Abstract
The E1B-55K protein plays an important role during human adenovirus type 5 productive infection. In the early phase of the viral infection, E1B-55K binds to and inactivates the tumor suppressor protein p53, allowing efficient replication of the virus. During the late phase of infection, E1B-55K is required for efficient nucleocytoplasmic transport and translation of late viral mRNAs, as well as for host cell shutoff. In an effort to separate the p53 binding and inactivation function and the late functions of the E1B-55K protein, we have generated 26 single-amino-acid mutations in the E1B-55K protein. These mutants were characterized for their ability to modulate the p53 level, interact with the E4orf6 protein, mediate viral late-gene expression, and support virus replication in human cancer cells. Of the 26 mutants, 24 can mediate p53 degradation as efficiently as the wild-type protein. Two mutants, R240A (ONYX-051) and H260A (ONYX-053), failed to degrade p53 in the infected cells. In vitro binding assays indicated that R240A and H260A bound p53 poorly compared to the wild-type protein. When interaction with another viral protein, E4orf6, was examined, H260A significantly lost its ability to bind E4orf6, while R240A was fully functional in this interaction. Another mutant, T255A, lost the ability to bind E4orf6, but unexpectedly, viral late-gene expression was not affected. This raised the possibility that the interaction between E1B-55K and E4orf6 was not required for efficient viral mRNA transport. Both R240A and H260A have retained, at least partially, the late functions of wild-type E1B-55K, as determined by the expression of viral late proteins, host cell shutoff, and lack of a cold-sensitive phenotype. Virus expressing R240A (ONYX-051) replicated very efficiently in human cancer cells, while virus expressing H260A (ONYX-053) was attenuated compared to wild-type virus dl309 but was more active than ONYX-015. The ability to separate the p53-inactivation activity and the late functions of E1B-55K raises the possibility of generating adenovirus variants that retain the tumor selectivity of ONYX-015 but can replicate more efficiently than ONYX-015 in a broad spectrum of cell types.
The E1B-55K protein plays an important role during the productive infection of human adenovirus type 5 (Ad5). In the early phase of infection, E1B-55K forms a stable complex with p53 (32) and inhibits p53-mediated transcriptional activation (38, 40). Furthermore, E1B-55K and another adenoviral protein, E4orf6, cooperate to relocate p53 to the cytoplasm for active degradation (25, 28, 35). This inactivation of p53 is critical for the productive replication of adenovirus, which requires cells to enter the S phase. During the late phase of infection with Ad5, viral mRNAs are selectively exported to the cytoplasm and are efficiently translated, while the nucleocytoplasmic transport of most host cell mRNAs is inhibited (2, 4, 24). This selective accumulation of viral mRNAs during the late phase of infection is mediated by a protein complex that includes E1B-55K and E4orf6 (6, 12, 21, 31). This complex actively shuttles between the nucleus and the cytoplasm, serving as a nucleocytoplasmic transporter for viral mRNAs (7). The E1B-55K protein has also been demonstrated to directly inhibit host cell protein synthesis (host cell shutoff), which ensures that the cellular resources are used for viral replication (1).
ONYX-015, originally named dl1520, is a mutant adenovirus that does not express the E1B-55K protein (3). This virus contains a stop codon immediately following the translation initiation codon ATG, plus a large deletion of the E1B-55K coding sequence. These mutations result in the complete abrogation of E1B-55K expression but do not alter the expression of the E1B-19K protein encoded by an overlapping open reading frame. Consequently, this virus lacks the ability to bind and inactivate p53, allowing it to replicate efficiently only in tumor cells that are defective in p53 function but not in normal cells where p53 function is normal (5, 15, 26). This forms the foundation of utilizing ONYX-015 as an anti-tumor agent. Phase III clinical trials with ONYX-015 in the treatment of human head-and-neck carcinomas in combination with chemotherapy are ongoing.
Lack of E1B-55K function in ONYX-015 results in defective cytoplasmic accumulation and translation of the viral late mRNAs, as well as diminished host cell shutoff, compromising the ability of the mutant virus to reproduce itself (13). Thus, it would be highly desirable to create an E1B-55K mutant that fails to bind and inactivate p53 yet retains the late functions. Such mutations would allow the virus to replicate selectively in cells that are deficient in p53 function without compromising the ability of the virus to replicate in tumor cells. Creation of such mutations will also allow us to further elucidate the mechanism of ONYX-015's tumor cell selectivity, as several groups have described in vitro studies showing that the host range specificity of ONYX-015 is independent of p53 gene status (9, 13, 14, 29). It has been argued that the loss of E1B-55K's RNA transport activity, rather than its p53 binding and inactivation function, may account for the tumor cell specificity of ONYX-015. Separating these two functions of E1B-55K would allow us to begin to distinguish these possibilities.
The regions of the E1B-55K protein responsible for p53 binding, viral mRNA transport, and host cell shutoff appear to overlap (Fig. 1). The region of E1B-55K that mediates its interaction with p53 has been mapped to amino acids 224 to 354 (19, 39). The same region appears to be critical for E1B-55K's ability to mediate mRNA transport. The regions required for E4orf6 binding (30), the regions required to bind E1B-AP5, a cellular protein implicated in nucleocytoplasmic transport (8), and the regions of E1B-55K that have putative RNA binding capability (17) all partially overlap with the region required for p53 binding. Thus far, all efforts to separate the p53 binding and inactivation and the late functions of the protein have been unsuccessful.
FIG. 1.
Amino acid sequence of the E1B-55K protein of Ad5. The region for p53 binding (19) is underlined, the putative RNA-binding domains (K. Leppard, personal communication) are indicated by the solid boxes, and the region required for transcription repression (P. E. Branton, personal communication) is defined by a broken box. Asterisks indicate amino acids that are mutated.
In this study, we have constructed a series of single-amino-acid substitution mutations in the p53-binding domain and the transcriptional repression domain of the E1B-55K protein. These mutations were recombined into an infectious virus (dl309) background, and the resulting mutant adenoviruses were characterized for their abilities to modulate the p53 level, interact with the E4orf6 protein, mediate viral late functions, and support virus replication in human cancer cells. Two E1B-55K mutants, R240A and H260A, appeared to lose the ability to inactivate p53 but retained, at least partially, the late functions of the wild-type protein. R240A fully restored the wild-type replication capacity of ONYX-015 in human cancer cells, while H260A did so only partially. The ability to separate the p53-inactivation activity and the late functions of E1B-55K raises the possibility of creating adenovirus variants that replicate more efficiently than ONYX-015 but retain the tumor selectivity of ONYX-015.
MATERIALS AND METHODS
Tumor cell lines and culturing conditions.
A549, 293, U2OS, DU145, and H1299 cells were obtained from the American Type Culture Collection. HCT116−/−, a derivative of HCT116 with p53 gene knock-out, is a generous gift from F. McCormick, University of California—San Francisco. All cells were grown as monolayer cultures in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 μg of nonessential amino acids per ml, 10 U of penicillin per ml, and 10 μg of streptomycin per ml. All adenovirus infections were performed in DMEM-high glucose supplemented with 2% FBS, 2 mM l-glutamine, 100 μg of nonessential amino acids per ml, 10 U of penicillin per ml, and 10 μg of streptomycin per ml.
Construction of E1B-55K alanine mutation viruses.
All E1B-55K mutants were generated using Stratagene's QuikChange Site-Directed Mutagenesis kit following the manufacturer's recommended protocol. For each mutation, a forward primer and a reverse primer were used. The mutations and their respective primers are summarized in Table 1. Briefly, 20 ng of pXC-1 was used as the template, and the cycling parameters were as follows: 1 cycle of 95°C for 30 s, and 16 cycles of 95°C for 30 s, 55°C for 1 min, and 68°C for 20 min. The parental DNA was digested by adding 10 U of DpnI to each sample and incubating it for 1 h at 37°C. Final products were transformed into XL-1 cells and confirmed by DNA sequencing. Viruses were constructed by cotransfecting pJM17 with plasmids containing the mutations (22) into 293 cells. Two rounds of plaque purification were done to rule out wild-type contamination. Mutations were confirmed by PCR followed by sequencing of the E1B-55K region. All viruses, including WtD, dl309, ONYX-015, and the mutant viruses created in this study, were propagated in 293 cells, purified on CsCl gradients (as described in reference 36), and quantified by plaque assays on 293 cells.
