Abstract
Infection with high-risk human papillomaviruses (HPV) is a major risk factor for development of cervical cancer. Expression of the HPV E6 and E7 oncoproteins increases in differentiating keratinocytes, resulting in inactivation of the p53 and retinoblastoma proteins, two important transcriptional regulators. We used cDNA microarrays to examine global alterations in gene expression in differentiating cervical keratinocytes after infection with retroviruses encoding HPV type 16 (HPV-16) E6 and E7. Expression of 80 cellular genes (approximately 4% of the genes on the array) was altered reproducibly by E6 and/or E7. Cluster analysis classified these genes into three functional groups: (i) interferon (IFN)-responsive genes, (ii) genes stimulated by NF-κB, and (iii) genes regulated in cell cycle progression and DNA synthesis. HPV-16 E6 or a dominant negative p53 protein downregulated multiple IFN-responsive genes. E6 decreased expression of IFN-α and -β, downregulated nuclear STAT-1 protein, and decreased binding of STAT-1 to the IFN-stimulated response element. E7 alone was less effective; however, coexpression of E6 and E7 downregulated IFN-responsive genes more efficiently than E6. The HPV-16 E6 protein also stimulated expression of multiple genes known to be inducible by NF-κB and AP-1. E6 enhanced expression of functional components of the NF-κB signal pathway, including p50, NIK, and TRAF-interacting protein, and increased binding to NF-κB and AP-1 DNA consensus binding sites. Secretion of interleukin-8, RANTES, macrophage inflammatory protein 1α, and 10-κDa IFN-γ-inducible protein were increased in differentiating keratinocytes by E6. Thus, high-level expression of the HPV-16 E6 protein in differentiating keratinocytes directly alters expression of genes that influence host resistance to infection and immune function.
Human papillomaviruses (HPVs) are small DNA tumor viruses that replicate in differentiating keratinocytes of the epidermis and anogenital tract (59). The E6 and E7 viral genes are expressed at low levels in proliferating basal cells, but transcription is activated as cells enter the terminal differentiation pathway (11, 16). E6 and E7 delay keratinocyte differentiation, reactivate host DNA synthesis, and stimulate cell cycle progression (60), allowing the virus to utilize host DNA synthetic enzymes to replicate its genome. HPVs induce benign warts and papillomas; however, infection with high-risk types (HPV-16, -18, -31, and -45) is a major risk factor for the development of cervical cancer (reviewed in reference 60). The E6 and E7 genes are particularly important because they are retained and expressed in most cervical carcinomas, and continued expression is required to maintain the malignant phenotype (55).
An early step in HPV-associated carcinogenesis is perturbation of cellular gene expression by the E6 and E7 oncoproteins. E6 binds to a number of cellular proteins and transactivates or represses gene expression (reviewed in reference 48). An important target for E6 is E6-associated protein (E6AP), a protein-ligase of the ubiquitin pathway of proteolysis. E6-E6AP complexes target the tumor suppressor protein p53 for degradation by the proteasome (51). p53 is a central transcription activator that regulates responses to stress and DNA damage. Loss of p53 leads to genetic instability and rapid malignant progression. The HPV-16 E7 protein binds to the retinoblastoma protein (pRb) (17) and members of the pRb family (reviewed in reference 31). Interaction occurs primarily with the hypophosphorylated form of pRb causing release of active E2F transcription factors, which in turn stimulate expression of genes involved in cell cycle progression and DNA synthesis (21). The E7 protein also binds to and alters the functions of other proteins, including AP-1 transcription factors (2). E6 and E7 exert overlapping effects on cell cycle control, and in combination, they efficiently immortalize human keratinocytes (39).
Because E6 and E7 interact with numerous cellular transcription regulators, these viral proteins have the potential to significantly modify keratinocyte gene expression. Recently, cDNA microarrays have been successfully used to study global patterns of gene expression in human cancer (1, 13, 14, 25, 47). For this methodology, labeled cDNAs from two samples are hybridized with a microarray containing spots for thousands of genes. Differences in gene expression are measured directly, and groups of differentially expressed genes can be clustered to identify common regulatory pathways. We used cDNA arrays to identify alterations in gene expression in cultures of human cervical keratinocytes infected with HPV-16 E6 and E7 retroviruses. Cervical keratinocytes are the natural target for HPV infection and the progenitors for cervical cancer. We examined how E6 and E7 influenced the pattern of gene expression in proliferating and differentiating cultures, as well as after treatment with tumor necrosis factor alpha (TNF-α), a factor that plays a role in cervical infection and inflammation. Our results show that E6 and E7 alter expression of a large number of cellular genes and that these genes can be grouped into functional categories, suggesting common regulatory pathways.
MATERIALS AND METHODS
Cell culture.
Primary cultures of human ectocervical keratinocytes were established from fresh cervical tissue obtained after hysterectomy due to fibroids or endometriosis (57). We used cell pools as well as cell preparations from single donors. Cells were maintained in serum-free MCDB153-LB medium (K-SFM; Life Technologies, Gaithersburg, Md.). Primary cultures were infected with high-titer retroviruses expressing the neomycin resistance gene only (pLXSN) or the HPV-16 E6, E7, or E6/E7 genes (27). Retroviruses encoding mutant E6 proteins with single amino acid exchanges (F2V, F125V, and L110Q) have been described elsewhere (35, 40). A retrovirus producer line expressing a dominant negative p53 (p53DN) miniprotein was established in the PA317 packaging cell line, using a recombinant construct kindly provided by Moshe Oren (26). After retrovirus infection, cells were always selected in G418 (200μg/ml for 2 to 4 days) and subcultured prior to extraction of RNA. Killing of noninfected cells is observed after 2 days in G418 and is complete by day 4. At this time point, all cells in the mock-infected control dish are dead. Longer exposure to 200 μg of G418/ml shows no detectable cytopathic effects. Western blots show that E7 is actively expressed under these conditions and that p53 is strongly diminished, suggesting that E6 is also expressed (40). For array analysis, all cell cultures, including the immortalized cell lines and those for the experiments using recombinant IFNs, were treated identically. After reaching confluency, keratinocyte cultures were rendered quiescent (growth arrested) by removal of growth factors including epidermal growth factor, bovine pituitary extract, insulin, hydrocortisone, triiodothyronine, and transferrin for 24 h. Quiescent cells were then induced to undergo terminal differentiation by adding 1.4 mM calcium for 24 h. In additional experiments, keratinocytes were treated with 300 to 1,000 U of recombinant alpha interferon (IFN-α) or IFN-β per ml, 500 U of IFN-γ per ml, 1 nM TNF-α, or 1 nM interleukin-1α (IL-1α) (all from R&D Systems, Minneapolis, Minn.). Treatment was for 4 to 6 h. For immortalization, keratinocytes were infected with HPV-16 E6/E7 retroviruses, and cells were seeded at low density (105 cells/150-mm-diameter dish) and passaged several times until growth ceased due to crisis or senescence (usually between 7 and 15 passages). Senescent cultures were maintained in complete medium until colonies of rapidly growing cells appeared as described in reference 40. After colonies reached a diameter of approximately 1 cm, selected colonies were picked and cultured for at least three more passages (eight population doublings) to confirm that cells were immortal.
RNA isolation and purification.
Confluent monolayers of differentiating, growth-arrested cells were washed with prewarmed phosphate-buffered saline (PBS) and lysed with Trizol reagent (Life Technologies), and residual genomic DNA was removed by digestion with RNase-free DNase I. DNase, proteins, and other contaminants were removed by two extractions with buffer-saturated phenol-chloroform, and RNA was precipitated with 1.5 volumes of isopropanol or lithium chloride. The precipitated RNA was centrifuged at 6,000 × g, washed with 70% ethanol, dried by evaporation, and resuspended in diethyl pyrocarbonate-treated water. RNA integrity was confirmed by electrophoresis, and samples were stored at −80°C until used. All RNA combinations used for array analysis were matched. For example, pLXSN controls and HPV-16 E6- and/or E7-infected cells were from the same donor(s) and treated identically (basal medium with 1.4 mM calcium for 24 h). The same applies to immortalized cell clones, which were compared to matching, pLXSN-infected primary cultures from the same donors or cell pools, cultured under identical growth conditions. This parameter is critical considering the high biological variability and age, gender, and racial differences between donors. We used a number of different single donors and, in about 50% of experiments, pools of three to five donors. A small number of arrays are duplicates using the same RNA preparations in independent experiments. These can easily be identified in Fig. 1 by their highly similar expression patterns.
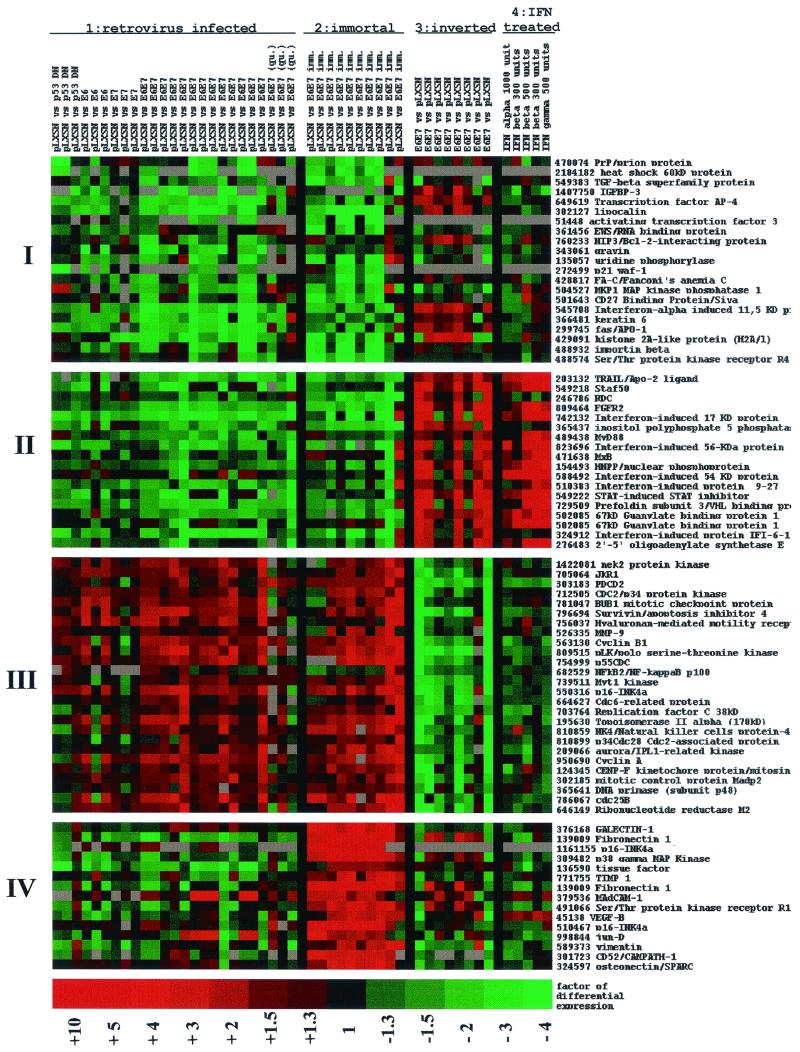
FIG. 1.
