Abstract
While the elements encoding enhancers and promoters have been relatively well studied, the full spectrum of insulator elements which bind the CCCTC binding factor (CTCF), is relatively poorly characterized. This is partly due to the genomic context of CTCF sites greatly influencing their roles and activity. Here we have developed an experimental system to determine the ability of minimal, consistently sized, individual CTCF elements to interpose between enhancers and promoters and thereby reduce gene expression during differentiation. Importantly, each element is tested in the identical location thereby minimising the effect of genomic context. We found no correlation between the ability of CTCF elements to block enhancer–promoter activity with the degree of evolutionary conservation; their resemblance to the consensus core sequences; or the number of CTCF core motifs harboured in the element. Nevertheless, we have shown that the strongest enhancer–promoter blockers include a previously described bound element lying upstream of the CTCF core motif. In addition, we found other uncharacterised DNaseI footprints located close to the core motif that may affect function. We have developed an assay of CTCF sequences which will enable researchers to sub-classify individual CTCF elements in a uniform and unbiased way.
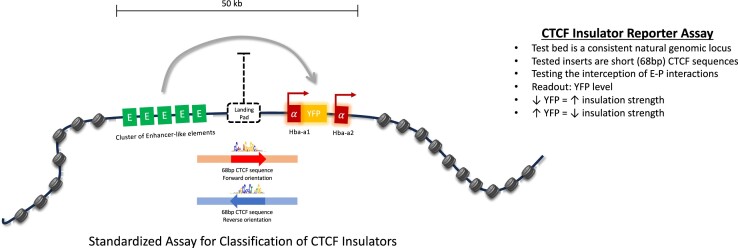
Graphical Abstract
Graphical Abstract.
Introduction
The regulation of gene expression is controlled by two general classes of cis-acting elements: enhancers and promoters. However, within each class there is considerable variation in activity. For example, individual promoters and enhancers can be considered as strong or weak, and as widely active or cell-type restricted. These features can to some extent be predicted from their sequence composition, ability to bind specific transcription factors and epigenetic signatures (1). However, there may be overlap between these classes of element: to some extent all enhancers may act as promoters and some promoters can act as enhancers (2–4). The activities of enhancers and promoters are modified by a third class of fundamental cis-acting elements referred to as CCCTC binding factor (CTCF) elements which have a wide range of activities including: blocking interactions between enhancers and promoters (5–7); facilitating interactions between enhancers and promoters (8–11); acting as barriers between active and inactive chromatin (12–14); contributing directly to activity of enhancers and promoters; and playing a key role in the three dimensional structure of the genome thereby influencing the physical proximity of enhancers and promoters (15–17). It is estimated that there are over 250 000 high confidence CTCF binding sites in the human genome, and around 50 000 sites are bound in individual cell types (14,18–20). However, other than binding CTCF proteins, relatively little is known about how to classify individual elements and to assess their ability to perform each aspect of their many potential activities. Previous studies derived from large datasets have correlated the activities of CTCF elements with features such as the core CTCF sequences, its evolutionary conservation, the amount of CTCF and cohesin enriched at the CTCF sites, the persistence of CTCF binding after CTCF depletion, DNA methylation at CTCF core motifs, nucleosome positioning, and the number of binding motifs in the CTCF element (18,19,21–29). However, the predictive values of these parameters have not been extensively tested on individual elements.
Assays of enhancers and promoters in terms of sequence, epigenetic modification and function are relatively well developed and standardized whereas assays of CTCF elements are less well characterised, in part because they have such diverse functions, and their role greatly depends on their genomic context. For example, for a CTCF element to block the activity of an enhancer on its cognate promoter it has to lie between such elements (5–7), whereas for CTCF elements to assist enhancer–promoter interactions they must lie within or close to the interacting sequences (8–10). We have recently investigated the roles of CTCF sites within and surrounding a ∼65 kb sub-TAD containing the mouse alpha-globin locus (Figure 1A) by investigating how deletion of individual sites and combinations of sites affect 3D structure of the sub-TAD and alpha-globin expression (30,31). None of these sites appear to influence the levels of alpha globin expression but two sites (HS-38 and HS-39) insulate neighbouring genes (Mpg, Rhbd1, Il9r) from the very strong cluster of enhancers controlling alpha-globin expression (31).
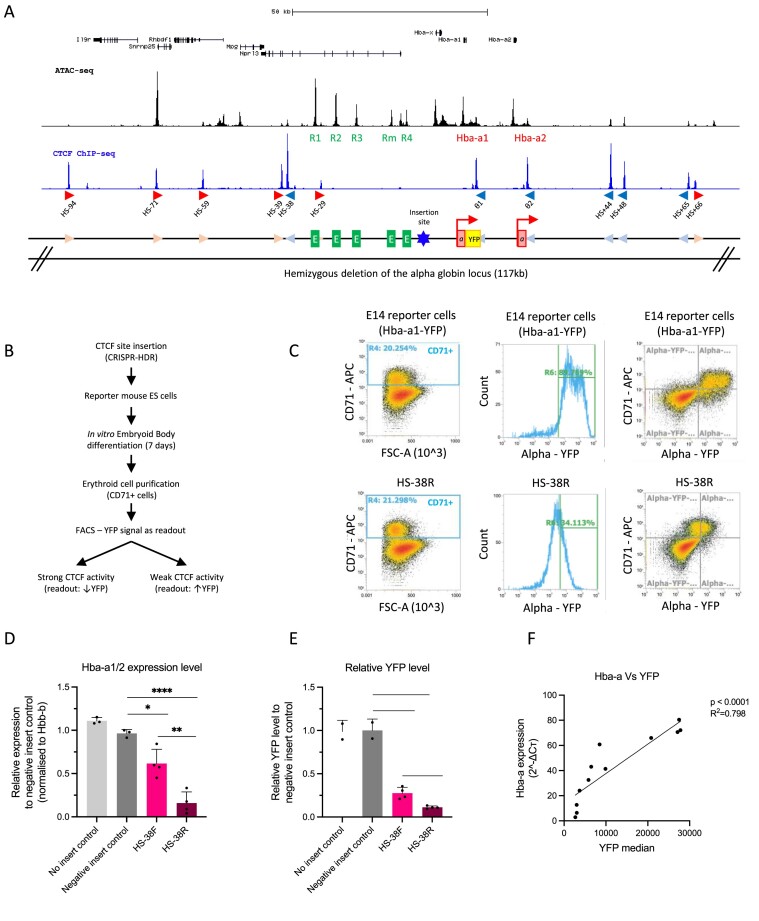
Figure 1.
An experimental system to evaluate the capacity of individual CTCF elements to block enhancer–promoter interactions. (A) Top, UCSC refseq genes at the genomic locus encompassing the alpha-globin locus. ATAC-seq track (black) shows the accessible regions with the alpha-globin enhancer-like elements (R1–R4) and genes (Hba-a1, Hba-a2) indicated. The blue track shows CTCF ChIP-seq with the peaks enriched over CTCF sites that are marked in red (forward orientation) and blue (reverse orientation) arrows as well as their corresponding label starting from the left with HS-94. Below a graphical representation of the alpha-globin locus indicating all three regulatory elements: CTCF sites in light blue and pink arrows, the enhancer-like elements in green, and the genes in red with the YFP tag in yellow and the red arrow indicating the transcriptionally active genes. The CTCF site of interest was inserted between the enhancer clusters and the promoters indicated by the blue star. An allele of the alpha-globin gene locus (117 kb) was hemizygously deleted in the reporter mouse ES cells (mESCs) as indicated. (B) The workflow starting with The CTCF site of interest inserted into the hemizygous alpha-globin locus of the reporter mESCs by the CRISPR-HDR method. The modified reporter mESCs were differentiated into EBs and the cells were harvested for analysis. The YFP level of the CD71+ cell population in the EBs were determined by FACS analysis and interpreted as the readout of the CTCF ability to block enhancer–promoter interaction as indicated. (C) Top panels, FACS plots to characterise the control E14 mESCs upon EB differentiation; left panel shows the proportion of CD71+ erythroid cells from the total EB population at day 7 of differentiation. Middle panel is a histogram showing the YFP signal (Hba-a expression) specifically in the CD71+ erythroid population (as gated in left panel). The right panel shows the same data plotted for the whole population based on two parameters, the CD71 + erythroid marker and YFP. All CD71+ cells isolated from the differentiated EBs representing the erythroid population exhibit high level of YFP, indicating high expression of the alpha-globin as expected. The lower panels represent the same FACS profile for mESCs with the inserted HS-38R site. (D) RT-qPCR data showing that insertion of the HS-38 sequence in both the forward and reverse orientation (pink and maroon columns) significantly represses the expression of the alpha-globin genes (Hba-a1/2) when compared to the ‘no insert’ and ‘negative insert’ controls. Data is normalised to the β-globin (Hbb-b). Bars indicate the standard deviation, the black dots represent single experiment, and the stars indicate the statistical significance resulting from unpaired, two-tailed t-tests, ∗P< 0.05; ∗∗P< 0.01; ∗∗∗P< 0.001; ∗∗∗∗P< 0.0001. (E) Same as in (D) except this is based on the YFP levels measured by FACS of the reporter cells compared to the ‘no insert’ and ‘negative insert’ controls. (F) The expression of the alpha-globin genes is positively and significantly correlated to the level of the YFP in the samples (Linear regression, P< 0.0001, R2= 0.798).
