Abstract
Influenza virus, the causative agent of the common flu, is a worldwide health problem with significant economic consequences. Studies of influenza virus biology have revealed elaborate mechanisms by which the virus interacts with its host cell as it inhibits the synthesis of cellular proteins, evades the innate antiviral response, and facilitates production of viral RNAs and proteins. With the advent of DNA array technology it is now possible to obtain a large-scale view of how viruses alter the environment within the host cell. In this study, the cellular response to influenza virus infection was examined by monitoring the steady-state mRNA levels for over 4,600 cellular genes. Infections with active and inactivated influenza viruses identified changes in cellular gene expression that were dependent on or independent of viral replication, respectively. Viral replication resulted in the downregulation of many cellular mRNAs, and the effect was enhanced with time postinfection. Interestingly, several genes involved in protein synthesis, transcriptional regulation, and cytokine signaling were induced by influenza virus replication, suggesting that some may play essential or accessory roles in the viral life cycle or the host cell's stress response. The gene expression pattern induced by inactivated viruses revealed induction of the cellular metallothionein genes that may represent a protective response to virus-induced oxidative stress. Genome-scale analyses of virus infections will help us to understand the complexities of virus-host interactions and may lead to the discovery of novel drug targets or antiviral therapies.
Although it has been nearly 7 decades since the isolation of human influenza virus (34), it remains a world health threat with large economic consequences (28, 43, 52). Although vaccine and drug strategies have managed to contain the spread of the disease and the severity of its symptoms, recent outbreaks, such as the one in Hong Kong in 1997, emphasize the need for continued research efforts for influenza prevention. An abundant but often overlooked source of potential antiviral targets are those cellular genes whose expression is most affected by viral infection. With DNA microarray technology it is now possible to measure the mRNA levels of thousands of cellular genes under a variety of experimental conditions. This approach is increasingly being used to monitor cellular gene expression in response to viral infections (5, 19, 20, 25, 30, 55, 59), expression of viral genes (21, 31, 58), or treatment with antiviral compounds such as interferon (12).
Influenza virus is a negative-stranded RNA virus that induces a profound inhibitory effect on the synthesis of cellular proteins. Much of this effect occurs at a posttranscriptional level, as viral RNAs are selectively translated while the initiation and elongation of cellular proteins are inhibited (15). On the other hand, viral proteins carry out a variety of functions within the nucleus, such as removing 5′ methyl caps from host cell mRNAs (50), blocking mRNA export (32), and inhibiting mRNA splicing (38) that could profoundly alter the steady-state levels of cellular mRNAs. Despite these characteristics, studies aimed at determining the effect of influenza virus infection on cellular mRNA levels have been limited to the analysis a few selected cellular mRNAs (3, 23, 27). A comprehensive large-scale analysis of host cell mRNAs during influenza virus infection has not been performed until now.
The expression of more than 4,500 cellular genes during the course of influenza virus infection was examined by cDNA microarrays. As a control to determine if viral replication was required to alter cellular gene expression, infections with an inactivated and replication-incompetent virus were performed. The pattern of gene expression was used to identify changes that were either dependent or independent of viral replication. At 4 h postinfection (p.i.), cellular genes were altered in both a replication-independent and a replication-dependent manner. However, as infection proceeded, changes in cellular mRNA levels were almost exclusively dependent on viral replication. These results suggest that early events in the viral life cycle are capable of inducing a change in host cell mRNA levels, possibly by attachment or fusion to the host cell. Although the relationships between cellular mRNA levels and cellular protein synthesis remain unclear, the findings are discussed in the context of host cell response and the viral replication cycle.
MATERIALS AND METHODS
Cell line and infection conditions.
HeLa cells were grown as monolayers in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 100 U of penicillin/ml, and 100 μg of streptomycin sulfate/ml at 37°C. When the cells were approximately 80% confluent, supernatant was removed and replaced with either medium alone (Dulbecco's modified Eagle's medium with 2% calf serum), medium containing untreated influenza virus (48), or medium containing inactivated virus (see below). The multiplicity of infection was approximately 50 PFU/cell. Viral attachment was carried out at 4°C for 45 min with gentle agitation. Cells were then incubated at 37°C for 4 or 8 h. At the indicated times, medium was removed, the cells were harvested, and the total RNA was extracted using the method of Chomczynski and Sacchi (7).
Inactivation procedures and agglutination assay.
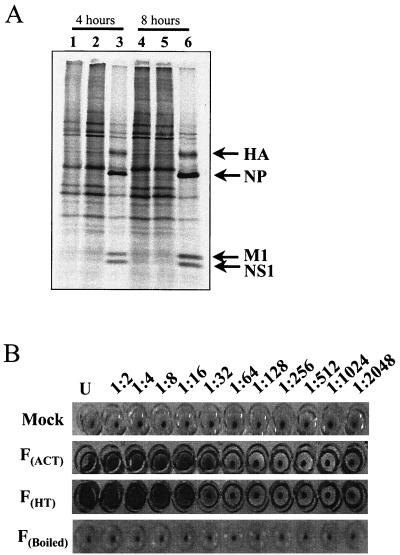
Heat inactivation of influenza virus stocks was performed by incubating the virus stock at 56°C for 90 min. When incubated under these conditions, the virus was rendered unable to inhibit host cell protein synthesis as monitored by [35S]methionine-cysteine labeling experiments (Fig. 1A). UV inactivation of influenza virus stocks was performed in a Stratalinker 2400 (Stratagene, La Jolla, Calif.) with increasing amounts of energy. Optimal inactivation was determined by [35S]methionine-cysteine labeling of infected cells to be 80 mJ (data not shown; see Fig. S1 at http://thor.csi.washington.edu/katzelab/papers/Geiss2000/index.htm). To determine if the viral hemagglutinin (HA) protein was still active after inactivation procedure, agglutination assays of chicken red blood cells were performed. Medium alone (for mock infection), untreated influenza virus, or inactivated virus was diluted (1:2 to 1:2,048) in ice-cold phosphate-buffered saline in a conical-bottom 96-well plate. An equal volume of chicken red blood cells was mixed with the dilutions and incubated at 4°C for 30 min. Loss of HA activity was monitored by the formation of a visible pellet of blood cells.
FIG. 1.
Effect of inactivation of influenza virus on the synthesis of host cell proteins and viral hemagglutination titers. (A) Protein extracts from mock-infected cells (lanes 1 and 4), cells infected with heat-inactivated virus (lanes 2 and 5), and cells infected with active influenza virus (lanes 3 and 6) were examined by [35S]methionine-cysteine pulse-labeling at 4 and 8 h p.i.. The relative positions of four viral proteins, HA, nucleoprotein (NP), membrane protein 1 (M1), and NS1, are indicated by arrows. (B) Comparison of HA titers. Serial dilutions of medium alone (mock), FACT virus, and FHT virus were mixed with chicken red blood cells to determine relative HA titers. As an additional negative control, an influenza virus preparation was heated in a boiling water bath for 90 min (boiled). All reactions were performed in triplicate in 96-well plates; one replica is shown here.
Analysis of cellular and viral protein synthesis.
The extent of influenza virus infection and the effectiveness of the inactivation procedure were monitored by examining host cell protein synthesis after infection. Mock- or virus-infected HeLa cells were labeled at the indicated times with [35S]methionine-cysteine (NEN, Boston, Mass.), and protein extracts were examined on 14% sodium dodecyl sulfate (SDS)-polyacrylamide gels as described previously (17).
RNA isolation, first-strand cDNA synthesis, and Northern blot analysis.
Total RNA preparations were performed essentially as described previously (7) with an additional phenol-chloroform (49:1) extraction. Poly(A)+ RNA was isolated from total RNA using an Oligotex mRNA purification kit (Qiagen, Valencia, Calif.). Northern blots were performed as described previously (1). Radioactive probes for Northern analysis were generated from double-stranded (ds) PCR products by either Ready-to-go beads (AP Biotech, Little Chalfont, Buckinghamshire, United Kingdom) or by amplification of the minus strand with Taq polymerase using strand-specific primers.