Cluster analysis of cellular gene expression. Data from 48 independent hybridization experiments examined multiple experimental conditions and different combinations of HPV-16 E6 and/or E7 genes or p53DN. Data were organized and displayed using cluster analyses (K-Means clustering algorithm). The four treatment groups are indicated at the top. Group 1 included cells expressing p53DN, E6, E7, or E6/E7 and maintained in the absence of growth factors with 1.4 mM Ca2+ to induce differentiation. Three experiments were performed using cells that were made quiescent but that were not induced to differentiate with calcium (qui). Group 2 included HPV-immortalized keratinocytes in Ca2+ to induce differentiation. Group 3 denotes a subset of experiments performed in parallel with group 1 but with inversion of Cy3 and Cy5 dyes as a control for labeling. Group 4 includes HPV-infected, differentiating keratinocytes treated with 300 to 1,000 of U IFN-α, -β, or -γ per ml. In groups 1 to 3, vector-only-infected cells are compared to HPV-infected cells; in group 4, E6/E7-expressing cells with and without IFN are compared. Clusters of differentially expressed mRNAs are shown on the left (I to IV); gene names and IMAGE clone IDs are given at the right. Color coding: green, downregulation of gene expression; red, induction; black, no significant change; grey, no data available.
cDNA synthesis and cDNA array hybridization.
The National Cancer Institute (NCI) oncochip glass arrays were manufactured and printed at the NCI microarray core facility (Advanced Technology Center, Gaithersburg, Md.). The oncochip contains 2,208 named cDNAs that are immobilized on poly-l-lysine-coated glass slides. Thirty to 50 μg of purified total RNA in 20 μl of diethyl pyrocarbonate-treated water was denatured at 70°C for 5 min and primed with 4 μg of oligo(dT)20 primer while cooling to room temperature. Reverse transcription was performed in a volume of 40 μl using 8 μl of first-strand reaction buffer, 4 μl of 0.1 M dithiothreitol, 4 μl of 10 mM deoxynucleoside triphosphate (dNTP), 2 μl of RNasin (Promega, Madison, Wis.), and 4 μl of 1 mM Cy3- or Cy5-dUTP (Amersham, Piscataway, N.J.). cDNA synthesis was completed at 42°C for 1 h. Remaining RNA was hydrolyzed by adding 10 μl of 1 M NaOH and 5 μl of 0.5 M EDTA and incubation for 0.5 1 h at 65°C or by adding DNase-free RNase. The reaction was quenched by adding 25 μl of 1 M Tris HCl (pH 7.4), and the probe was purified using Microcon YM-30 columns. Purified probes were combined and reduced to a volume of 15.6 μl, and 14 μg of cot-1 DNA, 14 μg of poly(A), 5.6 μg of yeast tRNA, and 4.4 μl of 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) were added to a final volume of 24 μl. Before hybridization, the mixture was heated to 100°C for 2 min and cooled to room temperature, and 1 μl of 10% sodium dodecyl sulfate (SDS) was added. Hybridization was performed overnight at 65°C. After hybridization, slides were washed in 2× SSC with 0.1% SDS for 2 min and then sequentially with 1× SSC, 0.2× SSC, and 0.05× SSC (1 min with each). Before drying by centrifugation, slides were briefly immersed in ultrapure water.
Scanning and analysis of cDNA array data.
Hybridized arrays were scanned at 10-μm resolution on a GenePix 4000 scanner (Axon Instruments, Inc., Foster City, Calif.) at variable photomultiplier tube voltage to obtain maximal signal intensities with <1% probe saturation. Resulting images were analyzed with the ArraySuite software (National Human Genome Research Institute, Bethesda, Md.). Differentially expressed genes were defined as reproducible outliers in consecutive arrays that exhibit Cy3/Cy5 ratios significantly different from 1.0 at a 99% confidence level (cutoff ratio in most experiments usually 1.6- to 1.7-fold). Array data (images and sample intensity files) were entered into the NCI microarray database (mAdb), and expression profiles and single data were retrieved from multiple arrays for cluster analysis using a high-stringency data filter (Cy3/Cy5 or Cy5/Cy3 ratios of >3-fold in at least 33.3% or more of all relevant arrays, fluorescence intensity of >500 in each channel unless that of the other channel was >2,000, and a spot size of >50 μm or 50 pixels). Data were processed with the Cluster program using the K-Means clustering algorithm and visualized using the TreeView software (18). Protocols and gene lists for the NCI oncochip can be downloaded from http://nciarray.nci.nih.gov.
RT-PCR.
For cDNA synthesis, 5 to 10 μg of purified total RNA were reverse transcribed for 1 h at 42°C in a volume of 50 μl containing 500 μM each dNTP, 10 μM dithiothreitol, 1.25 μM oligo(dT) primer (T16–18), 20 to 40 U of RNasin (Promega), and 75 U of Superscript Reverse Transcriptase II (Life Technologies) in 1× reverse transcription (RT) buffer. RT reaction mixtures were diluted 2.5- to 10-fold and stored at −70°C. PCRs were performed in 20 μl containing 2 μl of reaction product, 50 μM dNTPs, 0.2 to 0.5 μM each oligonucleotide primer, 1.75 mM magnesium chloride, and 1 U of AmpliTaq Gold (Perkin-Elmer Corp.). Amplification was limited to a total of 20 to 25 cycles to avoid saturation of the reaction. Each reaction was performed at least twice in independent experiments to confirm reproducibility. Oligonucleotide primers were designed using the Genetics Computer Group (Madison, Wis.) program package, available at http://bimas.dcrt.nih.gov.
ELISA analysis of cell-associated and secreted cytokines.
To measure cell-associated cytokines, cells were lysed in buffer containing 150 mM NaCl, 50 mM Tris-HCl (pH 7.5), 1.5 mM MgCl2, 1% glycerol, 1% Triton X-100, 5 mM EGTA, and protease inhibitors 4-(2-aminoethyl) benzenesulfonyl fluoride (AEBSF), leupeptin, and aprotinin (each at 100 μg/ml). Protein samples and standards were diluted with lysis buffer to a concentration of 0.25 to 0.5 μg of protein/μl. Total protein concentration was determined by the bicinchoninic acid protein assay (Pierce, Rockford, III.), and protein concentration was used for normalization of enzyme-linked immunosorbent assay (ELISA) data. Roughly, 1 mg of total protein equaled approximately 106 cells. To measure secreted cytokines, cell conditioned medium (5 ml/10-cm-diameter dish) was supplemented with 10 mM EDTA and the protease inhibitors AEBSF (1 mM) and aprotinin, pepstatin A, and leupeptin (each at 10 μg/ml) and then centrifuged for 5 min at 5,000 × g to remove cell debris. After sampling media, cells were trypsinized and counted in a Coulter Counter for normalization of ELISA data. Samples and standards were added to ELISA plates in duplicate or triplicate (200 μl/well) and incubated overnight at 4°C with constant agitation. Plates were washed four times, and the protocol was continued according to the manufacturer's recommendations. Levels of macrophage inflammatory protein 1α (MIP-1α), RANTES, vascular endothelial growth factor (VEGF), IL-8, granulocyte-macrophage colony-stimulating factor, TNF-α, and IL-6 were measured using Quantikine ELISA kits from R&D Systems. For the 10-kDa IFN-γ-induced protein (IP-10), matched antibody pairs and recombinant IP-10 were purchased from R&D Systems. IFN-α and IFN-β ELISA kits were purchased from Research Diagnostics.
Preparation of cytoplasmic and nuclear protein extracts.
Cells were washed with PBS, scraped from the plate, and snap-frozen in liquid nitrogen. For cytoplasmic extracts, frozen pellets were thawed in 4 volumes of buffer (10 mM HEPES [pH 7.9], 10 mM NaCl, 0.1 mM EDTA, 1 mM dithiothreitol, 5% [vol/vol] glycerol, 500 mM phenylmethylsulfonyl fluoride, 50 mM NaF, 100 mM sodium vanadate, 100 μg of leupeptin/ml, 4 μg of aprotinin/ml, 2 μg of pepstatin/ml, 2 μg of antipain/ml). After repeated pipetting, the lysate was centrifuged at 10,000 rpm for 20 min, and the supernatant (cytoplasmic fraction) was frozen at −70°C until use. For nuclear extracts, the pellet was resuspended in 2 volumes of the above buffer with 0.4 M NaCl and centrifuged again, and the supernatant (nuclear fraction) was collected and frozen at −70°C.
EMSA.
The Promega gel shift assay system containing oligonucleotides for SP1, AP-1, AP-2, CREB, NF-κB, Oct-1, and TFIID transcription factors was used for electrophoretic mobility shift assay (EMSA). Additional oligonucleotides (STAT-1, insulin response factor 1 [IRF-1], and SIE) for band shift experiments were purchased from Santa Cruz Biotechnology (San Diego, Calif.). Oligonucleotides were labeled using Escherichia coli polynucleotide kinase and [γ-32P]dATP (>3,000 Ci/mmol; Amersham) and purified by gel filtration; 10,000 cpm was used per reaction. Reactions were performed according to manufacturer's protocol, using 5 μg of nuclear protein/20 μl of reaction mixture. Supershift experiments were performed using p50, p52, and p65 NF-κB and p84/p91 STAT-1α antibodies (Santa Cruz Biotechnology). Reaction products were fractionated on polyacrylamide gels at room temperature; gels were dried on Whatman 3MM paper and analyzed by autoradiography.
Western blotting.
Cells were lysed in lysis buffer (10 mM Tris HCl, 150 mM NaCl, 1% deoxycholate, 1% Nonidet P-40, 0.1% SDS, 5 mM EDTA, 1% Triton X-100, 1 μg of leupeptin/ml, 1 mM phenylmethylsulfonyl fluoride, 0.5 mM orthovanadate) on ice for 5 min. Lysates were centrifuged at 15,000 rpm for 15 min to remove insoluble components. Equal amounts of protein lysates were separated by SDS-polyacrylamide gel electrophoresis on 10% or 4 to 15% gradient gels (Bio-Rad) and transferred to Immobilon-P polyvinylidene difluoride membranes (Millipore, Bedford, Mass.). The membranes were blocked with 5% dry milk in wash buffer (PBS, 0.05% Tween 20) for 1 h. Primary antibodies were incubated in PBS–1% bovine serum albumin–0.05% thimerosal or 5% dry milk overnight at 4°C, washed for 1 h with several changes of buffer, and incubated with anti-rabbit or anti-mouse secondary antibodies conjugated with horseradish peroxidase (Sigma Chemical Co.) for another hour. After a second wash as described above, the signal was visualized by enhanced chemiluminescence (ECL Plus; Amersham). Antibodies against human STAT-1 p84/91, phosphorylated STAT-1, RelA p65, NF-κB-1 (p50) and -2 (p52), and IRF-1 were purchased from Santa Cruz Biotechnology.
RNase protection assay.
RNase protection assays were performed using RiboQuant Multi-Probe RNase Protection Assay Systems hCK3 and hCK, an in vitro transcription kit, and an RPA kit (all Pharmingen, San Diego, Calif.). Reactions were performed following the standard protocol provided by the manufacturer. Purified, DNase-treated total RNA (10 or 20 μg per reaction) was incubated with in vitro-transcribed radioactive probe for 16 h at 56°C and then 15 min 37°C, followed by RNase treatment (RNase A-RNase T1 mix for 45 min at 30°C). Residual RNase was digested by proteinase K (15 min at 37°C), and protected fragments were precipitated with 75% ethanol–4 M ammonium acetate for 16 h at −70°C. RNA pellets were dissolved in 8 μl of loading buffer, and 4 μl was loaded on 6% polyacrylamide–50% urea gels. Dried gels were analyzed and signals were quantified using a STORM PhosphorImager (Molecular Dynamics, Sunnyvale, Calif.).