In addition to this observation, we inserted single or multiple copies of HS-38 into a non-coding region of the sub-TAD lying between the alpha-globin enhancers and promoters (insertion site in Figure 1A) (32). In this position HS-38 acted to partially block the interaction between the enhancers and promoters and significantly reduced the level of alpha-globin expression. Multiple insertions of HS-38 (2–4 copies) increasingly compromised enhancer–promoter interactions and further decreased alpha-globin expression. Importantly, we demonstrated that the blocking activity of HS-38 and accumulation of cohesin at this site was greater when the N-terminus of bound CTCF was orientated towards the enhancers rather than towards the promoters, consistent with the bound CTCF stalling translocation of cohesin from the enhancers to mediate loop extrusion (32–34). Some degree of enhancer–promoter blocking was also seen when the CTCF element was inserted in the opposite orientation (32). Consistent with previous conclusions, it is proposed that blocking the translocation of cohesin reduces the frequency of juxtaposition between enhancers and their cognate promoters within a TAD or sub-TAD (35,36).
In the past, CTCF elements have been analysed in the context of relatively large fragments of genomic DNA (1–4kb) (7,36). Here, we have engineered the alpha-globin cluster to enable a direct comparison and ranking of the extent to which individual, minimal (68 bp) CTCF elements, in an identical chromatin context, can compromise enhancer–promoter activity and thereby reduce alpha-globin expression during differentiation: just one of many activities of such sites. Here we have analysed a set of 18 CTCF elements whose roles have been previously determined by deletion from their natural genomic environment and/or analysed in other assays of CTCF elements.
Materials and methods
Construction of plasmids
To generate the CRISPR-Cas9 and chimeric guide RNA expression construct, the single guide-RNAs (sgRNAs) targeting the CTCF element insertion site were cloned into the pX335-U6-Chimeric_BB-CBh-hSpCas9n(D10A) (Addgene, 42335) vector as previously described (31). The pX335 vector was modified to contain a puromycin selection cassette. Sequences of the sgRNA are listed in Supplementary Table S1. Next, oligonucleotides corresponding to each CTCF element were cloned into the pROSA-TV2 vector (created by Prof. Benjamin Davies group) between the pair of BsaI sites using the Golden Gate Assembly Kit (NEB, E1601) and this generates the HDR donor template containing the inserted CTCF element. The pROSA-TV2 vector was designed to contain the 1.4 and 1.2 kb homology arms of the inserted site and a hygromycin selection cassette that is flanked by a pair of lox sites (loxP). Sequences of the homology arms are listed in Supplementary Table S2 and the inserted CTCF element oligonucleotides are listed in Table 1.
Table 1.
Genomic location and core-sequence based orientation of the tested CTCF elements
| Tested CTCF site | Chromosomal location (mm10) | CTCF core motif orientation in endogenous site | Inserted CTCF sequence in forward orientation (Core motif in bold) | Inserted CTCF sequence in reverse orientation (Core motif in bold) |
|---|---|---|---|---|
| HS-94 | chr11: 32182671–32182738 | Forward | CCCAGGAGGCTGCATTACCACAGA TGGACAGTAGAGGGAGACAG AGATCGTGAGTTGAAAATTAAAAG | CTTTTAATTTTCAACTCACGATCT CTGTCTCCCTCTACTGTCCA TCTGTGGTAATGCAGCCTCCTGGG |
| HS-71 | chr11: 32205080–32205147 | Forward | CACGCGAGTGGCAACGGCAGGCCC CAGGCAGCAGGAGGCGCCCG CCTCCCAGCGTCACCTTGTTGGGT | ACCCAACAAGGTGACGCTGGGAGG CGGGCGCCTCCTGCTGCCTG GGGCCTGCCGTTGCCACTCGCGTG |
| HS-59 | chr11: 32217005–32217072 | Forward | CCTTGGAGCAGAAGCCACAATAAA TGACCACGAGGTGGCGCCAA CTGCCAAAAAAGGCATCGAGGCAG | CTGCCTCGATGCCTTTTTTGGCAG TTGGCGCCACCTCGTGGTCA TTTATTGTGGCTTCTGCTCCAAGG |
| HS-39 | chr11: 32237185–32237252 | Forward | TCATGGATTCAAAGCCACTGAGGC CTGGCCACTGGGGGCGCCAT TCGCCATTAAAAGGTCCTGCTGGG | CCCAGCAGGACCTTTTAATGGCGA ATGGCGCCCCCAGTGGCCAG GCCTCAGTGGCTTTGAATCCATGA |
| HS-38 | chr11: 32238628–32238695 | Reverse | CACCTGGTAGTGCTATGCACCCTG AGGCCACCAGAGGGTAGCAG AGGCCTACCCAGGACCTCAGTTTG | CAAACTGAGGTCCTGGGTAGGCCT CTGCTACCCTCTGGTGGCCT CAGGGTGCATAGCACTACCAGGTG |
| HS-29 | chr11: 32247237–32247304 | Forward | CAAATTCCTGTGTCCCTCCAAATT GGTCCACTGGGTGGCACTTG GAGGCCTCAAGCCCTGAGCTGTGC | GCACAGCTCAGGGCTTGAGGCCTC CAAGTGCCACCCAGTGGACC AATTTGGAGGGACACAGGAATTTG |
| Theta 1 | chr11: 32286905–32286972 | Reverse | CTCAAAGACGTCCTGAAACACAAG AGGCCGCCAGGGGGCGCTGC ATCGTTCCAGGATGCCTAGGTGTT | AACACCTAGGCATCCTGGAACGAT GCAGCGCCCCCTGGCGGCCT CTTGTGTTTCAGGACGTCTTTGAG |
| Theta 2 | chr11: 32300010–32300077 | Reverse | CTCAAAGATGTCCTGAAACACAAG AGGCCGCCAGGGGGCGCTGC ATCGTTCCAGGATGCCTAGGTGTT | AACACCTAGGCATCCTGGAACGAT GCAGCGCCCCCTGGCGGCCT CTTGTGTTTCAGGACATCTTTGAG |
| HS + 44 | chr11: 32321366–32321433 | Reverse | GAAAAGCCTTGCACCTACTTATAG TGGCCTGCAGGGGGCGCCCC AGAGACCTCCTGCCCGGTAGTCAT | ATGACTACCGGGCAGGAGGTCTCT GGGGCGCCCCCTGCAGGCCA CTATAAGTAGGTGCAAGGCTTTTC |
| HS + 48 | chr11: 32324813–32324880 | Reverse | CTTGGAGCTGCGAAGTTCCGAGTC CCGCCACACGGGGGTGCTCG TCGCCTTCTTATATCTCCGGTAGC | GCTACCGGAGATATAAGAAGGCGA CGAGCACCCCCGTGTGGCGG GACTCGGAACTTCGCAGCTCCAAG |
| HS + 65 | chr11: 32341265–32341332 | Reverse | CCAGGTTGGAGCACTACATCAATC TCTCCTGCAGGTGGCGCCCT TCCGCGCGACTCACTCTTATGGAC | GTCCATAAGAGTGAGTCGCGCGGA AGGGCGCCACCTGCAGGAGA GATTGATGTAGTGCTCCAACCTGG |
| HS + 66 | chr11: 32343076–32343143 | Forward | GTGTGTGAATGCAGGTGCCTACAG TGGACAGAAGAGGGCGCCAG ATCCTCTGGAGCTGGAGTTTCAGG | CCTGAAACTCCAGCTCCAGAGGAT CTGGCGCCCTCTTCTGTCCA CTGTAGGCACCTGCATTCACACAC |
| mouse β 3′HS1 | chr7: 103807045–103807112 | Reverse | AGAGCTCTGAGGCATGTTCTCAGT CAACCTCAAGGGGGCAGTAT TGAGCTTGGAATTCAGTATCAACT | AGTTGATACTGAATTCCAAGCTCA ATACTGCCCCCTTGAGGTTG ACTGAGAACATGCCTCAGAGCTCT |
| HoxA5|6 | chr6: 52204630–52204697 | Reverse | TCGGAAGCGAAGCGATGCGCCCAG TCTCCAGCGGGTGGCGCTCG AGTCCGACTGAACGGCGGCAACGG | CCGTTGCCGCCGTTCAGTCGGACT CGAGCGCCACCCGCTGGAGA CTGGGCGCATCGCTTCGCTTCCGA |
| H19-Igf2-m3 | chr7: 142580737–142580804 | Reverse | TTATGTGCAACAAGGGAACGGATG CTACCGCGCGGTGGCAGCAT ACTCCTATATATCGTGGCCCAAAT | ATTTGGGCCACGATATATAGGAGT ATGCTGCCACCGCGCGGTAG CATCCGTTCCCTTGTTGCACATAA |
| Sox9-CBS1 | chr11: 111537815–111537882 | Forward | GAAAATATTGTGCAAGGCAACATG TTACCAGCAGGTGGCAGTCC AGTAGACTTCTACAGAACTGTGGC | GCCACAGTTCTGTAGAAGTCTACT GGACTGCCACCTGCTGGTAA CATGTTGCCTTGCACAATATTTTC |
| Sox9-CBS2 | chr11: 111536194–111536261 | Forward | AAGAAAAGGTGTACTCTCCGTTGT CTACCGCCAGATGGCAGCAT GCACACACCAATTAAAATTTGTTC | GAACAAATTTTAATTGGTGTGTGC ATGCTGCCATCTGGCGGTAG ACAACGGAGAGTACACCTTTTCTT |
| Epha4-Pax3-R4 | chr1: 78008893–78008960 | Reverse | TACGACAGAGGCTGCGTCTCACTA TGTCCATGCGGGGGCGCTCT TTTGCAGTCTCTCTGCAGACCTCA | TGAGGTCTGCAGAGAGACTGCAAA AGAGCGCCCCCGCATGGACA TAGTGAGACGCAGCCTCTGTCGTA |
| Negative control | chr11: 32274291–32274358 | nil | AATGTAATGAAAATATGGCTAATTA ATCTTTACATTCTGTCTTGGCCA AGATGCAGAGAAGCGACAGT | nil |
Tissue culture, transfection and drug selection
The mouse embryonic stem cell line (mESC), E14TG2a was used for the CTCF activity reporter assays. To facilitate the genetic modifications and downstream analysis, one wild-type copy of the alpha-globin locus was hemizygously deleted, and a yellow fluorescent protein (mVenus) was introduced at the end of exon 3 of Hba-a1 gene (37). The mESCs were maintained in Glasgow's Minimal Essential Medium (Gibco 21710-082) supplemented with 10% heat-inactivated foetal bovine serum (Gibco, 10270-106) 1 mM sodium pyruvate (Gibco 11360-039), 2 mM l-gluatamine (Gibco, 25030-024), 1× MEM (NE) AA (Gibco 11140-035), 0.1 mM beta-mercaptoethanol (Gibco, 31350-010) and 1 U/μl leukaemia inhibiting factors (Cell guidance system, LIF, GFM200) at 37°C humidified chamber with 5% CO2. Genetically modified mESC models with inserted CTCF elements were generated using the double nickase CRISPR-mediated homology-directed repair (HDR) strategy. The mESCs were co-transfected with the 1.66 μg of the two sgRNA nickase-Cas9 plasmids and 0.83 μg of the HDR plasmid using the Lipofectamine LTX and Plus Reagent (Invitrogen, 15338-100) according to manufacturer's instructions. Transfected cells underwent 1 μg/ml puromycin selection for 2 days and a subsequent 250 μg/ml hygromycin selection for 6 days. The positively selected cells were then transfected with 2.5 μg pCre-Pac vector using the Lipofectamine LTX and Plus Reagent (Invitrogen, 15338-100) which facilitated removal of the hygromycin-resistance cassette (Supplementary Figure S1 and Supplementary Figure S2). The insertion efficiency of this strategy ranges between 14.8% and 30.8%.
