregulation was detected in Kadaknath; however, it was less prominent than in Aseel, with the maximum expression of chIFITM1 at 6 hpi (2.79 folds) and 24 hpi (2.49 folds), chIFITM2 at 3 hpi (3.29 folds) and 6 hpi (2.30 folds), and chIFITM3 at 3 hpi (3.86 folds) to 24 hpi (4.15 folds), was observed respectively.
In cecal tonsil tissue, chIFITM1 showed consistent upregulation in Aseel, peaking at 12 hpi with a fold change of 9.47. A mild upregulation was observed for chIFITM2 at 3 hpi, while significant upregulation was noticed at 3 (2.97 folds) and 6 hpi (2.54 folds) for chIFITM3. In Kadaknath, strong and significant upregulation was observed throughout the study period for chIFITM1, with the highest fold change ranging from 2.13 to 8.67. chIFITM2 exhibited significant upregulation at 3 and 6 hpi with the fold change of 2.11 nd 2.42 folds, respectively. While chIFITM3 mRNA was abundant only at 24 hpi (5.69 folds).
In embryonic heart tissue, the chIFITM1 gene expression was undetectable in both breeds. Throughout the study, chIFITM2 showed significant and stable upregulation in both Aseel and Kadaknath. chIFITM3 also displayed stable upregulation across all time points in both breeds (2.08–3.29 folds). In lung tissue, chIFITM1 exhibited strong upregulation at 6 hpi (6.05 folds), 12 hpi (2.40 folds) and 24 hpi (3.12 folds) in Aseel. Whereas strong upregulation was noticed at 24 hpi (4.22 folds) and 48 hpi (6.17 folds) in Kadaknath. The chIFITM2 had significant upregulation at 24 hpi (6.40 folds) in Aseel. At the same time, abundant mRNA of chIFITM2 was found throughout the post-infection period in Kadaknath and it ranged from 5.23 to 17.91 folds. The chIFITM3 showed stable upregulation was noticed as 3.09–4.10 folds throughout the period of study in Aseel. Contrary to Aseel chIFITM3 gene expression was until 12 hpi in Kadaknath as 3.17 folds.
In spleen tissue, chIFITM1 gene expression was undetectable in both Aseel and Kadaknath. In Aseel, chIFITM2 displayed significant upregulation from 3 hpi to 12 hpi, with the highest expression as 3.51 folds. Similarly, in Kadaknath, chIFITM2 was significantly upregulated from 3 to 24 hpi, with the highest expression at 4.08 folds. The chIFITM3 showed stable expression throughout the period of study in Aseel, with a range of 2.09–3.53 folds, while in Kadaknath, it exhibited mild to moderate expression, with a range of 1.70–2.18 folds.
3.4.2. chIFITM5 gene expression
The gene expression profiling of the chIFITM5 gene was carried out in all five tissue samples in both chicken breeds. The results demonstrated significant (p < 0.01) upregulation of the chIFITM5 gene in chicken embryos at all time points, except in lung tissue. Specifically, in Aseel, the highest fold change of chIFITM5 gene expression was 8.70, 9.79, and 7.42 folds at 3 hpi in bursa, cecal tonsil, and spleen, respectively. While, at 24 hpi a fold change of 7.80 folds was noticed in heart tissue. Strong upregulation was observed at 24 hpi (6.02 folds) in the lung. In Kadaknath, the results showed a significant (p < 0.01) upregulation of the chIFITM5 gene in chicken embryos at all time points only in heart tissue. The highest fold change was noticed at 3 hpi, and 48 hpi and the same was maintained as 8.37–10.45 folds throughout the time points.
3.4.3. IFN-γ and Mx gene expression
The expression profiling of IFN-γ and Mx genes was included as immune positive genes. The results showed that the IFN-γ gene was significantly and consistently upregulated in most of the tissue samples in both breeds. The IFN-γ gene exhibited peak levels of 8.90 (6 hpi), 4.35 (6 hpi), 2.17 (24 hpi) 2.75 (3 hpi), and 2.47 (48 hpi), respectively in bursa, cecal tonsil, heart, lung and spleen in Aseel breed. While in Kadaknath, the peak levels were 2.76 (3 hpi), 9.36 (12 hpi), 5.49 (6 hpi), 2.51 (48 hpi) and 3.00 (3 hpi), respectively. The gene expression fold changes for the Mx gene were observed in Aseel chickens, with the highest expression at 24 hpi (6.23 folds), 3 hpi (2.27 folds), 12 hpi (2.62 folds), 12 hpi (4.25 folds), and 48 hpi (3.26 folds), respectively, in bursa, cecal tonsil, heart, lung, and spleen. While in Kadaknath, the highest expression was observed at 24 hpi (10.68 folds), 24 hpi (6.75 folds), 6 hpi (4.86 folds), 24 hpi (6.95 folds), and 3 hpi (7.77 folds), respectively.
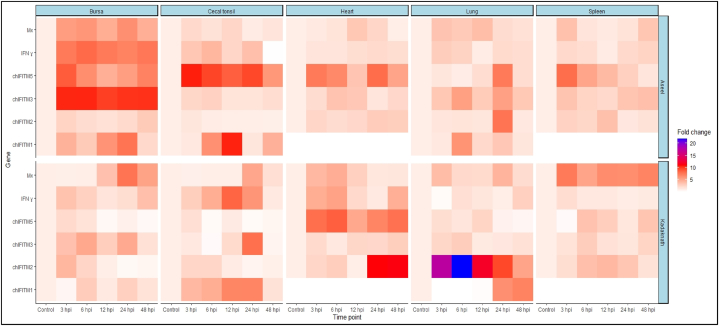
Fig. 1(a and b) shows marked variations in the expression of immune genes within breeds at various post-infection time points across all tissues in Aseel and Kadaknath. Fig. 2 depicts the heat map underscoring the notable disparities among breeds in terms of immune gene transcript levels at different post-infection stages.
