Abstract
U7 snRNA is part of the U7 snRNP complex, required for the 3′ end processing of replication-dependent histone pre-mRNAs in S phase of the cell cycle. Here, we show that U7 snRNA plays another function in inhibiting the expression of a subset of long terminal repeats of human endogenous retroviruses (HERV1/LTR12s) and LTR12-containing long intergenic noncoding RNAs (lincRNAs), both bearing sequence motifs that perfectly match the 5′ end of U7 snRNA. We demonstrate that U7 snRNA inhibits LTR12 and lincRNA transcription and propose a mechanism in which U7 snRNA hampers the binding/activity of the NF-Y transcription factor to CCAAT motifs within LTR12 elements. Thereby, U7 snRNA plays a protective role in maintaining the silencing of deleterious genetic elements in selected types of cells.
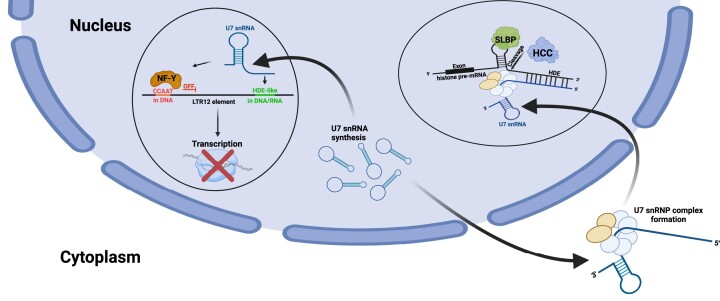
Graphical Abstract
Graphical Abstract.
Introduction
U7 snRNA (U7 small nuclear RNA) is synthetized in metazoan cells by RNA polymerase II (RNAP2) and reaches a length of 63 nucleotides in humans. The 3′ region of U7 snRNA (27 nucleotides) forms a stem−loop secondary structure required for its stability, while the central part is occupied by a noncanonical sequence at the Sm-binding site (5′-AUUUGUCUAG-3′). In the mature U7 snRNP (small nuclear ribonucleoprotein) complex, the Sm site is wrapped by the Sm/Lsm heptameric protein core, which consists of five proteins shared with other U snRNPs (SmB/B’, SmD3, SmE, SmF and SmG) and two unique proteins (Lsm10 and Lsm11) (1–3). U7 snRNA and proteins are assembled into the complex in the same maturation pathway as spliceosomal U snRNPs. Thus, after transcription, the U7 snRNA precursor is first exported to the cytoplasm, where it combines with the core proteins and is further processed. Next, the particle is reimported into the nucleus and remains mostly concentrated in subnuclear sites called histone locus bodies (HLBs) throughout the cell cycle (3–8).
In contrast to other U snRNPs, mostly involved in splicing, U7 snRNP is a key factor involved in the 3′ end processing of replication-dependent histone (RDH) pre-mRNAs (9). During this unique maturation event, which takes place in S phase of the cell cycle within microenvironment of HLBs, RDH pre-mRNAs undergo a single endonucleolytic cleavage after a specific stem−loop structure located at their 3′ ends, which is recognized by stem−loop binding protein (SLBP) (7,10–12). Downstream of the stem−loop structure, there is a conserved purine-rich sequence known as the histone downstream element (HDE), which is highly complementary to the 5′ end of U7 snRNA (5′-UUACAGCUCUUU-3′, with the core being the last 6 nucleotides) (5,13,14). The base-pair binding of U7 snRNP to the HDE aids in the recruitment of other factors involved in processing that form the histone cleavage complex (HCC) (15). Cleavage occurs between the 3′ stem−loop and the HDE and is catalyzed by the endonuclease CPSF73. Cleavage releases mature histone transcripts, and there is no polyadenylation step involved (6,12,16). Additionally, U7 snRNA has previously been described as a negative transcriptional regulator. U7 snRNA interacts with the transcription factor NF-Y and inhibits its binding to the promoter region of the MDR1 (multidrug resistance gene 1) gene encoding P-glycoprotein that confers the drug resistance of cancer cells via ATP-dependent extrusion of different drugs from the animal cells (17). Binding between NF-Y and U7 snRNA has been shown to be specific; however, the molecular mechanism of this interaction is still unclear (18).
NF-Y is an ubiquitously expressed heterotrimeric transcription factor (TF) consisting of the subunits NF-YA, NF-YB and NF-YC (19). It specifically recognizes the CCAAT motif in forward and reverse orientation at proximal promoters and cell-type-specific enhancers located away from transcription start sites (20,21). NF-YA contains DNA-binding and transactivation domains and is responsible for sequence-specific DNA contacts, while NF-YB/NF-YC contact DNA nonspecifically (19). Unlike most TFs, NF-Y is capable of binding to its DNA motif within closed, transcriptionally inactive chromatin, and therefore it has been characterized as a bifunctional TF, activator or repressor, associated with positive or negative histone marks (22).
Here, we propose another function of U7 snRNA in human cells related to its negative regulation of NF-Y, not linked to the cell cycle. We suggest that U7 snRNA inhibits the transcription of specific long terminal repeats (LTRs) of human endogenous retroviruses (HERVs) and long intergenic noncoding RNAs (lincRNAs), both of which contain U7 snRNA-complementary sequences and CCAAT motifs recognized by NF-Y.
HERVs belong to the ‘retrotransposon’ class of transposable elements (TEs), which constitute approximately half of the human genome. They originated from mobile DNA integration events that occurred in the past; however, TEs have now mostly lost the ability to undergo transposition (23). There are ∼500 000 copies of HERV loci in the human genome, and they account for ∼8% of the genome (24). A full-length HERV consists of two long terminal repeats (LTRs) and open reading frames (GAG, POL and ENV). However, 90% of them exist as solitary LTRs, which nevertheless may contain active regulatory sequences such as promoters, enhancers, splice sites, and polyadenylation signals. Therefore, despite being transposition-inactive, LTRs can regulate the expression of other genes (24,25). Moreover, many HERVs are transcribed to form long noncoding RNAs (lncRNAs), and most lncRNAs contain TE sequences with enrichment of LTR elements (25–27). LncRNAs are transcripts longer than 200 nucleotides, and almost half of them (∼15 000 genes) belong to the lincRNA class, whose sequences are located intergenically. LincRNAs can be transcribed by RNAP2, spliced, alternatively spliced, capped and polyadenylated. They play various roles in gene expression, in both the nucleus and the cytoplasm, as shown at the levels of epigenetics, transcription, and translation (28–33).
HERVs and lincRNAs are generally expressed in a spatiotemporally modulated manner, rather than constitutively. As both act in complex gene regulatory networks, aberrant levels of HERVs and lincRNAs are often associated with human diseases (28,34–37). Therefore, cells need to tightly control their expression. In this study, we show that U7 snRNA mediates this regulatory pathway through the transcription factor NF-Y. By this discovery, we describe a new function of U7 snRNA in the cell nucleus, in addition to its role in S phase of the cell cycle within the U7 snRNP complex. Furthermore, we suggest a novel mechanism of regulation of HERV LTR and lincRNA expression in human cells. In HEK293T, HeLa and SH-SY5Y cells, U7 snRNA blocks the expression of specific LTRs belonging to the HERV1 family – LTR12s and a subset of lincRNAs. In this mechanism, U7 snRNA interacts with complementary motifs present within LTR12s and lincRNAs containing LTR12 elements. This may in turn attenuate the activity and/or binding of the transcription factor NF-Y to the CCAAT motifs located in the same elements, leading to transcriptional inhibition. In this model, U7 snRNA plays a protective role in maintaining the silenced state of these genetic elements in numerous somatic cell types.
Materials and methods
Cell cultures and transfection
HEK293T, HeLa and SH-SY5Y cells were grown in Dulbecco's modified Eagle's medium with l-glutamine and 4.5 g/l glucose (DMEM; Lonza or Biowest) supplemented with 10% fetal calf serum (Gibco) and antibiotics (100 U/ml penicillin, 100 μg/ml streptomycin, 0.25 μg/ml amphotericin B (Sigma-Aldrich)) at 37°C in a humid atmosphere containing 5% CO2. For differentiation into neuron-like cells, SH-SY5Y cells were treated with 75 μM retinoic acid (USP, Tretinoin, 167400) for 10 days (38), with medium replacement every 3 days. The differentiation efficiency was analyzed by measuring the abundance of the MAP2 protein or the mRNA levels of two differentiation markers, RARB and MYC (39).
For U7 snRNA depletion, the chemically modified chimeric ASO (40) was introduced to the cells at a 100 nM concentration using Lipofectamine 2000, and GFP ASO was used as a control (Supplementary Table S1). For NF-Y or Lsm10 knockdown, HEK293T cells were transfected with 20 nM siRNA against NF-YA, NF-YB, NF-YC (Santa Cruz Biotechnology), 50 nM siRNA against Lsm10 (Merck, sequence from (41)) or corresponding concentration of universal negative control #1 siRNA (Merck) using Lipofectamine 2000 reagent (Thermo Scientific) according to the manufacturer's instructions. The cells were harvested and analyzed 48 h (NF-Y knockdown) or 72 h (Lsm10 knockdown) posttransfection; knockdown efficiencies were verified by RT−qPCR, Northern blotting and Western blotting. For overexpression experiments, HEK293T cells were transfected with pcDNA3.1(+)-LINC01554, pcDNA3.1(+)-ARRDC4-1 and pcDNA3.1(+)-ADCYAP1-2 plasmids using Lipofectamine 2000. Cells were harvested 24 h posttransfection. Overexpression efficiencies were verified by RT−qPCR.
HEK293T cells with mutations in HDE-like elements of lnc-ADCYAP1-2 (AP000829.1) (two out of four elements) and lnc-ARRDC4-1 (AC024651.2) (three elements) genes were generated according to (42) using SpCas9-2A-Puro (PX459) plasmids encoding the sgRNA 5′-CACACTTTCAGAATGTGACG-3′ or 5′-TTAGTGGGTTTGTAATCTCG-3′, a donor plasmid for HDR (homology-directed repair), and Lipofectamine 2000. Twenty-four hours after transfection puromycin selection at a concentration of 2 μg/ml was applied for 5 days. Clonal cell lines were isolated by serial dilution. Subsequently, colonies grown from single cells were picked, and gDNA was isolated for clone screening using the ExtractMe Genomic DNA Kit (Blirt). The selected clones were genotyped using DreamTaq DNA polymerase (Thermo Scientific) and primers located outside the region spanned by the homology arms to avoid false detection of residual repair template. Lnc-ARRDC4-1 HDE-mut cell line is heterozygous with only one allele carrying desired mutations in all three HDE-like motifs. In the case of the lnc-ADCYAP1-2 HDE-mut cell line, two HDE-like motifs located in the first exon of lnc-ADCYAP1-2 were edited in both alleles.
RNA extraction, cDNA synthesis, PCR and RT−qPCR
Total RNA from normal human tissues was obtained from Clontech (636643). Total RNA was isolated from cells using TRIzol reagent (0.8 M guanidine thiocyanate, 0.4 M ammonium thiocyanate, 0.1 M sodium acetate pH 5.0, 5% v/v glycerol, 38% v/v saturated acidic phenol) and the Direct-zol RNA MiniPrep Kit (including on-column DNase I treatment, Zymo Research, R2052) or according to the protocol for TRIzolTM Reagent (Invitrogen). For the latter samples, 10 μg of RNA was treated with 2 U TURBO DNase (Ambion) for 40–60 min at 37°C followed by standard phenol−chloroform extraction and ethanol precipitation. First-strand cDNA was synthesized in 20 μl reactions with 1–3 μg of RNA using 200 ng of random hexamer primers and 200 U of Superscript III reverse transcriptase (Thermo Scientific) according to the manufacturer's protocol. Semiquantitative PCR amplification was performed using DreamTaq DNA Polymerase (Thermo Scientific) and gene-specific oligonucleotide primer pairs. For qPCR, 1 μl of 2–5× diluted cDNA template, 0.2 μM primer mix, and 5 μl of SYBR Green PCR master mix (Applied Biosystems) were mixed in a 10 μl reaction with the following conditions: denaturation for 10 min at 95°C, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min (Applied Biosystems QuantStudio 6 Flex). For testing U7 snRNA levels in U7 KD cells, cDNA was synthesized in a coupled polyadenylation reverse transcription reaction as described in (43) (Figure 1B and C) or using random hexamers (Figure 1G and H, Supplementary Figures S1A, S5A, S8, S11). The statistical significance of the RT−qPCR results was determined by Student's t test and was calculated using relative values. The primers used for PCR and RT−qPCR are listed in Supplementary Table S2. The graphs were generated using tidyverse collection of R packages (https://cran.r-project.org/web/packages/tidyverse/index.html), along with ggbreak (https://cran.r-project.org/web/packages/ggbreak/index.html).