RESULTS
Microarray analysis of keratinocytes expressing HPV-16 E6, E7, or E6/E7.
We used the first version of the NCI human oncochip with 2,208 cDNAs to search for genes whose expression is reproducibly altered by E6 and/or E7. Cultures of human cervical keratinocytes were infected with retroviruses encoding HPV-16 E6, E7, E6/E7, or the empty vector (pLXSN) as a control and selected with G418 for stably expressing cells, and RNA was purified for array hybridization. In these experiments, we compared gene expression in vector-infected, differentiating cells to that in cells with one or both HPV oncoproteins. Cells were analyzed under different conditions by using medium that promoted either cell growth (data not included), quiescence (growth factor removal), or squamous differentiation (1.4 mM Ca2+). We also examined E6E7-expressing, differentiating cervical keratinocytes that were treated with IFN-α, -β, or -γ(300 to 1,000 U/ml for 6 h), as well as several independent, E6/E7-immortalized cervical cell lines. E6 and E7 stimulated very few changes in gene expression in rapidly growing cells; therefore, these experiments were excluded from further analyses. Similarly, infection with E6 and E7 oncogenes of HPV-6b, a low-risk HPV type associated with benign genital warts, resulted in few changes in cellular gene expression in cervical keratinocytes and were also excluded from this study. In contrast, HPV-16 E6 and E7 reproducibly altered expression of 80 to 90 genes in quiescent and/or differentiating cultures (approximately 4% of all genes on the array). HPV-16 E6/E7-immortalized cells also exhibited numerous, usually more pronounced alterations relative to normal keratinocytes. Gene expression patterns varied slightly in cultures isolated from different patients, and these differences probably represent true biological variations between donors since they were reproducible in multiple assays. For this reason, a large number of array experiments (total of 48 selected for this study) had to be performed and analyzed.
Cluster analysis of expression data.
Figure 1 represents an analysis of combined data from 48 independent hybridization experiments. Genes were considered to be differentially expressed if they were upregulated or downregulated more than threefold in at least one-third of all comparable experiments. Expression data were sorted using the K-Means clustering algorithm and displayed using the TreeView program (18). As expected, clustering reorganized all array experiments into treatment groups based on similarity in patterns of gene expression. These included (i) differentiating keratinocytes infected with HPV-16 E6, E7, E6/E7, or p53DN, (ii) E6/E7-immortalized cell lines, (iii) experiments in which the Cy3-Cy5 cDNA labeling was inverted as an internal control for cDNA labeling, and (iv) keratinocytes expressing E6/E7 and treated with 300 to 1,000 U of recombinant IFN-α, -β, or -γ.
Most importantly, the K-Means algorithm grouped the 80 differentially expressed genes into four distinct gene clusters (I to IV) based on similar patterns of expression (Fig. 1). Figure 1 also shows gene names (denoted by product encoded) and IMAGE clone identifier (ID) numbers; additional data (UniGene ID numbers, gene symbols, and average factors of differential mRNA expression) are included in Table 1. Several genes were represented more than once on the array and are usually located in the same cluster (e.g., p16 INK4a and p67 guanylate binding protein). These spots are composed of different portions of the same gene and serve as internal controls for the quality of hybridization and clustering. Clusters I and III contain mostly genes that are involved in cell cycle progression and DNA synthesis. However, they also contain genes whose differential expression indirectly correlates with changes in cell proliferation but have no functional role in cell cycle regulation (keratin 6, prion protein, heat shock protein 60, insulin-like growth factor binding protein 3 (IGFBP-3), gravin, etc.). Additionally, cluster III also contains a subset of genes inducible by the proinflammatory cytokine TNF-α and the transcription factor NF-κB (Table 2). Cluster II represents IFN-inducible genes. Last but not least, cluster IV shows genes that are over expressed exclusively in immortal cell lines in comparison to primary keratinocytes. Each cluster is discussed in detail below.
TABLE 1.
| Gene | Symbol | UniGene ID | Retrovirus-infected E6E7/vector | Immortalized/vector | Retrovirus-infected vector/E6E7 | IFN treatedb |
|---|---|---|---|---|---|---|
| Cluster I | ||||||
| Prion protein | PRNP | Hs.74621 | 0.57 | 0.42 | 1.31 | 0.88 |
| Heat shock protein 60 | HSPD1 | Hs.79037 | 0.27 | 0.30 | ||
| TGF-β | PLAB | Hs.116577 | 0.56 | 0.39 | 1.65 | 1.15 |
| IGFBP-3 | IGFBP3 | Hs.77326 | 0.21 | 0.46 | 1.91 | |
| Transcription factor AP-4 | TFAP4 | Hs.3005 | 0.24 | 0.59 | 2.20 | 0.85 |
| Lipocalin | LCN2 | Hs.204238 | 0.24 | 0.09 | 1.72 | 0.88 |
| Activating transcription factor 3 | ATF3 | Hs.460 | 0.41 | 0.24 | ||
| EWS/RNA binding protein | DDIT3 | Hs.279946 | 0.99 | 0.41 | 0.84 | |
| NIP3/Bcl-2-interacting protein | BNIP3 | Hs.79428 | 0.49 | 1.07 | 1.69 | 1.06 |
| Gravin | AKAP12 | Hs.788 | 0.27 | 0.85 | 0.84 | |
| Uridine phosphorylase | UP | Hs.77573 | 0.48 | 0.48 | 1.63 | 0.80 |
| p21 Waf-1 | CDKNIA | Hs.179665 | 0.47 | 0.47 | ||
| Fanconi's anemia C | FANCC | Hs.37953 | 0.53 | 0.39 | 1.09 | 0.91 |
| MKP-1 | DUSP1 | Hs.171695 | 0.45 | 0.45 | 1.14 | |
| CD27 binding protein/Siva | SIVA | Hs.112058 | 0.62 | 0.34 | 0.92 | 1.10 |
| α-IFN-induced 11.5-kDa protein | IFI27 | Hs.278613 | 0.20 | 0.45 | 2.98 | 0.87 |
| Keratin 6 | KRT6A | Hs.111758 | 0.26 | 0.56 | 1.28 | 0.95 |
| Fas/APO-1 | TNFRSF6 | Hs.82359 | 0.24 | 0.59 | 2.20 | 0.85 |
| Histone 2A-like protein | H2AFL | Hs.28777 | 0.28 | 0.23 | 2.01 | 1.07 |
| Importin beta | KPNB1 | Hs.180446 | 0.47 | 0.24 | 1.11 | 0.98 |
| Ser/Thr protein kinase receptor | Unknown | 0.73 | 0.21 | 0.86 | ||
| Cluster II | ||||||
| TRAIL/Apo-2 ligand | TNFSF10 | Hs.83429 | 0.33 | 0.43 | 2.59 | 11.01 |
| Staf50 | STAF50 | Hs.68054 | 0.45 | 0.65 | 2.66 | 2.69 |
| RDC1 | RDC1 | Hs.23016 | 0.66 | 0.26 | 2.06 | 0.84 |
| Fibroblast growth factor receptor 2 | FGFR2 | Hs.278581 | 0.35 | 0.35 | 3.39 | 3.13 |
| IFN-induced 17-kDa protein | ISG15 | Hs.833 | 0.18 | 0.32 | 3.83 | 6.27 |
| Inositol phosphatase IPP110 | INPP5D | Hs.155939 | 0.45 | 0.28 | 2.84 | 0.96 |
| MyD88 | MYD88 | Hs.82116 | 0.45 | 0.83 | 1.73 | 2.53 |
| IFN-induced 56-kDa protein | IFIT1 | Hs.20315 | 0.37 | 0.32 | 3.45 | 15.85 |
| MxB | MX2 | Hs.926 | 0.76 | 1.18 | 2.01 | 2.68 |
| HNPP/nuclear phosphoprotein | IFI75 | Hs.38125 | 0.57 | 0.53 | 3.24 | 2.86 |
| IFN-induced 54-kDa protein | IFI54 | Hs.169274 | 0.43 | 0.85 | 3.06 | 48.43 |
| IFN-induced protein 9-27 | IFITM1 | Hs.146360 | 0.43 | 0.81 | 1.99 | 1.81 |
| STAT-induced STAT inhibitor | SOCS1 | 0.45 | 0.65 | 2.66 | 2.69 | |
| Prefoldin 3/VHL binding protein 1 | VBP1 | Hs.198307 | 0.35 | 0.56 | 3.20 | 2.39 |
| 67-kDa guanylate binding protein 1 | GBP1 | Hs.62661 | 0.45 | 0.40 | 2.96 | 6.60 |
| IFN-induced protein IFI-6-16 | GIP3 | Hs.265827 | 0.24 | 0.36 | 2.36 | 1.01 |
| 2′-5′ Oligoadenylate synthetase E | OAS1 | Hs.82396 | 0.45 | 0.70 | 2.81 | 5.56 |
| Cluster III | ||||||
| Nek2 protein kinase | NEK2 | Hs.153704 | 2.32 | 1.99 | 0.52 | 0.96 |
| JkR1 | TACC3 | Hs.104019 | 2.04 | 1.99 | 0.39 | 1.03 |
| PDCD2 | PDCD2 | Hs.41639 | 1.53 | 2.41 | 0.29 | 0.52 |
| cdc2 p34 protein kinase | CDC2 | Hs.184572 | 1.73 | 2.24 | 0.48 | 0.98 |
| BUB1 mitotic checkpoint protein | BUB1 | Hs.98658 | 2.26 | 1.97 | 0.60 | 1.06 |
| Survivin/apoptosis inhibitor 4 | BIRC5 | Hs.1578 | 3.22 | 2.54 | 0.19 | 0.67 |
| Hyaluronan-receptor RHAMM | HMMR | Hs.72550 | 2.53 | 1.61 | 0.57 | 1.03 |
| Matrix metalloproteinase 9 | MMP9 | Hs.151738 | 1.41 | 1.89 | 0.37 | 1.08 |
| Cyclin B1 | CCNB1 | Hs.23960 | 2.51 | 2.96 | 0.36 | 0.63 |
| Polo-like kinase | PLK | Hs.77597 | 3.02 | 3.42 | 0.14 | 0.66 |
| p55CDC | CDC20 | Hs.82906 | 2.78 | 5.91 | 0.34 | 0.81 |
| NFκB2/NF-κB p100 | NFKB2 | Hs.73090 | 1.14 | 1.01 | 0.41 | 0.90 |
| Myt1 kinase | PKMYT1 | Hs.77783 | 1.55 | 1.90 | 0.32 | 0.83 |
| p16-INK4a clone IMAGE 550316 | CDKN2A | Hs.1174 | 2.91 | 3.38 | 0.33 | 0.77 |
| cdc6-related protein | CDC6 | Hs.69563 | 1.79 | 1.48 | 0.30 | 0.75 |
| Replication factor C 38 kDa | RFC3 | Hs.115474 | 1.42 | 1.82 | 0.45 | 0.56 |
| Topoisomerase IIα (170 kDa) | TOP2A | Hs.156346 | 2.83 | 3.58 | 0.19 | 0.75 |
| Natural killer cells protein 4 | NK4 | Hs.943 | 1.86 | 3.40 | 0.44 | 1.14 |
| cdc2-associated protein | CKS1 | Hs.77550 | 1.30 | 1.79 | 0.42 | 0.79 |
| aurora/IPL1-related kinase | STK15 | Hs.48915 | 1.90 | 2.83 | 0.38 | 0.60 |
| Cyclin A | CCNA2 | Hs.85137 | 2.67 | 2.89 | 0.38 | 0.78 |
| CENP-F kinetochore protein/mitosin | CENPF | Hs.77204 | 1.82 | 2.08 | 0.33 | 0.87 |
| Madp2 | MAD2L1 | Hs.79078 | 2.12 | 1.94 | 0.35 | 0.83 |
| DNA primase (subunit p48) | PRIM1 | Hs.82741 | 1.59 | 1.67 | 0.52 | 0.93 |
| cdc25B | CDC25B | Hs.153752 | 2.56 | 2.24 | 0.40 | 0.73 |
| Ribonucleotide reductase M2 | RRM2 | Hs.75319 | 2.03 | 2.92 | 0.29 | 0.79 |
| Cluster IV | ||||||
| Galectin-1 | LGALS1 | Hs.227751 | 1.38 | 5.85 | 0.71 | 0.88 |
| Fibronectin 1 | FN1 | Hs.287820 | 0.42 | 17.82 | 1.55 | 0.62 |
| p16-INK4a (clone IMAGE1161155) | CDKN2A | Hs.1174 | 1.04 | 10.37 | ||
| p38 gamma MAPK | MAPK12 | Hs.55039 | 0.63 | 8.90 | 1.02 | 0.76 |
| Tissue factor | F3 | Hs.62192 | 0.27 | 1.80 | 1.80 | 0.66 |
| TIMP 1 | TIMP1 | Hs.5831 | 0.70 | 3.22 | 0.67 | 1.01 |
| MAdCAM-1 mucosal addressin | MADCAM1 | Hs.102598 | 2.26 | 2.54 | 1.32 | 1.55 |
| Protein kinase receptor 1 | ACVR1 | Hs.150402 | 0.64 | 5.94 | 1.29 | 0.65 |
| VEGF-B | VEGFC | Hs.79141 | 0.84 | 2.87 | 0.72 | 2.31 |
| p16-INK4a (clone IMAGE 510467) | CDKN2A | Hs.1174 | 1.31 | 13.40 | 1.00 | 1.00 |
| JunD | JUND | Hs.2780 | 1.54 | 2.52 | 0.94 | 0.70 |
| Vimentin | VIM | Hs.2064 | 0.84 | 2.77 | 0.57 | 0.73 |
| CD52/CAMPATH-1 | CDW52 | Hs.276770 | 0.85 | 5.16 | 0.68 | 0.79 |
| Osteonectin/SPARC | SPARC | Hs.111779 | 1.15 | 3.66 | 0.61 | 0.87 |
Ratios represent average mRNA expression data from keratinocytes expressing HPV-16 E6/E7 oncogenes (13 experiments) or immortalized cells (10 experiments) compared to vector control (pLXSN), or vice versa (8 experiments). Values in bold indicate upregulated genes; those in italics indicate repressed genes (factors between >1.7 or <0.6). Gene names, symbols, and IDs are in accordance with the UniGene database.