Genotyping
Genetically modified mESCs that survived the antibiotic selections and Cre-recombinase vector transfection were single cell sorted in 96-well plate and grown as clonal colonies. Genomic DNA was extracted from individual colonies and were subjected to three sets of PCR screening (across the insert, left homology side spanning, and right homology side spanning) for the successful insertion of the CTCF elements to the target site and to eliminate possible concatemer integrations (Thermo Scientific, DreamTaq Green PCR Master Mix, K1081) (Supplementary Figure S3). Genotypes of positive clones were confirmed by Sanger sequencing. Sequences of the PCR screening primers are listed in Supplementary Table S1.
In vitro embryoid body (EB) differentiation
EB differentiation of mESCs were based on previously published protocol (37). In brief, mESCs were induced by passage into IMDM base media (Iscove's modified Dulbecco's medium (IMDM, Gibco, 31980030), supplemented with 15% heat-inactivated FBS, 1.4 × 10–4 M monothioglycerol (Sigma-Aldrich, MTG, M6145), 50 U/ml penicillin-streptomycin (Gibco, 15140122) and 1000 U/ml LIF (Cambridge Bioscience, GFM200-5)) at 48 h prior to differentiation. During primary plating, mESCs were trypsinized and plated in the EB differentiation medium (IMDM medium supplemented with 15% heat-inactivated FBS, 5% protein-free hybridoma medium (PFHM-II, Gibco, 12040-077), 2 mM L-glutamine, 50 μg/ml l-ascorbic acid (Sigma Aldrich, A4544), 3 × 10–4 M MTG and 300 μg/ml human transferrin (Roche, 10652202001)) in a triple vent petri 10 cm dishes (Thermo fisher, 101VR20) seeded at 1 × 104 cells for up to seven days. The dishes were gently shaken daily to disrupt any potential attachment to the dish. At day 7, EBs were harvested and disaggregated in 0.25% trypsin (Gibco, 25200-056) for 3 min at 37°C and neutralized with FBS. The disaggregated EBs were labelled with anti-mouse CD71-FITC antibody (eBioscience, 11-0711-85) (1:200 in staining buffer), washed, and further incubated with the MACS anti-FITC separation microbeads (Miltenyi, 130-048-701, 10 μl per 107 cells). The bead labelled CD71 + cells were then isolated by magnetic column separation (Miltenyi, LS Column, 130-042-401), according to the manufacturer's instructions.
Flow cytometry
Differentiated EB cells were disaggregated and stained with the anti-mouse CD71-APC antibody (eBioscience, 11-0711-80) (1:8000 in staining buffer) and Hoechst (Invitrogen, 33258) (1:10 000 in staining buffer) for 30 min at 4°C in dark. The level of YFP signal in the live CD71 positive cells was measured and used as a proxy for the level of the alpha-globin gene expression in cell line models. Stained cells were analysed using the Attune NxT Flow cytometer (Thermo fisher) and the Attune NxT software package.
RNA expression analysis
Total RNA was isolated from 1 × 107 EB-derived CD71+ cells using TRI reagent (Sigma, T9424) followed by RNA extraction and the on-column DNaseI digestion using the Direct-zol RNA miniprep kit, according to manufacturer's protocol (Zymo Research, R2050). The quantity and quality of the RNA are accessed by the Qubit RNA BR Assay (Invitrogen, Q10211) and the RNA ScreenTape (Agilent Technologies, 5067-5576), respectively. 1 μg of the total RNA was used for synthesising cDNA using the Superscript III first-strand synthesis SuperMix (Invitrogen, 11752-050) according to manufacturer's protocols. A no reverse transcriptase control was included to test for potential genomic DNA contamination in the sample. Quantitative real-time PCR was performed in triplicate on the 5× diluted cDNA using the fast SYBR green master mix (Thermo Fisher, 4385612) to assess the relative changes in gene expression. Primers used in the qPCR reaction are listed in Supplementary Table S1. The ΔΔCt method was used to quantify the RNA abundance of Hba-a relative to Hbb-b.
CTCF core binding motif occurrence
The occurrence of the CTCF core binding motif was determined by the ‘Find Individual Motif Occurrences’ FIMO (version 5.4.1) program in the MEME suite (38). The CTCF core motif position weight matrices used in the FIMO analysis were based on the HOCOMOCO database (CTCF.MOUSE.H11MO.0.A) (39). The inserted sequences (18 selected CTCF elements, 68 bp in length in their native orientation) were used as input sequences for the FIMO analysis with the zero-order background model. The p-value generated from the FIMO analysis evaluates the likelihood of observing the CTCF core motif occurrence in the input sequence by chance. When multiple CTCF core motifs were detected within the sequence, only the best match was retained.
Conservation of the CTCF element
The degree of evolutionary conservation of the 68bp inserted CTCF sequence was indicated by PhastCons conservation score. PhastCons is a hidden Markov model-based method that estimates the probability that each nucleotide belongs to a conserved element based on multiple alignment in 30 vertebrate species (40).
Enrichment level of CTCF and Rad21
The enrichment level of CTCF protein and RAD21 protein on individual CTCF binding sites were determined by re-analysing the previously published CTCF and RAD21 ChIP-sequencing data in mESCs and erythroid cells with the Lanceotron peak calling framework (LanceOtron.molbiol.ox.ac.uk) (41). Peak calling was performed using the bigwig files of the ChIP-seq dataset as the input, and the enrichment level of CTCF or RAD21 individual ChIP-seq peak is calculated based on the peak statistics (width, height, area) in Lanceotron.
Chromatin immunoprecipitation sequencing
Chromatin immunoprecipitation sequencing (ChIP-seq) was performed using the ChIP Assay Kit according to the manufacturer's instructions (Milliopore, 17-295). In brief, 5 × 106 purified CD71 positive erythroid cells derived from two biological replicates were subjected to crosslinking with 1% formaldehyde for 10 min and was quenched with 125 mM of glycine for 5 min. Chromatin fragmentation was performed using the Bioruptor Pico Sonicator (Diagenode) with 6 cycles of 30 s ON and 30 s OFF at 4°C to obtain an average fragment size of 200–500 bp. The fragmented chromatin was immunoprecipitated with 10μl of Rabbit Anti-CTCF antibody (Millipore, 07-729) at 4°C overnight on a rotator. The mix was incubated with Protein A agarose beads for 1 h at 4°C on a rotator prior to the washing steps and the elution. DNA sequencing libraries were prepared with the NEBNext Ultra II DNA library prep kit according to the manufacturer's instructions (New England Biolabs, E7645S) and sequenced on the Illumina NextSeq platform with the 75-cycle paired end kit (NextSeq 500/550 High Output Kit).
ChIP-seq data was analysed using an in-house pipeline. In brief, bowtie (version 1.2.3) was used to align the raw fastq files with the reference genome (an edited customed genome based on mm10 that includes the CTCF insertion sequence). The remaining unaligned reads are trimmed by Trimgalore (version 0.6.5) and flashed by using FLASh (version 1.2.11) before they were re-aligned to the reference genome by bowtie again. Samtools (version 1.10) was used to filter, sort, fixmate, remove PCR duplicates and index the mapped reads. The bamCoverage function of Deeptools (version 2.2.2) was used to create the bigwig file with normalisation set to CPM and smooth length of 300 bp.The bigwig files were visualized on the University of California Santa Cruz (UCSC) genome browser as individual tracks for comparison. All ChIP-seq experiments were performed at least in biological duplicate with similar results.