TABLE 1.
Primer sequences of the mutationsa
| Virus | Mutation | Primer sequences |
|---|---|---|
| ONYX-051 | R240A | GTTATTATGAATGTAGCGTTTACTGGCCCC, GGGGCCAGTAAACGCTACATTCATAATAAC |
| ONYX-052 | T255A | GTTTTCCTGGCCAATGCCAACCTTATCCTACAC, GTGTAGGATAAGGTTGGCATTGGCCAGGAAAAC |
| ONYX-053 | H260A | CCAACCTTATCCTAGCCGGTGTAAGCTTC, GAAGCTTACACCGGCTAGGATAAGGTTGG |
| ONYX-054 | C271A | GGGTTTAACAATACCGCCGTGGAAGCCTGG, CCAGGCTTCCACGGCGGTATTGTTAAACCC |
| ONYX-056 | R281A | CGATGTAAGGGTTGCGGGCTGTGCCTTTTAC, GTAAAAGGCACAGCCCGCAACCCTTACATCG |
| ONYX-057 | G282A | GTAAGGGTTCGGGCCTGTGCCTTTTAC, GTAAAAGGCACAGGCCCGAACCCTTAC |
| ONYX-058 | A284S | GTAAGGGTTCGGGGCTGTTCCTTTTACTGCTGCTGGAAGG, CCTTCCAGCAGCAGTAAAAGGAACAGCCCCGAACCCTTAC |
| ONYX-059 | F285L | GGTTCGGGGCTGTGCCTTATACTGCTGCTGGAAGGGG, CCCCTTCCAGCAGCAGTATAAGGCACAGCCCCGAACC |
| ONYX-060 | C288A | GGGCTGTGCCTTTTACTGCGCCTGGAAGGGGGTGGTGTG, CACACCACCCCCTTCCAGGCGCAGTAAAAGGCACAGCCC |
| ONYX-061 | W289F | GCTGTGCCTTTTACTGCTGCTTTAAGGGGGTGGTGTGTCGC, GCGACACACCACCCCCTTAAAGCAGCAGTAAAAGGCACAGC |
| ONYX-062 | W289A | GCTGTGCCTTTTACTGCTGCGCGAAGGGGGTGGTGTGTCG, CGACACACCACCCCCTTCGCGCAGCAGTAAAAGGCACAGC |
| ONYX-063 | K290A | CTGCTGCTGGGCGGGGGTGGTG, CACCACCCCCGCCCAGCAGCAG |
| ONYX-064 | R295A | TGGAAGGGGGTGGTGTGTGCCCCCAAAAGCAGGGCTTC, GAAGCCCTGCTTTTGGGGGCACACACCACCCCCTTCCA |
| ONYX-065 | K297A | GGTGGTGTGTCGCCCCGCAAGCAGGGCTTCAATTAAGAAATG, CATTTCTTAATTGAAGCCCTGCTTGCGGGGCGACACACCACC |
| ONYX-066 | K303A | CCAAAAGCAGGGCTTCAATTGCGAAATGCCTCTTTGAAAGGTG, CACCTTTCAAAGAGGCATTTCGCAATTGAAGCCCTGCTTTTGG |
| ONYX-067 | E308A | CTTCAATTAAGAAATGCCTCTTTGCAAGGTGTACCTTGGGTATCC, GGATACCCAAGGTACACCTTGCAAAGAGGCATTTCTTAATTGAAG |
| ONYX-068 | R309A | TCAATTAAGAAATGCCTCTTTGAAGCGTGTACCTTGGGTATCCTGTC, GACAGGATACCCAAGGTACACGCTTCAAAGAGGCATTTCTTAATTGA |
| ONYX-069 | E317A | TACCTTGGGTATCCTGTCTGCGGGTAACTCCAGGGTGCG, CGCACCCTGGAGTTACCCGCAGACAGGATACCCAAGGTA |
| ONYX-070 | G318A | TACCTTGGGTATCCTGTCTGAGGCTACCTCCAGGGTCCGCC, GGCGGACCCTGGAGGTAGCCTCAGACAGGATACCCAAGGTA |
| ONYX-071 | G318A-N | TACCTTGGGTATCCTGTCTGAGGCTAACTCCAGGGTGCGCC, GGCGCACCCTGGAGTTAGCCTCAGACAGGATACCCAAGGTA |
| ONYX-080 | E421A | CTAAGATATTGCTTGAGCCGGCGAGCATGTCCAAGGTGAAC, GTTCACCTTGGACATGCTCGCCGGCTCAAGCAATATCTTAG |
| ONYX-081 | K425A | GAGCCCGAGAGCATGTCCGCGGTGAACCTGAACGGGG, CCCCGTTCAGGTTCACCGCGGACATGCTCTCGGGCTC |
| ONYX-082 | D433A | GAACCTGAACGGGGTGTTTGCCATGACCATGAAGATCTGG, CCAGATCTTCATGGTCATGGCAAACACCCCGTTCAGGTTC |
| ONYX-083 | K440A | CCATGAAGATCTGGGCGGTGCTGAGGTAC, GTACCTCAGCACCGCCCAGATCTTCATGG |
| ONYX-084 | R443A | GGAAGGTGCTGGCGTACGATGAGACC, GGTCTCATCGTACGCCAGCACCTTCC |
| ONYX-085 | Y444A | GGAAGGTGCTGAGGGCCGATGAGACCCGC, GCGGGTCTCATCGGCCCTCAGCACCTTCC |
For each virus, the sequence of the forward primer is given first, and the sequence for the reverse primer is given second.
Western blot analysis.
A549 cells were infected with various viruses at a multiplicity of infection (MOI) of 10. At 24 h postinfection, the cells were lysed in sodium dodecyl sulfate (SDS) gel loading buffer (100 mM Tris-Cl [pH 6.8], 5 mM EDTA, 1% SDS, 5% β-mercaptoethanol). Proteins were fractionated by electrophoresis on Bio-Rad precast protein gels. After electrophoresis, the proteins were electrophoretically transferred to nylon membranes. Blots were then incubated with antibodies diluted in phosphate-buffered saline (PBS) containing 1% dry milk and 0.1% Tween-20 and were visualized by ECL (Amersham). Anti-p53 antibody DO-1 (Calbiochem) was diluted 1:1,000; anti-E1B-55K antibody 2A6 (33) and anti-E2A antibody B6-6 (generous gift from A. J. Levine, Rockefeller University) tissue culture supernatants were diluted 1:10; rabbit anti-fiber antibody (American Qualex) was used at 1:1,000.
Assays for protein interactions.
A549 cells were infected at an MOI of 10. At 24 h postinfection, cells were radiolabeled for 3 h at 37°C with methionine- and cysteine-free DMEM supplemented with 2% dialyzed FBS and 100 μCi of [35S]-express labeling mix (DuPont) per ml. Labeled cells were washed with cold saline, harvested, and solubilized in a solution containing 50 mM Tris-Cl (pH 8.0), 5 mM EDTA, 150 mM NaCl, 0.5% NP-40, 3 mM 2-mercaptoethanol, and 1× EDTA-free protease inhibitor mix (Boehringer Mannheim). Cell lysates were cleared by microcentrifugation for 10 min. Immunoprecipitations were carried out as described previously (34). Antibodies against p53 (mAb421) or E1B-55K (2A6) were used to bring down the protein complex. Proteins were separated by SDS–4 to 20% polyacrylamide gel electrophoresis (PAGE) (Bio-Rad) and visualized by autoradiography.