Data for cells treated with IFN-α and -β (four experiments).
TABLE 2.
Average expression data for a selection of genes in clusters I and III
| Gene | Differentiationa
|
TNF-αb
|
||
|---|---|---|---|---|
| Avg | SD | Avg | SD | |
| Cluster I | ||||
| Prion protein | 2.83 | 0.96 | NC | |
| MKP-1 | 3.03 | 1.20 | NC | |
| Fanconi's anemia C | NC | 2.12 | 0.95 | |
| Keratin 6 | 2.74 | 1.53 | NC | |
| SOCS-5 | 3.27 | 0.72 | NC | |
| Histone 2A-like protein | 5.10 | 1.64 | NC | |
| Tissue factor | 3.90 | 2.34 | NC | |
| Fas/APO-1, CD95 | NC | 2.30 | 0.12 | |
| Gravin | 17.78 | 1.53 | NC | |
| Uridine phosphorylase | 3.68 | 1.13 | NC | |
| TGF-β superfamily protein PLAB | 7.33 | 0.93 | NC | |
| Lipocalin | 6.54 | 3.85 | NC | |
| EWS/RNA binding protein (GADD 153) | 2.79 | 0.27 | NC | |
| Cluster III | ||||
| p55CDC | NC | 2.27 | ||
| NFκB2/NF-κB p100/p52 | NC | 2.75 | 0.77 | |
| Polo-like serine-threonine kinase | 0.59 | 0.02 | 3.15 | 1.46 |
| Cyclin B1 | 0.40 | 0.03 | NC | |
| JNK kinase 2/MAPK kinase | NC | 1.59 | ||
| Hyaluronan-mediated motility receptor RHAMM | NC | 2.15 | 0.36 | |
| Survivin/apoptosis inhibitor 4 | 0.35 | 0.23 | NC | |
| cdc2 p34 protein kinase | 0.48 | 0 | NC | |
| Matrix metalloprotease 9 | NC | 1.98 | 0.32 | |
| p16-INK4a, cyclin-dependent kinase inhibitor p16 | 0.65 | 0.18 | 4.40 | 0.88 |
| aurora/IPL1-related kinase | 0.45 | 0.16 | NC | |
| CENP-F kinetochore protein/mitosin | 0.41 | 0 | NC | |
| Cyclin A | 0.22 | 0 | NC | |
| cdc25B M-phase inducer phosphatase 2 | 0.42 | 0 | NC | |
| Topoisomerase IIα (170 kDa) | 0.51 | 0.2 | NC | |
| MAdCAM-1 mucosal adhesin | 0.26 | 0.66 | NC | |
| Insulin-induced protein 1 | 0.39 | 0.10 | NC | |
| Matrix metalloprotease 1 | 0.20 | 0 | NC | |
Differentially expressed after induction of keratinocyte differentiation (average of two experiments). Expression of these genes correlates with keratinocyte growth and proliferation.
Differentially expressed upon treatment with TNF-α (average of three experiments). These genes are responsive to NF-κB, which is induced by TNF-α.
E6 inhibits expression of IFN-inducible genes and IFN-α and -β.
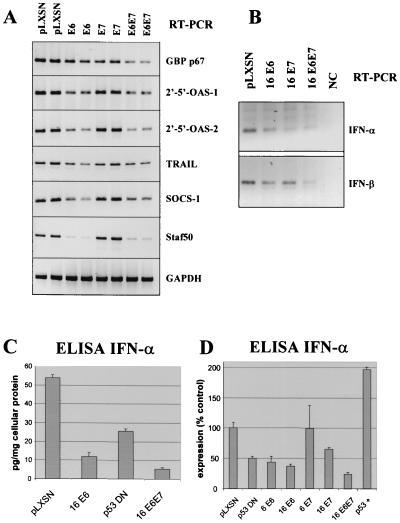
Genes in cluster II were consistently downregulated in HPV-16 E6- and E6/E7-expressing keratinocytes, and this effect was even more pronounced in their immortal derivatives. The IFN inducibility of genes in cluster II was confirmed by treatment of E6/E7-expressing, differentiating keratinocytes with 300 to 1,000 U of recombinant IFN-α or -β per ml (Table 1). Each IFN type strongly stimulated expression of genes in cluster II. Interestingly, E6 or E6/E7 decreased expression of most genes in cluster II more effectively than E7 alone. Decreased mRNA expression for some candidates in E6- or E6/E7-infected cells was confirmed by RT-PCR (Fig. 2A). For example, two isoforms of 2′-5′ oligoadenylate synthetase, a well-established IFN-inducible gene, were downregulated. Expression of both TRAIL (TNF family member 10) and the p67 guanylate binding protein were decreased. The highest factors of downregulation were observed for Staf50 and SOCS-1 (STAT-induced STAT inhibitor 1), genes originally identified as regulated by STAT-1. The STAT-1 transcription factor constitutes part of the IFN-stimulated gene factor 3 (ISGF-3) complex that activates IFN-regulated gene expression in response to IFN exposure (reviewed in references 12 and 24). Promoter sequences of genes in cluster II, if represented in GenBank, were analyzed using the PromoterScan program (courtesy of Dan Prestridge, Singapore University), and most contained one or multiple IFN-stimulated response elements (ISRE), which promote the expression of IFN-responsive genes.
FIG. 2.
RT-PCR and ELISA of differential expression of IFN-inducible genes in cluster II. (A) RT-PCR analysis of IFN-regulated genes after infection of keratinocytes with HPV E6 and/or E7. Duplicate data are from two independent experiments. 2′-5′-OAS, 2′-5′ oligoadenylate synthase. (B) RT-PCR analysis of IFN-α and -β RNAs using consensus primer pairs for IFN. NC indicates the negative control in which no enzyme was added to the RT reaction. (C) ELISA analysis of intracellular IFN-α from one representative experiment. Keratinocytes were infected with HPV-16 E6, E6/E7, or p53DN. (D) ELISA analysis of intracellular IFN-α from three independent experiments. Cells were infected with HPV-16 oncogenes or p53DN mutant as above, with low-risk HPV-6 E6 or E7, or with sense p53 (p53+). Bars depict the means of three experiments, and brackets indicate standard errors. Changes other than HPV-6 E7 were statistically significant (Fisher's exact test).
E6 and E6/E7 downregulate IFN-α and -β expression.
In contrast to E7, E6 decreased expression of RNAs for IFN-α and -β, as shown by RT-PCR analysis (Fig. 2B). IFN-α and -β RNAs were detected in both growing and differentiating keratinocytes, but differentiating cells expressed significant higher levels (data not shown). Secreted IFN-α or -β proteins were not detected by ELISA in conditioned medium of normal keratinocytes or retrovirus-infected cells. The inability to detect secreted IFN may be due to rapid sequestering of secreted IFN molecules by the extracellular matrix that is produced in abundance by keratinocytes in culture. Similar effects have been observed with TGF-β (40). However, intracellular IFN-α protein could be detected in cell lysates by ELISA (Fig. 2C and D). In this context, it is important to note that infection of human cervical keratinocytes with retrovirus vectors (pLXSN) and selection with G418 does not induce detectable changes in cellular IFN-α/β expression. In several control experiments, uninfected primary keratinocytes and pLXSN-infected cells expressed similar levels of IFN. The total amount of intracellular IFN-α ranged from 40 to 55 pg/mg of cellular protein in cultures from different individuals, and E6 or E6/E7 decreased levels to 0 to 10 pg/mg of protein (factors of 5 or greater). Infection of keratinocytes with a p53DN retrovirus (26) reduced levels of IFN-α only about twofold. Although the total amount of IFN-α produced was variable in different cultures, HPV-16 E6 and E6/E7 reproducibly lowered IFN-α levels in three independent experiments (Fig. 2D). The E7 protein was considerably less effective. Over expression of functional wild-type p53 protein induced IFN-α expression approximately twofold. When normal keratinocytes were treated with double-stranded RNA (dsRNA; a strong inducer of IFN production), IFN-α expression increased 5- to 10-fold; however, cells expressing E6/E7 were partially resistant to induction of IFNs by dsRNA (only two- to fivefold increase [data not shown]). We were not able to reproducibly detect the IFN-β protein using several commercially available ELISA test systems.