Fluorescently labeled cDNA probes were generated by reverse transcription as follows. Two micrograms of poly(A)+ RNA from mock- or influenza virus-infected HeLa cells, 2.5 ng of green fluorescent protein poly(A)+ RNA, 8 pmol of anchored dT primer, and 1 μg of random nonamers were combined in a 10.5-μl reaction volume. The solution was heated to 70°C for 10 min, chilled briefly on ice, and centrifuged. Reverse transcription was performed in a 20-μl reaction volume. Final concentrations were 1× first-strand buffer (Life Technologies, Rockville, Md.), 10 mM dithiothreitol, 100 nM dATP, dGTP, and dTTP, 50 nM nonlabeled dCTP, 50 nM FluoroLink-dCTP (either Cy3 or Cy5 labeled; AP Biotech), and 0.5 U of placental RNase inhibitor (Promega, Madison, Wis.)/μl. The contents were mixed and incubated at room temperature for 10 min. Superscript II RT (Life Technologies) was added (200 U), and the reaction mixtures were incubated at 42°C for 2 h. RNA was hydrolyzed with sodium hydroxide (0.25 N final concentration) for 15 min at 37°C. Samples were neutralized by addition of 2 M MOPS (morpholinepropanesulfonic acid) buffer to 0.4 M. Unincorporated nucleotides were removed using 96-well multiscreen-FB filter plates (Millipore, Bedford, Mass.) followed by G-50 ProbeQuant columns (AP Biotech).
Microarray construction, hybridization, and detection.
The human cDNA I.M.A.G.E. clones (36) used in this analysis were obtained from Research Genetics (Huntsville, Ala.) and consisted of a subset of the Homo sapiens 15K sequence verified set (UniGene Build 19, plates 1 to 44) plus a 384-well control plate. cDNA inserts for I.M.A.G.E clones and controls were PCR amplified and purified (see protocols at http://thor.csi.washington.edu/katzelab/papers/Geiss2000/index.htm). DNA pellets were suspended in a 50% solution of reagent D (AP Biotech) and deposited on 75- by 25-mm coated glass microscope slides (type VII; AP Biotech) with the use of a Molecular Dynamics (Sunnyvale, Calif.) Generation III microarray spotter. After spotting, microarrays were air dried, cross-linked at 450 mJ, and stored desiccated under liquid N2 until needed.
Prior to hybridization, microarray slides were pretreated for 20 min at 55°C in 5× SSC–0.2% SDS (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), rinsed briefly in deionized water, and dried with compressed air. Fluorescently labeled first-strand cDNAs were concentrated by drying and resuspended in 20 μl of 1× hybridization solution [5× SSC, 5× Denhardt's solution, 0.1% SDS, 50% formamide, 0.1 μg of Cot1 DNA/μl, and 20 μg of poly(A)72/ml]. The appropriate Cy3- and Cy5-labeled probes were combined (total hybridization volume, 40 μl), denatured by boiling, and applied to the slides under a 22- by 64-mm glass coverslip. Microarrays were hybridized at 42°C in a humidified chamber for 16 to 20 h. Following hybridization, slides were washed briefly in 1× SSC–0.2% SDS at 55°C to remove the coverslip and then washed once in 1× SSC–0.2% SDS for 10 min (55°C), twice in 0.1× SSC–0.2% SDS for 10 min (55°C°), twice in 0.1× SSC for 1 min (at room temperature), and once with deionized water for 10 s (at room temperature). Microarrays were dried with compressed air and scanned at 532 and 633 nm with an Avalanche dual-laser confocal scanner (Molecular Dynamics).
Data analysis and differentially expressed clone selection and control genes.
Each slide contained 4,608 cDNAs spotted in duplicate. Included in this number was a set of 384 selected cDNAs that were spotted on every slide. This set contained four influenza virus genes used as positive controls, nonhuman genes used as negative controls, and genes for a variety of selected transcription factors, ligands, and receptors chosen from the Research Genetic 15K human gene set. A complete list of “control” genes, including intensities, ratios, and errors, can be found at http://thor.csi.washington.edu/katzelab/papers/Geiss2000/index.htm. For each of the three infection conditions (Fig. 2), duplicate slides were hybridized with the same RNAs but with the fluorescent labels reversed to control for dye-specific effects as described previously (19). In addition, each infection was repeated independently for a total of four slides per condition (eight measurements per gene). Intensity values in Cy3 and Cy5 channels were extracted from each image, and the Cy3/Cy5 ratio was determined using Spot-on Image software. Data for all replicates were combined and normalized with custom software, Spot-on Unite. Briefly, Spot-on Unite normalizes the data, rejects outliers, and calculates the mean and standard deviation for each replicate measurement. The normalization method takes into account nonlinearities in the Cy3/Cy5 ratio as a function of intensity by fitting a second-order polynomial to Cy3/Cy5 ratio versus Cy3-plus-Cy5 intensity. This normalization procedure should also compensate for the amount of viral mRNA in the infected samples that are not present in cells infected with heat-inactivated virus or mock infection medium. Outlier rejection was then performed by removing each replicate data point one at a time and recalculating the mean and standard deviation. If the removal of a replicate point resulted in a twofold or larger reduction of the standard deviation, that point was removed from the calculation of the mean. The resulting mean and standard deviation calculations were then imported into the program Spot-on SELECT. A clone was considered differentially expressed if the signal (intensity minus background) was above 750 (at least three times the standard deviation of the background; see Fig. S2 at http://thor.csi.washington.edu/katzelab/papers/Geiss2000/index.htm) and the ratio plus and minus its standard deviation was at least 1.5-fold. A list of all genes differentially expressed in at least one experiment was generated for each time point. For each gene that was differentially expressed in at least one experiment, the mean intensity and standard deviation were extracted for all experiments (at each time point) using a program called Spot-on MERGE. Those genes that exhibited a consistent pattern of expression in both infections are presented in Results. Genes were then classified as either independent of or dependent on viral replication based on their pattern of expression. All image and data analysis programs were designed and written by E. Hammersmark and R. Bumgarner (unpublished data).
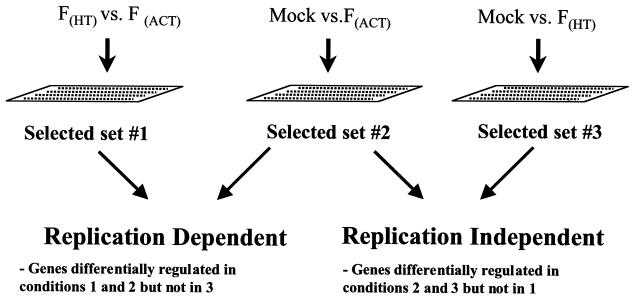
FIG. 2.
Schematic representation of experimental design and classification of differentially expressed genes. Microarray experiments were performed in a pairwise fashion with RNA from mock-infected cells (M), cells infected with FHT virus, and cells infected with FACT virus. A set of differentially expressed genes was generated for each of the three possible comparisons. The whole set of experiments was then repeated with RNA from independent infections. The lists of differentially expressed genes for each time point and condition were combined, and the pattern of expression for each gene was used to determine whether it was dependent on viral replication. A gene that is dependent on viral replication should be differentially regulated in the experiments depicted in the left and center panels but not during experiments depicted on the right, whereas a replication-independent gene should be differentially regulated under the conditions shown at center and right but not in FHT-versus-FACT experiments. The same pattern of expression had to be observed in both independent infections in order to be considered in this study (Table 1).
RESULTS
Generation of a replication-incompetent influenza virus that retains cell binding activity.
Before microarray analysis was performed it was necessary to generate an inactive viral preparation to compare with infected cells. For the purposes of these experiments, inactivated virus was defined as one that could not replicate or express viral mRNAs or proteins. Replication-deficient viruses were generated by either heat treatment or exposure to UV light. The effectiveness of the inactivation procedures was monitored by examining the rate of cellular protein synthesis with [35S]methionine-cysteine pulse-labeling of infected cells. As Fig. 1A shows, a 90-min treatment at 56°C was sufficient to inactivate influenza virus' ability to inhibit host cell protein synthesis at 4 and 8 h p.i. (compare heat-treated virus [FHT virus] [lanes 2 and 5] with mock infection [M] [lanes 1 and 4] and untreated influenza virus [FACT virus] [lanes 3 and 6]). In addition, viral proteins were not synthesized in cells infected with FHT virus but are clearly visible in extracts from influenza virus-infected cells (Fig. 1A). Moreover, Northern blots with probes specific for viral mRNA failed to detect transcription of viral genes in cells infected with heat-treated virus. However, the possibility that some viral mRNAs are transcribed below our detection limits remains has not been completely ruled out. The results were similar for cells that were infected with virus inactivated by 80 mJ of UV energy (data not shown; see Fig. S1 at http://thor.csi.washington.edu/katzelab/papers/Geiss2000/index.htm). These results indicated that inactivation procedures were sufficient to inhibit viral replication at both the 4- and 8-h time points.
