Abstract
Retroviral Gag expression is sufficient for capsid assembly, which occurs through interaction between distinct Gag domains. Human foamy virus (HFV) capsids assemble within the cytoplasm, although their budding, which mainly occurs in the endoplasmic reticulum, requires the presence of homologous Env. Yet little is known about the molecular basis of HFV Gag precursor assembly. Using fusions between HFV Gag and a nuclear reporter protein, we have identified a strong interaction domain in the N terminus of HFV Gag which is predicted to contain a conserved coiled-coil motif. Deletion within this region in an HFV provirus abolishes viral production through inhibition of capsid assembly.
While the genomic organization of foamy viruses (FVs) is that of a retrovirus, they present several very unusual features, such as the presence of infectious viral DNA in extracellular particles or Pol expression through a specific spliced transcript (16, 31, 33). In that sense, FVs are closely related to pararetroviruses, such as the hepatitis B virus (15).
For retroviruses, the Gag precursor is encoded by the full-length genomic mRNA and is cleaved by the viral protease yielding three mature products, the matrix, the capsid, and the nucleocapsid (13). For the so-called human foamy virus (HFV), Gag maturation has a distinct processing pathway: the 72-kDa Gag precursor is cleaved near its C-terminal end to yield a 68-kDa product, a cleavage required for efficient virus infectivity (23). Although some minor proteolytic cleavages occur within the 68-kDa product, the classical tripartite processing of Gag does not exist in HFV, and consequently only two high-molecular-weight proteins, of 72 and 68 kDa, are detected in infected cells or extracellular virions (14). Nucleic acid binding is performed through the C-terminal domain of HFV Gag, which contains three glycine/arginine-rich sequences, the GRI, -II, and -III boxes, instead of the canonical cysteine-histidine motifs observed in other retroviruses (27). While GRI interacts with nucleic acids (27, 32), GRII contains a nuclear localization sequence, targeting Gag to the nucleus (27). GRII is dispensable for infectivity but important for reaching a high proviral load in infected cells (20). Although GRIII is required for optimal viral infectivity, it was not assigned a specific role as yet. Its absence in the feline FV (FFV) and the equine FV (EFV) demonstrates its dispensability (28, 30). Finally, Gag was shown to target HFV preintegration complexes to the centrosome during the early steps of infection (26). Since Gag is not myristoylated, the basis of its membrane targeting and capsid assembly is not understood.
Retroviral Gag polyprotein expression is sufficient for capsid assembly (13). In type C retroviruses, Gag molecules assemble into capsids at the plasma membrane during virus budding. In type B and D retroviruses, capsids are preassembled within the cytoplasm prior to transport to the plasma membrane, where they exit by budding, even in the absence of Env. Assembly of FV capsids is similar to that of type B and D retroviruses, although their budding in the endoplasmic reticulum (ER) or at the plasma membrane requires the presence of homologous Env, implying that distinct intra- and intermolecular interactions control assemby and traffic of these structures (1, 2, 12, 24). Point mutations in Gag precursors lead to drastic changes in the morphogenic pathways for capsid assembly or in the site of budding (5, 11, 25).
In this report, using fusion proteins between HFV Gag and a nuclear reporter protein (the promyelocytic protein [PML]), we identify a Gag-Gag interaction domain in the N terminus of Gag, predicted to form a coiled-coil motif. Deletion of this domain in an infectious HFV clone completely abolishes viral capsid formation. Our results identify a strong and conserved Gag-Gag interaction domain that is implicated in capsid formation.
MATERIALS AND METHODS
Recombinant DNA.
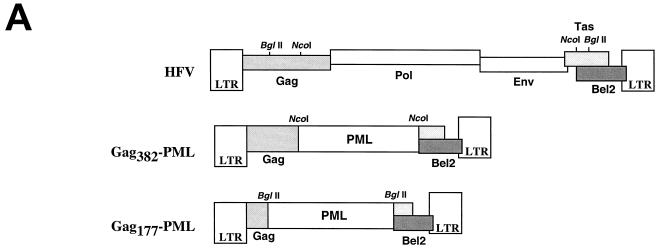
All HFV sequences were initially derived from pHFV13, the infectious molecular clone of HFV. Retroviral reporter constructs (see Fig. 1A) are deletion derivatives of pHFV13, made by digestion with either NcoI or BglII and insertion of a PML isoform cDNA (described in reference 9) in frame to the truncated gag gene.
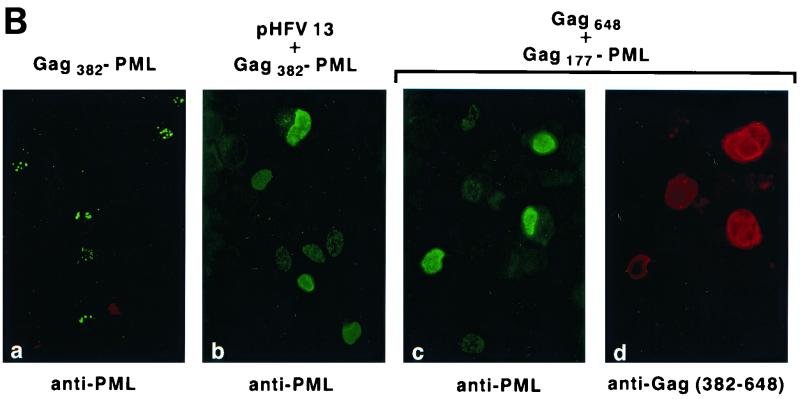
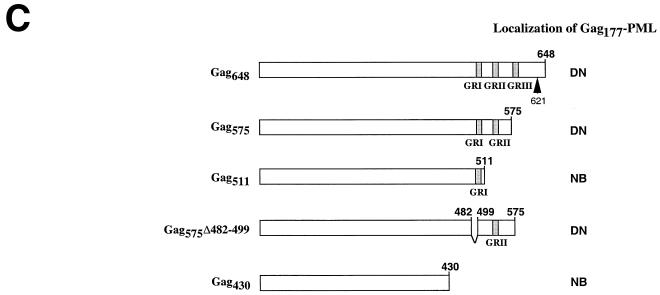
FIG. 1.
