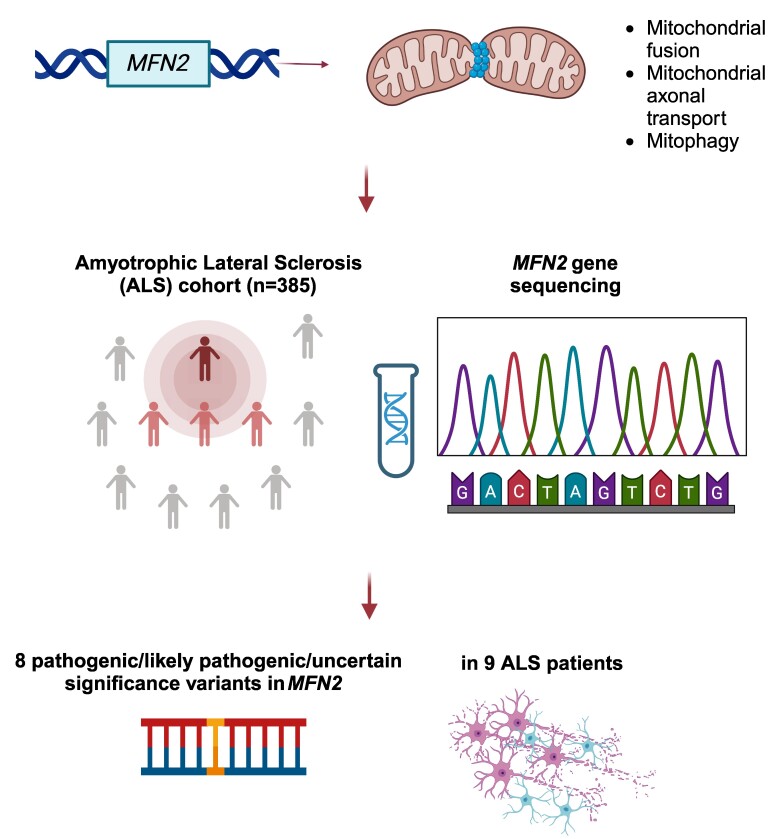
Abstract
The MFN2 gene encodes mitofusin 2, a key protein for mitochondrial fusion, transport, maintenance and cell communication. MFN2 mutations are primarily linked to Charcot–Marie–Tooth disease type 2A. However, a few cases of amyotrophic lateral sclerosis and amyotrophic lateral sclerosis/frontotemporal dementia phenotypes with concomitant MFN2 mutations have been previously reported. This study examines the clinical and genetic characteristics of an Italian cohort of amyotrophic lateral sclerosis patients with rare, non-synonymous MFN2 mutations. A group of patients (n = 385) diagnosed with amyotrophic lateral sclerosis at our Neurology Units between 2008 and 2023 underwent comprehensive molecular testing, including MFN2. After excluding pathogenic mutations in the main amyotrophic lateral sclerosis–related genes (i.e. C9orf72, SOD1, FUS and TARDBP), MFN2 variants were classified based on the American College of Medical Genetics and Genomics guidelines, and demographic and clinical data of MFN2-mutated patients were retrieved. We identified 12 rare, heterozygous, non-synonymous MFN2 variants in 19 individuals (4.9%). Eight of these variants, carried by nine patients (2.3%), were either pathogenic, likely pathogenic or variants of unknown significance according to the American College of Medical Genetics and Genomics guidelines. Among these patients, four exhibited a familial pattern of inheritance. The observed phenotypes included classic and bulbar amyotrophic lateral sclerosis, amyotrophic lateral sclerosis/frontotemporal dementia, flail arm, flail leg and progressive muscular atrophy. Median survival after disease onset was extremely variable, ranging from less than 1 to 13 years. This study investigates the prevalence of rare, non-synonymous MFN2 variants within an Italian cohort of amyotrophic lateral sclerosis patients, who have been extensively investigated, enhancing our knowledge of the underlying phenotypic spectrum. Further research is needed to understand whether MFN2 mutations contribute to motor neuron disease and to what extent. Improving our knowledge regarding the genetic basis of amyotrophic lateral sclerosis is crucial both in a diagnostic and therapeutic perspective.
Keywords: mitofusin 2, amyotrophic lateral sclerosis, genetics, mitochondria
Abati et al. reported nine patients diagnosed with amyotrophic lateral sclerosis and carrying eight rare variants within the MFN2 gene, identified among a cohort of 385 amyotrophic lateral sclerosis patients who underwent MFN2 screening. This study enhances the knowledge regarding the phenotypic spectrum underlying MFN2 alterations.
Graphical Abstract
Graphical Abstract.
Introduction
Amyotrophic lateral sclerosis is a devastating neurodegenerative disease characterized by the progressive loss of motor neurons (MNs) in the motor cortex and spinal cord.1,2 Sporadic amyotrophic lateral sclerosis, occurring without any family history, represents 85–90% of all cases, with median onset age ranging 58–63 years.1 Familial amyotrophic lateral sclerosis accounts for the remaining 10–15% cases, with a slightly younger age at onset (47–53 years).1,3 Causative mutations have been identified in more than 50 genes. However, most mutations are found in only four of them (i.e. C9orf72, TARDBP, FUS and SOD1), which account for almost 75% familial amyotrophic lateral sclerosis and 20% sporadic amyotrophic lateral sclerosis cases.1,4,5 Despite being the most common form of MN disease (MND) in adults, effective treatments for amyotrophic lateral sclerosis, particularly for sporadic forms, remain elusive. In this scenario, the molecular therapy approach is emerging as a compelling and viable strategy, bolstered by the recent approval of tofersen,6 which underscores the importance of identifying genetic causes in amyotrophic lateral sclerosis patients.
