Abstract
Ames dwarf mice (df/df) display delayed aging relative to their normal (N) siblings, living approximately 40–60 % longer. As such, investigating the mechanisms that enable these organisms to have extended lifespan is useful for the development of interventions to slow aging and deter age-related disease. Nonalcoholic fatty liver disease (NAFLD) is a condition that is characterized by the accumulation of excess adipose tissue in the liver. Previous studies highlight the potential of calorie restriction (CR) in promoting longevity, but little is known about its effects on the biomolecular processes that govern NAFLD. In this study, we examined the role of 6-month CR on genes regulating lipid metabolism in the livers of long-living df/df mice and their N littermates. Importantly, our findings showed significant downregulation of miR-34a-5p in N-CR mice and df/df mice regardless of dietary regimen. Alongside, our RT-PCR results indicated that downregulation of miR-34a-5p is correlated with the expression of metabolism-associated mRNAs involved in modulating the processes of de novo lipogenesis (DNL), fatty acid oxidation (FAO), very-low density lipoprotein transport (VLDL-T), and reverse cholesterol transport (RCT). To further verify the role of miR-34a-5p in regulating metabolic processes, we transfected the human liver cancer (HepG2) cell line with miR-34a mimic, and studied its effect on direct targets Sirt1, Ampk, and Ppara as well as downstream lipid transport regulating genes. Our findings suggest that CR and df/df life extending mutation are robust drivers of the miR-34a-5p signaling pathway and prevent the pathogenesis of age-related diseases by improving overall lipid homeostasis.
Keywords: Age-related diseases, miRNAs, mRNAs, Ames dwarf, Dietary intervention
1. Introduction
Nonalcoholic fatty liver disease (NAFLD) is an idiopathic metabolic disorder that affects roughly 83 million Americans, with an incidence rate of roughly 33–50 % in older adults (Perumpail et al., 2017; Pitisuttithum and Treeprasertsuk, 2022; Xia, 2023). Although the pathogenesis of this condition is poorly understood, it is believed to be influenced by unhealthy lifestyle choices such as excess energy intake (Perumpail et al., 2017; Barzilai et al., 2012). Presently, dietary intervention and physical activity are the only options that exist to stagnate the progression of this age-related illness (Arora et al., 2019). Calorie restriction (CR) is recognized as an effective intervention capable of significantly lengthening lifespan (Nisoli et al., 2005; Finley and Haigis, 2009; Fontana, 2009; Duregon et al., 2021). Alongside, CR has other benefits including decreased body weight, reduced cholesterol levels, and improved insulin sensitivity (Finley and Haigis, 2009; Fontana, 2009; Shintani et al., 2023). Notably, the effects of CR on aging are believed to be the product of changes in gene expression that may be influenced by metabolic processes including lipid trafficking (Nisoli et al., 2005). Previous studies elucidate the protective effects of calorie restriction (CR)-induced autophagy against cellular senescence, but little is known about its role in the biomolecular processes that regulate NAFLD (Cani et al., 2007). Unfortunately, compliance to any forms of CR regimens in developed countries is often futile with strict adherence by only 50 % of patients (Arora et al., 2019). Due to the indefinite commitment to these non-medicinal lifestyle changes, it is imperative to discover breakthrough therapies that may potentially mitigate NAFLD, enhance healthspan, and improve the quality of life.
Besides dietary intervention, there are known genetic mouse variants that carry significant protection from a variety of age-related diseases. Ames dwarf mice (df/df) are natural mutants of the prophet of pituitary factor-1 (PROP1) gene, resulting in pituitary hypoplasia (Hill et al., 2016; Noureddine, 2022). Consequently, they exhibit deficiencies in growth hormone (GH), prolactin (PRL), and thyroid stimulating hormone (TSH) (Bartke and Brown-Borg, 2004). As a byproduct of these distinct hormonal inadequacies relative to normal mice, df/df mice display improved memory, delayed aging, living 40–60 % longer (Hill et al., 2016; Noureddine, 2022). Alongside, df/df mutants are characterized with enhanced insulin signaling, which could minimize the risk of developing metabolic diseases (Bartke and Brown-Borg, 2004; Masternak et al., 2005). This makes the df/df genotype a valuable animal model by enabling the possibility of uncovering biomolecular processes that may extend life.
There exists significant emphasis on studying the development of pharmaceutical drugs that can regulate gene expression and biological pathways of interest (Shintani et al., 2023). However, in recent years, studying the role of small non-coding RNAs as major regulators of biomolecular pathways in different diseases including cancer, Alzheimer diseases, metabolic complications and aging has gained interest (Noureddine, 2022; O’Brien et al., 2018). MicroRNAs (miRNAs) are a group of ubiquitous, small noncoding RNAs about 25 nucleotides in length that regulate the process of post-transcriptional gene expression (O’Brien et al., 2018). These endogenous molecules bind to the 3′ untranslated region of messenger RNA’s (mRNAs) to block translation and prompt degradation of mRNA (O’Brien et al., 2018). As such, understanding the role of miRNAs in aging may prompt the discovery of treatments to combat age-associated diseases.
Recently, miR-34a has emerged as a proposed biomarker of liver disease with assumed influence on NAD-dependent deacetylase silent information T1 (SIRT1) and adenosine monophosphate-activated protein kinase (AMPK), coregulators of energy metabolism (Li et al., 2011; Ding et al., 2015; Li et al., 2015; Xu et al., 2021). Particularly, the SIRT1 gene is a facilitator of healthy aging and is known to inhibit oxidative stress and support DNA stability (Canto and Auwerx, 2009; Canto et al., 2009; Ding et al., 2017). AMPK is activated by high AMP to ATP ratio due to low cellular energy, inducing catabolism (Canto and Auwerx, 2009; Canto et al., 2009). Alongside, miR-34a has been reported to target peroxisome proliferator-activated receptor gamma coactivator-1 alpha (PGC1А) and peroxisome proliferator-activated receptor-a (PPARА), transcription factors that maintain the activity of fatty acid metabolizing genes (Li et al., 2015; Canto and Auwerx, 2009; Besse-Patin et al., 2019). As such, CR may improve lipid turnover by suppressing the expression of miR-34a making it a promising therapy to examine in understanding NAFLD.
In this study, we investigated how CR influences dysregulated metabolic pathways to curb the pathology of age-related diseases. We suppose CR restores lipid homeostasis by improved glucose sensitivity thereby promoting healthy fatty acid oxidation. Overall, we seek to understand the mechanism by which CR and the df/df genotype modulate age-related diseases in liver tissue and the role that miRNAs have in this process. Specifically, we focused on the regulation of miR-34a in the liver tissue in df/df and N mice subjected to 6 months of CR.
2. Methods
2.1. Experimental design and animals
Animal protocol was approved by the University of Central Florida Institutional Animal Care and Use Committee (IACUC). Normal wild-type (N) and Ames dwarf (df/df) littermates were housed under a 12-h light and dark cycle at approximately 20–24 °C with unlimited access to food (Rodent Laboratory Chow 5001; not autoclaved; 23.4 % protein, 4.5 % fat, 5.8 % crude fiber; LabDiet PMI Feeds, Inc., St. Louis, MO). At 3 months of age, male mice were separated into the following experimental groups (n = 8–11 per group): normal mice fed ad libitum (N-AL), normal mice subjected to 30 % calorie restriction (N-CR), Ames dwarf mice fed ad libitum (df/df-AL), and Ames dwarf mice subjected to 30 % calorie restriction (df/df-CR).
Animals selected for CR were introduced to the regimen with a gradual reduction of food intake during the first three weeks of the study. Specifically, 90 % food intake during the first week, 80 % intake during the second week, and 70 % intake thereafter. Food consumption was measured weekly in efforts to appropriately moderate 30 % calorie restriction relative to the ad libitum controls. At the age of 9 months (6 months of continuous CR), mice were fasted overnight and sacrificed via cervical dislocation under sedation with isoflurane. Liver tissue was harvested and snap frozen in liquid nitrogen with subsequent storage in −80 °C for future analysis.
2.2. Transfection of cell lines
Human hepatocellular carcinoma (HEPG2) cell line was generously donated from the laboratory of Dr. Shadab Siddiqi (University of Central Florida, Burnett School of Biomedical Sciences, Orlando, FL, USA) and cultured in 50:50 Dulbecco’s Modified Eagle Medium/Ham’s Nutrient Mixture F-12 (DMEM/F12; Corning, Manassas, VA, USA) supplemented with L-glutamine, 10 % fetal bovine serum (FBS; Corning, Woodland, CA, USA) and 1 % antibiotics (5000 units/ml penicillin and 5000μg/ml streptomycin; Gibco, Grand Island, NY, USA) at 37 °C with 5 % CO2.
At 80 % confluency, HEPG2 cells were washed in phosphate buffered saline (Corning, Manassas, VA, USA) and dissociated from the culture dish using 0.25 % trypsin-EDTA (Gibco, Grand Island, NY, USA). After trypsin inactivation, cells were pelleted via centrifugation at 125 ×g for 7 min and resuspended in antibiotic-free, FBS supplemented 50:50 DMEM/F12 media.
Using an automated cell counter (Corning CytoSmart), cells were counted and seeded at a density of 2.0*105 cells/ml with a final volume of 2.3 ml per well. After two days, cells were separated into two groups (control, mimic; n = 8 per group) and incubated in 100ul of transfection complexes consisting of 6 μl HiPerFect transfection reagent (Qiagen) in 100 μl serum-free DMEM/F12 media, with the mimic group incubated in 5 nM mimic of hsa-miR-34a-5p (Dharmacon). After three hours, 2.2 ml of antibiotic-free, FBS supplemented 50:50 DMEM/F12 was added to the cells. At 48 h, cells were collected in 500 μl of Trizol (Life Technologies, Carlsbad, CA, USA) for RNA extraction.
2.3. RNA extraction and cDNA synthesis
RNA extraction of mouse liver tissue samples was completed via manufacturer’s protocol for RNeasy® Mini Kit (Qiagen, Valencia, CA, USA). Briefly, tissues were homogenized in the Bullet Blender Homogenizer BBX24 (Next Advance, Averill Park, NY, USA) using 500 μl QIAzol® Lysis Reagent and 0.5 mm Zirconium oxide beads. Thereafter, an additional 400 μl of QIAzol® Lysis Reagent along with 100 μl gDNA eliminator solution and 180 μl of chloroform was added to the samples which were then shaken vigorously prior to centrifugation at 12,000 rcf for 15 min at 4 °C. Following centrifugation, the upper aqueous phase was collected. RNA was purified and eluted using RNeasy® mini spin column and 30 μl of RNase-free water. Concentrations were measured with BioTek Epoch microplate spectrophotometer and quality was ensured to have A260/A280 ≥ 1.9. RNA was stored at −80 °C until analysis.
RNA extraction of human hepatocytes was completed via the TRIzol method. Briefly, cell lysates were collected in 500 μl of TRIzol reagent and centrifuged with 100 μl chloroform for 15 min at 12,000rcf at 4 °C. The upper aqueous phase was transferred to a new tube and total RNA was precipitated using 250 μl of isopropanol. Samples were incubated for 10 min at −20 °C and subsequently centrifuged for 10 min at 12,000rcf at 4 °C. Supernatant was discarded and RNA pellet was washed in 500 μl of 75 % ethanol. Samples were vortexed briefly and centrifuged for 5 min at 7500rcf at 4 °C. Supernatant was removed and RNA was redissolved by air-drying the samples for 10 min and then resuspending the pellet in 20 μl of nuclease-free water. Thereafter, samples were incubated in a 60 °C heat block for 15 min. Concentrations were measured with BioTek Epoch microplate spectrophotometer and quality was ensured to have A260/A280 ≥ 1.9. RNA was stored at −80 °C until analysis.