(A) Schematic representation of the HFV provirus and Gag-PML retroviral constructs. Parts of the gag gene and the entire pol and env genes were replaced by the PML cDNA. (B) IF analysis demonstrating delocalization of Gag-PML fusion after transfection of the full-length infectious HFV molecular clone (pHFV13) or Gag-expressing vector (pGag648). NIH 3T3 cells stably expressing Tas (panels a and b) or COS6 (panels c and d) were transfected and analyzed by IF with the indicated antibodies. (C) Localization of the Gag177-PML construct upon expression of Gag648 or derived deletion mutants, followed by IF using an anti-PML antiserum. The locations of the three glycine/arginine-rich boxes (GRI, -II, and -III) are indicated by shaded regions. The arrow points to the viral protease cleavage site. The staining was diffuse nuclear (DN) or associated to NB.
The Gag expression plasmids depicted in Fig. 1C were generated by insertion of 2.54-kb Bsu36I (Gag648), 1.77-kb AvrII (Gag575), and 1.63-kb HinpI (Gag511) fragments into the SmaI site of the pSG5M eukaryotic expression vector (Libin Ma, Leiden, The Netherlands). A series of Gag deletion mutants was produced by PCR with specific primers to introduce an EcoRI site at the 5′ end and a BamHI site at the 3′ end. The resulting PCR product was ligated into the EcoRI- and BamHI-digested pSG5M expression vector. Additional Gag mutants were generated by site-directed mutagenesis using the appropriate primers (Tables 1 and 2).
TABLE 1.
Oligonucleotides used for PCR amplificationa
| Primer | Sequence |
|---|---|
| 1 | 5′-GCTTCAGTGACTCCTCCCTCTCAAGGACCAG-3′ |
| 2 | 5′-TGGTCCTTGAGAGGGAGGAGTCACTGAAGC-3′ |
| 3 | 5′-GCGAATTCCATGGACTACAAGGACGACGACGACAAGGCTTCAGGAAGTAATGTTGAAG-3′ |
| 4 | 5′-CGAAGCCATGCATGACGGGATCCTCATCATAACCCTGAGAAGCTGCTTAC-3′ |
| 5 | 5′-CGAAGCCATGCATGACGGGATCCTCATCATTGTAATATTAGTCGTACCATCT-3′ |
| 6 | 5′-CGAAGCCATGCATGACGGGATCCTCATCAAGGATCTCCTTGAACATAATGA-3′ |
| 7 | 5′-GACGACGACGACAAGTATAGTAGTTCTTACAG-3′ |
| 8 | 5′-CTGTAAGAACTACTATACTTGTCGTCGTCGTC-3′ |
| 9 | 5′-CTTACAGACCAGTAACATATGAAGTTGAAAC-3′ |
| 10 | 5′-GTTTCAACTTCATATGTTACTGGTCTGTAAG-3′ |
| 11 | 5′-TATGTACTACTAGTACTGTGGAATTCATGGCTTCAGGAAGTAAT-3′ |
| 12 | 5′-CGGGATCCTTGTCGTCGTCGTCCTTGTAGTCTAACCCTGAGAAGCTGCT-3′ |
| 13 | 5′-TTATGTACTACTAGTACTGTGGAATCCCATGGCAAGTTTGCCCTCTATAC-3′ |
| 14 | 5′-CGGGATCCTTGTCGTCGTCGTCCTTGTAGTCCTGTTGTAATGCAGTTAC-3′ |
| 15 | 5′-TTATGTACTACTAGTACTGTGGAATTCCATGGC TGCATTACAACAGCGTT-3′ |
| 16 | 5′-CGGGATCCTTGTCGTCGTCGTCCTTGTAGTCGTCCCTTTGATCTCC-3′ |
Added restriction sites are underlined, whereas the Flag sequence is in bold characters.
TABLE 2.
Constructs and primers useda
| Construct | Primers |
|---|---|
| Gag575Δ(482–499) | 1, 2 |
| Flag-Gag203 | 3, 4 |
| Flag-Gag123 | 3, 5 |
| Flag-Gag52 | 3, 6 |
| ∗Flag-Gag(123–203) | 7, 8 |
| ∗∗GagΔ(132–163) | 9, 10 |
| Gag203-Flag | 11, 12 |
| Gag(203–441)-Flag | 13, 14 |
| Gag(437–648)-Flag | 15, 16 |
Gag575 was used as a template except where indicated by a single or double asterisk, where FlagGag203 or pHFVΔEcoRI, respectively, was used as a template. Sites of deletion are indicated by the amino acid number at which the reading frame stops. Numbers within each mutant's designation represent the first and last amino acids which are preserved after deletion.
To create an HFV DNA clone with a deletion in Gag, the subclone pHFVΔEcoRI made by digestion of pHFV13 with EcoRI and religation was used as a template for site-directed mutagenesis using primers 9 and 10; the 2.8-kb EclXI-SwaI fragment was then cloned back into pHFV13 and named pHFVGagΔ(131–162).
Cell culture.
Cells were grown in Dulbecco's modified Eagle's medium with 10% fetal calf serum. G418 was added at concentrations of 1 and 0.5 mg/ml, respectively, to NIH 3T3 and BHK-21, stably expressing Tas, provided by A. Rethwilm. COS6, an SV40-transformed African green monkey kidney cell line, was purchased from ATCC.
Generation of an LTR-GFP indicator cell line (FV-activated GFP expression; FAG cells).