Lately, a growing number of genes have been linked to both Charcot–Marie–Tooth disease type 2 and amyotrophic lateral sclerosis (i.e. CHCHD10, GARS, FIG4, KIF5A, NEFH, VCP and SPG11),7 suggesting a genetic overlap between the two diseases, although further studies are needed to confirm this association. Interestingly, a few cases of occurrence of amyotrophic lateral sclerosis/frontotemporal dementia with MFN2 mutations have been reported in the literature.8,9 More recently, Russel et al.10 reported six rare MFN2 variants, which were classified as deleterious according to the VAAST (Variant Annotation, Analysis and Search Tool) and Phevor (Phenotype Driven Variant Ontological Re-ranking) software, within a cohort of 140 amyotrophic lateral sclerosis patients, and other 15 variants in the publicly available database ALSdb, composed of ∼2800 amyotrophic lateral sclerosis patients. Autosomal dominant and, to a lesser extent, recessive mutations in MFN2 are associated with Charcot–Marie–Tooth disease type 2A,11 the most frequent axonal subtype of Charcot–Marie–Tooth disease. Charcot–Marie–Tooth disease type 2A has been associated with early-onset, severe motor-predominant polyneuropathy presenting with muscle weakness, atrophy and sensory loss with mainly distal involvement.12,13 Moreover, Charcot–Marie–Tooth disease type 2A patients may manifest a wide spectrum of additional symptoms, such as optic atrophy, pyramidal tract alterations and cerebral white matter abnormalities.12-18 While there is still no cure for Charcot–Marie–Tooth disease type 2A, our group has been actively involved in the development of gene therapy approaches aimed at modulating mutant/wild-type MFN2 expression in preclinical models, as described in our recent work,19 an approach that could be applied to all MFN2-related diseases.
MFN2 is a mitochondrial transmembrane GTPase that plays a crucial role in several mitochondrial processes, including fusion, trafficking, turnover and organelle interactions, ensuring proper mitochondrial shape, function and distribution within cells.20,21 The mitochondrial fusion process involves the MFN2 heptad repeat domain and GTP hydrolysis, leading to the binding and oligomer formation of MFN2 and MFN1 proteins on distinct mitochondria, facilitating their fusion into a single mitochondrion. The importance of MFN2 is evident from studies led in Mfn2 knock-out (KO) mice, which do not survive past midgestation, and homozygous mutants that die postnatally.20,22 Intriguingly, in a mouse model of amyotrophic lateral sclerosis, the MFN2 protein was found to interact with TDP-43, a protein involved in the pathogenesis and neuropathology of frontotemporal dementia and amyotrophic lateral sclerosis.23
Here, we retrospectively reviewed our cohort of 385 amyotrophic lateral sclerosis patients to investigate the prevalence of rare, non-synonymous MFN2 variants. Our analysis led to the identification of eight different MFN2 variants, which were classified as either pathogenic, likely pathogenic or variants of unknown significance (VUS) according to the American College of Medical Genetics and Genomics (ACMG) criteria, in heterozygous state in nine individuals. The observed phenotypes ranged across a wide spectrum of MNDs, including classic amyotrophic lateral sclerosis, amyotrophic lateral sclerosis/frontotemporal dementia and restricted presentations.
Materials and methods
Data collection
We recruited a cohort of 385 patients with MND (amyotrophic lateral sclerosis, primary lateral sclerosis and progressive muscular atrophy) according to the El Escorial and Awaji-Shima criteria,24,25 among those diagnosed (n = 1600) in the MND Clinics of the Neurology Units of Fondazione IRCCS Ca′ Granda Ospedale Maggiore Policlinico of Milan and IRCCS Auxologico of Milan between January 2008 and July 2023. Neurological evaluation and medical chart review were performed by neurologists experienced in neuromuscular diseases. Gender, age and site of onset, family history of neuromuscular disorders, disease duration, presence of bulbar and respiratory involvement and presence of extra-motor features were recorded.
Genetic analysis
The extraction of genomic DNA from peripheral blood samples was performed using established protocols (Qiagen’s FlexiGene DNA Handbook).
Given the length of the study period, genetic analysis was performed with different methods, namely, Sanger sequencing, next-generation sequencing (NGS) MN gene panel, whole-exome sequencing (WES) and short-read whole-genome sequencing (WGS). All patients underwent analysis of the SOD1 gene, exon 6 of the TARDBP gene and exons 13-14-15 of the FUS gene, as previously described.26 Analysis of C9orf72 expansion was performed using the Asuragen AmplideX® polymerase chain reaction (PCR)/CE C9orf72 Kit, according to the manufacturer’s instructions.
In a subset of patients, a combination of PCR analysis and subsequent direct Sanger sequencing on an ABI Prism 3130 apparatus was conducted to analyse the coding sequence of MFN2 (NM_014874.3), as previously described.18 The specific primers employed are available upon request. In the other cases, NGS was run on Illumina platforms (MySeq and NextSeq 550), according to the manufacturer’s instructions.
All patients were screened for non-synonymous variants in MFN2. All the MFN2 variants detected were searched for within Franklin using the Genoox website (https://franklin.genoox.com) and screened in the Genome Aggregation Database (gnomAD). The categorization of identified variants into classes (i.e. ‘pathogenic’, ‘likely pathogenic’, ‘VUS’, ‘likely benign’ and ‘benign’) was carried out following the guidelines set forth by the ACMG criteria.27
In familial cases, co-segregation analyses were not possible because of the unavailability of genetic material of affected family members, except for Patient 4 (detailed segregation studies are described in Manini et al.26).
Statistical analysis
Descriptive statistics was performed where appropriate. Data are expressed in numbers (%) or median (range).
Protocol approval and informed consent
All patients or their legal representatives signed an informed consent form prior to enrolment. The study was approved by the local Institutional Review Board (Comitato Etico di Milano Area 2 Protocol Number 898_2020bis) and by the ethics committee of the IRCCS Instituto Auxologico Italiano (2023_01_31_01). The study was performed following relevant guidelines and regulations.
Results
MFN2-amyotrophic lateral sclerosis cohort overview
During the above-mentioned period, 385 amyotrophic lateral sclerosis patients underwent analysis of the MFN2 gene through either Sanger sequencing (n = 10), NGS panel (n = 88), WES (n = 239) or short-read WGS (n = 48). Among them, 23 (5.6%) had a positive family history of amyotrophic lateral sclerosis/frontotemporal dementia.
We identified 19 patients (4.9%) with 12 different non-synonymous variants in MFN2 (Table 1; Supplementary Table 1) in heterozygous state. None of our patients were homozygous or compound heterozygous for non-synonymous MFN2 variants. All the variants were absent or rare in public databases (gnomAD allele frequency < 0.01), except for the p.(His20Tyr) variant, which is present in both amyotrophic lateral sclerosis cases and controls in Project MinE data set.28,29
Table 1.