Complementary DNA (cDNA) for mRNA was generated with 1 μg of total RNA per reaction using iScriptTM cDNA Synthesis Kit (Bio-Rad, Hercules, CA, USA) in accordance with the manufacturer’s protocol. Samples were diluted 1:5 for qPCR. cDNA was stored in −20 °C.
Complementary DNA (cDNA) for miRNA was generated with 5 ng of total RNA per reaction using TaqMan Advanced miRNA cDNA Synthesis Kit (Applied Biosystems, Carlsbad, CA, USA) in accordance with the manufacturer’s protocol. Samples were diluted 1:10 for qPCR. cDNA was stored at −20 °C.
2.4. Quantitative real-time polymerase chain reaction (qPCR)
Real-time/Quantitative PCR (qPCR) was conducted on the Quant Studio 7 Flex System (Applied Biosystems, Foster City, CA, USA) for 40 cycles with an initial hold stage at 95 °C for 20s, followed by 40 cycles of 1) denaturing step at 95 °C for 1 s and 2) annealing/extension step at 60 °C for 20s per cycle, with ramp up of 2.63 °C/s and ramp down of 2.42 °C/s. A melt curve was obtained after each qPCR reaction was completed.
To determine mRNA levels, PCR reaction with SYBR-Green fluorescent dye was performed in duplicate with 2 μl of 1:5 dilution of mRNA cDNA template in 5 μl SYBR Green Master Mix, 0.2 μl of 10 μM forward primer, 0.2 μl of 10 μM reverse primer, and 12.6 μl nuclease- free water, for a final volume of 20 μl following previous protocols (King et al., 2024; Ashiqueali, 2024; Avila, 2023; Hense et al., 2022). Primer sequences are listed in Tables 1 and 2. Primers were manufactured by Integrated DNA Technologies. Transcript levels were normalized to the housekeeping gene β-2-microglobulin (β2M) and calculated as discussed in the statistics section.
Table 1.
Mus musculus Primers used for qPCR of in vivo liver tissue samples.
| Gene | Forward (5′ → 3′) | Reverse (5′ → 3′) | Function |
|---|---|---|---|
| β2m | AAGTATACTCACGCCACCCA | CAGGCGTATGTATCAGTCTC | Housekeeping gene |
| Sirt1 | CAGTGAGAAAATGCTGGCCTA | TTGGTGGTACAAACAGGTATTGA | Energy sensor activated by high NAD+/NADH ratio; associates with PPARα to stimulate fatty acid oxidation |
| Akt2 | GAGGACCTTCCATGTAGACT | CTCAGATGTGGAAGAGTCAC | Serine/threonine kinase that regulates glucose uptake by insulin |
| Glut2 | GGTCAGAACTACCTCTCTTTG | GTGTGTGTGGAATTGTCCTC | Plasma membrane glucose sensor that promotes glucose secretion during gluconeogenesis |
| Srebp1c | AAGACATGCTCCAGCTCATC | TACGCACGTCGAACGAAACT | Transcription factor activated by insulin; induces de novo lipogenesis |
| Pparg | GTCAGTGTCGGTTTCAG | CAGATCAGCAGACTCTGGGT | Nuclear receptor that promotes lipid uptake |
| Acc | GTCCCCAGGGATGAACCAATA | GCCATGCTCAACCAAAGTAGC | Enzyme; converts acetyl-CoA to malonyl CoA, generates fatty acids (FA); malonyl CoA inhibits beta oxidation by preventing FA from associating with carnitine which hampers them from travelling to the mitochondria |
| Scd1 | GAGGCCTGTACGGGATCATA | TGAGAGAAGAAGAAGCCACGG | Enzyme activated by Srebp1c, converts saturated FA to monounsaturated FA which can be synthesized into triglycerides |
| Pparα | GGGAAAGACCAGCAACC | TGGAAGAATCGGACCTCTGC | Transcription factor that activates genes involved with mitochondrial beta-oxidation |
| Cpt1 | CTCAGTGGGAGCGACTCTTCA | GGCCTCTGTGGTACACGACAA | Enzyme that creates carnitine so fatty acids can enter mitochondria to produce ATP; inhibited by malonyl CoA |
| Mttp | CTCTTGGCAGTGCTTTTTCTCT | GAGCTTGTATAGCCGCTCATT | Transfers triglycerides onto Apob100 in the endoplasmic reticulum |
| Apob | CGTGGGCTCCAGCATTCTA | TCACCAGTCATTTCTGCCTTTG | Apolipoprotein responsible for binding lipids and shuttling VLDL to the Golgi |
| Sar1a | GGGCCGTTGTAAGCATCAATA | TTCCAGATTTCTTGTAGAGTCCTAGGA | COPII coat protein that enables VLDL transport to the Golgi apparatus |
| Sar1b | GCTAAAAAGGCAGGGCTATGG | GGCCGCTGCTAATCGATGTA | COPII coat protein that enables VLDL transport to the Golgi apparatus |
| Sec22b | CATTGATAGCCGTGCTCGGAG | CGCATCTTCAAGTACTTCGCATC | Component of VTV that enables ER to Golgi delivery of VLDL |
| Apoa1 | CCACACCCTTCAGGATGAAAG | TGGCTCCCTGTCAGGAAGAC | Major component of HDL; moves HDL to liver for reverse cholesterol transport |
| Apoa4 | CAGCTGACCCCATACATCCAG | TCATCGAGGTGTGCAGGTTG | Component of HDL that mediates reverse cholesterol transport |
| Lxr | TGCCATCAGCATCTTCTCTG | GGCTCACCAGCTTCATTAGC | Nuclear receptor that functions as a cholesterol sensor and activates ATP-binding cassette transporters |
| Abcg5 | ATCCAACACCTCTATGCTAAATCAC | TACATTATTGGACCAGTTCAGTCAC | Activated by LXR and stimulates cholesterol efflux; transports cholesterol into the bile |
| Cyp7a1 | CCAGGGAGATGCTCTGTGTTC | ACCCAGACAGCGCTCTTTGAT | Enzyme that modulates cholesterol catabolism by generating bile acids |
Table 2.
Human primer sequences used for qPCR of samples from HEPG2 cells.
| Gene | Forward | Reverse |
|---|---|---|
| β2M | 5′ – GAGTATGCCTGCCGT – 3′ | 5′ – CGGCATCTTCAAACCTCCAT – 3′ |
| SIRT1 | 5′ – CTTCAGGTCAAGGGATGGTAT – 3′ | 5′ – GCGTGTCTATGTTCTGGGTAT – 3′ |
| AMPKa | 5′ – TGCGTGTACGAAGGAAGAATCC – 3′ | 5′ – TGTGACTTCCAGGTCTTGGAGTT – 3′ |
| SCD1 | 5′ – GCAGGACGATATCTCTAGCT – 3′ | 5′ – GTCTCCAACTTATCTCCTCCATTC – 3′ |
| PPARα | 5′ – TGAACGATCAAGTGACATTGC – 3′ | 5′ – TGGGAAGAGAAAGATATCG – 3′ |
| CPT1 | 5′ – TGAGGTGCTCTCGGAACCCT – 3′ | 5′ – CTCGCCTGCAATCATGTAGG – 3′ |
| APOB | 5′ – GGAGCTGCTGGACATTGCTA – 3′ | 5′ – ATGGCAGCTTTCTGGATCAT – 3′ |
| STX5A | 5′ – GGCTCCATCTTTCAGCAGTTGG – 3′ | 5′ – GTTGGAGGTGACAGACTGGAAG – 3′ |
| APOA1 | 5′ – CCCTGGGATCGAGTGAAGGA – 3′ | 5′ – CTGGGACACATAGTCTCTGCC – 3′ |
| APOE | 5′ – TGTCTGAGCAGGTGCAGGAG – 3′ | 5′ – TCCAGTTCCGATTTGTAGG –3′ |
| LXR | 5′ – AAGCCCTGCATGCCTACGT – 3′ | 5′ – TGCAGACGCAGTGCAAACA – 3′ |
| ABCG5 | 5′ – CCCAAGGGACTCCGGGGTCA – 3′ | 5′ – GACCCATGGACCCTCCGGGG – 3′ |
To quantify miRNA levels, real-time PCR was conducted with TaqMan Fast Advanced Master Mix (Applied Biosystems) and was performed in duplicate with 2.5 μl of 1:10 diluted miRNA cDNA template in 5 μl of 2× TaqMan Fast Advanced Master Mix, 0.5 μl of 20× miRNA primer (TaqMan Applied Biosystems), and 2 μl of nuclease-free water, for a final volume of 10 μl. The expression level of experimental miRNA was normalized to reference gene miR-16. The delta-delta Ct (2−ΔΔCt) method, explained in the statistical analysis section, was employed to determine the relative expression of each target of interest.
2.5. Statistical analysis
To analyze the relative expression of each gene we used the delta-delta Ct (2−ΔΔCt) method equation:
| (1) |
where Ct(experimental gene first sample) is the cycle of threshold (Ct) of the gene of interest of the first control sample (constant for each 96-well plate); Ct(experimental gene) is the Ct of the gene of interest of the analyzed sample; Ct(reference gene first control sample) is the Ct of the housekeeping gene in the first control sample (constant for each 96-well plate); Ct(reference gene) is the Ct of the housekeeping gene of the analyzed sample. Thereafter, the values of the N-AL group were averaged, and this number was used in the equation 2−ΔΔCt/average (2−ΔΔCt control sample) to calculate fold changes. Lastly, fold change outputs were averaged to calculate relative expression and this data was used to determine the effect of treatment and genotype on gene expression (Masternak et al., 2005).
Statistical analysis for tissue samples was performed by using two-way analysis of variance (2-way ANOVA) and Tukey’s post hoc multiple comparison of means. For PCR data from cells, t-test with Welch’s correction was performed. Outliers were detected via the Tukey interquartile rule, where values <1.5 times the first quartile or >1.5 times the third quartile are considered aberrations in the dataset. Significance was defined as p < 0.05 and all results represent mean ± standard error of the mean (SEM). Statistics were performed using Sigma Plot v14.0 (Systat Software, Palo Alto, CA, USA).
To evaluate the contribution of each gene to the overall difference among the different treatments and animal types, we performed principal component analysis (PCA) and visualized the results on PCA biplots. Statistical analyses for PCA biplot was performed using Stata MP 15 (StataCorp LLC, 2019) and R packages. All tests were two-tailed with a significance level of α (type I error) < 0.05.
3. Results
3.1. miRNA profiling reveals downregulation of candidate miR-34a in the livers of calorie restricted normal mice and long-living Ames dwarf mice
Through extensive literature and bioinformatics analysis, we identified miRNAs associated with fatty liver and evaluated their response to CR. Of the miRNAs that were studied, both miR-34a and miR-149 were most significantly downregulated in the livers of CR mice and df/df mice fed AL (Fig. 1, Table 3). Overall effects of 2-way ANOVA showed significance for genotypic (p < 0.01; p < 0.001, respectively), dietary (p < 0.01), and interactive terms, with less interactive effects noted between genotype and diet for miR-34a (p < 0.05) than for miR-149 (p < 0.001).
Fig. 1.