In order to determine if either inactivation procedure was destroying the virus' ability to interact with the cellular membrane, the activity of viral HA was examined. HA is the viral glycoprotein that mediates attachment and fusion of the virus with the host cell (reference 53 and references therein). Serial dilutions (1:2 to 1:2,048) of medium alone, untreated virus, FHT virus, and an aliquot of influenza virus that was boiled for 90 min (which presumably destroys the structure of the virion and was therefore used as a negative control) were incubated with chicken red blood cells to determine relative HA titer. The red blood cells in wells that contain sufficient HA activity will not form a pellet upon centrifugation. As Fig. 1B shows, the HA activity of FHT virus was very similar to that of untreated virus while medium alone and boiled virus showed no activity. The HA activity of UV-inactivated (FUVI) virus was also similar to that of untreated virus (data not shown; see Fig. S3 at http://thor.csi.washington.edu/katzelab/papers/Geiss2000/index.htm). We concluded that replication of inactivated virus is blocked at a step after interacting with the host cell since HA retained structural conformation and ability to interact with cells whereas a completely denatured sample (boiled) or medium alone did not. For the bulk of this work, data from FHT influenza virus was used as a primary source of inactivated virus. Results from cells infected with FUVI virus at 4 h p.i. were used to corroborate some of our findings.
Microarray analysis was used to identify replication-dependent and -independent changes in gene expression.
The major goal of this study was to identify global changes in host cell gene expression that occur during influenza virus infection. For all of this work, we utilized cDNA microarrays that were constructed at the University of Washington's Center for Expression Arrays. A distinct advantage of using an “in-house” microarray system is that multiple conditions can be compared in parallel and repeated as many times as necessary without the cost restraints of commercial microarray sources. In addition, controls for viral infection, fluorescent dye incorporation, and nonspecific hybridization can be routinely performed and statistically analyzed. This feature, along with experiments specifically designed to distinguish between replication-dependent and independent changes in gene expression (see below and Fig. 2), provides an extra degree of confidence in the interpretation of our large-scale gene expression data.
The use of a replication-incompetent virus that retains cell-binding activity had two advantages. First, it allowed us to compare cells infected with FHT virus with those infected with FACT virus (Fig. 2, left). Changes in gene expression observed in FHT-versus-FACT microarray experiments are likely due to viral replication, since FHT virus cannot replicate (Fig. 1A) but other factors (e.g., attachment) remain roughly constant. Second, RNA from cells infected with FHT virus can be compared to that of mock-infected cells (Fig. 2, right) to identify changes in gene expression that were totally independent of viral replication. Finally, since untreated influenza virus retains both its replication-dependent and -independent properties, the comparison of mock-infected cells with cells infected with active influenza virus (Fig. 2, center) should yield a composite of differentially regulated gene sets observed in the other two conditions. Therefore, changes in gene expression that are dependent upon virus replication are defined as those coordinately regulated in FHT-versus-FACT and M-versus-FACT experiments but not M-versus-FHT experiments. Accordingly, a gene that is regulated in the M-versus-FHT and M-versus-FACT conditions but not during FHT-versus-FACT experiments is defined as replication independent. This method should minimize potential artifacts and false positives due to experimental variation. However, a potential consequence of this conservative approach is that host cell genes regulated by inactivated influenza virus that are otherwise inhibited by viral replication will not be scored.
Microarray experiments were performed on cells infected at both 4 and 8 h under the three pairwise conditions described above. Fluorescently labeled first-strand cDNAs were generated (19) from poly(A)-selected RNA from mock-infected cells or cells infected with FHT virus, FUVI virus, or untreated influenza virus. The appropriate probes were combined and hybridized to replica slides containing 4,608 cDNAs (spotted in duplicate). To control for dye-specific incorporation effects and differences in the saturation curves for the two dyes, all experiments were done on duplicate slides where the labeling scheme was reversed. In addition, microarray experiments were repeated with mRNA from independent infections with a different viral stock. Therefore for each gene on the array, eight separate measurements (four from each infection) were made per time point (see Materials and Methods).
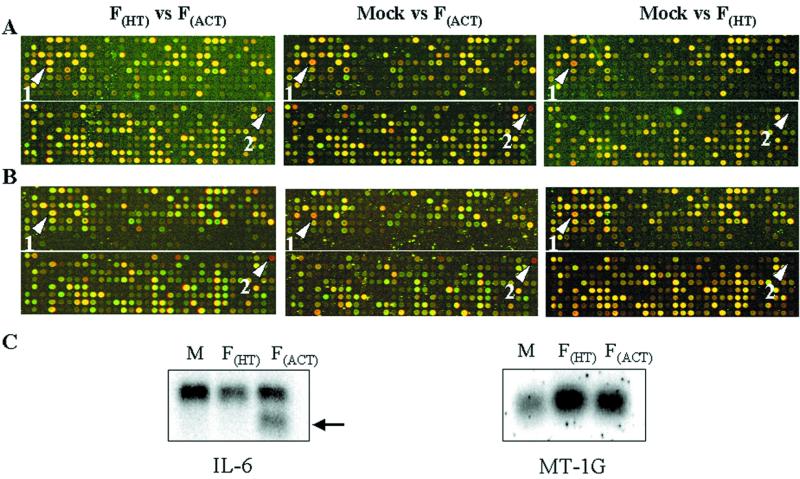
Hybridization signals were quantitated using the Spot-on software package, and a set of differentially expressed genes was generated for each condition and time point described in Materials and Methods. Host cell protein synthesis (Fig. 1A) and influenza virus gene expression (see Fig. S4 at http://thor.csi.washington.edu/katzelab/papers/Geiss2000/index.htm) was monitored for every experiment. An example of the microarray results is shown in Fig. 3A and B. Between one-third and two-thirds of the 4,608 cDNAs were above the minimal intensity value for any given experiment (see Table S1 at http://thor.csi.washington.edu/katzelab/papers/Geiss2000/index.htm). Ratios for genes below this threshold could not be measured accurately and therefore were eliminated from further consideration. Three general points were extracted from this analysis: (i) more changes in cellular gene expression were dependent on the presence of replication competent virus than not, (ii) inactivated virus alone was capable of affecting host cell gene expression, and (iii) more cellular genes were downregulated by influenza virus infection than were induced. The numbers of replication-dependent and -independent genes identified by the analysis described above are summarized in Table 1. The raw microarray images, quantitation results (pre- and postnormalization), and selected sets of differentially regulated genes are available at http://thor.csi.washington.edu/katzelab/papers/Geiss2000/index.htm and will be submitted to the first publicly available gene expression database.
FIG. 3.
Example of microarray results and determination of replication-dependent and -independent differentially expressed genes. False-color images for a portion of the microarray for the three pairwise conditions using FHT virus are shown in panels A (4 h p.i.) and B (8 h p.i.). Replica microarrays were hybridized with FHT virus (green) versus FACT virus (red), mock infection (green) versus FACT virus (red), and mock infection (green) versus FHT virus (red). A list of differentially expressed genes was generated for each set of samples using Spot-on software (see Materials and Methods). Examples of differentially expressed genes that are independent of or dependent on viral replication are shown (arrows 1 and 2, respectively). (C) RNA from mock-infected cells (M) or cells infected with FHT virus or FACT virus were run in 1% agarose gels under denaturing conditions and transferred to nylon membranes. Blots were hybridized with 32P-labeled DNA specific for IL-6 or metallothionein IG (MT-IG). Northern analysis was performed on RNA from the 4-h time point.
TABLE 1.
Summary of genes differentially expressed during influenza infection
| Classification | h p.i. | No. of genesa
|
||
|---|---|---|---|---|
| Upregulated | Downregulated | Total | ||
| Replication independent | 4 | 0 | 74 | 84 |
| Replication dependent | 4 | 2 | 59 | 61 |
| Total | 4 | 12 | 133 | 145 |
| Replication independent | 8 | 5 | 8 | 13 |
| Replication dependent | 8 | 37 | 292 | 329 |
| Total | 8 | 42 | 300 | 342 |
The number of differentially expressed genes represents the combination of two independent experiments, performed at different times with two separate preparations of viral stocks. Differentially expressed genes were defined by criteria described in Materials and Methods. Genes that did not exhibit a consistent pattern of expression in both infections were not included in these numbers.
Changes in cellular gene expression reveal pathways that may be important for viral replication.