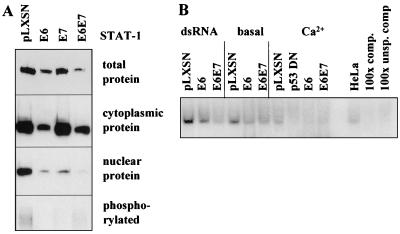
E6 decreases STAT-1 protein expression and binding to the ISRE.
RT-PCR demonstrated that E6 or/and E7 did not significantly alter expression of mRNAs encoding components of the IFN signal transduction pathway including IFN receptors R1 and R2, Jak kinases, STAT-1, STAT-2, and p48 (data not shown). Similarly, cDNA array experiments did not show any significant differences in mRNA expression for these genes. However, E6 reproducibly reduced total and cytoplasmic STAT-1 protein as detected by Western blotting (Fig. 3A). Both E6 and E7 reduced the levels of STAT-1 proteins that are translocated to the nucleus. E6 plus E7 decreased nuclear STAT-1 expression more effectively than E6 or E7 alone. E6 or E7 also reduced phosphorylated STAT-1 protein in comparison to controls infected with the empty vector (Fig. 3A, bottom panel). EMSA showed that E6 or E6/E7 reduced the amount of nuclear protein that bound to specific oligonucleotides containing the ISRE consensus sequence (Fig. 3B). Binding of STAT-1 to oligonucleotides was reduced by E6 under different culture conditions including the treatment with dsRNA to induce IFN-mediated gene transcription, use of basal medium to induce quiescence, or the addition of Ca2+ to induce squamous differentiation. Keratinocytes expressing p53DN (26) also showed slightly reduced specific DNA binding activity.
FIG. 3.
Western analysis showing altered expression and activity of STAT-1 in keratinocytes expressing E6 and E7. (A) Western analysis of total, cytoplasmic, nuclear, and phosphorylated STAT-1 protein in keratinocytes after infection with retroviruses containing HPV-16 E6 and/or E7. (B) EMSA for binding of STAT transcription factors to oligonucleotides containing the ISRE consensus sequence. Nuclear extracts were derived from cells infected with E6 and/or E7 or p53DN mutant and grown in the presence of dsRNA (which induces STAT activation), basal medium to induce quiescence (basal), or the same medium with Ca2+ to induce differentiation. HeLa, control nuclear extract from HeLa cell line; 100x comp., competition for specific DNA binding with a 100-fold molar excess of unlabeled oligonucleotide; 100x unsp. comp, unspecific competition with unrelated oligonucleotide.
E6 and E7 perturb expression of cell cycle regulatory genes.
Most genes in clusters I and III encode proteins that regulate cell cycle progression, DNA synthesis, or squamous differentiation. These genes were either strongly repressed (cluster I) or induced (cluster III) by E6 and/or E7 retroviruses or, more prominently, after immortalization (compare Fig. 1 and Table 1). Table 2 summarizes data from three independent experiments which show that expression of most genes in clusters I and III correlates with keratinocyte differentiation. Most genes in cluster I were upregulated during differentiation but downregulated by E6 and E7. Many of these genes were also strongly responsive to TGF-β, as shown previously (40). Differentiation-related genes include cytokeratin K6, tissue factor, gravin, lipocalin, and the TGF superfamily protein MIC-1. Cluster I also contains some proapoptotic genes, including Fas/APO-1, CD27 binding protein or Siva, and NIP3/Bcl-interacting protein. MKP-1 (mitogen-activated protein kinase [MAPK] phosphatase 1) and the EWS/RNA binding protein (CHOP or GADD 153) were previously identified as characteristic stress-inducible genes. In contrast, genes in cluster III were mostly decreased during differentiation but induced by E6 and E7. Many of these encode proteins that stimulate cell cycle progression including p55CDC (cdc42), polo-like kinase, cdc2 p34, and cdc2-associated protein, the cyclins A and B, MAPK kinase, and cdc25B. Other genes in cluster III encode proteins involved in DNA replication and mitosis including CENP-F, DNA primase, Madp2, topoisomerase IIα, and replication factor C.
Most interestingly, genes in cluster IV were significantly and strongly induced exclusively in HPV-16 E6/E7-immortalized keratinocytes that are derived from retrovirus-infected cells after extended in vitro culture (>20 passages or population doublings). These mRNAs are usually repressed in differentiation, and many encode cytoskeletal proteins (e.g., vimentin) or regulators of cell adhesion and extracellular matrix (TIMP-1, galectin-3, BM-40, or osteonectin [BM-40, SPARC], fibronectin, and MadCAM-1).
E6 and E7 enhance expression of genes regulated by NF-κB and AP-1 in differentiating cervical keratinocytes.
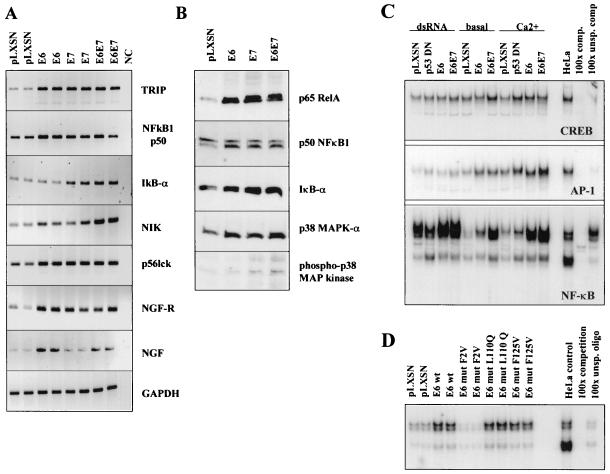
A subset of genes in cluster III were induced after treatment of keratinocytes with TNF-α (Table 2), a proinflammatory cytokine that activates the transcription factors NF-κB and AP-1. We confirmed that TNF-α stimulated expression of several genes in cluster III, including IL-8, NF-κB2/p52, matrix metalloproteinase 9, and others by RT-PCR (data not shown). We also found that E6, E7, or E6/E7 stimulated expression of several additional genes that are known to be involved in NF-κB activation (Fig. 4A). Additional RT-PCR analyses showed that E6/E7 characteristically increased mRNA expression of TRAF-interacting protein 1, p50 (NF-κB1), IκBα, and NF-κB-inducing kinase (NIK). p56 Lck is a tyrosine kinase in the Src family that is required for TNF-mediated activation of NF-κB and AP-1. Both nerve growth factor (NGF) and the 75-kDa NGF receptor are induced by E6, can protect from apoptosis, and contribute to NF-κB activation (23, 37). Western blot analysis showed induction of p65 RelA protein expression, one of two major functional subunits shown to be expressed in cervical keratinocytes (Fig. 4B). In contrast, p50 (NF-κB1) expression was basically unchanged. Supershift assays using antibodies against the p50, p52, and p65 subunits of NF-κB showed that cervical keratinocytes strongly express p65 (cRe1) and p50 but express little p52 (NF-κB2). Induction of mRNA and the corresponding proteins by HPV were also found for IκBα and NIK. IκB-α is an inhibitor of NF-κB activity, but this protein is a component of a large complex of proteins including NIK and IKK which are generally induced as a result of NF-κB activation (for example, by IL-1 and TNF-α). p38 MAPK is similarly induced by E6 and E7. MAPK is required for both NF-κB- and AP-1-dependent gene expression (6, 7). Levels of both total and phosphorylated p38 MAPK protein are induced by HPV-16 E6. Very similar results were observed for p44/p42 MAPK (data not shown).
FIG. 4.
RT-PCR and EMSA demonstrate that E6 increased expression of NF-κB-regulated genes and enhanced activity of NF-κB and AP-1. (A) RT-PCR analysis of several NF-κB-regulated genes after infection with HPV-16 E6 and/or E7. Duplicate data are from two independent experiments. (B) Western analysis of p65 RelA, p50 (NF-κB1), IκBα, and p38 MAPK proteins in keratinocytes after infection with retroviruses containing HPV-16 E6 and/or E7. (C) EMSA analysis for binding of CREB, AP-1, and NF-κB transcription factors in nuclear extracts to oligonucleotides with the respective DNA consensus sequences. Nuclear extracts were derived from cells growing in the presence of dsRNA (which induces STAT activation), and quiescent (basal) or differentiating keratinocytes (Ca2+). HeLa, control nuclear extract from HeLa cell line; 100x comp., competition for specific DNA binding with a 100-fold molar excess of unlabeled oligonucleotide; 100x unsp. comp, unspecific competition with unrelated oligonucleotide. D. EMSA analysis of NF-κB DNA binding activity in keratinocytes expressing wild-type HPV-16 E6 and mutants with variable p53-degrading activities. E6mut F2V does not bind and degrade p53 in vitro. Mutants L110Q and F125V retain p53 binding activity.
HPV-16 E6 induces NF-κB DNA binding.
We analyzed the activity of NF-κB in keratinocytes expressing E6 or E6/E7 by EMSA and detected activation of DNA binding for NF-κB and AP-1 by E6 (Fig. 4C). NF-κB was activated in both quiescent (basal) and differentiating (Ca2+-induced) keratinocytes. The high NF-κB activity in the presence of dsRNA, an inducer of cellular stress, was further increased by E6 (Fig. 4C). DNA binding activity of AP-1 was also induced by E6 and E6/E7, particularly under conditions of quiescence or keratinocyte differentiation. p53DN did not induce NF-κB activity as effectively as E6 or E6/E7. We also found that NF-kB activation was induced by several functionally impaired E6 mutants (35) that retain p53 binding activity (Fig. 4D). One E6 mutant (F2V) that does not bind p53 did not increase NF-κB binding. Supershift assays using antibodies against the p50 (NF-κB1), p52 (NF-κB2), and p65 (Re1A) subunits identified p50 and p65 as the major components of functional NF-κB in cervical keratinocytes (data not shown), with little p52 expression. In contrast to NF-κB, DNA binding activity of transcription factors CREB (Fig. 4C) and Oct-1 transcription factors (data not shown) did not change significantly in HPV-expressing keratinocytes.
HPV-16 E6 and E7 stimulate secretion of specific proinflammatory cytokines.
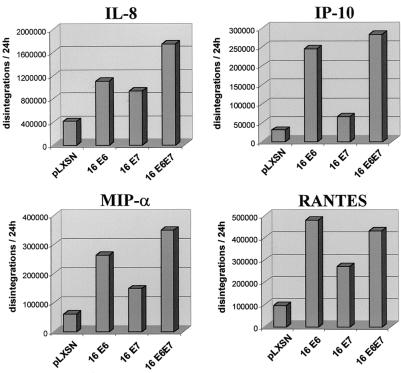
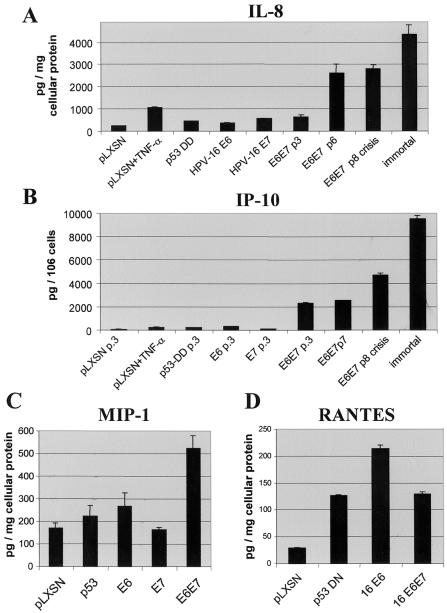
NF-κB and AP-1 activate expression of specific proinflammatory and immunoregulatory cytokines. Using the RNase protection assay, we examined whether E6 and/or E7 induced expression of selected cytokines (Fig. 5). E6 or mutant p53 induced IL-8, IP-10, MIP-1α, and RANTES. The promoters of each of these genes contain one or more binding sites for NF-κB or AP-1. We also examined conditioned medium from keratinocyte cultures by ELISA to determine whether E6 and/or E7 or p53DN altered cytokine secretion (Fig. 6). Secretion of IL-8 and IP-10 was increased directly after infection with E6 and/or E7. E6 but not E7 increased secretion of MIP-1α and RANTES. E6 and E7 stimulated expression of several other cytokine mRNAs but did not significantly increase secretion in vitro (IL-6, IL-12 p35 and p40 subunits, granulocyte-macrophage colony-stimulating factor, and VEGF-B and -C [data not shown]).