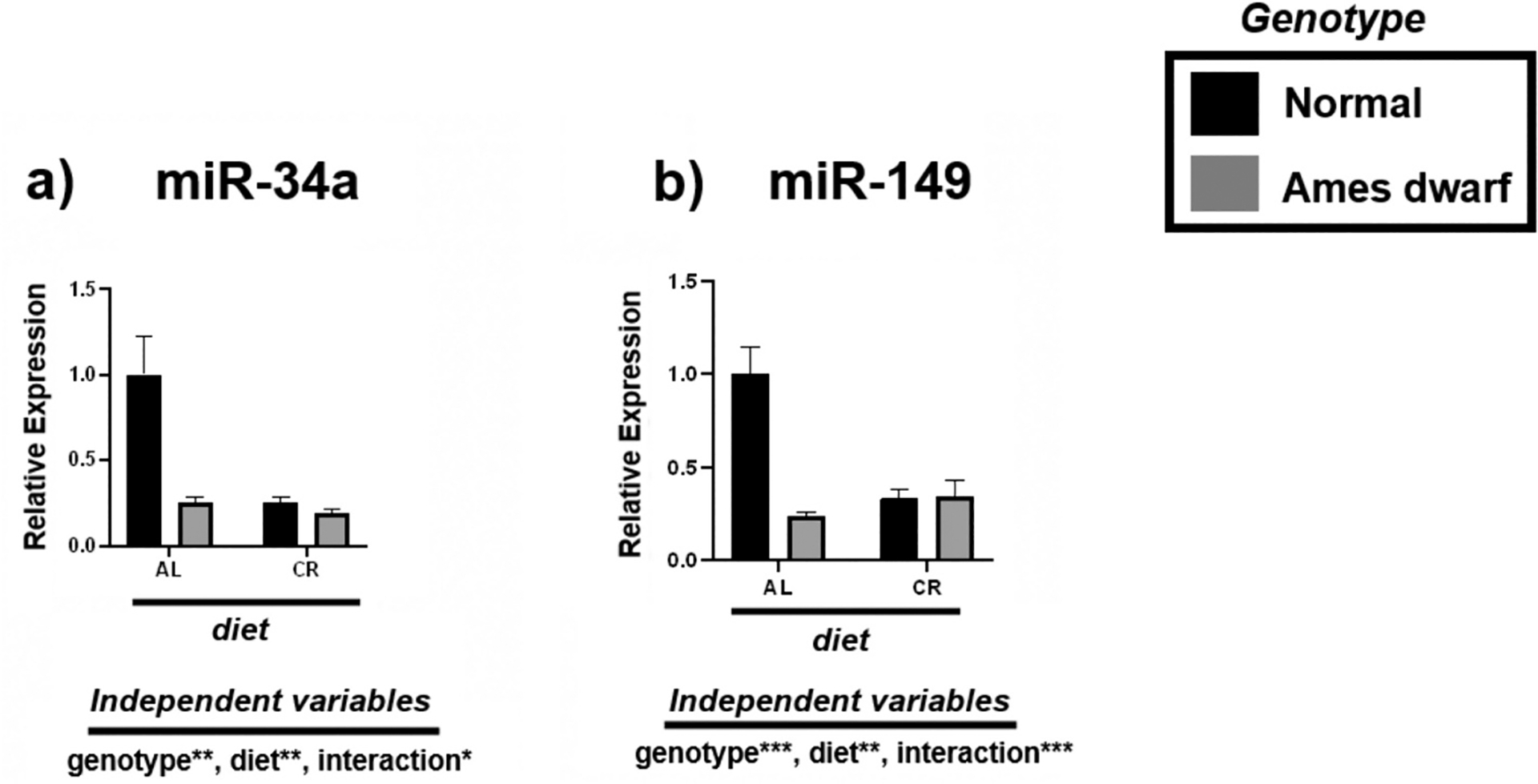
Expression of metabolism-associated miRNAs in the liver regulated by genotype (normal or Ames dwarf) and treatment (ad libitum (AL) or caloric restriction (CR)), with y-axis equal to miRNA expression relative to control miR-16, with 8–11 biological replicates. Two-way analysis of variance (ANOVA) was performed with Tukey’s post hoc multiple comparison test using the SigmaPlot v14.0 software. (*) p < 0.05; (**) p < 0.01; (***) p < 0.001; (****) p < 0.0001.
Table 3.
Tukey pairwise comparisons for Fig. 1.
| miR-34a | mirR-149 | |
|---|---|---|
| AL:Normal vs. AL:Ames dwarf | 0.0016 | <0.0001 |
| AL:Normal vs. CR:Normal | 0.0006 | <0.0001 |
| AL:Normal vs. CR:Ames dwarf | 0.0004 | 0.0001 |
| AL:Ames dwarf vs. CR:Normal | >0.9999 | 0.8729 |
| AL:Ames dwarf vs. CR:Ames dwarf | 0.9872 | 0.8756 |
| CR:Normal vs. CR:Ames dwarf | 0.9845 | >0.9999 |
3.2. Calorie restriction promotes Sirt1 activity and inhibits de novo lipogenesis
We investigated the expression of energy sensor Sirt1 and downstream de novo lipogenesis associated mRNAs including Cd36, Srebp1, Pparg, and Acc (Fig. 2). Sirt1 activity was upregulated in both N-CR mice and df/df mice regardless of diet with an overall significant genotypic (p < 0.05) and dietary (p < 0.01) effect noted by 2-way ANOVA (Fig. 2a). Interestingly, Cd36 was upregulated in both df/df-AL mice and df/df-CR mice with an overall effect of genotype evident (p < 0.0001) (Fig. 2b). CR appeared to suppress Srebp1 expression in both N and df/df mice (p < 0.01) providing an example of dietary, and not genotypic, mediated improvement to liver lipid trafficking homeostasis (Fig. 2c). Alongside, CR also mediated effects on Pparg in df/df mice with additive effects between genotype and diet evident (p < 0.0001) and contributing to increased expression of this mRNA (Fig. 2d, Table 4). Lastly, the expression of Acc was downregulated in both N-CR and df/df mice regardless of diet, with overall significance attributed to genotype (p < 0.05) (Fig. 2e).
Fig. 2.
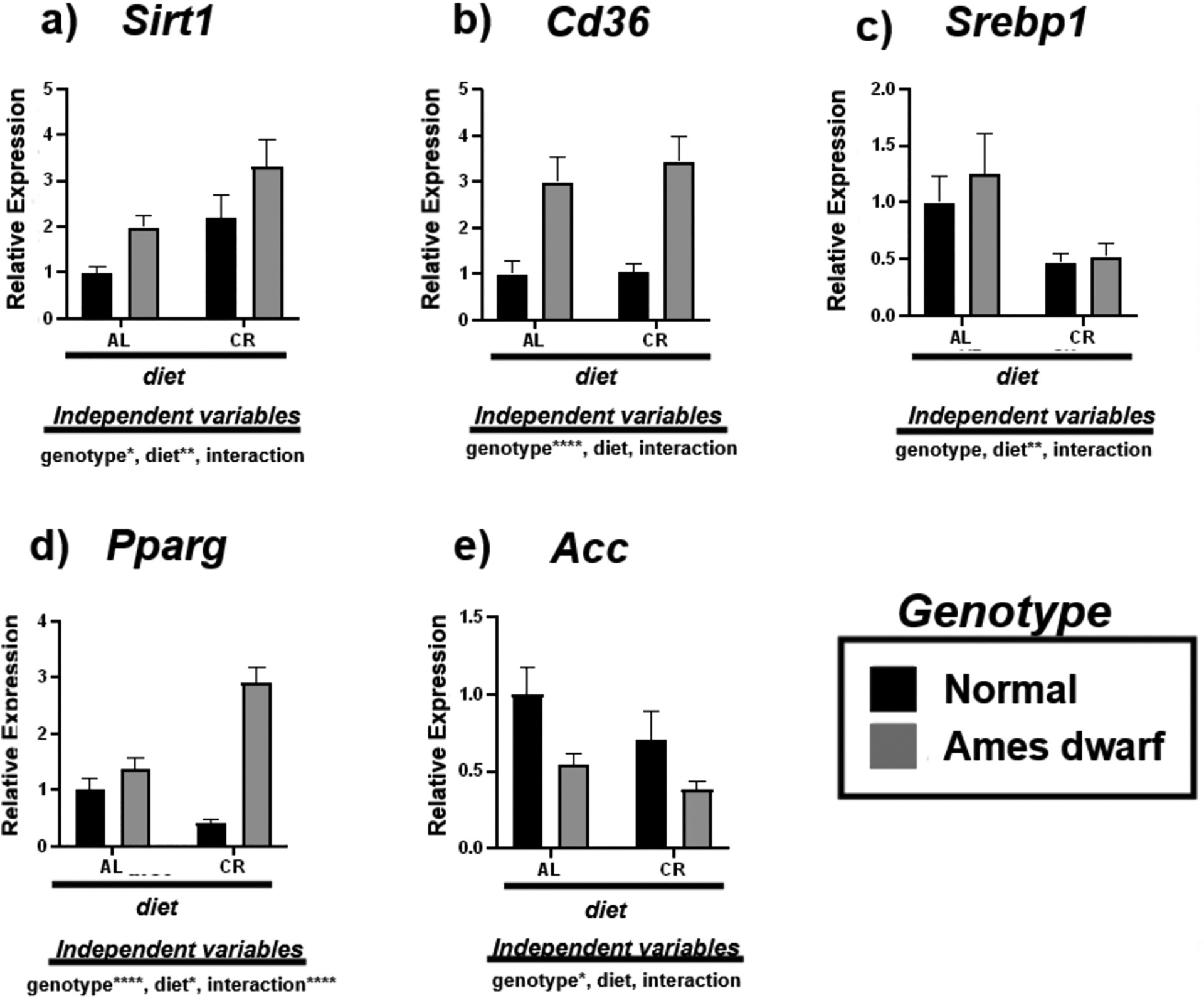
Expression of Sirt1 and de novo lipogenesis associated mRNAs in the liver regulated by genotype (normal or Ames dwarf) and treatment (ad libitum (AL) or caloric restriction (CR)), with y-axis equal to mRNA expression relative to control beta-2-microglobulin (β2M), with 8–11 biological replicates. Two-way analysis of variance (ANOVA) was performed with Tukey’s post hoc multiple comparison test using the SigmaPlot v14.0 software. (*) p < 0.05; (**) p < 0.01; (***) p < 0.001; (****) p < 0.0001.
Table 4.
Tukey pairwise comparisons for Fig. 2.
| Sirt1 | Cd36 | Srebp1 | Pparg | Acc | |
|---|---|---|---|---|---|
| AL:Normal vs. AL:Ames dwarf | 0.3033 | 0.0037 | 0.8532 | 0.5319 | 0.2181 |
| AL:Normal vs. CR:Normal | 0.1237 | 0.9998 | 0.2793 | 0.1096 | 0.4767 |
| AL:Normal vs. CR:Ames dwarf | 0.0012 | 0.0004 | 0.4549 | <0.0001 | 0.043 |
| AL:Ames dwarf vs. CR:Normal | 0.9817 | 0.0045 | 0.0844 | 0.0067 | 0.8989 |
| AL:Ames dwarf vs. CR:Ames dwarf | 0.1291 | 0.8675 | 0.171 | <0.0001 | 0.9171 |
| CR:Normal vs. CR:Ames dwarf | 0.2068 | 0.0004 | 0.9986 | <0.0001 | 0.4856 |
3.3. Calorie restriction and df/df genotype influence insulin signaling and fatty acid oxidation
We analyzed the expression of mRNAs involved in the process of insulin signaling (Glut2, Akt2) and fatty acid oxidation (Pgc1α, Pparα, Cpt1α) to further assess liver metabolic function in response to CR (Fig. 3). Glut2 was upregulated in response to genotype (p < 0.01) and CR (p < 0.05), with Akt2 significantly correlating with the df/df genotype (p < 0.01) (Fig. 3a–b). Alongside, we observed the effects of CR and df/df genotype on genes that regulate fatty acid oxidation. Our Tukey pairwise comparison results show an increase in Pgc1α in the N-CR mice (p = 0.0222) relative to the N-AL mice (Table 5) with 2-way ANOVA analysis revealing overall significance due to dietary regimen (p < 0.01) (Fig. 3c). In the df/df mice, Tukey pairwise analysis reveals that genotype and diet appear to influence the expression of Pgc1α in an additive, yet noninteractional manner (p = 0.0549) (Table 5). Additionally, we found no effects on Pparα due to CR of df/df genotype (Fig. 3d). CR upregulated the expression of Cpt1α in the livers of N-CR mice relative to N-AL mice (p = 0.006) (Table 5). 2-way ANOVA demonstrates interactive effects between CR and genotype (p < 0.05), suggesting both independent variables may have overlap in influencing the organism’s aerobic energy sources (Fig. 3e).