Viral replication resulted in the differential expression of 61 and 329 genes at the 4- and 8-h time points, respectively (Table 1). A partial list based on the levels of expression at the 8-h time point is presented in Table 2. A complete list is available at http://thor.csi.washington.edu/katzelab/papers/Geiss2000/index.htm. The cellular processes most affected by viral replication include transcriptional regulation (38 genes), interleukin and growth factor signaling (32 genes), mRNA processing (15 genes), protein synthesis (9 genes), and protein degradation (6 genes). While a large percentage of these genes (351) decreased in expression during influenza virus infection, the expression of 39 genes was upregulated by viral replication. As one might predict of changes in mRNA expression that were dependent on viral replication, 39 of the 61 genes regulated at the 4-h time point were also regulated at 8 h (Table 2 and see http://thor.csi.washington.edu/katzelab/papers/Geiss2000/index.htm).
TABLE 2.
Partial list of cellular genes that were dependent on viral replicationa
| Gene type | I.M.A.G.E. identification | Gene product | Fold change
|
|
|---|---|---|---|---|
| FHT vs FACT | M vs FACT | |||
| Protein synthesis | 303048 | Ribosomal protein S6 | 2.8+ | 1.8+ |
| X69150 | Ribosomal protein S18 | 2.6+ | 1.7+ | |
| 214565 | ESTs, ribosomal protein S14 | 2.7+ | 2.2+ | |
| 141854 | Eukaryotic translation elongation factor 1 gamma | 1.7+ | 1.9+ | |
| 562867 | Homolog of yeast ribosome biogenesis regulatory protein | 2.7+ | 2.0+ | |
| 897982d | Translation initiation factor eIF-3 p110 subunit | 5.8− | 4.0− | |
| 34849d | Eukaryotic translation elongation factor 2 | 4.2− | 4.0− | |
| 788511 | Ribosomal protein S6 kinase, 90 kDa, polypeptide 2 | 4.8− | 2.7− | |
| 40567de | Human homolog translational activator GCN1 | 8.5− | 4.7− | |
| Cytokine and growth factor signaling | 310406de | IL-6 | 18.2+ | 7.9+ |
| 811920 | IL-11 receptor alpha chain | 2.3− | 1.9− | |
| 289606 | IL-15 | 3.7− | 2.0− | |
| 897570 | Tumor necrosis factor type 1 receptor-associated protein | 5.1− | 2.4− | |
| 810444d | B94 protein | 2.4− | 1.8− | |
| 788185 | TRAIL receptor 2 | 2.4− | 1.8− | |
| 813184 | B12 protein | 2.4− | 1.8− | |
| 811900 | Lymphotoxin-beta receptor precursor | 3.7− | 2.6− | |
| 136821cd | TGF-β 1 precursor | 3.9− | 2.5− | |
| 768168b | Bone morphogenetic protein 6 precursor | 3.3− | 1.9− | |
| 898092d | Connective tissue growth factor | 5.2+ | 3.0+ | |
| 79712de | Insulin-like growth factor 2 receptor | 2.8− | 2.8− | |
| 898218c | Insulin-like growth factor-binding protein | 4.4− | 4.8− | |
| 85093de | KIAA0062 gene, similar to estrogen regulated LIV-1 protein | 6.8− | 3.4− | |
| 297122e | Epidermal growth factor receptor (v-erb-b) oncogene homolog | 4.6− | 4.6− | |
| 768496d | Cytokine receptor (EBI3) | 3.9− | 5.2− | |
| Transcription factors and DNA binding proteins | 898221 | Immediate-early response protein NOT | 2.7+ | 1.7+ |
| 812965 | v-myc avian myelocytomatosis viral oncogene homolog | 4.7+ | 4.2+ | |
| 770670 | Putative DNA binding protein A20 | 8.1+ | 3.7+ | |
| 146577 | TGF-β-inducible early protein | 2.6+ | 3.8+ | |
| 377320e | Zinc finger protein | 5.6− | 2.5− | |
| 771220 | Transcription factor p65, NF-κB | 4.3− | 3.2− | |
| 755821bd | Transcription factor 11 | 2.8− | 2.1− | |
| 725680d | Transcription factor ERF-1 | 11.7− | 2.0− | |
| 785816bd | Nuclear factor NF90 | 5.9− | 3.8− | |
| 40781d | Interleukin enhancer-binding factor | 4.3− | 3.1− | |
| 240367 | Transcriptional repressor (CTCF) | 3.8− | 2.6− | |
| 712840 | Transcription factor Stat5b | 2.6− | 1.9− | |
| 137387d | ESTs,f transcription factor AP-2 | 10.0− | 3.5− | |
| 161950d | ESTs, weakly similar to hairless protein | 4.5− | 2.8− | |
| 770910de | Epithelial cell-spectrum transcription factor ESE-1b | 5.2− | 3.7− | |
| 783681de | KIAA0312 gene, upstream regulatory element binding protein | 6.4− | 3.4− | |
| Processing and export of mRNA | 67074 | ESTs, pre-mRNA splicing factor RNA helicase PRP22 | 4.5− | 2.6− |
| 814119de | RNA helicase (HRH1) | 4.3− | 2.7− | |
| 741841 | CHL1 potential helicase (CHLR1) | 5.7− | 7.1− | |
| 809535 | Splicing factor, arginine/serine-rich 2 | 5.5− | 2.8− | |
| 811108 | Pre-mRNA splicing factor SF2, P32 subunit precursor | 3.6− | 2.5− | |
| 123400 | Human FUSE binding protein 2 (FBP2) | 3.3− | 2.1− | |
| 825224 | Export protein Rae1 (RAE1) | 4.2− | 3.2− | |
| 714426 | Cleavage stimulation factor, 3′ pre-RNA, subunit 2, 64 kDa | 2.8− | 1.8− | |
| 50188 | Zinc finger protein | 4.1− | 3.0− | |
| 823663d | Fragile X mental retardation syndrome-related protein (FXR2) | 4.3− | 2.1− | |
| 25588 | RNA-binding protein CUG-BP/hNab50 | 3.7− | 2.4− | |
| 811911d | Hlark | 3.5− | 2.5− | |
| 121621d | Heterogenous nuclear ribonucleoprotein U | 7.0− | 3.0− | |
| 309032 | Cleavage and polyadenylation specificity factor | 3.9− | 2.4− | |
| 366558 | RanGTPase activating protein 1 | 4.7− | 3.0− | |
| Ubiquitin pathway | 210405 | Proteasome activator hPA28 subunit beta | 1.9+ | 1.7+ |
| X63237 | Human ubiquitin | 2.6+ | 1.7+ | |
| 898262d | Ubiquitin activating enzyme E1 | 11.3− | 4.0− | |
| 134172 | ESTs, ubiquitin-conjugating enzyme E2-17 KD 11 | 3.7− | 1.9− | |
| 789232 | Antisecretory factor-1 | 1.9− | 3.0− | |
| 613126 | Isopeptidase T-3 (ISOT-3) | 3.0− | 2.7− | |
The gene names and fold changes are based on the results from the 8-h time point. The ratios were generated by taking the means of eight measurements at the two appropriate conditions, mock infection (M) versus FACT and FHT versus FACT (Fig. 3). A complete list of genes and the ratios for each individual experiment as well as the other condition (mock infection versus FHT) are available at http://thor.csi.washington.edu/katzelab/papers/Geiss2000/index.htm in Table S3.
The I.M.A.G.E clone was present in two copies on the array.
The clone was present twice on the array and regulated at both time points.
The cDNA was regulated in both FHT and FUVI experiments.
The mRNA was also differentially regulated in a replication-dependent manner at the 4-h time point.
ESTs, expressed sequence tags.
Influenza virus replication induced the expression of the interleukin 6 (IL-6) gene (present in two copies) at 4 h p.i. and 37 genes at 8 h p.i. Given that influenza virus generally represses the synthesis of cellular proteins (Fig. 1A) and gene expression (see below), upregulated genes are attractive candidates for future functional studies provided that their protein levels increase accordingly. IL-6 was the only gene upregulated by viral replication at the 4-h time point. Its mRNA expression levels increased almost eightfold at 8 h p.i. (Fig. 3B and Table 2). This proinflammatory cytokine is regulated by a variety of extracellular stresses, including bacterial and viral infections (2, 51). Northern blot analysis of the three RNA samples with an IL-6-specific probe detected two bands and confirmed the increase in IL-6 signal (lower band) only in cells infected with active influenza virus (Fig. 3C, left, lane F). The significance of the two transcripts is not known at this time, but one band may represent an alternatively spliced IL-6 product that has been detected in another system (29) which is not induced by infection. The induction of IL-6 mRNA is specific, since a similar increase in mRNAs encoding other interleukin genes present on the array was not upregulated by influenza virus. For example, mRNA levels for IL-15 and IL-11 as well as interleukin enhancer binding factor 3 (Table 2) were actually downregulated. Although IL-6 protein levels were not directly addressed in this study, the microarray data are consistent with reported IL-6 increases in cell lines (41, 57) and in mice (42). Finally, the induction of IL-6 mRNA was also observed in other microarray-based studies during cytomegalovirus (59), poliovirus (25), and coxsackievirus (55) infections. If the IL-6 protein expression verifies the expression levels in this system, it may hint at a potential role for IL-6 in the host's innate antiviral response.