Statistical analysis
Statistical analysis was carried out with Graphpad Prism (version 9). All the experiments were performed on three or four biological replicates (3–4 individual clones and three individual EB differentiations) with similar results, and standard deviation is shown for all measurements. Statistical analysis between control and target groups was performed using unpaired, two-tailed t-tests. Spearman correlation was performed for the FIMO P-value and CTCF enhancer–promoter blocking activity. Linear regression was performed to determine the relationship between the CTCF enhancer–promoter blocking activity and variables including the degree of conservation, levels of CTCF enrichment, and level of Rad21 enrichment. P-values are represented as NSP > 0.05; ∗P< 0.05; ∗∗P< 0.01; ∗∗∗P< 0.001; ∗∗∗∗P< 0.0001.
Results
An experimental system to evaluate the capacity of individual CTCF elements to block enhancer–promoter interactions.
Using the mouse alpha-globin locus, we developed an experimental system to quantify the strength and effectiveness of individual CTCF elements to block the interaction between enhancers and promoters during erythroid differentiation. The regulatory landscape of the mouse alpha-globin locus has been well-characterised and the protocol for erythroid differentiation is fully established (37). We have previously shown that the duplicated alpha-globin genes are regulated by a cluster of five enhancers (R1, R2, R3, Rm and R4) located 14–37 kb upstream of the Hba-a1 gene. The alpha-globin enhancers and genes are located within a ∼65kb sub-TAD (topologically associating domain) that is largely delimited by an array of convergently oriented CTCF binding sites (31,42,43) (Figure 1A).
We have recently shown that insertion of an 83 bp fragment spanning a previously characterised CTCF binding element (HS-38) between the alpha-globin enhancers and the alpha-globin promoters significantly reduced the interaction between these elements and reduced the level of transcription from the alpha-globin genes (32). This CTCF element acted in an orientation dependent manner, having a greater effect when the N-terminus of the bound CTCF protein was directed towards the enhancers lying upstream of the CTCF site. Based on this study, we engineered the alpha-globin locus to allow insertion of any CTCF bound element into a landing pad placed in a non-coding region between the enhancers and promoters (insertion site in Figure 1A) to determine the ability of each independent element to reduce the interaction between the enhancers and their cognate promoters. To standardise the comparison of CTCF elements we analysed uniformly sized sequences (68 bp) containing the 20 bp core CTCF motif, 24 bp upstream and 24 bp downstream to include all known DNA binding surfaces of CTCF.
In undifferentiated mESCs, CTCF elements of interest were inserted into a landing pad (insertion site) between the alpha-globin enhancers and the alpha-globin promoters using CRISPR-mediated homology-directed repair (HDR) (Supplementary Figure S1). To facilitate the CRISPR-HDR insertion and analysis of the engineered allele, we hemizygously deleted the wild-type copy of the alpha-globin locus (117 kb). In addition, we tagged the Hba-a1 gene with a yellow fluorescent protein (YFP), so that the YFP signal could be used as a proxy for the level of alpha-globin gene expression (Figure 1A) (37). These engineered mESCs were subsequently differentiated into erythroid cells in vitro within embryoid bodies (EBs) (37) since the interaction between the alpha-globin enhancers and their cognate promoters only occurs in differentiating erythroid cells. The levels of YFP in differentiated CD71 + erythroid cells were quantified by flow cytometry as a readout of the degree to which any CTCF element acted as an enhancer blocker (Figure 1B and C): high levels of YFP indicate low levels of insulation and low levels of YFP are associated with high levels of insulation.
To validate this as a means of testing how individual CTCF elements could block enhancer–promoter interactions, we initially inserted the HS-38 CTCF binding site into the landing pad. This element had previously been shown to effectively reduce enhancer–promoter interactions at the alpha-globin locus (32) where it normally insulates interaction between the strong alpha-globin enhancers and genes (Mpg, Rhbdf1 and Il9r) lying upstream of the cluster (31). Insertion of a 68bp element spanning the HS-38 CTCF element, in both the forward and reverse orientations exhibited significant reductions in alpha-globin gene expression when compared to the no insert control (the unmodified mESCs) and a negative insert control (insertion of the 68bp neutral sequence with no boundary activity) (Figure 1D). The reduction of alpha-globin expression was accurately reflected by the decreased level of YFP signal (Figure 1E) which is strongly and positively correlated with the level of alpha-globin RNA expression, assessed by RT-qPCR (P < 0.0001, R2= 0.798) (Figure 1F). Consistent with the work of Stolper and Tsang et al., we observed that the ability of the CTCF element to block an interaction between the enhancers and promoters was greater when the HS-38 site was inserted in the reverse orientation with the N-terminus of bound CTCF orientated towards the upstream enhancers (Figure 1D, P= 0.0047; Figure 1E, P= 0.0031). This observation may reflect the directional tracking of the cohesin complex from the enhancers to the promoters on the alpha-globin locus as previously discussed (32). In summary, we have shown that this engineered cell line and the associated YFP reporter faithfully and accurately reflect the ability of an individual minimal CTCF element to reduce the functional interaction between an enhancer and promoter as the cells undergo differentiation.
Analysing previously characterised CTCF elements
To investigate whether this experimental model could distinguish the degree to which individual CTCF elements might differ in their ability to interfere with enhancer–promoter interactions, we analysed previously characterised elements. Our previous work deleting individual boundary elements in the alpha-globin locus showed that HS-38 acts as a strong boundary which normally delimits the activity of the alpha-globin enhancers on genes lying upstream of the cluster (31). Deletion of other CTCF elements in the alpha-globin sub-TAD had variable effects on its 3D structure. Importantly, deletion of individual sites and combinations of sites from their natural positions in the alpha-globin sub-TAD had no discernible effects on alpha-globin expression (30).
Insertion of the twelve individual CTCF elements of the alpha-globin locus (Figure 1A and Table 1 in turn into the landing pad blocked the enhancer–promoter interaction to variable degrees (Figure 2A and B). The levels of CTCF binding at the inserted landing pad region determined by CTCF ChIP-seq, in three selected models, demonstrated the inserted 68bp CTCF binding sequence recapitulated the CTCF binding level of the respective endogenous CTCF sites (Supplementary Figure S4–S6). As the CTCF core binding motif is non-palindromic, we have also tested these CTCF sequences in different orientations. The orientation of the sequence is defined by the orientation of the CTCF core motif, irrespective of the orientation of the endogenous CTCF site in its native context (Table 1). Insertion of these CTCF binding sequences in both forward and reverse orientation in the reporter assay showed similar trends (Figure 2A and B) but again with greater effect when the sequence is reversely oriented and the N-terminus of bound CTCF was directed towards the enhancers (Supplementary Figure S7). These observations demonstrated that the experimental system established here can distinguish CTCF elements which display different degrees of insulation in an orientation-dependent manner.
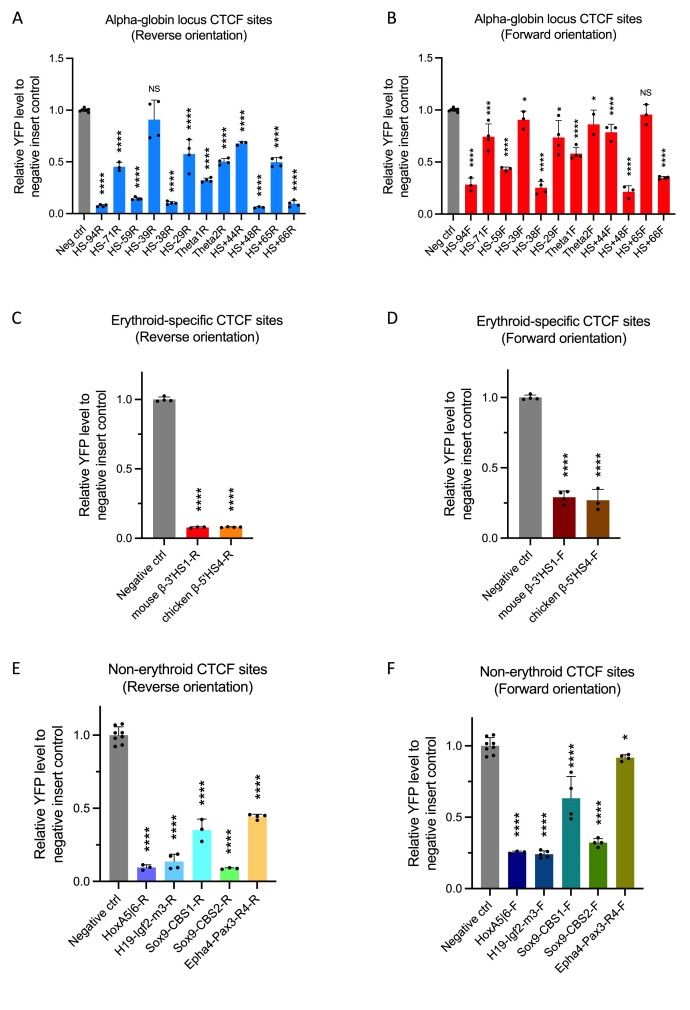
Figure 2.
Testing previously characterised CTCF elements for their insulation activity. (A, B) YFP levels derived from FACS analysis of EB cell populations derived from mESCs engineered to contain the indicated CTCF sites and plotted relative to a negative control mESC-derived signal. Nearly all CTCF sites in the alpha-globin locus blocked the enhancer–promoter interaction to variable degrees in both orientations. (C, D) Analysis as in panels above. Both β-globin locus CTCF binding sites in mouse (3′HS1) and chicken (5′HS4) significantly blocked the enhancer–promoter interaction. (E, F) Analysis as in panels above. CTCF binding elements from other non-erythroid specific loci also blocked the enhancer–promoter interaction and reduced alpha-globin expression to variable degrees. Bars indicate the standard deviation, the black dots represent single experiment, and the stars indicate the statistical significance resulting from unpaired, two-tailed t-tests, NS not significant; * P< 0.05; ** P< 0.01, *** P< 0.001, **** P< 0.0001.