List of rare, non-synonymous MFN2 variants detected in the amyotrophic lateral sclerosis cohort under investigation
| # | Chromosomal position | Nucleotide change | Amino acid change | ACMG | gnomAD AF | Previously reported | First description of the patient |
|---|---|---|---|---|---|---|---|
| 1 | 1:11989226 | c.58C>T | p.(His20Tyr) | VUS | 7.07e−5 | In CMT2A (Bombelli et al.51; Al-Harbi et al.52) | Present study |
| 2 | 1:11992626 | c.247T>C | p.(Ser83Pro) | LP | 0 | Not previously reported | Present study |
| 3 | 1:11997346 | c.524C>T | p.(Ala175Val) | LP | 7.95e−6 | Not previously reported | Present study |
| 4 | 1:11997403 | c.581A>C | p.(Asp194Ala) | LP | 0 | Not previously reported | Vinciguerra et al.9 |
| 5 | 1:12001423 | c.839G>A | p.(Arg280His) | P | 4.00e−6 | In CMT2A (Zuchner et al.11) | Abati et al.18 |
| 6 | 1:12001423 | c.839G>A | p.(Arg280His) | P | 4.00e−6 | In CMT2A (Zuchner et al.11) | Abati et al.18 |
| 7 | 1:12002028 | c.1085C>T | p.(Thr362Met) | P | 3.18e−5 | In CMT2A - Heterozygous state—mild phenotype (Chung et al.17) - Compound heterozygous state with either exon 5–6 deletion (Carr et al.53) or p.(Ala164Val) (Nicholson et al.54; Calvo et al.55)—severe and early-onset phenotype |
Present study |
| 8 | 1:12009690 | c.2168T>C | p.(Val723Ala) | VUS | 0 | Not previously reported | Present study |
| 9 | 1: 12011539 | c.2248C>T | p.(His750Tyr) | VUS | 0 | Not previously reported | Present study |
Variants are classified according to the nomenclature Nm_ for the cDNA and Np_ for the protein.
ACMG, American College of Medical Genetics and Genomics; AF, allele frequency; VUS, variants of unknown significance; LP, likely pathogenic; P, pathogenic; CMT2A, Charcot-Marie-Tooth disease type 2A.
Among our cohort, nine patients (2.3%), consisting of five males and four females, were heterozygous for eight different variants which were classified as either pathogenic (n = 2), likely pathogenic (n = 3) or VUS (n = 3) according to the ACMG guidelines and were all missense (Table 1). From this point on, these individuals will be referred to as MFN2-amyotrophic lateral sclerosis. Of these, five cases had a positive family history of amyotrophic lateral sclerosis/frontotemporal dementia, and four were classified as sporadic amyotrophic lateral sclerosis. None of the MFN2-amyotrophic lateral sclerosis patients tested positive for the pathogenic C9orf72 gene expansion, nor did they exhibit mutations in the SOD1 gene or the known mutational hotspots of the TARDBP (exon 6) and FUS (exons 13-14-15) genes.
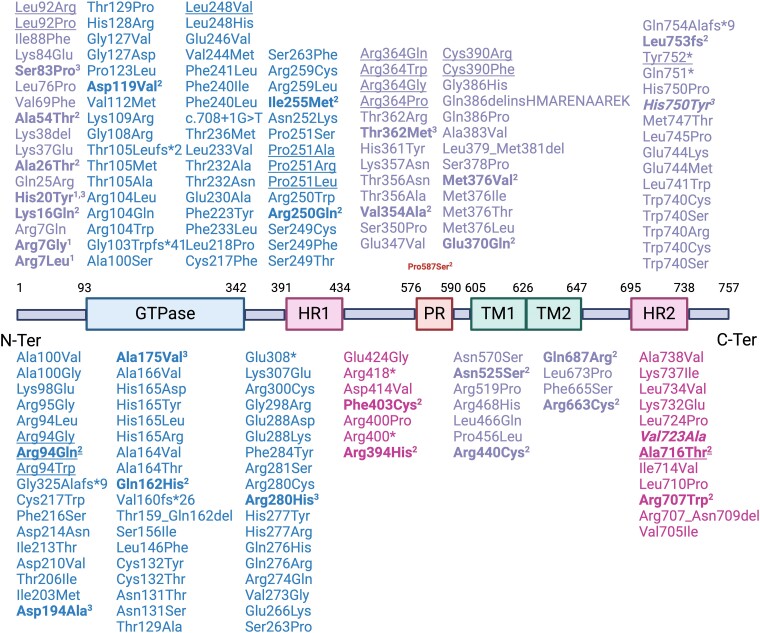
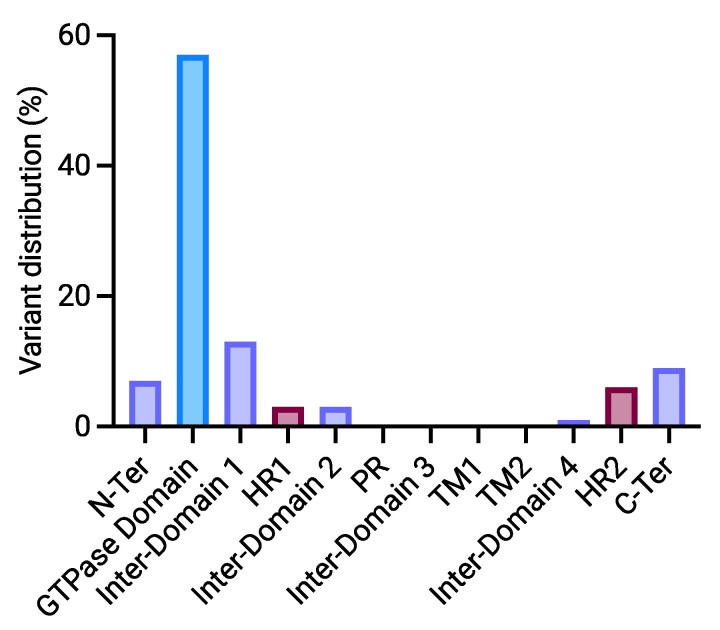
Tables 1 and 2 outline the demographic, genetic and clinical attributes of the MFN2-amyotrophic lateral sclerosis patients. Among the identified mutations in the MFN2 gene, three have already been reported in association with axonal hereditary motor and sensory neuropathy (HMSN), namely, the p.(Arg280His), the p.(His20Tyr) and the p.(Thr362Met). Conversely, the others have never been described in patients. Two unrelated patients carried the p.(Arg280His) mutation. The other variants were retrieved in one patient each. Localization at protein level of variants described so far in MFN2 is shown in Fig. 1, and distribution across MFN2 domains is shown in Fig. 2. Variants associated with amyotrophic lateral sclerosis appear to be scattered along the protein sequence rather than showing a specific domain distribution.
Table 2.