The mRNAs for several genes involved in the process of protein synthesis were also induced in a replication-dependent manner. These include mRNAs for the ribosomal subunit S6, translation elongation factor 1-gamma, a homolog of the recently identified ribosome biogenesis regulatory protein, and several other ribosomal proteins (Table 2). The upregulation of mRNAs for ribosomal proteins, especially S6 (13) and RRS1 (56), was intriguing given that viral mRNAs were selectively translated in infected cells, despite an overall inhibition of the synthesis of cellular proteins. Interestingly, not all the mRNAs for genes in this pathway are upregulated, indicating that influenza virus may recruit specific components of the translation machinery. The concomitant downregulation of translation factors by influenza virus could account for the similarity of polysome profiles in mock- and influenza virus-infected cells despite an obvious decrease in the synthesis of cellular proteins (26). It is also possible that some of these genes cooperate with additional cellular or viral proteins such as GRSF-1 (49) or nonstructural protein 1 (NS1) (11), respectively, in the selective translation of viral mRNAs. Of course, follow-up studies will be required to verify the protein levels, phosphorylation states, enzyme activities, and functional significance of key translation components to demonstrate a potential role in selective translation of influenza virus mRNAs.
Induction of cellular genes potentially involved in transcriptional regulation was also observed at the 8-h time point. These genes could have several roles in an infected cell such as maintaining the pool of capped RNA polymerase II transcripts for viral mRNA synthesis or controlling expression of other genes that mediate the cellular stress response. Two of these genes encode zinc finger proteins, A20 and the product of the transforming growth factor (TGF) β-inducible early gene, that are normally induced by tumor necrosis factor and TGF, respectively. This result is the opposite of what was observed for mRNAs encoding other transcriptional regulators that were shown to decrease during infection (see below and Table 2). The induction of A20 or TEIG amidst the broad downregulation of many other cellular genes may indicate that they are directly induced by viral components, such as viral dsRNAs. Indeed, the level of A20 mRNA has been shown to be upregulated by pI-pC treatment of GRE cells used as a model system for dsRNA signaling (G. K. Geiss and G. C. Sen, submitted for publication).
In contrast to the examples of cellular genes induced by influenza virus infection, the major cellular response was the downregulation of genes from a diverse set of cellular pathways. The number of downregulated cellular genes increased from 61 genes at 4 h p.i. to 329 at 8 h p.i.. The number of downregulated genes represents, on average, less than 4% of the total number of genes detected at the 4-h time point and less than 17% of the number detected at 8 h p.i., suggesting that a complete degradation of cellular mRNAs did not occur. However, the potential effects that influenza virus infection has on mRNA processing and stability, especially considering the removal of the 5′ cap and inhibition of cellular protein synthesis, could, in part, explain the downregulation of some genes, since uncapped messages are quickly degraded (24). Whether the mechanisms behind the decrease in steady-state mRNA levels in influenza virus-infected cells are due to transcriptional regulation, the degradation of aberrantly processed mRNAs, altered mRNA stability, or some combination of these events has not yet been addressed on a genome-wide scale.
The identities of the replication-dependent downregulated genes indicated that a broad array of cellular genes were affected. The classes with the highest number of genes downregulated include those involved in growth factor and cytokine signaling, those encoding DNA binding proteins, extracellular and cytoskeletal genes, and genes involved in mRNA processing and export. The latter class of genes is interesting in the context of influenza virus infection, given the role of the viral NS1 protein in inhibiting mRNA polyadenylation, splicing, and export of cellular mRNAs within the nucleus. Interestingly, a direct interaction between the influenza virus NS1 protein and cellular cleavage and polyadenylation specificity factor has recently been reported (45). Whether NS1-induced perturbations in mRNA processing are responsible for the decrease in cellular mRNA levels, potentially through its interactions with cellular cleavage and polyadenylation specificity factor, could be addressed with infections using mutant influenza viruses lacking the NS1 gene (16).
Cellular genes regulated independently of viral replication identify a potential antiviral pathway.
It is clear from the microarray data that the steady-state level of cellular mRNAs was also altered by events that do not require viral replication. Since heat-inactivated virus retains HA activity similar to that of untreated virus, it is likely that replication-independent steps of the viral life cycle, for example, attachment or fusion to the host cell, were responsible for inducing most of these changes. In contrast to the replication-dependent changes in gene expression, the number of replication-independent changes decreased dramatically over the course of infection, from 84 genes at 4 h p.i. (58% of all genes that change at that time point) to 13 at 8 h p.i. (4% of the genes regulated). This result is consistent with the notion that contact or fusion of the virus with the host cell transmits signals that alter host cell gene expression.
There were a total of 97 differentially regulated genes at both time points that did not require active viral replication. A partial list is presented in Table 3, based on their expression at the 4-h time point. A complete list is available at http://thor.csi.washington.edu/katzelab/papers/Geiss2000/index.htm. The cellular pathway genes affected include the metallothionein genes (five genes), genes involved in cell cycle progression (five genes), those encoding transcriptional regulators (eight genes), those involved in the ubiquitin/proteasome pathway (six genes), and those encoding various cellular kinases (three genes) (Table 3 and see http://thor.csi.washington.edu/katzelab/paper/Geiss2000/index.htm). As mentioned above, the majority of these genes were exclusively regulated at the 4-h time point. Only eight genes (four of the five metallothionein genes, the genes for flavin monooxygenase, cyclin A, and extracellular regulated kinase-3, and I.M.A.G.E. ID 299737) were differentially regulated at both time points.
TABLE 3.
Partial list of cellular genes that were regulated independently of viral replicationa
| Gene type | I.M.A.G.E. identification | Gene product | Fold change
|
|
|---|---|---|---|---|
| M vs FACT | M vs FHT | |||
| Metallothioneins | 232772bc | Metallothionein IB | 2.5+ | 2.2+ |
| 297392b | Metallothionein IL | 2.6+ | 2.3+ | |
| 202535b | Metallothionein IG | 3.2+ | 2.8+ | |
| 214162c | Metallothionein IH | 3.2+ | 2.7+ | |
| 129585bc | ESTS, metallothionein II | 3.1+ | 2.0+ | |
| Cell cycle related | 214572 | Cyclin-dependent kinase 6 | 1.7− | 1.7− |
| 950690b | Cyclin A | 2.6− | 2.0− | |
| 547058 | H. sapiens mRNA for cyclin G1 | 2.4− | 2.0− | |
| 898286 | Cell division cycle 2, G1 to S and G2 to M | 3.2− | 2.2− | |
| 814701 | Mitotic feedback control protein Madp2 homolog | 2.7− | 2.1− | |
| Transcriptional regulators | 77577 | FOS-related antigen 2 | 2.8+ | 2.0+ |
| 741067 | SWI/SNF complex 60-kDa subunit (BAF60b) | 1.9+ | 1.8+ | |
| 773188 | Human mRNA for EAR-1r | 2.0− | 1.8− | |
| 897761 | EST, similar to SNF2L1 | 2.4− | 2.3− | |
| 109265 | Human CACCC box-binding protein | 2.5− | 2.9− | |
| 795847 | v-jun avian sarcoma virus 17 oncogene homolog | 2.7− | 1.8− | |
| 144951 | ESTs, highly similar to zinc finger protein 91 | 2.4− | 1.6− | |
| 50614 | Human nuclear protein Skip | 1.8− | 1.6− | |
| Ubiquitin pathway | 788247 | Human CUL-2 (cul-2) | 2.9− | 2.0− |
| 760231 | H. sapiens mRNA for ubiquitin hydrolase | 2.4− | 1.8− | |
| 843094 | Human ubiquitin homology domain protein PIC1 | 1.7− | 1.8− | |
| 814246 | Proteasome component C8 | 2.3− | 2.1− | |
| 767049 | Proteasome subunit p42 | 3.8− | 1.9− | |
| 24085 | Tripeptidyl peptidase II | 2.0− | 2.1− | |
| Cellular kinases | 167032 | Protein kinase Cγ | 3.3+ | 2.2+ |
| 50506b | ERK3 mRNA | 2.2− | 1.8− | |
| 362853 | Human protein tyrosine kinase | 3.2− | 3.0− | |
The genes presented here are those that were differentially regulated in the absence of viral replication at 4 h p.i. The fold change is the mean of eight measurements under the two relevant conditions that define a replication-independent change in gene expression, mock infection (M) versus FACT and mock infection versus FHT (Fig. 3). A complete list of genes and the ratios for each individual experiment as well as the other condition (FHT versus FACT) are available at http://thor.csi.washington.edu/katzelab/papers/Geiss2000/index.htm in Table S2.