The GFP gene of the pGFP-C1 plasmid (Clontech) was replaced by a green fluorescent protein (GFP) sequence harboring the nuclear localization sequence (MAPKKKRK) of the T antigen of simian virus 40 (SV40) at the N-terminal end. This modified GFP gene was inserted at the NheI/BglII sites by PCR using the following primers: direct 5′-C TAG CTA GCC ATG GCC CCC AAG AAG AAG CGC AAG GTG AGC AAG GGC GAG GAG-3′ and reverse 5′-CAG TCC AGT GTT CTT CCT AAG CTG GAG ATC TGA TGC CGG-3′, where the nuclear localization sequence is underlined, the GFP sequences are in bold, and the NheI/BglII sites are in italic. The PCR product was digested with NheI and BglII and used to replaced the GFP gene in pGFP-C1, leading to the pGFPnls vector. The immediate-early promoter from cytomegalovirus was removed from pGFPnls by AseI-NheI digestion and replaced with the U3 region of the HFV long terminal repeat (LTR) obtained by PCR using the following primers: direct 5′-G TGG ATT AAT GCC ACT AGA AAC TAG GGA-3′ and reverse 5′-C TAG CTA GCG ACG CAG CGA GTA GT-3′, where the AseI or NheI site is underlined. This amplified product encompasses the transcriptional initiation start of HFV in the R region at 20 bp. All amplified products were sequenced with the ThermoSequenase sequencing kit (Amersham) before the cloning procedures. U3GFP cell lines were obtained after transfection of BHK-21 cells by the Lipofectin reagent (Gibco) with pU3GFPnls, which harbors the G418 resistance gene. These cells were maintained in the same medium supplemented with 500 μg of G418 (Gibco)/ml. Note that the sensitivity of the FAG assay is similar to that of the one described for the FAB cells (34).
Protein analysis.
Cells were transfected with the different constructs using Lipofect (Gibco-BRL) as specified by the manufacturer. Forty-eight hours posttransfection, protein expression was analyzed.
For immunofluorescence, cells were fixed with 4% paraformaldehyde at 4°C for 10 min and permeabilized with methanol at 4°C for 5 min. PML fusion proteins were revealed with rabbit polyclonal or mouse monoclonal antibodies. Proteins tagged with Flag (DYKDDDDK) were revealed with the anti-Flag M2 monoclonal antibody (Kodak). HFV proteins were detected by rabbit polyclonal anti-HFV antiserum or by anti-Gag382–648 antiserum raised in the laboratory. D11 is a mouse monoclonal antibody directed against Bet and is used at a dilution of 1/400.
For immunoprecipitation, 36 h posttransfection, cells were labeled with [35S]methionine-cysteine (75 μCi/ml, 1.175 Ci/mmol specific activity; Dupont NEN) for 7 h 30 min. Cells were harvested by scraping them into cold phosphate-buffered saline and then lysed in a solution containing 10 mM Tris-HCl (pH 7.5), 150 mM NaCl, 1% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate, and 1 mM phenylmethylsulfonyl fluoride for 30 min at 4°C. Nuclei were separated from the lysate by centrifugation at 12.000 × g for 5 min at 4°C and lysed in 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate buffer containing 0.85 M NaCl. For coimmunoprecipitation experiments, cytoplasmic and nuclear fractions were incubated overnight at 4°C with anti-HFV antiserum or specific anti-Gag antiserum. Protein A-Sepharose was then added for 1 h at 4°C. Immune complexes were centrifuged, washed four times in lysis buffer, and analyzed by sodium dodecyl sulfate–5 to 15% polyacrylamide gel electrophoresis, followed by autoradiography.
For Western blot analyses, transiently transfected cells were lysed directly in Laemmli buffer and proteins were resolved by sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis. The culture supernatant was cleared from cellular debris and centrifuged through a 20% sucrose cushion in a solution containing 100 mM NaCl, 10 mM Tris-HCl (pH 7.4), and 1 mM EDTA at 25,000 rpm for 3 h in a SW41 rotor at 4°C. The resulting pellets and cellular viral protein extracts were visualized by immunoblotting with rabbit anti-Gag antibodies and peroxidase-conjugated antibodies and revealed by enhanced chemiluminescence (Amersham).
Prediction of the coiled-coil motif was performed by the COILS program (http://www.ch.embnet.org/software/COILS_form.html).
Electron microscopy.
Monolayers were fixed in situ with 1.6% glutaraldehyde (Taab Laboratory Equipment Ltd., Reading, United Kingdom) in 0.1 M Sörensen phosphate buffer (pH 7.3 to 7.4) for 1 h at 4°C. Cells were scraped from their plastic substratum and centrifuged. The resulting pellets were successively postfixed with 2% aqueous osmium tetroxide for 1 h at room temperature, dehydrated in ethanol, and embedded in Epon. Ultrathin sections were collected on 200-mesh copper grids coated with Formvar and carbon and stained with uranyl acetate and lead citrate prior to being observed with a Philips 400 transmission electron microscope at 80 kV; magnification, ×2,800 to ×36,000.
RESULTS
HFV Gag-Gag interaction.
During the course of our studies aimed at characterizing the encapsidation signal of HFV, we generated HFV reporter constructs in which a PML cDNA (9) was substituted for the pol, env, and bel/tas sequences (Fig. 1A). The PML gene was originally identified through its fusion to the RARα gene in the t(15;17) translocation found in acute promyelocytic leukemia. PML has a speckled nuclear expression pattern as a consequence of its localization onto nuclear bodies (NBs), structures belonging to the nuclear matrix (15). This distinct speckled localization of PML makes it an attractive reporter, since very low levels of expression are readily detected by immunofluorescence (IF). In the Gag-PML retroviral construct, PML is expressed from the LTR promoter as a Gag-PML fusion protein that retains the first 382 amino acids of Gag (Fig. 1A). Since expression of this Gag382-PML protein is dependent on HFV LTR activity, we transiently transfected NIH 3T3-Tas cells, which stably express the HFV transactivator Tas (originally called Bel1). Our PML antibodies do not detect the endogenous PML protein in NIH 3T3, COS6, or BHK-21 cells, and therefore, expression of transfected Gag382-PML was monitored by IF. In transfected cells, antibodies to PML detect the Gag-PML fusion onto NBs (Fig. 1B, panel a), demonstrating that fusion to Gag does not alter the NB targeting of PML and therefore its folding.