Clinical features of the amyotrophic lateral sclerosis patients harbouring heterozygous, rare, non-synonymous MFN2 variants within the cohort under investigation
| # | Amino acid change | Family history | Sex | Age at onset (Ys) | Site of onset | Phenotype | Disease duration (Ys) | UMN signs | LMN signs | Bulbar/respiratory | Non-motor features |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | p.(His20Tyr) | sALS | M | 63 | UL | Flail arm | >1 | + | + | - | - |
| 2 | p.(Ser83Pro) | fALS | M | 68 | UL | Flail arm | 3 | + | + | + (NIV 17 months) | - |
| 3 | p.(Ala175Val) | sALS | F | 29 | UL | Classic | 12 | + | + | + (PEG 18 months) | - |
| 4 | p.(Asp194Ala) | fALS | F | 64 | LL | ALS-FTD | 3 | + | + | + | FTD |
| 5 | p.(Arg280His) | fALS | M | 58 | LL | PMA | 13 | - | + | + (NIV) | - |
| 6 | p.(Arg280His) | fALS | F | 69 | LL | PMA | 8 | - | + | - | - |
| 7 | p.(Thr362Met) | sALS | M | 56 | LL | Classic | >3a | + | + | - | Sensory symptoms (distal tingling paresthesias) Urinary symptoms (polyuria, incontinence) |
| 8 | p.(Val723Ala) | sALS | M | 65 | LL | Classic | >1a | + | + | + (NIV 10 months) | Sensory symptoms (distal tingling paresthesias) |
| 9 | p.(His750Tyr) | fALS | F | 51 | Bulbar | Bulbar | >1a | + | + | - | - |
ALS, amyotrophic lateral sclerosis; fALS, familial amyotrophic lateral sclerosis; sALS, sporadic amyotrophic lateral sclerosis; Ys, years; UMN, upper motor neuron; LMN, lower motor neuron; M, male; F, female; NIV, non-invasive ventilation; PEG, percutaneous endoscopic gastrostomy; FTD, frontotemporal dementia; PMA, progressive muscular atrophy; UL, upper limbs; LL, lower limbs.
aFollow-up not available.
Figure 1.
Schematic representation of the MFN2 protein structure and genetic variants identified so far. MFN2 variants detected in patients and their positions along the mitofusin 2 protein, with data sourced from the Leiden Open Variation Database (LOVD) and https://www.progettomitofusina2.com/portfolio-articoli/database-mutazioni-mfn2/. Superscripts indicate the following: 1variants included in the Project MinE database, 2variants previously described by Russell et al.10 and 3variants identified in this study. Underlined variants have been detected in Charcot–Marie–Tooth disease type 2A patients with a predominant motor phenotype. Variants in bold have been associated with a MND phenotype. Variants in italics are classified as VUS according to the ACMG.
Figure 2.
Distribution of variants across MFN2 domains. C-Ter, C-terminal; HR, heptad repeat; N-Ter, N-terminal; PR, proline-rich; TM, transmembrane.
The median age of onset was 63 years (range 29.1–69.2 years). Most cases (88.8%) had a spinal onset; among them, five patients first experienced symptoms in the lower limbs, while three initially presented with upper limb involvement. In one individual, the disease began with pseudobulbar palsy. We observed significant variability in both the progression and the duration of the disease among the remaining patients (Table 2). Disease duration spanned from less than 1 to 13 years. Notably, two patients have been living with the disease for over a decade (Patients 3 and 5). Patients displayed a huge variety of clinical phenotypes. Two had flail limb syndrome, three had the classical amyotrophic lateral sclerosis phenotype, one had bulbar-onset symptoms and two had progressive muscular atrophy [both patients carrying the p.(Arg280His) mutation]. As the disease progressed, bulbar symptoms emerged in five patients (Patients 2, 3, 4, 5 and 8), while one patient developed frontotemporal dementia (Patient 4). Five patients displayed non-motor symptoms, specifically subjective sensory (Patients 7 and 8) and urinary symptoms (Patient 7), as detailed in Table 2. Six individuals (Patients 1, 3, 4, 5, 6 and 8) exhibited alterations in motor nerve conduction studies, though clinical sensory symptoms and signs were absent and neurophysiological sensory involvement was either absent (Patient 6) or very mild (Patients 4). In five individuals (Patients 1, 4, 5, 7 and 9), motor evoked potentials were available, revealing pathological alterations.
A detailed description of the clinical and neurophysiological characteristics of each patient is provided in the Supplementary material (clinical vignettes; Supplementary Table 2).
Discussion
Although not consolidated yet, there is a growing body of evidence8,9 suggesting an association between variants in the MFN2 and the amyotrophic lateral sclerosis/frontotemporal dementia spectrum. Moreover, a biological interaction between MFN2 and TDP-43, the main protein involved in amyotrophic lateral sclerosis pathogenesis, has been described in an amyotrophic lateral sclerosis mouse model.23 Here, we reported a retrospective study on a large Italian cohort of amyotrophic lateral sclerosis patients aimed at investigating the presence of MFN2 variants in patients with amyotrophic lateral sclerosis/frontotemporal dementia. Among the 385 amyotrophic lateral sclerosis patients, negative for pathogenic variants in the four most common amyotrophic lateral sclerosis–associated genes, we identified 9 individuals (2.3%) with 8 MFN2 rare, heterozygous, missense mutations, which were classified as pathogenic, likely pathogenic or VUS according to the ACMG guidelines. Three of them have already been reported in association with axonal HMSN, namely, the p.(His20Tyr), the p.(Arg280His) and the p.(Thr362Met).
The first report of a patient with co-occurrence of Charcot–Marie–Tooth disease type 2A and amyotrophic lateral sclerosis associated with an MFN2 mutation dates back to 2011.8 A second report was published in 2023 by Vinciguerra et al.,9 detailing one amyotrophic lateral sclerosis/frontotemporal dementia patient with the p.(Asp194Ala) mutation that is now included in our study (Patient 4). In addition, Russell et al.10 recently performed WGS on 140 amyotrophic lateral sclerosis patients, identifying 21 previously unknown MFN2 mutations within this cohort. However, to the best of our knowledge, this is the first study to provide a clinical, neurophysiological and molecular characterization of multiple patients affected by amyotrophic lateral sclerosis harbouring MFN2 variants and the largest work to assess their prevalence.