Fig. 1.
Expression levels of immune genes in embryonic tissues of (a) Aseel and (b) Kadaknath.
Fig. 2.
Heatmap illustrating tissue specific and breed specific immune gene expression patterns against NDV infection in Aseel and Kadaknath chicken embryos.
4. Discussion
4.1. Characterization of chicken IFITM (chIFITM) locus
The chIFITM locus was found to be flanked by the ATHL1 and B4GALNT4 genes [27,28]. The identified SNVs were located in both coding and non-coding regions of the locus. Similarly, it was found that chicken cell lines such as HD11, DF1, and OU-2 cells were a total of 27 SNVs identified in the coding and noncoding regions of the chIFITM gene based on Gallus gallus_5 reference genome, indicating relatively low sequence diversity or variation [10]. Whereas a limited number of IFITM gene variations were present in a total of 206 samples from different chicken breeds, with 10, 15, 7, and 18 SNVs, respectively, in the European, Commercial, Pirbright, and local chicken breeds [28]. The findings highlighted the genetic diversity present in the chIFITM locus among Aseel and Kadaknath chicken breeds. The presence of SNVs in both coding and non-coding regions suggests potential functional implications, as variations in non-coding regions can also affect gene expression and regulation. The differences observed between the two reference genomes indicated breed-specific genetic variations within the chIFITM locus. According to the published data, the chicken genome architecture confirms that the gene order in the chicken IFITM locus is on the q arm of chromosome 5q, viz., B4GALNT4-chIFITM3-chIFITM1-chIFITM2-chIFITM5-ATHL1 [27].
4.1.1. IR-chIFITM gene variants
The chIFITM1 gene exhibited the highest number of variants compared to the chIFITM2 and chIFITM3 genes. It was located at the LOC422993 locus on chromosome 5q [27,56]. In contrast to the Kadaknath breed, a lower number of variants were present in chIFITM1 genes of the European breed as only two variants (g.13141G > A and g.14105C > T) and only one variant (g.13141G > A) was present in the commercial chicken breeds [28]. On the other hand, eight SNVs in the chIFITM1 gene were present in various laboratory cell lines (DF1, CEF, HD11 and DT40) [10]. Among these SNVs, one was a synonymous mutation located in the exon region, while the remaining seven were intronic mutations. The research findings highlight the potential variability in the number and type of variants observed in different chicken populations. It was discovered that two specific SNPs within the chicken IFITM1 gene exhibited a significant correlation with susceptibility to H7N9 virus disease [32]. A significant two haplotypes rs77537847, and rs11246062 were found in huIFITM1 of the Korean population with ulcerative colitis [37]. However, no such reports were found till date for the association of SNPs against ND virus infection in chickens.
Next to the chIFITM1 gene, the chIFITM2 gene was found to have higher genetic variations in the chicken population. A cytosine insertion was observed in the chIFITM2 non-coding region of the Assel chicken. In line with our findings, Whitehead also discovered a 3bp (TCC) insertion in 5′ UTR in chicken cell lines [10], and similar insertion resulted in frameshift mutation in the chIFITM2 exonic region of the indigenous chicken breed Onigbaogbe/Rose [28]. However, varying numbers of SNVs were identified: 3 in European (g.14960G > T) [28], 8 in Commercial [28], 2 in Pirbright [28]. and 10 in indigenous chicken [28], and a total of 13 SNVs in the laboratory chicken cell lines [10]. Additionally, two frameshift mutations were observed in the indigenous chicken breeds as g.16243-16244 TG > T and g.16269A > ACC. This suggests that the chIFITM2 gene might have a higher level of genetic diversity, and variability in different chicken populations, and it was described that the SNP rs1059091-A/G in the huIFITM2 gene were highly expressive in the Jurkat E6-1 cell population [57].
The chIFITM3 gene was situated at the LOC770612 locus on chromosome 5q and flanked at the 5′ end by the B4GALNT4 genes and at the 3′ end by the chIFITM1 genes [27,28,56]. The chIFITM3 gene had the least number of variations, one unique SNV in each breed was observed in the untranslated region of exon. Similarly, it was discovered that two SNVs and two INDELs in the HD-1 chicken cell line [10]. Comparatively lower numbers of SNVs were reported as 2 in European (g.18814 T > C, g.18862C > T), 1 in Commercial (g.18862C > T), 1 in Pirbright (g.18862C > T), and 3 in Indigenous (g.18862C > T, g.19572G > A and g.19576-19577 AC > A) chicken breeds [28]. This indicates a lower level of genetic variation or diversity in the chIFITM3 gene among the chicken populations when compared to the chIFITM1 gene. In contract, thirteen single nucleotide polymorphisms (SNPs) and one insertion/deletion was present in the IFITM3 gene of Dekalb White and Ross chicken breeds [12]. It was found that the variants rs12252 [31] and rs6598045 [33,58] within the huIFITM3 gene have a notable correlation with the severity of IAV infection and mortality in COVID-19. These findings indicate a potential role for genetic variations in the IFITM3 gene in modulating the susceptibility and severity of the infection. However, up to date, there were no reports indicates an association of such SNVs with chicken diseases.
Overall, the results showed the presence of genetic differences between the Aseel and Kadaknath chicken breeds. The presence of SNVs in coding and non-coding regions of the chIFITM1, chIFITM2 and chIFITM3 genes might have a significant impact on the expression pattern of these genes and the identified SNVs provide valuable insights into the genetic diversity of these antiviral genes and they may have a potential implication for disease resistance and immunity in chickens.