Initial experiments were performed on CTCF elements derived from the mouse alpha-globin sub-TAD. A more extensive set of CTCF elements was tested to ensure that the experimental system was suitable for analysing a broader range of elements. To this end, we first analysed the well-characterised erythroid-specific 3′HS1 CTCF element from the mouse β-globin locus (13), and showed that insertion of this site exhibited a prominent reduction in the level of YFP (Figure 2C and D). A similar effect was observed when we inserted the erythroid specific 5′HS4 CTCF element of the chicken β-globin locus (5) (Figure 2C and D), showing that the experimental system can quantify the activities of boundary elements even from other species.
We next tested CTCF binding elements from non-erythroid specific loci, including previously characterised CTCF elements located at the H19-Igf2, HoxA and Sox9-Kcnj2 loci (6,12,44,45), and a less prominent CTCF binding site located at the Epha4-Pax3 TAD boundary (46). This showed that insertion of the CTCF elements from the H19-Igf2, HoxA and Sox9 loci strongly and significantly diminished the YFP level in the experimental system, while insertion of the less prominent CTCF site from the Epha4-Pax3 TAD boundary reduced the YFP level to a lesser extent (Figure 2E and F).
Nearly all these diverse CTCF elements blocked the enhancer–promoter interaction and reduced alpha-globin expression to variable degrees and again all had a greater effect when they are reversely oriented; where the N-terminus of the bound CTCF protein was directed towards the enhancers (Figure 2, Supplementary Figures S7 and S8). Together, these observations showed that the experimental system established here could quantify the activities of a wide variety of CTCF elements contained within small fragments of 68 bp, including the proposed DNA contacts of CTCF, and reflect their varied ability to block enhancer–promoter activities.
Correlating the structure and function of individual CTCF elements
Previous studies of CTCF elements have correlated their activities with a variety of associated features including the sequence of the core CTCF binding element, its evolutionary conservation, the amount of CTCF and cohesin bound in ChIP assays, and their persistence following acute depletion of CTCF protein (18,21–25). While these parameters derived from large datasets provide general trends, their predictive values have not been extensively tested on individual elements at the identical genomic position where all other variables are minimised.
First, by performing the FIMO analysis we found no correlation between the enhancer–promoter blocking activity of the CTCF elements tested in this study and their resemblance to the consensus CTCF core motif described as position-specific scoring matrices (Figure 3A and Supplementary Table S3) (Pearson correlation, P= 0.84, R2= 0.0026). The degree of conservation of the CTCF element was also not correlated with their ability to alter enhancer–promoter activity as tested (Figure 3B and Supplementary Table S3) (linear regression, P= 0.2171, R2= 0.0935).
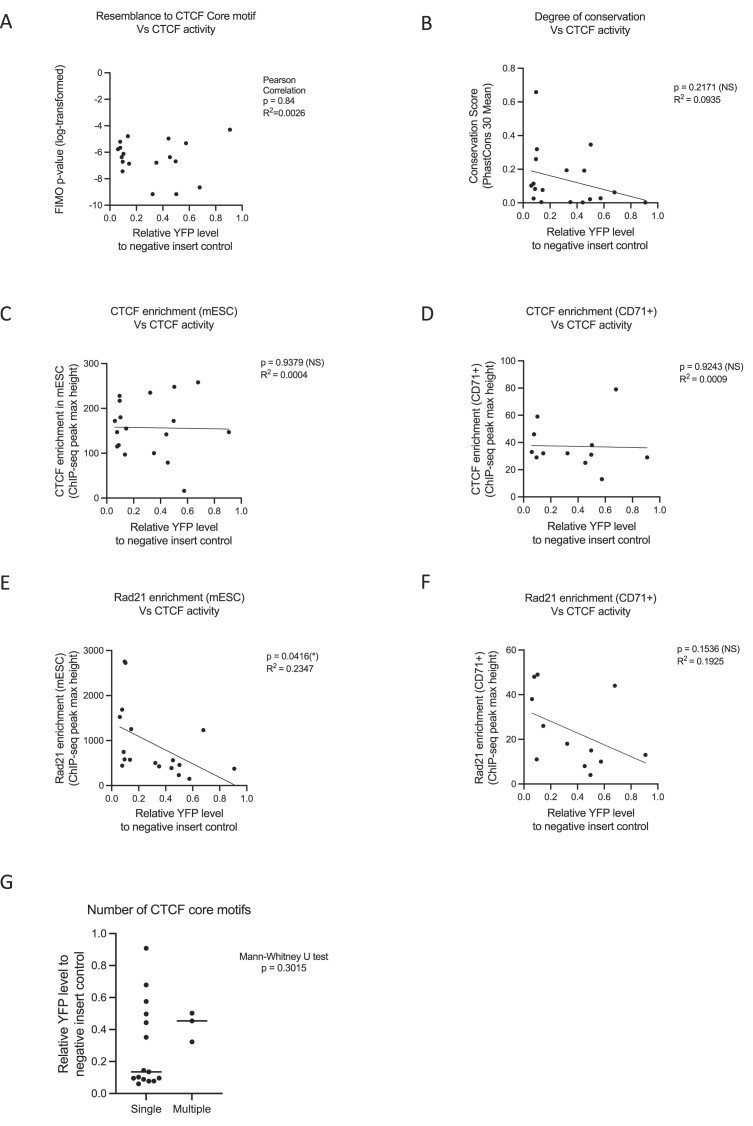
Figure 3.
Correlating binding activity and sequence characteristics with insulation function of individual CTCF elements. (A) There is no correlation between the core CTCF binding sequences (p-value of the FIMO analysis) with their ability to alter enhancer promoter activity (Spearman correlation, P= 0.65). (B) There is no correlation between the conservation of the inserted CTCF binding (PhastCons Vert30 genome conservation score) with their ability to block enhancer–promoter interaction (linear regression, P= 0.2171, R2= 0.0935). (C) There is no significant correlation between CTCF activity and CTCF enrichment in the mESCs (linear regression, P= 0.9379, R2= 0.0004). (D) There is also no correlation between CTCF activity and CTCF enrichment when only focused on the erythroid-specific alpha-globin CTCF binding sites in the CD71+ erythroid cells isolated from EBs (linear regression, P= 0.9243, R2= 0.0009). (E) CTCF binding sites with stronger enhancer–promoter blocking activity showed a higher level of Rad21 enrichment in mESCs (linear regression, P= 0.0416, R2= 0.2347). (F) The correlation between the activities of the alpha-globin locus CTCF binding sites and enrichment of cohesin in CD71+ cells isolated from the differentiated EBs was not statistically significant (linear regression, P= 0.1536, R2= 0.1925). (G) The presence of more than one overlapping core motif in the three tested CTCF sites (labeled as ‘multiple’) did not manifest as a stronger insulation effect as shown by the YFP levels when comparing the ‘multiple’ to ‘single’ (CTCF sites with only one core CTCF motif) datasets.
Second, we investigated the relationship between the effect of each CTCF element and the levels of enrichment seen at these sites in their native context in the genome obtained from CTCF ChIP-seq data. In mESCs, we observed no significant correlation between activity and CTCF enrichment (Figure 3C and Supplementary Table S3) (linear regression, P= 0.9379, R2= 0.0004). Similarly, there was also no such correlation when we only focused on the erythroid-specific alpha-globin CTCF binding sites in the CD71+ erythroid cells isolated from EBs (Figure 3D and Supplementary Table S3) (linear regression, P= 0.9243, R2= 0.0009). One limitation of this correlation study is that the enhancer–promoter blocking activity of the CTCF sequence is measured based on its insertion at the landing pad region, while the CTCF protein enrichment levels of individual sites are taken from their endogenous CTCF sites. Although the levels of CTCF protein binding at the insertion site appear similar to the levels of CTCF bound at endogenous sites in three insertion models (Supplementary Figure S4–S6), we cannot be certain that this holds true for all insertion models. Therefore, our observations suggest that high levels of CTCF enrichment at their normal genomic positions may not necessarily predict their ability to block enhancer–promoter interactions.
Next, we interrogated the relationship between the enhancer–promoter blocking activity of CTCF elements and cohesin enrichment estimated from RAD21 ChIP-seq data. Interestingly, we observed that CTCF binding sites with stronger activity (i.e. lower relative YFP levels) showed a higher level of RAD21 enrichment in mESCs (Figure 3E and Supplementary Table S3) (linear regression, P= 0.0416, R2= 0.2347). As it has been shown that the loading and the enrichment level of cohesin may be tissue-specific (47,48), we have also correlated the activities of the alpha-globin locus CTCF binding sites and enrichment of cohesin in CD71 + cells isolated from the differentiated EBs. Although not statistically significant, we observed a trend that stronger CTCF elements (i.e. lower relative YFP level) tend to have higher levels of cohesin enrichment in their native loci (Figure 3F and Supplementary Table S3) (linear regression, P= 0.1536, R2= 0.1925). However, the levels of cohesin enrichment are likely to be affected by the position of each element in the native locus.
A recent study proposed that certain CTCF binding sites may contain multiple CTCF core motifs, either in parallel or in opposite orientation, which could potentially enhance their insulation activity (29). In this study, we have identified the presence of additional CTCF core motifs within three of the tested CTCF sites, namely HS-71, theta-1 and theta-2 from the alpha-globin locus (Supplementary Figure S9 and Supplementary Table S4). Despite the presence of the additional CTCF core motifs, our findings indicate that the insulation strength of these CTCF sites does not increase compared to CTCF sites harbouring a single core motif (see Figure 3G; Mann–Whitney U test, P= 0.3015).