Fig. 3.
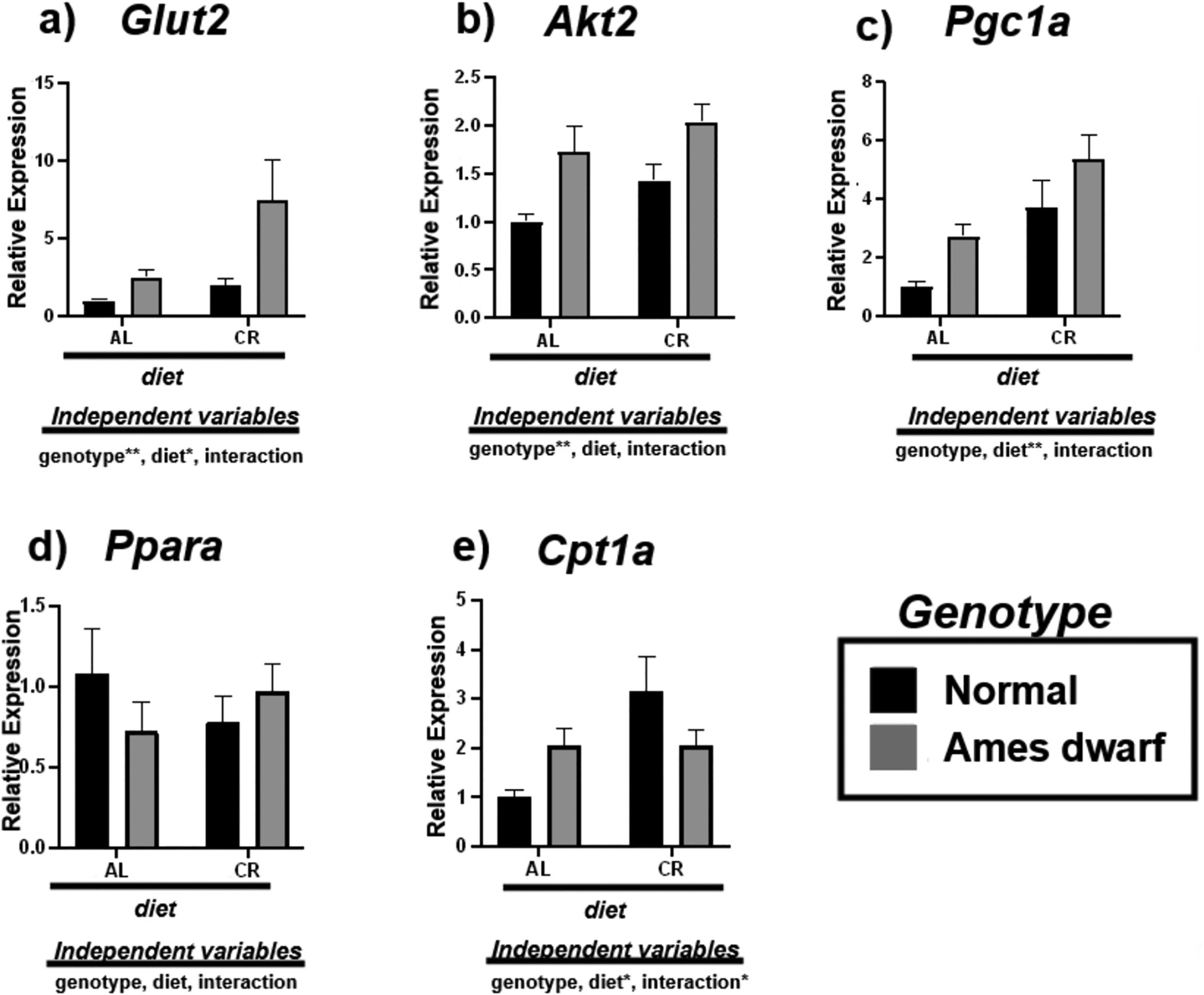
Expression of insulin associated (a,b) and fatty acid oxidation (c-e) mRNAs in the liver regulated by genotype (normal or Ames dwarf) and treatment (ad libitum (AL) or caloric restriction (CR)), with y-axis equal to mRNA expression relative to control beta-2-microglobulin (β2M), with 8–11 biological replicates. Two-way analysis of variance (ANOVA) was performed with Tukey’s post hoc multiple comparison test using the SigmaPlot v14.0 software. (*) p < 0.05; (**) p < 0.01; (***) p < 0.001; (****) p < 0.0001.
Table 5.
Tukey pairwise comparisons for Fig. 3.
| Glut2 | Akt2 | Pgc1a | Ppara | Cpt1a | |
|---|---|---|---|---|---|
| AL:Normal vs. AL:Ames dwarf | 0.8438 | 0.041 | 0.2727 | 0.6509 | 0.3755 |
| AL:Normal vs. CR:Normal | 0.9339 | 0.3088 | 0.0222 | 0.7155 | 0.006 |
| AL:Normal vs. CR:Ames dwarf | 0.0049 | 0.0028 | 0.0002 | 0.985 | 0.4126 |
| AL:Ames dwarf vs. CR:Normal | 0.9914 | 0.6532 | 0.7497 | 0.9973 | 0.346 |
| AL:Ames dwarf vs. CR:Ames dwarf | 0.0585 | 0.6671 | 0.0549 | 0.8718 | >0.9999 |
| CR:Normal vs. CR:Ames dwarf | 0.0166 | 0.1159 | 0.3008 | 0.9239 | 0.3768 |
3.4. Calorie restriction influences VLDL transport in the Ames dwarf liver
We investigated the effect of CR on the VLDL trafficking mRNAs Apob, Sar1α, Sar1b, Cideb, Sec22b, and Stx5α in the livers of N and df/df mice (Fig. 4, Table 6). The df/df genotype upregulates the expression of Apob (p < 0.05) while diet appears to upregulate the expression of Cideb (p < 0.05) (Fig. 4a, d). Despite this assumed biogenesis of VLDL, CR appears to facilitate VLDL maturation by promoting the expression of COPII protein Sar1α (p < 0.05) (Fig. 4b). No effect was observed on Sar1b (Fig. 4c). This is further supported by the upregulation of vesicle membrane SNARE (v-SNARE) Sec22b due to interactive effects of diet and genotype (p < 0.05) along with VTV-Golgi fusion protein Stx5α due to diet (p < 0.05) in the df/df-CR mice (Fig. 4e–f).
Fig. 4.
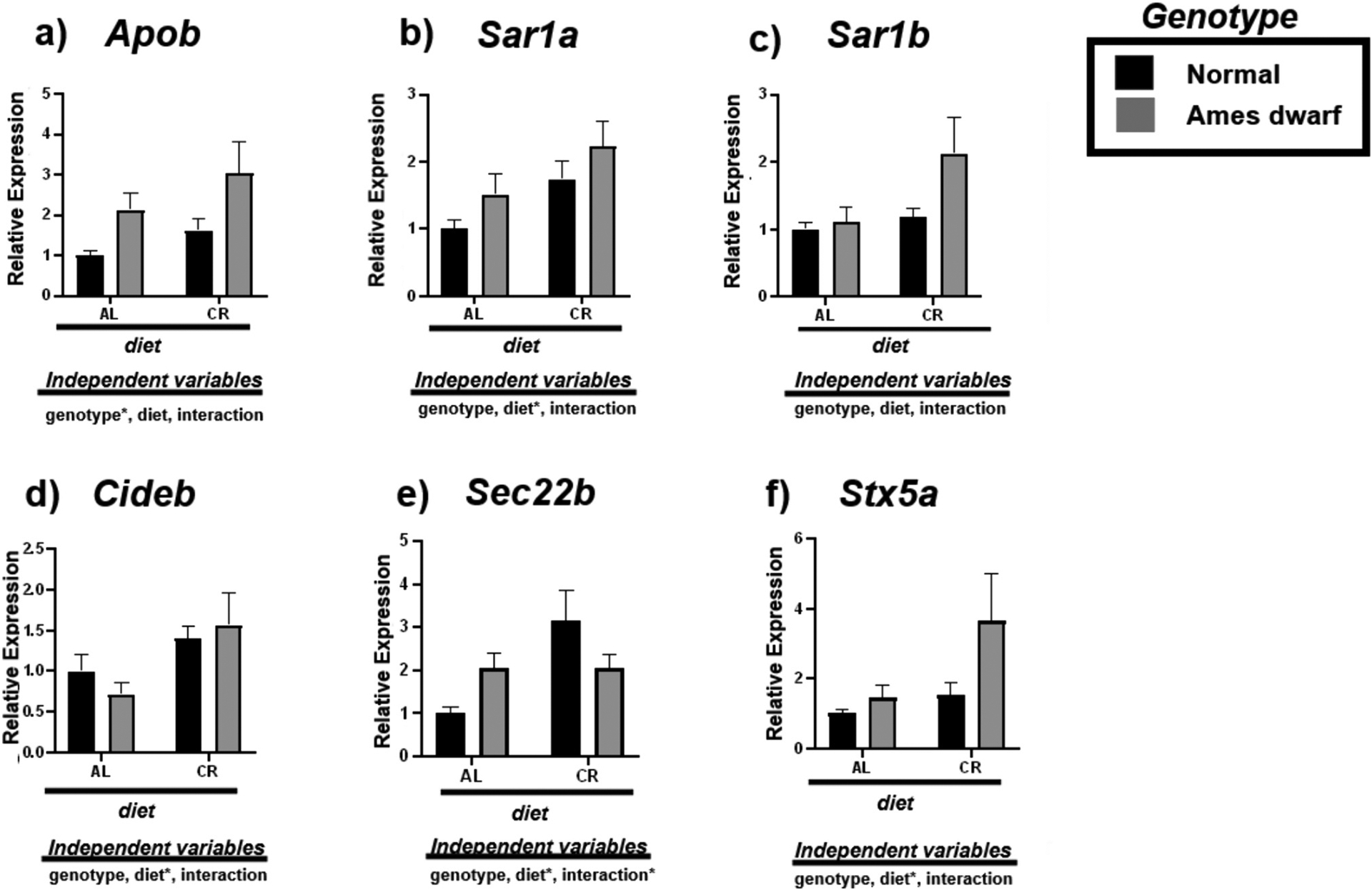
Expression of very-low-density lipoprotein transport associated mRNAs in the liver regulated by genotype (normal or Ames dwarf) and treatment (ad libitum (AL) or caloric restriction (CR)), with y-axis equal to mRNA expression relative to control beta-2-microglobulin (β2M), with 8–11 biological replicates. Two-way analysis of variance (ANOVA) was performed with Tukey’s post hoc multiple comparison test using the SigmaPlot v14.0 software. (*) p < 0.05; (**) p < 0.01; (***) p < 0.001; (****) p < 0.0001.
Table 6.
