Abstract
BPHE-1 cells, which harbor 50 to 200 viral episomes, encapsidate viral genome and generate infectious bovine papillomavirus type 1 (BPV1) upon coexpression of capsid proteins L1 and L2 of BPV1, but not coexpression of BPV1 L1 and human papillomavirus type 16 (HPV16) L2. BPV1 L2 bound in vitro via its C-terminal 85 residues to purified L1 capsomers, but not with intact L1 virus-like particles in vitro. However, when the efficiency of BPV1 L1 coimmunoprecipitation with a series of BPV1 L2 deletion mutants was examined in vivo, the results suggested that residues 129 to 246 and 384 to 460 contain independent L1 interaction domains. An L2 mutant lacking the C-terminal L1 interaction domain was impaired for encapsidation of the viral genome. Coexpression of BPV1 L1 and a chimeric L2 protein composed of HPV16 L2 residues 1 to 98 fused to BPV1 L2 residues 99 to 469 generated infectious virions. However, inefficient encapsidation was seen when L1 was coexpressed with either BPV1 L2 with residues 91 to 246 deleted or with BPV1 L2 with residues 1 to 225 replaced with HPV16 L2. Impaired genome encapsidation did not correlate closely with impairment of the L2 proteins either to localize to promyelocytic leukemia oncogenic domains (PODs) or to induce localization of L1 or E2 to PODs. We conclude that the L1-binding domain located near the C terminus of L2 may bind L1 prior to completion of capsid assembly, and that both L1-binding domains of L2 are required for efficient encapsidation of the viral genome.
Papillomaviruses are nonenveloped double-stranded DNA tumor viruses. Their capsid comprises 360 molecules of the major capsid protein L1, arranged as 72 pentamers, or capsomers, in a T=7d icosahedral surface lattice (2). Expression of L1 protein results in the self-assembly of virus-like particles (VLPs), which have the size, shape, and conformational epitopes of virion capsids (14). Bovine papillomavirus type 1 (BPV1) virions in the presence of low ionic strength and dithiothreitol (DTT) (18) and human papillomavirus type 11 (HPV11) and HPV33 VLPs in the presence of reducing agents (21, 25) are disassembled into capsomers. Recombinant HPV11 L1 protein with a Cys-to-Gly mutation in the C terminus of L1 forms pentamers but cannot assemble into capsid-like structures (18). Taken together, the data imply that both ionic and disulfide bonds mediate interpentamer binding in the papillomavirus capsid.
Virions also contain L2, the minor capsid protein (5). The number of L2 molecules per capsid has been estimated at between 12 (30) and 36 (5) molecules per virion. If L2 is coexpressed with L1, the L2 protein is coassembled into VLPs with a stoichiometry similar to that seen in authentic virions (15). Three-dimensional reconstruction of cryo-electron micrographs of quench-frozen BPV virions has revealed the capsid architecture to 9-Å resolution. This analysis detected a protein density within the central cavity of the pentavalent capsomers, suggesting that L2 may be associated with these 12 vertex capsomers (30).
Rodent fibroblasts maintain the BPV1 genome at 50 to 200 episomes/cell (17, 33). These cells express the nonstructural viral proteins, but no virus is produced because the cells do not express the capsid proteins (1). However, expression of L1 and L2 in trans causes encapsidation of viral episomes and formation of infectious virions (26, 36, 37). L2 is not absolutely required for generation of pseudovirions in vitro (29) or in vivo (31). However, L2 enhances DNA encapsidation in vivo by >50-fold (23, 26, 36, 37). DNA encapsidation may also be enhanced by nucleotides 1506 to 1625 of the BPV1 genome (34), as well as by E2 (a virally encoded transcription/replication factor) in some systems that generate pseudovirions (35) but not in others (31). L2 colocalizes with the promyelocytic leukemia protein (PML) in subnuclear domains called PML oncogenic domains (PODs) or nuclear domain-10 (3). Further, while BPV1 E2 and L1 exhibit a diffuse nuclear localization in its absence, L2 causes E2 (both the full-length E2TA and short repressor form E2TR) and L1 to traffic to PODs (3, 10). This localization partially, or completely, overlaps with the site of HPV11 DNA replication (27). Interestingly, overexpressed HPV11 E2 is associated with the nuclear matrix (38), and HPV5 E2 is associated with RNA splicing factors in subnuclear foci (16). L2 binds directly to two regions of BPV1 E2 in vitro and attenuates E2-mediated transcription but not viral replication (10).
In the present study, we have sought to characterize the interaction between L1 and L2 during virion formation and specifically to determine (i) which domains of L2 mediate its binding to L1; (ii) at what stage during capsid assembly L2 binds to L1; and (iii) whether the L1 interaction domains in L2 contribute to virion assembly.
MATERIALS AND METHODS
Generation of vectors and recombinant viruses.
The full-length BPV1 L2 gene and a PCR-amplified fragment (L2Δ384-469) comprising L2 nucleotides 1 to 1149 were inserted between the BamHI and EcoRI sites of Bluescript II SK(−) phagemid (Stratagene Cloning Systems, La Jolla, Calif.), downstream from the T3 RNA polymerase promoter, to generate Bluescript II pSK(−)L2 and Bluescript II pSK(−)L2Δ384-469, respectively. The baculovirus transfer vectors containing the BPV1 L1 or L2 genes were constructed as previously described. The baculovirus transfer vector was constructed by subcloning the XbaI-HindIII minor capsid gene fragment into pFastBac1. Recombinant baculovirus was generated using the BAC-TO-BAC Baculovirus Expression System under the manufacturer's recommended reaction conditions (Gibco BRL Life Technologies, Gaithersburg, Md.). Deletions were introduced into L2 by PCR as described previously (22, 24), using the following oligonucleotides: L2Δ1-88 employed CGCGAGATCTACCATGGGATCCAGAGCTGTAAC and GCGCAGATCTTTAGGCATGTTTCCG; L2Δ173-469 employed CGCGAGATCTACCATGAGTGCACGAAAAAGAG and GCGCAGATCTTTAAACCGCTATGTCCTCG; L2Δ247-469 employed CGCGAGATCTACCATGAGTGCACGAAAAAGAG and GCGCAGATCTTTAGGCAATACTGCGGGGCGT; L2Δ395-469 employed CGCGAGATCTACCATGAGTGCACGAAAAAGAG and GCGCAGATCTTTACTGAGTTGGAATGAGGC; and L2Δ461-469 employed CGCGAGATCTACCATGAGTGCACGAAAAAGAG and GCGCAGATCTTTACAACAAGGAGGGATGC. The oligonucleotides were used to PCR amplify BPV L2 fragments which, after BglII digestion, were cloned into the BamHI site of pSFV-1. Deletions L2Δ91-129 and L2Δ91-246 were amplified using 5′CGCGGGATCCGCCCCTGCAATAGTC and CGCGGGATCCTCTAAATCACGTGGC, respectively, with 3′ oligonucleotide GTAGTGTCATCGATAAC. The PCR fragments were cloned into pSFV-1.BPV1 L2 using BamHI and ClaI. Chimeras H98B and H225B were generated using 5′ oligonucleotides CGCGGGGCCCTAGTATAGGTGCGGGC and GCGCCCTAGGAGAAAACATTGAACTGAC, respectively, and 3′ oligonucleotide CGTTTGCGTAGGGATGTAAT to amplify a fragment from pSFV-1.BPV1 L2. Chimera H98B was generated by cloning an ApaI and XmaI-digested PCR fragment into pSFV-1.HPV16 L2. Chimera H225B was generated by cloning an AvrII- and XmaI-digested PCR fragment into pSFV-1.HPV16 L2. All constructs were confirmed by automated fluorescence sequence analysis (Seqwright). Recombinant Semliki Forest virus (SFV) expressing mutant L2 proteins was constructed as described previously (23). The recombinant pSFV-1 clones containing either the L1 or mutant L2 genes and pHelper-2 plasmid were linearized using SpeI (or NruI for pSFV-1.NruI-based clones). The DNAs were phenol-chloroform extracted and ethanol precipitated. To generate SFV RNA, 1 μg of each linearized pSFV-1 clone and 1 μg of pHelper-2 were resuspended in 100-μl reaction mixtures containing 1 mM ATP, 1 mM CTP, 1 mM UTP, 0.5 mM GTP, 1 mM RNA capping analog m7G(5′)ppp(5′)G, 5 mM DTT, 100 U of human placental ribonuclease inhibitor, and 75 U of SP6 RNA polymerase in 1× SP6 reaction buffer. The reactions were incubated for 1 h at 37°C, and 2.5 μl was analyzed on a 0.8% agarose gel to assess the integrity of the SFV RNAs. A total of 107 BHK21 cells released into suspension by trypsin treatment was mixed with 5 μg of SFV RNA and 5 μg of Helper-2 RNA in 0.8 ml of serum-free Glasgow's minimal essential medium (GMEM). After transfer to a cuvette, the cells were electroporated (225 V, 800 μF, low ohms; Life Technologies Electroporator) twice and plated out in 25 ml of complete GMEM (5% fetal calf serum, 10% tryptose-phosphate broth, 10 mM HEPES [pH 7.4], 1× nonessential amino acids, 100 U of penicillin/ml, and 100 μg of streptomycin/ml in GMEM). After incubation for 24 h the medium was harvested, clarified by centrifugation (1,000 × g, 10 min), aliquoted, and stored at −80°C.