FIG. 5.
RNase protection analysis of chemokine gene expression. Cervical keratinocytes were infected with empty vector (pLXSN) or E6, E7, and E6/E7 retroviruses and induced to undergo squamous differentiation. Raw data were normalized using glyceraldehyde-3-phosphate dehydrogenase and L32 expression. Data were analyzed by densitometry using a PhosphorImager.
FIG. 6.
ELISA analysis of chemokine secretion. Keratinocytes were infected with HPV-16 E6 and/or E7 retroviruses or p53DN mutant. Cells were induced to undergo differentiation, and some cultures were treated with TNF-α. ELISA was performed on supernatants of E6/E7-infected cultures; in other experiments, cells were examined after several (three to eight) subpassages or after immortalization. Data are averages of four individual ELISA readings from experiments and were normalized for absolute cell number or total cellular protein.
DISCUSSION
Infection with high-risk HPV is a major risk factor for the development of cervical cancer (60). In infected epithelia and low-grade cervical intraepithelial neoplasias (CIN I), expression of E6 and E7 mRNAs is increased as keratinocytes enter the terminal differentiation pathway or progress to high-grade dysplasia (CIN II and III) and cancer (16). We used retroviruses to express high levels of the E6 and E7 oncoproteins in cultures of differentiating cervical keratinocytes and directly examined resultant alterations in gene expression using cDNA arrays. Approximately 80 genes out of 2,208 on the chip (4%) were reproducibly increased or decreased more than threefold by E6 and/or E7. These genes fell into several functional groups providing insights into pathways by which E6 and E7 deregulate differentiation, cell cycle progression, and the host response to viral infection.
The most obvious result was that E6/E7 strongly decreased expression of IFN-responsive genes, as shown in cluster II. Similar or even more pronounced results were observed in cells immortalized by E6 and E7. Efficient production of IFNs in virally infected and/or immortalized cells is an essential aspect of the host defense and is regulated by a number of cellular transcription factors. The transcription factor complex ISGF-3 was originally identified as a critical mediator of IFN signaling. ISGF-3 is formed by association of STAT-1, STAT-2, and p48 proteins. Upon stimulation of the IFN receptor, the complex translocates to the nucleus, binds to ISREs, and activates expression of IFNs and IFN-inducible genes (12, 24). Transgenic animals that carry a disruption in STAT-1, p48, or IFN receptor genes exhibit severe defects in virus-induced IFN-α/β gene expression (28). Our observations that E6 and E7 downregulate expression of IFNs and IFN-responsive genes confirm and extend recent work by Chang et al. (8) that showed that immortalized foreskin keratinocytes transfected with the complete HPV-31 genome have reduced expression of multiple IFN-responsive genes. Our cluster analysis determined that the relative effectiveness of oncoproteins was E6/E7 > E6 > E7. Thus, in particular under conditions promoting differentiation, E6 appears to be primarily responsible for most but not all changes. Furthermore, most IFN-responsive genes were downregulated by infection with the retrovirus encoding a p53DN miniprotein (26), suggesting that E6-mediated degradation of p53 contributes to this response but is not exclusively responsible. We found that E6 decreased constitutive expression of IFN-α and -β mRNAs and diminished the intracellular level of IFN-α 5- to 10-fold. IFN-α production was also inhibited by the p53DN mutant, although not as effectively as by E6 or E6/E7. We also showed that E6 decreased the level of cytoplasmic and nuclear STAT-1, an important component of the ISGF-3 complex which binds to ISRE and activates expression of IFN and IFN-inducible genes. This downregulation of cytoplasmic and nuclear STAT-1 appears to occur at the posttranscriptional level, as arrays and RT-PCR analyses did not show a strong reduction of STAT-1 mRNA in response to E6/E7. Downregulation and inactivation of STAT-1 might directly inhibit IFN production in E6-expressing cells. It would be interesting to determine whether STAT-1 protein is targeted for degradation by E6-E6AP complexes, as is observed for p53 protein. Ronco et al. have shown that the HPV-16 E6 protein binds and blocks transcriptional activity of IRF-3 another important activator of IFN-responsive gene expression (50). In addition, Li et al. used HT1080 cells to show that E6 binds to and diminishes the activity of Tyk2 a kinase involved in activation of STAT-1 and STAT-2 (34). Thus, E6 has been shown to target IFN signaling by at least two different types of interactions. Our observation that E6 downregulates multiple IFN-responsive genes directly demonstrates the biological relevance of these interactions in the natural target of HPV infection, primary ectocervical keratinocytes.
HPV-16 E6 or E6/E7 decreased expression of multiple IFN-responsive genes. However, E7 alone was less effective and diminished expression of only a subset of genes. Recently, Barnard et al. showed that E7 binds to p48, a component of IGSF3, and abrogates signaling mediated by IFN-α in HaCaT cells (3). Furthermore, they recently showed that E7 is able to inhibit the antiproliferative and antiviral functions of IFN-α (4). E7 also binds and inhibits transactivation by IRF-1, an important component of IFN signaling (44, 46). Thus, E7 targets at least two components of the IFN signal pathway. Surprisingly, our own results showed that E7 alone did not effectively downregulate the majority of IFN-responsive genes in differentiating human cervical keratinocytes. However, coexpression of E6 and E7 was more effective than E6 alone in inhibiting IFN-responsive genes. These results suggest that E6 is more effective than E7. However, we have previously shown that high-level expression of E7 in differentiating cervical keratinocytes sensitizes cells to apoptosis (29). Thus, it is possible that some cells (approximately 5% in our previous study) that express very high levels of E7 are eliminated before the analysis. This might explain, in part, why E7 had reduced effectiveness for downregulation of IFN-responsive genes. IFNs provide an important first line of defense against viral infection. IFNs stimulate expression of several intracellular proteins with direct antiviral activity. IFNs also enhance expression of major histocompatibility class I and II proteins, which mediate immune recognition of viral antigens. Thus, the ability of E6 and E7 to interfere with expression of IFN-responsive genes may allow circumvention of the host response to HPV-infected keratinocytes.
A hallmark of HPV infection is reactivation of host DNA synthesis and cell cycle progression in differentiating keratinocytes. This allows a small DNA tumorvirus to utilize host enzymes to replicate its own DNA. Cluster I contains genes that are downregulated by E6 and E7 and that promote keratinocyte differentiation or growth arrest. Some of these genes are regulated by p53 (reviewed in reference 19), which is targeted for degradation by HPV-16 E6 (51). p53-inducible genes that are downregulated by HPV in cluster I include the Fas/APO-1 receptor, p21 Waf-1, p53-induced genes 8 and 10, Fanconi's anemia antigen C, IGFBP-3, EWS/GADD 153 or CHOP, the TNF-related proapoptotic ligand TRAIL, and the TRAIL receptors R2 and R3/decoy receptor DcR3 (not included in array and cluster analysis but confirmed by RT-PCR). Most p53-inducible genes play a role in regulation of cellular responses to genotoxic stress. E6 and/or E7 also repressed expression of genes typically induced during keratinocyte differentiation or in inflammation. For instance, tissue factor/thromboplastin is expressed by keratinocytes and induced during differentiation. GADD 153, heat shock protein 60, gravin, and lipocalin are induced under pathological conditions and inflammation (10). MAPK phosphatase 1 (MKP-1) is a dual-specificity protein phosphatase induced in immediate-early gene responses to growth factors and during cellular stress. MKP-1 selectively inactivates stress-activated protein kinase 3 in vitro by dephosphorylation (22). Cluster I also contains proapoptotic genes, including Fas/APO-1, CD27 binding protein or Siva, and NIP3. In general, genes in cluster I encode proteins that regulate various aspects of host response to cellular stress. Downregulation of these genes could contribute to increased resistance of keratinocytes to environmental or host stress factors.
The mRNAs found in cluster III were induced by HPV-16 E6 and E7. Many represent proteins that stimulate cell cycle progression or are functional in mitosis (15), including BUB1, CENP-F/mitosin, Madp2, p55CDC, polo-like kinase 1, cdc2 p34, and cdc2-associated protein, cdc6, the cyclins A and B, and cdc25B. Other genes in cluster III encode proteins involved in DNA synthesis and repair, including ribonucleotide reductase M2, DNA primase, topoisomerase IIα, and several subunits of replication factor C. These are representative of a large number of genes that are induced by the E2F transcription factor family, not all of which were included in the first version of the NCI oncochip. Active E2F factors are released upon binding of HPV-16 E7 protein to hypophosphorylated pRB and directly induce expression of genes involved in DNA synthesis (S phase) and cell cycle progression. Many of these genes are strongly repressed by TGF-β, one of the most potent growth suppressors known that strongly inhibits keratinocyte proliferation. TGF-β itself is efficiently repressed by HPV-16, as shown previously (40). Previous work over the past decade has demonstrated that HPV-16 E6 and E7 proteins enhance expression of many of the genes located in cluster III (53), thus confirming the accuracy of the cDNA array analysis.
A fraction of genes in cluster III encode proteins known as induced by the transcription factors NF-κB and/or AP-1. Because of apparently similar expression profiles, the clustering software could not distinguish between cell cycle regulatory genes and NF-κB-inducible genes in cluster III. We used additional microarray experiments to show that these genes were induced by TNF-α (Table 2). TNF-α activates both NF-κB and AP-1 and stimulates expression of many inflammatory genes such as IL-6 and IL-8. However, TNF-α does not significantly effect cell cycle regulation and proliferation of normal, primary keratinocytes (reviewed in reference 43). Using EMSA, we show that E6 significantly induced DNA binding activity for NF-κB and AP-1, consistent with the hypothesis that these transcription factors regulate a subset of E6-induced genes. We also observed increased mRNA and/or protein expression of a number of genes that are necessary for or involved in NF-κB activation. These include p38 MAPK, which is required for NF-κB dependent gene expression (6, 7). Also induced were NIK, NF-κB1 and -2 (p50 and p52), TRAF-interacting protein 1, p56 Lck (36), NGF (37), and the 75-kDa NGF receptor (23). Transcriptional activation of NF-κB and AP-1 in HPV-immortalized mouse keratinocytes has been reported recently (32). In contrast, Patel et al. have reported that E6 reduces NF-κB activity in SAOS and U20S cells, which are derived from osteosarcomas, a mesenchymal tissue (45). The explanation for this difference between this study and our results is unclear; however, one possibility is that E6 affects NF-κB activation differently in differentiating cervical keratinocytes than in osteosarcoma cells.