Figure 1.
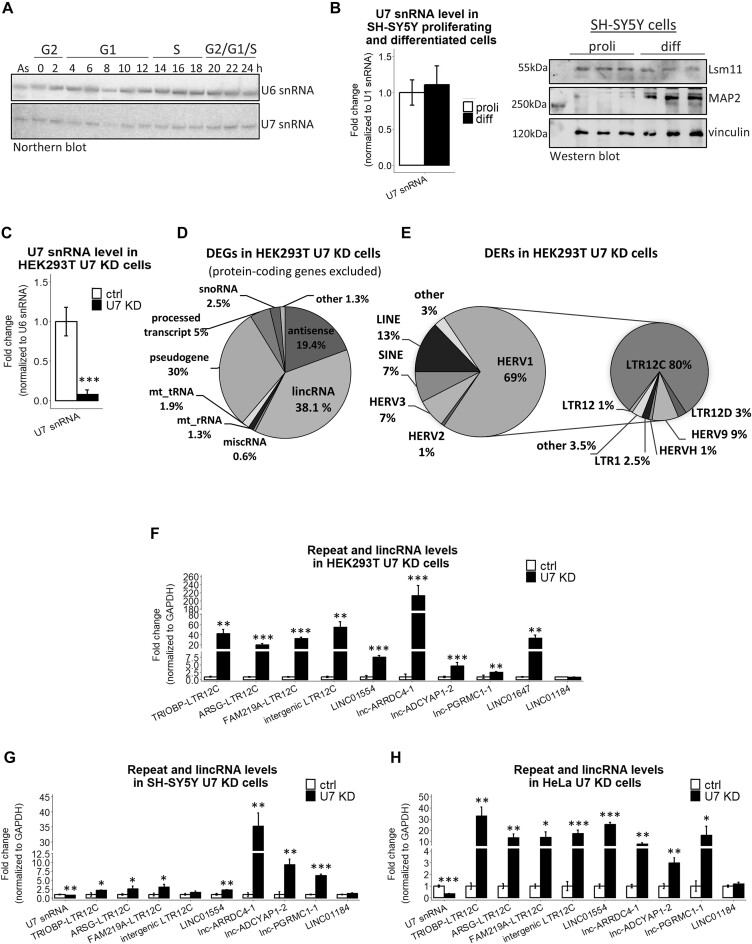
The expression level of U7 snRNA is inversely correlated with the expression of LTR12s and lincRNAs. (A) The levels of U7 snRNA in particular phases of the cell cycle were analyzed by Northern blotting, and U6 snRNA served as a loading control. The synchronization of HeLa cells was performed as described in (Figure 3A in (43)), and cell cycle phases at the indicated time points after release from nocodazole block are presented at the top. (B) U7 snRNA was detected by RT-qPCR, U1 snRNA levels were used for normalization. The level of Lsm11 protein was checked by Western blotting and immunodetection in proliferating (proli) and differentiated (diff) SH-SY5Y cells. MAP2 was used as a differentiation marker, and vinculin was used as a loading control. (C) The U7 snRNA knockdown efficiency in HEK293T cells treated with ASO targeting U7 snRNA (U7 KD) in comparison to control ASO (ctrl) was checked by RT−qPCR. U6 snRNA levels were used for normalization. (D, E) Pie charts showing differentially expressed genes (DEGs) (D) and differentially expressed repeats (DERs) (E), with special consideration of the HERV1 class in U7 KD cells. For clarity, protein-coding genes were omitted from the DEG analysis. (F–H) The expression levels of selected LTR12s and lincRNAs were analyzed in HEK293T (F), proliferating SH-SY5Y (G) and HeLa (H) cell lines, ctrl cells and U7 KD cells by RT−qPCR. LINC01184 was not identified as a DEG in the RNA-seq data and was used as a negative control. GAPDH levels were used for normalization. Data are presented as means ± SDs (n = 3). P values were calculated using two-sided Student's t test, and statistical significance was defined as follows: *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
RNA immunoprecipitation
Nuclear extracts from ∼6 × 106 HEK293T cells, prepared as described in (43), were subjected to RNA immunoprecipitation (RIP) using 2 μg of anti-NF-YA (Santa Cruz Biotechnology, sc-17753) or mouse IgG (Santa Cruz Biotechnology, sc-2025). Protein extracts were incubated with 25 μl of Dynabeads Protein G (Invitrogen) for 2 h at 4°C. After washing the immunoprecipitated complexes 3× with TBS-0.05% NP-40, coprecipitated RNAs were eluted from the beads with TRIzol, precipitated with ethanol, treated with TURBO DNase, and used for cDNA synthesis with hexamer primers followed by qPCR, as described above. As an input, 10% of the extract used for IP was directly added to TRIzol.
Northern blotting
For the Northern blot analysis, 30 μg of total RNA was used to detect U7 snRNA. RNA electrophoresis, blot transfer and hybridization were performed as previously described (44). The U6 snRNA hybridization signal was used as a loading control. The sequences of the hybridization probes are listed in Supplementary Table S1.
Metabolic labeling of nascent RNA with 4sU and nascent RNA purification
Metabolic labeling of newly transcribed RNA was performed as described in (45) with some modifications. Cells were treated with 500 μM 4sU (POL-AURA, PA-03-6085-N#100MG) for 15 min. Nascent RNAs were eluted twice with 100 μl of fresh 0.1 M dithiothreitol (DTT) and precipitated overnight with 2 μl of GlycoBlue (15 mg/ml), 20 μl of 3 M sodium acetate pH 5.2 and 600 μl of ice-cold ethanol. The precipitated RNA was centrifuged for 30 min at 16 000×g. The pellet was washed twice with 75% ethanol, centrifuged for 10 min at 16 000×g, air-dried and resuspended in RNase-free water. The RNA obtained was reverse transcribed into cDNA as described above. The levels of newly transcribed RNAs were analyzed by RT−qPCR, as described above.
Western blotting and immunodetection
For histone extraction, cells were lysed on ice in TEB buffer (PBS containing 0.5% Triton X-100, 2 mM PMSF, 0.02% sodium azide), centrifuged and washed with TEB buffer. Then, the pellet was resuspended in 0.2 N HCl and incubated overnight at 4°C with gentle stirring. Protein concentration in the supernatant obtained was determined using Bradford assay. Protein extraction, SDS-polyacrylamide gel electrophoresis (SDS−PAGE) and immunodetection were performed as described in (46) using the following antibodies: anti-actin (MP Biomedicals, 691001), anti-NF-YA (Santa Cruz Biotechnology, sc-17753), anti-NF-YB (Santa Cruz Biotechnology, sc-376546), anti-EWSR1 (Santa Cruz Biotechnology, sc-48404), anti-Lsm11 (Abcam, ab201159), anti-MAP2 (Cell Signaling, 8707), anti-vinculin (Thermo Scientific, MA5-11690), anti-H2A (Santa Cruz Biotechnology, sc-10807), anti-H3 (Santa Cruz Biotechnology, sc-10809), anti-H4 (Santa Cruz Biotechnology, sc-8658-R) and anti-Lsm10 (St John's Laboratory, STJ196051).
Plasmid construction
Plasmid-based donor repair templates for CRISPR were prepared in the pGEM-T Easy vector (Promega). Homology arms (each approximately 800–1000 bp) flanking the sites of alterations (HDE-like motifs) were amplified from the genomic DNA using CloneAmp HiFi PCR Premix (Clontech), A-tailed with DreamTaq DNA polymerase and ligated into the pGEM-T Easy vector. The mutagenesis of the HDE-like motifs and PAM sequence within the donor plasmid for HDR was performed using the QuikChange II Site-Directed Mutagenesis Kit (Agilent Technologies). pcDNA3.1(+)-ARRDC4-1 and pcDNA3.1(+)-ADCYAP1-2 expression vectors were constructed by cloning lnc-ARRDC4-1 and lnc-ADCYAP1-2 sequences into the pcDNA3.1(+) vector by using the NheI, XbaI and NheI, ApaI restriction sites, respectively. The pcDNA3.1(+) vector containing the LINC01554 sequence was kindly provided by Professor Xin-Yuan Guan (47). The primers used for the construction of all vectors are listed in Supplementary Table S2.
Chromatin immunoprecipitation
Chromatin immunoprecipitation was performed as described in (39) with some modifications. A total of 225 μl of chromatin (∼5 × 106 cells equivalent), diluted in a 1:5 ratio using dilution buffer, was used for IP with NF-YA or NF-YB antibodies. IP with only beads served as a negative control. One tenth of the chromatin used for immunoprecipitation was used as an input.
Flow cytometry
The synchronization of HeLa cells in G2/M was performed by the addition of 200 ng/ml nocodazole (Sigma−Aldrich) to the medium for 18 h. After synchronization, the cells were washed twice with PBS and then collected 2, 4, 6, 8, 10, 12, 14, 16, 18, 20, 22 and 24 h after release, as described in (43). For cytofluorometric analysis, the cells were stained with propidium iodide as described in (48). Cell cycle profiles were analyzed with a Guava easyCyte System (Merck Millipore) flow cytometer, and the data were processed with InCyte Software (utilities from guavaSoft 3.1.1).
Total RNA-seq library preparation
One microgram of RNA was first subjected to the rRNA depletion step using RiboCop rRNA Depletion Kit, and next, libraries were prepared using the SENSE Total RNA-Seq Library Preparation Kit (Lexogen) following the manufacturer's instructions. An Agilent High Sensitivity DNA Kit (Agilent) was used to assess library quality on an Agilent Bioanalyzer 2100, and the libraries were quantified using a Qubit dsDNA HS Assay Kit (Invitrogen). RNA sequencing was performed for 125 bp paired-end reads using an Illumina HiSeq 2500 platform at the Lexogen NGS facility.
Bioinformatic analysis
Differential expression of repeats and HDE-like motifs: a quality check was performed with FastQC v0.11.5 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) and read processing and quality control were performed with BBDUK2 v37.02 from the BBMAP package (sourceforge.net/projects/bbmap/). Then, reads were mapped against the human genome version GRCh38 using STAR v2.5.3a_modified (49). Here, due to the nature of repetitive elements, the reads were required to map uniquely (–outFilterMultimapNmax 1 parameter was applied). Additionally, human genome annotations in a GTF format from ENSEMBL 109 were used to improve the quality of read mapping. The resulting BAM files were then input to the featureCounts v1.5.0-p1 program from Subread software (50) to calculate the raw expression values of the repeats. Finally, the calculated expression values were subjected to differential expression analysis using DESeq2 v1.34.0 from R/Bioconductor (51), requiring the adjusted P value to be below 0.05.
Differential expression of genes: after checking read quality with FastQC, the reads were subjected to processing with BBDUK2. Additionally, reads that mapped to human ribosomal RNAs were discarded using Bowtie 2 v2.3.5.1 (52). Then, SALMON was used to estimate the expression values of genes and transcripts, which was followed by the assessment of differential gene expressions using DESeq2.