In attempts to pseudotype the Gag-PML construct with HFV structural proteins, we have cotransfected this construct with an infectious molecular HFV clone, pHFV13. To our surprise, coexpression with pHFV13 led to diffuse nuclear PML staining (Fig. 1B, panel b). In contrast to several other viruses (4), HFV does not alter the structure of NBs and consequently the location of endogenous or transfected PML (data not shown). Hence, our observations suggested that HFV infection specifically alters the NB targeting of the Gag382-PML through interactions with its Gag moiety. When Gag-PML fusion proteins were expressed from an SV40 promoter rather than from the HFV LTR, a similar HFV-induced delocalization was observed, allowing the subsequent use of naive cells rather than Tas-expressing ones. Since a similar HFV-induced delocalization was observed with a shorter Gag177-PML (data not shown), the N-terminal region of Gag is required for delocalization of Gag-PML fusion proteins.
To identify the HFV gene product(s) responsible for the altered localization of Gag-PML, we cotransfected Gag177-PML with expression vectors encoding the full-length HFV Gag648, Env, or Bet proteins. Delocalization of the Gag177-PML fusion was exclusively obtained following expression of the Gag648 protein (Fig. 1B, panels c and d), suggestive for the existence of specific Gag-Gag interactions.
During HFV infection, the Gag precursor is detected both in the nucleus and in the cytoplasm of infected cells, as a p72/p68 doublet. The C terminus of Gag648 contains glycine/arginine-rich sequences (GR boxes) involved in nucleic acid binding (GRI) and nuclear localization (GRII) (Fig. 1C). To determine whether the GR boxes are required for Gag177-PML delocalization, GR deletion mutants were derived from the Gag648 expression vector. As summarized in Fig. 1C, all constructs harboring GRII delocalized Gag177-PML from nuclear bodies, even in the absence of GRI. These results demonstrate that Gag-PML delocalization by Gag requires the presence of Gag residues 511 to 575 (including GRII), which likely bind distinct nuclear components preventing the NB targeting of the Gag-PML fusion protein. Interaction between this region and putative nuclear components is necessarily stronger than that driving Gag-PML onto NBs. This nuclear anchor, which remains to be characterized, together with the nuclear localization signal function harbored by GRII, is likely to play a role in the nuclear accumulation of Gag.
Mapping of sequences involved in Gag-Gag interaction.
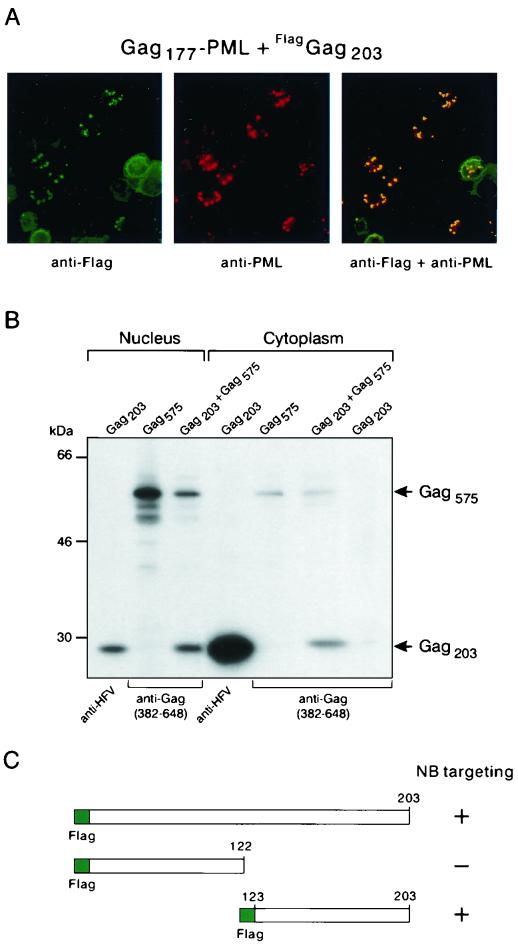
To determine which part of Gag interacts with Gag177-PML fusion, three constructs were generated, expressing Gag from amino acids 1 to 203, 203 to 441, and 437 to 648, all tagged with a C-terminal Flag sequence. This particular partition of Gag was made according to protease cleavage sites that were initially predicted (18). COS6 cells were transfected with these expression vectors together with Gag177-PML. Only the construct expressing Gag203 led to NB-associated anti-Flag staining (Fig. 2A and data not shown), demonstrating the existence of an interaction between Gag203 and Gag177-PML.
FIG. 2.
Gag-Gag interaction involves the N-terminal domain. (A) IF analysis. Transfected COS6 cells were subjected to double immunofluorescence labeling with the mouse monoclonal anti-Flag M2 (revealed by fluorescein isothiocyanate) and rabbit polyclonal anti-PML antibodies (revealed with Texas red). (B) Coimmunoprecipitation assay. [S35]methionine-cysteine-labeled viral proteins were immunoprecipitated from cytoplasmic and nuclear extracts with the indicated antibodies. (C) Interaction of Gag mutants with Gag177-PML onto NBs was assessed by IF using the anti-Flag M2 antibody: the plus sign corresponds to an efficiency similar to that of Gag203, and the minus sign corresponds to an absence of interaction.