Binding of E4orf6 by various E1B-55K mutants was examined by immunoprecipitation followed by Western blotting. A549 cells were infected and lysed as described above, except that cells were not labeled with radioisotope. Cleared cell extracts were immunoprecipitated with the anti-E1B-55K antibody 2A6. The precipitated materials were separated by SDS-PAGE (a 4 to 20% gradient gel) and analyzed by Western blotting either with (i) a polyclonal antibody against E4orf6, 1807-3 (a generous gift from P. E. Branton, McGill University), or with (ii) rat anti-E1B-55K antibody (a generous gift from C. O'Shea, University of California—San Francisco).
In vitro binding experiments were performed essentially as described earlier (37). A549 cells were infected and lysed as described above, except that cells were not labeled with radioisotope. Cleared cell extracts were mixed with in vitro-labeled p53 (generated and labeled with [35S]methionine by transcription and translation in vitro [TNT T3-system; Promega]). The mixtures were incubated at 30°C for 30 min, followed by immunoprecipitation with the monoclonal anti-E1B-55K antibody 2A6 and four washing steps with the lysis buffer. p53 coprecipitated along with E1B-55K protein was resolved on a 10% precast protein gel (Bio-Rad) and was detected by fluorography.
Indirect immunofluorescent staining.
A549 and U2OS cells were seeded on Lab-Tek II chamber slides the day before infection to roughly 60% confluency. Cells were then infected at an MOI of 10 with the following viruses: dl309, ONYX-015, ONYX-051 (R240A), ONYX-052 (T255A), and ONYX-053 (H260A). Twenty-four hours postinfection cells were washed with PBS and fixed for 30 min at room temperature using 4% formaldehyde in PBS. Cells were permeabilized and blocked with PBS supplemented with 0.1% Triton X-100, 0.05% Tween-20, and 10% goat serum for 30 min at room temperature and then incubated for 1 h at room temperature with properly diluted primary antibodies. All antibodies were diluted in the permeabilization and blocking solution. p53 antibody DO-1 (Calbiochem) was diluted 1:100, anti-E1B-55K antibody 9C10 (Calbiochem) was diluted 1:100, and the rabbit anti-E4orf6 antibody 1807-3 was diluted 1:500. Antigens were visualized with Alexa Fluor 594 (red)- or Alexa Fluor 488 (green)-coupled secondary antibody (Molecular Probes).
Cold-sensitivity assay.
Infections of H1299 cells were performed at two temperatures, 32 and 39°C. Infections at 32°C were performed 1 h after the temperature shift from 39°C. All infections were at an MOI of 5, and infected cells were incubated at 32 and 39°C. Four days after infection, cells and culture media were harvested, pooled, and freeze-thawed three times to release virus particles. Viral yields were determined by enzyme-linked immunosorbent assay (L. Hawkins, Y. Wang, and T. Hermiston, unpublished data) on 293 cell monolayers.
Assay for protein synthesis during the late phase of virus infection.
A549 cells were either mock infected or infected with various adenovirus mutants at an MOI of 10. At 24 h postinfection cells were labeled with [35S]methionine-cysteine for a 3-h period. Labeled cells were washed with cold saline, harvested, and solubilized in a solution containing 50 mM Tris-Cl (pH 8.0), 5 mM EDTA, 150 mM NaCl, 0.5% NP-40, 3 mM β-mercaptoethanol, and 1× EDTA-free protease inhibitor mix (Boehringer Mannheim). Cleared cell extracts were resolved by SDS-PAGE (4 to 20% gradient gel) and visualized by autoradiography. The same amount of radioactivity was loaded in each lane.
MTT assay.
DU145 and U2OS cells were seeded into 96-well plates at a density of 2.5 × 103 cells/well using DMEM-high glucose supplemented with 2% FBS, 2 mM l-glutamine, 100 μg of nonessential amino acids per ml, 10 U of penicillin per ml, and 10 μg of streptomycin per ml. The cells were infected 24 h after being seeded with the E1B-55K mutant viruses (dl309 and ONYX-015 were included as controls). All infections were carried out in quadruplet with serial threefold dilutions of the viruses. A total of 10 dilutions were used for each virus, starting at an MOI of 30 and ending at an MOI of 1.5 × 10−3. Cells that were mock infected were used as negative controls. Infected cells were incubated at 37°C and monitored daily. Colorimetric reactions were performed when significant cytopathic effect was observed in the cells infected with the wild-type virus, dl309, at the median MOI of 0.1. For most cell lines we have tested, this occurred at 4 to 7 days postinfection, allowing the viruses to go through multiple rounds of infection and spread. In the experiments reported here, MTT reactions were carried out at 6 days postinfection using Promega's CellTiter 96 Non-Radioactive Cell Proliferation assay according to the manufacturer's instructions. Cell killing curves from such MTT assays demonstrated a Poisson distribution.
RESULTS
To separate the p53 binding and inactivation function from the other activities of E1B-55K, a series of single-amino-acid mutations in the p53-binding region (19) and the region required for transcriptional repression (P. E. Branton, personal communication) were generated (Fig. 1 and Table 1). In each case, one native amino acid was changed to Ala (Table 1). In addition, four mutations (Ala 284 to Ser, Phe 285 to Leu, Cys 288 to Ala, Trp 289 to Phe) were generated in the putative RNA-binding region of the protein. It was shown previously that some of these mutations can modulate the RNA-binding activity of the E1B-55K protein (K. Leppard, personal communications). In one case (ONYX-070), two amino acid changes were introduced inadvertently (Gly 318 to Ala and Asn 319 to Thr). All mutations were recombined into infectious virus (dl309 background), and the resulting recombinant viruses were characterized for their ability to produce stable 55K protein, modulate p53 degradation, mediate viral late-gene expression, and support virus replication. All the E1B-55K mutations and the resulting viruses are summarized in Table 1.
Steady-state level of p53.
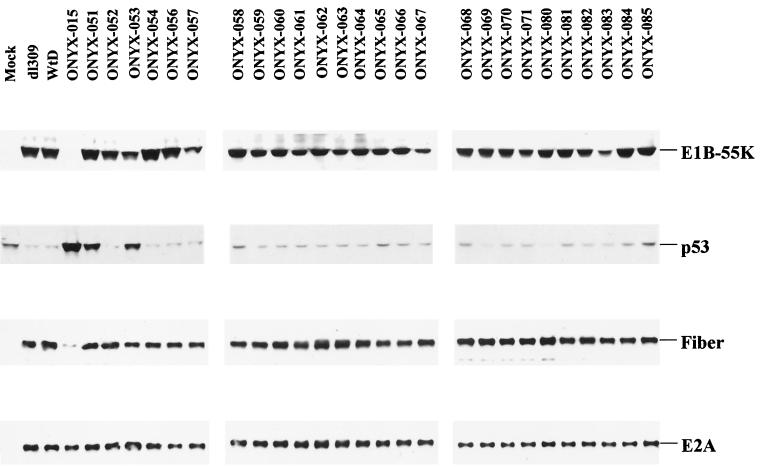
E1B-55K cooperates with E4orf6 to target p53 for active degradation. In order to identify E1B-55K mutants that were incapable of this activity, we examined the steady-state level of p53 in infected A549 cells by Western blot analysis (Fig. 2). In cells infected with dl309 or WtD, the p53 level was significantly reduced due to the targeted degradation mediated by E1B-55K and E4orf6. In contrast, cells infected with ONYX-015 exhibited an increase in the p53 level. Most viral mutants caused a decrease in the level of p53. However, cells infected with ONYX-051 and -053 (expressing E1B-55K mutants R240A and H260A, respectively) displayed a significant increase in p53 levels compared to mock-infected A549 cells. All of the point-mutant adenoviruses accumulated E1B-55K protein to levels comparable to WtD or dl309 upon infection (Fig. 2). These observations raised the possibility that E1B-55K mutants R240A and H260A may fail to bind p53 and/or E4orf6.
