Abstract
The multifunctional herpes simplex virus type 1 (HSV-1) protein IE63 (ICP27) interacts with the essential pre-mRNA splicing factor, spliceosome-associated protein 145 (SAP145), and in infected cells IE63 and SAP145 colocalize. This interaction was reduced or abrogated completely using extracts from cells infected with IE63 viral mutants, with mutations in IE63 KH and Sm homology domains, which do not exhibit host shutoff or inhibit splicing. In the presence of IE63, splicing in vitro was inhibited prior to the first catalytic step and the B/C complex formed during splicing was shifted up in mobility and reduced in intensity. With the use of splicing extracts, IE63 and SAP145 both comigrated with the B/C complex, suggesting that they interact within this complex to inhibit B/C complex formation or conversion. The inhibition of splicing may facilitate the export of viral or cellular transcripts, possibly via other protein partners of IE63. These data provide important new insights into how IE63 influences pre-mRNA processing during HSV-1 infection.
Herpes simplex virus type 1 (HSV-1) is a nuclear replicating virus with a large double-stranded DNA genome that encodes some 80 gene products (30). During lytic infection, viral genes are expressed in a temporal manner (48) and are classified as immediate-early (IE), early, or late. The five IE genes do not require prior viral protein synthesis for their expression; four regulate early and late viral gene expression (11, 13, 15, 29, 37, 50, 51, 54, 67), while one subverts the host cytotoxic T-lymphocyte response (20).
IE63 (ICP27), an HSV-1 RNA binding protein (21, 34, 52) essential throughout infection (50), promotes accumulation of a subset of viral early and late mRNAs and is necessary for the switch from early to late virus gene expression (29). IE63 is required for efficient viral DNA replication and stimulates the accumulation of certain viral early and late mRNAs (16, 29, 31, 32, 45, 50, 56, 63, 65). Itself not a transcriptional activator, IE63 is required for expression of viral early and late genes at both transcriptional and posttranscriptional levels. At the posttranscriptional level, IE63 has effects on the processing of pre-mRNA and mediates nucleocytoplasmic transport of transcripts from intronless viral genes which form the majority of viral RNAs (52). IE63 inhibits splicing of cellular pre-mRNAs to mediate host cell shutoff (18), redistributes splicing factors (39, 53), and increases RNA 3′ processing at weak virus poly(A) sites (31). In addition, IE63 shuttles between the nucleus and the cytoplasm (35, 40, 52, 59).
IE63 contains several functional domains (see Fig. 3B). The N terminus contains an acidic activation domain essential for enhancing early gene expression and hence viral DNA replication (46), a leucine-rich nuclear export signal which promotes nuclear shuttling (52), nuclear and nucleolar localization signals (33), and an RGG box involved in RNA binding (34). Along with a putative zinc finger (66), IE63 contains three KH-like domains homologous to RNA binding regions of hnRNP K (60) and an Sm domain (60), a motif found in certain spliceosomal proteins which potentiates protein-protein interactions (19, 22). Mutations in the KH1 and KH3 domains have shown their requirement for virus growth, up-regulation of certain late transcripts, and IE63 shuttling (47, 59–61), while the Sm domain has been associated with host shutoff (59–61). However, the KH-like domains in IE63 have not been shown to bind RNA and are not fully typical of KH domains, as they lack an invariant GXXG motif.
FIG. 3.
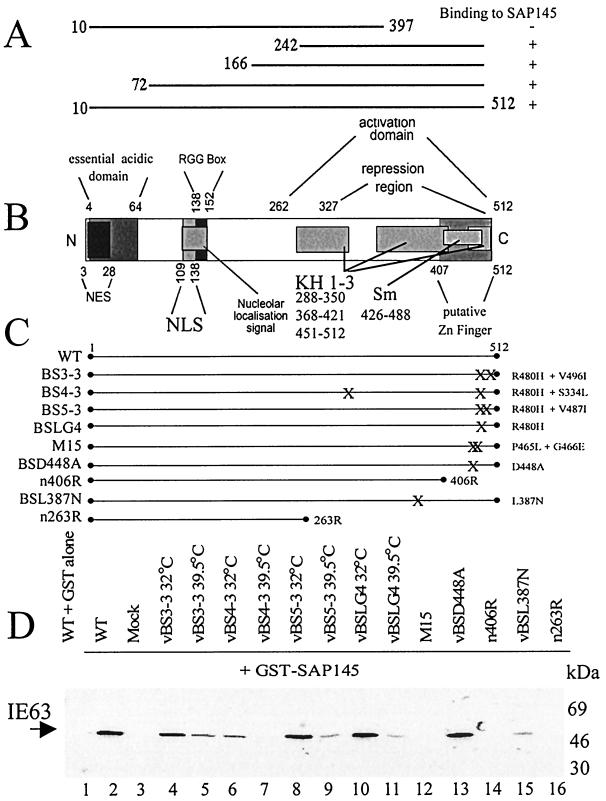
Mapping the interaction of IE63 with SAP145. (A) Truncations used in the yeast two-hybrid assay to map the IE63 region involved in interaction with SAP145; results of the interactions are shown as plus or minus signs. (B) Schematic representation (not to scale) of IE63 protein showing the different functional regions as described in reference 67. NES, leucine-rich nuclear export signal; NLS, nuclear localization signal; RGG box, arginine-rich RNA binding region; KH 1-3, hnRNP K homology domains; Sm, homology domain present in certain Sm proteins. (C) HSV-1 IE63 mutant viruses used to study the interaction with SAP145. X's indicate the locations of amino acid substitutions which are described on the right-hand side of the schematic; truncations also are shown, and the last amino acid is indicated. Mutation R480H in mutants BS3-3, BS4-3, BS5-3, and BSLG4 leads to a ts phenotype; the second mutations in these viruses are intragenic suppressors of this phenotype (see text). (D) Extracts from HeLa cells infected with viruses shown in panel C were mixed with GST alone (lane 1) or GST-SAP145 (lanes 2 to 16); bound proteins were separated by SDS-polyacrylamide gel electrophoresis, transferred to nitrocellulose, and analyzed by Western blotting using IE63 antiserum.
While IE63-mediated host shutoff is not essential for expression of virus proteins, the abilities of IE63 to shut off host synthesis and to inhibit splicing are correlated (17, 18). As well as contributing to host cell shutoff, inhibition of splicing could uncouple the splicing and mRNA export pathways and facilitate nuclear export of transcripts from viral intronless genes which may contain cryptic splice sites, allowing them to bypass the normal stringent cellular requirement for splicing prior to nuclear export (8).
Splicing specificity is determined by multiple RNA-RNA and RNA-protein interactions involving sequences at both 5′ and 3′ splice sites, at the branch site, and within the exon. During splicing, successive recognition of these elements occurs as spliceosomal complexes E, A, B, and C assemble on pre-mRNA in a stepwise manner (43). In the commitment complex E, U1 snRNP is bound to the 5′ splice site and splicing factors U2AF and SF1 are attached to the polypyrimidine tract upstream of the 3′ splice site and the branch site, respectively. As U2 snRNP binds and complex E is converted to presplicing complex A, a short helix forms between a single-stranded region in U2 snRNA and the intron branch site which requires both the 12S snRNP and splicing factors SF3a and SF3b (3). The next step is binding of the U4/U5/U6 tri-snRNP at the 5′ splice site to form the B complex. Just prior to splicing, conformational changes within complex B destabilize the association of U4 and transform complex B into complex C. Spliceosome-associated protein 145 (SAP145), an essential component of the SF3b subunit, can bind to pre-mRNA (62) and is implicated in the tethering of U2 snRNA to the branch site (7). The splicing reaction occurs in two stages (38, 49). In the first step, pre-mRNA is cleaved at the 5′ splice site, generating a linear first exon and an intron-second exon RNA species in a lariat formation (the lariat-second exon). In the second step, the 3′ splice site is cleaved and concomitantly the exons are ligated together, generating the spliced mRNA.