The mRNA was also differentially regulated in a replication-independent manner at the 8-h time point.
The cDNA was regulated in both FHT and FUVI experiments.
The mRNAs that most consistently increased in a replication-independent manner were members of the metallothionein gene family (encoding metallothioneins IB, IG, IH, IL, and II) (Table 3). All five cDNAs representing metallothionein genes on this array were induced at the early time point, and four continue to be regulated at the later time point. Northern blot analysis with a radiolabeled metallothionein IG probe detected a single band and confirmed the increased mRNA levels in cells infected with FHT and active influenza virus (Fig. 3C, right). However, due to the high sequence homology among the metallothionein mRNAs, an increase in a single species cannot be distinguished on these cDNA microarrays. Although the exact physiological role of metallothioneins has not been fully elucidated, they are induced by a variety of extracellular stimuli, including IL-6, heavy metals, oxidative stress, and bacterial endotoxins (10, 14, 22, 35, 37, 47). Indeed, influenza virus infections with either active or inactive viral preparations have previously been shown to induce oxidative stress (6, 8). If protein levels confirm the expression data, metallothionein induction may represent a host response to attachment-induced oxidative stress. It will be extremely interesting to examine the outcome of influenza virus infection in cell lines by inducing metallothionein gene expression (by treatment with Zn or IL-6) before or after infection or in metallothionein knockout mice (44). With regard to the former, the clinical administration of Zn at the onset of illness has been shown to reduce the severity and longevity of cold-like symptoms (18, 40) and has been shown to reduce herpes simplex virus infectivity in vitro (33).
Nearly all of the genes regulated independently of viral replication were downregulated (82 of 97), most of them exclusively at the 4-h time point (74 of 82). Among the pathways most influenced were those involved in cell cycle progression, protein degradation, and transcriptional regulation. Binding of influenza virus to the cell membrane may initiate a cellular response that “sets up” the host environment for viral infection or induces extracellular stress that results in the downregulation of these genes. For example, influenza virus may require a specific stage of the cell cycle or downregulation of the ubiquitin pathway in order to achieve maximum replication efficiency or synthesis of viral proteins. Interestingly, several members of the protein degradation pathway were upregulated by viral replication during later times of infection (Table 2), suggesting that increased expression of viral proteins during replication eventually activates the protein degradation process. Finally, downregulation of these genes does not likely represent a nonspecific effect of cell death, since inactivated virus does not inhibit host cell protein synthesis (Fig. 1A) or significantly reduce cell viability (data not shown).
Gene expression analysis of UV-inactivated virus confirms the replication-dependent and -independent changes in cellular gene expression.
To provide additional evidence that changes in steady-state mRNA levels were due to infection, microarray experiments were repeated with FUVI virus at 4 h p.i. as described above. We compared the expression level for genes differentially expressed in the first set of experiments to those observed during the FUVI microarray experiments. There were 13 genes that were altered in a replication-independent manner and 142 genes whose changes were dependent on viral replication at 4 h p.i.. Four of the 13 replication-independent genes (encoding metallothioneins IB, IH, and II and alanine-glyoxylate aminotransferase) were differentially regulated in the same manner as FHT virus. Similarly, 96 (4 up and 92 down) of the 142 replication-dependent genes were also differentially regulated in UV experiments. These include genes for IL-6 (two copies), connective tissue growth factor, and transcription factors and genes involved in mRNA processing (Table 2). In addition, flavin monooxygenase and metallothionein IG mRNA levels also increased moderately but not enough to meet our strict cutoff criteria. These data in conjunction with previous findings in other studies support the hypothesis that the changes in expression of the metallothionein genes and others are due to the presence of viral components.
There were also significant differences in mRNA levels that were specific to the method of inactivation. First, the absence of replication-independent downregulated genes and a increase in the number of replication-dependent genes suggests that the kinetics of infection were increased slightly in UV experiments, resulting in a gene expression profile more similar to that of 8-h FHT virus (Table 2). In addition, there were a number of genes that were differentially regulated exclusively by UV and not FHT virus and others that were regulated in the opposite direction, suggesting that they are regulated by factors that were affected differently by the two distinct inactivation procedures.
DISCUSSION
Our efforts to control for nonviral factors that may alter host cell gene expression support the idea that the majority of changes in mRNA abundance identified here are due to influenza virus infection. We used a DNA microarray-based approach to show that influenza virus infection affects the steady-state mRNA levels for a wide variety of cellular genes in both a replication-dependent and a replication-independent manner. The mechanisms driving the observed changes in mRNA abundance for each gene are not known and are probably due to both altered mRNA stability and transcriptional regulation. We have attempted to minimize potential problems by (i) requiring a gene to be regulated similarly in two of the three relevant pairwise conditions, (ii) performing duplicate infections and selecting only genes that were consistently regulated, and (iii) preparing inactivated virus by two different mechanisms. This approach minimizes the selection of genes whose expression patterns vary from experiment to experiment and the potential effects of the method of inactivation. The phenomenon of genes being regulated consistently under all conditions is almost certainly due to virus-mediated events, since a nonviral factor would have to resist heat and UV inactivation in order to induce a similar pattern of expression in both experiments. Finally, although some of the findings reported here are corroborated by published reports, it is important to note that replication-dependent changes in gene expression occur in the presence of an inhibition of cellular protein synthesis. Therefore, it will be critical to verify that genes whose steady-state mRNA levels increase during infection do, in fact, escape the virus-induced inhibition of protein synthesis.
The number of DNA array-based studies aimed at elucidating host cell gene expression during viral and bacterial infections has increased substantially in the past 2 years (9, 39). A comparative analysis of influenza virus infections with gene expression studies on genetically unrelated viruses revealed some interesting similarities and differences with influenza virus. For instance, poliovirus and coxsackievirus also induce mRNAs encoding ribosomal components (25, 55), while cytomegalovirus induces expression of IL-6 (see above), and metallothionein I RNA is upregulated in coxsackievirus infections. In addition, both influenza virus and cytomegalovirus downregulate the expression of genes in the insulin and TGF pathways. On the other hand, the differences in these viral systems is exemplified by the finding that cytomegalovirus infection induces a large set of interferon-induced genes which is not seen in these influenza virus infections or experiments on papillomavirus-expressing cell lines in which the interferon response was specifically repressed (5). As mentioned above, the differences observed in these large-scale studies are likely due to a number of factors. For instance, the lack of an interferon-induced gene expression in these experiments might be attributed to influenza virus' ability to block interferon signaling via the viral NS1 protein (4, 16, 54) or the presence or absence of exogenous interferon in the viral preparation. Alternatively, the timing of the experiments may not be the optimal system for observing changes in this classic antiviral system, since this HeLa cell line is at least partially responsive to exogenous interferon treatment (G. K. Geiss and M. G. Katze, unpublished data).
Microarray-based gene expression studies are especially well suited for comparing the cellular response to viral infections, viral components (purified proteins and dsRNA), and treatment with various cytokines or antiviral compounds such as those approved for influenza treatment and prevention. The cellular response to influenza virus and other viral infections will ultimately be defined by comparing the results from different viruses, host cells, and infection kinetics with the present knowledge base and future functional analysis. In addition, highly parallel and genome-wide comparisons might allow us to determine which cellular genes are essential to virus or host cell survival. This study is the first in what we anticipate will be an ongoing effort to define global gene expression patterns in response to different influenza virus strains, mutants, proteins, and host cell types. Furthermore, recent advances in reverse genetic techniques (46) will allow a systematic analysis of wild-type influenza virus genes or mutants and their influence on cellular gene expression. The information obtained from this and related studies will revolutionize our view of influenza virus infection and its influence on host cell biology and may eventually lead to novel therapeutic targets.
