Abstract
Autonomous parvovirus minute virus of mice (MVM) DNA replication is strictly dependent on cellular factors expressed during the S phase of the cell cycle. Here we report that MVM DNA replication proceeds in specific nuclear structures termed autonomous parvovirus-associated replication bodies, where components of the basic cellular replication machinery accumulate. The presence of DNA polymerases α and δ in these bodies suggests that MVM utilizes partially preformed cellular replication complexes for its replication. The recruitment of cyclin A points to a role for this cell cycle factor in MVM DNA replication beyond its involvement in activating the conversion of virion single-stranded DNA to the duplex replicative form.
Autonomous parvoviruses and other members of the Parvoviridae family are unique among animal viruses in having linear single-stranded DNA genomes. The termini of their approximately 5,000-nucleotide genomes contain palindromic sequences which fold into stable hairpin structures and serve as primers for viral DNA synthesis. The replicative cycle, which takes place in the nucleus, is initiated by the synthesis of the cDNA strand leading to the formation of double-stranded replicative-form DNA. This reaction, also known as conversion, is exclusively dependent on cellular factors. In addition, the subsequent amplification reactions require the activity of the virus-encoded major nonstructural protein NS1 (7).
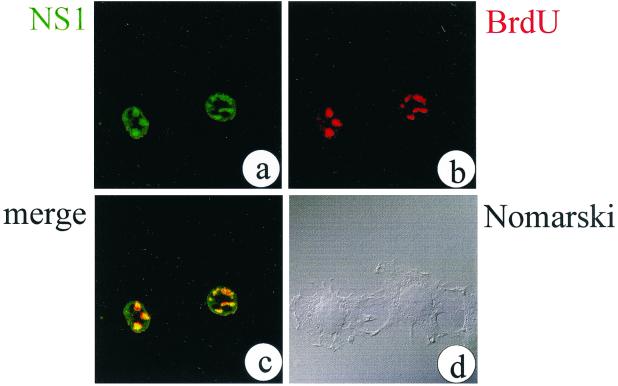
Several cellular factors which are essential for DNA replication of the prototype autonomous parvovirus minute virus of mice (MVM) have been identified through cell fractionation and complementation assays in vitro replication systems (5, 8, 20). However, very little is known about the subnuclear organization of MVM DNA replication in vivo. In contrast to viruses with double-stranded DNA genomes that replicate in close proximity to preformed nuclear structures known as promyelocytic leukemia (PML) bodies (for review see reference 18), autonomous parvovirus H-1 was shown to induce characteristic nuclear structures, termed H-1 parvovirus-associated replication (PAR) bodies, which are unrelated to PML bodies (9). Similar structures were also characterized in Aleutian mink disease virus-infected cells (21, 22). In this study, we aimed to determine whether MVM also establishes PAR body-like structures in the nuclei of infected cells and to analyze the subnuclear distribution of cellular factors assumed to be involved in MVM DNA replication in vivo. To this end, A9 mouse cells were infected with MVMp at a multiplicity of infection of 10 PFU per cell. At 15 h postinfection, cells were labeled for 20 min with bromodeoxyuridine (BrdU) at a concentration of 10 μM and then immediately fixed in 1% formaldehyde for 10 min at room temperature. BrdU incorporation into replicated viral DNA was detected with a BrdU-specific antibody (Becton Dickinson) without prior denaturation, thus excluding the detection of chromosomal DNA replication (21). Simultaneously, NS1 was detected with the NS1-specific SP8 antibody (11). Analysis by confocal microscopy (LSM510 UV; Zeiss, Jena, Germany) revealed the accumulation of NS1 in specific nuclear bodies (Fig. 1a) which were also found to be the sites of ongoing viral DNA replication, as indicated by BrdU incorporation (Fig. 1b and c). At this time postinfection, no signs of virus-induced cytotoxicity were visible (Fig. 1d). From these data, we concluded that MVM DNA replication proceeds in specific nuclear structures similar to the ones previously described for other autonomous parvoviruses (9, 21, 22). We would therefore like to propose the more general term autonomous PAR (APAR) bodies for these virus-induced structures.
FIG. 1.
MVM DNA replication colocalizes with NS1 in APAR bodies in the nuclei of infected A9 cells. Representative confocal optical sections through the nuclei of infected cells are shown. NS1 was localized with the SP8 polyclonal antiserum and a fluorescein isothiocyanate (FITC)-conjugated secondary antibody (a). Replication was monitored by incorporation of BrdU and indirect immunofluorescence using a BrdU-specific antibody and a tetramethyl rhodamine isothiocyanate (TRITC)-conjugated secondary antibody (b). In a merged image, colocalized structures from panels a and b appear yellow (c). By phase-contrast microscopy (Nomarski), the cells show no obvious sign of NS1-induced cytotoxicity at the time of fixation (15 h postinfection) (d).