Preparative purification of intact and disassembled VLPs.
Preparation of intact particles was performed as described previously (15). Disassembled particles were prepared by dialyzing 200 μl of intact particles at 4°C against 250 ml of disassembly buffer (10 mM Tris-HCl [pH 7.4], 3 mM DTT, 1 mM EDTA) containing 1 pellet of Complete protease inhibitor cocktail tablets (Roche). The dialysate was clarified by centrifugation at 14,000 rpm for 15 min at 4°C. Protein concentrations were determined with a colorimetric protein assay (Bio-Rad Laboratories, Hercules, Calif.). For transmission electron microscopy, samples were spotted on carbon-coated grids, negatively stained with 1% uranyl acetate, and examined with a Philips electron microscope (model EM 400T) at 36,000× magnification as previously described.
Cell-free L1-L2 binding assay.
In vitro transcription of nonlinearized DNA at 25 μg/ml with T3 RNA polymerase (Promega Corp., Madison, Wis.) was performed for 120 min at 37°C in the recommended reaction buffer (Gibco BRL Corp.). Transcription products were precipitated with sodium acetate and ethanol, dissolved in water to a concentration of 35 μg/ml, and then diluted 25-fold in the TNT rabbit reticulocyte lysate system (Promega Corp.) containing [35S]cysteine (>1,000 Ci/mmol; Amersham) for translation for 90 min at 30°C. Twenty-five microliters from the in vitro translation reaction mixture was incubated with 4 μg of purified particles in 100 μl of phosphate-buffered saline (PBS) for 2 h at 30°C. Samples were diluted to 500 μl with radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris [pH 8.0], 150 mM NaCl, 1.0% NP-40, 0.5% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS]) containing 1 mM phenylmethylsulfonyl fluoride, 1 U of Trasylol/ml, and 0.67 μl of rabbit preimmune serum for 1 h of continuous rotation at 4°C. This was followed by an additional 30 min of continuous rotation at 4°C in the presence of 30 μl of protein A-Sepharose CL-4B beads (Pharmacia Biotech, Uppsala, Sweden) suspended in RIPA buffer. After centrifugation at 16,000 × g for 10 s, the supernatant was immunoprecipitated with 0.30 μl of rabbit polyclonal antiserum to L1, essentially as described previously (4). Nonradiolabeled full-length L2 and six overlapping L2 peptides were used to block in vitro-translated L2 binding to L1. Generation of the recombinant bacteria expressing these proteins, induction of protein expression, and purification were performed as described previously (24). Rabbit polyclonal antisera to L1, L2, peptide B, corresponding to amino acids 45 to 173 of L2, and peptide F, corresponding to amino acids 384 to 469 of L2, were generated as described previously (24).
Immunofluorescent localization of BPV proteins.
As described previously (3), recombinant SFV stocks expressing L1 and L2 were rendered infectious by incubation with 0.5 mg of chymotrypsin A4 (Boehringer Mannheim) per ml for 30 min on ice and treatment with 0.5 mg of aprotinin (Sigma) per ml. Activated virus, diluted 1:100 in Dulbecco's modification of PBS (D-PBS), was added to BPHE-1 cells on glass coverslips for 1 h at 37°C and then replaced with medium. After 6 h, the cells were washed in PBS and fixed for 10 min in 1% paraformaldehyde–PBS. After blocking in 200 mM glycine, the cells were permeabilized with 0.1% Brij 58, and all incubations were performed at 4°C. L2 was detected with rabbit antiserum to full-length L2 (24); L1 was detected with monoclonal antibody (MAb) 5B6 (24); and E2 was detected with MAb B201 (E. Androphy, Tufts Medical Center). Hybridoma supernatants were diluted 1:100, and rabbit polyclonal serum was diluted 1:1,000. Secondary antibody (fluoroscein isothiocyanate [FITC] or Texas Red labeled) was used at 5 μg/ml and mounted with Fluoromount mounting fluid (Southern Biotechnology Associates) on a glass slide. Fluorescence was examined using a Bio-Rad MRC 1024 laser-scanning confocal system attached to a Zeiss Axioplan microscope. All images were acquired with a Zeiss 63× N.A. 1.4 Planapo objective. Control slides with isotype-matched, irrelevant MAbs or preimmune serum were used to establish that fluorescence between the green and red channels did not overlap and that immunofluorescent labeling was specific. The images were arranged and pseudocolored using Adobe Photoshop.
Coimmunoprecipitation of L1 and L2 mutants.
A total of 4 × 106 BHK21 cells were coinfected with equivalent titers of recombinant SFV expressing L1 and mutant L2. The cells were harvested 24 h postinfection by scraping and collected by centrifugation (600 × g, 10 min). The cell pellet was resuspended in 1 ml of lysis buffer (1% NP-40, 0.5 M NaCl, 50 mM Tris-HCl [pH 8], and Complete [Roche] protease inhibitors) at 4°C. The lysates were sheared by sonication (Branson Sonifier 250, ice-cold water bath; maximum setting; 30 s) and clarified by centrifugation (10,000 × g, 10 min, 4°C). Sequential immunoprecipitations were performed for 1 h each with a 50-μl packed volume of protein A-Sepharose per tube precoupled to 5 μl of (i) preimmune rabbit antiserum, (ii) rabbit anti-BPV L1 VLP (14), and (iii) rabbit anti-BPV1 L2-6His (24). The immunoprecipitates were washed three times with 1 ml of lysis buffer, and the bound proteins were eluted by boiling in gel sample buffer. The immunoprecipitates were subjected to SDS–10% polyacrylamide gel electrophoresis (PAGE) and Western blotting. Coimmunoprecipitated capsid proteins were detected with 1 μg of L2-specific MAb C6 (20) or MAb 3A10 (12) per ml, peroxidase-linked anti-mouse immunoglobulin G antibody (1:10,000), and chemiluminescent substrate (Kirkegaard & Perry Laboratories).
Analysis of genome encapsidation.
A total of 107 BPHE-1 cells were coinfected with recombinant SFVs expressing wild-type L1 alone or with each deletion mutant of L2. The BPHE-1 cells were harvested 30 h postinfection by scraping into the medium and collected by centrifugation. The cells were washed in PBS and lysed on ice by sonication (Branson sonifier, 1 min at power level 7) in 1 ml of PBS containing 1% (vol/vol) Nonidet P-40, 10 μg of aprotinin/ml, and 100 U of DNase I/ml. The lysates were clarified by centrifugation (10,000 × g, 10 min, 4°C). Preimmune serum (10 μl) and protein A-Sepharose (50-μl packed volume) were added to the clarified extract, and the sample was tumbled at 4°C for 1 h. The beads were removed by centrifugation for 1 min at 2,000 × g. The supernatant was transferred, and 10 μl of rabbit anti-BPV1 L1/L2 VLP serum and protein A-Sepharose (50-μl packed volume) were added. The sample was tumbled at 4°C for 1 h, and the beads were recovered by centrifugation at 2,000 × g for 1 min. The beads were washed three times with 1 ml of D-PBS containing 1% (vol/vol) Nonidet P-40, 10 μg of aprotinin/ml, and 100 U of DNase I and RNase A per ml and incubated for 1 h at 37°C to allow digestion of accessible DNA. The beads were then washed twice more in buffer lacking nuclease and aprotinin. Encapsidated DNA was released from the beads by resuspension in 400 μl of 50 mM Tris-HCl (pH 8), 10 mM EDTA, 1 mM DTT, and 100 μg of proteinase K/ml. After 15 min of incubation at 37°C to allow digestion of the capsid proteins, the sample was centrifuged for 1 min at 2,000 × g. The supernatant was transferred and extracted using buffered phenol and then chloroform. DNA was precipitated from the supernatant by addition of 20 μg of glycogen, 40 μl of 3 M sodium acetate (pH 5.2), and 1 ml of ethanol and cooling to −20°C overnight. The DNA was recovered by centrifugation (16,000 × g, 10 min), washed with 70% ethanol, and resuspended in Tris-EDTA. The DNA (uncut) was separated on a 0.8% agarose–Tris-acetate-EDTA gel and transferred to a Nytran+ membrane. BPV1 DNA was detected by Southern blotting. Biotinylated probe was prepared by random priming of the EcoRI-BamHI large fragment of BPV-pML. The probe was detected using strepavidin-peroxidase and enhanced chemiluminescence according to the manufacturer's instructions (Pierce).