4.1.2. chIFITM5 gene variants
The chIFITM5 gene was located after the chIFITM2 gene and before ATHL1 on chromosome 5 [27,56]. Among the identified SNVs in Kadaknath three were found to be new variations. Four SNVs such as g.1556674A > G, g.1556686T > C, g.1556705A > C, and g.1556716T > C were discovered in the non-coding region of the chIFITM5 gene in all chicken cell lines [10]. Whereas two SNVs in European, one in commercial and one in Pirbright chicken breeds were also reported [28]. These breed-specific SNPs indicate genetic diversity and variation within the chIFITM5 gene among different chicken populations. Studies have shown that a single recurring mutation, c.-14C > T [59] and c.-9C > A [60], in the huIFITM5 gene is linked to Osteogenesis Imperfecta Type V in humans. However, no such findings have been reported for the chIFITM5 gene in chicken breeds.
4.2. Prediction of IR-chIFITM protein and comparison
The predicted amino acid sequences were evaluated and identified that there was no structural alteration in these protein sequences among the breeds. This infers that the synonymous mutations observed in chIFITM1, coupled with the lack of SNVs in the coding regions of chIFITM2 and chIFITM3 genes, may not cause functional change of these proteins between the breeds.
The SNVs primarily occurred in the UTRs and introns of the chIFITM genes. The UTRs are non-coding regions of the gene that flank the coding sequence and play a role in regulating gene expression [61]. The SNVs in the UTR regions and introns did not alter the amino acid sequence of chIFITM proteins. Hence, the observed SNVs likely did not have an impact on the protein's primary structure [62]. However, it is important to note that SNVs in UTR regions and introns can still have functional implications. They may affect the regulation of gene expression, mRNA stability, or splicing patterns, which can indirectly influence protein production [61,63].
The transition and transversion changes at codon 14 and 112 of Kadaknath chIFITM1 results in similar amino acids viz., serine and arginine respectively with respect to that of reference sequence. These amino acids were reported to have a critical involvement in innate and adaptive immunity [64,65]. These residual amino acids may be conserved for regulation of these protein functions. In line with our observations, we interestingly identified that 8 of the SNVs were synonymous or silent substitutions, potentially leaving the proteins function unchanged [28]. Whereas, it was found that IFITM3 contains three lysine amino acids which serve as ubiquitination sites [25]. Overall, the analysis indicated that the chIFITM protein sequences in both Kadaknath and Aseel chickens remained largely conserved, with most of the genetic variations occurring in noncoding regions and introns.
4.3. In ovo embryonic chIFITM gene expression against NDV
In ovo study revealed several significant findings regarding the impact of various factors on immune gene expression in chicken embryos against NDV. First, the breed factor was found to have a significant influence on immune gene expression and the findings highlight the intricate interplay among genetic factors, tissue types and developmental time points in shaping the immune gene expression response to NDV infection in chicken embryos. Similar kind of study had been carried out for gene expression in ducks and chickens [40], Goosling [57] and RIR chicken [10].
4.3.1. IR-chIFITM gene expression
In comparison to the control uninfected embryo of the Aseel and Kadaknath chicken breeds at various times of post-infection, a general trend of gene upregulation in all organs of infected chicken embryos was found. A significant and plentiful amount of chIFITM1 mRNA was detected in the cecal tonsils of both breeds and in bursa of Aseel. Similarly gene expression pattern was studied in various organs such as bursa [24] and cecal tonsil of RIR chicken against Lyssa virus [24] and lung tissue of duck against IAV [40], cecal tissue of gosling against Tembusu virus [25], intestine tissues of RIR against H9N2 Influenza Viruses [10] and lung of transgenic chicken H5N1 Influenza Viruses [14]. In concordance to our study, chIFITM2 gene expression was specifically observed in all tissues of RIR chicken [56], lung tissue of duck [24,40] and in lung and liver of RIR [10].
In the current study, the chIFITM3 gene consistently exhibited higher expression levels in the bursa of Aseel chickens compared to all other organs, while a stable expression was noticed in Kadaknath breed. Similar expression was noticed in the thymus, spleen, bursa of Fabricius, cecal tonsil, gastrointestinal tract, trachea, bone marrow, brain, muscle, heart, liver, kidney, lung, and skin of Rhode Island Red (RIR) chickens against Lyssaviruses [56], lung tissue of duck against Influenza virus [24,40], in pulmonary epithelium of human against Human Influenza Virus [66], in respiratory organs of gosling in Tembusu Virus Infection [25]. In addition, the bursa of yellow feathered broilers also expressed strongly chIFITM3 gene against Avian Reo virus [17] and lung and liver tissues of RIR against H9N2 Influenza Viruses [10]. Apart from the viral infection studies, it was also noticed that heightened expression of the IFITM3 gene in the spleen and lungs of swine during an inflammatory response induced by lipopolysaccharide (LPS) [67]. The bursa serves as a primary immune organ, while the spleen and cecal tonsil function as secondary immune organs. On the other hand, the lung and heart are vital organs responsible for essential physiological processes. These varying roles and functions among the tissues may contribute to the observed differences in gene expression patterns. It suggests that the expression of chIFITM genes may be tissue specific [26,68] and vary across different chicken breeds.
Overall, these results demonstrate that the expression patterns of IR-chIFITM genes vary across different tissues and time points in response to NDV infection in Aseel and Kadaknath chicken breeds. The observed differences highlight the complex and dynamic nature of the immune response to viral infections in chickens.
4.3.2. chIFITM5 gene expression
The chIFITM5 gene exhibited significant upregulation throughout all time points, in bursa, cecal tonsils and heart. However, in the lung, the chIFITM5 gene did not show substantial up-regulation, displaying low fold changes. On the other hand, in Kadaknath chickens, the chIFITM5 gene exhibited strong expression specifically in the heart tissue. The gene transcribed in the majority of tissues and were activated specifically in response to infections [69]. The IFITM5 gene was known for bone-specific expression [70], which was also upregulated in response to the NDV infection in various tissues. Similarly, a significant upregulation of IFITM5 genes in duck lung tissue was noticed following highly pathogenic IAV infection [24]. The expression levels of IFITM5 were found to be increased by approximately 5-fold at one day post infection, indicating the induction of these genes during the infection [24].