We show that IE63 interacts with the essential splicing factor SAP145 and that, in HSV-1-infected cells, IE63 and SAP145 colocalize. The IE63-SAP145 interaction was reduced or abrogated completely using extracts from cells infected with IE63 viral mutants that fail to shut off host protein synthesis and which map in the KH and Sm homology domains. From in vitro splicing reactions in the presence of IE63, pre-mRNA splicing was inhibited prior to the first catalytic step and both IE63 and SAP145 comigrated with the B/C complex formed during splicing. These data strongly suggest that the association of IE63 with SAP145 acts to inhibit splicing. This inhibition could facilitate the nuclear export of RNAs and contribute to host cell shutoff.
MATERIALS AND METHODS
Plasmids and antisera.
For transient expression of IE63, plasmid pCMV63 (4) was used. IE63 constructs used in the yeast two-hybrid screen have been described previously (67). Anti-IE63 antiserum H1113 was a mouse monoclonal antibody (1) supplied by the Goodwin Institute for Cancer Research. Anti-SAP145 antiserum was a rabbit antibody (55) kindly provided by R. Reed. Glutathione S-transferase (GST) fusion protein constructs pGEX145 from R. Reed (55), pGEX27 from S. Rice (32), and pGEXK from K. Bomsztyk (36) have been described previously. The pre-mRNA transcript used for splicing was transcribed from pBSAD1 (23).
Cells and viruses.
HeLa cells were grown in Dulbecco's modified Eagle's medium supplemented with 2.5% fetal calf serum, 2.5% newborn calf serum, 100 U of penicillin/ml, and 100 μg of streptomycin/ml. Stocks of wild-type (wt) HSV-1 strain 17+; the IE63 insertion mutant HSV-1 27-lacZ (58); and the HSV-1 mutant viruses vBSLG4, vBS3-3, vBS4-3, and vBS5-3 (59); vBSL387N and vBSD448A (60); n263R and n406R (44); and M15 (47) were grown as described previously (31) in BHK cells or in the IE63-complementing cell line 2-2, a derivative of Vero cells that expresses IE63 under the control of its own promoter. Complementing cells were maintained in Dulbecco's modified Eagle's medium supplemented with 500 μg of G418 (Geneticin; Gibco BRL) per ml. The mutations present in each virus are shown in Fig. 3C.
Infection of cells and preparation of extracts.
Ninety percent confluent HeLa cell monolayers (4 × 107 cells) were infected with HSV-1 wt or IE63 mutant virus at a multiplicity of 10 PFU/cell or left uninfected (mock infected). Growth of temperature-sensitive (ts) viruses was carried out at 32 and 39.5°C, and growth of wt virus and other mutants and mock infection were carried out at 37°C. After 1 h of absorption, medium was added and cells were left for 16 h. For preparation of cell extracts, monolayers were washed with phosphate-buffered saline (PBS), and cells were lysed by suspension in 1 ml of cell extract buffer (50 mM HEPES, 50 mM NaCl, 0.1% Nonidet NP-40 [pH 7.5]) containing a protease inhibitor cocktail (Boehringer Mannheim). Extracts were sonicated on ice, cell debris was pelleted, and the protein concentration was determined by the Bradford assay (Bio-Rad). For immunofluorescence, 13-mm-diameter coverslips were seeded at 0.5 × 105 HeLa cells per well in 1 ml of normal HeLa medium and incubated overnight at 37°C prior to infection at a multiplicity of 10 PFU/cell. Alternatively, HeLa cells were transfected with the IE63-expressing plasmid pCMV63, a procedure in which 5 × 106 cells were transfected by electroporation with 20 μg of DNA and left for 24 h before extracts were prepared as described above or cells were fixed for immunofluorescence assay.
For splicing assays, nuclear extracts were prepared from 80% confluent mock-, wt-, or 27-lacZ-infected HeLa cells. After being washed with PBS, cells were resuspended in 1 packed cell volume of 10 mM HEPES–1.5 mM MgCl2–10 mM KCl–1 mM dithiothreitol (pH 8.0) and left on ice for 15 min. The suspension was then passaged 10 times through a 25-gauge needle and microcentrifuged at full speed for 20 s. The nuclear pellet was then resuspended in 2/3 packed cell volume of 20 mM HEPES–1.5 mM MgCl2–25% glycerol–420 mM NaCl–0.2 mM EDTA–1 mM dithiothreitol–0.5 mM phenylmethylsulfonyl fluoride (pH 8.0), and mixed on ice, and nuclear debris was pelleted.
Fusion protein expression and pull-down assays.
GST, GST-SAP145, GST-IE63, and GST-K fusion proteins were expressed, bound onto glutathione beads, and purified as described previously (4). An equal amount of each fusion protein was mixed with wt virus-, mutant virus-, or mock-infected cell extract. For wt-, 27-lacZ-, and mock-infected cell extracts, 100 μg of total protein extract was added, while for mutant virus extracts an amount of IE63 equal to that found in 100 μg of wt-infected cell extract (as determined by Western blotting) was used. After washing, bound proteins were removed from the beads by boiling, loaded onto sodium dodecyl sulfate (SDS)-polyacrylamide gels, and transferred to nitrocellulose for analysis by Western blotting.
Immunoprecipitation and Western blotting.
One hundred micrograms of cell extract was mixed with 5 μl of anti-SAP145 rabbit antibody in 50 μl of binding buffer (100 mM Tris HCl, 5 mM EDTA, 1% Triton X-100 [pH 7.4]) for 3 h at 4°C; 75 μl of protein A-Sepharose was then added, and mixing was done for 1 h as described previously (4). After pelleting of the beads and multiple washes, the bound proteins were assayed for CK2 activity or eluted by boiling, separated by SDS-polyacrylamide gel electrophoresis, and detected by Western blotting. Western blotting was performed as described in reference 4, using primary antibody dilutions of 1:2,000 for IE63 and of 1:1,000 for SAP145.
CK2 activity assay.
Following washing, proteins that coimmunoprecipitated with SAP145 were resuspended in 30 μl of CK2 reaction buffer (50 mM Tris, 20 mM MgCl2 [pH 8.2]) containing 10 μCi of [α-32P]ATP per reaction mixture, either with or without 0.1 mM CK2 peptide substrate (Arg-Arg-Arg-Glu-Glu-Glu-Thr-Glu-Glu-Glu), and CK2 activity was detected as described previously (67).
Yeast two-hybrid screen and mapping of the regions of IE63 involved in the p32 interaction.
The yeast two-hybrid screen was performed using IE63 amino acids 10 to 512 as bait and target plasmids containing a HeLa cell cDNA library (Clontech) as described previously (67). Mapping the IE63 regions involved in the interaction utilized the truncated IE63 constructs described in reference 67; these were mated into yeast cells transfected with the SAP145 clone identified from the library screen, using a β-galactosidase (β-Gal) filter assay. Results from each cotransformation represented an analysis of some 200 individual colonies. The time taken for the positive cells to turn blue varied between experiments, depending on factors such as colony size. However, a positive interaction (+) was identified by the presence of blue colonies seen after 3 h on β-Gal filter assays which, compared to interactive standards assayed by liquid β-Gal assays, had activities of between 10 and 50 U, while standards showing lack of interaction had activities of <1 U. Filter assays and liquid β-Gal assays were performed according to the manufacturer's instructions (protocol PT3024-1; Clontech).
Indirect immunofluorescence.