In summary, despite correlations found in genome-wide analyses, none of the commonly found associations fully predict the behaviour of individual CTCF elements in the sensitive assay used here to detect perturbations in the interactions between enhancers and promoters and the consequent changes in the levels of gene expression.
Sequences flanking the core motif contribute to the strength of the CTCF element to perturb enhancer–promoter interactions
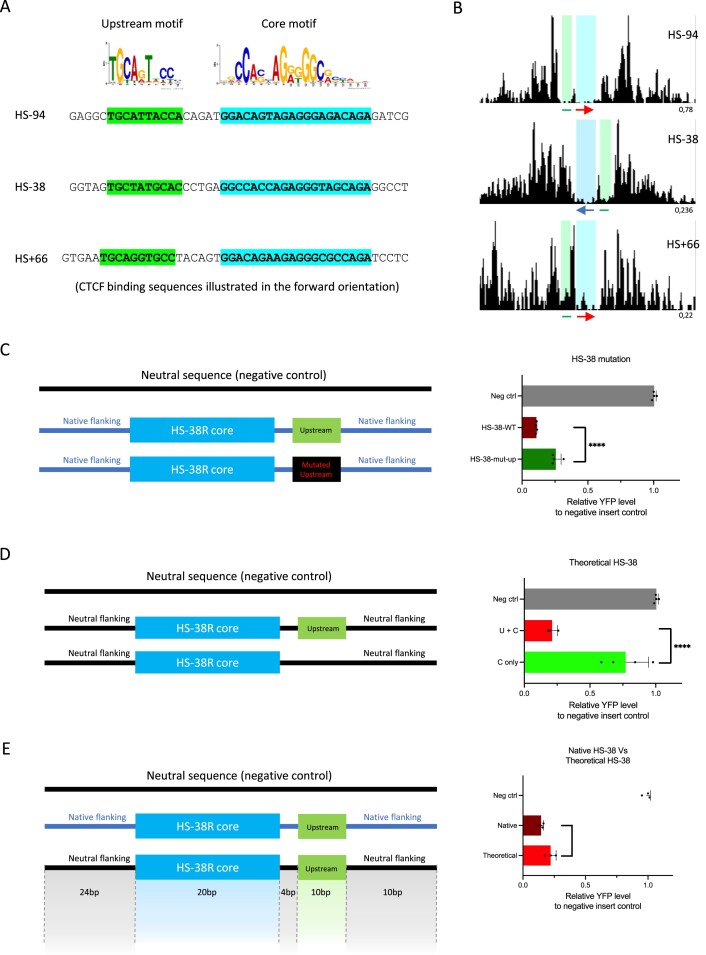
Although we found no predictive value for the CTCF blocking activity by analysing the core CTCF motif, it has been previously shown that sequences immediately flanking the CTCF core motif may contribute to the functional role of a CTCF binding site. For instance, a phylogenetically conserved upstream motif has been discovered at 15% of all CTCF binding sites (21,49). The upstream motif was shown to stabilise CTCF occupancy by interacting with the zinc fingers of the CTCF protein at its C terminus (22,50). We scanned all the sites that we have tested and identified this upstream motif at HS-94, HS-38, and HS + 66 sites from the alpha-globin locus (Figure 4A). High resolution DNaseI footprinting we have published previously showed that the upstream regions of these sites are indeed bound by proteins in erythroid cells (31) (Figure 4B). Consistently, the sites associated with this upstream motif (HS-94, HS-38 and HS + 66) exhibited very strong blocking activities compared to other CTCF binding sites analysed in this study (Figure 2A and B).
Figure 4.
Sequences flanking the core motif contribute to the strength of the CTCF element in perturbing enhancer–promoter interactions. (A) The upstream and core motif sequences highlighted in green and blue respectively on the alpha-globin HS-94, HS-38 and HS + 66 CTCF binding sites. (B) DNaseI footprinting data of the HS-94, HS-38 and HS + 66 CTCF binding sites. Red arrow, forward oriented CTCF core motif; blue arrow, reverse oriented CTCF core motif; green line, known upstream motif; blue highlighted area, DNaseI footprints occupied by the CTCF core motifs; green highlighted area, DNaseI footprint occupied by the upstream motifs). (C) YFP levels derived from FACS analysis of EB cell populations derived from mESCs engineered to contain the indicated engineered CTCF sites (HS-38 wildtype and HS-38 with a mutated upstream motif) and plotted relative to a negative control mESC-derived signal. Mutation of the upstream motif of the native HS-38 site (dark green bar) has significantly increased the relative YFP level when compared to the wildtype (maroon bar). (D) Analysis same as above. The relative YFP level was remarkably increased when the upstream motif was absent in the theoretical HS-38 site (light green bar). (E) Analysis same as above. The insulation ability of the native HS-38 (maroon) is higher than that of the theoretical HS-38 site (light red). Bars indicate the standard deviation, the black dots represent single experiment, and the stars indicate the statistical significance resulting from unpaired, two-tailed t-tests, **** P< 0.0001, * P< 0.05.
To further explore the effect of the upstream motif on boundary strength, we synthesised artificial CTCF sites based on the alpha-globin HS-38 sequence and tested them in the CTCF activity reporter assay. First, we mutated the upstream motif of the native HS-38 sequence while keeping the core motif intact. Mutation of the upstream motif significantly increased the relative level of YFP (indicating reduced insulator activity) when compared to the wildtype HS-38 site (Figure 4C and Supplementary Table S5). Next, we investigated how the core and upstream motif sequences alone affect insulator strength without the influence of any native flanking sequences on HS-38. To achieve this, we synthesised two ‘theoretical’ CTCF sites: one containing the HS-38 core motif plus the upstream motif and another with the core motif only, both embedded in a neutral sequence derived from the alpha-globin locus with no detected CTCF binding activity. Again, the relative level of YFP was remarkably increased when the upstream motif was absent, indicating a significant reduction of the insulator strength (Figure 4D and Supplementary Table S5). These results show that the upstream motif sequence positively contributes to the insulator strength of CTCF binding sites. To confirm the underlying mechanism of the upstream motif, we have performed CTCF ChIP-seq in the theoretical HS-38 models. Interestingly, we observed almost no binding of CTCF in the absence of the upstream motif (Supplementary Figure S10). This suggests that the upstream motif sequences may strengthen the binding of the CTCF proteins on the element.
Of interest, the newly synthesised ‘theoretical’ HS-38 site did not fully recapitulate the boundary strength of the ‘native’ HS-38 site. Even though the ‘native’ and newly synthesised ‘theoretical’ sites contain exactly the same core and upstream motif sequences (a total of 30 bp), the boundary strength of the native HS-38 site is significantly stronger (Figure 4E and Supplementary Table S5) (P= 0.023). CTCF ChIP-seq revealed that the inserted ‘native’ HS-38 site binds CTCF protein at similar level as the ‘endogenous’ HS-38 site. However, the ‘theoretical’ HS-38 site recruits much less CTCF protein than the inserted ‘native’ HS-38 site and the ‘endogenous’ HS-38 site (Supplementary Figure S11). This data suggests that the sequences immediately flanking the core and upstream motifs (38 bp to the left and to the right of the core and upstream motifs) in the ‘native’ site could also contribute to the boundary activity.
It therefore appears that at least one common motif immediately flanking the core CTCF motif in the small (68 bp) sequences studied here can modify the activity of the element. From our previously published DNaseI footprinting data, we have observed additional DNaseI footprints both upstream and downstream of some CTCF sites such as the HS-59, HS + 44, HS + 48 and HS + 65 on the alpha-globin locus (31) (Supplementary Figure S12). It remains to be seen if such sites may bind other proteins associated with insulator activity.
Discussion
To compare the ability of CTCF elements to insulate interactions between enhancers and promoters, we analysed uniformly sized sequences (68 base pairs) containing the core CTCF motif (20 bp) and 24 bp flanking upstream and downstream from 18 well-characterised elements. We show that regardless of their role in their natural genomic locations, nearly all CTCF elements analysed in this fixed, identical genomic location block enhancer–promoter interaction to different extents. Notably, we observed that the effect of the CTCF elements was consistently greater when the N-terminus of the CTCF protein was orientated towards the enhancer, consistent with findings from our parallel study (32). This observed orientation bias may be due to the local topology effect of the alpha-globin locus. We have shown that cohesin complexes may be loaded and extruded from the alpha-globin enhancer cluster. Therefore, they may be more effectively impeded by the N-terminus of the CTCF protein binding to the reverse oriented inserted CTCF site, facing the enhancer cluster (34,51,52).
The alpha-globin locus represents an appropriate genomic region for establishing the insulator reporter assay to evaluate the insulation activity of individual CTCF elements during erythroid differentiation. Notably, our investigation into the two Sox9-Kcnj2 CTCF binding sites reveals intriguing insights. While a previous study utilizing larger fragments (2.3–2.5 kb) only reported minimal activities for these CTCF sites in a boundary reporter assay located at the Sox2 locus (36), our assay reveals a substantial insulation activity for these same CTCF sites. This indicates that the alpha-globin based experimental system may offer greater sensitivity in quantifying CTCF-element activities, allowing for a wider spread of blocking activity to be identified in a larger pool of tested sites.
The reporter assay described here effectively discriminates between the strength of previously validated strong and weak CTCF sites reported in other studies. This suggests that our reporter assay provides an unbiased platform to evaluate the insulation activity of minimal CTCF elements. By keeping the genomic context surrounding the CTCF insertion site constant, the only variable is the inserted CTCF sequence. Thus, the measured CTCF insulation activity is a direct reflection of the impact of the inserted CTCF sequence alone. It is important to note that one potential limitation of this reporter assay may be in its reliance on YFP as the readout for the insulation activity. YFP levels exhibit a sigmoidal rather than a linear relationship, which implies that extreme YFP values may have more predictive power than the intermediate YFP values. However, a direct assessment of transcriptional output could be performed (e.g. RT-qPCR) if needed.