FIG. 2.
Effects of E1B-55K mutations on p53 accumulation and viral gene expression. A549 cells were either mock infected (mock) or infected with dl309, WtD, ONYX-015, or viruses harboring various E1B-55K mutants. All infections were performed at an MOI of 10. Cell extracts were prepared at 24 h postinfection and were separated by SDS-PAGE. Steady-state levels of E1B-55K mutants, p53, E2A, and fiber were determined by Western blotting with monoclonal antibodies 2A6, DO-1, B6-6, and a rabbit polyclonal anti-fiber antibody, respectively. Blots were visualized with ECL, as described in Materials and Methods.
Binding of selected E1B-55K mutants with p53 and E4orf6.
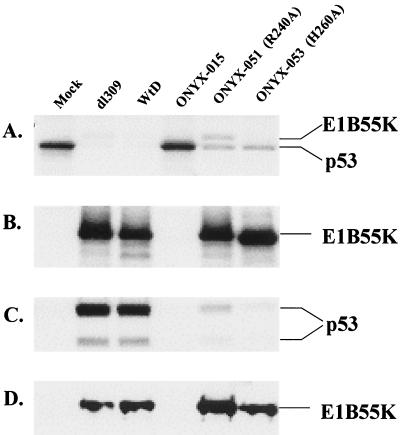
To directly examine the ability of E1B-55K mutants R240A and H260A to interact with p53, we performed an immunoprecipitation experiment using 35S-labeled cell extracts from infected A549 cells (Fig. 3). Anti-p53 antibody mAb421 coprecipitated small amounts of wild-type E1B-55K protein from dl309- and WtD-infected cells as expected (Fig. 3A). E1B-55K mutant R240A was also coprecipitated, indicating that it is capable of binding p53. Coprecipitation of H260A E1B-55K could be detected only after prolonged exposure, suggesting that this mutant interacts with p53 rather poorly. Incubation with the anti-E1B-55K antibody 2A6 immunoprecipitated E1B-55K proteins (Fig. 3B); however, it is difficult to determine whether p53 was coprecipitated, as the proteins share similar molecular weights and the E1B-55K protein was much more abundant. It is interesting to note that the amounts of p53 in ONYX-051 (R240A)- and -053 (H260A)-infected cells as detected by immunoprecipitation were lower than those in mock-infected cells, whereas by Western analysis, the p53 levels were higher in ONYX-051- and -053-infected cells. This is likely due to the fact that Western blot analysis detects the steady-state level of p53, whereas immunoprecipitation only detects newly synthesized p53. We hypothesize that R240A and H260A were able to mediate host cell shutoff (see below), causing p53 to be synthesized at a reduced rate compared to that of mock-infected cells. The slow rate of synthesis for p53 and its higher steady-state level in cells that are infected with ONYX-051 and -053 again suggest that these viruses have lost their ability to target p53 for degradation.
FIG. 3.
Coimmunoprecipitation of p53 by E1B-55K mutants. (A and B) Coimmunoprecipitation experiment. A549 cells were either mock infected (mock) or infected with various virus mutants at an MOI of 10. At 24 h postinfection, cells were metabolically labeled with [35S]methionine-cysteine for a 3-h period. Cell extracts were immunoprecipitated with anti-p53 antibody Ab421 (A) or anti-E1B-55K antibody 2A6 (B) and analyzed by SDS-PAGE as described in Materials and Methods. E1B-55K mutant R240A was expressed from ONYX-051, and H260A was expressed from ONYX-053. (C and D) In vitro-translated (and 35S-labeled) p53 was mixed with lysates prepared from infected A549 cells at 24 h post infection and immunoprecipitated with 2A6 anti-E1B-55K antibody. Immune complexes were separated by SDS-PAGE and visualized either directly by autoradiography (C) or by Western blotting with 2A6 antibody (D).
Because of the variability in the level of p53 in infected cell extracts (see Fig. 2), it is impossible to determine the relative binding efficiency of various E1B-55K mutant proteins to p53 from the coimmunoprecipitation experiment described above. In order to more accurately compare the efficiencies of binding of different E1B-55K mutant proteins to p53, we used an in vitro binding assay (37). Lysates of A549 cells infected with various adenoviruses were incubated with in vitro-translated 35S-labeled p53, and the anti-E1B-55K antibody 2A6 was used for immunoprecipitation. It was apparent from this experiment (Fig. 3C) that R240A (ONYX-051) and H260A (ONYX-053) bound p53 with significantly lower affinity than the wild-type E1B-55K protein. This inefficient precipitation of p53 was not due to lower levels of mutant E1B-55K protein in cells infected with ONYX-051 or -053; Western analysis confirmed that a similar or slightly higher level of E1B-55K protein was present in cell extracts derived from ONYX-051- and -053-infected cells (Fig. 3D). The inefficient binding of R240A and H260A to p53 may explain, at least in part, why p53 was not efficiently targeted for degradation in cells infected with ONYX-051 and -053.
Wild-type E1B-55K protein forms a complex with E4orf6 protein during adenovirus infection (6, 23). It has been suggested that this complex plays an important role in targeting p53 for active degradation (25, 28, 35) and in the regulation of the nuclear export of late viral mRNAs and cellular mRNAs (7, 12). Thus, it is important to determine if the mutations we introduced into E1B-55K changed the ability of this protein to complex with E4orf6. We examined this interaction by coimmunoprecipitation with an antibody against E1B-55K followed by Western analysis for E4orf6 (Fig. 4). The E1B-55K mutant R240A binds E4orf6 as well as the wild-type protein. However, the H260A mutant binds E4orf6 only very weakly. It is possible that the reduced binding to E4orf6 by H260A may also contribute to the lack of p53 degradation in ONYX-053 infection. T255A (ONYX-052) was included as a control, mainly because this mutation lies between the two mutations that disrupt binding to p53. Interestingly, T255A completely lost its ability to bind E4orf6 yet is still capable of degrading p53, as shown in Fig. 2. These results suggest that E1B-55K and E4orf6 may not need to form a stable complex to target p53 for active degradation.
FIG. 4.
Binding of E4orf6 by the E1B-55K mutants. A549 cells were either mock infected (mock) or infected with indicated virus mutants at an MOI of 10. At 24 h postinfection cells were lysed; cleared cell extracts were immunoprecipitated with anti-E1B-55K antibody 2A6. The precipitated materials were separated by SDS-PAGE and analyzed by Western blotting with a rat polyclonal antibody against E4orf6 (A) or with a rat anti-E1B-55K polyclonal antibody (B). (C) The level of E4orf6 expression in the infected cells by straight Western analysis using a polyclonal anti-E4orf6 antibody. dl355* is a dl309 derivative lacking the E4orf6 gene.
Immunofluorescent staining of p53 and E1B-55K.