Gag-Gag interaction was also assessed by biochemical analyses. In these experiments, Gag575 alone (which harbors both GRI and GRII boxes) was used rather than Gag177-PML, since PML is a nuclear matrix protein whose extraction requires stringent conditions that would disrupt protein complexes. COS6 cells were transfected with expression vectors for Gag203, Gag575, or both; labeled with [35S]methionine-cysteine; and separated into cytoplasmic and nuclear fractions. Cell extracts were then immunoprecipitated with either an antibody that recognizes all HFV proteins or one specific antiserum directed against the C-terminal region of Gag (amino acids 382 to 648) interacting solely with Gag575 and not with Gag203, as shown in the last lane of Fig. 2B. In both the cytoplasmic and the nuclear fractions, Gag203 was readily coimmunoprecipitated with Gag575 using the antibody directed against the C-terminal moiety of Gag (Fig. 2B). This implied that a significant fraction of the two proteins interacts and confirmed the existence of an interaction domain in the N terminus of HFV Gag. Moreover, this experiment proved that such an interaction occurs outside of a Gag-PML fusion protein context.
To precisely map the sequences within Gag203 that are implicated in Gag-Gag interaction, Flag-Gag203 deletion mutants were generated and tested for their ability to interact with Gag177-PML, using the same assay as that shown in Fig. 2A. While deletion of residues 123 to 203 abolished Gag-Gag interaction, Gag123–203 totally colocalized with Gag177-PML onto NBs (Fig. 2C), as already was observed with Gag203 (Fig. 2A), suggesting that the association between both molecules is stable. These results implied that the region encompassing amino acids 123 to 203 mediates Gag-Gag interaction.
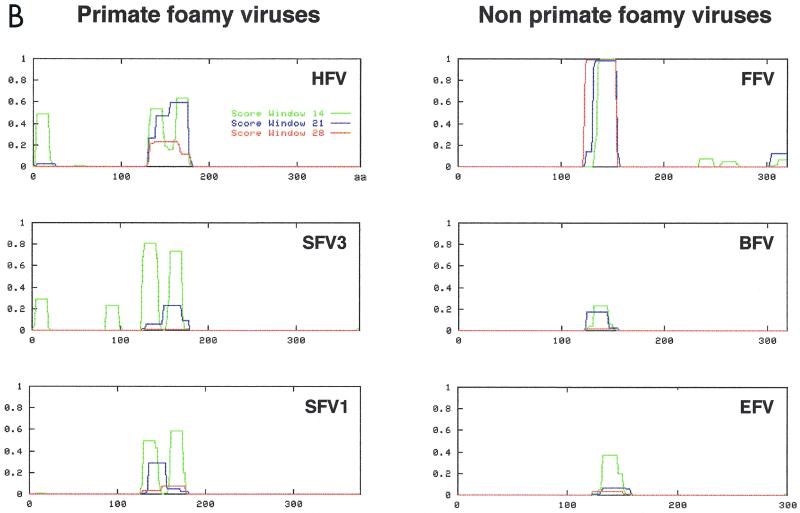
Careful examination of the HFV domain between amino acids 123 and 203 revealed the presence of regularly spaced hydrophobic residues (Fig. 3A), suggestive for the presence of a coiled-coil motif, which was confirmed by analysis of this region with the COILS program (Fig. 3B). Although FV Gag proteins show little similarity (29), alignment of this region from six foamy virus isolates (HFV, simian FV type 3, simian FV type 1, FFV, bovine FV [BFV], and EFV) revealed the conservation of this putative coiled-coil motif in all but the two nonprimate FVs, BFV and EFV (Fig. 3B).
FIG. 3.
(A) Multiple amino acids alignment shows that hydrophobic residues are conserved among FVs. Conserved residues are shown in boldface. (B) Prediction of coiled-coil motifs in N termini of Gag proteins from FVs by the COILS program (Matrix: MTIDK, weight of 2.5 for position N).
Biological role of the Gag-Gag interaction.
Based on the prediction of the COILS program, we generated a deletion spanning residues 131 to 162 in Gag203. Since this deletion abolished its interaction with Gag177-PML on NBs, a similar mutation was introduced into the infectious pHFV13 clone, leading to pHFVGagΔ(131–162). This mutant and the wild-type proviral clones were transfected in Tas-expressing BHK-21 cells. Two days after transfection, virus production was determined by exposing BHK-21 GFP indicator cells (see Materials and Methods) to cell supernatants from these transfected cells. Fluorescence observed at 24 h postinfection revealed that no infectious virus was produced with the Gag mutant clone (<0.5 fluorescent-cell-forming units [FCFU]/ml), whereas the wild-type provirus led to significant viral production (9.3 × 104 FCFU/ml). Trans-complementation of the mutant virus clone with a full-length HFV Gag648 expression vector [pHFVGagΔ(131–162)+pGag648] partially restored viral infectivity (3 × 104 FCFU/ml) as expected (values are averaged from three independent experiments) (35). Hence, Gag-Gag interaction is essential for viral infectivity.