Infected, transfected, or mock-infected HeLa cell monolayers were fixed for 10 min at 20°C with 2% sucrose–5% formaldehyde in PBS. After being washed three times in PBS, cells were permeabilized for 10 min at 20°C with 0.5% NP-40–10% sucrose in PBS. After further washes, primary antibody diluted to the appropriate concentration (IE63, 1:100, and SAP145, 1:150) in PBS with 1% calf serum was added for 60 min at 20°C. Cells were once again washed before incubation with secondary antibody coupled to fluorescein isothiocyanate or Cy 5 (Sigma) diluted 1:100 in PBS was carried out for 30 min at 20°C. After a final wash, cells were examined with a Zeiss LSM 510 confocal microscope system with two lasers giving excitation at 488 nm (fluorescein isothiocyanate) and 633 nm (Cy 5) and a Zeiss Axioplan microscope using a 63× oil-immersion objective lens, numerical aperture 1.4. Data were processed with LSM 510 software and then exported for preparation using Photoshop.
Splicing.
Splicing assays were performed using uniformly labeled, capped pre-mRNAs incubated with wt-, 27-lacZ-, or mock-infected nuclear extracts using the in vitro splicing conditions described previously (25) in which the pre-mRNA was the adenovirus major late precursor transcribed from Sau3A-digested plasmid pBSAD1. Splicing products were separated on 10% polyacrylamide–8 M urea denaturing gels and exposed to X-Omat film (Kodak) overnight. Splicing complexes were prepared and separated on nondenaturing agarose-polyacrylamide composite gels (26) and visualized by autoradiography overnight or transferred to nitrocellulose under standard transfer conditions prior to Western blotting for IE63 and SAP145.
RESULTS
SAP145 interacts with IE63.
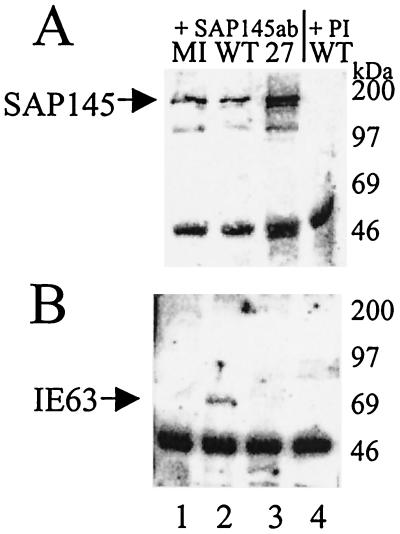
To identify cellular proteins capable of interacting with IE63 in the yeast two-hybrid assay, we screened a HeLa cell library fused to the GAL4 activation domain and expressed in pGADGH using IE63 amino acids 10 to 512 as bait. Eighty-two clones fulfilled the criteria for interaction of gene products from a total of 2.3 × 106 transformants screened. These were sequenced and checked against the GenBank-EMBL database. For one clone, containing an insert of approximately 900 bp, a 499-bp fragment had 89.4% homology with the 2,839-bp human SAP145 gene. Confirmation of the interaction was provided by coimmunoprecipitation from virus-infected cells and GST pull-down assays. With anti-SAP145 serum, SAP145 was immunoprecipitated from extracts of cells infected with wt or 27-lacZ and from mock-infected cells (Fig. 1A, lanes 1 to 3). IE63 was coimmunoprecipitated with SAP145 from wt-infected cell extracts only (Fig. 1B, lane 2); however, neither SAP145 nor IE63 was precipitated by the preimmune sera (Fig. 1A and B, lanes 4). The intense lower band present in each track is the heavy chain of the antibodies used.
FIG. 1.
Coimmunoprecipitation of IE63 and SAP145 in vivo using antibodies directed against SAP145. HSV-1-infected (WT), 27-lacZ-infected, or mock-infected (MI) HeLa cell extracts were immunoprecipitated with SAP145 antiserum. Aliquots of the precipitated proteins were separated by SDS-polyacrylamide gel electrophoresis, transferred to nitrocellulose, and analyzed by Western blotting using SAP145 antiserum or IE63 monoclonal antibody. (A) Immunoprecipitates obtained with SAP145 antiserum (lanes 1 to 3) or preimmune (PI) serum (lane 4), Western blotted with SAP145 antiserum. (B) Immunoprecipitates obtained with SAP145 antiserum (lanes 1 to 3) or preimmune serum (lane 4), Western blotted with IE63 antiserum.
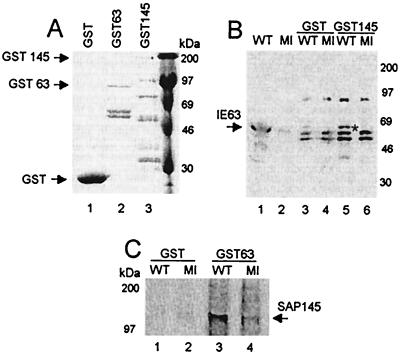
Recombinant GST-IE63 and GST-SAP145 proteins were expressed in Escherichia coli along with the control GST protein alone (Fig. 2A), and these were used in pull-down assays. GST-SAP145 was capable of pulling down IE63 from wt-infected cell extract (Fig. 2B, lane 5), whereas the control GST alone brought down an insignificant background amount (Fig. 2B, lane 3). Conversely, GST-IE63 pulled down SAP145 from both wt- and mock-infected extracts (Fig. 2C, lanes 3 and 4); again, GST alone did not bring down significant levels of SAP145 (Fig. 2C, lanes 1 and 2). The difference shown in the quantity of interacting SAP145 from wt- and mock-infected extracts (Fig. 2C, compare lanes 3 and 4) was not consistently found and is not thought to be significant.
FIG. 2.
In vitro interaction between GST-IE63 and SAP145 and between GST-SAP145 and IE63. HSV-1-infected (WT) or mock-infected (MI) HeLa cell extracts were mixed with GST-IE63 (GST63), GST-SAP145 (GST145), or GST alone. Bound proteins were separated by SDS-polyacrylamide gel electrophoresis, transferred to nitrocellulose, and analyzed by Western blotting using SAP145 antiserum or IE63 monoclonal antibody. (A) Coomassie blue staining of the GST fusion proteins used in the pull-down assays. (B) Proteins binding to GST (lanes 3 and 4) or GST-SAP145 (lanes 5 and 6) following Western blotting with IE63 antiserum. The cell extracts used (lanes 1 and 2) were loaded with 25% of the amount used for the pull-downs. (C) Proteins binding to GST (lanes 1 and 2) or GST-IE63 (lanes 3 and 4), following Western blotting with SAP145 antiserum.
Regions of IE63 involved in SAP145 binding.
To identify the region(s) of IE63 involved in the interaction with SAP145, both the yeast two-hybrid system and mutant IE63 viruses were used. The yeast two-hybrid mapping was performed using large truncations of IE63 cloned into the GAL4 DNA binding domain which were mated with the SAP145 clone pulled out of the initial yeast two-hybrid screen. Results indicated that the IE63 region(s) responsible for interaction was contained within amino acids 397 to 512 at the C-terminal region (Fig. 3A). A panel of IE63 mutant viruses containing amino acid substitutions within this region (Fig. 3C) was used to further characterize the region of interaction with SAP145 using the GST pull-down assay.
Mutant vBSLG4 contains a single mutation within an overlapping region of the Sm and KH3 domains (R480H); this gives rise to a ts phenotype defective in both host shutoff and synthesis of viral proteins. Defective growth at the nonpermissive temperature (39.5°C) reflects the failure to synthesize viral late proteins. Three unique second-site intragenic suppressor mutants of the ts phenotype also were used; they contain the R480H mutation plus mutations in KH1 S334L (vBS4-3), KH2 V496I (vBS3-3), or KH3 V487I (vBS5-3). At the nonpermissive temperature (39.5°C), revertants are restored in their ability to synthesize late proteins and hence are non-ts for growth but still fail to inhibit host cell protein synthesis. At the permissive temperature, vBSLG4 behaves like the wt while the revertants appear to show intermediate levels of host shutoff (59). The other mutants used were truncations of IE63 (n406R and n263R), a virus with two mutations in the overlapping Sm/KH3 domain (M15), a virus with a mutation solely in the Sm region (vBSD448A), and one with an amino acid substitution in the KH2 domain (L387N).