Within our cohort, age at onset and disease duration were widely variable, in line with literature data,2 highlighting the unpredictable nature of amyotrophic lateral sclerosis progression. Indeed, despite median survival in amyotrophic lateral sclerosis spanning between 2 and 4 years, individual courses may show high variability, especially for atypical phenotypes.2 Clinical symptom presentation in MFN2-amyotrophic lateral sclerosis patients was quite heterogeneous, with the majority exhibiting spinal onset, although upper limb onset and pseudobulbar palsy were also reported. Notably, five patients developed bulbar symptoms during the disease, and one developed frontotemporal dementia. Sensory deficits were observed in five patients, emphasizing the need for a comprehensive clinical assessment of amyotrophic lateral sclerosis patients, including sensory evaluation. Some patients also presented with a concomitant alteration of motor nerve conduction studies. Interestingly, the phenotypic presentation within the family of Patient 4 was diverse, as the proband’s son presented with Charcot–Marie–Tooth disease type 2A with additional mood disturbance. As previously stated, some of the variants reported in this amyotrophic lateral sclerosis cohort have already been described in Charcot–Marie–Tooth disease type 2A patients. Taken all together, these observations might suggest the presence of an MFN2-related clinical spectrum, possibly influenced by other genetic modifiers. A growing body of evidence has already demonstrated the presence of an overlap between hereditary neuropathy and MND at a genetic level, with several genes (i.e. CHCHD10, GARS, FIG4, KIF5A, NEFH, VCP and SPG11) associated with both amyotrophic lateral sclerosis and Charcot–Marie–Tooth disease type 2.7 These considerations notwithstanding, the significance of MFN2 variants in the context of amyotrophic lateral sclerosis pathogenesis is still unknown and requires further investigations.
Different lines of evidence might suggest the possibility of a link between MFN2 and amyotrophic lateral sclerosis on both molecular and cellular levels. In their previously mentioned work, Russell et al.10 employed both in vitro and in vivo models to understand the specific biological consequences of the novel MFN2 variants disclosed in their amyotrophic lateral sclerosis cohort. To assess the impact on mitochondrial morphology, the 21 MFN2 mutations identified were introduced into Mfn2 KO mouse embryonic fibroblasts (MEFs). Remarkably, none of them could restore normal morphology in Mfn2 KO MEFs. Some of these mutations also exhibited reduced membrane potential compared to wild-type MFN2 expression, indicating compromised mitochondrial function. These findings suggest that MFN2-driven mitochondrial dysfunction might potentially lead to MND in vivo. Mitochondria are indeed believed to play a significant role in amyotrophic lateral sclerosis development, as evidenced by mitochondrial abnormalities observed both in vitro and in vivo in multiple amyotrophic lateral sclerosis rodent models.30 Swollen mitochondria with unusual cristae organization have been detected early in the disease, even before symptom onset in SOD1 mice.31 Inhibiting mitochondrial fission through Drp1 has shown promise in slowing amyotrophic lateral sclerosis progression.32 As mentioned above, amyotrophic lateral sclerosis patients with diverse clinical phenotypes, including amyotrophic lateral sclerosis/frontotemporal dementia, pure amyotrophic lateral sclerosis and flail arm syndrome, and patients with axonal Charcot–Marie–Tooth disease have been found to carry missense mutations in the CHCHD10 gene, encoding the mitochondrial protein CHCHD10.7,30,33,34 Interestingly, these mutations have been linked to altered mitochondrial cristae structure, impaired stress response and disrupted mitochondrial dynamics, despite no significant changes in canonical fusion and fission protein levels, including MFN2.35 The robust body of evidence supporting mitochondrial dysfunction in various neurodegenerative diseases, such as Alzheimer’s and Parkinson’s diseases, underscores the significance of understanding mitochondrial dynamics.36,37 The interplay among key regulators of mitochondrial fission and fusion, like mitofusins, adds weight to the role of mitochondrial dynamics in the pathogenesis of these disorders.36
Studies using mouse models to explore mutations in mitochondrial fusion and fission regulators, including Mfn2, have demonstrated that they lead to mitochondrial depletion in neurites and synapses, resulting in the loss of MN synapses.38 Interestingly, amyotrophic lateral sclerosis patients exhibit inclusions containing TDP-43, which are also found in mitochondria within damaged MNs.38,39 Russell et al.10 also explored protein aggregation within the CNS in a Mfn2 KO zebrafish model, showing that both heterozygous and homozygous Mfn2 loss-of-function mutations led to increased TDP-43 levels in the hindbrain and cerebellum. Additionally, TDP-43 overexpression was able to rescue the insufficiency of Parkin and its downstream effectors, including MFN2, secondary to progranulin silencing in fibroblasts derived from healthy controls.40
TDP-43 has already been shown to negatively affect mitochondrial function and was found in mitochondria.23,41-44 Mutant TDP-43 was reported to disrupt mitochondria in mice, leading to impaired transport and membrane potential in neurons.41,44 Overexpressing Mfn2 in these mice restored normal mitochondria and membrane potential. TDP-43 bound mitochondrial mRNAs, especially with amyotrophic lateral sclerosis–causing mutations.42,43 Knocking down TARDBP reduced MFN2 and blocked its mitochondrial association, improving neuromuscular function in mutant mice.42,43 In Drosophila, overexpressing Marf was able to mitigate TDP-43-induced disease.45
The main genetic cause of amyotrophic lateral sclerosis is a hexanucleotide repeat expansion within C9orf72.46-48 Recent research demonstrated that the encoded C9orf72 protein plays a role in maintaining the respiratory chain of mitochondria.49 C9orf72 was found to reside in the inner mitochondrial membrane, and its loss caused decreased oxidative phosphorylation and ATP production under stress conditions.50 Dysfunctional mitochondrial transport and irregular mitochondrial respiration were observed in MNs differentiated from samples of patients with C9orf72 repeat expansion, potentially involving TDP-43-like effects on mitochondrial mRNAs.50
Overall, our findings contribute to a deeper insight into amyotrophic lateral sclerosis genetics and its overlap with other neurological diseases like Charcot–Marie–Tooth disease type 2. We hypothesize that MFN2-related clinical syndromes might form a spectrum of MN/axonal degeneration. Functional studies and larger cohorts are needed to further elucidate the role of MFN2 in MNDs. The creation of big consortia, together with the application of artificial intelligence tools for the analysis of large data sets, may help reach this goal. Furthermore, considering that MFN2 is a druggable target in neurodegeneration, these results are also interesting from a therapeutic perspective.
In conclusion, the present cohort, together with findings from other works, suggests that MFN2 variants may play a role in amyotrophic lateral sclerosis pathology, highlighting the relevance of mitochondria and MFN2 in neuronal health.