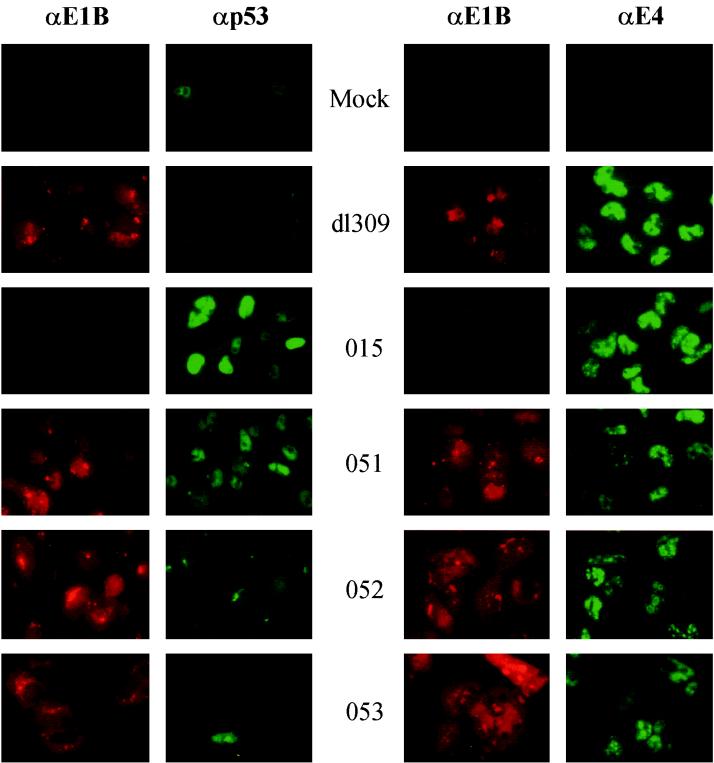
Next, we examined whether E1B-55K mutants R240A, T255A, and H260A are able to relocate p53 to the cytoplasm (Fig. 5). In mock-infected A549 cells, p53 was detected exclusively in nuclei. In cells infected with the wild-type virus dl309, p53 was present at a very low level, exclusively in the cytoplasm, and colocalized with the E1B-55K protein. Infection with ONYX-015 resulted in an elevated level of p53 localized in cell nuclei, as expected. In A549 cells infected with the R240A mutant virus (ONYX-051), the p53 level was elevated (compare ONYX-051 and mock-infected cells) in nuclei, as in the case of ONYX-015 infection. This is consistent with the fact that R240A binds p53 very weakly. Infection with the T255A mutant virus (ONYX-052) showed results similar to those for dl309, indicating that T255A can relocate p53 to the cytoplasm for degradation. Unexpectedly, cells that were infected with H260A virus (ONYX-053) showed variable p53 levels. Approximately two-thirds of the infected cells had undetectable levels of p53, whereas the rest showed high levels of p53 in nuclei. While the reason for this inconsistency is unclear, it is possible that this may reflect the cell cycle status of the cells when they were infected. Interestingly, this phenomenon was observed only in A549 cells. In U2OS cells (which harbor wild-type p53), p53 was present at high levels in nuclei when infected with ONYX-015, -051, and -053 but at low levels when infected with dl309 and ONYX-052 (data not shown).
FIG. 5.
Indirect immunofluorescent staining of adenovirus- and mock-infected (mock) cells. A549 cells grown on chamber slides were infected with dl309 or ONYX-015, -051, -052, or -053 at an MOI of 10 or were mock infected. At 24 h postinfection the cells were fixed, permeabilized, and analyzed by indirect immunofluorescent staining using the E1B-55K-specific monoclonal antibody 9C10 (αE1B), p53-specific monoclonal antibody DO-1 (αp53), and E4orf6-specific polyclonal antibody 1807-3 (αE4). Representative fields are shown for all cases.
Analysis of late functions of the E1B-55K mutants.
Forming a complex with E4orf6, E1B-55K actively facilitates nuclear export of the late viral mRNAs while concomitantly inhibiting the nuclear export of the cellular mRNAs. E1B-55K may also be involved in the active translation of late viral mRNAs (13). In the absence of E1B-55K, nuclear export and translation of late viral mRNAs do not occur efficiently, resulting in lower expression of the late viral proteins than of the wild-type virus (2, 6, 12, 20). To examine whether our E1B-55K mutant viruses expressed late proteins efficiently, we examined the synthesis of the adenovirus fiber protein. Previous work indicated that the mRNA for fiber protein depends on the E1B-55K/E4orf6 complex for efficient nuclear export and expression (24). Whereas the ability of ONYX-015 to express fiber protein was severely compromised compared to that of wild-type virus, all other mutant adenoviruses expressed fiber protein at a level similar to that of the wild-type virus (Fig. 2). As a control, we examined the expression of E2A, an early protein involved in viral DNA replication. The E2A protein level was uniform among all samples, suggesting that the difference in fiber expression was not due to variance in viral DNA replication. Thus, the E1B-55K protein of ONYX-051 and -053 was unable to inactivate p53 yet retained the ability to promote high levels of structural protein synthesis.
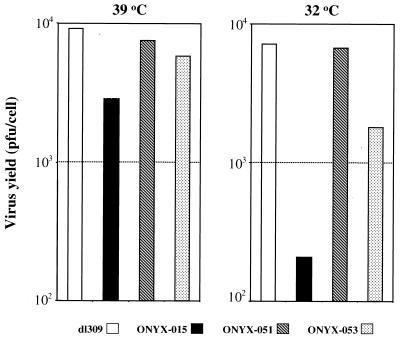
Several groups have reported that the replication of E1B-55K mutant adenoviruses is temperature dependent (9, 13, 16, 21). Whereas replication of E1B-55K mutant adenoviruses is equivalent to that of wild-type virus at 39°C, at 32°C replication of E1B-55K mutant viruses is significantly reduced compared to that of wild-type virus. This cold-sensitive phenotype of the E1B-55K-defective viruses presumably reflects the more severe defects in mRNA transport or late-gene expression at the colder temperature (9, 13). To determine whether ONYX-051 (R240A) and -053 (H260A) have a cold-sensitive phenotype, we performed the cold-sensitivity assay, using dl309 and ONYX-015 as controls (Fig. 6). As expected, all viruses replicated similarly at 39°C. The yield of dl309 was approximately fourfold higher than that of ONYX-015, and the yields of ONYX-051 and -053 fell between. However, at 32°C the ONYX-015 yield was reduced nearly 35-fold compared to that of dl309, consistent with previous reports. Replication of ONYX-051 was essentially identical to that of dl309, while replication of ONYX-053 was only slightly reduced (fourfold). These results indicated that mutant R240A can completely rescue the cold-sensitive phenotype of the E1B-55K-defective adenovirus, whereas H260A is partially functional.
FIG. 6.
Replication of dl309, ONYX-015, ONYX-051, and ONYX-053 as a function of time postinfection in p53-null H1299 cells at 32 and 39°C. H1299 cells were infected and maintained at two temperatures, 32 and 39°C. Infections at 32°C were performed 1 h after the temperature shift from 39°C. All infections were performed at an MOI of 5. Infected cells were incubated at 32 and 39°C, respectively. Ninety-six hours postinfection cells and culture media were harvested, pooled, and freeze-thawed three times to release virus particles. Viral yields were determined by enzyme-linked immunosorbent assay on a 293 cell monolayer. Total viral yields were divided by the number of cells at the time of infection to determine viral production per cell. Results are the average from two independent experiments that are highly consistent with each other.
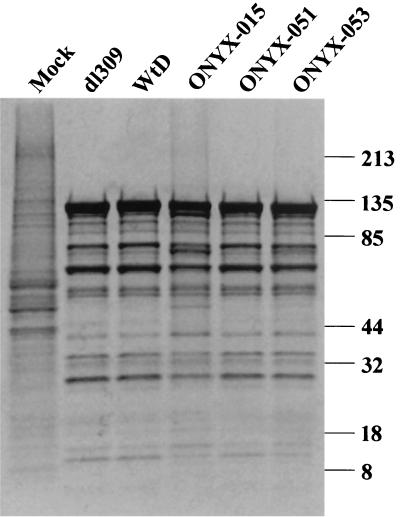
When total protein expression from infected cells was compared (Fig. 7), it was clear that expression of late viral proteins was lower in ONYX-015-infected cells than in dl309- and WtD-infected cells. At the same time, de novo synthesis of the cellular proteins was substantially reduced in cells infected with dl309 and WtD (compared to mock infection), presumably due to host cell shutoff. In ONYX-015-infected cells, however, shutoff of host protein synthesis was deficient, as evidenced by the smeared background in the lane labeled ONYX-015. The protein synthesis profile in cells infected with ONYX-051 and -053 was similar to that in cells infected with wild-type viruses dl309 and WtD. This observation suggested that mutants R240A and H260A are capable of modulating mRNA trafficking in favor of late viral mRNA nuclear export and promoting host cell shutoff.