In contrast to our result, Whitehead reported that there was no significant upregulation of the chIFITM5 gene observed at any time points of post-infection in the lung tissue of RIR [10]. However, significantly high levels of chIFITM5 expression were noticed in the liver at 24 and 48 hpi and the intestine at 24 hpi [10]. In contrast, it was observed that chIFITM5 expression levels were not consistent across any of the tissues [71]. These findings suggest that the regulation and function of the chIFITM5 gene may vary between different tissues and chicken breeds, potentially playing distinct roles in development, immune response or other biological processes specific to each tissue and breed.
The highest upregulation of chIFITM genes was observed at specific time points in each tissue, but there were still notable up-regulations observed at other time points as well. It is important to note that the expression patterns of chIFITM genes appeared to be both tissue-specific and breed-specific [10]. Indeed, it is still unclear if certain tissues are able to induce chIFITM expression to a greater or lesser extent and whether this is temporally regulated as the embryo develops. The relationship between tissue-specific induction of chIFITM gene expression and sequential regulation during embryonic development remains to be elucidated. Further studies are essential to ascertain the relative capabilities of distinct tissues to modulate chIFITM gene expression and to differentiate how this regulatory mechanism evolves throughout embryogenesis.
4.3.3. IFN γ and Mx gene expression
The findings of our study revealed that the IFN γ gene exhibited consistent, and significant upregulation in all of the tissue samples from both Aseel and Kadaknath breeds. However, there were some notable differences between the breeds and tissues. The findings of the studies are consistent with our research regarding the production of IFN γ in chickens infected with virulent NDV. A very high level of IFN γ expression was noticed in the lymphoid tissues such as thymus, bursa, cecal tonsils, and spleen of NDV infected chickens [[72], [73], [74], [75]]. Taken together, these findings provide further support for the induction of IFN γ in various lymphoid tissues in response to NDV infection, highlighting the dynamic nature of the immune response in different organs and stages of infection.
Similarly, the observation was made that the Mx gene exhibited high expression levels in the bursa and spleen of Kadaknath chicken embryos. Our findings align with the results presented in Whitehead study, where significant upregulation of the Mx gene was observed in the lung, liver tissues, and intestine of Rhode Island Red chicken embryos following H9N2 infection [10].
These findings indicate that both the IFN γ and Mx genes play a critical role in the immune response of chickens, as their expression levels were upregulated in multiple tissues. The upregulation of these genes suggests activation of the immune system, possibly in response to an infectious agent or other immune stimuli. Further investigation and functional studies were needed to better understand the specific roles of these genes in chicken immunity and their potential implications for disease resistance in the Aseel and Kadaknath breeds.
5. Conclusion
The investigation focused on establishing a unique dataset of SNVs and INDEL for the chIFITM locus specific to Aseel and Kadaknath chicken breeds, and the analysis revealed that chIFITM1 had the most SNVs, followed by chIFITM2, with chIFITM3 exhibiting the least variations. However, the presence of SNVs in IR-chIFITM genes of Aseel and Kadaknath indicates substantial genetic diversity in these chicken populations. Despite this diversity, the coding sequences of all chIFITM genes exhibit a high degree of conservation, indicating a robust preservation of genetic information. Genetic variations in chIFITM genes could impact gene regulation, splicing, and various aspects of gene expression, mRNA stability, and translation efficiency. The resilient and consistent expression of the chIFITM3 gene across diverse tissues provides compelling support for its potent antiviral efficacy against viral infections. The results, suggests the need for further in-vitro, and in-vivo association studies to understand disease resistance and antiviral protein expression, especially in the context of UTR variants in chIFITM3 (g.1814938G > A and g.1815386A > C), for effective breeding strategies.
Data availability statement
The original contributions presented in the study will be provided on request.
Ethics statement
The experiment was conducted with the approval of the Institutional Bio-safety Committee (IBSC) (Approval Lr. No. 1764/VCRI-NKL/IBSC/2022 dated May 11, 2022 of the Dean, VCRI, Namakkal) and Institutional Animal Ethical committee (IAEC) (Project proposal No. 09/VCRI-NKL/2023 dated September 08, 2023) of TANUVAS-Veterinary College and Research Institute, Namakkal, Tamil Nadu, India.
Funding
The financial assistance for this research was provided by the Tamil Nadu Veterinary and Animal Sciences University (TANUVAS) in Chennai, Tamil Nadu, India is greatly appreciated. (USO. No. 20652/A1/2020 and No.273/A1/2021, dated February 26, 2021 of Registrar, TANUVAS, Chennai – 51, India.)