Supplementary Material
Acknowledgements
We wish to thank Progetto Mitofusina 2 Onlus and Associazione Amici del Centro Dino Ferrari for their support. The authors would like to thank the ‘Project MinE ALS Sequencing Consortium’. A.R. acknowledges ‘Aldo Ravelli Center for Neurotechnology and Experimental Brain Therapeutics’, Università degli Studi di Milano. The authors acknowledge the Italian Ministry of Health [‘Piano Nazionale Complementare Ecosistema Innovativo della Salute - Hub Life Science-Diagnostica Avanzata (HLS-DA)’ - PNC-E3-2022-23683266 - ‘INNOVA’], the Italian Ministry of Education and Research - MUR (‘Dipartimenti di Eccellenza’ Programme 2023–27 - Dept. of Pathophysiology and Transplantation, Università degli Studi di Milano) and the European Reference Network (ERN) Euro-NMD for their support. Graphical abstract and figures were created with Biorender.com.
Contributor Information
Elena Abati, Dino Ferrari Centre, Department of Pathophysiology and Transplantation (DEPT), Università degli Studi di Milano, 20122 Milan, Italy; Neurology Unit, Fondazione IRCCS Ca’ Granda Ospedale Maggiore Policlinico, 20122 Milan, Italy.
Delia Gagliardi, Dino Ferrari Centre, Department of Pathophysiology and Transplantation (DEPT), Università degli Studi di Milano, 20122 Milan, Italy.
Arianna Manini, Dino Ferrari Centre, Department of Pathophysiology and Transplantation (DEPT), Università degli Studi di Milano, 20122 Milan, Italy.
Roberto Del Bo, Dino Ferrari Centre, Department of Pathophysiology and Transplantation (DEPT), Università degli Studi di Milano, 20122 Milan, Italy.
Dario Ronchi, Dino Ferrari Centre, Department of Pathophysiology and Transplantation (DEPT), Università degli Studi di Milano, 20122 Milan, Italy.
Megi Meneri, Neurology Unit, Fondazione IRCCS Ca’ Granda Ospedale Maggiore Policlinico, 20122 Milan, Italy.
Francesca Beretta, Department of Neurosciences, Psychology, Drug Research and Child Health (NEUROFARBA), Università degli Studi di Firenze, 50139 Firenze, Italy.
Annalisa Sarno, Dino Ferrari Centre, Department of Pathophysiology and Transplantation (DEPT), Università degli Studi di Milano, 20122 Milan, Italy.
Federica Rizzo, Neurology Unit, Fondazione IRCCS Ca’ Granda Ospedale Maggiore Policlinico, 20122 Milan, Italy.
Edoardo Monfrini, Dino Ferrari Centre, Department of Pathophysiology and Transplantation (DEPT), Università degli Studi di Milano, 20122 Milan, Italy; Neurology Unit, Fondazione IRCCS Ca’ Granda Ospedale Maggiore Policlinico, 20122 Milan, Italy.
Alessio Di Fonzo, Neurology Unit, Fondazione IRCCS Ca’ Granda Ospedale Maggiore Policlinico, 20122 Milan, Italy.
Maria Teresa Pellecchia, Department of Medicine, Surgery and Dentistry ‘Scuola Medica Salernitana’, Neuroscience Section, Università degli Studi di Salerno, 84081Salerno, Italy.
Alberto Brusati, Department of Brain and Behavioral Sciences, Università degli Studi di Pavia, 27100 Pavia, Italy.
Vincenzo Silani, Dino Ferrari Centre, Department of Pathophysiology and Transplantation (DEPT), Università degli Studi di Milano, 20122 Milan, Italy; Department of Neurology and Laboratory of Neuroscience, IRCCS Istituto Auxologico Italiano, 20149 Milan, Italy.
Giacomo Pietro Comi, Dino Ferrari Centre, Department of Pathophysiology and Transplantation (DEPT), Università degli Studi di Milano, 20122 Milan, Italy; Neurology Unit, Fondazione IRCCS Ca’ Granda Ospedale Maggiore Policlinico, 20122 Milan, Italy.
Antonia Ratti, Department of Neurology and Laboratory of Neuroscience, IRCCS Istituto Auxologico Italiano, 20149 Milan, Italy; Department of Medical Biotechnology and Translational Medicine, Università degli Studi di Milano, 20133 Milan, Italy.
Federico Verde, Dino Ferrari Centre, Department of Pathophysiology and Transplantation (DEPT), Università degli Studi di Milano, 20122 Milan, Italy; Department of Neurology and Laboratory of Neuroscience, IRCCS Istituto Auxologico Italiano, 20149 Milan, Italy.
Nicola Ticozzi, Dino Ferrari Centre, Department of Pathophysiology and Transplantation (DEPT), Università degli Studi di Milano, 20122 Milan, Italy; Department of Neurology and Laboratory of Neuroscience, IRCCS Istituto Auxologico Italiano, 20149 Milan, Italy.
Stefania Corti, Dino Ferrari Centre, Department of Pathophysiology and Transplantation (DEPT), Università degli Studi di Milano, 20122 Milan, Italy; Neuromuscular and Rare Diseases Unit, Department of Neuroscience, Fondazione IRCCS Ca’ Granda Ospedale Maggiore Policlinico, Milan 20122 Milan, Italy.
Supplementary material
Supplementary material is available at Brain Communications online.
Funding
This study was funded by the Italian Ministry of Health grant GR-2018–12365358 to F.R. (2018–21), by the Fondazione Telethon grant GGP19002 to S.C., by the Istituto di Ricovero e Cura a Carattere Scientifico (IRCCS) Fondazione Ca’ Granda Ospedale Maggiore Policlinico Ricerca Corrente 2020 to G.P.C. and by the Italian Ministry of Health (grants RF-2021-12374238 to N.T. and RF-2018-12367768 to V.S. and A.R.).
Competing interests
V.S. received compensation for consulting services and/or speaking activities from AveXis, Cytokinetics, Italfarmaco, Liquidweb S.r.l., Novartis Pharma AG and Zambon Biotech SA and receives or has received research support from the Italian Ministry of Health, AriSLA and E-Rare Joint Transnational Call. He is member of the editorial boards of Amyotrophic Lateral Sclerosis and Frontotemporal Degeneration, European Neurology, American Journal of Neurodegenerative Diseases, Frontiers in Neurology and Exploration of Neuroprotective Therapy. N.T. received compensation for consulting services from Amylyx Pharmaceuticals, Biogen, Italfarmaco and Zambon Biotech SA. He is an associate editor for Frontiers in Aging Neuroscience. The other authors report no competing interests.