In this study, we found no correlation between the ability of CTCF elements to block enhancer–promoter activity with their resemblance to the consensus CTCF core motif; the degree of evolutionary conservation; or the number of CTCF core motif harboured in the CTCF elements. Our limited observations also suggest that the levels of CTCF and cohesin enrichment at the natural genomic positions of the CTCF element may not necessarily reflect their ability to block enhancer–promoter interactions. Nevertheless, using DNaseI footprinting we have shown that three of the strongest enhancer–promoter blockers include a previously described bound element lying upstream of the core CTCF binding sequence (21,49). In addition, we found other uncharacterized footprints located close to the core sequence that may affect function. This suggests that other proteins binding near to CTCF sites may contribute to insulator activity.
Altogether, the analysis of our 126 engineered models (encompassing the naturally occurring 18 CTCF sites, generated in both orientations, in addition to the mutants and artificial versions, each derived in three independent mESC clones) demonstrate that the insulator assay located at the alpha-globin locus is a reliable and sensitive platform to quantify the activities of universal boundary elements in a non-biased manner. Together these findings show that, as with enhancers and promoters, using the approach outlined here, it will ultimately be possible to sub-classify CTCF elements in an unbiased way by sequence analysis independently of their roles in their natural genomic environment.
Supplementary Material
Acknowledgements
We are very grateful to Lars Hanssen for initiating the work on CTCF insertion between the alpha-globin enhancers and promoters; Helena Francis, Andrew King and Danuta Jeziorksa for establishing the YFP-tagged hemizygous mESCs cell line; Helena Francis for the YFP reporter validation as a proxy for alpha-globin transcriptional output; Lance Hentges for his advice on using LanceOtron; Prof. Jim Hughes, Prof. James Davies, Dr Robert Beagrie and all the members of the Higgs lab for helpful comments on the manuscript; the Flow Cytometry Facility at the Weatherall Institute for Molecular Medicine (WIMM) for helping with the FACS sorting experiments.
Notes
Present address: Rosa J. Stolper, Wellcome Centre for Human Genetics, Nuffield Department of Medicine, University of Oxford, Old Road Campus, Oxford OX3 7BN, UK.
Present address: Benjamin Davies, Francis Crick Institute, London NW1 1AT, UK.
Contributor Information
Felice H Tsang, Chinese Academy of Medical Sciences Oxford Institute, Nuffield Department of Medicine, University of Oxford, Old Road Campus, Oxford OX3 7BN, UK; MRC Weatherall Institute of Molecular Medicine, Radcliffe Department of Medicine, University of Oxford, John Radcliffe Hospital, Oxford OX3 9DS, UK.
Rosa J Stolper, MRC Weatherall Institute of Molecular Medicine, Radcliffe Department of Medicine, University of Oxford, John Radcliffe Hospital, Oxford OX3 9DS, UK.
Muhammad Hanifi, MRC Weatherall Institute of Molecular Medicine, Radcliffe Department of Medicine, University of Oxford, John Radcliffe Hospital, Oxford OX3 9DS, UK.
Lucy J Cornell, MRC Weatherall Institute of Molecular Medicine, Radcliffe Department of Medicine, University of Oxford, John Radcliffe Hospital, Oxford OX3 9DS, UK.
Helena S Francis, MRC Weatherall Institute of Molecular Medicine, Radcliffe Department of Medicine, University of Oxford, John Radcliffe Hospital, Oxford OX3 9DS, UK.
Benjamin Davies, Wellcome Centre for Human Genetics, Nuffield Department of Medicine, University of Oxford, Old Road Campus, Oxford OX3 7BN, UK.
Douglas R Higgs, MRC Weatherall Institute of Molecular Medicine, Radcliffe Department of Medicine, University of Oxford, John Radcliffe Hospital, Oxford OX3 9DS, UK; Chinese Academy of Medical Sciences Oxford Institute, Nuffield Department of Medicine, University of Oxford, Old Road Campus, Oxford OX3 7BN, UK.
Mira T Kassouf, MRC Weatherall Institute of Molecular Medicine, Radcliffe Department of Medicine, University of Oxford, John Radcliffe Hospital, Oxford OX3 9DS, UK.
Data availability
ChIP-sequencing data generated for this study have been deposited in the Gene Expression Omnibus (GEO) under accession code GSE240678. Previously published ChIP-seq, DNaseI-seq and ATAC-seq data reanalysed here are available under the following accession codes: GSE97871, GSE30203, GSE90994.
All other data supporting the findings of this study are available from the corresponding author on reasonable request.
Supplementary data
Supplementary Data are available at NAR Online.
Funding
Chinese Academy of Medical Sciences (CAMS) Innovation Fund for Medical Science (CIFMS), China [2018-I2M-2-002]. Medical Research Council (MRC) Programme grant [MR/T014067/1]. Funding for open access charge: University of Oxford.
Conflict of interest statement. None declared.
References
- 1. Oudelaar A.M., Higgs D.R.. The relationship between genome structure and function. Nat. Rev. Genet. 2021; 22:154–168. [DOI] [PubMed] [Google Scholar]
- 2. Andersson R., Sandelin A.. Determinants of enhancer and promoter activities of regulatory elements. Nat. Rev. Genet. 2020; 21:71–87. [DOI] [PubMed] [Google Scholar]
- 3. Mikhaylichenko O., Bondarenko V., Harnett D., Schor I.E., Males M., Viales R.R., Furlong E.E.M.. The degree of enhancer or promoter activity is reflected by the levels and directionality of eRNA transcription. Genes Dev. 2018; 32:42–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nguyen T.A., Jones R.D., Snavely A.R., Pfenning A.R., Kirchner R., Hemberg M., Gray J.M.. High-throughput functional comparison of promoter and enhancer activities. Genome Res. 2016; 26:1023–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bell A.C., West A.G., Felsenfeld G.. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell. 1999; 98:387–396. [DOI] [PubMed] [Google Scholar]
- 6. Bell A.C., Felsenfeld G.. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature. 2000; 405:482–485. [DOI] [PubMed] [Google Scholar]
- 7. Valadez-Graham V., Razin S.V., Recillas-Targa F.. CTCF-dependent enhancer blockers at the upstream region of the chicken alpha-globin gene domain. Nucleic Acids Res. 2004; 32:1354–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kubo N., Ishii H., Xiong X., Bianco S., Meitinger F., Hu R., Hocker J.D., Conte M., Gorkin D., Yu M.et al.. Promoter-proximal CTCF binding promotes distal enhancer-dependent gene activation. Nat. Struct. Mol. Biol. 2021; 28:152–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Monahan K., Rudnick N.D., Kehayova P.D., Pauli F., Newberry K.M., Myers R.M., Maniatis T.. Role of CCCTC binding factor (CTCF) and cohesin in the generation of single-cell diversity of protocadherin-alpha gene expression. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:9125–9130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zhang X., Zhang Y., Ba Z., Kyritsis N., Casellas R., Alt F.W.. Fundamental roles of chromatin loop extrusion in antibody class switching. Nature. 2019; 575:385–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lee J., Krivega I., Dale R.K., Dean A.. The LDB1 Complex Co-opts CTCF for Erythroid Lineage-Specific Long-Range Enhancer Interactions. Cell Rep. 2017; 19:2490–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Narendra V., Rocha P.P., An D., Raviram R., Skok J.A., Mazzoni E.O., Reinberg D.. CTCF establishes discrete functional chromatin domains at the Hox clusters during differentiation. Science. 2015; 347:1017–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Splinter E., Heath H., Kooren J., Palstra R.J., Klous P., Grosveld F., Galjart N., de Laat W.. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 2006; 20:2349–2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cuddapah S., Jothi R., Schones D.E., Roh T.Y., Cui K., Zhao K.. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 2009; 19:24–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Tang Z., Luo O.J., Li X., Zheng M., Zhu J.J., Szalaj P., Trzaskoma P., Magalska A., Wlodarczyk J., Ruszczycki B.et al.. CTCF-mediated human 3D genome architecture reveals chromatin topology for transcription. Cell. 2015; 163:1611–1627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Narendra V., Bulajic M., Dekker J., Mazzoni E.O., Reinberg D.. CTCF-mediated topological boundaries during development foster appropriate gene regulation. Genes Dev. 2016; 30:2657–2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lupianez D.G., Kraft K., Heinrich V., Krawitz P., Brancati F., Klopocki E., Horn D., Kayserili H., Opitz J.M., Laxova R.et al.. Disruptions of topological chromatin domains cause pathogenic rewiring of gene-enhancer interactions. Cell. 2015; 161:1012–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim T.H., Abdullaev Z.K., Smith A.D., Ching K.A., Loukinov D.I., Green R.D., Zhang M.Q., Lobanenkov V.V., Ren B.. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007; 128:1231–1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang H., Maurano M.T., Qu H., Varley K.E., Gertz J., Pauli F., Lee K., Canfield T., Weaver M., Sandstrom R.et al.. Widespread plasticity in CTCF occupancy linked to DNA methylation. Genome Res. 2012; 22:1680–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Fang C., Wang Z., Han C., Safgren S.L., Helmin K.A., Adelman E.R., Serafin V., Basso G., Eagen K.P., Gaspar-Maia A.et al.. Cancer-specific CTCF binding facilitates oncogenic transcriptional dysregulation. Genome Biol. 2020; 21:247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Schmidt D., Schwalie P.C., Wilson M.D., Ballester B., Goncalves A., Kutter C., Brown G.D., Marshall A., Flicek P., Odom D.T.. Waves of retrotransposon expansion remodel genome organization and CTCF binding in multiple mammalian lineages. Cell. 2012; 148:335–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nakahashi H., Kieffer Kwon K.R., Resch W., Vian L., Dose M., Stavreva D., Hakim O., Pruett N., Nelson S., Yamane A.et al.. A genome-wide map of CTCF multivalency redefines the CTCF code. Cell Rep. 2013; 3:1678–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wendt K.S., Yoshida K., Itoh T., Bando M., Koch B., Schirghuber E., Tsutsumi S., Nagae G., Ishihara K., Mishiro T.et al.. Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature. 2008; 451:796–801. [DOI] [PubMed] [Google Scholar]