CRediT authorship contribution statement
Malarmathi Muthusamy: Writing – review & editing, Writing – original draft, Project administration, Methodology, Formal analysis, Data curation, Conceptualization. Murali Nagarajan: Writing – review & editing, Project administration, Conceptualization. Sivakumar Karuppusamy: Writing – review & editing, Supervision, Project administration. Kannaki T. Ramasamy: Writing – review & editing, Resources, Methodology. Amutha Ramasamy: Writing – review & editing, Resources, Project administration. Ramya Kalaivanan: Writing – review & editing, Resources, Project administration. Gopala Krishna Murthy Thippicettipalayam Ramasamy: Writing – review & editing, Supervision, Methodology, Formal analysis. Thiruvenkadan Aranganoor Kannan: Writing – review & editing, Writing – original draft, Supervision, Resources, Project administration, Methodology, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
The authors gratefully acknowledge the support provided by the Tamil Nadu Veterinary and Animal Sciences University (TANUVAS), Chennai, Tamil Nadu, India and Theomics International Pvt Ltd, Bengaluru, Karnataka 560038, for Sequencing facilities and Support.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e37729.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Friedman R.L., Manly S.P., McMahon M., Kerr I.M., Stark G.R. Transcriptional and posttranscriptional regulation of interferon-induced gene expression in human cells. Cell. 1984;38:745–755. doi: 10.1016/0092-8674(84)90270-8. [DOI] [PubMed] [Google Scholar]
- 2.Reid L.E., Brasnett A.H., Gilbert C.S., Porter A.C., Gewert D.R., Stark G.R., Kerr I.M. A single DNA response element can confer inducibility by both alpha- and gamma-interferons. Proc. Natl. Acad. Sci. U. S. A. 1989;86:840–844. doi: 10.1073/pnas.86.3.840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brass A.L., Huang I.-C., Benita Y., John S.P., Krishnan M.N., Feeley E.M., Ryan B.J., Weyer J.L., van der Weyden L., Fikrig E., Adams D.J., Xavier R.J., Farzan M., Elledge S.J. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell. 2009;139:1243–1254. doi: 10.1016/j.cell.2009.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Savidis G., Perreira J.M., Portmann J.M., Meraner P., Guo Z., Green S., Brass A.L. The IFITMs inhibit Zika virus replication. Cell Rep. 2016;15:2323–2330. doi: 10.1016/j.celrep.2016.05.074. [DOI] [PubMed] [Google Scholar]
- 5.Ren L., Du S., Xu W., Li T., Wu S., Jin N., Li C. Current progress on host antiviral factor IFITMs. Front. Immunol. 2020;11 doi: 10.3389/fimmu.2020.543444. https://www.frontiersin.org/articles/10.3389/fimmu.2020.543444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Z., Liu J., Li M., Yang H., Zhang C. Evolutionary dynamics of the interferon-induced transmembrane gene family in vertebrates. PLoS One. 2012;7 doi: 10.1371/journal.pone.0049265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Webb A.E., Gerek Z.N., Morgan C.C., Walsh T.A., Loscher C.E., Edwards S.V., O'Connell M.J. Adaptive evolution as a predictor of species-specific innate immune response. Mol. Biol. Evol. 2015;32:1717–1729. doi: 10.1093/molbev/msv051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.TenOever B.R. The evolution of antiviral defense systems. Cell Host Microbe. 2016;19:142–149. doi: 10.1016/j.chom.2016.01.006. [DOI] [PubMed] [Google Scholar]
- 9.Muñoz-Moreno R., Cuesta-Geijo M.Á., Martínez-Romero C., Barrado-Gil L., Galindo I., García-Sastre A., Alonso C. Antiviral role of IFITM proteins in African swine fever virus infection. PLoS One. 2016;11 doi: 10.1371/journal.pone.0154366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Whitehead T.J. Characterisation of chicken interferon-inducible transmembrane proteins: locus architecture, gene expression and viral restriction. 2018. https://discovery.ucl.ac.uk/id/eprint/10058159 Doctoral thesis (Ph. D)
- 11.Smith S.E., Gibson M.S., Wash R.S., Ferrara F., Wright E., Temperton N., Kellam P., Fife M. Chicken interferon-inducible transmembrane protein 3 restricts influenza viruses and Lyssaviruses in vitro. J. Virol. 2013;87:12957–12966. doi: 10.1128/jvi.01443-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim Y.-C., Jeong Min-Ju. Byung-Hoon Jeong, Genetic characteristics and polymorphisms in the chicken interferon-induced transmembrane protein (IFITM3) gene. Vet. Res. Commun. 2019;43:203–214. doi: 10.1007/s11259-019-09762-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fife M., Moore J. IFITM knockdown/knockout technology for vaccine production. Ann Clin Trials V accines R es. 2017;1:46. [Google Scholar]
- 14.Rohaim M.A., Al-Natour M.Q., Abdelsabour M.A., El Naggar R.F., Madbouly Y.M., Ahmed K.A., Munir M. Transgenic chicks expressing interferon-inducible transmembrane protein 1 (IFITM1) restrict highly pathogenic H5N1 influenza viruses. Int. J. Mol. Sci. 2021;22:8456. doi: 10.3390/ijms22168456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lanz C., Yángüez E., Andenmatten D., Stertz S., Correction for Lanz, et al. Swine interferon-inducible transmembrane proteins potently inhibit influenza A virus replication. J. Virol. 2015;89:2988. doi: 10.1128/JVI.03533-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu J., Qian P., Wu Q., Liu S., Fan W., Zhang K., Wang R., Zhang H., Chen H., Li X. Swine interferon-induced transmembrane protein, sIFITM3, inhibits foot-and-mouth disease virus infection in vitro and in vivo. Antivir. Res. 2014;109:22–29. doi: 10.1016/j.antiviral.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang, Y. X Q., L. C., D. H., W. C Distribution of IFITM3 in yellow-feathered broilers and inhibition of avian reovirus multiplication by IFITM3. Braz. J. Poult. Sci. 2018;20:377–386. doi: 10.1590/1806-9061-2017-0662. [DOI] [Google Scholar]