Generation of infectious BPV.
The system used to generate infectious BPV1 in vitro has been described previously (23). Briefly, 107 BPHE-1 cells were coinfected with recombinant SFVs expressing wild-type L1 alone or with each deletion mutant of L2. The BPHE-1 cells were harvested 30 h postinfection by scraping into the medium and collected by centrifugation. The cells were resuspended in 2 ml of D-PBS and lysed by sonication (Branson sonifier; 1 min at power level 7) on ice. Monolayers of C127C mouse fibroblasts in 60-mm-diameter dishes were incubated with 1 ml of extract for 1 h at 37°C. The cells were washed and the medium was replaced with Dulbecco's MEM containing 10% fetal calf serum, 100 U of penicillin/ml, and 100 μg of streptomycin/ml. The C127C cells were cultured for 3 weeks and then stained with 0.5% (wt/vol) methylene blue and 0.25% (wt/vol) carbol fuchsin in methanol.
RESULTS AND DISCUSSION
Coimmunoprecipitation of BPV1 L1 and deletion mutants of L2.
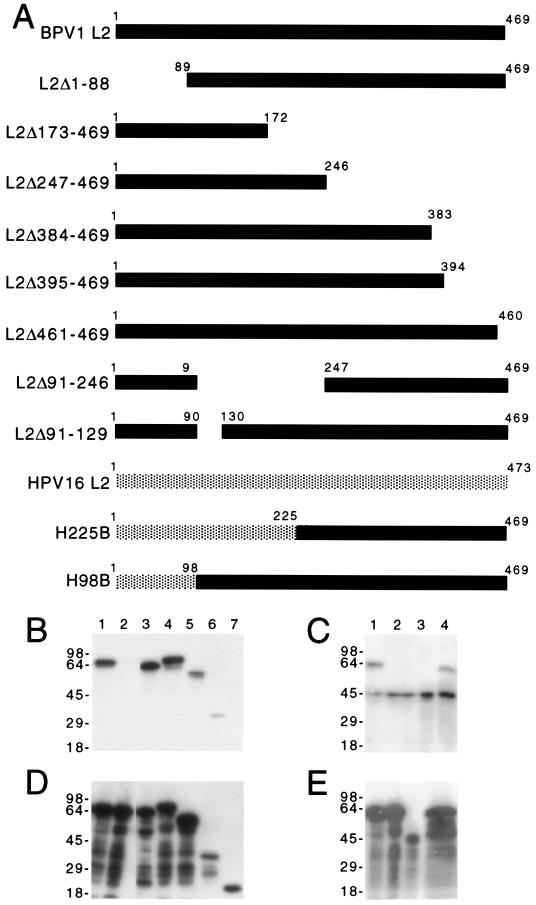
To determine regions of L2 that interact with L1, a series of deletion mutants of the BPV1 L2 gene was generated by PCR and cloned into vector pSFV-1 (19) (Fig. 1A). Defective recombinant SFVs encoding these deletion mutants were generated (23). Coexpression of L1 and L2 leads to their coassembly into VLPs with a stoichiometry similar to that of virions obtained from bovine papillomas (23). We therefore examined the interaction of wild-type BPV1 L1 with seven of the L2 deletion mutants in BHK21 cells that had been coinfected with recombinant SFV that express L1 and mutant L2 (19, 23). Sequential immunoprecipitations were performed using preimmune rabbit serum first, then rabbit anti-BPV1 L1 VLPs (14), and finally rabbit anti-BPV1 L2–six-His (24). By SDS-PAGE and Western blotting with L2-specific antibody, no L2 was detected upon immunoprecipitation with preimmune rabbit serum (results not shown) or when L2 was expressed alone and immunoprecipitated by rabbit anti-BPV1 L1 VLPs (Fig. 1B, lane 2). All mutants expressed high levels of L2 (Fig. 1D and E). The extra immunoreactive band at ∼50 kDa in Fig. 1C likely results from cross-reactivity of the secondary antibody with the heavy chain of the rabbit antibody used to immunoprecipitate L1. When the L2 deletion mutants were coexpressed with L1, only L2Δ173-469 failed to coimmunoprecipitate with L1 (Fig. 1B, lane 7) despite its high level expression (Fig. 1D, lane 7). L2Δ395-469, L2Δ247-469 (Fig. 1B, lanes 5 and 6), and L2Δ91-246 (Fig. 1C, lane 3) coimmunoprecipitated with L1, although with reduced efficiency compared to full-length L2. The remaining L2 deletion mutants (L2Δ1-88, L2Δ461-469, and L2Δ91-129) coimmunoprecipitated with L1 to the same extent as wild-type L2 (Fig. 1B and C). Taken together, the data imply that L2 may contain two independent L1 interaction domains. One L1 interaction domain is located near the C terminus of L2, between residues 395 and 460, and the second is located between residues 129 and 246 of L2. Consistent with this possibility, studies of L2-specific MAb binding to L1 and L2 VLPs suggest that both these regions of L2 are internal to the capsid (20, 32).
FIG. 1.
Coimmunoprecipitation of BPV1 L1 and L2 deletion mutants. (A) Schematic diagram of BPV1 L2 deletion mutants and chimeras with HPV16 L2. (B and D) BHK21 cells were infected with recombinant SFV expressing BPV1 L1 (lanes 1 and 3 to 7) and full-length L2 (lanes 1 and 2), L2Δ1-88 (lane 3), L2Δ461-469 (lane 4), L2Δ395-469 (lane 5), L2Δ247-469 (lane 6), or L2-173–469 (lane 7). (C and E) BHK21 cells were infected with recombinant SFV expressing BPV1 L1 (lanes 1, 3, and 4) and full-length L2 (lanes 1 and 2), L2Δ91-246 (lane 3), or L2Δ91-129 (lane 4). After incubation for 24 h, the cells were harvested and sequential immunoprecipitation was performed using preimmune serum first (results not shown), then rabbit antiserum to L1 VLPs (B and C), and finally rabbit antiserum to full-length L2 (D and E). The presence of L2 in immunoprecipitates was determined by Western blotting using L2-specific MAb C6 (B and C) or 3A10 (D and E).
L2 binds in vitro to L1 capsomers, but not to intact VLPs.
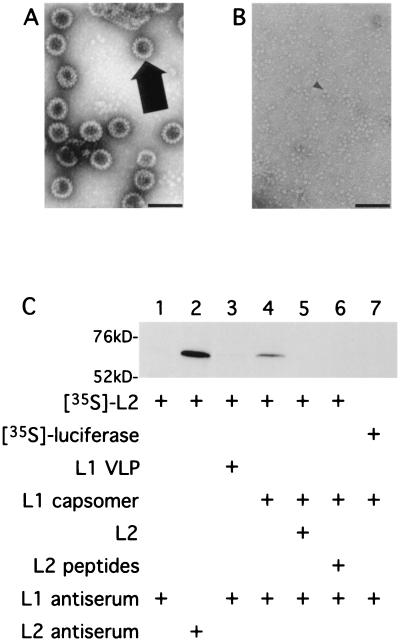
Since many viruses inject their genome into preformed capsid structures (7, 11) and L2 is required for efficient papillomavirus DNA encapsidation (23), we sought to determine if L2 interacts with intact L1 capsids or with capsomers. To produce capsomers, L1 VLPs were purified from Sf9 insect cells infected with recombinant baculovirus expressing the L1 gene (Fig. 2A) and dialyzed against a disassembly buffer. As expected (18, 21, 25), this procedure resulted in the disassembly of intact VLPs into capsomers, as determined by electron microscopy (Fig. 2B) and by gel filtration (Superose 6 sizing column) (data not shown).