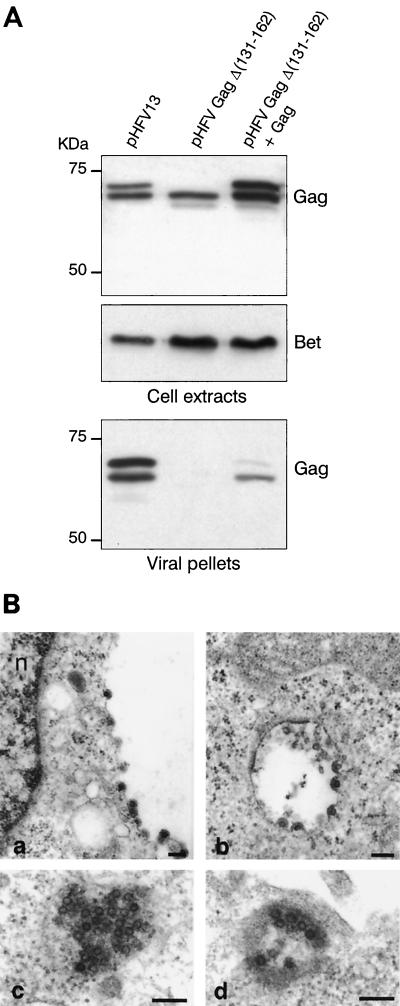
To test whether the lack of infectivity of the HFV Gag mutant clone is due to either impairment of capsid formation, virus particle egress, or release of a defective virus, supernatants from transfected cells were centrifuged through a sucrose cushion and the resulting pellets were analyzed by Western blotting (WB) and compared to the corresponding cellular extracts. Cell extracts from wild-type or mutant provirus-transfected cells showed no major difference in the level of intracellular p72/p68 Gag expression; anti-Bet antibodies (D11) also revealed similar levels of p62 Bet in all transfected cell extracts, demonstrating that, as expected, deletion of Gag did not impair viral gene expression (Fig. 4A). Strikingly, when the viral pellets were analyzed by WB, no Gag protein was detected in the supernatant of mutant transfected cells, showing that this mutation abolished virus release (Fig. 4). Cotransfection of the mutant HFV clone with a full-length Gag-expressing vector partially restored virus release to the supernatant as expected. Yet in that case, a p68 protein was mainly detected. It is most likely that this represents the cleaved p72 fragment rather than the full-size Gag polyprotein derived from the mutated virus. The smaller amount of Gag (and hence of virus) retrieved in that case indicates that the deleted Gag polyprotein has some dominant-negative effects on capsid assembly, suggestive for the existence of other interaction interfaces in Gag as reported previously (3, 10). One possible explanation for the almost exclusive presence of p68 could then be that GagΔ (131–163) selectively binds p72 Gag.
FIG. 4.
(A) Western blot analysis of cell extracts and virus pellets from cells transfected either with pHFVGagΔ(131–162) or with pHFV13 clones. Antibodies used are either a rabbit polyclonal anti-Gag382–648 antibody or a monoclonal anti-Bet antiserum (D11). Mutation of Gag completely abolishes virus release from transfected cells, whereas cotransfection with a wild-type Gag (pGag648) partially restores virus production. (B) Electron micrographs of BHK-21 cells 48 h after transfection with pHFV13 (panels a and c) and pHFVGagΔ(131– 162) trans-complemented with Gag648 (panels b and d). Images represent budding of virus particles (panels a and b) or clusters of capsids (panels c and d). The bars correspond to 200 nm (a and b) or 400 nm (c and d).
The absence of viruses in the supernatant of pHFVGagΔ(131–162)-transfected cells could be due to the impairment of capsid formation or of virus exit. Hence, cells transfected with wild-type or mutant proviral DNAs were analyzed by electron microscopy. In HFV-expressing cells, capsids and budding viruses were observed in a large number of cells. In contrast, in cells transfected by the virus harboring the mutated Gag, no viral capsid could be observed, despite a similar transfection efficiency. Again, trans-complementation of the mutant virus with a Gag-expressing vector (Gag648) led to the appearance of viral particles (Fig. 4B). Therefore, Gag-Gag interaction through its 131 to 162 amino acid domain is absolutely required for capsid formation.
DISCUSSION
This report identifies a strong Gag-Gag interaction domain at the N terminus of HFV Gag that is required for intracellular capsid assembly. This interaction domain was first demonstrated by the analysis of nuclear fluorescence patterns, diffuse or onto NBs, of Gag molecules and derivative mutants in the presence of a Gag-PML fusion protein. Interaction was further confirmed by coimmunoprecipitation experiments. Finally, we showed using different approaches that the domain spanning residues 131 to 162 was necessary for the formation of viral capsids.
Interaction and subsequent multimerization of Gag molecules play a crucial role in capsid assembly (13). Depending on the virus, multiple strong dimerization interfaces have been described along the Gag protein, especially within the matrix, the C-terminal globular domain of the capsid, and the nucleocapsid sequence (3, 8). Multiple and sequential interactions of Gag molecules allow the precise concentration and folding of precursors to form capsids. While the mechanism of capsid assembly has been intensively studied for other retroviruses, little is known about FV capsid formation, in particular on Gag domains involved in this step of the viral cycle. Complementation analyses of Rous sarcoma virus and HFV Gag revealed the presence of two putative interaction domains (the I domains) in the C terminus of HFV Gag overlapping the GRI box (3). Since we have shown that the mutated clone pHFVGagΔ(131–162) is defective for capsid assembly, these I domains alone do not seem to be sufficient to form viral capsids. More recently, Eastman et al. have reported that mutation of a conserved Arg residue at position 50 in HFV Gag prevents formation of extracellular virions, presumably by disrupting capsid assembly. This amino acid lies within a motif resembling the morphogenetic signal of the type D retrovirus, Mason-Pfizer monkey virus (5, 25, 28). The sequential involvement of these characterized determinants (I domains, the Gag region between residues 131 to 162, and the Arg50 residue), which could be necessary to trigger capsid assembly, remains to be elucidated.
Here we show that the region mediating Gag-Gag interaction contains a conserved motif which could form a coiled coil (19). This sequence appears to function as a whole, since point mutants in the conserved residues of the putative coiled coil failed to abolish the interaction (data not shown). Note in that respect that deletions that were similar in size did not abrogate capsid formation in other viruses (11, 36). Such a domain was found in primate and nonprimate FVs. However, in BFV and EFV, the probability of forming a coiled coil is low. Interestingly, these two viruses also lack the ER retrieval dilysine motif in the Env cytoplasmic tails, which is responsible for HFV budding at the ER. In line with this observation, EFV was shown to bud mainly at the plasma membrane (28).