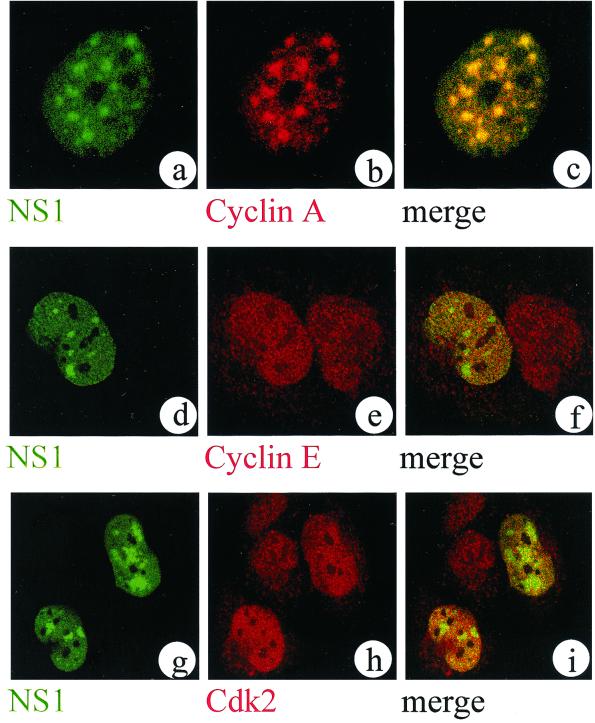
MVM DNA replication starts only after host cells enter the S phase of the cell cycle. Recently, it was shown that in vitro the conversion reaction is activated by the cell cycle factor cyclin A, whose production is induced at the G1/S transition (1). Cyclin A is involved in the regulation of the S and G2 phases (25) and has been reported to be required for chromosomal (16) and simian virus 40 (10, 12) DNA replication. In order to test whether cyclin A is present in APAR bodies, we performed double immunofluorescence labeling and confocal microscopical analysis of MVM-infected A9 cells. Cyclin A, which is homogeneously distributed throughout the nuclei of mock-infected cells and absent from nucleoli (reference 2 and data not shown), was indeed found to massively accumulate together with NS1 in the APAR bodies of infected cells (Fig. 2a to c). Even if one assumes that the subnuclear location at which the conversion reaction takes place predetermines the site of APAR body formation, a sole role for cyclin A in the conversion reaction is difficult to reconcile with such a strong accumulation of cyclin A in APAR bodies at 15 h postinfection, when conversion is thought to be complete. This result could indicate that the continuous presence of cyclin A is also required for subsequent NS1-dependent replicative steps. One can furthermore speculate that the sequestration of this factor is responsible for the previously described parvovirus-induced cell cycle arrest (23, 24) because deprivation of cyclin A may lead to a failure of cdk1 activation in the G2 phase, thus preventing cell cycle progression.
FIG. 2.
Cyclin A, but not cyclin E or cdk2, accumulates in APAR bodies. Representative confocal images of nuclei from double-labeled MVM-infected A9 (a to c) or NBE (d to i) cells are shown. NS1 was detected with FITC using the NS1-specific 3D9 antibody (a generous gift from N. Salomé and D. Pintel) (a, d, and g). Images of cyclin A (b), cyclin E (e), and cdk2 (h) were detected with TRITC in the same confocal plane as shown in the left column using the respective primary antisera for cyclin A (a generous gift from M. Pagano), cyclin E (Ab-1; Neomarkers, Fremont, Calif.), and cdk2 (Ab-3; Neomarkers). The superimposition of both channels from the left and middle columns reveals either the colocalization (yellow signal [c]) or the lack of colocalization (distinct red and green signals [f and i]) of the respective factors in relationship to APAR bodies.
Besides cyclin A, cyclin E is involved in the regulation of the G1/S transition and is known to play a role in chromosomal DNA replication (14, 16). In contrast to cyclin A, however, cyclin E did not accumulate in APAR bodies (Fig. 2d to f). Since the cyclin E-specific antibody best recognized the human protein, in these experiments we infected human NBE cells and verified that MVMp indeed induces the formation of APAR bodies in this human cell line, as detected by NS1-specific (Fig. 2d) and BrdU-specific (data not shown) antibodies. The failure to detect an accumulation of cyclin E does not exclude the presence of minute amounts, of this factor in APAR bodies. Our data are, however, in agreement with recent in vitro experiments showing no contribution of cyclin E or cdk2 in the initiation of conversion (1).