FIG. 2.
In vitro binding of L2 to L1 pentamers, but not to intact VLPs. BPV1 L1 VLPs disassemble into component capsomers after dialysis in disassembly buffer. BPV1 L1 VLPs purified from recombinant baculovirus-infected insect cells was left intact or was disassembled via dialysis against disassembly buffer (10 mM Tris-HCl [pH 7.4], 1 mM EDTA, 3 mM DTT) at 4°C overnight. Samples of intact BPV1 L1 VLPs (A) and disassembled BPV1 L1 VLPs (B) were examined via electron microscopy at 36,000× magnification (bar, 100 nm). Intact VLPs and pentamer are indicated with large and small arrows, respectively. (C) In vitro-transcribed L2 mRNA was translated in rabbit reticulocyte lysate supplemented with [35S]cysteine. The in vitro translation product was incubated with purified L1 VLPs or capsomer and then immunoprecipitated and subjected to SDS-PAGE and autoradiography. Lane 1, in vitro-translated L2 immunoprecipitated with anti-L1 serum; lane 2, in vitro-translated L2 immunoprecipitated with anti-L2 serum; lane 3, in vitro-translated L2 incubated with intact L1 VLPs and then immunoprecipitated with anti-L1 serum; lane 4, in vitro-translated L2 incubated with disassembled L1 VLPs and then immunoprecipitated with anti-L1 serum; lane 5, in vitro-translated L2 incubated with disassembled L1 VLPs and nonradiolabeled L2 with a C-terminal six-histidine tag and then immunoprecipitated with anti-L1 serum; lane 6, in vitro-translated L2 incubated with disassembled L1 VLPs and nonradiolabeled overlapping L2 peptides (A through F) with C-terminal six-histidine tags; lane 7, in vitro-translated luciferase incubated with disassembled L1 VLPs and then immunoprecipitated with anti-L1 serum. Molecular masses (in kilodaltons) are shown.
To examine whether L2 could interact with capsids or with capsomers, BPV1 L2 translated in rabbit reticulocyte lysate supplemented with [35S]cysteine was incubated with intact or disassembled L1 VLPs and immunoprecipitated with rabbit polyclonal antiserum that recognizes intact and disassembled L1, and the immunoprecipitates were subjected to SDS-PAGE (Fig. 2C). In vitro-translated L2 was not efficiently coprecipitated by anti-L1 serum in the presence of intact L1 VLPs or, as a control, in the absence of L1 (lanes 1 and 3). Upon extended exposure, a trace amount of in vitro-translated L2 coprecipitated with VLPs (lane 3), but this signal probably results from free capsomers contaminating the VLP preparation (Fig. 2A). In the presence of disassembled L1 VLPs, by contrast, in vitro-translated L2 was efficiently coprecipitated by anti-L1 serum (lane 4). This coprecipitation was specifically blocked by nonradiolabeled L2 produced in Escherichia coli (lane 5) or by a set of overlapping L2 peptides (A to F) that together encompass the full L2 protein (lane 6). Peptides A through F correspond to the following L2 amino acids: A, 1 to 88; B, 45 to 173; C, 130 to 257; D, 216 to 340; E, 300 to 425; and F, 384 to 469 (24). As additional specificity controls, in vitro-translated L2 was not coprecipitated by antipolyoma serum in the presence of disassembled polyoma VP1 VLPs (data not shown), and radiolabeled in vitro-translated luciferase was not coimmunoprecipitated by anti-L1 serum in the presence of disassembled L1 VLPs (lane 7). The finding that L2 bound to capsomers, but not to intact L1 VLPs, implies that L2 binds to L1 prior to completion of capsid assembly.
Determination of the L2 region that mediates binding to L1 in vitro.
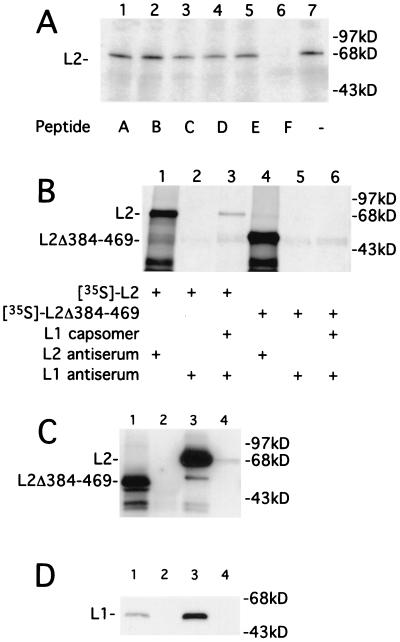
Since the set of six overlapping L2 peptides (A through F, described in reference 24) were able to block coimmunoprecipitation of radiolabeled L2 in the presence of disassembled L1 (Fig. 2C, lane 6), the peptides were tested individually in an in vitro binding assay for their ability to block L2 binding (Fig. 3A). Only peptide F, corresponding to the C-terminal 85 amino acids of L2 (amino acids 384 to 469), blocked L2 binding (Fig. 3A, lane 6). To exclude the possibility that the F domain blocks L1 binding by forming an inactive hetero-oligomer with full-length L2, L2 and peptide F were coincubated and then immunoprecipitated with a rabbit antiserum raised against peptide B (residues 45 to 173 of BPV1 L2) (24). However, there was no evidence of coprecipitation of peptide F with full-length L2 in the immunoprecipitated complexes when probed by Western blotting with a rabbit antiserum raised against peptide F (results not shown). To confirm that the L1-binding domain lies within the C-terminal 85 amino acids of L2, an L2 C-terminal deletion mutant lacking amino acids 384 to 469 (L2Δ384-469) was subjected to in vitro transcription and translation. In vitro binding of L2Δ384-469 to disassembled L1 VLPss was dramatically reduced when compared to binding by full-length L2 (Fig. 3B). Thus, in vitro binding of L2 to L1 requires sequences found at or near the C terminus of L2. This location is consistent with these sequences corresponding to the more C terminal domain of the two L1 interacting domains identified in Fig. 1.
FIG. 3.
L2 C-terminal 85 amino acids are required for L1 binding in vitro, but not in vivo. (A) Six overlapping peptides of L2 (A through F), expressed as six-His fusion proteins in E. coli, were tested for blocking of L1 binding to in vitro-translated L2. In vitro-translated 35S-labeled L2 and disassembled L1 VLPs were incubated either with peptides A through F (lanes 1 to 6) or buffer (lane 7). Samples were immunoprecipitated with L1 antiserum, and immunoprecipitates were subjected to SDS-PAGE and autoradiography. (B) In vitro-translated 35S-labeled L2 and L2Δ384-469 were immunoprecipitated with anti-L2 serum (lanes 1 and 4) or immunoprecipitated with anti-L1 serum in either the presence (lanes 3 and 6) or absence (lanes 2 and 5) of disassembled L1 VLPs. Immunoprecipitates were subjected to SDS-PAGE and autoradiography. The positions of L2 and L2Δ384-469 are indicated. Molecular masses (in kilodaltons) are shown. (C) Four micrograms each of intact L1/L2Δ384-469 VLPs (lane 1), in vitro-disassembled L1/L2Δ384-469 VLPs (lane 2), intact L1/L2 VLPs (lane 3), and in vitro-disassembled L1/L2 VLPs (lane 4) were immunoprecipitated with MAb 5B6 (lanes 1 to 4) and then subjected to SDS-PAGE and analyzed by Western blotting using rabbit antiserum to BPV1 L2 residues 45 to 173 and peroxidase-conjugated protein A. The positions of L2 and L2Δ384-469 are indicated. (D) Four micrograms each of purified L1/L2Δ384-469 VLPs (lanes 3 and 4) were immunoprecipitated with rabbit antiserum to L2 residues 45 to 173 (lanes 1 and 3) or preimmune serum (lanes 2 and 4) and then subjected to SDS-PAGE and analyzed by Western blotting with BPV1 L1 mouse monoclonal antibody 837 and peroxidase-conjugated protein A. The position of L1 is indicated.