Human repetitive elements were downloaded from UCSC Genome Browser via Table Browser utility (track: RepeatMasker, genome: GRCh38, format: BED). The coordinates of HERVs, LTR12s and other repetitive elements were then obtained by parsing the file using scripting languages. HDE-like elements were identified using a custom Python script that scanned both strands of the human genome (GRCh38), allowing up to two mismatches compared to the following consensus motif: AAGAGCTGTAACACT.
HDE-like motif frequency assessment: the human genome (GRCh38) was scanned in search for HDE-like motifs, resulting in finding 21 168 of them. Scanning for a shuffled genomic sequence of HDE-like motif (the same nucleotide composition but a different order) led to the identification of only 5 489 such motifs.
HDE-like conservation analysis: phyloP scores for the human genome were downloaded from (http://hgdownload.cse.ucsc.edu/goldenpath/hg38/phyloP100way/hg38.phyloP100way.bw), converted into a wig format with bigWigToWig utility (http://hgdownload.cse.ucsc.edu/admin/exe/), and finally to a BED format with wig2bed program from BEDOPS package (53), which resulted in a table that stores conservation scores at a single nucleotide resolution. Next, a custom Python script was used to calculate and compare conservation scores assigned to HDE-like elements (mean = 0.064), LTR12 elements (mean= −0.068) and exons of protein-coding genes (mean = 1.08) as a positive control. A Mann–Whitney test showed that the differences between each group are statistically significant (P value ≤ 10e-5).
Analysis of RNA-seq read coverage around HDE-like elements: the BAM files resulting from uniquely mapped RNA-seq reads were processed with bamCoverage from deepTools v2.5.7 (54) to obtain local coverage near those HDE-like elements located inside LTR12 classes of repetitive elements. The results were then parsed and plotted with a custom Python script using the Matplotlib library (https://matplotlib.org/).
Strand-specificity analysis: expression values of U7 snRNA-dependent LTR12 elements were calculated with RSEM 1.2.30 (55) using a –forward-prob 0 parameter to account for RNA-seq data strandedness. The calculations were performed twice - with LTR12 sequences in a sense orientation as well as using their reverse complement variants. This gave insight into profiles of their expression and numbers of mapped reads in both sense and antisense orientations.
Results
U7 snRNA regulates the expression of LTR12s and lincRNAs in human cells
The function of U7 snRNA is closely connected to the replication phase of the cell cycle, where it plays a crucial role in the 3′ end processing of RDH pre-mRNAs. However, the unique components of the U7 snRNP complex are detectable throughout the cell cycle, including U7 snRNA (Figure 1A) (56,57) and Lsm11 (43). In nondividing, differentiated cells, U7 snRNA, Lsm11 (Figure 1B) (58,59) and Lsm10 (58) are also present, despite the fact that these cells are no longer capable of cell division. These results suggest that U7 snRNA/snRNP plays other roles in a cell. To address this possibility, we knocked down U7 snRNA in HEK293T cells (U7 KD cells) using an antisense oligonucleotide. The knockdown efficiency reached 90%, as verified by RT−qPCR and Northern blotting, and resulted in inefficient 3′ end processing of RDH transcripts (Figure 1C, Supplementary Figure S1A−C). At the same time, histone protein levels remained unchanged in U7 KD cells (Supplementary Figure S1D). This is consistent with the high stability of the histone proteins and previously reported results showing that histone mRNA levels do not correspond with histone levels and suggesting that the U7 snRNP-mediated control of histone production is not the only mechanism that guarantees an adequate supply of these proteins during the S phase of the cell cycle (41). Thereafter, we analyzed the transcriptomes of the cells by high-throughput sequencing (RNA-seq) and found that U7 snRNA depletion resulted in the deregulation of hundreds of genes (DEGs, differentially expressed genes), mainly protein-coding genes and lncRNAs, with predominance of the lincRNA class (Figure 1D, Supplementary Table S3). Subsequently, using parameters provided in Materials and Methods, we uniquely mapped sequencing reads to genomic regions annotated as repeated sequences and we observed the differential expression of 250 regions annotated as repeat elements (DERs, differentially expressed repeats), with enrichment of the HERV1 family. Further analysis revealed that 84% of these HERV1s are solitary LTR12 elements, with overrepresentation of LTR12C (Figure 1E, Supplementary Table S4). Interestingly, transposable element derived sequences are also present in the majority of human lncRNAs, with specific enrichment of HERV LTRs (27,60,61). Guided by these findings, we analyzed the composition of lincRNAs affected by U7 snRNA knockdown (hereafter referred to as U7-dependent for simplicity) and we found that 50% of them contained LTR12 elements of the HERV1 family.
Among U7-dependent DERs and DEGs, all LTR12s and almost 90% of lincRNAs were upregulated, suggesting an inhibitory effect of U7 snRNA on these genomic regions in wild-type cells. These results were confirmed for randomly selected examples by RT−qPCR (Figure 1F). The same observations were made in SH-SY5Y and HeLa cell lines with U7 snRNA knockdown, suggesting a more common phenomenon, regardless of the cell type used (Figure 1G, H).
Furthermore, we noticed that U7-dependent repeats are either transcribed as relatively short independent RNA molecules from intergenic regions of the genome (in total they account for ∼55% of DERs) (Figure 2A) or are located within other genes (mainly in introns) (Figure 2B). In the case of intragenic repeats, the hosts were mainly protein-coding genes (52%) and lincRNAs (39%). By comparing the U7-dependent DERs with U7-dependent DEGs, we found that in only 22% of cases the activation of intragenically located repeats was accompanied by elevated expression of the host protein-coding gene transcripts. This observation was in consistence with previous results of RT−qPCR showing the specific upregulation of LTR12C elements located in introns of TRIOBP, ARSG and FAM219A genes in HEK293T U7 KD cells (Figure 1F, Supplementary Figure S2), with little or no change in the relative abundance of mature mRNAs and different regions of the host introns (Supplementary Figure S3). This suggests that such LTR12 elements may serve as promoters that drive the expression of short intronic RNAs, independently from their host genes. In the case of U7-dependent lincRNAs, the expression of either the LTR12 element or the full-length transcript was upregulated in U7 KD cells, as exemplified in Figure 2C.
Figure 2.
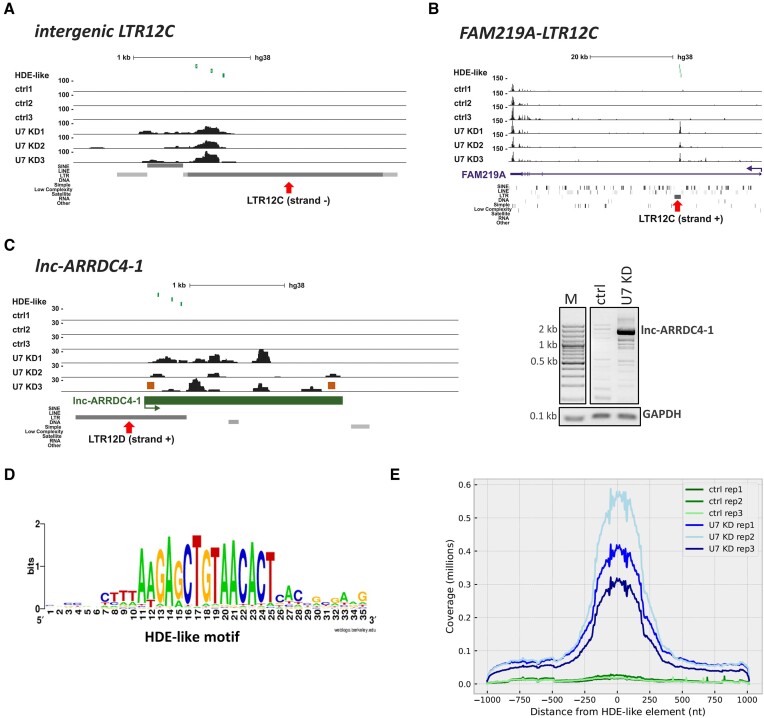
U7 snRNA regulates the expression of LTR12s and lincRNAs containing HDE-like motifs. (A–C) UCSC Genome Browser visualization of RNA-seq data from HEK293T ctrl and U7 KD cells (three biological replicates are shown) of the intergenic LTR12C element (A), the intragenic LTR12C element in FAM219A (B) and the LTR12-containing lincRNA lnc-ARRDC4-1 (C). Peaks within data tracks illustrate reads mapped to particular genomic regions. The positions of HDE-like motifs are shown in the top track as green vertical lines. The red arrows indicate the position of the LTR12 elements within the gene structures. The expression of full-length lnc-ARRDC4-1 was checked by semiquantitative PCR using primers depicted as orange boxes, and GAPDH was used for normalization (C, on the right). (D) Consensus sequence of the HDE-like motif present within HERVs in the context of the 10 nucleotides flanking the motif on both sides. (E) Graph showing the distribution of reads around HDE-like motifs within LTR12 elements present in the human genome. The Y axis represents the cumulative number of reads assigned to a corresponding position around the HDE-like elements, which are placed at position ‘0′.
U7 snRNA acts through HDE-like sequence motifs located in LTR12s and LTR12-containing lincRNAs
The above results imply that U7 snRNA may play a role in maintaining the silencing of transposable element derived sequences, such as LTR12s, in specific cells. Since small RNAs operate mainly via specific base pairing with target RNAs, we screened U7-dependent repeats to search for U7 snRNA-complementary regions (the entire U7 snRNA sequence was used as a query). This resulted in the identification of a sequence motif of 15 nucleotides in length exhibiting high complementarity (1–2 mismatches allowed) to the 5′ end of U7 snRNA (nucleotides 2–16 of U7 snRNA) (Figure 2D). Due to the analogy of this motif to the one in RDH genes, we called it the ‘HDE-like motif’. However, it should be pointed out that this motif is not present in any histone pre-mRNAs, since it is longer than the HDE. Further analysis showed that the HDE-like motif is present within the sequence of all U7-dependent LTR12s or is located in close proximity (from 1 bp to 15 kb, most frequently closer than 1kb) to the vast majority of differentially expressed repeats from other classes, such as LINEs (long interspersed nuclear elements) or SINEs (short interspersed nuclear elements). Similarly, U7-depenedent lincRNAs contain HDE-like motifs embedded within their LTRs. Importantly, in the majority of these lincRNAs, HDE-like motifs are present in several copies, predominantly at the beginning of the genes (in the first exons or the first introns) and perfectly or almost perfectly (1–2 mismatches) matched the 5′ end of U7 snRNA (Table 1). Notably, within the human genome, the HDE-like motif is overrepresented in repeated sequences, with a considerable prevalence of LTR12 elements of the HERV1 family (81% of all HERVs) (Table 2). Our further bioinformatic analyses revealed that HDE-like motifs in the total number of 21 168 copies (with 0 (3 150 copies), 1 (6 900 copies) and 2 (11 118 copies) mismatches) are present in the genome at frequencies higher than expected by chance and show higher evolutionary conservation than the rest of the sequence of LTR12 elements in the human genome. In summary, the high abundance of HDE-like motifs in the human genome is due to its prevalence in repetitive elements of the LTR12 class and supports its biological relevance.
Table 1.