FIG. 7.
Protein expression during the late phase of adenovirus infection. A549 cells were either mock infected (mock) or infected with various adenovirus mutants at an MOI of 10. At 24 h postinfection, cells were metabolically labeled with [35S]methionine-cysteine for a 3-h period. Cell extracts were resolved by SDS-PAGE (4 to 20% gradient gel). The positions of the molecular mass markers are indicated at the right.
Taken together, these results suggest that the E1B-55K mutant R240A may entirely maintain the normal late functions of the wild-type protein, whereas mutant H260A appeared to be at least partially active in the late functions.
Analysis of cytolytic activity in human cancer cells.
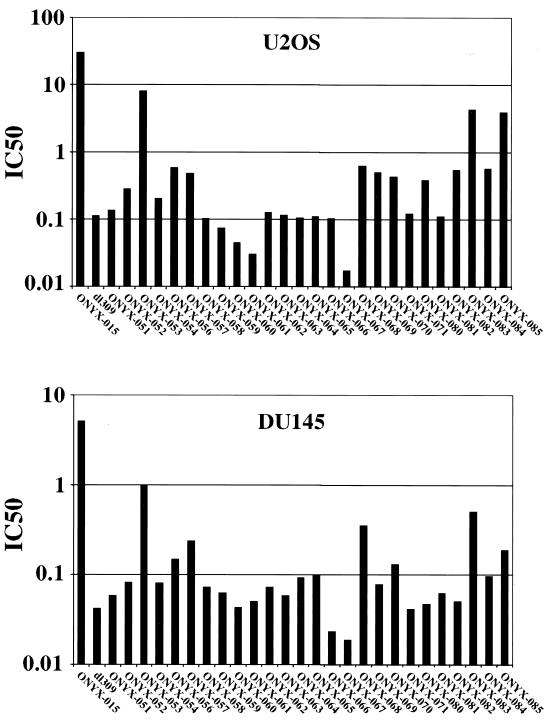
Each E1B-55K mutant virus was tested for its ability to replicate in and to kill human tumor cells. Although A549 cells were used to characterize the functional interaction between p53 and various 55K point mutants, this cell line has been reported to be rather permissive for the replication of the 55K-mutant viruses (18). Therefore, we used two other human cancer cell lines, U2OS and DU145, to examine the oncolytic activity of our mutant viruses. A quantitative assay, the MTT assay, was used for this analysis (see Materials and Methods). The MOI at which 50% of the cells were killed was determined and plotted (Fig. 8). In both cell lines, ONYX-015 was attenuated compared to its wild-type counterpart dl309. Among the viruses that were generated for this study, most (including ONYX-051) were comparable to dl309 in their ability to infect and kill tumor cells, though some (ONYX-053, -083, and -085) were significantly less potent than the wild-type virus but were more active than ONYX-015. In the case of ONYX-053, its tumor cytolytic activity was 35- to 100-fold lower than that of dl309 but was more active than ONYX-015 by a factor of four- to fivefold.
FIG. 8.
Cytolytic activity in tumor cells. DU145 and U2OS cells were seeded into 96-well plates at a density of 2.5 × 103 cells/well. Twenty-four hours after seeding, cells were infected with serial threefold dilutions of E1B-55K mutant viruses, ranging from an MOI of 30 to an MOI of 1.5 × 10−3. dl309 and ONYX-015 were included as controls. MTT assays were performed 6 days after infection as described in Materials and Methods. The MOIs that resulted in 50% cell killing were defined as IC50 and were plotted for each virus. Results from one of the two independent experiments are shown here. Similar results were obtained in the other experiment.
DISCUSSION
We have constructed and analyzed a series of single-amino-acid substitution mutations of the adenovirus E1B-55K protein in an effort to separate the p53 binding and inactivation function and late functions of this protein. Analysis of each mutant indicated that all of the E1B-55K proteins were expressed in infected cells. Of the 26 mutants, 24 mediated p53 degradation as efficiently as the wild-type protein. Two mutants, R240A (ONYX-051) and H260A (ONYX-053), failed to degrade p53 (Fig. 2). In fact, p53 levels in A549 cells infected with ONYX-051 and ONYX-053 were noticeably higher than that in the mock-infected A549 cells, although they were slightly lower than that in cells infected with ONYX-015. In in vitro assays, R240A and H260A mutant E1B-55K proteins associated with p53 with a greatly reduced affinity compared to the wild-type protein (Fig. 3). H260A also lost the ability to bind the E4orf6 protein, while R240A was fully functional in this interaction (Fig. 4). Immunofluorescent staining experiments confirmed that p53 was present at high levels in the nuclei of cells that were infected with ONYX-051 (Fig. 5). In cells that were infected with ONYX-053, p53 levels varied (Fig. 5), a phenomenon that deserves further detailed analysis. Both ONYX-051 (R240A) and ONYX-053 (H260A) expressed adenoviral fiber protein efficiently (Fig. 2), mediated shutoff of host cell protein synthesis (Fig. 7), and did not display the cold-sensitive growth phenotype (Fig. 6). Finally, ONYX-051 (R240A) replicated efficiently in human cancer cells, while ONYX-053 (H260A) did so only partially (Fig. 8).
Taken together, these data suggest that we have successfully isolated two E1B-55K mutants that are defective in p53 binding and inactivation yet retain many of the late functions of the wild-type protein which are essential for efficient replication of adenovirus. These results suggest that these distinct E1B-55K functions are separable. The implication of the functional separation is twofold. First, these E1B-55K mutants will allow us to better investigate the molecular mechanisms of the tumor-selective adenovirus ONYX-015. Second, by incorporating these mutations, we can create adenoviruses that maintain the tumor selectivity of ONYX-015 but replicate more efficiently in tumor cells than ONYX-015. It is gratifying that both ONYX-051 and -053 showed higher cytolytic activity in tumor cells than ONYX-015 (Fig. 8). We are currently testing whether these mutant viruses retained the tumor cell selectivity of ONYX-015.
It was hypothesized that deletion of the E1B-55K gene in ONYX-015 allows this virus to replicate efficiently only in tumor cells that are defective in p53 function but not in normal cells where p53 function is normal (5). This hypothesis has gained support from a number of in vitro and in vivo studies (15, 27). Recently, it was reported (26) that the loss of p14ARF function facilitates ONYX-015 replication. This finding demonstrated for the first time that the integrity of the p53 functional pathway rather than the p53 gene itself may determine the outcome of ONYX-015 infection. Nonetheless, several groups have described in vitro studies showing that the host range specificity of ONYX-015 is independent of p53 gene status (9, 13, 14, 29). It has been suggested that the loss of E1B-55K's RNA transport activity rather than its p53 binding and inactivation function may account for the tumor cell specificity of ONYX-015. The mutant viruses we created in this study, particularly ONYX-051 and -053, can be used to explore some of these differences.
Grand et al. (11) reported that the region of E1B-55K essential for p53 interaction lies between amino acids 216 and 235. This domain only partially overlaps the p53-binding region identified by Kao et al. and Yew et al. (19, 39), which maps to amino acids 224 to 354. The reason for this apparent discrepancy is not clear. Since Grand et al. used fusion proteins between Ad2 E1B-55K and Ad12 E1B-55K in their analysis, it is possible that additional domains in the Ad12 E1B-55K protein may be involved in p53 binding. Our results indicate that amino acids 240 and 260 are critical for p53 binding and inactivation, consistent with the results of Kao et al. Further studies are necessary to identify the minimal essential region of the E1B-55K protein for p53 interaction.