- 24. Khoury A., Achinger-Kawecka J., Bert S.A., Smith G.C., French H.J., Luu P.L., Peters T.J., Du Q., Parry A.J., Valdes-Mora F.et al.. Constitutively bound CTCF sites maintain 3D chromatin architecture and long-range epigenetically regulated domains. Nat. Commun. 2020; 11:54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Luan J., Xiang G., Gomez-Garcia P.A., Tome J.M., Zhang Z., Vermunt M.W., Zhang H., Huang A., Keller C.A., Giardine B.M.et al.. Distinct properties and functions of CTCF revealed by a rapidly inducible degron system. Cell Rep. 2021; 34:108783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kreibich E., Kleinendorst R., Barzaghi G., Kaspar S., Krebs A.R.. Single-molecule footprinting identifies context-dependent regulation of enhancers by DNA methylation. Mol. Cell. 2023; 83:787–802. [DOI] [PubMed] [Google Scholar]
- 27. Fu Y., Sinha M., Peterson C.L., Weng Z.. The insulator binding protein CTCF positions 20 nucleosomes around its binding sites across the human genome. PLoS Genet. 2008; 4:e1000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Barisic D., Stadler M.B., Iurlaro M., Schubeler D.. Mammalian ISWI and SWI/SNF selectively mediate binding of distinct transcription factors. Nature. 2019; 569:136–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Chang L.H., Ghosh S., Papale A., Luppino J.M., Miranda M., Piras V., Degrouard J., Edouard J., Poncelet M., Lecouvreur N.et al.. Multi-feature clustering of CTCF binding creates robustness for loop extrusion blocking and Topologically Associating Domain boundaries. Nat. Commun. 2023; 14:5615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Harrold C.L., Gosden M.E., Hanssen L.L.P., Stolper R.J., Downes D.J., Telenius J.M., Biggs D., Preece C., Alghadban S., Sharpe J.A.et al.. A functional overlap between actively transcribed genes and chromatin boundary elements. 2020; bioRxiv doi:01 July 2020, preprint: not peer reviewed 10.1101/2020.07.01.182089. [DOI]
- 31. Hanssen L.L.P., Kassouf M.T., Oudelaar A.M., Biggs D., Preece C., Downes D.J., Gosden M., Sharpe J.A., Sloane-Stanley J.A., Hughes J.R.et al.. Tissue-specific CTCF-cohesin-mediated chromatin architecture delimits enhancer interactions and function in vivo. Nat. Cell Biol. 2017; 19:952–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Stolper R.J., Tsang F.H., Georgiades E., Hanssen L.L., Downes D.J., Harrold C.L., Hughes J.R., Beagrie R.A., Davies B., Kassouf M.et al.. Loop extrusion by cohesin plays a key role in enhancer-activated gene expression during differentiation. 2023; bioRxiv doi:07 September 2023, preprint: not peer reviewed 10.1101/2023.09.07.556660. [DOI]
- 33. Rao S.S., Huntley M.H., Durand N.C., Stamenova E.K., Bochkov I.D., Robinson J.T., Sanborn A.L., Machol I., Omer A.D., Lander E.S.et al.. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014; 159:1665–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. de Wit E., Vos E.S., Holwerda S.J., Valdes-Quezada C., Verstegen M.J., Teunissen H., Splinter E., Wijchers P.J., Krijger P.H., de Laat W.. CTCF binding polarity determines chromatin looping. Mol. Cell. 2015; 60:676–684. [DOI] [PubMed] [Google Scholar]
- 35. Chakraborty S., Kopitchinski N., Zuo Z., Eraso A., Awasthi P., Chari R., Mitra A., Tobias I.C., Moorthy S.D., Dale R.K.et al.. Enhancer-promoter interactions can bypass CTCF-mediated boundaries and contribute to phenotypic robustness. Nat. Genet. 2023; 55:280–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Huang H., Zhu Q., Jussila A., Han Y., Bintu B., Kern C., Conte M., Zhang Y., Bianco S., Chiariello A.M.et al.. CTCF mediates dosage- and sequence-context-dependent transcriptional insulation by forming local chromatin domains. Nat. Genet. 2021; 53:1064–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Francis H.S., Harold C.L., Beagrie R.A., King A.J., Gosden M.E., Blayney J.W., Jeziorska D.M., Babbs C., Higgs D.R., Kassouf M.T.. Scalable in vitro production of defined mouse erythroblasts. PLoS One. 2022; 17:e0261950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Grant C.E., Bailey T.L., Noble W.S.. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011; 27:1017–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kulakovskiy I.V., Vorontsov I.E., Yevshin I.S., Sharipov R.N., Fedorova A.D., Rumynskiy E.I., Medvedeva Y.A., Magana-Mora A., Bajic V.B., Papatsenko D.A.et al.. HOCOMOCO: towards a complete collection of transcription factor binding models for human and mouse via large-scale ChIP-Seq analysis. Nucleic Acids Res. 2018; 46:D252–D259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Siepel A., Bejerano G., Pedersen J.S., Hinrichs A.S., Hou M., Rosenbloom K., Clawson H., Spieth J., Hillier L.W., Richards S.et al.. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 2005; 15:1034–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hentges L.D., Sergeant M.J., Cole C.B., Downes D.J., Hughes J.R., Taylor S.. LanceOtron: a deep learning peak caller for genome sequencing experiments. Bioinformatics. 2022; 38:4255–4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hay D., Hughes J.R., Babbs C., Davies J.O.J., Graham B.J., Hanssen L., Kassouf M.T., Marieke Oudelaar A.M., Sharpe J.A., Suciu M.C.et al.. Genetic dissection of the alpha-globin super-enhancer in vivo. Nat. Genet. 2016; 48:895–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Oudelaar A.M., Beagrie R.A., Gosden M., de Ornellas S., Georgiades E., Kerry J., Hidalgo D., Carrelha J., Shivalingam A., El-Sagheer A.H.et al.. Dynamics of the 4D genome during in vivo lineage specification and differentiation. Nat. Commun. 2020; 11:2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hark A.T., Schoenherr C.J., Katz D.J., Ingram R.S., Levorse J.M., Tilghman S.M.. CTCF mediates methylation-sensitive enhancer-blocking activity at the H19/Igf2 locus. Nature. 2000; 405:486–489. [DOI] [PubMed] [Google Scholar]
- 45. Despang A., Schopflin R., Franke M., Ali S., Jerkovic I., Paliou C., Chan W.L., Timmermann B., Wittler L., Vingron M.et al.. Functional dissection of the Sox9-Kcnj2 locus identifies nonessential and instructive roles of TAD architecture. Nat. Genet. 2019; 51:1263–1271. [DOI] [PubMed] [Google Scholar]
- 46. Anania C., Acemel R.D., Jedamzick J., Bolondi A., Cova G., Brieske N., Kuhn R., Wittler L., Real F.M., Lupianez D.G.. In vivo dissection of a clustered-CTCF domain boundary reveals developmental principles of regulatory insulation. Nat. Genet. 2022; 54:1026–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Liu N.Q., Maresca M., van den Brand T., Braccioli L., Schijns M., Teunissen H., Bruneau B.G., Nora E.P., de Wit E.. WAPL maintains a cohesin loading cycle to preserve cell-type-specific distal gene regulation. Nat. Genet. 2021; 53:100–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kagey M.H., Newman J.J., Bilodeau S., Zhan Y., Orlando D.A., van Berkum N.L., Ebmeier C.C., Goossens J., Rahl P.B., Levine S.S.et al.. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010; 467:430–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Boyle A.P., Song L., Lee B.K., London D., Keefe D., Birney E., Iyer V.R., Crawford G.E., Furey T.S.. High-resolution genome-wide in vivo footprinting of diverse transcription factors in human cells. Genome Res. 2011; 21:456–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Yin M., Wang J., Wang M., Li X., Zhang M., Wu Q., Wang Y.. Molecular mechanism of directional CTCF recognition of a diverse range of genomic sites. Cell Res. 2017; 27:1365–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li Y., Haarhuis J.H.I., Sedeno Cacciatore A., Oldenkamp R., van Ruiten M.S., Willems L., Teunissen H., Muir K.W., de Wit E., Rowland B.D.et al.. The structural basis for cohesin-CTCF-anchored loops. Nature. 2020; 578:472–476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pugacheva E.M., Kubo N., Loukinov D., Tajmul M., Kang S., Kovalchuk A.L., Strunnikov A.V., Zentner G.E., Ren B., Lobanenkov V.V.. CTCF mediates chromatin looping via N-terminal domain-dependent cohesin retention. Proc. Natl. Acad. Sci. U.S.A. 2020; 117:2020–2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
ChIP-sequencing data generated for this study have been deposited in the Gene Expression Omnibus (GEO) under accession code GSE240678. Previously published ChIP-seq, DNaseI-seq and ATAC-seq data reanalysed here are available under the following accession codes: GSE97871, GSE30203, GSE90994.
All other data supporting the findings of this study are available from the corresponding author on reasonable request.