HDE-like motif occurrence in the selected U7-dependent lincRNAs
| Gene ID | Gene name | Length | Exons | HDE-like motifs (number, localization, size, sequence similarity to U7 snRNA) |
|---|---|---|---|---|
| ENSG00000230392 | lnc-PGRMC1-1 | 2907 nt | 4 | 5 in exon 4: 2(15 nt, 100%), 2(15 nt, 93%), 1(15 nt, 87%);4 in intron 1: 3(15 nt, 100%), 1(15 nt, 87%, strand -) |
| ENSG00000263567 | lnc-SUZ12-3 | 505 nt | 2 | 3 in exon 1: 1(15 nt, 100%), 1(15 nt, 93%), 1(15 nt, 87%); 4 additional in close downstream region of the gene |
| ENSG00000259870 | lnc-ARRDC4-1 | 2098 nt | 1 | 3 in exon 1: 1(15 nt, 93%), 2(15 nt, 87%) |
| ENSG00000249790 | lnc-RIMKLB-6–3 | 1972 nt | 2 | 3 in exon 1: 2(15 nt, 100%), 1(15 nt, 93%); 3 additional immediately downstream of the gene |
| ENSG00000263551 | lnc-ADCYAP1-2 | 1132 nt | 8 | 2 in exon 1: (15 nt, 93%);2 in intron 1: 1(15 nt, 93%, strand -), 1(15 nt, 87%, strand -) |
| ENSG00000236882 | LINC01554 | 2052 nt | 4 | 3 in exon 1: 1(15 nt, 100%), 1(15 nt, 93%), 1(15 nt, 87%); 1 intron 3: 1(15 nt, 87%, strand -) |
| ENSG00000235643 | LINC01647 | 1351 nt | 3 | 3 in exon 1: 1(15 nt, 100%), 2(15 nt, 93%) |
Gene names according to LNCipedia. Sequence similarity to U7 snRNA is perfect (100%) or includes 1 (93%) or 2 (87%) mismatches.
Table 2.
HDE-like motif distribution across the human genome (hg38)
| HDE-like motifs present in the genome | HDE-like motifs embedded in all repeats | HDE-like motifs embedded in HERVs | HDE-like motifs embedded in LTR12s | |
|---|---|---|---|---|
| Number | 21 168 | 16 890 | 14 449 | 11 685 |
| Relative to the genome | 100% | 79.79% | 68.26% | 55.20% |
| Relative to repeats | 100% | 85.55% | 69.18% | |
| Relative to HERVs | 100% | 80. 87% |
The analysis encompasses 15 nucleotide HDE-like motifs (with sequence presented in Figure 2D) with 0, 1 or 2 mismatches allowed.
Subsequently, we asked how many HDE-like motifs are activated after U7 snRNA depletion. First, we used our RNA-seq data and calculated the expression values of HDE-like motifs across the genome. This analysis revealed that 4157 of 21 168 HDE-like motifs are expressed in HEK293T cells, with 3336 motifs expressed exclusively in U7 KD cells, and only 269 with expression confined to control cells. Next, we performed differential expression analysis to learn how many of them respond to U7 snRNA depletion. We found that 527 HDE-like motifs exhibit a statistically significant increase in expression in U7 KD compared to control cells, with only 12 motifs being downregulated (adjusted P value < 0.05). Out of the 527 upregulated HDE-like motifs, 525 overlap LTR12 elements in a sense orientation (+/+ or −/− genomic strands). In general, the distribution of the sequencing reads mapped across all LTR12s containing HDE-like motifs confirmed the higher accumulation of RNA-seq reads around HDE-like motifs in U7 KD cells in comparison to control cells (Figure 2E). The distribution also indicates that HDE-like elements are predominantly transcribed as relatively short intergenic or intragenic transcripts (100–250 nucleotides) from the LTR12 promoters, as exemplified in Supplementary Figure S2. The analysis of stranded RNA-seq data revealed that the reads map predominantly to transcripts in a sense orientation (i.e. the orientation consistent with LTR12 annotations in the genome) in both U7 KD cells and control cells (Supplementary Table S5). This means that U7 snRNA depletion actually resulted in upregulation of LTR12 transcripts containing HDE-like motifs and not their reverse complement variants. Admittedly, in some cases the reads were mapped to the antisense transcripts (bearing a sequence that is reverse complement to the HDE-like element), but their abundance was lower compared to sense transcripts. Altogether, this provides evidence that U7 snRNA regulates the expression of LTR12 elements and its depletion leads to the generation of transcripts that contain a U7 snRNA complementary sequence (i.e. the HDE-like motif).
Considering that lincRNAs can act as molecular sponges (e.g. for miRNAs) (29,31,32,37), we examined whether lincRNAs can regulate the accessibility of U7 snRNA. We observed that the levels of the selected lincRNAs remained unchanged in differentiated cells in comparison to proliferating cells (Supplementary Figure S4), although in the nondividing cells the activity of U7 snRNP is supposed to be dispensable. Furthermore, the overexpression of selected lincRNAs did not change the level of U7 snRNA or influence the efficiency of the 3′ end processing of RDH pre-mRNAs (Supplementary Figure S5A-B), contradicting the hypothesis that the level or accessibility of U7 snRNA is regulated by lincRNAs and indicating it is U7 snRNA that regulates the expression of other genes.
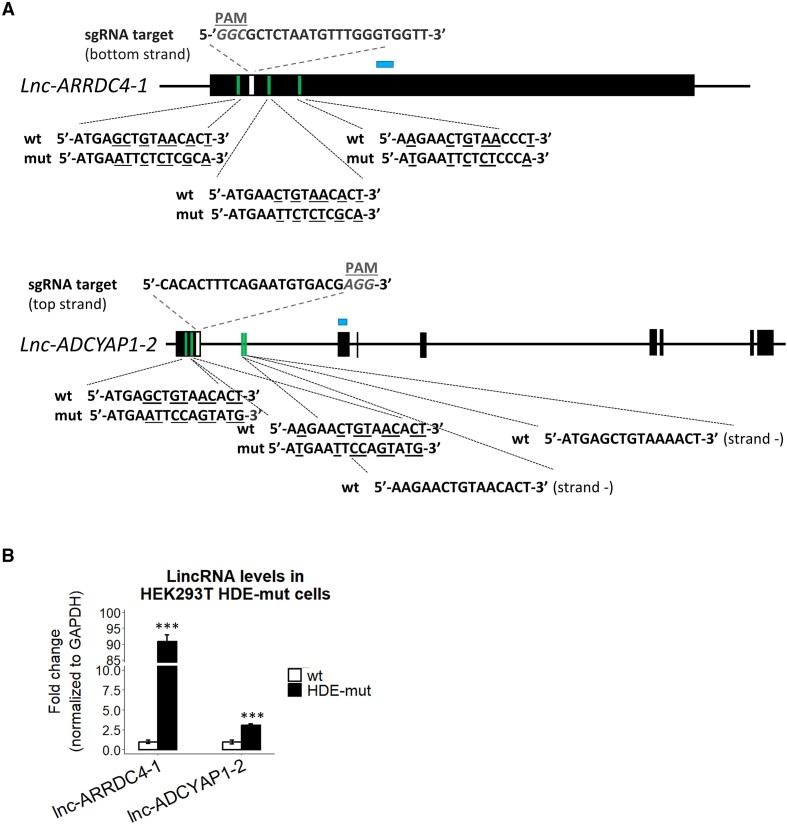
Based on the results mentioned above, we hypothesized that U7 snRNA directly regulates LTR12s and LTR12-containing lincRNAs and does it through base pairing with HDE-like motifs. To test this hypothesis, we used CRISPR−Cas9 technology based on homology directed repair and generated cells in which HDE-like motifs in the lnc-ARRDC4-1 and lnc-ADCYAP1-2 genes were mutated (hereafter called lnc-ARRDC4-1 HDE-mut and lnc-ADCYAP1-2 HDE-mut cells, respectively) (Figure 3A, Supplementary Figure S6A). As shown in Figure 3B, in both types of HDE-mut cells, the corresponding lincRNAs were upregulated regardless of the presence of U7 snRNA in the cells, confirming that this negative regulation relies on a base-pairing interaction between U7 snRNA and HDE-like motifs within the lincRNA sequences. Lnc-ADCYAP1-2 HDE-mut cells were homozygous, however lnc-ARRDC4-1 HDE-mut cell line used in the analysis was heterozygous with only one allele carrying the desired mutations in all three HDE-like motifs. Using primers that specifically amplify either the WT or mutated version of lnc-ARRDC4-1, we found that the mutated allele is predominantly expressed and is responsible for the increased level of this lincRNA in lnc-ARRDC4-1 HDE-mut cells (Supplementary Figure S6B).
Figure 3.
U7 snRNA controls the expression of LTR12s and lincRNAs via HDE-like motifs. (A) Schematic representation of lnc-ARRDC4-1 and lnc-ADCYAP1-2 (isoform ENST00000670380.1). The black boxes represent exons, and the lines depict introns. The positions of HDE-like motifs and sgRNA target sequences used for the generation of HEK293T lnc-ARRDC4-1 HDE-mut and lnc-ADCYAP1-2 HDE-mut cells using the CRISPR−Cas9 system are indicated in green and white, respectively. Nucleotides mutated in the HDE-like motifs are underlined. HEK293T lnc-ARRDC4-1 HDE-mut cells are heterozygous, with one allele corresponding to the wild-type versions of HDE-like motifs and the other carrying desired mutations within HDE-like motifs. Two HDE-like motifs located within the intron of the lnc-ADCYAP1-2 gene (in antisense orientation) were not edited. PAM, protospacer-adjacent motif. (B) The expression levels of the respective lincRNAs in lnc-ARRDC4-1 HDE-mut and lnc-ADCYAP1-2 HDE-mut cell lines were analyzed by RT−qPCR. The locations of the amplified qPCR products in the expression analysis are shown as blue boxes in (A). GAPDH levels were used for normalization. Data are presented as means ± SDs (n = 3). P values were calculated using two-sided Student's t test, and statistical significance was defined as follows: ***P ≤ 0.001.
Next, we addressed the question of whether U7 snRNA alone or the U7 snRNP complex regulates lincRNA and LTR12 expression. To answer this question, we knocked down Lsm10, one of the unique proteins of the U7 snRNP protein core. However, the reduction in Lsm10 mRNA and protein levels to ∼70% (followed by the increase of unprocessed replication-dependent histone pre-mRNAs) did not result in the expected upregulation of U7-dependent LTR12s and lincRNAs (Supplementary Figure S7A-C). This result suggests that U7 snRNA may regulate the expression of HDE-like-containing lincRNAs and LTR12s without the entire U7 snRNP complex.
U7 snRNA regulates the transcription of LTR12s and LTR12-containing lincRNAs through the transcription factor NF-Y
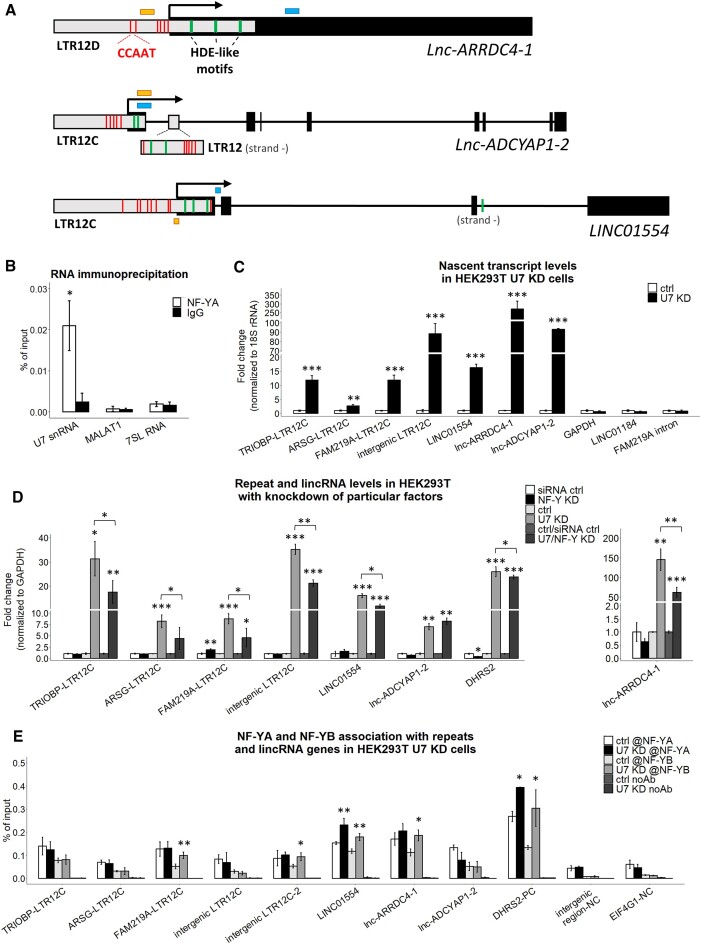
U7 snRNA has been reported to inhibit the activity of the transcription factor NF-Y which is capable of activating or repressing transcription (18). NF-Y is composed of three subunits, NF-YA, NF-YB and NF-YC, all of which are required for binding to a CCAAT motif (19,62–66). CCAAT motifs have been found in tissue-specific enhancers and within proximal promoter regions of the genes. Interestingly, it has been reported that the LTR12 class of transposable elements is abundant in CCAAT motifs as well (20). Likewise, U7-dependent LTR12s and LTR12-containing lincRNAs are rich in CCAAT motifs, usually located near HDE-like motifs within LTR12 elements (Figure 4A). These results suggest a mechanism in which U7 snRNA regulates the levels of LTR12s and lincRNAs by inhibiting their transcription mediated by NF-Y. This was further supported by the RIP experiment in which we confirmed NF-Y:U7 snRNA interaction in HEK293T cells (Figure 4B).