C-terminal L1-binding domain is not necessary in vivo for incorporation of L2 within VLPss.
To determine if the C-terminal 85 amino acids of L2 are necessary for L2 to associate with L1 in vivo, Sf9 cells were doubly infected with baculovirus encoding L1 and either L2Δ384-469 or full-length L2, and VLPss were purified (15). Both L2 and L2Δ384-469 copurified with L1 in VLPss. There was no visible difference in VLPs preparations and their disassembly using either BPV L1 only (Fig. 2A and B), BPV1 L1/L2, or L1/L2Δ384–469 (data not shown) as assessed by transmission electron microscopy. When purified L1/L2Δ384-469 and L1/L2 VLPss were immunoprecipitated with L1-specific antibody, both L2 proteins coprecipitated with L1, although L2Δ384-469 did so ∼3-fold less efficiently than did full-length L2 (Fig. 3C, lanes 1 and 3), consistent with the results obtained without VLPs purification (Fig. 1B). Analogous results were obtained when the purified VLPss were immunoprecipitated with antiserum to residues 45 to 172 of BPV1 L2, which neutralizes BPV infection and therefore presumably recognizes L2 epitopes exposed on the surface of intact virions and VLPs (Fig. 3D, lanes 1 and 3) (24). By contrast, when disassembled VLPss were subjected to L1 immunoprecipitation, full-length L2, but not L2Δ384-469, coprecipitated with L1 (Fig. 3C, lanes 2 and 4). The finding for full-length L2 is consistent with a previous report that L2 remains associated with pentamers upon disassembly of HPV33 L1 and L2 VLPss (25). Since a subset of L2 interactions with L1 are salt labile in vitro (25), perhaps the weak coimmunoprecipitation of L2 with capsomers reflects the relatively harsh washing conditions used (RIPA buffer). The results also suggest that the C terminus of L2 is required to form an interaction with L1 that is stable in vitro. However, we cannot exclude the possibility that L2Δ384-469 is more prone to degradation than L2 upon disassembly of these insect cell-derived VLPss, although L2Δ384-469 and full-length L2 exhibited similar stability upon translation in vitro (Fig. 3B). The higher stability of the association of L2Δ384-469 with VLPss compared to capsomers may be due to retention of the L2 within the closed structure of the VLPss. Consistent with this idea, antibody studies suggest that L2 is predominantly located within the capsid rather than exposed on the surface (9, 13, 20, 24).
Role of the C-terminal L1 interaction domain of L2 in genome encapsidation.
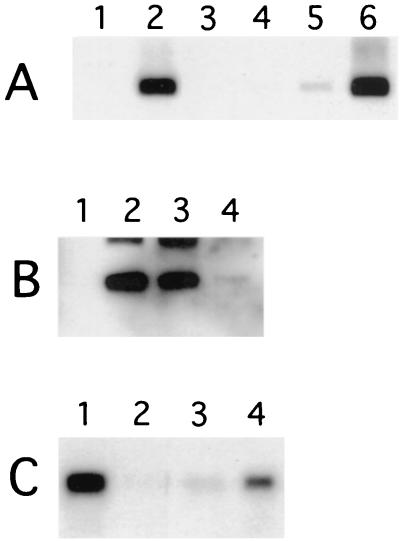
To assess the contribution to virion assembly of the L1 interaction domain at the C terminus of L2, we exploited a culture system that generates infectious papillomavirus (23). BPHE-1 cells harbor 50 to 200 episomal copies of the BPV1 genome per cell (33), but they produce no virus because the L1 and L2 genes are not expressed. However, expression of L1 and L2 in these cells via recombinant defective SFV vectors results in the generation of infectious BPV. BPHE-1 cells were coinfected with recombinant SFV expressing BPV1 L1 and mutant L2 (23). Thirty hours postinfection, the cells were harvested, the capsid proteins were immunoprecipitated and treated with DNase I to eliminate nonencapsulated genomes, and the quantity of DNase I-resistant BPV1 genome present in the immunoprecipitates was assessed by Southern blotting (Fig. 4). As expected, DNase I-resistant BPV genomes were recovered from immunoprecipitates of BPHE-1 cells expressing both L1 and L2 of BPV1, but not from cells expressing BPV1 L1 and HPV16 L2, which do not coassemble into VLPss, or from cells expressing only BPV1 L1 (Fig. 4A, lane 1, and Fig. 4C, lanes 1 and 2). The predominant size of the BPV1 genome in the immunoprecipitates derived from BPHE-1 cells expressing L1 and L2 was consistent with the supercoiled form of BPV1 DNA.
FIG. 4.
Analysis of BPV1 genome encapsidation in BPHE-1 cells coexpressing BPV1 L1 and L2 deletion mutants or HPV16-BPV1 chimeras. (A) BPHE-1 cells were coinfected with recombinant SFV expressing BPV1 L1 (lanes 1 to 6) and L2 (lane 2), L2Δ173-469 (lane 3), L2Δ247-469 (lane 4), L2Δ395-469 (lane 5), or L2Δ461-469 (lane 6). (B) BPHE-1 cells were coinfected with recombinant SFV expressing BPV1 L1 (lanes 1 to 4) and L2 (lane 2), L2Δ91-129 (lane 3), or L2Δ91-246 (lane 4). (C) BPHE-1 cells were coinfected with recombinant SFV expressing BPV1 L1 (lanes 1 to 4) and L2 (lane 1), HPV16 L2 (lane 2), H225B (lane 3), or H98B (lane 4). Thirty hours postinfection, the cells were harvested and the capsid proteins were immunoprecipitated from lysates using rabbit anti-BPV1 VLPs. The immunoprecipitates were treated with DNase I to eliminate nonencapsulated genomes, and the quantity of DNase I-resistant BPV1 genome present in the immunoprecipitates was assessed by Southern blotting. The predominant size of BPV1 DNA in immunoprecipitates derived from BPHE-1 cells expressing L1 and L2 is consistent with the supercoiled form of the viral genome.
When the ability of BPV1 L2 mutants to substitute for full-length L2 was assessed, equivalent quantities of the BPV1 genome were found to be encapsidated by full-length BPV1 L2 and L2Δ461-469 (Fig. 4A, lanes 2 and 6). However, BPV1 genome encapsidation was dramatically reduced for L2Δ395-469 and L2Δ247-469 and was absent from L2Δ173-469 (Fig. 4A, lanes 3 to 5). These results closely correlate with the relative efficiency with which the L2 mutants coprecipitated with VLPss (Fig. 1B, lanes 4 to 7). As the L2Δ395-469 mutant encapsidated the viral genome less efficiently than the L2Δ461-469 mutant or wild-type L2, the data suggest that the L1 interaction domain at the C terminus of L2 is necessary for efficient viral genome encapsidation. In addition, the C terminus of L2 may contain residues required to interact with BPV1 episomes in vivo (26), perhaps to the identified packaging enhancement sequence between nucleotides 1506 and 1625 (34).
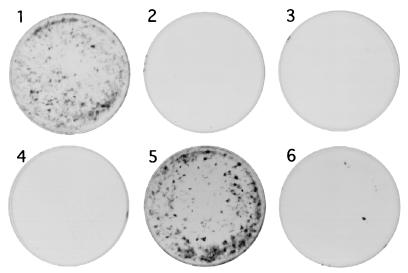
BPV1 infectivity was absent from crude extracts of BPHE-1 cells expressing L1 and L2Δ395-469 (Fig. 5, plate 2), L2Δ247-469, or L2Δ173-469 (results not shown), as assayed by focal transformation of mouse C127C cells (6). As we have recently demonstrated that the C-terminal 9 amino acids are necessary for infection but not virion assembly (R. B. S. Roden et al., unpublished data), no infectious particles would have been expected using these larger C-terminal deletions.
FIG. 5.
Ability of BPV1 L2 deletion mutants and HPV16-BPV1 chimeras to generate infectious virions when coexpressed in BPHE-1 cells. BPHE-1 cells were coinfected with recombinant SFV expressing BPV1 L1 (plates 1 to 6) and L2 (plate 1), L2Δ395-469 (plate 2), L2-91–246 (plate 3), HPV16 L2 (plate 4), H98B (plate 5), or H225B (plate 6). Thirty hours postinfection, the cells were harvested and lysed by sonication. Mouse C127C monolayers in 60-mm-diameter petri dishes were infected with lysates and maintained for 3 weeks in DMEM containing 10% FCS. The plates were stained with 0.5% (wt/vol) methylene blue and 0.25% (wt/vol) carbol fuchsin in methanol to highlight transformed foci.