Binding to SAP145 was assessed using the GST pull-down assay, and equal amounts of each mutant protein were added as determined by Western blotting of whole-cell extracts prior to incubation with GST-SAP145 (data not shown). As previously, GST-SAP145 pulled down IE63 from wt-infected cell extract, while GST alone did not (Fig. 3D, compare lanes 1 and 2). In the SAP145 pull-downs, the viral mutants were divided into three groups, those in which IE63 still interacted normally, those in which binding of SAP145 to IE63 was reduced, and those in which IE63 no longer bound SAP145. At the permissive temperature, mutant vBSD448A (Fig. 3D, lane 13) and mutants vBS3-3, vBS5-3, and vBSLG4 (Fig. 3D, lanes 4, 8, and 10, respectively) showed wt SAP145 binding, while at the nonpermissive temperature, mutants vBS3-3, vBS5-3, and vBSLG4 (Fig. 3D, lanes 5, 9, and 11, respectively) along with mutant vBSL387N (Fig. 3D, lane 15) exhibited reduced binding. Interestingly, in contrast to the other mutants containing the R480H mutation, vBS4-3 showed reduced binding at the permissive temperature (Fig. 3D, lane 6) and binding was not detected at the nonpermissive temperature (Fig. 3D, lane 7). Other viruses in which SAP145 binding was completely disrupted were M15, n406R, and n263R (Fig. 3D, lanes 12, 14, and 16, respectively).
Due to the overlapping of KH3 and Sm domains, the relative contribution of each was difficult to assess. All the mutants in the overlap region showed reduced interaction. When more than one amino acid within this region was substituted (M15) or a mutation in KH1 was combined with the R480H mutation (vBS4-3), the interaction was completely disrupted. A mutation in the KH2 region alone (vBSL387N) considerably reduced SAP145 binding. The mutation solely in the Sm region, and not overlapping KH3, did not affect the interaction. As expected, the large deletion mutants completely disrupted the interaction.
IE63 is required for the association of SAP145 with hnRNP K and CK2.
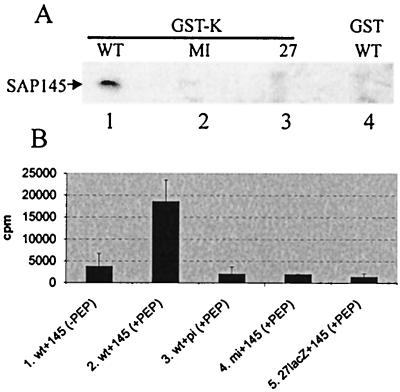
To determine if SAP145 was associated with other IE63 partner proteins, and whether IE63 was interacting with these partners simultaneously, GST-hnRNP K fusion protein was used in pull-down assays with wt-, 27-lacZ-, and mock-infected cell extracts (Fig. 4A, lanes 1 to 3). SAP145 bound to GST-hnRNP K but only using wt-infected cell extracts (Fig. 4A, lane 1), and SAP145 from wt-infected cell extracts did not bind GST alone (Fig. 4A, lane 4). Therefore, hnRNP K can associate with SAP145 but only in the presence of IE63, and IE63 must be associated with hnRNP K and SAP145 simultaneously. Under the conditions used here, CK2 was previously detected in hnRNP K coimmunoprecipitations but only when IE63 was present (67). Thus, the SAP145-hnRNP K association in the presence of IE63 could occur as an indirect interaction via CK2. To examine this, anti-SAP145 serum was used as before (Fig. 1B) to bring down SAP145 from wt-, 27-lacZ-, and mock-infected cell extracts, and CK2 activity of immunoprecipitates was assayed using the peptide assay. CK2 activity was coimmunoprecipitated with SAP145 only in the presence of IE63 (Fig. 4B, lane 2), and assays performed in the absence of peptide or in the presence of peptide with mock-infected or 27-lacZ-infected cell extracts or with preimmune sera did not result in any activity (Fig. 4B, lanes 1, 3, 4, and 5). Hence, similar to its association with hnRNP K, CK2 was found with SAP145 only in the presence of IE63, and IE63 must be able to form a complex with CK2 and SAP145 simultaneously.
FIG. 4.
SAP145 interacts with hnRNP K and CK2 only in the presence of IE63. (A) HSV-1-infected (WT), 27-lacZ-infected (27), or mock-infected (MI) HeLa cell extracts were mixed with GST-hnRNP K (GST-K, lanes 1 to 3) or GST alone (lane 4). Bound proteins were separated by SDS-polyacrylamide gel electrophoresis, transferred to nitrocellulose, and analyzed by Western blotting using SAP145 antiserum. (B) Following immunoprecipitation with anti-SAP145 antiserum, as shown in Fig. 1, samples were analyzed for CK2 activity using a peptide assay. CK2 assays were performed with immunoprecipitates generated by preimmune (pi) serum (lane 3) and SAP145 (145) antiserum (lanes 1, 2, 4, and 5) from HSV-1 wt-infected (wt), 27-lacZ-infected (27lacZ), or mock-infected (mi) HeLa cell extracts and in the absence (lane 1) or presence (lanes 2 to 5) of peptide substrate.
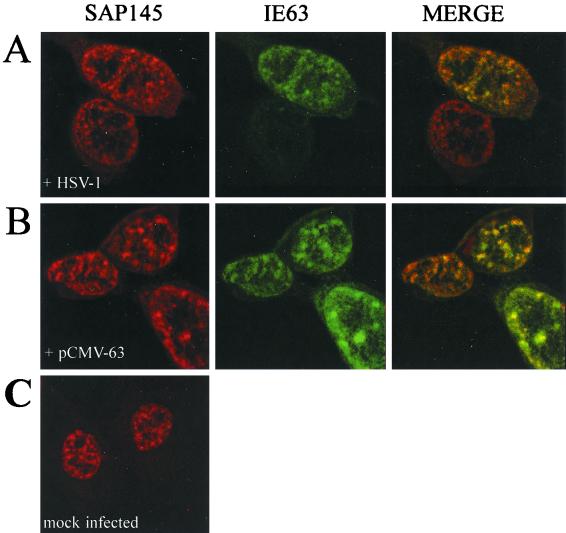
SAP145 and IE63 colocalize in HSV-infected cells.
During HSV-1 infection, the splicing components examined so far redistribute within the nucleus to give a characteristic clumped pattern (28); IE63 is responsible for this effect and colocalizes with the redistributed factors (39, 53). SAP145 is a splicing factor, and its location in infected cells was of interest. Confocal microscopy revealed that, in wt-infected cells (Fig. 5A) and in IE63-transfected cells (Fig. 5B), IE63 colocalizes with SAP145 in a speckled distribution similar to that seen for other snRNP proteins. The nuclear distribution of SAP145 shown in the non-IE63-expressing cells in Fig. 5A and in uninfected cells (Fig. 5C) was speckled, whereas two other splicing factors examined in cells subsequently infected with HSV-1 were more diffusely spread (39, 53). Splicing factors exhibit different levels of punctate staining (24) that can vary under different growth conditions, reflecting the dynamic state of the nucleus, where factors move at different rates from inactive storage sites to active sites.
FIG. 5.
IE63 colocalizes with SAP145 in HSV-1-infected cells. Immunofluorescence assay was performed on HeLa cells using anti-IE63 serum and anti-SAP145 serum. (A) HSV-1 wt virus-infected cells. (B) pCMV-63-transfected cells. (C) Mock-infected cells.