ACKNOWLEDGMENTS
This work was supported by Public Health Service grants AI-22646, AI-41629, and RR-00166 from the National Institutes of Health. Mahru C. An was a recipient of a Howard Hughes Fellowship and an Early Identification Program Presidential Scholarship.
We acknowledge M. J. Korth and other members of the Katze laboratory for critical reading and review of the manuscript and A. B. van 't Wout for helpful discussions and for construction and design of the control plate. We also thank Mary Claire King's laboratory for access to the human cDNA I.M.A.G.E clones and the University of Washington's Center for Expression Arrays for providing microarray services and software.
REFERENCES
- 1.Agy B M, Wambach M, Foy K, Katze M G. Expression of cellular genes in CD4 positive lymphoid cells infected by the human immunodeficiency virus, HIV-1: evidence for a host protein synthesis shut-off induced by cellular mRNA degradation. Virology. 1990;177:251–258. doi: 10.1016/0042-6822(90)90478-a. [DOI] [PubMed] [Google Scholar]
- 2.Appelberg R. Protective role of interferon gamma, tumor necrosis factor alpha and interleukin-6 in Mycobacterium tuberculosis and M. avium infections. Immunobiology. 1994;191:520–525. doi: 10.1016/S0171-2985(11)80458-4. [DOI] [PubMed] [Google Scholar]
- 3.Beloso A, Martinez C, Valcarcel J, Santaren J F, Ortin J. Degradation of cellular mRNA during influenza virus infection: its possible role in protein synthesis shutoff. J Gen Virol. 1992;73:575–581. doi: 10.1099/0022-1317-73-3-575. [DOI] [PubMed] [Google Scholar]
- 4.Bergmann M, Garcia-Sastre A, Carnero E, Pehamberger H, Wolff K, Palese P, Muster T. Influenza virus NS1 protein counteracts PKR-mediated inhibition of replication. J Virol. 2000;74:6203–6206. doi: 10.1128/jvi.74.13.6203-6206.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chang Y E, Laimins L A. Microarray analysis identifies interferon-inducible genes and Stat-1 as major transcriptional targets of human papillomavirus type 31. J Virol. 2000;74:4174–4182. doi: 10.1128/jvi.74.9.4174-4182.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Choi A M, Knobil K, Otterbein S L, Eastman D A, Jacoby D B. Oxidant stress responses in influenza virus pneumonia: gene expression and transcription factor activation. Am J Physiol. 1996;271:L383–L391. doi: 10.1152/ajplung.1996.271.3.L383. [DOI] [PubMed] [Google Scholar]
- 7.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 8.Coppola J A, Cole M D. Constitutive c-myc oncogene expression blocks mouse erythroleukemia cell differentiation but not commitment. Nature. 1986;320:760–763. doi: 10.1038/320760a0. [DOI] [PubMed] [Google Scholar]
- 9.Cummings C A, Relman D A. Using DNA microarrays to study host-microbe interactions. Emerg Infect Dis. 2000;6:513–525. doi: 10.3201/eid0605.000511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De S K, McMaster M T, Andrews G K. Endotoxin induction of murine metallothionein gene expression. J Biol Chem. 1990;265:15267–15274. [PubMed] [Google Scholar]
- 11.de la Luna S, Fortes P, Beloso A, Ortin J. Influenza virus NS1 protein enhances the rate of translation initiation of viral mRNAs. J Virol. 1995;69:2427–2433. doi: 10.1128/jvi.69.4.2427-2433.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Der S D, Zhou A, Williams B R, Silverman R H. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci USA. 1998;95:15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dufner A, Thomas G. Ribosomal S6 kinase signaling and the control of translation. Exp Cell Res. 1999;253:100–109. doi: 10.1006/excr.1999.4683. [DOI] [PubMed] [Google Scholar]
- 14.Durnam D M, Hoffman J S, Quaife C J, Benditt E P, Chen H Y, Brinster R L, Palmiter R D. Induction of mouse metallothionein-I mRNA by bacterial endotoxin is independent of metals and glucocorticoid hormones. Proc Natl Acad Sci USA. 1984;81:1053–1056. doi: 10.1073/pnas.81.4.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gale M, Jr, Katze M G. Molecular mechanisms of interferon resistance mediated by viral-directed inhibition of PKR, the interferon-induced protein kinase. Pharmacol Ther. 1998;78:29–46. doi: 10.1016/s0163-7258(97)00165-4. [DOI] [PubMed] [Google Scholar]
- 16.Garcia-Sastre A, Egorov A, Matassov D, Brandt S, Levy D E, Durbin J E, Palese P, Muster T. Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology. 1998;252:324–330. doi: 10.1006/viro.1998.9508. [DOI] [PubMed] [Google Scholar]
- 17.Garfinkel S M, Katze M G. Translational control by influenza virus. Selective and cap-dependent translation of viral mRNAs in infected cells. J Biol Chem. 1992;267:9383–9390. [PubMed] [Google Scholar]
- 18.Garland L M, Hagmeyer K O. The role of zinc lozenges in treatment of the common cold. Ann Pharmacother. 1998;32:63–69. doi: 10.1345/aph.17128. [DOI] [PubMed] [Google Scholar]
- 19.Geiss G K, Bumgarner R E, An M C, Agy M B, van 't Wout A B, Hammersmark E, Carter V S, Upchurch D, Mullins J I, Katze M G. Large-scale monitoring of host cell gene expression during HIV-1 infection using cDNA microarrays. Virology. 2000;266:8–16. doi: 10.1006/viro.1999.0044. [DOI] [PubMed] [Google Scholar]
- 20.Haberberger T C, Kupfer K, Murphy J E. Profiling of genes which are differentially expressed in mouse liver in response to adenoviral vectors and delivered genes. Gene Ther. 2000;7:903–909. doi: 10.1038/sj.gt.3301181. [DOI] [PubMed] [Google Scholar]
- 21.Han J, Yoo H Y, Choi B H, Rho H M. Selective transcriptional regulations in the human liver cell by hepatitis B viral X protein. Biochem Biophys Res Commun. 2000;272:525–530. doi: 10.1006/bbrc.2000.2801. [DOI] [PubMed] [Google Scholar]
- 22.Hernandez J, Hidalgo J. Endotoxin and intracerebroventricular injection of IL-1 and IL-6 induce rat brain metallothionein-I and -II. Neurochem Int. 1998;32:369–373. doi: 10.1016/s0197-0186(97)00096-x. [DOI] [PubMed] [Google Scholar]
- 23.Inglis S C. Inhibition of host protein synthesis and degradation of cellular mRNAs during infection by influenza and herpes simplex virus. Mol Cell Biol. 1982;2:1644–1648. doi: 10.1128/mcb.2.12.1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jacobson A, Peltz S W. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu Rev Biochem. 1996;65:693–739. doi: 10.1146/annurev.bi.65.070196.003401. [DOI] [PubMed] [Google Scholar]
- 25.Johannes G, Carter M S, Eisen M B, Brown P O, Sarnow P. Identification of eukaryotic mRNAs that are translated at reduced cap binding complex eIF4F concentrations using a cDNA microarray. Proc Natl Acad Sci USA. 1999;96:13118–13123. doi: 10.1073/pnas.96.23.13118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Katze G M, DeCorato D, Krug R M. Cellular mRNA translation is blocked at both initiation and elongation after infection by influenza virus or adenovirus. J Virol. 1986;60:1027–1039. doi: 10.1128/jvi.60.3.1027-1039.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katze G M, Krug R M. Metabolism and expression of RNA polymerase II transcripts in influenza virus-infected cells. Mol Cell Biol. 1984;4:2198–206. doi: 10.1128/mcb.4.10.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Keavey S. Preparing for the next influenza outbreak—or (inevitably) pandemic. J Acad Physicians Assist. 1999;12:28–30. , 33–4, 37–40. [PubMed] [Google Scholar]
- 29.Kestler D P, Agarwal S, Cobb J, Goldstein K M, Hall R E. Detection and analysis of an alternatively spliced isoform of interleukin-6 mRNA in peripheral blood mononuclear cells. Blood. 1995;86:4559–4567. [PubMed] [Google Scholar]