It was reported (19, 39) that a four-amino-acid insertional mutation at amino acid 380 of the E1B-55K protein (S380) reduced its interaction with p53, while the ability of the virus to replicate was not compromised. Furthermore, this mutant virus (S380) resembles the wild-type virus with respect to late viral protein synthesis and host cell shutoff. Thus, S380 appears to be very similar to ONYX-051 and -053. It should be noted that the characterization of this mutant, S380, was performed in HeLa cells, where human papillomavirus oncoproteins exist and may complicate the analysis due to interactions between human papillomavirus 18 E6 protein and p53. Recently, Gabler et al. reported that S380 failed to bind E1B-AP5, suggesting that this mutant may be defective in mediating late viral mRNA transport (8). Further characterization of this mutant with respect to its interaction with E4orf6, its subcellular localization, its activity in mRNA transport, and its ability to rescue the cold-sensitive phenotype would allow ONYX-051 and -053 and S380 to be directly compared.
Two of the E1B-55K mutant proteins we have tested, T255A (ONYX-052) and H260A (ONYX-053), are defective in binding the E4orf6 protein, as determined by the coimmunoprecipitation experiment (Fig. 4). No interaction between the E1B-55K T255A mutant and E4orf6 could be detected. However, in cells that were infected with ONYX-052, p53 was clearly exported to the cytoplasm (Fig. 5) and degraded (Fig. 2 and 5). These results suggest that E1B-55K and E4orf6, both shown to be necessary for p53 degradation (25, 28, 35), may not need to be in a stable complex to target p53 for degradation. Moreover, T255A appeared to be active in mediating fiber expression (Fig. 2) and did not display the cold-sensitive phenotype (Fig. 6), suggesting that its function in mRNA trafficking during the late phase of adenovirus infection was intact. Similarly, H260A bound E4orf6 poorly yet was still capable of mediating efficient fiber expression and other late functions. These observations are in contrast to previous literature, which suggests that E1B-55K and E4orf6 need to interact to modulate mRNA transport. It is possible that other cellular or viral proteins may interact with these adenoviral proteins and keep them in the same complex (10, 23). It was reported recently (19a) that the E1B-55K protein itself is a highly active shuttle protein capable of shuttling independent of E4orf6. Whether this finding supports our observations remains to be tested.
Phase I and II clinical trials with the head-and-neck cancer patients have indicated that ONYX-015 replicates in and destroys tumor cells at the exclusion of surrounding normal tissue. In combination with standard chemotherapy, intratumoral injection of ONYX-015 resulted in tumor regression in more than two-thirds of the patients treated. However, the deletion of E1B-55K in ONYX-015 has compromised the replication capability of this virus. Compared to wild-type virus, ONYX-015 is attenuated in a number of tumor cell lines (9, 13), possibly limiting its potential as an anticancer drug. One can imagine that virus mutants that have improved replication capacity yet retain the tumor selectivity of ONYX-015 would provide more efficient treatment for cancer patients. In this regard, ONYX-051 and -053 and viruses like them may serve as candidates for clinical development. Additional studies are underway in our laboratory to determine the replication capacity of these viruses in the presence and absence of normal p53 activity and to compare their replication efficiency in normal primary cells and tumor cells.
ACKNOWLEDGMENTS
We thank Adam Sampson-Johannes, Sandy McCoy, and Wen Yan for excellent technical support, Josh Watanabe for virus production, and Jaina Sumortin for DNA sequencing. We also thank Tom Alber, Matthias Dobbelstein, Leisa Johnson, and Keith N. Leppard for helpful discussions, Philip E. Branton for the E4orf6 antibodies, Clodaugh O'Shea for the rat anti-E1B-55K antibodies, and Allan Balmain and Tom Dubensky for critical reading of the manuscript.
REFERENCES
- 1.Babich A, Feldman L T, Nevins J R, Darnell J E, Weinberger C. Effect of adenovirus on metabolism of specific host mRNAs: transport control and specific translational discrimination. Mol Cell Biol. 1983;3:1212–1221. doi: 10.1128/mcb.3.7.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Babiss L E, Ginsberg H S, Darnell J E J. Adenovirus E1B proteins are required for accumulation of late viral mRNA and for effects on cellular mRNA translation and transport. Mol Cell Biol. 1985;5:2552–2558. doi: 10.1128/mcb.5.10.2552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barker D D, Berk A J. Adenovirus proteins from both E1B reading frames are required for transformation of rodent cells by viral infection and DNA transfection. Virology. 1987;156:107–121. doi: 10.1016/0042-6822(87)90441-7. [DOI] [PubMed] [Google Scholar]
- 4.Beltz G A, Flint S J. Inhibition of HeLa cell protein synthesis during adenovirus infection. Restriction of cellular messenger RNA sequences to the nucleus. J Mol Biol. 1979;131:353–373. doi: 10.1016/0022-2836(79)90081-0. [DOI] [PubMed] [Google Scholar]
- 5.Bischoff J R, Kirn D H, Williams A, Heise C, Horn S, Muna M, Ng L, Nye J A, Sampson-Johannes A, Fattaey A, McCormick F. An adenovirus mutant that replicates selectively in p53-deficient human tumor cells. Science. 1996;274:373–376. doi: 10.1126/science.274.5286.373. [DOI] [PubMed] [Google Scholar]
- 6.Bridge E, Ketner G. Interaction of adenoviral E4 and E1b products in late gene expression. Virology. 1990;174:345–353. doi: 10.1016/0042-6822(90)90088-9. [DOI] [PubMed] [Google Scholar]
- 7.Dobbelstein M, Roth J, Kimberly W T, Levine A J, Shenk T. Nuclear export of the E1B 55-kDa and E4 34-kDa adenoviral oncoproteins mediated by a rev-like signal sequence. EMBO J. 1997;16:4276–4284. doi: 10.1093/emboj/16.14.4276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gabler S, Schutt H, Groitl P, Wolf H, Shenk T, Dobner T. E1B 55-kilodalton-associated protein: a cellular protein with RNA-binding activity implicated in nucleocytoplasmic transport of adenovirus and cellular mRNAs. J Virol. 1998;72:7960–7971. doi: 10.1128/jvi.72.10.7960-7971.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goodrum F D, Ornelles D A. p53 status does not determine outcome of E1B 55-kilodalton mutant adenovirus lytic infection. J Virol. 1998;72:9479–9490. doi: 10.1128/jvi.72.12.9479-9490.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goodrum F D, Shenk T, Ornelles D A. Adenovirus early region 4 34-kilodalton protein directs the nuclear localization of the early region 1B 55-kilodalton protein in primate cells. J Virol. 1996;70:6323–6335. doi: 10.1128/jvi.70.9.6323-6335.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Grand R J, Parkhill J, Szestak T, Rookes S M, Roberts S, Gallimore P H. Definition of a major p53 binding site on Ad2E1B58K protein and a possible nuclear localization signal on the Ad12E1B54K protein. Oncogene. 1999;18:955–965. doi: 10.1038/sj.onc.1202358. [DOI] [PubMed] [Google Scholar]
- 12.Halbert D N, Cutt J R, Shenk T. Adenovirus early region 4 encodes functions required for efficient DNA replication, late gene expression, and host cell shutoff. J Virol. 1985;56:250–257. doi: 10.1128/jvi.56.1.250-257.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harada J N, Berk A J. p53-independent and -dependent requirements for E1B–55K in adenovirus type 5 replication. J Virol. 1999;73:5333–5344. doi: 10.1128/jvi.73.7.5333-5344.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hay J G, Shapiro N, Sauthoff H, Heitner S, Phupakdi W, Rom W N. Targeting the replication of adenoviral gene therapy vectors to lung cancer cells: the importance of the adenoviral E1b–55kD gene. Hum Gene Ther. 1999;10:579–590. doi: 10.1089/10430349950018652. [DOI] [PubMed] [Google Scholar]