Figure 4.
U7 snRNA regulates the transcription of LTR12s and LTR12-containing lincRNAs. (A) Schematic representation of lnc-ARRDC4-1, lnc-ADCYAP1-2 (isoform ENST00000670380.1) and LINC01554 (isoform ENST00000436592.5). The black boxes represent exons, the gray boxes illustrate LTR12 elements, and the lines represent introns. The positions of the 5′ ends of the genes are indicated by arrows. Positions of the HDE-like and CCAAT motifs (within LTR elements only) are indicated in green and red, respectively. The locations of the amplified qPCR products in the 4sU and ChIP experiments are shown as blue and yellow boxes, respectively. (B) The interaction between U7 snRNA and NF-YA subunit was analyzed using RNA immunoprecipitation followed by RT-qPCR in HEK293T wild-type cells. The abundantly expressed RNAs, MALAT1 and 7SL RNA, were used as negative controls. (C) The levels of newly transcribed LTR12s and lincRNAs in HEK293T U7 snRNA knockdown cells (U7 KD) compared to ctrl cells (ctrl) were tested by the 4sU assay followed by RT−qPCR. The levels of GAPDH, LINC01184 and the FAM219A intron fragment (outside LTR12) were used as negative controls. 18S rRNA was used for normalization. (D) The expression levels of selected LTR12s and lincRNAs in HEK293T cells with combined depletion of U7 snRNA and all NF-Y subunits. GAPDH levels were used for normalization. DHRS2 was used as a positive control. (E) NF-YA and NF-YB binding to LTR12s and lincRNA gene regions containing CCAAT sequences near HDE-like motifs in HEK293T ctrl and U7 KD cells was analyzed by ChIP followed by qPCR. Empty beads were used as a negative IP control. The bars represent the recovery, expressed as a percentage of the input (the relative amount of immunoprecipitated DNA compared to the input DNA after qPCR analysis). DHRS2 was used as a positive control (PC). An intergenic region and the fragment of EIF4G1 gene deprived of CCAAT were used as negative controls (NC). Statistical significance was calculated to assess the association of NF-Y subunits within selected genomic locations between U7 KD and ctrl cells. @, antibody. Data are presented as means ± SDs (n = 3). P values were calculated using two-sided Student's t test, and statistical significance was defined as follows: *P ≤ 0.05; **P ≤ 0.01; ***P ≤ 0.001.
To determine whether this is indeed the case, we measured the transcriptional activity of these genes by 4-thiouridine (4sU) assay. As shown in Figure 4C, the levels of newly synthesized U7-dependent LTR12 RNAs and lincRNAs were significantly higher in U7 KD cells than in control cells. Next, we examined the role of the transcription factor NF-Y in this regulation. First, we knocked down all three subunits of NF-Y in wild-type cells (NF-Y KD) and found no significant alterations in the expression levels of U7-dependent LTR12s and lincRNAs (Figure 4D). In turn, they were highly activated in U7 KD cells, in which NF-Y subunits were present (U7 KD). Then, we measured the levels of U7-dependent LTR12s and lincRNAs in cells with depletion of U7 snRNA followed by the depletion of the whole NF-Y complex (U7/NF-Y KD) and we determined that in most cases their synthesis declined under these conditions compared to the levels observed in U7 KD cells (Figure 4D, Supplementary Figure S8). This finding supports our conclusion that the transcriptional activity of NF-Y on LTR12 and lincRNA genes is unleashed after U7 snRNA knockdown. DHRS2 was used as a positive control in the experiment since it is known to be positively regulated by NF-Y and driven by the LTR12 promoter (67). In addition, to better understand the role of NF-Y in the repression of U7-dependent genes, we performed chromatin immunoprecipitation (ChIP) using antibodies against NF-Y subunits A and B. Primer pairs were designed to amplify gene regions containing CCAAT sequences near HDE-like motifs or the intergenic region and the EIF4G1 gene fragment devoid of CCAAT motifs as negative controls. As shown in Figure 4E, we observed the binding of NF-YA and NF-YB to the majority of LTR12s and lincRNAs within regions containing CCAAT motifs, which is consistent with previously published results (20). Intriguingly, in some cases (FAM219A-LTR12C, intergenic LTR12C-2, LINC01554, lnc-ARRDC4-1), this binding increased in the absence of U7 snRNA, as in the case of the LTR12 promoter region of a positive control (DHRS2-PC). In other cases, such obvious changes were not detected, although corresponding transcription was increased (Figure 4C). We speculate that this could have been due to the partial or full saturation of the given region by NF-Y. When the region is partially saturated by NF-Y (presumably in the case of FAM219A-LTR12C, intergenic LTR12C-2, LINC01554, lnc-ARRDC4-1), after U7 snRNA knockdown extra NF-Y subunits can bind to unoccupied motifs and eventually all of them are activated. In turn, when all CCAAT motifs in a given region are occupied, only NF-Y activity is affected after U7 snRNA knockdown, not the amount, therefore there was no effect on NF-Y binding in the ChIP assay. In summary, these results confirm the role of U7 snRNA as an inhibitor of LTR12 and lincRNA transcription through inhibition of the binding/activity of the transcription factor NF-Y (Figure 5).
Figure 5.
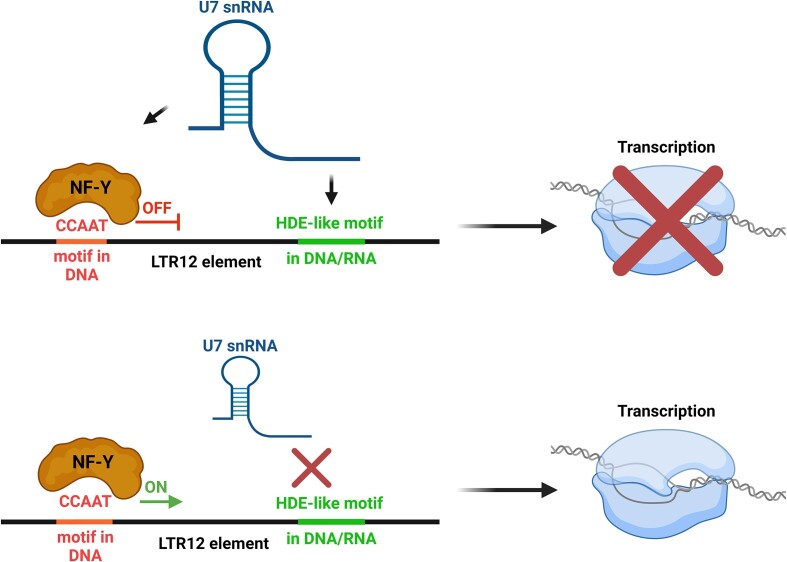
Mechanism of the regulation of LTR12 and LTR12-containing lincRNA expression by U7 snRNA. In wild-type cells, U7 snRNA undergoes base pairing with the HDE-like motif(s) present in the LTR12 element and attenuates the binding/activity of the transcription factor NF-Y, which recognizes the CCAAT motif located nearby. The transcription of LTR12s and LTR12-containing lincRNAs is inhibited (upper panel). In particular conditions (U7 snRNA deficiency, mutations in the HDE-like motif) or cell types, the interaction of U7 snRNA and the HDE-like motif is abolished, and NF-Y activates transcription of LTR12s and LTR12-containing lincRNAs (lower panel). Created with BioRender.com.
The key question arises what is the biological relevance of LTR12 and lincRNA silencing via U7 snRNA. In general, the expression of both HERVs and lincRNAs is tightly regulated and can differ depending on the type of cells and tissues or the stage of development and diseases (31,68,69). According to our results, U7-dependent LTR12 and lincRNA expression is repressed in some cell lines, such as HEK293T, HeLa, and SH-SY5Y cells (Figure 1F–H). However, when we analyzed their levels using the genotype-tissue expression (GTEx) portal, it appeared that their expression predominates in the testis (Supplementary Figures S9 and S10). Furthermore, we supported this trend by examining the expression levels of selected U7-dependent LTR12s and lincRNAs in multiple normal human tissues. In the case of lnc-ARRDC4-1 and LINC01647, testis expression was predominant, whereas TRIOBP-LTR12C, lnc-ADCYAP1-2, and intergenic LTR12C were highly expressed in the testis, cerebellum, or placenta (Supplementary Figure S11). In the same set of samples, the relative levels of U7 snRNA were more stable, with a prominent decrease observed only in the heart and small intestine (Supplementary Figure S11). The results obtained are in consistence with the previously reported data showing DNA demethylation in a subset of LTR12s in primordial germ cells (PGCs) and transcriptional activation of LTR12C class in spermatogonial stem cells (SSCs) and early differentiated spermatogonia (70–72). In summary, U7-dependent LTR12s and lincRNAs might be relevant in primordial germ cells and during early spermatogenesis in the testes, with an adverse role in other types of tissues.
Discussion
U7 snRNA regulates LTR12 and lincRNA expression through a novel mode of action
The novel function of U7 snRNA in human cells proposed here can be exerted outside of S phase, in addition to its role in RDH gene expression. This is in line with the observation that U7 snRNA is present not only in proliferating but also in terminally differentiated cells. We showed that U7 snRNA functions as a transcriptional regulator of various genes/regions that possess a CCAAT motif by a mechanism involving NF-Y, as previously suggested (Figure 5) (18). Our ChIP experiment revealed NF-Y occupancy at U7-dependent LTR12 elements. This is in agreement with previously published data that identified LTR12s as one of the most predominant classes containing NF-Y binding sites in the human genome (40% of all sites are found within MLT1 and LTR12 families of LTRs) (20). However, as it was reported in K562 cells, although LTR12s are frequently bound by NF-Y, they appear to be inactive as they are located within closed chromatin regions (20). We propose two possible scenarios that can eventually result in the activated expression of LTR12 elements upon U7 snRNA knockdown: (i) when all CCAAT motifs available within certain LTR12s are occupied by NF-Y, U7 snRNA deficiency may result solely in the liberating the transcriptional activity of the already bound NF-Y; (ii) U7 snRNA depletion may cause binding of extra NF-Y to unoccupied CCAAT motifs, affecting not only the activity, but also the pool of NF-Y associated with certain loci.