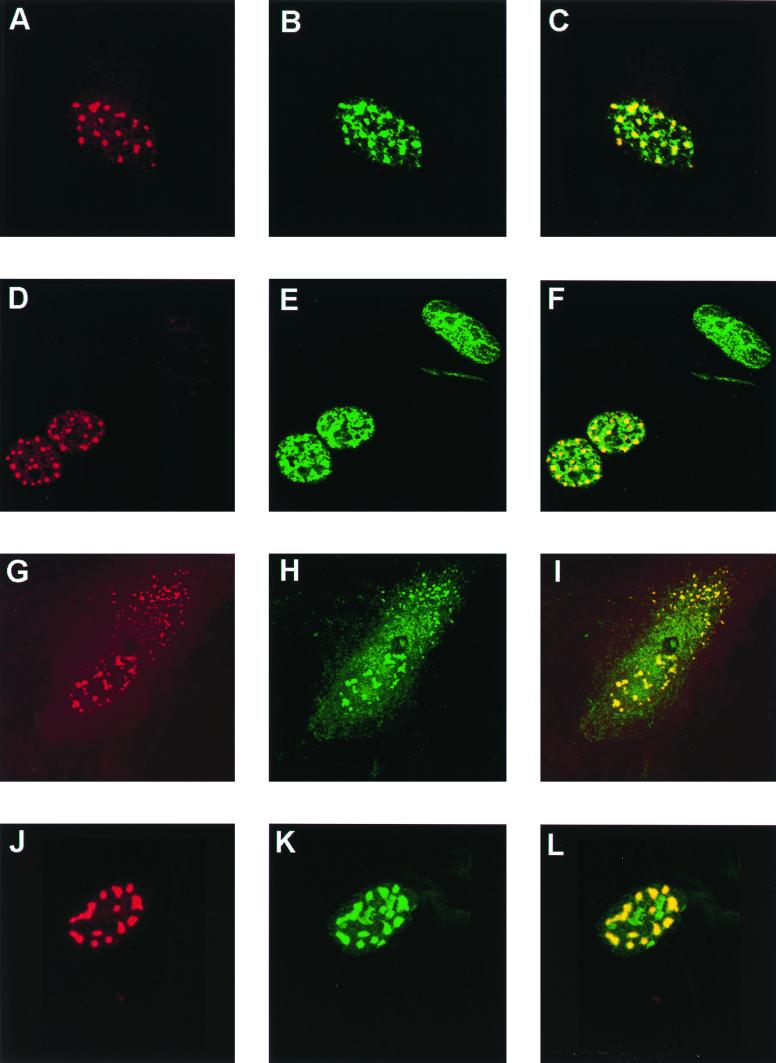
Our previous studies in BPHE-1 cells had demonstrated that (i) L2 is targeted to PODs in the absence of other papillomavirus proteins and DNA; and (ii) L1 and E2 (a sequence-specific papillomavirus DNA-binding protein) are targeted to PODs in the presence, but not in the absence, of L2 (3). In addition, Zhao et al. observed that E2 expression enhanced in vivo DNA encapsidation 6- to 10-fold (35), and direct interaction between L2 and E2 in vitro has recently been demonstrated (10). Further, Swindle et al. found partial or complete overlap of PML with HPV11 E1, E2, the host replication factor RP-A, and bromodeoxyuridine incorporation and the viral replication origin, consistent with viral DNA amplification at this subnuclear domain (28). To exclude the possibility that the POD-related L2 functions were compromised by deletion of the C-terminal L1 interaction domain, thus preventing viral genome encapsidation, the intracellular localization of L2Δ395-469 and L2Δ247-469 with respect to PML (data not shown), L1 (Fig. 6), and E2 (Fig. 7) were examined by immunofluorescent staining. L2Δ395-469 trafficked to PODs, colocalized with PML (data not shown), and recruited E2, as did full-length L2 (Fig. 6A to C and 7A to C). Therefore the inability of L2Δ395-469 to encapsidate the BPV1 genome when coexpressed with L1 does not result from improper subnuclear localization of L2 or impairment of its recruitment of E2. We also determined that L2Δ395-469 caused L1 to traffic to PODs. After examining many coexpressing cells, it is our impression that the colocalization was to a lesser degree than that with L1 and full-length L2 (Fig. 6) or L2Δ460-469 (results not shown). This result is consistent with the deletion of the L1 interaction domain at the C terminus of L2. However, this conclusion must be considered tentative, as the degree of colocalization of L1 and L2 was quite variable, even for wild-type L2 (as previously reported), and could not be quantified.
FIG. 6.
Immunolocalization of BPV1 L2 deletion mutants and an HPV16-BPV1 chimera with respect to BPV1 L1 in SFV-infected BPHE-1 cells. The cells were coinfected with the L1 and various L2 SFVs, fixed, and stained with antiserum against the L2 protein, detected with goat anti-rabbit Texas Red (panels A, D, G, and J), and MAb 5B6 directed against the L1 protein, detected with FITC-labeled goat anti-mouse antibody (panels B, E, H, and K). The digital superimposition of the two images is shown in panels C, F, I, and L. Colocalization is evident in the merged image as shown in yellow. The distribution of proteins for wild-type L2 is shown in panels A, B, and C. L2Δ395-469 is shown in panels D to F. L2Δ91-246 is shown in panels G to I. Chimera H225B is shown in panels J to L.
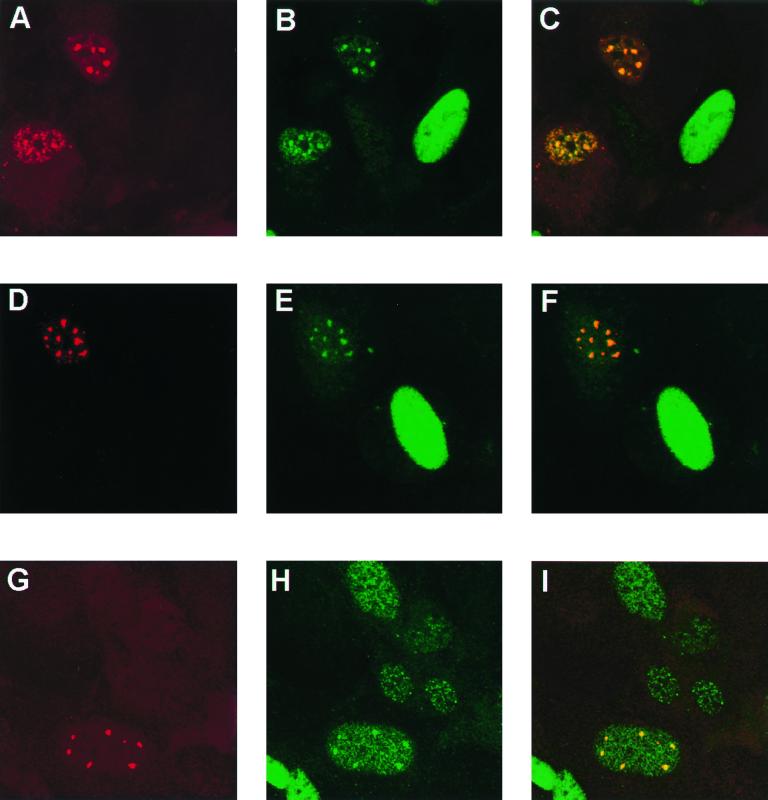
FIG. 7.
Immunolocalization of BPV1 L2 deletion mutants with respect to endogenous E2 in SFV-infected BPHE-1 cells. The cells were infected with the deletion mutant L2 SFVs and colocalized with respect to the endogenous E2 expressed in the BPHE-1 cells. L2 was detected with the rabbit polyclonal antiserum and Texas Red-labeled goat anti-rabbit antibody (A, D, and G). E2 was detected with MAb B201 and FITC-labeled goat anti-mouse antibody (B, E, and H). The merged images are shown in panels C, F, and I. The cells infected with wild-type L2 are shown in panels A to C. L2Δ395-469 is shown in panels D to F. L2Δ91-246 is shown in panels G to I.