IE63 halts splicing, alters splicing complexes, and is found in the B/C but not the A complex.
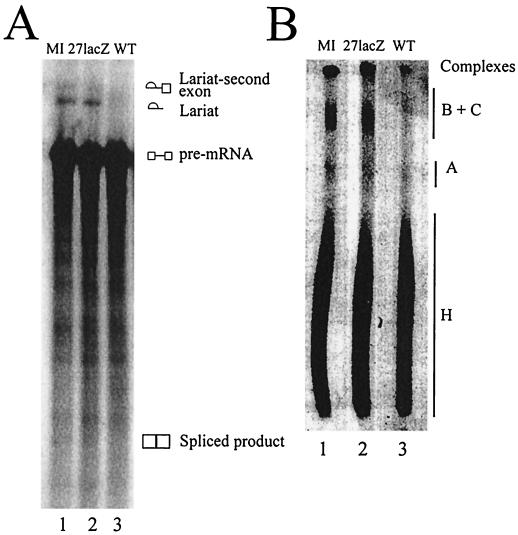
As SAP145 is an essential component of the splicing factor SF3b subunit, we wished to examine the interaction of IE63 with SAP145 in the context of pre-mRNA splicing in vitro. IE63 expression was necessary for the inhibition of pre-mRNA splicing in vitro using nuclear extracts from HSV-1-infected cells (18). To confirm this, we performed in vitro splicing reactions using nuclear extracts from cells infected with wt and 27-lacZ viruses and from mock-infected cells. As expected, with wt-infected cell extracts splicing was inhibited as shown by the lack of lariat product; in addition, the intermediate second exon-lariat structure was not detected (Fig. 6A, lane 3). In contrast, with mock-infected and 27-lacZ-infected cell extracts the splicing products of the free lariat and the lariat-second exon RNAs were seen (Fig. 6A, lanes 1 and 2). The splicing reaction was further studied by examining the protein complexes formed. In reactions performed with mock-infected and 27-lacZ-infected cell extracts, presplicing complex A, active splicing complex B/C, and associated complex H were all observed (Fig. 6B, lanes 1 and 2). However, when wt-infected cell extract was used, complex A was reduced in intensity, complex B/C was reduced to a greater extent with its mobility shifted upward on the gel (Fig. 6B, lane 3), and complex H formed normally.
FIG. 6.
IE63 inhibits splicing in vitro and alters splicing complex formation. In vitro splicing assays were performed using adenovirus major late precursor mRNA with nuclear extracts from HSV-1 wt-infected (lanes 3), 27-lacZ-infected (lanes 2), or mock-infected (lanes 1) HeLa cells. The products of the splicing reaction were run on gels as follows. (A) A 10% polyacrylamide–8 M urea denaturing gel, to separate the RNA products formed. The pre-mRNA was 32P labeled, and products were visualized by exposure to X-ray film overnight. (B) A nondenaturing agarose-polyacrylamide composite gel to separate the protein-RNA complexes formed. Complexes bound to labeled RNA were visualized by exposure to X-ray film overnight.
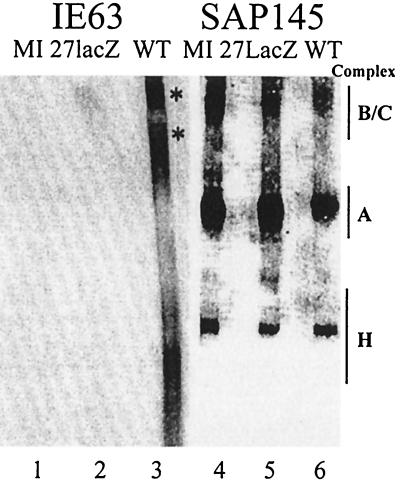
The presence of IE63 and SAP145 in splicing complexes was examined by Western blotting. SAP145 was detected in all three complexes using all the extracts examined (Fig. 7, lanes 4 to 6); however, the amount was reduced in intensity in the A and B/C complexes formed with wt-infected cell extract, and the reduction was again more dramatic in complex B/C than in complex A (Fig. 7, compare lanes 4 and 6). IE63 was not detected in complex A, was seen in the complex H region which comprises RNA-protein complexes that form on RNAs irrespective of whether they are splicing substrates (25) and mainly contains hnRNP proteins (5), and also was present as two larger bands. Interestingly, the upper IE63 band corresponded in mobility to that of the splicing B/C complex, and within this complex, the upper IE63 band and SAP145 migrated at the same rate, while within the H complex they did not (Fig. 7, compare lane 3 with lanes 4 to 6). IE63 was capable of forming complexes with RNA that appear unrelated to splicing, as suggested by its presence in the H complex and as a larger band that did not comigrate with SAP145.
FIG. 7.
IE63 and SAP145 proteins are found in splicing complex B/C. Duplicate samples of the protein complexes, as formed in the results shown in Fig. 6B, were transferred to nitrocellulose and assayed by Western blotting using SAP145 (lanes 4 to 6) or IE63 antiserum (lanes 1 to 3). Complexes were formed using HSV-1 wt-infected (WT), 27-lacZ-infected, or mock-infected (MI) HeLa cell nuclear extracts. The locations of the complexes as detected by autoradiography are indicated on the right-hand side.
DISCUSSION
The interaction of IE63 with SAP145 was identified using the yeast two-hybrid assay and confirmed using GST pull-downs and coimmunoprecipitation from infected cells. Further, no other HSV-1 proteins were required, as GST-IE63 associated with SAP145 with the use of extracts from uninfected cells. The SAP145 and IE63 proteins colocalized in infected cells under conditions in which IE63 was present in clumps with other redistributed snRNP components (39). As previous studies using viruses with mutations in the Sm and KH domains of IE63 showed redistribution of snRNPs, similar to wt infection (53), interaction with SAP145 was unlikely to be the cause of snRNP redistribution.
IE63 associated with its SAP145, hnRNP K, and CK2 partners simultaneously, forming a complex that was not seen in uninfected cells. These interactions could occur by IE63 directly binding each partner. Alternatively, IE63 could potentiate the modification of one or more partners, allowing them to interact with each other or to bind via an unknown intermediate. Recruitment of CK2 to the complex could phosphorylate proteins not normally associated with this kinase, as seen for hnRNP K and p32 partners (4, 67). However, we have no evidence that CK2 in the complex can phosphorylate SAP145, which lacks CK2 consensus phosphorylation sites (data not shown). The ability of IE63 protein to oligomerize (67, 68) could facilitate its interaction with several proteins, and this provides an explanation of its multifunctional activities. Interestingly, while IE63 can interact with hnRNP K, CK2, and p32 simultaneously (4, 67), the IE63-SAP145 and IE63-p32 interactions appear to be mutually exclusive (data not shown), and the region of IE63 (amino acids 241 to 397) identified as interacting with p32 (4) includes the KH1 and KH2 domains that also facilitate the SAP145 interaction. We consider that different conditions could favor the interaction of IE63 with one or another partner, and the binding of one partner could then exclude binding of another. The selective interaction of IE63 with a particular partner protein could reflect timing postinfection, different cellular locations, or alternative protein subfractions.
At the nonpermissive temperature, mutants vBSLG4, vBS3-3, vBS4-3, and vBS5-3 do not shut off host protein synthesis and fail to process β-actin pre-mRNA (59). Reduced SAP145 binding by IE63 was seen at the nonpermissive temperature with these mutants. At the permissive temperature, intermediate levels of host shutoff were observed and host shutoff with vBS5-3 appeared greatest (59). At the permissive temperature, the IE63-SAP145 interaction with vBS5-3 was similar to that for wt, while the vBS4-3 level was considerably reduced. IE63 inhibits splicing, and host shutoff is correlated with inhibition of splicing (18). As SAP145 is a constitutive splicing factor involved in tethering pre-mRNA to the U2 snRNP complex (7), we propose that this association, probably along with other factors, acts to shut off host synthesis via inhibition of splicing. Host shutoff by IE63 need not reflect exclusively the inhibition of splicing, as competition for cellular factors, such as those involved in mRNA export, also could cause this effect.