Both p53 and NF-κB regulate the cellular response to stress. In contrast to p53, which is proapoptotic, NF-κB can efficiently protect keratinocytes from apoptosis (9). Recent studies have shown that NF-κB and p53 directly compete for binding to the transcriptional coactivator CBP/p300. Therefore, degradation of p53 by E6 could result in an activation of NF-κB as shown in our work. Furthermore, cross-transcriptional interference between p53 and RelA may occur due to direct interaction between these two transcription factors which is mediated by their dimerization/tetramerization domains and results in inhibition of each other's transcriptional activity (30, 56). The postulated cross talk between p53 and NF-κB is consistent with our observation that wild-type E6 and E6 mutants that degrade p53 also induce NF-κB activity, in contrast to a mutant that binds p53 insufficiently (E6mut F2V). Constitutive activation of NF-κB and AP-1 transcription factors occur regularly during mouse skin carcinogenesis (5), and elevated NF-κB activity is typically detected in HPV-immortalized cell lines (32). In contrast, inhibitors of NF-κB and AP-1 activation block neoplastic transformation (33). Thus, NF-κB and AP-1 activation occurs commonly during carcinogenesis, and increased activity may contribute to HPV-induced transformation.
Expression of E6 and E7 in differentiating keratinocytes stimulated secretion of several cytokines including IL-8, IP-10, MIP-1α, and RANTES. IL-8 and IP-10 are members of the CXC family of chemokines that exert inflammatory, growth-promoting, and/or chemotactic activity. CXC chemokines are induced by stressful stimuli such as wound healing (20), inflammation, or exposure to UV light (54). IP-10 is chemotactic for monocytes and T lymphocytes and promotes adhesion to endothelial cells. IL-8 stimulates leukocyte chemotaxis. RANTES is a CC chemokine that is chemotactic for memory T lymphocytes and activates naive T cells (49). Elevated RANTES and IP-10 occur in psoriasis and cutaneous hypersensitivity reactions. Both IL-8 and IP-10 were more strongly induced in HPV-immortalized cell lines (data not shown). The potential biological significance of increased chemokine production in differentiating keratinocytes after high-level E6/E7 expression is unclear. Most chemokines are not normally expressed in resting cells but are rapidly induced in response to various inflammatory and mitogenic stimuli. Induction of chemokine secretion might reflect activation by NF-κB or AP-1 transcription factors due to cell stress induced by E6 and/or E7. Increased chemokine production might facilitate recognition and elimination of HPV-infected cells in vivo. However, chemokines such as IL-8 can also stimulate tumor proliferation or angiogenesis (38, 42).
Cluster IV contains a number of genes that are characteristically induced in immortal cell lines established from retrovirus-infected keratinocytes. Infection of keratinocytes with high-risk HPV E6 and E7 results in hyperproliferation, in dysregulation of the cell cycle, and eventually in increasing genetic instability and acquisition of somatic mutations. In vivo, these events can eventually lead to malignant progression and cervical cancer. In vitro, neoplastic progression is reflected in immortalization of primary cells. Immortalization requires long-term cell culture and survival of cellular senescence or crisis and involves massive losses and/or amplifications of genetic material. HPV-immortalized cell lines show highly dysplastic differentiation and growth properties and acquire many characteristics shared by preneoplastic, high-grade CIN lesions (58). Therefore, we were particularly interested in identification of genes induced in immortalized cell lines independently of early responses to the E6 and E7 oncogenes. Many genes in cluster IV encode components of the cytoskeleton or the extracellular matrix such as vimentin, osteonectin (SPARC or BM-40), and fibronectin. Interestingly, increased production of soluble and cell surface-associated fibronectin or thrombospondin is not specific for HPV-16-immortalized keratinocytes, since these genes are also characteristically increased in adenovirus and simian virus 40-immortalized cells (52). Upregulation of fibronectin gene expression and extracellular matrix formation are consistent characteristics of malignantly converted human keratinocytes. Fibronectin and other genes in cluster IV, including galectin-1 and osteonectin/SPARC, might therefore be useful as potential molecular markers that indicate an increased risk for malignant progression of HPV-associated lesions (41) and could be highly interesting as novel diagnostic tools for early detection of preneoplastic cervical lesions.
ACKNOWLEDGMENTS
We thank Eliot Androphy and Yun Liu for providing HPV-16 E6 mutants, Moshe Oren for the p53 miniprotein used for constructing the p53DN retrovirus, and John Sunwoo for assistance with the EMSAs. Human cervical tissue was supplied by the Cooperative Human Tissue Network.
REFERENCES
- 1.Alizadeh A A, Eisen M B, Davis R E, Ma C, Lossos I S, Rosenwald A, Boldrick J C, Sabet H, Tran T, Yu X, Powell J I, Yang L, Marti G E, Moore T, Hudson J, Jr, Lu L, Lewis D B, Tibshirani R, Sherlock G, Chan W C, Greiner T C, Weisenburger D D, Armitage J O, Warnke R, Staudt L M. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 2.Antinore M J, Birrer M J, Patel D, Nader L, McCance D J. The human papillomavirus type 16 E7 gene product interacts with and trans-activates the AP1 family of transcription factors. EMBO J. 1996;15:1950–1960. [PMC free article] [PubMed] [Google Scholar]
- 3.Barnard P, McMillan N A. The human papillomavirus E7 oncoprotein abrogates signaling mediated by interferon-alpha. Virology. 1999;259:305–313. doi: 10.1006/viro.1999.9771. [DOI] [PubMed] [Google Scholar]
- 4.Barnard P, Payne E, McMillan N A. The human papillomavirus E7 protein is able to inhibit the antiviral and anti-growth functions of interferon-alpha. Virology. 2000;277:411–419. doi: 10.1006/viro.2000.0584. [DOI] [PubMed] [Google Scholar]
- 5.Budunova I V, Perez P, Vaden V R, Spiegelman V S, Slaga T J, Jorcano J L. Increased expression of p50-NF-κB and constitutive activation of NF-κB transcription factors during mouse skin carcinogenesis. Oncogene. 1999;18:7423–7431. doi: 10.1038/sj.onc.1203104. [DOI] [PubMed] [Google Scholar]
- 6.Carter A B, Hunninghake G W. A constitutive active MEK→ ERK pathway negatively regulates NF-κB-dependent gene expression by modulating TATA-binding protein phosphorylation. J Biol Chem. 2000;275:27858–27864. doi: 10.1074/jbc.M003599200. [DOI] [PubMed] [Google Scholar]
- 7.Carter A B, Knudtson K L, Monick M M, Hunninghake G W. The p38 mitogen-activated protein kinase is required for NF-κB-dependent gene expression. The role of TATA-binding protein (TBP) J Biol Chem. 1999;274:30858–30863. doi: 10.1074/jbc.274.43.30858. [DOI] [PubMed] [Google Scholar]
- 8.Chang Y E, Laimins L A. Microarray analysis identifies interferon-inducible genes and Stat-1 as major transcriptional targets of human papillomavirus type 31. J Virol. 2000;74:4174–4182. doi: 10.1128/jvi.74.9.4174-4182.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chaturvedi V, Qin J Z, Denning M F, Choubey D, Diaz M O, Nickoloff B J. Apoptosis in proliferating, senescent, and immortalized keratinocytes. J Biol Chem. 1999;274:23358–23367. doi: 10.1074/jbc.274.33.23358. [DOI] [PubMed] [Google Scholar]
- 10.Cowland J B, Borregaard N. Molecular characterization and pattern of tissue expression of the gene for neutrophil gelatinase-associated lipocalin from humans. Genomics. 1997;45:17–23. doi: 10.1006/geno.1997.4896. [DOI] [PubMed] [Google Scholar]
- 11.Crish J F, Bone F, Balasubramanian S, Zaim T M, Wagner T, Yun J, Rorke E A, Eckert R L. Suprabasal expression of the human papillomavirus type 16 oncoproteins in mouse epidermis alters expression of cell cycle regulatory proteins. Carcinogenesis. 2000;21:1031–1037. doi: 10.1093/carcin/21.5.1031. [DOI] [PubMed] [Google Scholar]
- 12.Darnell J E., Jr STATs and gene regulation. Science. 1997;277:1630–1635. doi: 10.1126/science.277.5332.1630. [DOI] [PubMed] [Google Scholar]
- 13.DeRisi J, Penland L, Brown P O, Bittner M L, Meltzer P S, Ray M, Chen Y, Su Y A, Trent J M. Use of a cDNA microarray to analyse gene expression patterns in human cancer. Nat Genet. 1996;14:457–460. doi: 10.1038/ng1296-457. [DOI] [PubMed] [Google Scholar]
- 14.Diehn M, Alizadeh A A, Brown P O. Examining the living genome in health and disease with DNA microarrays. JAMA. 2000;283:2298–2299. [PubMed] [Google Scholar]
- 15.Duensing S, Lee L Y, Duensing A, Basile J, Piboonniyom S, Gonzalez S, Crum C P, Munger K. The human papillomavirus type 16 E6 and E7 oncoproteins cooperate to induce mitotic defects and genomic instability by uncoupling centrosome duplication from the cell division cycle. Proc Natl Acad Sci USA. 2000;97:10002–10007. doi: 10.1073/pnas.170093297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Durst M, Glitz D, Schneider A, zur Hausen H. Human papillomavirus type 16 (HPV 16) gene expression and DNA replication in cervical neoplasia: analysis by in situ hybridization. Virology. 1992;189:132–140. doi: 10.1016/0042-6822(92)90688-l. [DOI] [PubMed] [Google Scholar]
- 17.Dyson N, Guida P, Munger K, Harlow E. Homologous sequences in adenovirus E1A and human papillomavirus E7 proteins mediate interaction with the same set of cellular proteins. J Virol. 1992;66:6893–6902. doi: 10.1128/jvi.66.12.6893-6902.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eisen M B, Spellman P T, Brown P O, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.El Deiry W S. Regulation of p53 downstream genes. Semin Cancer Biol. 1998;8:345–357. doi: 10.1006/scbi.1998.0097. [DOI] [PubMed] [Google Scholar]
- 20.Engelhardt E, Toksoy A, Goebeler M, Debus S, Brocker E B, Gillitzer R. Chemokines IL-8, GROα, MCP-1, IP-10, and Mig are sequentially and differentially expressed during phase-specific infiltration of leukocyte subsets in human wound healing. Am J Pathol. 1998;153:1849–1860. doi: 10.1016/s0002-9440(10)65699-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Foster S A, Galloway D A. Human papillomavirus type 16 E7 alleviates a proliferation block in early passage human mammary epithelial cells. Oncogene. 1996;12:1773–1779. [PubMed] [Google Scholar]