Tukey pairwise comparisons for Fig. 4.
| Apob | Sar1a | Sar1b | Cideb | Sec22b | Stx5a | |
|---|---|---|---|---|---|---|
| AL:Normal vs. AL:Ames dwarf | 0.3018 | 0.59 | 0.9931 | 0.8488 | 0.4689 | 0.962 |
| AL:Normal vs. CR:Normal | 0.7275 | 0.2158 | 0.97 | 0.5881 | 0.3322 | 0.94 |
| AL:Normal vs. CR:Ames dwarf | 0.0163 | 0.0195 | 0.0417 | 0.3486 | 0.6049 | 0.0364 |
| AL:Ames dwarf vs. CR:Normal | 0.8464 | 0.9408 | 0.9985 | 0.2001 | 0.9993 | 0.9999 |
| AL:Ames dwarf vs. CR:Ames dwarf | 0.5271 | 0.3443 | 0.1025 | 0.0999 | 0.0765 | 0.1328 |
| CR:Normal vs. CR:Ames dwarf | 0.1298 | 0.6064 | 0.123 | 0.9619 | 0.0387 | 0.1333 |
3.5. Calorie restriction and df/df genotype influence reverse cholesterol transport
To examine cholesterol homeostasis, we assessed the expression of mRNAs involved with modulating reverse cholesterol transport including Apoa1, Apoa4, Apoe, Lxr, Abcg5, and Cyp7a1 (Fig. 5). Interestingly, df/df mice appear to be characterized by less Apoa1 (p < 0.001) and Apoa4 (p < 0.01) (Fig. 5a–b). However, Apoe expression was significantly upregulated due to CR in both N and df/df mice (p < 0.05) (Fig. 5c). Alongside, CR appears to stimulate Lxr (p < 0.05) (Fig. 4d), inducing upregulation of Abcg5 in the livers of both N-CR compared to N-AL mice (p = 0.0497) and df/df-CR relative to df/df-AL mice (p = 0.0002) (Table 7). 2-way ANOVA further reveals that diet remarkably contributes to this evident increase in expression of Abcg5 (p < 0.0001) (Fig. 5e). Lastly, Cyp7a1 is upregulated due to both df/df genotype (p < 0.05) and CR (p < 0.05) indicating reduced cholesterol deposition in the liver (Fig. 5f).
Fig. 5.
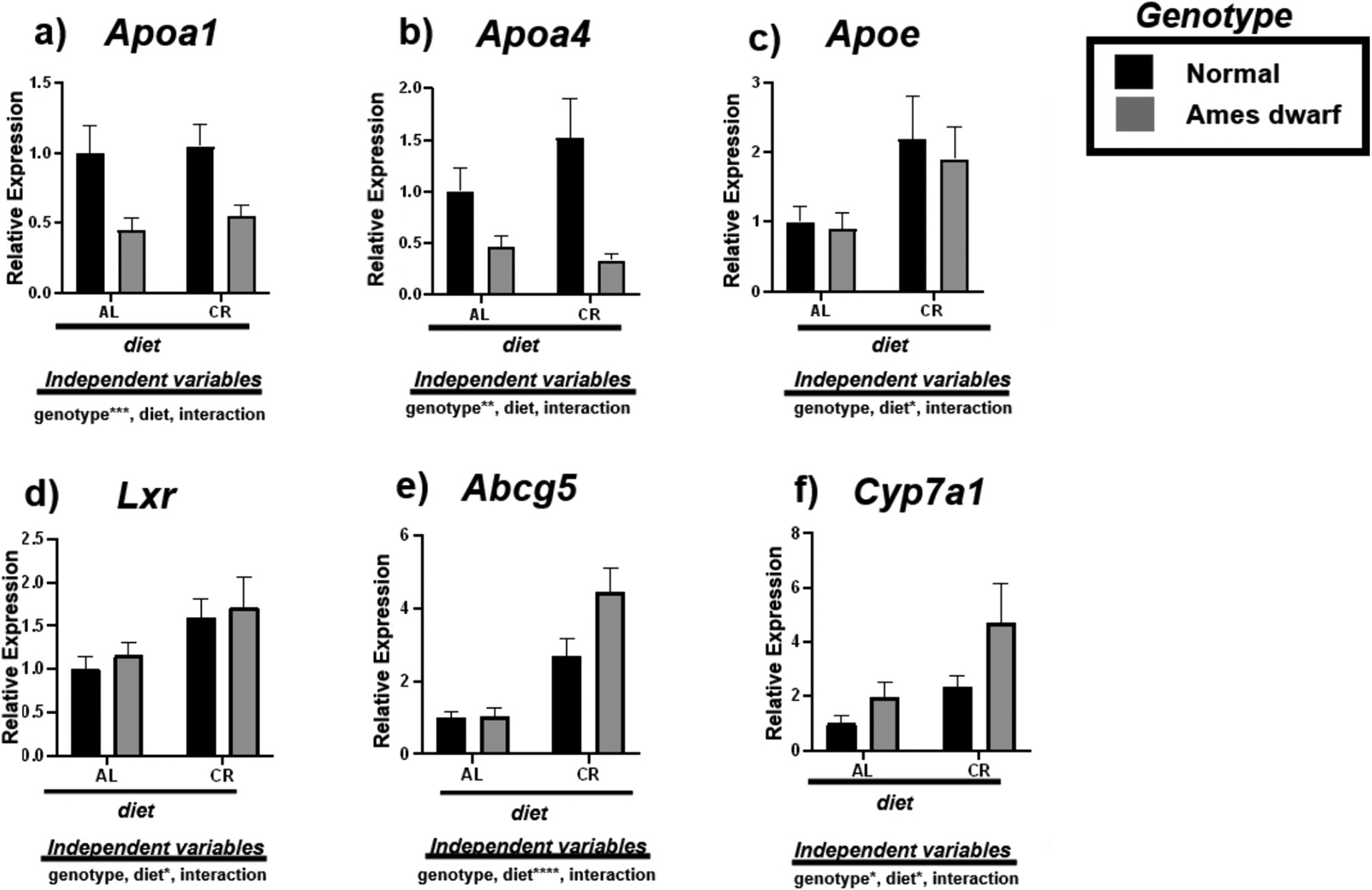
Expression of reverse cholesterol transport associated mRNAs in the liver regulated by genotype (normal or Ames dwarf) and treatment (ad libitum (AL) or caloric restriction (CR)), with y-axis equal to mRNA expression relative to control beta-2-microglobulin (β2M), with 8–11 biological replicates. Two-way analysis of variance (ANOVA) was performed with Tukey’s post hoc multiple comparison test using the SigmaPlot v14.0 software. (*) p < 0.05; (**) p < 0.01; (***) p < 0.001; (****) p < 0.0001.
Table 7.
Tukey pairwise comparisons for Fig. 5.
| Apoa1 | Apoa4 | Apoe | Lxr | Abcg5 | Cyp7a1 | |
|---|---|---|---|---|---|---|
| AL:Normal vs. AL:Ames dwarf | 0.0622 | 0.4542 | 0.9985 | 0.9723 | >0.9999 | 0.8415 |
| AL:Normal vs. CR:Normal | 0.9947 | 0.4255 | 0.1948 | 0.2821 | 0.0497 | 0.6256 |
| AL:Normal vs. CR:Ames dwarf | 0.1394 | 0.2402 | 0.4716 | 0.1729 | <0.0001 | 0.0123 |
| AL:Ames dwarf vs. CR:Normal | 0.0323 | 0.0287 | 0.2053 | 0.6055 | 0.0947 | 0.9892 |
| AL:Ames dwarf vs. CR:Ames dwarf | 0.9699 | 0.984 | 0.4542 | 0.4282 | 0.0002 | 0.1187 |
| CR:Normal vs. CR:Ames dwarf | 0.0772 | 0.0088 | 0.9666 | 0.9799 | 0.0433 | 0.1714 |
3.6. Principal Component Analysis (PCA) biplot demonstrates overlapping ellipses for calorie restricted mice and long-living Ames dwarf mice
PCA analysis demonstrated that PCA1 (treatment) contributed to 39.2 % of variance while PCA2 (genotype) contributed to 11.6 % of variance (Fig. 6). Both the N-CR and df/df-CR groups overlap with df/df-AL group but not with the N-AL group (Fig. 6). Alongside, df/df AL mice exhibit a wide, horizontal ellipse across the x-axis while df/df CR mice exhibit a narrower ellipse rotating to the left (Fig. 6).
Fig. 6.
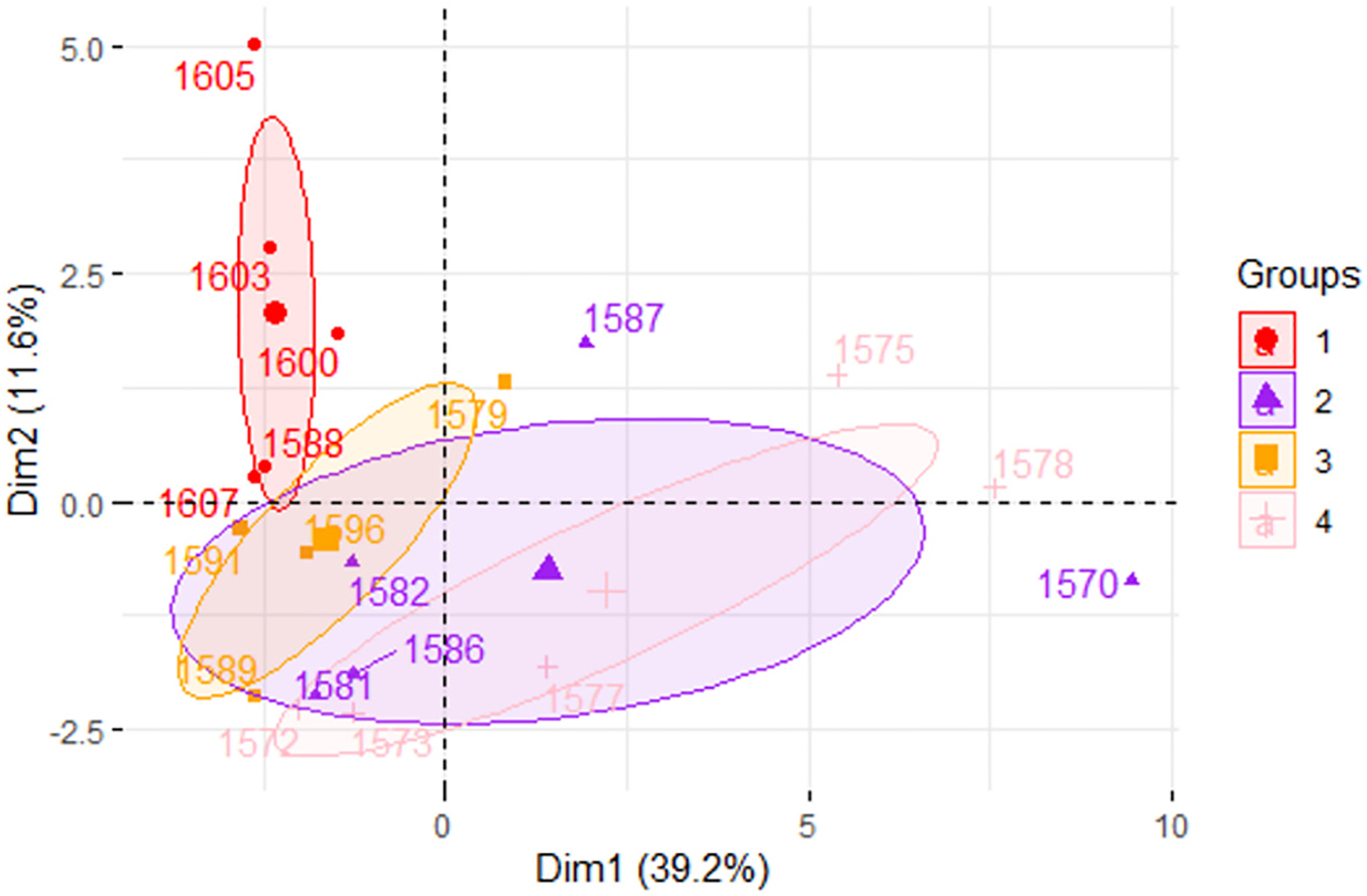
Principal component analysis biplot demonstrating the effects of dietary treatment (PCA1) and genotype (PCA2). Group 1 (red) represents normal mice under AL diet. Group 2 (purple) represents Ames dwarf mice under AL diet. Group 3 (orange) represents normal mice under CR diet. Group 4 (light pink) represents Ames dwarf mice under CR diet.