In this study, two types of domains have been examined, Sm and KH-like. Sm domains allow protein-protein interactions of Sm proteins associated with U1, U2, U4, and U5 spliceosomal RNAs (19, 22). Similar domains are present in Sm-like proteins, which in yeast may be involved in mRNA decay (64), and these potentiate interaction with each other and with U6 snRNA. As SAP145 does not contain an Sm motif, any interaction involving the IE63 Sm domain would not be via a direct Sm-Sm interaction. The D448A mutant in the Sm region alone did not affect the IE63-SAP145 interaction. Unlike the R480H suppressor mutants, which fail to shut off throughout infection, the D448A mutation acts to delay shutoff (60), and extracts from cells infected with this virus were made at a later time when shutoff is apparent. The overlap of Sm and KH3 domains in the other mutants of this region makes it difficult to assess the relative contributions of these two regions. A striking observation was that mutation in either one or more of the KH-like domains reduced or destroyed the IE63-SAP145 interaction. KH domains commonly are involved in RNA binding. Either RNA could potentiate the IE63-SAP145 association by influencing IE63 structure to facilitate contact, or the interaction could be via RNA.
In hnRNP K, KH domains exhibit discrete and independent nucleic acid binding activities (10, 57). In the context of IE63, selectivity of RNA processing and transport could occur through individual KH domains binding distinct viral RNAs (61). Thus, a subpopulation of transcripts containing introns could interact with KH2 or KH1, allowing splicing to be inhibited by the IE63-SAP145 interaction. Selective inhibition of splicing via RNA binding could explain how certain viral transcripts, such as that from the late HSV-1 UL15 gene (2, 9), which contain one or more exons in all herpesvirus genomes sequenced to date, are still spliced during infection. This mechanism would enable the expression of proteins from alternatively spliced viral RNAs as seen for the IE110 gene (6, 14).
Analysis of the products of a splicing reaction using extracts from cells infected with wt virus demonstrated, in agreement with previous data (18), that the lariat-second exon RNA structure did not form, which implies that splicing is inhibited before the first catalytic step. Splicing complex A formed normally, although with reduced intensity; however, the B/C complex was more drastically reduced and shifted up in mobility. Normally, as the first catalytic step occurs, complex B is converted into complex C. The reduction in B/C intensity with wt HSV-1-infected cell extracts is most likely due to complex C not forming (the band present being complex B) but could reflect a reduction in both complexes. The B/C mobility shift suggested the presence of an additional protein(s), and Western blotting revealed the presence of IE63 within this complex but not in the A complex; in the B/C complex, the upper IE63 band migrated similarly to the SAP145-containing band. These data are consistent with a model whereby, at the point of complex B/C formation or conversion, IE63 interacts with SAP145 within the spliceosome and inhibits splicing before the first catalytic step. As IE63 is not found in complex A, pre-mRNA could be bound normally in this complex. The reduction in complex A with wt-infected cell extracts, however, suggests that IE63 also could affect earlier splicing events, such as by sequestering splicing components before they enter the spliceosome, and the interaction with p32 could occur at this point. Further experiments are under way to elucidate the mechanism(s) of splicing inhibition exerted by IE63.
IE63 induces splicing-independent transport of RNAs from the intron-containing α-globin gene irrespective of the presence of introns and, possibly, and to a lesser extent, of transcripts from the HSV-1 ICP0 gene (12). Normally, intron-containing messages need to be spliced prior to export (27). In the presence of IE63, a transcript rescue mechanism (8), analogous to that proposed for human immunodeficiency virus Rev (42), which blocks splicing via the SAP145 interaction to prevent nonproductive interactions with the spliceosome, could enhance RNA 3′ processing and export. In this scheme, the inhibition of splicing would enable certain intron-containing transcripts, or transcripts with cryptic splice sites such as intronless viral RNAs, to access export pathways, with any effects on host cell shutoff being a secondary consequence. As IE63 is proposed to bind viral late gene transcripts in the nucleus and shuttle with them into the cytoplasm to facilitate their expression (52, 59), blocking splicing could facilitate direct IE63-mediated RNA export or RNAs could access alternative pathways.
ACKNOWLEDGMENTS
We thank John McLauchlan for comments on the manuscript.
This work was supported by an award to J.B.C. from the Medical Research Council (G9826324) and sequencing provision was provided by an award from the Wellcome Trust (04674/2/96). Other support was provided by U.S. Public Health Service grant AI-33952 to S.J.S.
REFERENCES
- 1.Ackermann M, Braun D K, Pereira L, Roizman B. Characterization of herpes simplex virus 1 α proteins 0, 4, and 27 with monoclonal antibodies. J Virol. 1984;52:108–118. doi: 10.1128/jvi.52.1.108-118.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baines J D, Roizman B. The cDNA of UL15, a highly conserved herpes simplex virus 1 gene, effectively replaces the two exons of the wild-type virus. J Virol. 1992;66:5621–5626. doi: 10.1128/jvi.66.9.5621-5626.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Behrens S-E, Tyc K, Kastner B, Reichelt J, Lührmann R. Small nuclear ribonucleoprotein (RNP) U2 contains numerous additional proteins and has a bipartite RNP structure under splicing conditions. Mol Cell Biol. 1993;13:307–319. doi: 10.1128/mcb.13.1.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bryant H E, Matthews D A, Wadd S, Scott J E, Kean J, Graham S, Russell W C, Clements J B. Interaction between herpes simplex virus type 1 IE63 protein and cellular protein p32. J Virol. 2000;74:11322–11328. doi: 10.1128/jvi.74.23.11322-11328.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calvio C, Neubauer G, Mann M, Lamond A I. Identification of hnRNP P2 as TLS/FUS using electrospray mass spectrometry. RNA. 1995;1:724–733. [PMC free article] [PubMed] [Google Scholar]
- 6.Carter K L, Roizman B. Alternatively spliced mRNAs predicted to yield frame-shift proteins and stable intron 1 RNAs of the herpes simplex virus regulatory gene α0 accumulate in the cytoplasm of infected cells. Proc Natl Acad Sci USA. 1996;93:12535–12540. doi: 10.1073/pnas.93.22.12535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Champion-Arnaud P, Reed R. The prespliceosome components SAP49 and SAP145 interact in a complex implicated in tethering U2 snRNP to the branch site. Genes Dev. 1996;8:1974–1983. doi: 10.1101/gad.8.16.1974. [DOI] [PubMed] [Google Scholar]
- 8.Cheung P, Ellison K S, Verity R, Smiley J R. Herpes simplex virus ICP27 induces cytoplasmic accumulation of unspliced polyadenylated α-globin pre-mRNA in infected HeLa cells. J Virol. 2000;74:2913–2919. doi: 10.1128/jvi.74.6.2913-2919.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costa R H, Draper K G, Kelly T J, Wagner E K. An unusual spliced herpes simplex virus type 1 transcript with sequence homology to Epstein-Barr virus DNA. J Virol. 1985;54:317–328. doi: 10.1128/jvi.54.2.317-328.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dejgaard K, Leffers H. Characterisation of the nucleic acid binding activity of KH domains. Different properties of different domains. Eur J Biochem. 1996;241:425–431. doi: 10.1111/j.1432-1033.1996.00425.x. [DOI] [PubMed] [Google Scholar]