- 22.Franklin C C, Kraft A S. Conditional expression of the mitogen-activated protein kinase (MAPK) phosphatase MKP-1 preferentially inhibits p38 MAPK and stress-activated protein kinase in U937 cells. J Biol Chem. 1997;272:16917–16923. doi: 10.1074/jbc.272.27.16917. [DOI] [PubMed] [Google Scholar]
- 23.Gentry J J, Casaccia-Bonnefil P, Carter B D. Nerve growth factor activation of nuclear factor κB through its p75 receptor is an anti-apoptotic signal in RN22 schwannoma cells. J Biol Chem. 2000;275:7558–7565. doi: 10.1074/jbc.275.11.7558. [DOI] [PubMed] [Google Scholar]
- 24.Gilmour K C, Reich N C. Signal transduction and activation of gene transcription by interferons. Gene Expr. 1995;5:1–18. [PMC free article] [PubMed] [Google Scholar]
- 25.Golub T R, Slonim D K, Tamayo P, Huard C, Gaasenbeek M, Mesirov J P, Coller H, Loh M L, Downing J R, Caligiuri M A, Bloomfield C D, Lander E S. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–537. doi: 10.1126/science.286.5439.531. [DOI] [PubMed] [Google Scholar]
- 26.Gottlieb E, Haffner R, von Ruden T, Wagner E F, Oren M. Down-regulation of wild-type p53 activity interferes with apoptosis of IL-3-dependent hematopoietic cells following IL-3 withdrawal. EMBO J. 1994;13:1368–1374. doi: 10.1002/j.1460-2075.1994.tb06390.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Halbert C L, Demers G W, Galloway D A. The E7 gene of human papillomavirus type 16 is sufficient for immortalization of human epithelial cells. J Virol. 1991;65:473–478. doi: 10.1128/jvi.65.1.473-478.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harada H, Matsumoto M, Sato M, Kashiwazaki Y, Kimura T, Kitagawa M, Yokochi T, Tan R S, Takasugi T, Kadokawa Y, Schindler C, Schreiber R D, Noguchi S, Taniguchi T. Regulation of IFN-α/β genes: evidence for a dual function of the transcription factor complex ISGF3 in the production and action of IFN-α/β. Genes Cells. 1996;1:995–1005. doi: 10.1046/j.1365-2443.1996.870287.x. [DOI] [PubMed] [Google Scholar]
- 29.Iglesias M, Yen K, Gaiotti D, Hildesheim A, Stoler M H, Woodworth C D. Human papillomavirus type 16 E7 protein sensitizes cervical keratinocytes to apoptosis and release of interleukin-1α. Oncogene. 1998;17:1195–1205. doi: 10.1038/sj.onc.1202054. [DOI] [PubMed] [Google Scholar]
- 30.Ikeda A, Sun X, Li Y, Zhang Y, Eckner R, Doi T S, Takahashi T, Obata Y, Yoshioka K, Yamamoto K. p300/CBP-dependent and -independent transcriptional interference between NF-κB Re1A and p53. Biochem Biophys Res Commun. 2000;272:375–379. doi: 10.1006/bbrc.2000.2786. [DOI] [PubMed] [Google Scholar]
- 31.Jones D L, Munger K. Interactions of the human papillomavirus E7 protein with cell cycle regulators. Semin Cancer Biol. 1996;7:327–337. doi: 10.1006/scbi.1996.0042. [DOI] [PubMed] [Google Scholar]
- 32.Li J J, Rhim J S, Schlegel R, Vousden K H, Colburn N H. Expression of dominant negative Jun inhibits elevated AP-1 and NF-κB transactivation and suppresses anchorage independent growth of HPV immortalized human keratinocytes. Oncogene. 1998;16:2711–2721. doi: 10.1038/sj.onc.1201798. [DOI] [PubMed] [Google Scholar]
- 33.Li J J, Westergaard C, Ghosh P, Colburn N H. Inhibitors of both nuclear factor-κB and activator protein-1 activation block the neoplastic transformation response. Cancer Res. 1997;57:3569–3576. [PubMed] [Google Scholar]
- 34.Li S, Labrecque S, Gauzzi M C, Cuddihy A R, Wong A H, Pellegrini S, Matlashewski G J, Koromilas A E. The human papilloma virus (HPV)-18 E6 oncoprotein physically associates with Tyk2 and impairs Jak-STAT activation by interferon-alpha. Oncogene. 1999;18:5727–5737. doi: 10.1038/sj.onc.1202960. [DOI] [PubMed] [Google Scholar]
- 35.Liu Y, Chen J J, Gao Q, Dalal S, Hong Y, Mansur C P, Band V, Androphy E J. Multiple functions of human papillomavirus type 16 E6 contribute to the immortalization of mammary epithelial cells. J Virol. 1999;73:7297–7307. doi: 10.1128/jvi.73.9.7297-7307.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Manna S K, Sah N K, Aggarwal B B. Protein tyrosine kinase p56lck is required for ceramide-induced but not tumor necrosis factor-induced activation of NF-κB, AP-1, JNK, and apoptosis. J Biol Chem. 2000;275:13297–13306. doi: 10.1074/jbc.275.18.13297. [DOI] [PubMed] [Google Scholar]
- 37.Marconi A, Vaschieri C, Zanoli S, Giannetti A, Pincelli C. Nerve growth factor protects human keratinocytes from ultraviolet-B-induced apoptosis. J Investig Dermatol. 1999;113:920–927. doi: 10.1046/j.1523-1747.1999.00773.x. [DOI] [PubMed] [Google Scholar]
- 38.Metzner B, Hofmann C, Heinemann C, Zimpfer U, Schraufstatter I, Schopf E, Norgauer J. Overexpression of CXC-chemokines and CXC-chemokine receptor type II constitute an autocrine growth mechanism in the epidermoid carcinoma cells KB and A431. Oncol Rep. 1999;6:1405–1410. doi: 10.3892/or.6.6.1405. [DOI] [PubMed] [Google Scholar]
- 39.Munger K, Phelps W C, Bubb V, Howley P M, Schlegel R. The E6 and E7 genes of the human papillomavirus type 16 together are necessary and sufficient for transformation of primary human keratinocytes. J Virol. 1989;63:4417–4421. doi: 10.1128/jvi.63.10.4417-4421.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nees M, Geoghegan J M, Munson P, Prabhu V, Liu Y, Androphy E, Woodworth C D. Human papillomavirus type 16 E6 and E7 proteins inhibit differentiation-dependent expression of transforming growth factor-β2 in cervical keratinocytes. Cancer Res. 2000;60:4289–4298. [PubMed] [Google Scholar]
- 41.Nees M, van Wijngaarden E, Bakos E, Schneider A, Durst M. Identification of novel molecular markers which correlate with HPV-induced tumor progression. Oncogene. 1998;16:2447–2458. doi: 10.1038/sj.onc.1201785. [DOI] [PubMed] [Google Scholar]
- 42.Norgauer J, Metzner B, Schraufstatter I. Expression and growth-promoting function of the IL-8 receptor beta in human melanoma cells. J Immunol. 1996;156:1132–1137. [PubMed] [Google Scholar]
- 43.Pahl H L. Activators and target genes of Rel/NF-κB transcription factors. Oncogene. 1999;18:6853–6866. doi: 10.1038/sj.onc.1203239. [DOI] [PubMed] [Google Scholar]
- 44.Park J S, Kim E J, Kwon H J, Hwang E S, Namkoong S E, Um S J. Inactivation of interferon regulatory factor-1 tumor suppressor protein by HPV E7 oncoprotein. Implication for the E7-mediated immune evasion mechanism in cervical carcinogenesis. J Biol Chem. 2000;275:6764–6769. doi: 10.1074/jbc.275.10.6764. [DOI] [PubMed] [Google Scholar]
- 45.Patel D, Huang S M, Baglia L A, McCance D J. The E6 protein of human papillomavirus type 16 binds to and inhibits co-activation by CBP and p300. EMBO J. 1999;18:5061–5072. doi: 10.1093/emboj/18.18.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Perea S E, Massimi P, Banks L. Human papillomavirus type 16 E7 impairs the activation of the interferon regulatory factor-1. Int J Mol Med. 2000;5:661–666. doi: 10.3892/ijmm.5.6.661. [DOI] [PubMed] [Google Scholar]
- 47.Perou C M, Sorlie T, Eisen M B, Van de R M, Jeffrey S S, Rees C A, Pollack J R, Ross D T, Johnsen H, Akslen L A, Fluge O, Pergamenschikov A, Williams C, Zhu S X, Lonning P E, Borresen-Dale A L, Brown P O, Botstein D. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 48.Rapp L, Chen J J. The papillomavirus E6 proteins. Biochim Biophys Acta. 1998;1378:F1–F19. doi: 10.1016/s0304-419x(98)00009-2. [DOI] [PubMed] [Google Scholar]
- 49.Raychaudhuri S P, Jiang W Y, Farber E M, Schall T J, Ruff M R, Pert C B. Upregulation of RANTES in psoriatic keratinocytes: a possible pathogenic mechanism for psoriasis. Acta Derm Venereol. 1999;79:9–11. doi: 10.1080/000155599750011615. [DOI] [PubMed] [Google Scholar]
- 50.Ronco L V, Karpova A Y, Vidal M, Howley P M. Human papillomavirus 16 E6 oncoprotein binds to interferon regulatory factor-3 and inhibits its transcriptional activity. Genes Dev. 1998;12:2061–2072. doi: 10.1101/gad.12.13.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scheffner M, Werness B A, Huibregtse J M, Levine A J, Howley P M. The E6 oncoprotein encoded by human papillomavirus types 16 and 18 promotes the degradation of p53. Cell. 1990;63:1129–1136. doi: 10.1016/0092-8674(90)90409-8. [DOI] [PubMed] [Google Scholar]
- 52.Sheibani N, Rhim J S, Allen-Hoffmann B L. Malignant human papillomavirus type 16-transformed human keratinocytes exhibit altered expression of extracellular matrix glycoproteins. Cancer Res. 1991;51:5967–5975. [PubMed] [Google Scholar]
- 53.Steinmann K E, Pei X F, Stoppler H, Schlegel R, Schlegel R. Elevated expression and activity of mitotic regulatory proteins in human papillomavirus-immortalized keratinocytes. Oncogene. 1994;9:387–394. [PubMed] [Google Scholar]
- 54.Venner T J, Sauder D N, Feliciani C, McKenzie R C. Interleukin-8 and melanoma growth-stimulating activity (GRO) are induced by ultraviolet B radiation in human keratinocyte cell lines. Exp Dermatol. 1995;4:138–145. doi: 10.1111/j.1600-0625.1995.tb00237.x. [DOI] [PubMed] [Google Scholar]
- 55.von Knebel Doeberitz D M, Rittmuller C, zur Hausen H, Durst M. Inhibition of tumorigenicity of cervical cancer cells in nude mice by HPV E6–E7 anti-sense RNA. Int J Cancer. 1992;51:831–834. doi: 10.1002/ijc.2910510527. [DOI] [PubMed] [Google Scholar]
- 56.Webster G A, Perkins N D. Transcriptional cross talk between NF-κB and p53. Mol Cell Biol. 1999;19:3485–3495. doi: 10.1128/mcb.19.5.3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Woodworth C D, Bowden P E, Doniger J, Pirisi L, Barnes W, Lancaster W D, DiPaolo J A. Characterization of normal human exocervical epithelial cells immortalized in vitro by papillomavirus types 16 and 18 DNA. Cancer Res. 1988;48:4620–4628. [PubMed] [Google Scholar]
- 58.Woodworth C D, Waggoner S, Barnes W, Stoler M H, DiPaolo J A. Human cervical and foreskin epithelial cells immortalized by human papillomavirus DNAs exhibit dysplastic differentiation in vivo. Cancer Res. 1990;50:3709–3715. [PubMed] [Google Scholar]
- 59.zur Hausen H, de Villiers E M. Human papillomaviruses. Annu Rev Microbiol. 1994;48:427–447. doi: 10.1146/annurev.mi.48.100194.002235. [DOI] [PubMed] [Google Scholar]
- 60.zur Hausen H, Rosl F. Pathogenesis of cancer of the cervix. Cold Spring Harbor Symp Quant Biol. 1994;59:623–628. doi: 10.1101/sqb.1994.059.01.071. [DOI] [PubMed] [Google Scholar]