3.7. Transfection of HepG2 cells with candidate miR-34a mimic reveals dysregulation of fatty acid oxidation and cholesterol transport
After selecting miR-34a as a candidate biomarker, we investigated if this molecule is directly involved in the regulation of SIRT1, AMPKА, PPARА, and downstream lipid metabolic genes (Fig. 7). Transfection efficiency was assessed by qPCR and indicated that there was significant upregulation of miR-34a after 48-h of transfection with mimic (p < 0.0001) (Fig. 7a). Surprisingly, transfection with miR-34a mimic displayed no significant change in the expression of energy sensors SIRT1 and AMPKА although a decreasing trend was noted (Fig. 7b–c). However, there was significant downregulation of direct target PPARА (p < 0.01) (Fig. 7d). Interestingly, reduced de novo lipogenesis was evident by significantly decreased SCD1 levels between control and mimic cells (p < 0.01) (Fig. 7e). Furthermore, less fatty acid oxidation was also observed with significantly decrease in expression of CPT1А (p < 0.05) (Fig. 7f). A significant decrease in APOB (p < 0.05) was seen alongside a significant decrease in SCD1 (p < 0.01) (Fig. 7g). Alongside, there was also a decrease in COPII transport and fusion protein STX5А (p < 0.01) (Fig. 7h). Lastly, there was a significant decrease in expression of high-density lipoprotein (HDL) component APOA1 (P < 0.01), lipid transporter APOE (p < 0.01), cholesterol sensor LXR (p < 0.05), and cholesterol transporter ABCG5 (p < 0.05) (Fig. 7i–l).
Fig. 7.
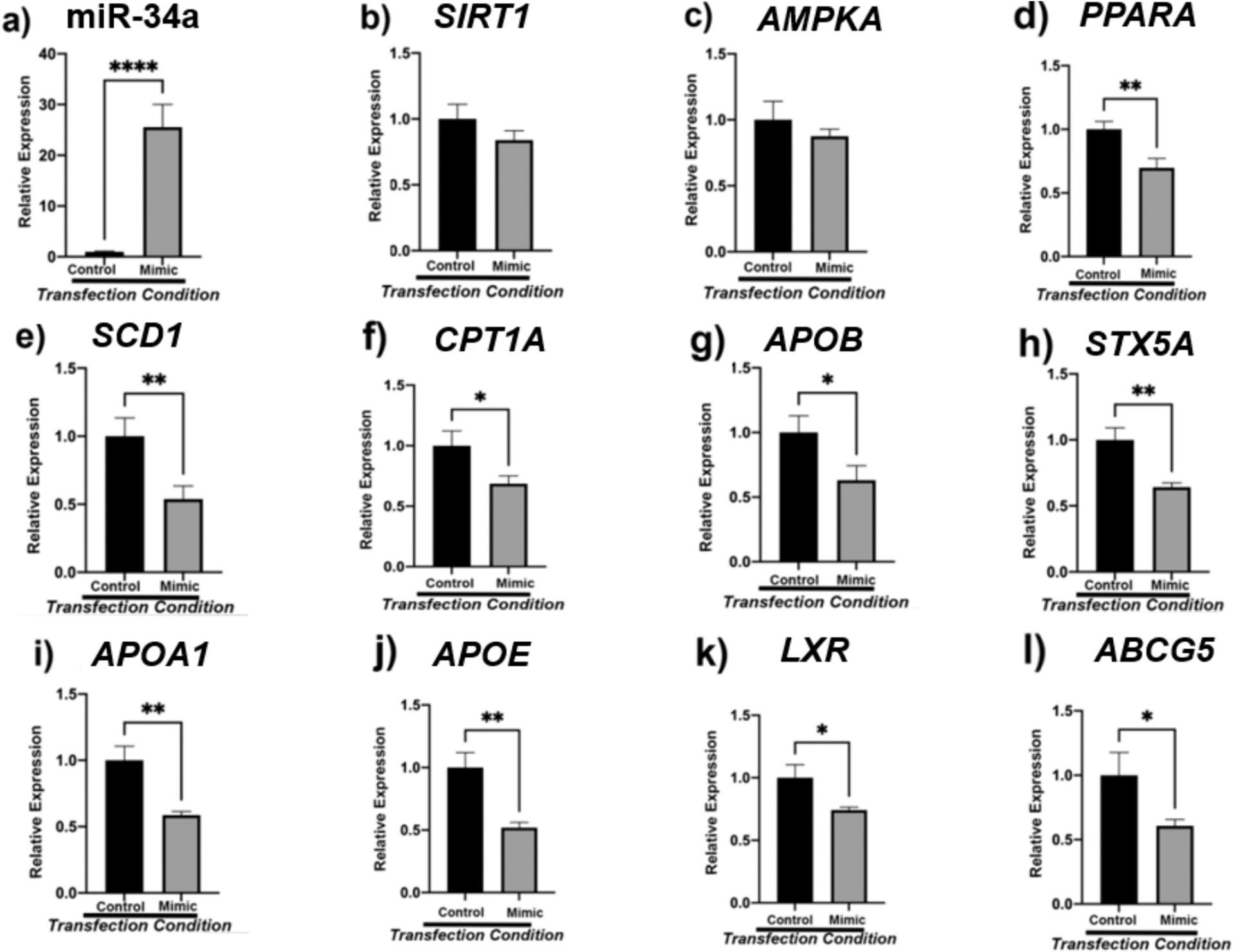
Human hepatocellular carcinoma cells (HEPG2) were transfected with 5 nM miR-34a for 48 h. miR-34a transfection efficiency was verified via qPCR (7a). Thereafter, the expression of energy sensing (7b-c), lipogenesis associated (7e), fatty acid oxidation (7d, 7f), lipid trafficking (7 g-h), and reverse cholesterol transport (7i-l) mRNAs was assessed. β2M served as internal control, with n = 16 biological replicates for both control and mimic groups. One-way analysis of variance (ANOVA) was performed with Welch’s correction using the SigmaPlot v14.0 software. (*) p < 0.05; (**) p < 0.01; (***) p < 0.001; (****) p < 0.0001.
4. Discussion
NAFLD is a multifactorial condition that is influenced by the consequences of nutritional overload which include imbalance between triglyceride retention and excretion. The absence of drug therapies to help combat NAFLD is a major disadvantage as adherence to lifestyle modifications such as diet and exercise is ill-fated with only 50 % compliance rate (Arora et al., 2019). Therefore, understanding the epigenetic regulation of CR on miRNAs and downstream metabolic processes may lead to the discovery of novel remedies that could curtail the pathogenesis of this idiopathic disease. In the present study, we investigated the effects of CR on metabolic signaling pathways in the livers of long-living df/df mice and thereby aimed to elucidate the mechanisms by which both diet and genotype impact the progression of this age-associated illness.
Several studies have demonstrated that individuals with obesity are 3.5 times more likely to develop NAFLD and are more vulnerable to other chronic age-related diseases (Romero-Gomez et al., 2023). As such, nutritional strategies offer a promising avenue to modulate liver health and overall vitality. The human body relies on dietary lipids, carbohydrates, and proteins as a source of fuel for cellular functions (Rui, 2014). Specifically, these macromolecules are broken down into their chemical building blocks by the digestive system and subsequently absorbed into the blood stream (Rui, 2014). This nutrient-rich blood, containing fatty acids, glucose, and amino acids, is transported to the liver via the hepatic portal vein and expended during cellular respiration (Rui, 2014). Thus, the liver is a vital metabolic instrument that plays a central role in controlling energy balance (Rui, 2014).
Due to fluctuations in nutrient availability during the anabolic and catabolic states, different oxidation pathways are made readily available to fuel the energy demands of cells. To elaborate, the consumption of food triggers postprandial glucose abundance thereby eliciting insulin secretion (Sanders and Griffin, 2016). Insulin accelerates the absorption of blood glucose by the plasma membrane glucose transporters (Glut2) of the hepatocytes and encourages the synthesis of glycogen and fatty acids (Sanders and Griffin, 2016). Leftover glucose is shuttled to the peripheral tissues to enable various cellular functions. Meanwhile, during the post-absorptive state glucagon induces lipolysis and gluconeogenesis due to limited glucose reservoirs. Fatty acid oxidation is the preferred route of energy production and allows the cells to carry out their life-sustaining functions (Finley and Haigis, 2009; Rui, 2014).
The accumulation of excess adipose deposits due to hypercaloric diet impairs key metabolic signaling pathways and predisposes individuals to imminent liver injury (Cani et al., 2007). To elaborate, the etiology of NAFLD is attributed to long-term overnutrition which subsequently induces de novo lipogenesis, mitochondrial dysfunction, compromised very low-density lipoprotein (VLDL) assembly and maturation, and diminished cholesterol efflux provoking inflammation, oxidative stress, and damage in the endoplasmic reticulum (ER) (Rui, 2014; Vacca et al., 2015). Metabolic reprogramming through dietary modifications can induce potent epigenetic changes to extend lifespan and improve healthspan.
In particular, the etiology of NAFLD is associated with the over-consumption of carbohydrates and fats (Kargulewicz et al., 2014). This results in anabolic signaling and subsequent alterations in nutrient sensing pathways including the activation of mammalian target of rapamycin (Mtor), a transcript known to accelerate cellular aging (Melnik, 2012; Howell and Manning, 2011). Several studies have shown that the Western diet, rich in processed foods and refined sugars, can stimulate the Igf1-Mtor-Srebp1 signal transduction pathway thereby inducing lipogenesis and shortening lifespan (Fekete et al., 2022; Melnik et al., 2011).
To mitigate the effects of aging, different dietary interventions such as the Mediterranean diet, the Okinawan (centenarian) diet, intermittent fasting, and the ketogenic diet have been largely explored, with these approaches able to improve cellular health, support healthy aging, and extend lifespan (Fekete et al., 2022). Therefore, CR is a potential cure-all that could counteract the detrimental effects of nutritional overload by mimicking fasting, inducing the hydrolysis of triglycerides, and influencing the expression of powerful lipid trafficking genes and proteins.
miRNAs have promising properties to be therapeutic owing to their small size and are powerful endogenous biomarkers with the capacity to broadly regulate physiology dependent on the sequence of mRNA they complement (O’Brien et al., 2018). They work by silencing post-transcriptional gene expression and play an important role in regulating several physiological processes including cell metabolism (O’Brien et al., 2018). miRNA profiling in the livers of AL and CR animals revealed significant downregulation of miR-34a in N-CR, df/df-AL, and df/df-CR mice suggesting the overlapping yet different mechanisms by which diet and genotype influence lipid trafficking, evidenced by significant interaction terms for some genes, gene alignment along both axes, and the shift of the centroid/ellipse-size between treatment groups. In particular, df/df-AL mice depicted a wide ellipse across the x-axis that was narrowed and shifted vertically after 30 % CR, demonstrating the potential combined influence of both genetics and dietary intervention on regulating liver health and homeostasis. Alongside N-CR mice also exhibited an ellipse that shifted across the x-axis and that did not overlap with the N-AL animals but that did overlap with the df/df-AL animals, showing that nutritional intervention can have impacts on liver health. Further, literature and bioinformatic search suggested that this miRNA has been implicated in fatty liver by allegedly downregulating the expression of Sirt1, Ampk, and Pparα ultimately resulting in increased DNL and impaired VLDL transport, FAO, and RCT (Li et al., 2011; Ding et al., 2015; Li et al., 2015; Xu et al., 2021).