The assay outlined here to detect protein-protein interactions in vivo using NB targeting of PML as an end point could be of general interest. Indeed, using NB targeting of a protein through interaction with another protein fused to PML can be a sensitive way to demonstrate protein-protein interaction within the nucleus. It can also be used to compare the strengths of different intranuclear targeting signals.
For exogenous retroviruses, cellular expression of Gag is sufficient for capsid assembly and release of virus-like particles. FV assembly and egress are unique among the retroviral family (17). In particular, budding of preassembled viral capsids requires the expression of the cognate FV envelope, reminiscent of the hepadnavirus assembly pathway. Although the results presented in this report bring new insights into the formation of FV capsids, several points remain to be elucidated, one of the most intriguing being the targeting of Gag to the membranes of the ER. For intracisternal A type particles, which, like HFV, lack an N-terminal myristoylation signal, a stretch of hydrophobic amino acids at the N terminus of Gag directs virus budding to the ER (21). However, it has already been demonstrated that this is not the case for HFV capsids, which never bud at any internal membrane in the absence of Env (12). Whether the Gag domain characterized in this study is involved in Gag-Env interactions remains to be investigated.
ACKNOWLEDGMENTS
Ali Saïb and Hugues de Thé should be considered senior coauthors.
We warmly thank J. Lasneret, F. Rocher, and A. Janin for electron microscopy studies and LPH (Laboratoire Photo de l'Institut d'Hématologie) for the photographic work.
H. de Thé is supported by Université Paris 7 and Assistance Publique.
REFERENCES
- 1.Baldwin D N, Linial M L. The roles of Pol and Env in the assembly pathway of human foamy virus. J Virol. 1998;72:3658–3665. doi: 10.1128/jvi.72.5.3658-3665.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baldwin D N, Linial M L. Proteolytic activity, the carboxy terminus of Gag, and the primer binding site are not required for Pol incorporation into foamy virus particles. J Virol. 1999;73:6387–6393. doi: 10.1128/jvi.73.8.6387-6393.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowzard J B, Bennett R P, Krishna N K, Ernst S M, Rein A, Wills J W. Importance of basic residues in the nucleocapsid sequence for retrovirus Gag assembly and complementation rescue. J Virol. 1998;72:9034–9044. doi: 10.1128/jvi.72.11.9034-9044.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chelbi-Alix M K, Quignon F, Pelicano L, Koken M H M, de Thé H. Resistance to virus infection conferred by the interferon-induced promyelocytic leukemia protein. J Virol. 1998;72:1043–1051. doi: 10.1128/jvi.72.2.1043-1051.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Choi G, Park S, Choi B, Hong S, Lee J, Hunter E, Rhee S S. Identification of a cytoplasmic targeting/retention signal in a retroviral Gag polyprotein. J Virol. 1999;73:5431–5437. doi: 10.1128/jvi.73.7.5431-5437.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conte M R, Klikova M, Hunter E, Ruml T, Matthews S. The three-dimensional solution structure of the matrix protein from the type D retrovirus, the Mason-Pfizer monkey virus, and implications for the morphology of retroviral assembly. EMBO J. 1997;16:5819–5826. doi: 10.1093/emboj/16.19.5819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Conte M R, Matthews S. Retroviral matrix proteins: a structural perspective. Virology. 1998;246:191–198. doi: 10.1006/viro.1998.9206. [DOI] [PubMed] [Google Scholar]
- 8.Dawson L, Yu X F. The role of nucleocapsid of HIV-1 in virus assembly. Virology. 1998;251:141–157. doi: 10.1006/viro.1998.9374. [DOI] [PubMed] [Google Scholar]
- 9.de The H, Lavau C, Marchio A, Chomienne C, Degos L, Dejean A. The PML-RAR alpha fusion mRNA generated by the t(15; 17) translocation in acute promyelocytic leukemia encodes a functionally altered RAR. Cell. 1991;66:675–684. doi: 10.1016/0092-8674(91)90113-d. [DOI] [PubMed] [Google Scholar]
- 10.Eastman S W, Benki S, Baldwin D N, Linial M L. Proceedings of the 2000 Cold Spring Harbor Meeting on Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 2000. Domains of foamy virus Gag involved in particle assembly and morphogenesis; p. 114. [Google Scholar]
- 11.Facke M, Janetzko A, Shoeman R L, Krausslich H G. A large deletion in the matrix domain of the human immunodeficiency virus gag gene redirects virus particle assembly from the plasma membrane to the endoplasmic reticulum. J Virol. 1993;67:4972–4980. doi: 10.1128/jvi.67.8.4972-4980.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fischer N, Heinkelein M, Lindemann D, Enssle J, Baum C, Werder E, Zentgraf H, Muller J G, Rethwilm A. Foamy virus particle formation. J Virol. 1998;72:1610–1615. doi: 10.1128/jvi.72.2.1610-1615.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freed E O. HIV-1 gag proteins: diverse functions in the virus life cycle. Virology. 1998;251:1–15. doi: 10.1006/viro.1998.9398. [DOI] [PubMed] [Google Scholar]
- 14.Giron M L, Colas S, Wybier J, Rozain F, Emanoil-Ravier R. Expression and maturation of human foamy virus Gag precursor polypeptides. J Virol. 1997;71:1635–1639. doi: 10.1128/jvi.71.2.1635-1639.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koken M H, Puvion-Dutilleul F, Guillemin M C, Viron A, Linares-Cruz G, Stuurman N, de Jong L, Szostecki C, Calvo F, Chomienne C, et al. The t(15;17) translocation alters a nuclear body in a retinoic acid-reversible fashion. EMBO J. 1994;13:1073–1083. doi: 10.1002/j.1460-2075.1994.tb06356.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lecellier C, Saib A. Foamy viruses: between pararetroviruses and retroviruses. Virology. 2000;271:1–8. doi: 10.1006/viro.2000.0216. [DOI] [PubMed] [Google Scholar]
- 17.Linial M L. Foamy viruses are unconventional retroviruses. J Virol. 1999;73:1747–1755. doi: 10.1128/jvi.73.3.1747-1755.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lochelt, M., and R. Flugel. The molecular biology of human and primate spuma retroviruses, p. 239–292. In J. Levy (ed.), The Retroviridae. Plenum Press, New York, N.Y.