As shown in Supplementary Figure S7A, in contrast to the role of U7 snRNA in the expression of the RDH genes, the mechanism of U7-dependent LTR12 and lincRNA expression may depend on U7 snRNA alone, rather than the canonical U7 snRNP with the Sm/Lsm ring. In wild-type cells, we hypothesize that after synthesis a pool of U7 snRNA can remain in the nucleus where it hybridizes with HDE-like motifs, thereby attenuating NF-Y transcriptional activity. Another pool of U7 snRNA is transported to the cytoplasm and assembled into the U7 snRNP complex. Next, U7 snRNP is reimported to the nucleus, where together with other proteins from the histone cleavage complex it participates in the 3′ end processing of RDH gene transcripts within HLBs. In this process, SLBP stabilizes the base-pair interaction of U7 snRNA with HDE motifs located in the 3′UTRs of histone pre-mRNAs, which are not perfectly complementary to each other. Interestingly, it has been reported that SLBP activity is not necessary for stable interaction in the case of perfect complementarity between U7 snRNA and the HDE motif (73). In the novel mechanism of regulation described here, most HDE-like motifs perfectly match U7 snRNA (Table 1); therefore, there is no need for additional stabilizing factors. Furthermore, HDE-like motifs are located mainly at the beginning of the lincRNA transcripts, in the first exon or intron, and we demonstrated that the U7 snRNA-mediated regulation of these genes and LTR12s relies on transcriptional inhibition, which is the next argument confirming a novel mode of action of U7 snRNA alone, apart from the U7 snRNP complex that triggers cleavage. According to our calculation, ∼500 HDE-like elements were upregulated after U7 snRNA knockdown, suggesting that such a number of elements must engage U7 snRNA-dependent mechanism for their repression at a given time. The number of U7 snRNA copies required could be even lower in the case where multiple HDE-like motifs are present in close proximity in a single LTR12 locus and/or due to chromatin topology, and thus not all of them have to be saturated with U7 snRNA for efficient inhibition of the LTR12 region. Therefore, even considering that abundance of U7 snRNA is very low, reaching 1 × 103 to 2 × 104 molecules per cell (5,6,74,75), it is still possible that the number of U7 snRNA copies present in the cell is high enough to fill both U7 snRNA pools: one involved in the inhibition of LTR12 expression, and the other which is a component of the U7 snRNP complex.
The link between U7 snRNA and transposable elements has been described in murine embryonic stem cells (ES) and in preimplantation embryos. It has been observed that the removal of the 5′ fragment of tRNA-Gly-GCC (tRF-GG) resulted in the derepression of ∼50 genes regulated by the LTR of the endogenous retroelement MERVL (mouse endogenous retrovirus L) (76,77). This was paralleled by decreased synthesis of U7 snRNA/snRNP, which in turn led to decreased expression of histones at the transcript and protein levels. Although the detailed mechanism was not described, the authors suggested that eventually the lower supply of histones would cause chromatin changes and transcriptional activation of the MERVL element, in turn stimulating the expression of MERVL-linked genes in murine ES cells and preimplantation embryos (76). In contrast, we demonstrated that U7 snRNA depletion does not influence histone protein levels in HEK293T cells (Supplementary Figure S1D), which is consistent with the results obtained by Ideue et al. (41). This is additional evidence supporting a distinctness of the mode of action described here, which is based on the direct regulation of LTR12s by U7 snRNA via base-pair complementarity with HDE-like motifs (which are absent in the MERVL sequence) and involves the transcription factor NF-Y.
U7 snRNA acts as a transcription inhibitor to ensure tissue-dependent expression of LTR12s and lincRNAs
HERVs and lincRNAs are present in thousands of copies in the human genome, and both are important components of gene expression regulatory networks. In the case of HERVs, this function is mostly related to regulatory sequences present in LTR regions, and most HERVs exist as solo LTRs. Therefore, despite being transpose-inactive, HERVs are essential repositories of transcription factor-binding sites, such as CCAAT motifs recognized by NF-Y, which may serve as promoters or enhancers (24,25). In turn, lincRNAs can function both in the nucleus and in the cytoplasm at the DNA, RNA, and protein levels (29–33,78).
As mentioned, the expression of both HERVs and lincRNAs is tightly regulated. HERVs are mostly silenced in somatic cells, including nondividing, terminally differentiated cells; however, they are highly expressed in germ cells, during embryogenesis, cell transformation, in neuronal precursors and pluripotent stem cells (PSCs) (68,69). Similarly, lincRNAs are expressed primarily in a cell type-specific, tissue-specific, developmental stage-specific, or disease state-specific manner, with overrepresentation in the brain and testis tissues, suggesting involvement in neurogenesis and reproductive functions (31). In other kinds of cells, their expression should be restricted or may lead to human diseases. According to GTEx data, U7-dependent LTR12s and lincRNAs display cell type specificity as well, being predominantly restricted to testis (Supplementary Figures S9-S11). This observation is consistent with the reported extensive chromatin remodeling and widespread transcription, including TE-driven genes and intergenic regions, during spermatogenesis in vertebrates (79,80). NF-Y has been shown to participate in the activation of LTR12 elements (67,81), and NF-Y binding sites are considerably enriched in LTR12C, LTR12D, and LTR12E elements, which are upregulated in hSSCs and oocytes (70). Furthermore, NF-Y binding motifs are frequently found near DMRT1 binding sites, a transcription factor essential for human gonadogenesis (70,82). The close localization of HDE-like motifs and sites recognized by these two pioneer transcription factors, NF-Y and DMRT1, suggests their possible cooperation in the specific activation of LTR12C transcription in hSSCs. These findings demonstrate that LTR12 silencing, essential for genome stability in somatic cells, and their specific derepression during spermatogenesis may require multiple distinct mechanisms and one of them may involve the interplay between NF-Y and U7 snRNA.
Exaptation of LTR elements into promoters and enhancers is a widespread phenomenon in mammalian genomes that contributes to the evolution of gene regulation (83). According to our RNA-seq results, U7 snRNA depletion caused a significant upregulation of some LTR12-driven protein-coding genes, such as RAE1, DHRS2, CGREF1, SEMA4D, CSF3, TNFRSF10B, IRGM, ACSBG1, DNAH3, NPC1, DPEP2, TPH2, DLEU1, OFCC1, PCDH17, ETV7, SH2D4B, TCN2, RPL3L, WSCD1, DENND3 (Supplementary Table S3) (67,72,84–87), from which RAE1, DHRS2, CGREF1 and SEMA4D are known to be positively regulated by NF-Y (67,84). In all of these genes, LTR12 elements along with HDE-like motifs are located within promoters/alternative promoters, suggesting U7-dependent control of transcription. Tissue-specific expression analysis using GTEx, single-cell transcriptomic data (88) deposited in the Human Protein Atlas (proteinatlas.org), and results published by Iouranova et al. (72) and Brind’Amour et al. (87) disclosed that these genes exhibited enhanced expression in early embryo, oocytes, testis and/or at different stages of the spermatogenesis. For instance, Sema4D is involved in ovary follicular development in mice, whereas Rae1, NPC1 and Dnah3 have already been linked to sperm development (89–93). In addition, Beyer et al. suggested that cell death and immunity-related genes under LTR12 control, such as TNFRSF10B and IRGM, may increase the apoptotic potential or defense mechanisms in germ cells, conferring enhanced germ cell protection in the light of the long lifetime and fertility of male individuals (85). Importantly, Drosophila U7 snRNA null mutants displayed reproductive phenotypes; males and females are viable but sterile due to compromised histone pre-mRNA processing (94). In summary, all these findings reinforce the essential role of LTR12 promoters/elements in the gamete formation (72,87), and implicate an essential role of U7 snRNA in the regulation of these hominoid-specific LTR12 elements. Finally, we do not rule out the possibility that U7 snRNA is not only engaged in the inhibition, but it may also mediate the reactivation process of this specific class of HERV1 elements. Given that U7 snRNA levels do not differ considerably between human tissues (Supplementary Figure S11), NF-Y-mediated transcription activation in testes might be triggered by U7 snRNA displacement from these elements.
Very recent studies identified LTR12s as enhancers with oncogenic potential in acute myeloid leukemia and liver cancer cells (95,96). Importantly, some of these liver cancer-specific LTR12s are upregulated in HEK293T U7 KD cells, suggesting the role of the U7 snRNA- LTR12 module not only in developmental processes, but also in carcinogenesis.
Limitations of the study
As mentioned above, ∼80% of annotated human lncRNAs have been reported to contain TE sequences, with enrichment of LTR elements (25,27,60,61), and we observed a similar phenomenon in this study: half of U7-dependent lincRNAs contained LTR12 elements, and 39% of intragenic U7-dependent DERs were encoded by sequences within lincRNAs. Importantly, some genomic regions found to be affected by U7 snRNA deficiency had representatives in both U7-dependent lincRNAs and U7-dependent repeats. For example, lnc-PGRMC1-1 (AC004835.1) contained four repeats showing upregulated expression upon U7 snRNA knockdown, and the expression of the entire gene was also shown to be activated in DESeq2 analysis. It must be pointed out that we cannot rule out the possibility that some lincRNAs listed as DEGs are in fact included in the list only due to differential expression of the embedded LTR12(s) and vice versa. This ambiguity probably results from the imperfection of the algorithm used, which can sometimes identify a gene as a DEG based on the sequencing reads mapping only to one region of the gene (e.g. TE elements). Therefore, it must be further verified in each single case whether the whole lincRNA is upregulated under U7 snRNA knockdown conditions. However, when LTR12 elements are located at the beginning of a gene, they can act as transcriptional activators of the gene; indeed, we observed reads covering the entire gene for the majority of U7-dependent lincRNAs with LTR12 elements located in the first exon or the first intron.
In this report, we also showed that 69% of DERs belong to the HERV1 family with LTR12s containing HDE-like motifs, representing the most prevalent class of U7-dependent repeats. Additionally, our manual inspection of activated repeats of other types revealed that approximately 80% of them were located in close proximity to HDE-like motifs present in neighboring LTR12s. Since the DER analysis in U7 KD cells was performed using only uniquely mapped sequencing reads, we assume that some loci activated by U7 snRNA deficiency could have escaped identification due to the high similarity of LTR12 elements in the human genome. Thus, we believe that the novel U7 snRNA-mediated mechanism of LTR12 silencing described in this report may actually make a bigger contribution to cell fitness.
Supplementary Material
Acknowledgements
We thank Magdalena Maslon for help with the 4sU assay, Kishor Gawade for help with RNA-seq data, and Bartosz Kwiatkowski for help with graph preparation. We also thank Prof. Wojciech Makalowski for stimulating and fruitful discussions about transposable elements. A graphical abstract was created with BioRender.com.
Authors contributions: conceptualization, K.D.R., P.P., A.S.; methodology, P.P., M.W.S., K.D.R.; investigation, P.P., M.W.S., K.D.R., R.S., A.S.; funding acquisition, K.D.R.; resources: K.D.R., I.M., E.W.; supervision, K.D.R.; writing, K.D.R., P.P., with feedback from all the authors.
Contributor Information
Patrycja Plewka, Department of Gene Expression, Laboratory of RNA Processing, Institute of Molecular Biology and Biotechnology, Faculty of Biology and Center for Advanced Technology, Adam Mickiewicz University, Poznan, Poland.
Michal W Szczesniak, Institute of Human Biology and Evolution, Faculty of Biology, Adam Mickiewicz University, Poznan, Poland.
Agata Stepien, Department of Gene Expression, Laboratory of RNA Processing, Institute of Molecular Biology and Biotechnology, Faculty of Biology and Center for Advanced Technology, Adam Mickiewicz University, Poznan, Poland.
Robert Pasieka, Department of Gene Expression, Laboratory of RNA Processing, Institute of Molecular Biology and Biotechnology, Faculty of Biology and Center for Advanced Technology, Adam Mickiewicz University, Poznan, Poland.
Elzbieta Wanowska, Institute of Human Biology and Evolution, Faculty of Biology, Adam Mickiewicz University, Poznan, Poland.
Izabela Makalowska, Institute of Human Biology and Evolution, Faculty of Biology, Adam Mickiewicz University, Poznan, Poland.
Katarzyna Dorota Raczynska, Department of Gene Expression, Laboratory of RNA Processing, Institute of Molecular Biology and Biotechnology, Faculty of Biology and Center for Advanced Technology, Adam Mickiewicz University, Poznan, Poland.