The significance of the interaction term for miR-149 suggests this miRNA has overlapping roles to regulate both the Ames dwarf and calorie restricted liver, while miR-34a has weak significance for the interaction term, which may suggest less overlap in the mechanism of CR and genotype on liver metabolism and homeostasis. miR-34a appears to be regulated by both genotype and diet; namely, while CR significantly downregulated the expression of miR-34a in the liver, Ames dwarf mice had a significant reduction in this microRNA regardless of dietary regimen suggesting that there are different mechanisms by which CR and genotype influence the expression of this miRNA. Studying the difference in mechanism by which diet and Ames dwarf genotype influence liver homeostasis may provide insights for preventing age-related diseases and is why we chose to further investigate miR-34a.
Sirt1 was studied due to its role as a major regulator and sensor of energy metabolism in the liver (Canto and Auwerx, 2009; Canto et al., 2009; Ding et al., 2017; Canto, 2013; Cantó et al., 2009). Sirt1 is also a direct target of miR-34a and has been shown to have protective effects when overexpressed in the liver (Li et al., 2015). Lipid, glucose, and oxidative metabolic processes are regulated by Sirt1 to retain homeostasis and prevent the pathogenesis of NAFLD (Canto and Auwerx, 2009). CR and Ames dwarf genotype have been shown to have significant effects on liver energy metabolism such as inducing changes in the NAD+/NADH ratio in the CR model or suppressing oxidative stress in the df/df liver (Canto and Auwerx, 2009; Canto et al., 2009). We hypothesized that CR and df/df genotype would correspondingly exert increased expression of Sirt1 and Ampk. Nutrient deficiency due to CR increased the cellular NAD+/NADH ratio and prompted increased expression of Sirt1 to signal the need for energy. In the df/df model, upregulation of Sirt1 in AL diet relative to normal mice may be attributed to small body size and suggests that minimized oxidative stress may contribute to the prolonged longevity of these mice. Coupled with CR, df/df mice exhibited higher levels of Sirt1 than both df/df-AL and N-CR animals although no interactive effects suggesting potential synergistic, additive functions of diet and genotype on energy sensing were noted.
To examine the effects of diet and genotype on DNL, we observed fatty acid uptake by quantifying the expression of the Cd36 plasma membrane translocase. Cd36 is a plasma membrane protein that facilitates the uptake and accumulation of fats in the liver (Rada et al., 2020). We found upregulation of Cd36 translocase in df/df mice regardless of diet indicating the requirement of fatty acid fuel use in their livers, perhaps due to their small body size and serious need for energy. In the normal mice, CR surprisingly does not influence Cd36 levels and indicates that genotypic rather than dietary effects regulate this gene and components of lipid metabolism. Pparg is a nuclear receptor that initiates the transcription of Srebp1, a protein which is involved with lipogenesis (Vacca et al., 2015; Nassir et al., 2015; Pettinelli and Videla, 2011). Our findings suggest that PParg is upregulated by df/df genotype, and we maintain that this may be due to high cellular demands resulting from their small body size, further evident by the interactive effects noted between CR and genotype. Further, CR decreased the expression of Srebp1 which suggests inhibition of DNL and supports the idea that low cellular energy encourages lipid trafficking. Suppression of Acc, an mRNA involved in the process of triglyceride retention by preventing the conversion of acetyl CoA to malonyl CoA, due to genotype demonstrates that df/df AL mice have a healthy balance of lipid import and export despite increased expression of Cd36 and Pparg (Sanders and Griffin, 2016). The Pparg mRNA represents a unique trend in this study in that its interaction term was significant, suggesting a synergy between diet and genotype.
To confirm the oxidation of these fatty acids, we explored the expression of insulin signaling genes Glut2, Akt2, and Pgc1α. Pgc1α influences both fatty acid oxidation and the insulin signaling pathway by regulating the ratio of insulin receptor substrates 1 and 2 (IRS1 and IRS2) in the hepatocytes. Increased IRS2 expression inhibits gluconeogenesis and prompts the activation of Akt (Besse-Patin et al., 2019; Hatting et al., 2018). During CR, glucagon induces gluconeogenesis to secrete glucose for energy in the peripheral tissues, while dwarf genotype has been shown to increase deacetylation of Pgc1α (Besse-Patin et al., 2019). Alongside, we looked at the mRNA expression of Glut2 and Akt2, mRNAs central to glucose uptake and metabolism (Besse-Patin et al., 2019). We studied Glut2 because it is important for the transport of glucose and Akt2 due to its response to insulin. Specifically, the upregulation of Pgc1α is believed to activate Pparα and initiate the transcription of Cpt1, a gene that generates carnitine to transport fatty acids to the mitochondria to undergo beta oxidation. Dysregulation of these processes is involved with hepatic steatosis (Ding et al., 2015; Li et al., 2015; Vacca et al., 2015).
We found that Pgc1α regulates gluconeogenesis and the hepatic response to CR by stimulating lipid catabolism, preventing fatty liver and insulin resistance, suggesting that these treatments may increase insulin sensitivity and enable a response to gluconeogenesis. The small size of the df/df mice may cause them to rely on glucose for energy and this process is augmented under long-term CR as seen by elevated levels of Glut2 in df/df-CR mice. Gluconeogenesis prompted activation of Akt2 in the df/df liver. In conjunction, gluconeogenesis is regulated by Pgc1α and the expression of this gene was upregulated regardless of diet but appeared to be heightened under CR. Lastly, Cpt1 expression was increased in the CR livers, with an interactive effect between genotype and diet evident. As such, the small size of the df/df mice may prompt lipid catabolism under CR due to the activation of gluconeogenesis.
Further, hepatic ER to Golgi lipid transport is facilitated by apolipoproteins and plays an important role in controlling intracellular trafficking (Tiwari and Siddiqi, 2012). Namely, with the assistance of microsomal triglyceride transfer protein (Mttp), Apob undergoes lipid extension in the endoplasmic reticulum and is developed into VLDL particles. VLDL cargo is docked to the Golgi apparatus by Sec22b-Stx5a complexes, released from the liver, and converted into potentially proatherogenic low density lipoproteins (LDL) (Tiwari and Siddiqi, 2012). Given the deleterious effects of LDL, the liver must efficiently remove VLDL and process cholesterol. We hypothesized that lipid trafficking and metabolism would be altered in response to CR and df/df genotype. We observed that df/df mice had increased levels of Apob but exhibited improved VLDL transport by CR suggesting efficient transport to the Golgi apparatus and subsequent secretion from hepatocytes.
While the expression of Apoa1 and Apoa4 was downregulated in df/df mice relative to N mice, CR upregulated the expression of Apoe and consequently improved cholesterol efflux and bile acid biosynthesis thereby improving reverse cholesterol transport. Alongside, dietary intervention increased the expression of Lxr, Abcg5, and Cyp7a1 suggesting decreased liver absorption of cholesterol, increased cholesterol efflux, and increased bile salt synthesis (Yu et al., 2002). Blood plasma from these mice could provide more useful information of HDL to LDL balance. High HDL could indicate increasing RCT in liver, and low LDL could indicate either impaired VLDL transport or decreased retention of fats in the liver. HDL to LDL ratio would give a better overall perspective on the overall cholesterol homeostasis of these mice.
To verify the influence of miR-34a on these processes, we transfected the HepG2 cancer cell line with 5 nM of mimic for miR-34a. We hypothesized that upregulation of miR-34a in HepG2 cells would correspondingly decrease the expression of SIRT1 and AMPK. Alongside, we maintained that the expression of this endogenous molecule would prompt hepatic steatosis and increased cholesterol deposition. Namely, we speculated that miR-34a would increase DNL (CD36, SREBP1, PPARG, ACC, SCD1), inhibit VLDL transport (APOB, MTTP, SAR1A, SAR1B, SEC22B, STX5A), decrease fatty acid oxidation (CPT1A, PPARA, PGC1A), and impair reverse cholesterol transport (APOA1, APOA4, APOE). Studying the associated genes with longevity and lipid trafficking in vitro using mimic for miR-34a enabled us to validate our findings. Our findings indicate that miR-34a impairs fatty acid oxidation and reverse cholesterol transport as evident by decreased expression of PPARA, CPT1A, APOA1, APOA4, LXR, and ABCG5. Although SIRT1 and AMPK in cells were not significantly downregulated, a downward trend was noted. Alongside, the upregulation of this miRNA did not induce DNL or hinder VLDL transport. Overall, this data indicates that miR34a is involved with lipid retention and metabolic processes in the liver thereby influencing the pathogenesis of NAFLD.
Our study reveals that CR improves metabolic processes in the Ames dwarf liver via downregulation of miR-34a and subsequent improvement of fatty acid oxidation and cholesterol efflux. Although DNL and VLDL transport are also mediated, these processes do not appear to be influenced by the regulation of miR-34a. It appears that miR-34a may inhibit the retention of fats in the liver but may also have downstream effects and thus influence the development of fatty liver. Alternatively, since there was decreased DNL in the cells following transfection with miR-34a there may also be less need for VLDL generation and transport. Overall, these results indicate that miR-34a is involved with fatty acid oxidation and cholesterol homeostasis, which may result in the pathogenesis of fatty liver. Given these findings, we speculate that CR and df/df genotype increased insulin sensitivity, improved VLDL and reverse cholesterol transport while increasing fatty acid oxidation to improve liver function.
Due to its implications on the Sirt1 signaling pathway, miR-34a-5p not only disrupts metabolic homeostasis, but also potentially influences cellular senescence. Particularly, Sirt1, known for its pivotal role in aging and metabolism, is activated by CR and notably elevated in long-living Ames dwarf mice. This suggests that miR-34a-5p may accelerate aging as further indicated by studies demonstrating its abundance in aged skeletal muscle, vasculature, and lens epithelial cells (Fulzele et al., 2019; Raucci et al., 2021; Wang et al., 2022). Concurrently, its interaction with other miRNAs such as miR-149, which was found to be downregulated by CR and life-extending mutation in our study and shown to be elevated in a high fat diet (HFD) induced mouse model of NAFLD, suggests a complex network of miRNA interactions that influence metabolic health and aging (Xiao et al., 2016). As such, this intricacy underscores the need for future studies to further elucidate these pathways and explore miR-34a-5p as a novel therapeutic target for managing age-related diseases.
Calorie restriction (CR) is regarded as the only intervention to significantly extend lifespan in various animal models (Most et al., 2017). This is particularly due to its modulation of important nutrient signaling pathways that are dysregulated in many age-related diseases including diabetes, fatty liver disease, cardiovascular disease, and neurodegenerative conditions (Dilova et al., 2007). Specifically, studies of the centenarian community in Okinawa have demonstrated that these long-living humans eat 17 % fewer calories than Japanese adults and 40 % fewer calories than American adults (Most et al., 2017). As such, the CALERIE (Comprehensive Assessment of Long-Term Effects of Reducing Intake of Energy) clinical trials were started by the National Institute of Aging to further explore the impacts of CR in healthy and overweight humans (Most et al., 2017; Dorling et al., 2021; van Beek et al., 2013). Overall, these studies demonstrate that CR can reduce alkaline phosphatase (ALT) and gamma-glutamyl transferase (GGT), improve insulin sensitivity, and decrease LDL-cholesterol and blood pressure (Most et al., 2017; Dorling et al., 2021). Improvement of parameters related to liver health in these studies shows that CR holds great promise in treating metabolic conditions like NAFLD. This information also elucidates the importance of understanding natural therapies to find biomarkers that can prevent the progression of age-related diseases such as NAFLD.