- 19.Lupas A. Predicting coiled-coil regions in proteins. Curr Opin Struct Biol. 1997;7:388–393. doi: 10.1016/s0959-440x(97)80056-5. [DOI] [PubMed] [Google Scholar]
- 20.Meiering C D, Comstock K E, Linial M L. Multiple integrations of human foamy virus in persistently infected human erythroleukemia cells. J Virol. 2000;74:1718–1726. doi: 10.1128/jvi.74.4.1718-1726.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mietz J A, Grossman Z, Lueders K K, Kuff E L. Nucleotide sequence of a complete mouse intracisternal A-particle genome: relationship to known aspects of particle assembly and function. J Virol. 1987;61:3020–3029. doi: 10.1128/jvi.61.10.3020-3029.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morikawa Y, Zhang W H, Hockley D J, Nermut M V, Jones I M. Detection of a trimeric human immunodeficiency virus type 1 Gag intermediate is dependent on sequences in the matrix protein, p17. J Virol. 1998;72:7659–7663. doi: 10.1128/jvi.72.9.7659-7663.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pfrepper K I, Lochelt M, Rackwitz H R, Schnolzer M, Heid H, Flugel R M. Molecular characterization of proteolytic processing of the Gag proteins of human spumavirus. J Virol. 1999;73:7907–7911. doi: 10.1128/jvi.73.9.7907-7911.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pietschmann T, Heinkelein M, Heldmann M, Zentgraf H, Rethwilm A, Lindemann D. Foamy virus capsids require the cognate envelope protein for particle export. J Virol. 1999;73:2613–2621. doi: 10.1128/jvi.73.4.2613-2621.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rhee S S, Hunter E. A single amino acid substitution within the matrix protein of a type D retrovirus converts its morphogenesis to that of a type C retrovirus. Cell. 1990;63:77–86. doi: 10.1016/0092-8674(90)90289-q. [DOI] [PubMed] [Google Scholar]
- 26.Saïb A, Puvion-Dutilleul F, Schmid M, Périès J, de Thé H. Nuclear targeting of incoming human foamy virus Gag proteins involves a centriolar step. J Virol. 1997;71:1155–1161. doi: 10.1128/jvi.71.2.1155-1161.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schliephake A W, Rethwilm A. Nuclear localization of foamy virus Gag precursor protein. J Virol. 1994;68:4946–4954. doi: 10.1128/jvi.68.8.4946-4954.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tobaly-Tapiero J, Bittoun P, Neves M, Guillemin M, Lecellier C, Puvion-Dutilleul F, Gicquel B, Zientara S, Giron M L, de Thé H, Saïb A. Isolation and characterization of an equine foamy virus (EFV) J Virol. 2000;74:4064–4073. doi: 10.1128/jvi.74.9.4064-4073.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang G, Mulligan M J. Comparative sequence analysis and predictions for the envelope glycoproteins of foamy viruses. J Gen Virol. 1999;80:245–254. doi: 10.1099/0022-1317-80-1-245. [DOI] [PubMed] [Google Scholar]
- 30.Winkler I, Bodem J, Haas L, Zemba M, Delius H, Flower R, Flugel R M, Lochelt M. Characterization of the genome of feline foamy virus and its proteins shows distinct features different from those of primate spumaviruses. J Virol. 1997;71:6727–6741. doi: 10.1128/jvi.71.9.6727-6741.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu S F, Baldwin D N, Gwynn S R, Yendapalli S, Linial M L. Human foamy virus replication: a pathway distinct from that of retroviruses and hepadnaviruses. Science. 1996;271:1579–1582. doi: 10.1126/science.271.5255.1579. [DOI] [PubMed] [Google Scholar]
- 32.Yu S F, Edelmann K, Strong R K, Moebes A, Rethwilm A, Linial M L. The carboxyl terminus of the human foamy virus Gag protein contains separable nucleic acid binding and nuclear transport domains. J Virol. 1996;70:8255–8262. doi: 10.1128/jvi.70.12.8255-8262.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu S F, Sullivan M D, Linial M L. Evidence that the human foamy virus genome is DNA. J Virol. 1999;73:1565–1572. doi: 10.1128/jvi.73.2.1565-1572.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu S F, Linial M L. Analysis of the role of the bel and bet open reading frames of human foamy virus by using a new quantitative assay. J Virol. 1993;67:6618–6624. doi: 10.1128/jvi.67.11.6618-6624.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zemba M, Wilk T, Rutten T, Wagner A, Flugel R F, Lochelt M. The carboxy-terminal p3Gag domain of the human foamy virus Gag precursor is required for efficient virus infectivity. Virology. 1998;247:7–13. doi: 10.1006/viro.1998.9234. [DOI] [PubMed] [Google Scholar]
- 36.Zhang W H, Hockley D J, Nermut M V, Morikawa Y, Jones I M. Gag-Gag interactions in the C-terminal domain of human immunodeficiency virus type 1 p24 capsid antigen are essential for Gag particle assembly. J Gen Virol. 1996;77:743–751. doi: 10.1099/0022-1317-77-4-743. [DOI] [PubMed] [Google Scholar]