- 11.DeLuca N A, McCarthy A M, Schaffer P A. Isolation and characterization of deletion mutants of herpes simplex virus type 1 in the gene encoding immediate-early regulatory protein ICP4. J Virol. 1985;56:558–570. doi: 10.1128/jvi.56.2.558-570.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ellison K S, Rice S A, Verity R, Smiley J R. Processing of α-globin in cells infected with herpes simplex virus type 1 ICP27 mutants. J Virol. 2000;74:7307–7319. doi: 10.1128/jvi.74.16.7307-7319.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Everett R D. Trans-activation of transcription by herpes virus products: requirement for two HSV-1 immediate-early polypeptides for maximum activity. EMBO J. 1984;3:3135–3141. doi: 10.1002/j.1460-2075.1984.tb02270.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Everett R D, Cross A, Orr A. A truncated form of herpes-simplex virus type 1 immediate-early protein Vmw110 is expressed in a cell-type dependent manner. Virology. 1993;197:751–756. doi: 10.1006/viro.1993.1651. [DOI] [PubMed] [Google Scholar]
- 15.Gelman I H, Silverstein S. Identification of immediate early genes from herpes simplex virus that transactivate the virus thymidine kinase gene. Proc Natl Acad Sci USA. 1985;82:5265–5269. doi: 10.1073/pnas.82.16.5265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hardwicke A M, Vaughan P J, Sekulovich R E, O'Conner R, Sandri-Goldin R M. The regions important for the activator and repressor functions of herpes simplex virus type 1 α protein ICP27 map to the C-terminal half of the molecule. J Virol. 1989;63:4590–4602. doi: 10.1128/jvi.63.11.4590-4602.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hardwicke A M, Sandri-Goldin R M. The herpes simplex virus regulatory protein ICP27 contributes to the decrease in cellular mRNA levels during infection. J Virol. 1994;68:4797–4810. doi: 10.1128/jvi.68.8.4797-4810.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hardy W R, Sandri-Goldin R M. Herpes simplex virus inhibits host cell splicing, and regulatory protein ICP27 is required for this effect. J Virol. 1994;68:7790–7799. doi: 10.1128/jvi.68.12.7790-7799.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hermann H, Fabrizio P, Raker V A, Foulaki K, Hornig H, Brahms H, Luhrmann R. SnRNP Sm proteins share two evolutionarily conserved motifs which are involved in Sm protein-protein interactions. EMBO J. 1995;14:2076–2088. doi: 10.1002/j.1460-2075.1995.tb07199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hill A, Jurgovic P, York I, Bennink J, Yewdell J, Ploegh H I, Johnson D. Herpes simplex virus turns off the TAP to evade host immunity. Nature. 1995;375:411–415. doi: 10.1038/375411a0. [DOI] [PubMed] [Google Scholar]
- 21.Ingram A, Phelan A, Dunlop J, Clements J B. Immediate early protein IE63 of herpes simplex virus type 1 binds RNA directly. J Gen Virol. 1996;77:1847–1851. doi: 10.1099/0022-1317-77-8-1847. [DOI] [PubMed] [Google Scholar]
- 22.Kambach C, Walke S, Young R, Avis J M, de la Fortelle E, Raker V A, Li R, Luhrmann J, Nagai K. Crystal structures of two Sm protein complexes and their implications for the assembly of the spliceosomal snRNPs. Cell. 1999;96:375–387. doi: 10.1016/s0092-8674(00)80550-4. [DOI] [PubMed] [Google Scholar]
- 23.Konarska M M, Sharp P A. Interactions between small nuclear ribonucleoprotein particles in formation of spliceosomes. Cell. 1987;49:763–774. doi: 10.1016/0092-8674(87)90614-3. [DOI] [PubMed] [Google Scholar]
- 24.Lamond A I, Earnshaw W C. Structure and function in the nucleus. Science. 1998;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- 25.Lamond A I, Konarska M M, Sharp P A. A mutational analysis of spliceosome assembly: evidence for splice site collaboration during spliceosome formation. Genes Dev. 1987;1:532–543. doi: 10.1101/gad.1.6.532. [DOI] [PubMed] [Google Scholar]
- 26.Lamond A I, Sproat B, Ryder U, Hamm J. Probing the structure and function of U2 Sn RNP with antisense oligonucleotides made of 2′-Ome RNA. Cell. 1989;58:383–390. doi: 10.1016/0092-8674(89)90852-0. [DOI] [PubMed] [Google Scholar]
- 27.Legrain P, Seraphin B, Rosbash M. Early commitment of yeast pre-mRNA to the spliceosome pathway. Mol Cell Biol. 1988;8:3755–3760. doi: 10.1128/mcb.8.9.3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Martin T E, Barghusen S C, Leser G P, Spear P G. Redistribution of the nuclear ribonucleoprotein antigens during herpes simplex virus infection. J Cell Biol. 1987;105:2069–2082. doi: 10.1083/jcb.105.5.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCarthy A M, McMahan L, Schaffer P A. Herpes simplex virus type 1 ICP27 deletion mutants exhibit altered patterns of transcription and are DNA deficient. J Virol. 1989;66:18–27. doi: 10.1128/jvi.63.1.18-27.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McGeoch D J, Schaffer P A. Herpes simplex virus. In: O'Brien S J, editor. Genetic maps. 6th ed. Vol. 1. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 147–156. [Google Scholar]
- 31.McGregor F, Phelan A, Dunlop J, Clements J B. Regulation of herpes simplex virus type 1 poly(A) site usage and the action of the immediate-early protein IE63 in the early-late switch. J Virol. 1996;70:1931–1940. doi: 10.1128/jvi.70.3.1931-1940.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McMahan L, Schaffer P A. The repressing and enhancing functions of herpes simplex virus regulatory protein ICP27 map to the C-terminal regions and are required to modulate viral gene expression very early in infection. J Virol. 1990;64:3471–3485. doi: 10.1128/jvi.64.7.3471-3485.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mears W E, Lam V, Rice S A. Identification of nuclear and nucleolar localization signals in the herpes simplex virus regulatory protein ICP27. J Virol. 1995;69:935–947. doi: 10.1128/jvi.69.2.935-947.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mears W E, Rice S A. The RGG box motif of the herpes simplex virus ICP27 protein mediates an RNA-binding activity and determines in vivo methylation. J Virol. 1996;70:7445–7453. doi: 10.1128/jvi.70.11.7445-7453.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mears W E, Rice S A. The herpes simplex virus immediate-early protein ICP27 shuttles between nucleus and cytoplasm. Virology. 1998;242:128–137. doi: 10.1006/viro.1997.9006. [DOI] [PubMed] [Google Scholar]
- 36.Michelotti E F, Michelotti G A, Aronsohn A I, Levens D. Heterogeneous nuclear ribonucleoprotein K is a transcription factor. Mol Cell Biol. 1996;16:2350–2360. doi: 10.1128/mcb.16.5.2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Hare P, Hayward G S. Evidence for a direct role for both the 175,000- and 110,000-molecular-weight immediate-early proteins of herpes simplex virus in the transactivation of delayed-early promoters. J Virol. 1985;53:751–760. doi: 10.1128/jvi.53.3.751-760.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Padgett R A, Konarska M, Grabowski P J, Hardy S F, Sharp P A. Lariat RNAs as intermediates and products in the splicing of messenger RNA precursors. Science. 1984;225:898–903. doi: 10.1126/science.6206566. [DOI] [PubMed] [Google Scholar]