- 30.Khodarev N N, Advani S J, Gupta N, Roizman B, Weichselbaum R R. Accumulation of specific RNAs encoding transcriptional factors and stress response proteins against a background of severe depletion of cellular RNAs in cells infected with herpes simplex virus 1. Proc Natl Acad Sci USA. 1999;96:12062–12067. doi: 10.1073/pnas.96.21.12062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kleines M, Finke K, Ritter K, Schaade L. Induction of growth rate and transcriptional modulation of growth promoters and growth inhibitors in epithelial cells by EBV-LMP1. Virus Res. 2000;68:63–69. doi: 10.1016/s0168-1702(00)00152-0. [DOI] [PubMed] [Google Scholar]
- 32.Krug R M. The regulation of export of mRNA from nucleus to cytoplasm. Curr Opin Cell Biol. 1993;5:944–949. doi: 10.1016/0955-0674(93)90074-z. [DOI] [PubMed] [Google Scholar]
- 33.Kumel G, Schrader S, Zentgraf H, Daus H, Brendel M. The mechanism of the antiherpetic activity of zinc sulphate. J Gen Virol. 1990;71:2989–2997. doi: 10.1099/0022-1317-71-12-2989. [DOI] [PubMed] [Google Scholar]
- 34.Lamb R A, Krug R M. Orthomyxoviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Vol. 1. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 1353–1395. [Google Scholar]
- 35.Lee D K, Carrasco J, Hidalgo J, Andrews G K. Identification of a signal transducer and activator of transcription (STAT) binding site in the mouse metallothionein-I promoter involved in interleukin-6-induced gene expression. Biochem J. 1999;337:59–65. [PMC free article] [PubMed] [Google Scholar]
- 36.Lennon G, Auffray C, Polymeropoulos M, Soares M B. The I.M.A.G.E. Consortium: an integrated molecular analysis of genomes and their expression. Genomics. 1996;33:151–152. doi: 10.1006/geno.1996.0177. [DOI] [PubMed] [Google Scholar]
- 37.Liu J, Liu Y P, Sendelbach L E, Klassen C D. Endotoxin induction of hepatic metallothionein is mediated through cytokines. Toxicol Appl Pharmacol. 1991;109:235–240. doi: 10.1016/0041-008x(91)90171-a. [DOI] [PubMed] [Google Scholar]
- 38.Lu Y, Qian X Y, Krug R M. The influenza virus NS1 protein: a novel inhibitor of pre-mRNA splicing. Genes Dev. 1994;8:1817–1828. doi: 10.1101/gad.8.15.1817. [DOI] [PubMed] [Google Scholar]
- 39.Manger I D, Relman D A. How the host ‘sees’ pathogens: global gene expression responses to infection. Curr Opin Immunol. 2000;12:215–218. doi: 10.1016/s0952-7915(99)00077-1. [DOI] [PubMed] [Google Scholar]
- 40.Marshall S. Zinc gluconate and the common cold. Review of randomized controlled trials. Can Fam Phys. 1998;44:1037–1042. [PMC free article] [PubMed] [Google Scholar]
- 41.Matsukura S, Kokubu F, Noda H, Tokunaga H, Adachi M. Expression of IL-6, IL-8, and RANTES on human bronchial epithelial cells, NCI-H292, induced by influenza virus A. J Allergy Clin Immunol. 1996;98:1080–1087. doi: 10.1016/s0091-6749(96)80195-3. [DOI] [PubMed] [Google Scholar]
- 42.Matsuo K, Iwasaki T, Asanuma H, Yoshikawa T, Chen Z, Tsujimoto H, Kurata T, Tamura S S. Cytokine mRNAs in the nasal-associated lymphoid tissue during influenza virus infection and nasal vaccination. Vaccine. 2000;18:1344–1350. doi: 10.1016/s0264-410x(99)00401-6. [DOI] [PubMed] [Google Scholar]
- 43.Meltzer M I, Cox N J, Fukuda K. The economic impact of pandemic influenza in the United States: priorities for intervention. Emerg Infect Dis. 1999;5:659–671. doi: 10.3201/eid0505.990507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Michalska A E, Choo K H. Targeting and germ-line transmission of a null mutation at the metallothionein I and II loci in mouse. Proc Natl Acad Sci USA. 1993;90:8088–8092. doi: 10.1073/pnas.90.17.8088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nemeroff E M, Barabino S M, Li Y, Keller W, Krug R M. Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′ end formation of cellular pre-mRNAs. Mol Cell. 1998;1:991–1000. doi: 10.1016/s1097-2765(00)80099-4. [DOI] [PubMed] [Google Scholar]
- 46.Palese P, Zheng H, Engelhardt O G, Pleschka S, Garcia-Sastre A. Negative-strand RNA viruses: genetic engineering and applications. Proc Natl Acad Sci USA. 1996;93:11354–11358. doi: 10.1073/pnas.93.21.11354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Palmiter R D. The elusive function of metallothioneins. Proc Natl Acad Sci USA. 1998;95:8428–8430. doi: 10.1073/pnas.95.15.8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Park Y W, Katze M G. Translational control by influenza virus. Identification of cis-acting sequences and trans-acting factors which may regulate selective viral mRNA translation. J Biol Chem. 1995;270:28433–28439. doi: 10.1074/jbc.270.47.28433. [DOI] [PubMed] [Google Scholar]
- 49.Park Y W, Wilusz J, Katze M G. Regulation of eukaryotic protein synthesis: selective influenza viral mRNA translation is mediated by the cellular RNA-binding protein GRSF-1. Proc Natl Acad Sci USA. 1999;96:6694–6699. doi: 10.1073/pnas.96.12.6694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Plotch S J, Bouloy M, Ulmanen I, Krug R M. A unique cap(m7GpppXm)-dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell. 1981;23:847–858. doi: 10.1016/0092-8674(81)90449-9. [DOI] [PubMed] [Google Scholar]
- 51.Ramshaw I A, Ramsay A J, Karupiah G, Rolph M S, Mahalingam S, Ruby J C. Cytokines and immunity to viral infections. Immunol Rev. 1997;159:119–135. doi: 10.1111/j.1600-065x.1997.tb01011.x. [DOI] [PubMed] [Google Scholar]
- 52.Snacken R, Kendal A P, Haaheim L R, Wood J M. The next influenza pandemic: lessons from Hong Kong, 1997. Emerg Infect Dis. 1999;5:195–203. doi: 10.3201/eid0502.990202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Steinhauer D A. Role of hemagglutinin cleavage for the pathogenicity of influenza virus. Virology. 1999;258:1–20. doi: 10.1006/viro.1999.9716. [DOI] [PubMed] [Google Scholar]
- 54.Talon J, Horvath C M, Polley R, Basler C F, Muster T, Palese P, Garcia-Sastre A. Activation of interferon regulatory factor 3 is inhibited by the influenza A virus NS1 protein. J Virol. 2000;74:7989–7996. doi: 10.1128/jvi.74.17.7989-7996.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Taylor L A, Carthy C M, Yang D, Saad K, Wong D, Schreiner G, Stanton L W, McManus B M. Host gene regulation during coxsackievirus B3 infection in mice: assessment by microarrays. Circ Res. 2000;87:328–334. doi: 10.1161/01.res.87.4.328. [DOI] [PubMed] [Google Scholar]
- 56.Tsuno A, Miyoshi K, Tsujii R, Miyakawa T, Mizuta K. RRS1, a conserved essential gene, encodes a novel regulatory protein required for ribosome biogenesis in Saccharomyces cerevisiae. Mol Cell Biol. 2000;20:2066–2074. doi: 10.1128/mcb.20.6.2066-2074.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Visseren F L, Verkerk M S, Bouter K P, Diepersloot R J, Erkelens D W. Interleukin-6 production by endothelial cells after infection with influenza virus and cytomegalovirus. J Lab Clin Med. 1999;134:623–630. doi: 10.1016/s0022-2143(99)90103-8. [DOI] [PubMed] [Google Scholar]
- 58.Wagner A, Doerks A, Aboud M, Alonso A, Tokino T, Flugel R M, Lochelt M. Induction of cellular genes is mediated by the Bel1 transactivator in foamy virus-infected human cells. J Virol. 2000;74:4441–4447. doi: 10.1128/jvi.74.10.4441-4447.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu H, Cong J-P, Mamtora G, Gingeras T, Shenk T. Cellular gene expression altered by human cytomegalovirus: global monitoring with oligonucleotide arrays. Proc Natl Acad Sci USA. 1998;95:14470–14475. doi: 10.1073/pnas.95.24.14470. [DOI] [PMC free article] [PubMed] [Google Scholar]