Data availability
The data underlying this article will be shared on reasonable request to the corresponding author.
References
- 1. Hardiman O, Al-Chalabi A, Chio A, et al. Amyotrophic lateral sclerosis. Nat Rev Dis Primers. 2017;3:17071. [DOI] [PubMed] [Google Scholar]
- 2. Feldman EL, Goutman SA, Petri S, et al. Amyotrophic lateral sclerosis. Lancet. 2022;400(10360):1363–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chiò A, Logroscino G, Traynor BJ, et al. Global epidemiology of amyotrophic lateral sclerosis: A systematic review of the published literature. Neuroepidemiology. 2013;41(2):118–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Al-Chalabi A, Van Den Berg LH, Veldink J. Gene discovery in amyotrophic lateral sclerosis: Implications for clinical management. Nat Rev Neurol. 2017;13(2):96–104. [DOI] [PubMed] [Google Scholar]
- 5. Al-Chalabi A, Fang F, Hanby MF, et al. An estimate of amyotrophic lateral sclerosis heritability using twin data. J Neurol Neurosurg Psychiatry. 2010;81(12):1324–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Miller TM, Cudkowicz ME, Genge A, et al. Trial of antisense oligonucleotide tofersen for SOD1 ALS. N Engl J Med. 2022;387(12):1099–1110. [DOI] [PubMed] [Google Scholar]
- 7. Gentile F, Scarlino S, Falzone YM, et al. The peripheral nervous system in amyotrophic lateral sclerosis: Opportunities for translational research. Front Neurosci. 2019;13(JUN):601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Marchesi C, Ciano C, Salsano E, et al. Co-occurrence of amyotrophic lateral sclerosis and Charcot-Marie-Tooth disease type 2A in a patient with a novel mutation in the mitofusin-2 gene. Neuromuscul Disord. 2011;21(2):129–131. [DOI] [PubMed] [Google Scholar]
- 9. Vinciguerra C, Di Fonzo A, Monfrini E, et al. Case report: Asp194Ala variant in MFN2 is associated with ALS-FTD in an Italian family. Front Genet. 2023;14:1235887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Russell KL, Downie JM, Gibson SB, et al. MFN2 Influences Amyotrophic Lateral Sclerosis Pathology. bioRxiv 466517. 10.1101/2021.10.30.466517, 31 October 2021, preprint: not peer reviewed. MFN2Influences amyotrophic lateral sclerosis pathology. [DOI]
- 11. Züchner S, Mersiyanova IV, Muglia M, et al. Mutations in the mitochondrial GTPase mitofusin 2 cause Charcot-Marie-Tooth neuropathy type 2A. Nat Genet. 2004;36(5):449–451. [DOI] [PubMed] [Google Scholar]
- 12. Feely SME, Laura M, Siskind CE, et al. MFN2 mutations cause severe phenotypes in most patients with CMT2A. Neurology. 2011;76(20):1690–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pipis M, Feely SME, Polke JM, et al. Natural history of Charcot-Marie-Tooth disease type 2A: A large international multicentre study. Brain. 2020;143(12):3589–3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhu D, Kennerson ML, Walizada G, Züchner S, Vance JM, Nicholson GA. Charcot-Marie-Tooth with pyramidal signs is genetically heterogeneous: Families with and without MFN2 mutations. Neurology. 2005;65(3):496–497. [DOI] [PubMed] [Google Scholar]
- 15. Verhoeven K, Claeys KG, Züchner S, et al. MFN2 mutation distribution and genotype/phenotype correlation in Charcot-Marie-Tooth type 2. Brain. 2006;129(8):2093–2102. [DOI] [PubMed] [Google Scholar]
- 16. Pareyson D, Marchesi C. Diagnosis, natural history, and management of Charcot-Marie-Tooth disease. Lancet Neurol. 2009;8(7):654–667. [DOI] [PubMed] [Google Scholar]
- 17. Chung KW, Kim SB, Park KD, et al. Early onset severe and late-onset mild Charcot-Marie-Tooth disease with mitofusin 2 (MFN2) mutations. Brain. 2006;129(Pt 8):2103–2118. [DOI] [PubMed] [Google Scholar]
- 18. Abati E, Manini A, Velardo D, et al. Clinical and genetic features of a cohort of patients with MFN2-related neuropathy. Sci Rep. 2022;12(1):6181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rizzo F, Bono S, Ruepp MD, et al. Combined RNA interference and gene replacement therapy targeting MFN2 as proof of principle for the treatment of Charcot-Marie-Tooth type 2A. Cell Mol Life Sci. 2023;80(12):373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen H, Detmer SA, Ewald AJ, Griffin EE, Fraser SE, Chan DC. Mitofusins Mfn1 and Mfn2 coordinately regulate mitochondrial fusion and are essential for embryonic development. J Cell Biol. 2003;160(2):189–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chandhok G, Lazarou M, Neumann B. Structure, function, and regulation of mitofusin-2 in health and disease. Biol Rev Camb Philos Soc. 2018;93(2):933–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stavropoulos F, Georgiou E, Schiza N, et al. Mitofusin 1 overexpression rescues the abnormal mitochondrial dynamics caused by the mitofusin 2 K357T mutation in vitro. J Peripher Nerv Syst. 2023;28(3):329–340. [DOI] [PubMed] [Google Scholar]
- 23. Davis SA, Itaman S, Khalid-Janney CM, et al. TDP-43 interacts with mitochondrial proteins critical for mitophagy and mitochondrial dynamics. Neurosci Lett. 2018;678:8–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Brooks BR, Miller RG, Swash M, Munsat TL. El Escorial revisited: Revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotroph Lateral Scler Other Motor Neuron Disord. 2000;1(5):293–299. [DOI] [PubMed] [Google Scholar]
- 25. de Carvalho M, Dengler R, Eisen A, et al. Electrodiagnostic criteria for diagnosis of ALS. Clin Neurophysiol. 2008;119(3):497–503. [DOI] [PubMed] [Google Scholar]