- 39.Phelan A, Carmo-Fonseca M, McLauchlan J, Lamond A I, Clements J B. A herpes simplex virus type 1 immediate-early gene product, IE63, regulates small nuclear ribonucleoprotein distribution. Proc Natl Acad Sci USA. 1993;90:9056–9060. doi: 10.1073/pnas.90.19.9056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Phelan A, Clements J B. Herpes simplex virus type 1 immediate-early protein IE63 shuttles between nuclear compartments and the cytoplasm. J Gen Virol. 1997;78:3327–3331. doi: 10.1099/0022-1317-78-12-3327. [DOI] [PubMed] [Google Scholar]
- 41.Phelan A, Clements J B. Posttranscriptional regulation in herpes simplex virus. Semin Virol. 1998;8:309–318. [Google Scholar]
- 42.Pollard V W, Malim M H. The HIV Rev protein. Annu Rev Microbiol. 1998;52:491–532. doi: 10.1146/annurev.micro.52.1.491. [DOI] [PubMed] [Google Scholar]
- 43.Reed R. Mechanisms of fidelity in pre-mRNA splicing. Curr Opin Cell Biol. 2000;12:340–345. doi: 10.1016/s0955-0674(00)00097-1. [DOI] [PubMed] [Google Scholar]
- 44.Rice S A, Knipe D M. Gene-specific transactivation by herpes simplex virus type 1 α protein ICP27. J Virol. 1988;62:3814–3823. doi: 10.1128/jvi.62.10.3814-3823.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rice S A, Knipe D M. Genetic evidence for two distinct transactivation functions of the herpes simplex virus α protein ICP27. J Virol. 1990;64:1704–1715. doi: 10.1128/jvi.64.4.1704-1715.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rice S A, Lam V, Knipe D M. The acidic amino-terminal region of herpes simplex virus type 1 alpha protein ICP27 is required for an essential lytic function. J Virol. 1993;67:1778–1787. doi: 10.1128/jvi.67.4.1778-1787.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rice S A, Lam V. Amino acid substitution mutations in the herpes simplex virus ICP27 protein define an essential gene regulation function. J Virol. 1994;68:823–833. doi: 10.1128/jvi.68.2.823-833.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roizman B, Sears A. The human herpes viruses: biology, pathogenesis and treatment. New York, N.Y: Raven Press; 1993. [Google Scholar]
- 49.Ruskin B, Krainer A R, Maniatis T, Green M R. Excision of an intact intron as a novel lariat structure during pre-mRNA splicing in vitro. Cell. 1984;38:317–331. doi: 10.1016/0092-8674(84)90553-1. [DOI] [PubMed] [Google Scholar]
- 50.Sacks W R, Greene C C, Aschman D P, Schaffer P A. Herpes simplex virus type 1 ICP27 is an essential regulatory protein. J Virol. 1985;55:796–805. doi: 10.1128/jvi.55.3.796-805.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sacks W R, Schaffer P A. Deletion mutants in the gene encoding the herpes simplex virus type 1 immediate-early protein ICP0 exhibit impaired growth in cell culture. J Virol. 1987;61:829–839. doi: 10.1128/jvi.61.3.829-839.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sandri-Goldin R M. ICP27 mediates HSV RNA export by shuttling through a leucine-rich nuclear export signal and binding viral intronless RNAs through an RGG motif. Genes Dev. 1998;12:868–879. doi: 10.1101/gad.12.6.868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sandri-Goldin R M, Hibbard M K, Hardwicke M A. The C-terminal repressor region of herpes simplex virus type 1 ICP27 is required for the redistribution of small nuclear ribonucleoprotein particles and splicing factor SC35; however, these alterations are not sufficient to inhibit host cell splicing. J Virol. 1995;69:6063–6076. doi: 10.1128/jvi.69.10.6063-6076.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sears A E, Halliburton I W, Meignier B, Silver S, Roizman B. Herpes simplex virus 1 mutant deleted in the α22 gene: growth and gene expression in permissive and restrictive cells and establishment of latency in mice. J Virol. 1985;55:338–346. doi: 10.1128/jvi.55.2.338-346.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seghezzi W, Chua K, Shanahan F, Gozani O, Reed R, Lees E. Cyclin E associates with components of the pre-mRNA splicing machinery in mammalian cells. Mol Cell Biol. 1998;18:4526–4536. doi: 10.1128/mcb.18.8.4526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sekulovich R E, Leary K, Sandri-Goldin R M. The herpes simplex virus type 1 α protein ICP27 can act as a trans-repressor or a trans-activator in combination with ICP4 and ICP0. J Virol. 1988;62:4510–4522. doi: 10.1128/jvi.62.12.4510-4522.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Siomi H, Choi M C, Siomi M C, Nussbaum R L, Dreyfuss G. Essential role for KH domains in RNA binding: impaired RNA binding by a mutation in the KH domain of FMR1 that causes fragile X syndrome. Cell. 1994;77:33–39. doi: 10.1016/0092-8674(94)90232-1. [DOI] [PubMed] [Google Scholar]
- 58.Smith I L, Hardwicke M A, Sandri-Goldin R M. Evidence that the herpes simplex virus immediate-early protein ICP27 acts post-transcriptionally during infection to regulate gene expression. Virology. 1992;186:74–86. doi: 10.1016/0042-6822(92)90062-t. [DOI] [PubMed] [Google Scholar]
- 59.Soliman T M, Sandri-Goldin R M, Silverstein S J. Shuttling of the herpes simplex virus type 1 regulatory protein ICP27 between the nucleus and the cytoplasm mediates the expression of late proteins. J Virol. 1997;71:9188–9197. doi: 10.1128/jvi.71.12.9188-9197.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Soliman T M, Silverstein S J. Herpesvirus mRNAs are sorted for export via Crm-1-dependent and -independent pathways. J Virol. 2000;74:2814–2825. doi: 10.1128/jvi.74.6.2814-2825.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Soliman T M, Silverstein S J. Identification of an export control sequence and a requirement for the KH domains in ICP27 from herpes simplex virus type 1. J Virol. 2000;74:7600–7609. doi: 10.1128/jvi.74.16.7600-7609.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Staknis D, Reed R. Direct interactions between pre-mRNA and six U2 small nuclear ribonucleoproteins during spliceosome assembly. Mol Cell Biol. 1994;14:2994–3005. doi: 10.1128/mcb.14.5.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Su L, Knipe D M. Herpes simplex virus α protein ICP27 can inhibit or augment viral gene transactivation. Virology. 1989;170:496–504. doi: 10.1016/0042-6822(89)90441-8. [DOI] [PubMed] [Google Scholar]
- 64.Tharun S, He W, Mayes A E, Lennertz P, Beggs J D, Parker R. Yeast Sm-like proteins function in mRNA decapping and decay. Nature. 2000;404:515–518. doi: 10.1038/35006676. [DOI] [PubMed] [Google Scholar]
- 65.Uprichard S L, Knipe D M. Herpes simplex ICP27 mutant viruses exhibit reduced expression of specific DNA replication genes. J Virol. 1996;70:1969–1980. doi: 10.1128/jvi.70.3.1969-1980.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vaughan P J, Thibault K J, Hardwicke M A, Sandri-Goldin R M. The herpes simplex virus immediate early protein ICP27 encodes a potential metal binding domain and binds zinc in vitro. Virology. 1992;189:377–384. doi: 10.1016/0042-6822(92)90720-a. [DOI] [PubMed] [Google Scholar]
- 67.Wadd S, Bryant H E, Filhol O, Scott J E, Hsieh T, Everett R, Clements J B. The multifunctional herpes simplex virus IE63 protein interacts with heterogeneous ribonucleoprotein K and with casein kinase 2. J Biol Chem. 1999;274:28991–28998. doi: 10.1074/jbc.274.41.28991. [DOI] [PubMed] [Google Scholar]
- 68.Watson R J, Clements J B. A herpes simplex virus type 1 function required for early and late virus RNA synthesis. Nature. 1980;285:329–330. doi: 10.1038/285329a0. [DOI] [PubMed] [Google Scholar]
- 69.Zhi Y, Sciabica K F, Sandri-Goldin R M. Self-interaction of the herpes simplex virus type 1 regulatory protein ICP 27. Virology. 1999;257:341–351. doi: 10.1006/viro.1999.9698. [DOI] [PubMed] [Google Scholar]