Acknowledgements
SAA, DW, SS, MMM conceptualized the project. SAA, XS, AS, DW, AG, MMM, SS developed the methodology. SAA, DW, SAN, CGCG performed the experiments. SAA, XS, DW, AG, AS, SAN, SS, MMM were responsible for visualization. SS, MMM acquired funding. SS, MMM supervised the project. SAA wrote the original draft of the manuscript. All authors were responsible for editing and reviewing the manuscript.
Funding
This work was supported by the National Institute on Aging of the National Institutes of Health R56 AG074499 (MM) and the National Institutes of Health RO1 DK-125596 (SS).
Footnotes
CRediT authorship contribution statement
Sarah A. Ashiqueali: Writing – original draft, Visualization, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. Xiang Zhu: Writing – review & editing, Methodology, Formal analysis, Data curation. Denise S. Wiesenborn: Writing – review & editing, Visualization, Methodology, Investigation, Conceptualization. Adam Gesing: Writing – review & editing, Visualization, Methodology, Investigation, Conceptualization. Augusto Schneider: Writing – review & editing, Visualization, Methodology, Investigation, Conceptualization. Sarah A. Noureddine: Writing – review & editing, Investigation. Christian G. Correa-Garcia: Writing – review & editing, Investigation. Michal M. Masternak: Writing – review & editing, Visualization, Supervision, Methodology, Investigation, Funding acquisition, Conceptualization. Shadab A. Siddiqi: Writing – review & editing, Visualization, Supervision, Methodology, Investigation, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare that no conflict of interest exists.
References
- Arora C, et al. , 2019. Lifestyle intervention framework for obese patients with non-alcoholic fatty liver disease - a tool for health professionals in resource constraint settings. Cureus 11 (10), e5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashiqueali SA, et al. , 2024. Fisetin modulates the gut microbiota alongside biomarkers of senescence and inflammation in a DSS-induced murine model of colitis. Geroscience 46, 3085–3103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avila BM, et al. , 2023. Effect of calorie restriction on redox status during chemically induced estropause in female mice. Geroscience 46 (2), 2139–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartke A, Brown-Borg H, 2004. Life extension in the dwarf mouse. Curr. Top. Dev. Biol 63, 189–225. [DOI] [PubMed] [Google Scholar]
- Barzilai N, et al. , 2012. The critical role of metabolic pathways in aging. Diabetes 61 (6), 1315–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Beek JH, et al. , 2013. The genetic architecture of liver enzyme levels: GGT. ALT and AST. Behav Genet 43 (4), 329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Besse-Patin A, et al. , 2019. PGC1A regulates the IRS1:IRS2 ratio during fasting to influence hepatic metabolism downstream of insulin. Proc. Natl. Acad. Sci. USA 116 (10), 4285–4290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cani PD, et al. , 2007. Metabolic endotoxemia initiates obesity and insulin resistance. Diabetes 56 (7), 1761–1772. [DOI] [PubMed] [Google Scholar]
- Canto C, Auwerx J, 2009. PGC-1alpha, SIRT1 and AMPK, an energy sensing network that controls energy expenditure. Curr. Opin. Lipidol 20 (2), 98–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto C, et al. , 2009. AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature 458 (7241), 1056–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantó C, G.-H. Z, Feige JN, Lagouge M, Noriega L, Milne JC, Elliott PJ, Puigserver P, Auwerx J, AMPK regulates energy expenditure by modulating NAD+ metabolism and SIRT1 activity. Nature, 2009. 458: p. 1056–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canto CAJ, 2013. PGC-1alpha, SIRT1 and AMPK, an energy sensing network that controls energy expenditure. Curr. Opin. Lipidol 20 (2), 98–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilova I, Easlon E, Lin SJ, 2007. Calorie restriction and the nutrient sensing signaling pathways. Cell. Mol. Life Sci 64 (6), 752–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J, et al. , 2015. Effect of miR-34a in regulating steatosis by targeting PPARalpha expression in nonalcoholic fatty liver disease. Sci. Rep 5, 13729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding RB, Bao J, Deng CX, 2017. Emerging roles of SIRT1 in fatty liver diseases. Int. J. Biol. Sci 13 (7), 852–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorling JL, et al. , 2021. Effect of 2 years of calorie restriction on liver biomarkers: results from the CALERIE phase 2 randomized controlled trial. Eur. J. Nutr 60 (3), 1633–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duregon E, et al. , 2021. Intermittent fasting: from calories to time restriction. Geroscience 43 (3), 1083–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fekete M, et al. , 2022. Nutrition strategies promoting healthy aging: from improvement of cardiovascular and brain health to prevention of age-associated diseases. Nutrients 15 (1). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley LW, Haigis MC, 2009. The coordination of nuclear and mitochondrial communication during aging and calorie restriction. Ageing Res. Rev 8 (3), 173–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana L, 2009. The scientific basis of caloric restriction leading to longer life. Curr. Opin. Gastroenterol 25 (2), 144–150. [DOI] [PubMed] [Google Scholar]
- Fulzele S, Mendhe B, Khayrullin A, Johnson M, Kaiser H, Liu Y, Isales C, Hamrick M, 2019. Musle-derived miR-34a increases with age in circulating extracellular vesicles and induces senescence of bone marrow stem cells. Aging 11, 1791–1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatting M, et al. , Insulin regulation of gluconeogenesis. Ann. N. Y. Acad. Sci, 2018. 1411(1): p. 21–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hense JD, et al. , 2022. Senolytic treatment reverses obesity-mediated senescent cell accumulation in the ovary. Geroscience 44 (3), 1747–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill CM, et al. , 2016. Long-lived hypopituitary Ames dwarf mice are resistant to the detrimental effects of high-fat diet on metabolic function and energy expenditure. Aging Cell 15 (3), 509–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell JJ, Manning BD, 2011. mTOR couples cellular nutrient sensing to organismal metabolic homeostasis. Trends Endocrinol. Metab 22 (3), 94–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kargulewicz A, Stankowiak-Kulpa H, Grzymislawski M, 2014. Dietary recommendations for patients with nonalcoholic fatty liver disease. Prz Gastroenterol 9 (1), 18–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King TL, et al. , 2024. Chronological and reproductive aging-associated changes in resistance to oxidative stress in post-reproductive female mice. Geroscience 46 (1), 1159–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, et al. , 2011. Increased expression of miR-34a and miR-93 in rat liver during aging, and their impact on the expression of Mgst1 and Sirt1. Mech. Ageing Dev 132 (3), 75–85. [DOI] [PubMed] [Google Scholar]
- Li X, et al. , 2015. Isocaloric pair-fed high-carbohydrate diet induced more hepatic steatosis and inflammation than high-fat diet mediated by miR-34a/SIRT1 Axis in mice. Sci. Rep 5, 16774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masternak M, Al-Regaiey K, Lim M, Bonkowski M, Panici J, Przybylski G, Bartke A, 2005. Calorie restriction results in decreased expression of peroxisome proliferator-activated receptor superfamily in muscle of Normal and long-lived growth hormone receptor/binding protein knockout mice. Journal of Gerontology: Biological Sciences 60A, 1238–1245. [DOI] [PubMed] [Google Scholar]
- Melnik B, 2012. Dietary intervention in acne: attenuation of increased mTORC1 signaling promoted by Western diet. Dermatoendocrinol 4 (1), 20–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melnik BC, John SM, Schmitz G, 2011. Over-stimulation of insulin/IGF-1 signaling by western diet may promote diseases of civilization: lessons learnt from laron syndrome. Nutr. Metab. (Lond.) 8, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Most J, et al. , 2017. Calorie restriction in humans: an update. Ageing Res. Rev 39, 36–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassir F, Rector S, Hammoud G, Ibdah J, 2015. Pathogenesis and prevention of hepatic steatosis. Gastroenterol. Hepatol 11 (3), 167–175. [PMC free article] [PubMed] [Google Scholar]
- Nisoli E, Tonello C, Cardile A, Cozzi V, Bracale R, Tedesco L, Falcone S, Valerio A, Cantoni O, Clementi E, Moncada S, Carruba M, 2005. Calorie restriction promotes mitochondrial biogenesis by inducing the expression of eNOS. Science 310, 314–317. [DOI] [PubMed] [Google Scholar]
- Noureddine S, et al. , 2022. GH deficiency confers protective advantages against Alzheimer’s disease through rescued miRNA expression profile in APP/PS1 mice. Geroscience 44 (6), 2885–2893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Brien J, et al. , 2018. Overview of MicroRNA biogenesis, mechanisms of actions, and circulation. Front Endocrinol (Lausanne) 9, 402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perumpail BJ, et al. , 2017. Clinical epidemiology and disease burden of nonalcoholic fatty liver disease. World J. Gastroenterol 23 (47), 8263–8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettinelli P, Videla LA, 2011. Up-regulation of PPAR-gamma mRNA expression in the liver of obese patients: an additional reinforcing lipogenic mechanism to SREBP-1c induction. J. Clin. Endocrinol. Metab 96 (5), 1424–1430. [DOI] [PubMed] [Google Scholar]
- Pitisuttithum P, Treeprasertsuk S, 2022. Nonalcoholic fatty liver disease (NAFLD) among older adults. Portal Hypertension & Cirrhosis 1 (3), 184–191. [Google Scholar]
- Rada P, et al. , 2020. Understanding lipotoxicity in NAFLD pathogenesis: is CD36 a key driver? Cell Death Dis. 11 (9), 802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raucci A, et al. , 2021. MicroRNA-34a: the bad guy in age-related vascular diseases. Cell. Mol. Life Sci 78 (23), 7355–7378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Gomez M, et al. , 2023. Nutrition could prevent or promote non-alcoholic fatty liver disease: an opportunity for intervention. BMJ 383, e075179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rui L, 2014. Energy metabolism in the liver. Compr. Physiol 4 (1), 177–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders FW, Griffin JL, 2016. De novo lipogenesis in the liver in health and disease: more than just a shunting yard for glucose. Biol. Rev. Camb. Philos. Soc 91 (2), 452–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani T, et al. , 2023. Calorie restriction mimetic drugs could favorably influence gut microbiota leading to lifespan extension. Geroscience 45 (6), 3475–3490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari S, Siddiqi SA, 2012. Intracellular trafficking and secretion of VLDL. Arterioscler. Thromb. Vasc. Biol 32 (5), 1079–1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vacca M, et al. , 2015. Fatty acid and glucose sensors in hepatic lipid metabolism: implications in NAFLD. Semin. Liver Dis 35 (3), 250–261. [DOI] [PubMed] [Google Scholar]
- Wang S, et al. , 2022. MiR-34a-5p negatively regulates oxidative stress on Lens epithelial cells by silencing GPX3 - a novel target. Curr. Eye Res 47 (5), 727–734. [DOI] [PubMed] [Google Scholar]
- Xia M, et al. , 2023. DNA methylation age acceleration contributes to the development and prediction of non-alcoholic fatty liver disease. Geroscience. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, et al. , 2016. miR-149 controls non-alcoholic fatty liver by targeting FGF-21. J. Cell. Mol. Med 20 (8), 1603–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y, et al. , 2021. Hepatocyte miR-34a is a key regulator in the development and progression of non-alcoholic fatty liver disease. Mol Metab 51, 101244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, et al. , 2002. Overexpression of ABCG5 and ABCG8 promotes biliary cholesterol secretion and reduces fractional absorption of dietary cholesterol. J. Clin. Invest 110 (5), 671–680. [DOI] [PMC free article] [PubMed] [Google Scholar]