- 18.Steyn A., Keep S., Bickerton E., Fife M. The characterization of chIFITMs in avian coronavirus infection in vivo, ex vivo and in vitro. Genes. 2020;11:918. doi: 10.3390/genes11080918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H., Ni R., Wang K., Tian Y., Gong H., Yan W., Tang Y., Lei C., Wang H., Yang X. Chicken interferon-induced transmembrane protein 1 promotes replication of coronavirus infectious bronchitis virus in a cell-specific manner. Vet. Microbiol. 2022;275 doi: 10.1016/j.vetmic.2022.109597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Malarmathi M., Murali N., Selvaraju M., Sivakumar K., Gowthaman V., Raghavendran V.B., Raja A., Peters S.O., Thiruvenkadan A.K. In vitro characterization of chIFITMs of Aseel and Kadaknath chicken breeds against Newcastle disease virus infection. Biology. 2023;12:919. doi: 10.3390/biology12070919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Friedlová N., Zavadil Kokáš F., Hupp T.R., Vojtěšek B., Nekulová M. IFITM protein regulation and functions: far beyond the fight against viruses. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.1042368. https://www.frontiersin.org/articles/10.3389/fimmu.2022.1042368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hickford D., Frankenberg S., Shaw G., Renfree M.B. Evolution of vertebrate interferon inducible transmembrane proteins. BMC Genom. 2012;13:155. doi: 10.1186/1471-2164-13-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hornick A.L., Li N., Oakland M., McCray P.B., Sinn P.L. Human, pig, and mouse interferon-induced transmembrane proteins partially restrict pseudotyped lentiviral vectors. Hum. Gene Ther. 2016;27:354–362. doi: 10.1089/hum.2015.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blyth G.A.D., Chan W.F., Webster R.G., Magor K.E. Duck interferon-inducible transmembrane protein 3 mediates restriction of influenza viruses. J. Virol. 2016;90:103–116. doi: 10.1128/JVI.01593-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang A., Sun L., Wang M., Jia R., Zhu D., Liu M., Sun K., Yang Q., Wu Y., Chen X., Cheng A., Chen S. Identification of IFITM1 and IFITM3 in Goose: gene structure, expression patterns, and immune reponses against Tembusu virus infection. BioMed Res. Int. 2017;2017 doi: 10.1155/2017/5149062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lu G., Ou J., Cai S., Lai Z., Zhong L., Yin X., Li S. Canine interferon-inducible transmembrane protein is a host restriction factor that potently inhibits replication of emerging canine influenza virus. Front. Immunol. 2021;12 doi: 10.3389/fimmu.2021.710705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bassano I., Ong S.H., Lawless N., Whitehead T., Fife M., Kellam P. Accurate characterization of the IFITM locus using MiSeq and PacBio sequencing shows genetic variation in Galliformes. BMC Genom. 2017;18:419. doi: 10.1186/s12864-017-3801-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bassano I., Ong S.H., Sanz-Hernandez M., Vinkler M., Kebede A., Hanotte O., Onuigbo E., Fife M., Kellam P. Comparative analysis of the chicken IFITM locus by targeted genome sequencing reveals evolution of the locus and positive selection in IFITM1 and IFITM3. BMC Genom. 2019;20:272. doi: 10.1186/s12864-019-5621-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okuzaki Y., Kidani S., Kaneoka H., Iijima S., Nishijima K.-I. Characterization of chicken interferon-inducible transmembrane protein-10. Biosci. Biotechnol. Biochem. 2017;81:914–921. doi: 10.1080/09168451.2016.1274639. [DOI] [PubMed] [Google Scholar]
- 30.Moffatt P., Gaumond M.-H., Salois P., Sellin K., Bessette M.-C., Godin E., de Oliveira P.T., Atkins G.J., Nanci A., Thomas G. Bril: a novel bone-specific modulator of mineralization. J. Bone Miner. Res. Off. J. Am. Soc. Bone Miner. Res. 2008;23:1497–1508. doi: 10.1359/jbmr.080412. [DOI] [PubMed] [Google Scholar]
- 31.Xuan Y., Wang L.N., Li W., Zi H.R., Guo Y., Yan W.J., Chen X.B., Wei P.M. IFITM3 rs12252 T>C polymorphism is associated with the risk of severe influenza: a meta-analysis. Epidemiol. Infect. 2015;143:2975–2984. doi: 10.1017/S0950268815000278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Layton D.S., Butler J., Stewart C., Stevens V., Payne J., Rootes C., Deffrasnes C., Walker S., Shan S., Gough T.J., Cowled C., Bruce K., Wang J., Kedzierska K., Wong F.Y.K., Bean A.G.D., Bingham J., Williams D.T. H7N9 bearing a mutation in the nucleoprotein leads to increased pathology in chickens. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.974210. https://www.frontiersin.org/articles/10.3389/fimmu.2022.974210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kim Y.-C., Jeong B.-H. Strong correlation between the case fatality rate of COVID-19 and the rs6598045 single nucleotide polymorphism (SNP) of the interferon-induced transmembrane protein 3 (IFITM3) gene at the population-level. Genes. 2021;12:42. doi: 10.3390/genes12010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim Y.-C., Jeong B.-H. No correlation of the disease severity of influenza A virus infection with the rs12252 polymorphism of the interferon-induced transmembrane protein 3 gene. Intervirology. 2017;60:69–74. doi: 10.1159/000479087. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y.-H., Zhao Y., Li N., Peng Y.-C., Giannoulatou E., Jin R.-H., Yan H.-P., Wu H., Liu J.-H., Liu N., Wang D.-Y., Shu Y.-L., Ho L.-P., Kellam P., McMichael A., Dong T. Interferon-induced transmembrane protein-3 genetic variant rs12252-C is associated with severe influenza in Chinese individuals. Nat. Commun. 