Data availability
The total RNA-seq data are available at NCBI GEO under GSE247500 accession number.
Supplementary data
Supplementary Data are available at NAR Online.
Funding
Polish National Science Centre [UMO-2015/19/B/NZ1/00233 to K.D.R.]. Funding for open access charge: Polish National Science Centre grant [UMO-2018/30/E/NZ2/00295].
Conflict of interest statement. None declared.
References
- 1. Pillai R.S., Grimmler M., Meister G., Will C.L., Luhrmann R., Fischer U., Schumperli D.. Unique Sm core structure of U7 snRNPs: assembly by a specialized SMN complex and the role of a new component, Lsm11, in histone RNA processing. Genes Dev. 2003; 17:2321–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pillai R.S., Will C.L., Luhrmann R., Schumperli D., Muller B.. Purified U7 snRNPs lack the Sm proteins D1 and D2 but contain Lsm10, a new 14 kDa Sm D1-like protein. EMBO J. 2001; 20:5470–5479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Schumperli D., Pillai R.S.. The special Sm core structure of the U7 snRNP: far-reaching significance of a small nuclear ribonucleoprotein. Cell. Mol. Life Sci. 2004; 61:2560–2570. [DOI] [PubMed] [Google Scholar]
- 4. Azzouz T.N., Pillai R.S., Dapp C., Chari A., Meister G., Kambach C., Fischer U., Schumperli D.. Toward an assembly line for U7 snRNPs: interactions of U7-specific lsm proteins with PRMT5 and SMN complexes. J. Biol. Chem. 2005; 280:34435–34440. [DOI] [PubMed] [Google Scholar]
- 5. Mowry K.L., Steitz J.A.. Identification of the human U7 snRNP as one of several factors involved in the 3' end maturation of histone premessenger RNA’s. Science. 1987; 238:1682–1687. [DOI] [PubMed] [Google Scholar]
- 6. Dominski Z., Marzluff W.F.. Formation of the 3' end of histone mRNA: getting closer to the end. Gene. 2007; 396:373–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ghule P.N., Dominski Z., Yang X.C., Marzluff W.F., Becker K.A., Harper J.W., Lian J.B., Stein J.L., van Wijnen A.J., Stein G.S.. Staged assembly of histone gene expression machinery at subnuclear foci in the abbreviated cell cycle of human embryonic stem cells. Proc. Natl. Acad. Sci. USA. 2008; 105:16964–16969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Frey M.R., Matera A.G.. Coiled bodies contain U7 small nuclear RNA and associate with specific DNA sequences in interphase human cells. Proc. Natl. Acad. Sci. U.S.A. 1995; 92:5915–5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Harris M.E., Bohni R., Schneiderman M.H., Ramamurthy L., Schumperli D., Marzluff W.F.. Regulation of histone mRNA in the unperturbed cell cycle: evidence suggesting control at two posttranscriptional steps. Mol. Cell. Biol. 1991; 11:2416–2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Muller B., Schumperli D.. The U7 snRNP and the hairpin binding protein: key players in histone mRNA metabolism. Semin. Cell Dev. Biol. 1997; 8:567–576. [DOI] [PubMed] [Google Scholar]
- 11. Wang Z.F., Whitfield M.L., Ingledue T.C. 3rd, Dominski Z., Marzluff W.F.. The protein that binds the 3' end of histone mRNA: a novel RNA-binding protein required for histone pre-mRNA processing. Genes Dev. 1996; 10:3028–3040. [DOI] [PubMed] [Google Scholar]
- 12. Romeo V., Schumperli D.. Cycling in the nucleus: regulation of RNA 3' processing and nuclear organization of replication-dependent histone genes. Curr. Opin. Cell Biol. 2016; 40:23–31. [DOI] [PubMed] [Google Scholar]
- 13. Schaufele F., Gilmartin G.M., Bannwarth W., Birnstiel M.L.. Compensatory mutations suggest that base-pairing with a small nuclear RNA is required to form the 3' end of H3 messenger RNA. Nature. 1986; 323:777–781. [DOI] [PubMed] [Google Scholar]
- 14. Marz M., Mosig A., Stadler B.M., Stadler P.F.. U7 snRNAs: a computational survey. Genom. Proteom. Bioinf. 2007; 5:187–195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sun Y., Zhang Y., Aik W.S., Yang X.C., Marzluff W.F., Walz T., Dominski Z., Tong L.. Structure of an active human histone pre-mRNA 3'-end processing machinery. Science. 2020; 367:700–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Yang X.C., Sabath I., Debski J., Kaus-Drobek M., Dadlez M., Marzluff W.F., Dominski Z.. A complex containing the CPSF73 endonuclease and other polyadenylation factors associates with U7 snRNP and is recruited to histone pre-mRNA for 3'-end processing. Mol. Cell. Biol. 2013; 33:28–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schinkel A.H., Mayer U., Wagenaar E., Mol C.A., van Deemter L., Smit J.J., van der Valk M.A., Voordouw A.C., Spits H., van Tellingen O.et al.. Normal viability and altered pharmacokinetics in mice lacking mdr1-type (drug-transporting) P-glycoproteins. Proc. Natl. Acad. Sci. U.S.A. 1997; 94:4028–4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Higuchi T., Anzai K., Kobayashi S.. U7 snRNA acts as a transcriptional regulator interacting with an inverted CCAAT sequence-binding transcription factor NF-Y. Biochim. Biophys. Acta. 2008; 1780:274–281. [DOI] [PubMed] [Google Scholar]
- 19. Romier C., Cocchiarella F., Mantovani R., Moras D.. The NF-YB/NF-YC structure gives insight into DNA binding and transcription regulation by CCAAT factor NF-Y. J. Biol. Chem. 2003; 278:1336–1345. [DOI] [PubMed] [Google Scholar]
- 20. Fleming J.D., Pavesi G., Benatti P., Imbriano C., Mantovani R., Struhl K.. NF-Y coassociates with FOS at promoters, enhancers, repetitive elements, and inactive chromatin regions, and is stereo-positioned with growth-controlling transcription factors. Genome Res. 2013; 23:1195–1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Oldfield A.J., Yang P.Y., Conway A.E., Cinghu S., Freudenberg J.M., Yellaboina S., Jothi R.. Histone-fold domain protein NF-Y promotes chromatin accessibility for cell type-specific master transcription factors. Mol. Cell. 2014; 55:708–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ceribelli M., Dolfini D., Merico D., Gatta R., Vigano A.M., Pavesi G., Mantovani R.. The histone-like NF-Y is a bifunctional transcription factor. Mol. Cell. Biol. 2008; 28:2047–2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bannert N., Kurth R.. The evolutionary dynamics of human endogenous retroviral families. Annu. Rev. Genomics Hum. Genet. 2006; 7:149–173. [DOI] [PubMed] [Google Scholar]
- 24. Ali A., Han K., Liang P.. Role of transposable elements in gene regulation in the Human genome. Life (Basel). 2021; 11:118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Thompson P.J., Macfarlan T.S., Lorincz M.C.. Long terminal repeats: from parasitic elements to building blocks of the transcriptional regulatory repertoire. Mol. Cell. 2016; 62:766–776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Babarinde I.A., Ma G., Li Y., Deng B., Luo Z., Liu H., Abdul M.M., Ward C., Chen M., Fu X.et al.. Transposable element sequence fragments incorporated into coding and noncoding transcripts modulate the transcriptome of human pluripotent stem cells. Nucleic Acids Res. 2021; 49:9132–9153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Fort V., Khelifi G., Hussein S.M.I.. Long non-coding RNAs and transposable elements: a functional relationship. Biochim. Biophys. Acta Mol. Cell Res. 2021; 1868:118837. [DOI] [PubMed] [Google Scholar]
- 28. Dhanoa J.K., Sethi R.S., Verma R., Arora J.S., Mukhopadhyay C.S.. Long non-coding RNA: its evolutionary relics and biological implications in mammals: a review. J. Anim. Sci. Technol. 2018; 60:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kazimierczyk M., Kasprowicz M.K., Kasprzyk M.E., Wrzesinski J.. Human long noncoding RNA interactome: detection, characterization and function. Int. J. Mol. Sci. 2020; 21:1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Quinn J.J., Chang H.Y.. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016; 17:47–62. [DOI] [PubMed] [Google Scholar]
- 31. Ransohoff J.D., Wei Y., Khavari P.A.. The functions and unique features of long intergenic non-coding RNA. Nat. Rev. Mol. Cell Biol. 2018; 19:143–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Statello L., Guo C.J., Chen L.L., Huarte M.. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021; 22:96–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mattick J.S., Amaral P.P., Carninci P., Carpenter S., Chang H.Y., Chen L.L., Chen R., Dean C., Dinger M.E., Fitzgerald K.A.et al.. Long non-coding RNAs: definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 2023; 24:430–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jansz N., Faulkner G.J.. Endogenous retroviruses in the origins and treatment of cancer. Genome Biol. 2021; 22:147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mao J., Zhang Q., Cong Y.S.. Human endogenous retroviruses in development and disease. Comput. Struct. Biotechnol. J. 2021; 19:5978–5986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Plewka P., Raczynska K.D.. Long intergenic noncoding RNAs affect biological pathways underlying autoimmune and neurodegenerative disorders. Mol. Neurobiol. 2022; 59:5785–5808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pasieka R., Zasonski G., Raczynska K.D.. Role of long intergenic noncoding RNAs in cancers with an overview of MicroRNA binding. Mol. Diagn. Ther. 2022; 27:29–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Simpson P.B., Bacha J.I., Palfreyman E.L., Woollacott A.J., McKernan R.M., Kerby J.. Retinoic acid evoked-differentiation of neuroblastoma cells predominates over growth factor stimulation: an automated image capture and quantitation approach to neuritogenesis. Anal. Biochem. 2001; 298:163–169. [DOI] [PubMed] [Google Scholar]
- 39. Gadgil A., Walczak A., Stepien A., Mechtersheimer J., Nishimura A.L., Shaw C.E., Ruepp M.D., Raczynska K.D.. ALS-linked FUS mutants affect the localization of U7 snRNP and replication-dependent histone gene expression in human cells. Sci. Rep. 2021; 11:11868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ideue T., Hino K., Kitao S., Yokoi T., Hirose T.. Efficient oligonucleotide-mediated degradation of nuclear noncoding RNAs in mammalian cultured cells. RNA. 2009; 15:1578–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ideue T., Adachi S., Naganuma T., Tanigawa A., Natsume T., Hirose T.. U7 small nuclear ribonucleoprotein represses histone gene transcription in cell cycle-arrested cells. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:5693–5698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., Zhang F.. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 2013; 8:2281–2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Raczynska K.D., Ruepp M.D., Brzek A., Reber S., Romeo V., Rindlisbacher B., Heller M., Szweykowska-Kulinska Z., Jarmolowski A., Schumperli D.. FUS/TLS contributes to replication-dependent histone gene expression by interaction with U7 snRNPs and histone-specific transcription factors. Nucleic Acids Res. 2015; 43:9711–9728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kruszka K., Pacak A., Swida-Barteczka A., Stefaniak A.K., Kaja E., Sierocka I., Karlowski W., Jarmolowski A., Szweykowska-Kulinska Z.. Developmentally regulated expression and complex processing of barley pri-microRNAs. BMC Genomics [Electronic Resource]. 2013; 14:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dolken L., Ruzsics Z., Radle B., Friedel C.C., Zimmer R., Mages J., Hoffmann R., Dickinson P., Forster T., Ghazal P.et al.. High-resolution gene expression profiling for simultaneous kinetic parameter analysis of RNA synthesis and decay. RNA. 2008; 14:1959–1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gawade K., Plewka P., Hafner S.J., Lund A.H., Marchand V., Motorin Y., Szczesniak M.W., Raczynska K.D.. FUS regulates a subset of snoRNA expression and modulates the level of rRNA modifications. Sci. Rep. 2023; 13:2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zheng Y.L., Li L., Jia Y.X., Zhang B.Z., Li J.C., Zhu Y.H., Li M.Q., He J.Z., Zeng T.T., Ban X.J.et al.. LINC01554-Mediated glucose metabolism reprogramming suppresses tumorigenicity in hepatocellular carcinoma via downregulating PKM2 expression and inhibiting akt/mTOR signaling pathway. Theranostics. 2019; 9:796–810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Brzek A., Cichocka M., Dolata J., Juzwa W., Schumperli D., Raczynska K.D.. Positive cofactor 4 (PC4) contributes to the regulation of replication-dependent canonical histone gene expression. BMC Mol. Biol. 2018; 19:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R.. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013; 29:15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Liao Y., Smyth G.K., Shi W.. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014; 30:923–930. [DOI] [PubMed] [Google Scholar]