Central domain of L2 functions in L1 binding and viral genome encapsidation.
Two strategies were adopted to explore the role of the second L1-binding domain of L2 in viral genome encapsidation. In the first, BPV1 L1 was coexpressed with L2Δ91-129 or with L2Δ91-246 in BPHE-1. Wild-type levels of genome encapsidation were observed when BPV1 L1 was coexpressed with L2-91-129 (Fig. 4B, lane 3), consistent with the efficient coimmunoprecipitation of L1 and L2Δ91-129 (Fig. 1C, lane 4). By contrast, viral genome encapsidation was drastically reduced when BPV1 L1 and L2Δ91-246, which coimmunoprecipitated only weakly (Fig. 1C, lane 3), were coexpressed (Fig. 4B, lane 4). Coexpression of L1 and L2Δ91-246 in BPHE-1 failed to produce infectious virions, consistent with the poor function of this mutant in genome encapsidation and L1 binding (Fig. 5, plate 3). L2Δ91-246 functioned similarly to full-length L2 with respect to POD localization and its redistribution of endogenous E2 to PODs (Fig. 7), suggesting that impairment of these functions does not account for inefficient genome encapsidation by this mutant. However, this mutant also appeared to cause localization of L1 to PODs to a lesser degree than full-length L2 (Fig. 6). This is consistent with the low efficiency of coimmunoprecipitation of L2Δ91-246 with L1 (Fig. 1C, lane 3) and suggests that L1 localization to PODs results from its direct interaction with L2.
In a second approach to determine the role of the central L1 interaction domain of L2 in genome encapsidation, we took advantage of the inability of HPV16 L2 to coimmunoprecipitate with BPV1 L1. To further define this L1 interaction domain of BPV1 L2, two chimeras were constructed: chimera H98B, which comprised residues 1 to 98 of HPV16 fused in frame with residues 99 to 469 of BPV1 L2, and chimera H225B, which comprised residues 1 to 225 of HPV16 fused in frame with residues 226 to 469 of BPV1 L2. As expected from the presence of the L1 interaction of BPV1 L2 at the C terminus of each chimera, both H98B and H225B coimmunoprecipitated with BPV1 L1 (not shown). However, H225B failed to promote encapsidation of the BPV1 genome when coexpressed with BPV1 L1 in BPHE-1 cells, whereas H98B demonstrated significant activity (Fig. 4C, lanes 3 and 4). The chimeras were also tested for their ability to generate infectious virions when coexpressed with BPV1 L1 in BPHE-1 cells. When the focus-forming activity on C127C monolayers was examined, extracts of BPHE-1 cells expressing L1 and H225B resulted in only a few foci, whereas H98B generated many hundreds (Fig. 5, plates 5 and 6). H98B induced approximately half as many foci as wild-type BPV1 L2, although the plates shown in Fig. 5 were deliberately overloaded to allow detection of low-level virion production (as for H225B). Notably, H98B expression was about twofold less than BPV1 L2 expression (results not shown). Overall, the results with the chimeric L2s suggest that there is a type-restricted interaction of L1 with a BPV1 L2 domain that includes amino acids 99 to 225, which correlates with the earlier results defining an L1 interaction domain located between L2 residues 129 and 246.
Since H225B functioned poorly in virion assembly, the subcellular localization of this L2 chimeric protein was examined. H225B trafficked to PODs as well as BPV1 L2 (Fig. 6) and HPV16 L2 (results not shown). Furthermore, H225B caused BPV1 E2 to traffic to PODs in the same manner as BPV1 L2 (data not shown). H225B also caused BPV1 L1 to colocalize in PODs (Fig. 6), consistent with the presence of the C-terminal L1 interaction domain of BPV1 L2 at the C terminus of this chimera. The immunostaining studies suggest that the poor functioning of H225B in virion assembly results from neither improper subnuclear localization nor inability to cause E2 and L1 to localize in PODs. Interestingly, BPV1 L1 colocalized with H225B to an even greater degree than did wild-type BPV1 L2. We speculate that H225B is partially functional, causing accumulation of L1 in PODs, but the BPV1 L1-H225B complex is unable to encapsidate the viral DNA and form virions that can exit PODs. By contrast, wild-type BPV1 L2 brings L1 to PODs, thereby forming virions that then can exit the PODs, perhaps accounting for the stronger colocalization with BPV1 L1 and H225B compared to that with BPV1 L2.
In summary, we have demonstrated that the C-terminal region of L2 (residues 384 to 460) interacts with L1 both in vivo and in vitro. This domain interacts in vitro with L1 pentamers, but not intact L1 VLPs, suggesting that L2 may be incorporated into virions prior to completion of capsid assembly. L2 possesses a second independent L1 interaction domain located between residues 129 and 246. This interaction was detected in vivo, but not in vitro. Both L1 interaction domains are necessary for L2 to efficiently encapsidate the BPV1 genome in BPHE-1 cells.
ACKNOWLEDGMENTS
We are most grateful to the late Jian Zhou for providing monoclonal antibody C6, to A. Bennett Jenson for monoclonal antibody 3A10, and to Elliot Androphy for monoclonal antibody B201. We thank Carl Olson for the bovine papilloma. We are also grateful to Jon Yewdell, Jack Bennink, and the Laboratory of Viral Diseases (NIH) for the use of their confocal microscope.
This work was supported by National Cancer Institute intramural funding, the Richard TeLinde endowment, the Cancer Research Institute (RBSR), and the Cancer Research Foundation of America (RBSR).
REFERENCES
- 1.Baker C C, Howley P M. Differential promoter utilization by the bovine papillomavirus in transformed cells and productively infected wart tissues. EMBO J. 1987;6:1027–1035. doi: 10.1002/j.1460-2075.1987.tb04855.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker T S, Newcomb W W, Olson N H, Cowsert L M, Olson C, Brown J C. Structures of bovine and human papillomaviruses. Analysis by cryoelectron microscopy and three-dimensional image reconstruction. Biophys J. 1991;60:1445–1456. doi: 10.1016/S0006-3495(91)82181-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Day P M, Roden R B, Lowy D R, Schiller J T. The papillomavirus minor capsid protein, L2, induces localization of the major capsid protein, L1, and the viral transcription/replication protein, E2, to PML oncogenic domains. J Virol. 1998;72:142–150. doi: 10.1128/jvi.72.1.142-150.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delos S E, Cripe T P, Leavitt A D, Greisman H, Garcea R L. Expression of the polyomavirus minor capsid proteins VP2 and VP3 in Escherichia coli: in vitro interactions with recombinant VP1 capsomeres. J Virol. 1995;69:7734–7742. doi: 10.1128/jvi.69.12.7734-7742.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doorbar J, Gallimore P H. Identification of proteins encoded by the L1 and L2 open reading frames of human papillomavirus la. J Virol. 1987;61:2793–2799. doi: 10.1128/jvi.61.9.2793-2799.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dvoretzky I, Shober R, Chattopadhyay S K, Lowy D R. A quantitative in vitro focus assay for bovine papilloma virus. Virology. 1980;103:369–375. doi: 10.1016/0042-6822(80)90195-6. [DOI] [PubMed] [Google Scholar]
- 7.Fujisawa H, Morita M. Phage DNA packaging. Genes Cells. 1997;2:537–545. doi: 10.1046/j.1365-2443.1997.1450343.x. [DOI] [PubMed] [Google Scholar]
- 8.Grande M A, van der Kraan I, van Steensel B, Schul W, de The H, van der Voort H T, de Jong L, van Driel R. PML-containing nuclear bodies: their spatial distribution in relation to other nuclear components. J Cell Biochem. 1996;63:280–291. doi: 10.1002/(sici)1097-4644(19961201)63:3<280::aid-jcb3>3.0.co;2-t. [DOI] [PubMed] [Google Scholar]