2013;4:1418. doi: 10.1038/ncomms2433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Everitt A.R., Clare S., Pertel T., John S.P., Wash R.S., Smith S.E., Chin C.R., Feeley E.M., Sims J.S., Adams D.J., Wise H.M., Kane L., Goulding D.A., Digard P., Anttila V., Baillie J.K., Walsh T.S., Hume D.A., Palotie A., Xue Y., Colonna V., Tyler-Smith C., Dunning J., Gordon S.B., Smyth R.L., Openshaw P., Dougan G., Brass A.L., Kellam P. IFITM3 restricts the morbidity and mortality associated with influenza. Nature. 2012;484:519–523. doi: 10.1038/nature10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mo J.-S., Na K.-S., Yu J.-I., Chae S.-C. Identification of the polymorphisms in IFITM1 gene and their association in a Korean population with ulcerative colitis. Immunol. Lett. 2013;156:118–122. doi: 10.1016/j.imlet.2013.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seo G.S., Lee J.K., Yu J.I., Yun K.J., Chae S.C., Choi S.C. Identification of the polymorphisms in IFITM3 gene and their association in a Korean population with ulcerative colitis. Exp. Mol. Med. 2010;42:99–104. doi: 10.3858/emm.2010.42.2.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu-yang Z., Pei-yu B., Chuan-tao Y., Wei Y., Hong-wei M., Kang T., Chun-mei Z., Ying-feng L., Xin W., Ping-zhong W., Chang-xing H., Xue-fan B., Ying Z., Zhan-sheng J. Interferon-induced transmembrane protein 3 inhibits Hantaan virus infection, and its single nucleotide polymorphism rs12252 influences the severity of hemorrhagic fever with renal syndrome. Front. Immunol. 2017;7:535. doi: 10.3389/fimmu.2016.00535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith J., Smith N., Yu L., Paton I.R., Gutowska M.W., Forrest H.L., Danner A.F., Seiler J.P., Digard P., Webster R.G., Burt D.W. A comparative analysis of host responses to avian influenza infection in ducks and chickens highlights a role for the interferon-induced transmembrane proteins in viral resistance. BMC Genom. 2015;16:574. doi: 10.1186/s12864-015-1778-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jaiswal G., Kumar S., Prasad Y. Immunocompetence traits and their inheritance pattern in Kadaknath native chicken. Indian J. Anim. Res. 2014;48:509. doi: 10.5958/0976-0555.2014.00021.1. [DOI] [Google Scholar]
- 42.Radhika R., Thiagarajan D., Veeramani P., Karthickeyan S.M.K. Aseel, Kadaknath and white Leghorn chicken immune response to variation in sheep red blood cell. Int. J. Pure Appl. Biosci. 2017;5:335–340. doi: 10.18782/2320-7051.5299. [DOI] [Google Scholar]
- 43.Kanakachari M., Chatterjee R.N., Reddy M.R., Dange M., Bhattacharya T.K. Indian Red Jungle fowl reveals a genetic relationship with South East Asian Red Jungle fowl and Indian native chicken breeds as evidenced through whole mitochondrial genome sequences. Front. Genet. 2023;14 doi: 10.3389/fgene.2023.1083976. https://www.frontiersin.org/articles/10.3389/fgene.2023.1083976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ramakrishnan S., Annamalai A., Sachan S., Kumar A., Sharma B.K., Govindaraj E., Chellappa M.M., Dey S., Krishnaswamy N. Synergy of lipopolysaccharide and resiquimod on type I interferon, pro-inflammatory cytokine, Th1 and Th2 response in chicken peripheral blood mononuclear cells. Mol. Immunol. 2015;64:177–182. doi: 10.1016/j.molimm.2014.11.013. [DOI] [PubMed] [Google Scholar]
- 45.Noorai R.E., Shankar V., Freese N.H., Gregorski C.M., Chapman S.C. Discovery of genomic variations by whole-genome resequencing of the North American Araucana chicken. PLoS One. 2019;14 doi: 10.1371/journal.pone.0225834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Warren W.C., Hillier L.W., Tomlinson C., Minx P., Kremitzki M., Graves T., Markovic C., Bouk N., Pruitt K.D., Thibaud-Nissen F., Schneider V., Mansour T.A., Brown C.T., Zimin A., Hawken R., Abrahamsen M., Pyrkosz A.B., Morisson M., Fillon V., Vignal A., Chow W., Howe K., Fulton J.E., Miller M.M., Lovell P., Mello C.V., Wirthlin M., Mason A.S., Kuo R., Burt D.W., Dodgson J.B., Cheng H.H. A new chicken genome assembly provides insight into avian genome structure. G3 GenesGenomesGenetics. 2017;7:109–117. doi: 10.1534/g3.116.035923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gallus gallus genome assembly GRCg6a, NCBI (n.d.). https://www.ncbi.nlm.nih.gov/data-hub/assembly/GCF_000002315.5/(accessed January 10, 2024).
- 48.Gallus gallus genome assembly bGalGal1.mat.broiler.GRCg7b, NCBI (n.d.). https://www.ncbi.nlm.nih.gov/data-hub/assembly/GCF_016699485.2/(accessed April 30, 2023).
- 49.Freese N.H., Norris D.C., Loraine A.E. Integrated genome browser: visual analytics platform for genomics. Bioinformatics. 2016;32:2089–2095. doi: 10.1093/bioinformatics/btw069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. 1000 genome Project data processing subgroup, the sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ruden D., Cingolani P., Patel V., Coon M., Nguyen T., Land S., Lu X. Using Drosophila melanogaster as a model for genotoxic chemical mutational studies with a new program, SnpSift. Front. Genet. 2012;3 doi: 10.3389/fgene.2012.00035. https://www.frontiersin.org/articles/10.3389/fgene.2012.00035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Danecek P., Auton A., Abecasis G., Albers C.A., Banks E., DePristo M.A., Handsaker R.E., Lunter G., Marth G.T., Sherry S.T., McVean G., Durbin R. 1000 genomes Project analysis group, the variant call format and VCFtools. Bioinformatics. 2011;27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pfaf M.W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 2001;29:45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods San Diego Calif. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 56.Smith S.E., Gibson M.S., Wash R.S., Ferrara F., Wright E., Temperton N., Kellam P., Fife M. Chicken interferon-inducible transmembrane protein 3 restricts influenza viruses and Lyssaviruses in vitro. J. Virol. 