- 26. Manini A, Casiraghi V, Brusati A, et al. Association of the risk factor UNC13A with survival and upper motor neuron involvement in amyotrophic lateral sclerosis. Front Aging Neurosci. 2023;15:1067954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Richards S, Aziz N, Bale S, et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17(5):405–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. van der Spek RAA, van Rheenen W, Pulit SL, Kenna KP, van den Berg LH, Veldink JH. The project MinE databrowser: Bringing large-scale whole-genome sequencing in ALS to researchers and the public. Amyotroph Lateral Scler Frontotemporal Degener. 2019;20(5–6):432–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Van Rheenen W, Pulit SL, Dekker AM, et al. Project MinE: Study design and pilot analyses of a large-scale whole-genome sequencing study in amyotrophic lateral sclerosis. Eur J Hum Genet. 2018;26(10):1537–1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bannwarth S, Ait-El-Mkadem S, Chaussenot A, et al. A mitochondrial origin for frontotemporal dementia and amyotrophic lateral sclerosis through CHCHD10 involvement. Brain. 2014;137(Pt 8):2329–2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tafuri F, Ronchi D, Magri F, Comi GP, Corti S. SOD1 misplacing and mitochondrial dysfunction in amyotrophic lateral sclerosis pathogenesis. Front Cell Neurosci. 2015;9(AUGUST):336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Joshi AU, Saw NL, Vogel H, Cunnigham AD, Shamloo M, Mochly-Rosen D. Inhibition of Drp1/Fis1 interaction slows progression of amyotrophic lateral sclerosis. EMBO Mol Med. 2018;10(3):e8166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Auranen M, Ylikallio E, Shcherbii M, et al. CHCHD10 variant p.(Gly66Val) causes axonal Charcot-Marie-Tooth disease. Neurol Genet. 2015;1(1):e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ronchi D, Riboldi G, Del Bo R, et al. CHCHD10 mutations in Italian patients with sporadic amyotrophic lateral sclerosis. Brain. 2015;138(Pt 8):e372. [DOI] [PubMed] [Google Scholar]
- 35. Shammas MK, Huang TH, Narendra DP. CHCHD2 and CHCHD10-related neurodegeneration: Molecular pathogenesis and the path to precision therapy. Biochem Soc Trans. 2023;51(2):797–809. [DOI] [PubMed] [Google Scholar]
- 36. Gao J, Wang L, Liu J, Xie F, Su B, Wang X. Abnormalities of mitochondrial dynamics in neurodegenerative diseases. Antioxidants (Basel). 2017;6(2):25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Manini A, Abati E, Comi GP, Corti S, Ronchi D. Mitochondrial DNA homeostasis impairment and dopaminergic dysfunction: A trembling balance. Ageing Res Rev. 2022;76:101578. [DOI] [PubMed] [Google Scholar]
- 38. Mou Y, Dein J, Chen Z, Jagdale M, Li XJ. MFN2 deficiency impairs mitochondrial transport and downregulates motor protein expression in human spinal motor neurons. Front Mol Neurosci. 2021;14:727552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Neumann M, Sampathu DM, Kwong LK, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314(5796):130–133. [DOI] [PubMed] [Google Scholar]
- 40. Gaweda-Walerych K, Walerych D, Berdyński M, Buratti E, Zekanowski C. Parkin levels decrease in fibroblasts with progranulin (PGRN) pathogenic variants and in a cellular model of PGRN deficiency. Front Mol Neurosci. 2021;14:676478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Xu YF, Gendron TF, Zhang YJ, et al. Wild-type human TDP-43 expression causes TDP-43 phosphorylation, mitochondrial aggregation, motor deficits, and early mortality in transgenic mice. J Neurosci. 2010;30(32):10851–10859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Wang W, Wang L, Lu J, et al. The inhibition of TDP-43 mitochondrial localization blocks its neuronal toxicity. Nat Med. 2016;22(8):869–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang W, Arakawa H, Wang L, et al. Motor-Coordinative and cognitive dysfunction caused by mutant TDP-43 could be reversed by inhibiting its mitochondrial localization. Mol Ther. 2017;25(1):127–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang W, Li L, Lin WL, et al. The ALS disease-associated mutant TDP-43 impairs mitochondrial dynamics and function in motor neurons. Hum Mol Genet. 2013;22(23):4706–4719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Khalil B, Cabirol-Pol MJ, Miguel L, Whitworth AJ, Lecourtois M, Liévens JC. Enhancing Mitofusin/Marf ameliorates neuromuscular dysfunction in Drosophila models of TDP-43 proteinopathies. Neurobiol Aging. 2017;54:71–83. [DOI] [PubMed] [Google Scholar]
- 46. Renton AE, Majounie E, Waite A, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72(2):257–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. DeJesus-Hernandez M, Mackenzie IR, Boeve BF, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72(2):245–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Gagliardi D, Costamagna G, Taiana M, et al. Insights into disease mechanisms and potential therapeutics for C9orf72-related amyotrophic lateral sclerosis/frontotemporal dementia. Ageing Res Rev. 2020;64:101172. [DOI] [PubMed] [Google Scholar]
- 49. Wang T, Liu H, Itoh K, et al. C9orf72 regulates energy homeostasis by stabilizing mitochondrial complex I assembly. Cell Metab. 2021;33(3):531–546.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mehta AR, Gregory JM, Dando O, et al. Mitochondrial bioenergetic deficits in C9orf72 amyotrophic lateral sclerosis motor neurons cause dysfunctional axonal homeostasis. Acta Neuropathol. 2021;141(2):257–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bombelli F, Stojkovic T, Dubourg O, et al. Charcot-Marie-Tooth disease type 2A: From typical to rare phenotypic and genotypic features. JAMA Neurol. 2014;71(8):1036–1042. [DOI] [PubMed] [Google Scholar]
- 52. Al-Harbi TM, Abdulmana SO, Bashir S, Dridi W. Novel MFN2 missense mutation induces hereditary axonal motor and sensory neuropathy in a Saudi Arabian family. J Clin Neuromuscul Dis. 2019;21(1):25–29. [DOI] [PubMed] [Google Scholar]
- 53. Carr AS, Polke JM, Wilson J, et al. MFN2 deletion of exons 7 and 8: Founder mutation in the UK population. J Peripher Nerv Syst. 2015;20(2):67–71. [DOI] [PubMed] [Google Scholar]
- 54. Nicholson GA, Magdelaine C, Zhu D, et al. Severe early-onset axonal neuropathy with homozygous and compound heterozygous MFN2 mutations. Neurology. 2008;70(19):1678–1681. [DOI] [PubMed] [Google Scholar]
- 55. Calvo J, Funalot B, Ouvrier RA, et al. Genotype-phenotype correlations in Charcot-Marie-Tooth disease type 2 caused by mitofusin 2 mutations. Arch Neurol. 2009;66(12):1511–1516. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article will be shared on reasonable request to the corresponding author.