- 51. Love M.I., Huber W., Anders S.. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014; 15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Langmead B., Salzberg S.L.. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012; 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Neph S., Kuehn M.S., Reynolds A.P., Haugen E., Thurman R.E., Johnson A.K., Rynes E., Maurano M.T., Vierstra J., Thomas S.et al.. BEDOPS: high-performance genomic feature operations. Bioinformatics. 2012; 28:1919–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Ramirez F., Dundar F., Diehl S., Gruning B.A., Manke T.. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014; 42:W187–W191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Li B., Dewey C.N.. RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinf. 2011; 12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bond U., Yario T.A.. The steady state levels and structure of the U7 snRNP are constant during the human cell cycle: lack of cell cycle regulation of histone mRNA 3' end formation. Cell Mol. Biol. Res. 1994; 40:27–34. [PubMed] [Google Scholar]
- 57. Hoffmann I., Birnstiel M.L.. Cell cycle-dependent regulation of histone precursor mRNA processing by modulation of U7 snRNA accessibility. Nature. 1990; 346:665–668. [DOI] [PubMed] [Google Scholar]
- 58. Lyons S.M., Cunningham C.H., Welch J.D., Groh B., Guo A.Y., Wei B., Whitfield M.L., Xiong Y., Marzluff W.F.. A subset of replication-dependent histone mRNAs are expressed as polyadenylated RNAs in terminally differentiated tissues. Nucleic Acids Res. 2016; 44:9190–9205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Larson D.E., Hoffmann I., Zahradka P., Birnstiel M.L., Sells B.H.. Histone H4 mRNA levels are down-regulated by 3' RNA processing during terminal differentiation of myoblasts. Biochim. Biophys. Acta. 1992; 1131:139–144. [DOI] [PubMed] [Google Scholar]
- 60. Kannan S., Chernikova D., Rogozin I.B., Poliakov E., Managadze D., Koonin E.V., Milanesi L.. Transposable element insertions in long intergenic non-coding RNA genes. Front. Bioeng. Biotechnol. 2015; 3:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Kapusta A., Kronenberg Z., Lynch V.J., Zhuo X., Ramsay L., Bourque G., Yandell M., Feschotte C.. Transposable elements are major contributors to the origin, diversification, and regulation of vertebrate long noncoding RNAs. PLoS Genet. 2013; 9:e1003470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Maity S.N., de Crombrugghe B.. Role of the CCAAT-binding protein CBF/NF-Y in transcription. Trends Biochem. Sci. 1998; 23:174–178. [DOI] [PubMed] [Google Scholar]
- 63. Sinha S., Maity S.N., Lu J., de Crombrugghe B.. Recombinant rat CBF-C, the third subunit of CBF/NFY, allows formation of a protein-DNA complex with CBF-A and CBF-B and with yeast HAP2 and HAP3. Proc. Natl. Acad. Sci. U.S.A. 1995; 92:1624–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Gurtner A., Manni I., Piaggio G.. NF-Y in cancer: impact on cell transformation of a gene essential for proliferation. Biochim. Biophys. Acta Gene Regul. Mech. 2017; 1860:604–616. [DOI] [PubMed] [Google Scholar]
- 65. Mantovani R. The molecular biology of the CCAAT-binding factor NF-Y. Gene. 1999; 239:15–27. [DOI] [PubMed] [Google Scholar]
- 66. Mantovani R. A survey of 178 NF-Y binding CCAAT boxes. Nucleic Acids Res. 1998; 26:1135–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kronung S.K., Beyer U., Chiaramonte M.L., Dolfini D., Mantovani R., Dobbelstein M.. LTR12 promoter activation in a broad range of human tumor cells by HDAC inhibition. Oncotarget. 2016; 7:33484–33497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wang T., Doucet-O’Hare T.T., Henderson L., Abrams R.P.M., Nath A.. Retroviral elements in Human evolution and neural development. J. Exp. Neurol. 2021; 2:1–9. [PMC free article] [PubMed] [Google Scholar]
- 69. Xiang Y., Liang H.. The regulation and functions of endogenous retrovirus in embryo development and stem cell differentiation. Stem Cells Int. 2021; 2021:6660936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Guo J., Grow E.J., Yi C., Mlcochova H., Maher G.J., Lindskog C., Murphy P.J., Wike C.L., Carrell D.T., Goriely A.et al.. Chromatin and single-cell RNA-seq profiling reveal dynamic signaling and metabolic transitions during Human spermatogonial stem cell development. Cell Stem Cell. 2017; 21:533–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Guo J., Grow E.J., Mlcochova H., Maher G.J., Lindskog C., Nie X., Guo Y., Takei Y., Yun J., Cai L.et al.. The adult human testis transcriptional cell atlas. Cell Res. 2018; 28:1141–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Iouranova A., Grun D., Rossy T., Duc J., Coudray A., Imbeault M., de Tribolet-Hardy J., Turelli P., Persat A., Trono D.. KRAB zinc finger protein ZNF676 controls the transcriptional influence of LTR12-related endogenous retrovirus sequences. Mob. DNA. 2022; 13:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Streit A., Koning T.W., Soldati D., Melin L., Schumperli D.. Variable effects of the conserved RNA hairpin element upon 3' end processing of histone pre-mRNA in vitro. Nucleic Acids Res. 1993; 21:1569–1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Soldati D., Schumperli D.. Structural and functional characterization of mouse U7 small nuclear RNA active in 3' processing of histone pre-mRNA. Mol. Cell. Biol. 1988; 8:1518–1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Grimm C., Stefanovic B., Schumperli D.. The low abundance of U7 snRNA is partly determined by its Sm binding site. EMBO J. 1993; 12:1229–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Boskovic A., Bing X.Y., Kaymak E., Rando O.J.. Control of noncoding RNA production and histone levels by a 5' tRNA fragment. Genes Dev. 2020; 34:118–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Sharma U., Conine C.C., Shea J.M., Boskovic A., Derr A.G., Bing X.Y., Belleannee C., Kucukural A., Serra R.W., Sun F.et al.. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science. 2016; 351:391–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Perry R.B., Ulitsky I.. The functions of long noncoding RNAs in development and stem cells. Development. 2016; 143:3882–3894. [DOI] [PubMed] [Google Scholar]
- 79. Davis M.P., Carrieri C., Saini H.K., van Dongen S., Leonardi T., Bussotti G., Monahan J.M., Auchynnikava T., Bitetti A., Rappsilber J.et al.. Transposon-driven transcription is a conserved feature of vertebrate spermatogenesis and transcript evolution. EMBO Rep. 2017; 18:1231–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Soumillon M., Necsulea A., Weier M., Brawand D., Zhang X., Gu H., Barthes P., Kokkinaki M., Nef S., Gnirke A.et al.. Cellular source and mechanisms of high transcriptome complexity in the mammalian testis. Cell Rep. 2013; 3:2179–2190. [DOI] [PubMed] [Google Scholar]
- 81. Yu X., Zhu X., Pi W., Ling J., Ko L., Takeda Y., Tuan D.. The long terminal repeat (LTR) of ERV-9 human endogenous retrovirus binds to NF-Y in the assembly of an active LTR enhancer complex NF-Y/MZF1/GATA-2. J. Biol. Chem. 2005; 280:35184–35194. [DOI] [PubMed] [Google Scholar]
- 82. Zarkower D., Murphy M.W.. DMRT1: an ancient sexual regulator required for Human gonadogenesis. Sex. Dev. 2022; 16:112–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Cohen C.J., Lock W.M., Mager D.L.. Endogenous retroviral LTRs as promoters for human genes: a critical assessment. Gene. 2009; 448:105–114. [DOI] [PubMed] [Google Scholar]
- 84. Jung Y.D., Lee H.E., Jo A., Hiroo I., Cha H.J., Kim H.S.. Activity analysis of LTR12C as an effective regulatory element of the RAE1 gene. Gene. 2017; 634:22–28. [DOI] [PubMed] [Google Scholar]
- 85. Beyer U., Kronung S.K., Leha A., Walter L., Dobbelstein M.. Comprehensive identification of genes driven by ERV9-LTRs reveals TNFRSF10B as a re-activatable mediator of testicular cancer cell death. Cell Death Differ. 2016; 23:64–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Brocks D., Schmidt C.R., Daskalakis M., Jang H.S., Shah N.M., Li D., Li J., Zhang B., Hou Y., Laudato S.et al.. DNMT and HDAC inhibitors induce cryptic transcription start sites encoded in long terminal repeats. Nat. Genet. 2017; 49:1052–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Brind’Amour J., Kobayashi H., Richard Albert J., Shirane K., Sakashita A., Kamio A., Bogutz A., Koike T., Karimi M.M., Lefebvre L.et al.. LTR retrotransposons transcribed in oocytes drive species-specific and heritable changes in DNA methylation. Nat. Commun. 2018; 9:3331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Karlsson M., Zhang C., Mear L., Zhong W., Digre A., Katona B., Sjostedt E., Butler L., Odeberg J., Dusart P.et al.. A single-cell type transcriptomics map of human tissues. Sci. Adv. 2021; 7:eabh2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Volpi S., Bongiorni S., Fabbretti F., Wakimoto B.T., Prantera G.. Drosophila rae1 is required for male meiosis and spermatogenesis. J. Cell Sci. 2013; 126:3541–3551. [DOI] [PubMed] [Google Scholar]
- 90. Jeganathan K.B., van Deursen J.M.. Differential mitotic checkpoint protein requirements in somatic and germ cells. Biochem. Soc. Trans. 2006; 34:583–586. [DOI] [PubMed] [Google Scholar]
- 91. Vedelek V., Bodai L., Grezal G., Kovacs B., Boros I.M., Laurinyecz B., Sinka R.. Analysis of drosophila melanogaster testis transcriptome. BMC Genomics [Electronic Resource]. 2018; 19:697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Regev A., Goldman S., Shalev E.. Semaphorin-4D (Sema-4D), the Plexin-B1 ligand, is involved in mouse ovary follicular development. Reprod. Biol. Endocrinol. 2007; 5:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Wang C., Ma Z., Scott M.P., Huang X.. The cholesterol trafficking protein NPC1 is required for Drosophila spermatogenesis. Dev. Biol. 2011; 351:146–155. [DOI] [PubMed] [Google Scholar]
- 94. Godfrey A.C., Kupsco J.M., Burch B.D., Zimmerman R.M., Dominski Z., Marzluff W.F., Duronio R.J.. U7 snRNA mutations in Drosophila block histone pre-mRNA processing and disrupt oogenesis. RNA. 2006; 12:396–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Deniz O., Ahmed M., Todd C.D., Rio-Machin A., Dawson M.A., Branco M.R.. Endogenous retroviruses are a source of enhancers with oncogenic potential in acute myeloid leukaemia. Nat. Commun. 2020; 11:3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Karttunen K., Patel D., Xia J., Fei L., Palin K., Aaltonen L., Sahu B.. Transposable elements as tissue-specific enhancers in cancers of endodermal lineage. Nat. Commun. 2023; 14:5313. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The total RNA-seq data are available at NCBI GEO under GSE247500 accession number.