2013;87:12957–12966. doi: 10.1128/JVI.01443-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wu W.-L., Grotefend C.R., Tsai M.-T., Wang Y.-L., Radic V., Eoh H., Huang I.-C. Δ20 IFITM2 differentially restricts X4 and R5 HIV-1. Proc. Natl. Acad. Sci. USA. 2017;114:7112–7117. doi: 10.1073/pnas.1619640114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gholami M., Sakhaee F., Sotoodehnejadnematalahi F., Zamani M.S., Ahmadi I., Anvari E., Fateh A. Increased risk of COVID-19 mortality rate in IFITM3 rs6598045 G allele carriers infected by SARS-CoV-2 delta variant. Hum. Genom. 2022;16:60. doi: 10.1186/s40246-022-00434-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cho T.-J., Lee K.-E., Lee S.-K., Song S.J., Kim K.J., Jeon D., Lee G., Kim H.-N., Lee H.R., Eom H.-H., Lee Z.H., Kim O.-H., Park W.-Y., Park S.S., Ikegawa S., Yoo W.J., Choi I.H., Kim J.-W. A single recurrent mutation in the 5′-UTR of IFITM5 causes osteogenesis imperfecta type V. Am. J. Hum. Genet. 2012;91:343–348. doi: 10.1016/j.ajhg.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hanagata N. IFITM5 mutations and osteogenesis imperfecta. J. Bone Miner. Metabol. 2016;34:123–131. doi: 10.1007/s00774-015-0667-1. [DOI] [PubMed] [Google Scholar]
- 61.Steri M., Idda M.L., Whalen M.B., Orrù V. vol. 9. Wiley Interdiscip. Rev. RNA; 2018. p. e1474. (Genetic Variants in mRNA Untranslated Regions). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brody T. In: Clin. Trials. second ed. Brody T., editor. Academic Press; Boston: 2016. Chapter 19 - biomarkers; pp. 377–419. [DOI] [Google Scholar]
- 63.Barrett L.W., Fletcher S., Wilton S.D. Regulation of eukaryotic gene expression by the untranslated gene regions and other non-coding elements. Cell. Mol. Life Sci. 2012;69:3613–3634. doi: 10.1007/s00018-012-0990-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rodriguez P.C., Ochoa A.C., Al-Khami A.A. Arginine metabolism in myeloid cells shapes innate and adaptive immunity. Front. Immunol. 2017;8 doi: 10.3389/fimmu.2017.00093. https://www.frontiersin.org/articles/10.3389/fimmu.2017.00093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wu Q., Chen X., Li J., Sun S. Serine and metabolism regulation: a novel mechanism in antitumor immunity and senescence. Aging Dis. 2020;11:1640. doi: 10.14336/AD.2020.0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sun H., Pu J., Wei Y., Sun Y., Hu J., Liu L., Xu G., Gao W., Li C., Zhang X., Huang Y., Chang K.-C., Liu X., Liu J. Highly pathogenic avian influenza H5N6 viruses exhibit enhanced affinity for human type sialic acid receptor and in-contact transmission in model ferrets. J. Virol. 2016;90:6235–6243. doi: 10.1128/JVI.00127-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Li H.-P., Chen P.-G., Liu F.-T., Zhu H.-S., Jiao X.-Q., Zhong K., Guo Y.-J., Zha G.-M., Han L.-Q., Lu W.-F., Wang Y.-Y., Yang G.-Y. Characterization and anti-inflammation role of swine IFITM3 gene. Oncotarget. 2017;8:73579–73589. doi: 10.18632/oncotarget.20568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tanaka S.S., Yamaguchi Y.L., Tsoi B., Lickert H., Tam P.P.L. IFITM/Mil/fragilis family proteins IFITM1 and IFITM3 play distinct roles in mouse primordial germ cell homing and repulsion. Dev. Cell. 2005;9:745–756. doi: 10.1016/j.devcel.2005.10.010. [DOI] [PubMed] [Google Scholar]
- 69.Siegrist F., Ebeling M., Certa U. The small interferon-induced transmembrane genes and proteins. J. Interferon Cytokine Res. 2011;31:183–197. doi: 10.1089/jir.2010.0112. [DOI] [PubMed] [Google Scholar]
- 70.Mäkitie R.E., Pekkinen M., Morisada N., Kobayashi D., Yonezawa Y., Nishimura G., Ikegawa S., Mäkitie O. A novel IFITM5 variant associated with phenotype of osteoporosis with calvarial doughnut lesions: a case report. Calcif. Tissue Int. 2021;109:626–632. doi: 10.1007/s00223-021-00878-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Cai S., Zheng Z., Cheng J., Zhong L., Shao R., Zheng F., Lai Z., Ou J., Xu L., Zhou P., Lu G., Zhang G. Swine interferon-inducible transmembrane proteins potently inhibit African swine fever virus replication. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.827709. https://www.frontiersin.org/articles/10.3389/fimmu.2022.827709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ecco R., Brown C., Susta L., Cagle C., Cornax I., Pantin-Jackwood M., Miller P.J., Afonso C.L. In vivo transcriptional cytokine responses and association with clinical and pathological outcomes in chickens infected with different Newcastle disease virus isolates using formalin-fixed paraffin-embedded samples. Vet. Immunol. Immunopathol. 2011;141:221–229. doi: 10.1016/j.vetimm.2011.03.002. [DOI] [PubMed] [Google Scholar]
- 73.Susta L., Cornax I., Diel D.G., Garcia S.C., Miller P.J., Liu X., Hu S., Brown C.C., Afonso C.L. Expression of interferon gamma by a highly virulent strain of Newcastle disease virus decreases its pathogenicity in chickens. Microb. Pathog. 2013;61–62:73–83. doi: 10.1016/j.micpath.2013.05.009. [DOI] [PubMed] [Google Scholar]
- 74.Kristeen-Teo Y.W., Yeap S.K., Tan S.W., Omar A.R., Ideris A., Tan S.G., Alitheen N.B. The effects of different velogenic NDV infections on the chicken bursa of Fabricius. BMC Vet. Res. 2017;13:151. doi: 10.1186/s12917-017-1071-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brown C., Zhang J., Pantin-Jackwood M., Dimitrov K., Ferreira H.L., Suarez D. In situ cytokine gene expression in early stage of virulent Newcastle disease in chickens. Vet. Pathol. 2022;59:75–81. doi: 10.1177/03009858211045945. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study will be provided on request.