- 15.Heise C, Sampson-Johannes A, Williams A, McCormick F, Von Hoff D D, Kirn D H. ONYX-015, an E1B gene-attenuated adenovirus, causes tumor-specific cytolysis and antitumoral efficacy that can be augmented by standard chemotherapeutic agents. Nat Med. 1997;3:639–645. doi: 10.1038/nm0697-639. [DOI] [PubMed] [Google Scholar]
- 16.Ho Y S, Galos R, Williams J. Isolation of type 5 adenovirus mutants with a cold-sensitive host range phenotype: genetic evidence of an adenovirus transformation maintenance function. Virology. 1982;122:109–124. doi: 10.1016/0042-6822(82)90381-6. [DOI] [PubMed] [Google Scholar]
- 17.Horridge J J, Leppard K N. RNA-binding activity of the E1B 55-kilodalton protein from human adenovirus type 5. J Virol. 1998;72:9374–9379. doi: 10.1128/jvi.72.11.9374-9379.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Imler J L, Chartier C, Dreyer D, Dieterle A, Sainte-Marie M, Faure T, Pavirani A, Mehtali M. Novel complementation cell lines derived from human lung carcinoma A549 cells support the growth of E1-deleted adenovirus vectors. Gene Ther. 1996;3:75–84. [PubMed] [Google Scholar]
- 19.Kao C C, Yew P R, Berk A J. Domains required for in vitro association between the cellular p53 and the adenovirus 2 E1B 55K proteins. Virology. 1990;179:806–814. doi: 10.1016/0042-6822(90)90148-k. [DOI] [PubMed] [Google Scholar]
- 19a.Kratzer F, Rosorius O, Heger P, Hirschmann N, Dobner T, Hauber J, Stauber R H. The adenovirus type 5 E1B-55K oncoprotein is a highly active shuttle protein and shuttling is independent of E4orf6, p53 and Mdm2. Oncogene. 2000;19:850–857. doi: 10.1038/sj.onc.1203395. [DOI] [PubMed] [Google Scholar]
- 20.Leppard K N. Selective effects on adenovirus late gene expression of deleting the E1b 55K protein. J Gen Virol. 1993;74:575–582. doi: 10.1099/0022-1317-74-4-575. [DOI] [PubMed] [Google Scholar]
- 21.Leppard K N, Shenk T. The adenovirus E1B 55kd protein influences mRNA transport via an intranuclear effect on RNA metabolism. EMBO J. 1989;8:2329–2336. doi: 10.1002/j.1460-2075.1989.tb08360.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McGrory W J, Bautista D S, Graham F L. A simple technique for the rescue of early region 1 mutations into infectious human adenovirus type 5. Virology. 1988;163:614–617. doi: 10.1016/0042-6822(88)90302-9. [DOI] [PubMed] [Google Scholar]
- 23.Ornelles D A, Shenk T. Localization of the adenovirus early region 1B 55-kilodalton protein during lytic infection: association with nuclear viral inclusions requires the early region 4 34-kilodalton protein. J Virol. 1991;65:424–429. doi: 10.1128/jvi.65.1.424-429.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pilder S, Moore M, Logan J, Shenk T. The adenovirus E1B–55K transforming polypeptide modulates transport or cytoplasmic stabilization of viral and host cell mRNAs. Mol Cell Biol. 1986;6:470–476. doi: 10.1128/mcb.6.2.470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Querido E, Marcellus R C, Lai A, Charbonneau R, Teodoro J G, Ketner G, Branton P E. Regulation of p53 levels by the E1B 55-kilodalton protein and E4orf6 in adenovirus-infected cells. J Virol. 1997;71:3788–3798. doi: 10.1128/jvi.71.5.3788-3798.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ries S J, Brandts C H, Chung A S, Biederer C H, Hann B C, Lipner E M, McCormick F, Korn M W. Loss of p14ARF in tumor cells facilitates replication of the adenovirus mutant dl1520 (ONYX-015) Nat Med. 2000;6:1128–1133. doi: 10.1038/80466. [DOI] [PubMed] [Google Scholar]
- 27.Rogulski K R, Freytag S O, Zhang K, Gilbert J D, Paielli D L, Kim J H, Heise C C, Kirn D H. In vivo antitumor activity of ONYX-015 is influenced by p53 status and is augmented by radiotherapy. Cancer Res. 2000;60:1193–1196. [PubMed] [Google Scholar]
- 28.Roth J, Konig C, Wienzek S, Weigel S, Ristea S, Dobbelstein M. Inactivation of p53 but not p73 by adenovirus type 5 E1B 55-kilodalton and E4 34-kilodalton oncoproteins. J Virol. 1998;72:8510–8516. doi: 10.1128/jvi.72.11.8510-8516.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rothmann T, Hengstermann A, Whitaker N J, Scheffner M, zur Hausen H. Replication of ONYX-015, a potential anticancer adenovirus, is independent of p53 status in tumor cells. J Virol. 1998;72:9470–9478. doi: 10.1128/jvi.72.12.9470-9478.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rubenwolf S, Schutt H, Nevels M, Wolf H, Dobner T. Structural analysis of the adenovirus type 5 E1B 55-kilodalton-E4orf6 protein complex. J Virol. 1997;71:1115–1123. doi: 10.1128/jvi.71.2.1115-1123.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sarnow P, Hearing P, Anderson C W, Halbert D N, Shenk T, Levine A J. Adenovirus early region 1B 58,000-dalton tumor antigen is physically associated with an early region 4 25,000-dalton protein in productively infected cells. J Virol. 1984;49:692–700. doi: 10.1128/jvi.49.3.692-700.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sarnow P, Ho Y S, Williams J, Levine A J. Adenovirus E1b-58 kd tumor antigen and SV40 large tumor antigen are physically associated with the same 54 kd cellular protein in transformed cells. Cell. 1982;28:387–394. doi: 10.1016/0092-8674(82)90356-7. [DOI] [PubMed] [Google Scholar]
- 33.Sarnow P, Sullivan C A, Levine A J. A monoclonal antibody detecting the adenovirus type 5-E1b–58Kd tumor antigen: characterization of the E1b–58Kd tumor antigen in adenovirus-infected and -transformed cells. Virology. 1982;120:510–517. doi: 10.1016/0042-6822(82)90054-x. [DOI] [PubMed] [Google Scholar]
- 34.Shen Y, Shenk T. Relief of p53-mediated transcriptional repression by the adenovirus E1B 19-kDa protein or the cellular Bcl-2 protein. Proc Natl Acad Sci USA. 1994;91:8940–8944. doi: 10.1073/pnas.91.19.8940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steegenga W T, Riteco N, Jochemsen A G, Fallaux F J, Bos J L. The large E1B protein together with the E4orf6 protein target p53 for active degradation in adenovirus infected cells. Oncogene. 1998;16:349–357. doi: 10.1038/sj.onc.1201540. [DOI] [PubMed] [Google Scholar]
- 36.Tollefson A E, Hermiston T W, Wold W S M. Preparation and titration of CsCl-banded adenovirus stock. Methods Mol Med. 1998;20:1–9. doi: 10.1385/1-59745-166-5:223. [DOI] [PubMed] [Google Scholar]
- 37.Wienzek S, Roth J, Dobbelstein M. E1B 55-kilodalton oncoproteins of adenovirus type 5 and 12 inactivate and relocalize p53, but not p51 or p73, and cooperate with E4orf6 proteins to destabilize p53. J Virol. 2000;74:193–202. doi: 10.1128/jvi.74.1.193-202.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yew P R, Berk A J. Inhibition of p53 transactivation required for transformation by adenovirus early 1B protein. Nature. 1992;357:82–85. doi: 10.1038/357082a0. [DOI] [PubMed] [Google Scholar]
- 39.Yew P R, Kao C C, Berk A J. Dissection of functional domains in the adenovirus 2 early 1B 55K polypeptide by suppressor-linker insertional mutagenesis. Virology. 1990;179:795–805. doi: 10.1016/0042-6822(90)90147-j. [DOI] [PubMed] [Google Scholar]
- 40.Yew P R, Liu X, Berk A J. Adenovirus E1B oncoprotein tethers a transcriptional repression domain to p53. Genes Dev. 1994;8:190–202. doi: 10.1101/gad.8.2.190. [DOI] [PubMed] [Google Scholar]