- 9.Heino P, Skyldberg B, Lehtinen M, Rantala I, Hagmar B, Kreider J W, Kirnbauer R, Dillner J. Human papillomavirus type 16 capsids expose multiple type-restricted and type-common antigenic epitopes. J Gen Virol. 1995;76:1141–1153. doi: 10.1099/0022-1317-76-5-1141. [DOI] [PubMed] [Google Scholar]
- 10.Heino P, Zhou J, Lambert P F. Interaction of the papillomavirus transcription replication factor, E2, and the viral capsid protein, L2. Virology. 2000;276:304–314. doi: 10.1006/viro.2000.0342. [DOI] [PubMed] [Google Scholar]
- 11.Homa F L, Brown J C. Capsid assembly and DNA packaging in herpes simplex virus. Rev Med Virol. 1997;7:107–122. doi: 10.1002/(sici)1099-1654(199707)7:2<107::aid-rmv191>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 12.Jin X W, Cowsert L M, Pilacinski W P, Jenson A B. Identification of L2 open reading frame gene products of bovine papillomavirus type 1 using monoclonal antibodies. J Gen Virol. 1989;70:1133–1140. doi: 10.1099/0022-1317-70-5-1133. [DOI] [PubMed] [Google Scholar]
- 13.Kawana K, Yoshikawa H, Taketani Y, Yoshiike K, Kanda T. Common neutralization epitope in minor capsid protein L2 of human papillomavirus types 16 and 6. J Virol. 1999;73:6188–6190. doi: 10.1128/jvi.73.7.6188-6190.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kirnbauer R, Booy F, Cheng N, Lowy D R, Schiller J T. Papillomavirus L1 major capsid protein self-assembles into virus-like particles that are highly immunogenic. Proc Natl Acad Sci USA. 1992;89:12180–12184. doi: 10.1073/pnas.89.24.12180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kirnbauer R, Taub J, Greenstone H, Roden R, Durst M, Gissmann L, Lowy D R, Schiller J T. Efficient self-assembly of human papillomavirus type 16 L1 and L1–L2 into virus-like particles. J Virol. 1993;67:6929–6936. doi: 10.1128/jvi.67.12.6929-6936.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lai M C, Teh B H, Tarn W Y. A human papillomavirus E2 transcriptional activator. The interactions with cellular splicing factors and potential function in pre-mRNA processing. J Biol Chem. 1999;274:11832–11841. doi: 10.1074/jbc.274.17.11832. [DOI] [PubMed] [Google Scholar]
- 17.Law M F, Lowy D R, Dvoretzky I, Howley P M. Mouse cells transformed by bovine papillomavirus contain only extrachromosomal viral DNA sequences. Proc Natl Acad Sci USA. 1981;78:2727–2731. doi: 10.1073/pnas.78.5.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li M, Beard P, Estes P A, Lyon M K, Garcea R L. Intercapsomeric disulfide bonds in papillomavirus assembly and disassembly. J Virol. 1998;72:2160–2167. doi: 10.1128/jvi.72.3.2160-2167.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liljestrom P, Garoff H. A new generation of animal cell expression vectors based on the Semliki Forest virus replicon. Bio/Technology. 1991;9:1356–1361. doi: 10.1038/nbt1291-1356. [DOI] [PubMed] [Google Scholar]
- 20.Liu W J, Gissmann L, Sun X Y, Kanjanahaluethai A, Muller M, Doorbar J, Zhou J. Sequence close to the N-terminus of L2 protein is displayed on the surface of bovine papillomavirus type 1 virions. Virology. 1997;227:474–483. doi: 10.1006/viro.1996.8348. [DOI] [PubMed] [Google Scholar]
- 21.McCarthy M P, White W I, Palmer-Hill F, Koenig S, Suzich J A. Quantitative disassembly and reassembly of human papillomavirus type 11 viruslike particles in vitro. J Virol. 1998;72:32–41. doi: 10.1128/jvi.72.1.32-41.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roden R B, Armstrong A, Haderer P, Christensen N D, Hubbert N L, Lowy D R, Schiller J T, Kirnbauer R. Characterization of a human papillomavirus type 16 variant-dependent neutralizing epitope. J Virol. 1997;71:6247–6252. doi: 10.1128/jvi.71.8.6247-6252.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Roden R B, Greenstone H L, Kirnbauer R, Booy F P, Jessie J, Lowy D R, Schiller J T. In vitro generation and type-specific neutralization of a human papillomavirus type 16 virion pseudotype. J Virol. 1996;70:5875–5883. doi: 10.1128/jvi.70.9.5875-5883.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roden R B, Weissinger E M, Henderson D W, Booy F, Kirnbauer R, Mushinski J F, Lowy D R, Schiller J T. Neutralization of bovine papillomavirus by antibodies to L1 and L2 capsid proteins. J Virol. 1994;68:7570–7574. doi: 10.1128/jvi.68.11.7570-7574.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sapp M, Volpers C, Muller M, Streeck R E. Organization of the major and minor capsid proteins in human papillomavirus type 33 virus-like particles. J Gen Virol. 1995;76:2407–2412. doi: 10.1099/0022-1317-76-9-2407. [DOI] [PubMed] [Google Scholar]
- 26.Stauffer Y, Raj K, Masternak K, Beard P. Infectious human papillomavirus type 18 pseudovirions. J Mol Biol. 1998;283:529–536. doi: 10.1006/jmbi.1998.2113. [DOI] [PubMed] [Google Scholar]
- 27.Swindle C S, Engler J A. Association of the human papillomavirus type 11 E1 protein with histone H1. J Virol. 1998;72:1994–2001. doi: 10.1128/jvi.72.3.1994-2001.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Swindle C S, Zou N, Van Tine B A, Shaw G M, Engler J A, Chow L T. Human papillomavirus DNA replication compartments in a transient DNA replication system. J Virol. 1999;73:1001–1009. doi: 10.1128/jvi.73.2.1001-1009.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Touze A, Coursaget P. In vitro gene transfer using human papillomavirus-like particles. Nucleic Acids Res. 1998;26:1317–1323. doi: 10.1093/nar/26.5.1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Trus B L, Roden R B, Greenstone H L, Vrhel M, Schiller J T, Booy F P. Novel structural features of bovine papillomavirus capsid revealed by a three-dimensional reconstruction to 9 Å resolution. Nat Struct Biol. 1997;4:413–420. doi: 10.1038/nsb0597-413. [DOI] [PubMed] [Google Scholar]
- 31.Unckell F, Streeck R E, Sapp M. Generation and neutralization of pseudovirions of human papillomavirus type 33. J Virol. 1997;71:2934–2939. doi: 10.1128/jvi.71.4.2934-2939.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Volpers C, Sapp M, Snijders P J, Walboomers J M, Streeck R E. Conformational and linear epitopes on virus-like particles of human papillomavirus type 33 identified by monoclonal antibodies to the minor capsid protein L2. J Gen Virol. 1995;76:2661–2667. doi: 10.1099/0022-1317-76-11-2661. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Y L, Lewis A, Jr, Wade-Glass M, Schlegel R. Levels of bovine papillomavirus RNA and protein expression correlate with variations in the tumorigenic phenotype of hamster cells. J Virol. 1987;61:2924–2928. doi: 10.1128/jvi.61.9.2924-2928.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhao K N, Frazer I H, Jun Liu W, Williams M, Zhou J. Nucleotides 1506–1625 of bovine papillomavirus type 1 genome can enhance DNA packaging by L1/L2 capsids. Virology. 1999;259:211–218. doi: 10.1006/viro.1999.9714. [DOI] [PubMed] [Google Scholar]
- 35.Zhao K N, Hengst K, Liu W J, Liu Y H, Liu X S, McMillan N A, Frazer I H. BPV1 E2 protein enhances packaging of full-length plasmid DNA in BPV1 pseudovirions. Virology. 2000;272:382–393. doi: 10.1006/viro.2000.0348. [DOI] [PubMed] [Google Scholar]
- 36.Zhao K N, Sun X Y, Frazer I H, Zhou J. DNA packaging by L1 and L2 capsid proteins of bovine papillomavirus type 1. Virology. 1998;243:482–491. doi: 10.1006/viro.1998.9091. [DOI] [PubMed] [Google Scholar]
- 37.Zhou J, Stenzel D J, Sun X Y, Frazer I H. Synthesis and assembly of infectious bovine papillomavirus particles in vitro. J Gen Virol. 1993;74:763–768. doi: 10.1099/0022-1317-74-4-763. [DOI] [PubMed] [Google Scholar]
- 38.Zou N, Lin B Y, Duan F, Lee K Y, Jin G, Guan R, Yao G, Lefkowitz E J, Broker T R, Chow L T. The hinge of the human papillomavirus type 11 E2 protein contains major determinants for nuclear localization and nuclear matrix association. J Virol. 2000;74:3761–3770. doi: 10.1128/jvi.74.8.3761-3770.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]