Cyclin A forms an active complex with cdk2 at the beginning of the S phase. This complex is assumed to be involved in the regulation of DNA replication, since both cyclin A and cdk2 were found to be associated with replication foci in mammalian cells (3) as well as with replicating DNA in the simian virus 40 in vitro replication system (13). We therefore attempted to detect cdk2 in APAR bodies. Using several different cdk2-specific antibodies, cdk2 could not be shown to accumulate in APAR bodies (Fig. 2g to i). The observed accumulation of cyclin A in APAR bodies (Fig. 2b) would then point to a structural role for cyclin A in these bodies, independent of complex formation with cdk2. However, it is still possible that only catalytic amounts too small to be detected by the method used here are required for MVM DNA replication in APAR bodies.
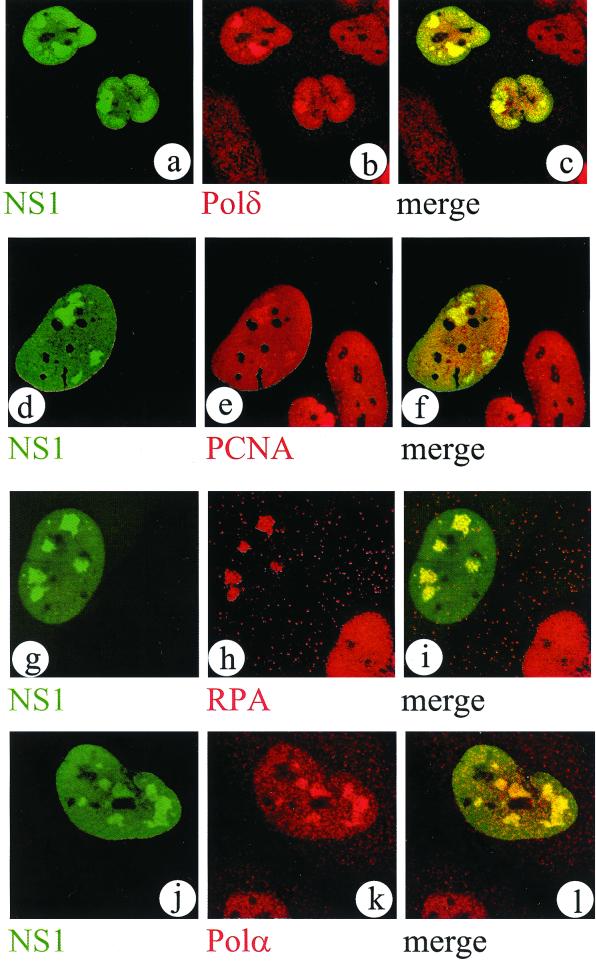
Parvovirus DNA replication proceeds according to a leading-strand-synthesis mechanism and has been shown to be dependent on proliferating cell nuclear antigen (PCNA) in vitro (5). These and other lines of evidence point to DNA polymerase δ as the enzyme responsible for parvovirus DNA replication (6). Here, we show that DNA polymerase δ indeed accumulates in APAR bodies in MVM-infected NBE cells (Fig. 3a to c) and therefore at the sites of ongoing MVM DNA replication. DNA polymerase δ requires for processive DNA synthesis the presence of the cofactor PCNA, which is also involved in other processes, like DNA recombination and repair (15). PCNA is distributed throughout the nucleus except the nucleoli, but it specifically accumulates in the APAR bodies of MVM-infected NBE cells (Fig. 3d to f). This finding corroborates earlier in vitro data demonstrating that PCNA is indispensable for MVM DNA replication (1, 5).
FIG. 3.
Cellular replication factors DNA polymerases δ and α, PCNA, and RPA accumulate in APAR bodies. Representative confocal images of nuclei from double-labeled MVM-infected NBE cells are shown. NS1 was detected with FITC using the SP8 antiserum (a, d, g, and j). Replication factors were detected with TRITC in the same confocal plane as shown in the left column, using the respective antibodies against DNA polymerase δ (Transduction Laboratories) (b), PCNA (PC10; Upstate Biotechnology) (e), RPA (Ab-1; Oncogene) (h), and DNA polymerase α (SJK132-20 [26]) (k), respectively. A merged image of both channels from the left and center columns provides evidence for colocalization of these factors in APAR bodies (c, f, i, and l).
The single-strand-DNA-binding protein replication protein A (RPA) is required for MVM DNA replication in vitro (5) and cannot be replaced by other proteins with single-strand-DNA-binding activity (J. Christensen, personal communication). Furthermore, a direct interaction between the 70-kDa subunit of RPA and NS1 was demonstrated in vitro (4). In agreement with these in vitro findings we have observed that RPA massively accumulates at the sites of ongoing parvovirus DNA replication in infected cells (Fig. 3g to i), which strongly suggests its involvement in this process in vivo.
Surprisingly, DNA polymerase α was also found at the sites of MVM DNA replication (Fig. 3j to l). So far, there is no evidence for a contribution of DNA polymerase α to parvovirus DNA replication, since neutralizing antibodies against this enzyme do not impair viral DNA replication in vitro (5, 19). Furthermore, the presence of preformed hairpin primers makes a DNA polymerase α-dependent primase activity dispensable. However, it was shown that DNA polymerase α can be isolated together with DNA polymerase δ and the PCNA loading factor, replication factor C, from mammalian cells as a stable complex that is replication competent when template DNA, PCNA, and nucleotides are added (17). It could be demonstrated that in this complex, DNA polymerase α is active on single-stranded-DNA templates only when primer synthesis is a precondition for replication. On a primer-template junction, as present in parvovirus DNA, the DNA-binding affinity of replication factor C is higher than that of DNA polymerase α, thereby rendering the latter inactive (17, 27). Since DNA polymerase α is required for chromosomal DNA replication, it is tempting to speculate that MVM utilizes cellular replication complexes that are at least partially preformed, exploiting only the DNA polymerase δ activity and thus evading the need for an energy-consuming dissociation of preformed cellular complexes. This would again highlight the opportunistic and minimalistic strategies used by these viruses.
In summary, the data presented here show for the first time the formation of APAR bodies in MVM-infected cells and the accumulation of various cellular replication factors in these structures. Our data support the idea that the basic cellular replication machinery is used by MVM for viral DNA replication, including DNA polymerase δ, PCNA, RPA, and cyclin A. DNA polymerase α, which is part of the cellular replication machinery, was unexpectedly found to accumulate in APAR bodies but is most likely not active in parvovirus DNA replication. For cyclin A, it will be of interest to identify the role this factor plays in parvovirus DNA replication, in addition to its role in the activation of the conversion reaction in vitro.
Acknowledgments
We acknowledge the generous gift of antibodies from D. Pintel and N. Salomé (University of Missouri—Columbia) and antiserum from M. Pagano (New York University, New York, N.Y.). The expert support of H. Spring (DKFZ, Heidelberg, Germany) with acquisition of data by confocal microscopy is gratefully acknowledged.
REFERENCES
- 1.Bashir T, Horlein R, Rommelaere J, Willwand K. Cyclin A activates the DNA polymerase delta-dependent elongation machinery in vitro: a parvovirus DNA replication model. Proc Natl Acad Sci USA. 2000;97:5522–5527. doi: 10.1073/pnas.090485297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brenot-Bosc F, Gupta S, Margolis R L, Fotedar R. Changes in the subcellular localization of replication initiation proteins and cell cycle proteins during G1-to S-phase transition in mammalian cells. Chromosoma. 1995;103:517–527. doi: 10.1007/BF00355316. [DOI] [PubMed] [Google Scholar]
- 3.Cardoso M C, Leonhardt H, Nadal-Ginard B. Reversal of terminal differentiation and control of DNA replication: cyclin A and Cdk2 specifically localize at subnuclear sites of DNA replication. Cell. 1993;74:979–992. doi: 10.1016/0092-8674(93)90721-2. [DOI] [PubMed] [Google Scholar]
- 4.Christensen J. Binding of the major non-structural protein (NS1) of MVM to the large subunit of human replication protein is involved in origin unwinding. Infect Dis Rev. 2000;2:152. [Google Scholar]
- 5.Christensen J, Cotmore S F, Tattersall P. A novel cellular site-specific DNA-binding protein cooperates with the viral NS1 polypeptide to initiate parvovirus DNA replication. J Virol. 1997;71:1405–1416. doi: 10.1128/jvi.71.2.1405-1416.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cossons N, Faust E A, Zannis-Hadjopoulos M. DNA polymerase delta-dependent formation of a hairpin structure at the 5′ terminal palindrome of the minute virus of mice genome. Virology. 1996;216:258–264. doi: 10.1006/viro.1996.0058. [DOI] [PubMed] [Google Scholar]
- 7.Cotmore S F, Tattersall P. The autonomously replicating parvoviruses of vertebrates. Adv Virus Res. 1987;33:91–174. doi: 10.1016/s0065-3527(08)60317-6. [DOI] [PubMed] [Google Scholar]
- 8.Cotmore S F, Tattersall P. High-mobility group 1/2 proteins are essential for initiating rolling-circle-type DNA replication at a parvovirus hairpin origin. J Virol. 1998;72:8477–8484. doi: 10.1128/jvi.72.11.8477-8484.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cziepluch C, Lampel S, Grewenig A, Grund C, Lichter P, Rommelaere J. H-1 parvovirus-associated replication bodies: a distinct virus-induced nuclear structure. J Virol. 2000;74:4807–4815. doi: 10.1128/jvi.74.10.4807-4815.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D'Urso G, Marraccino R L, Marshak D R, Roberts J M. Cell cycle control of DNA replication by a homologue from human cells of the p34cdc2 protein kinase. Science. 1990;250:786–791. doi: 10.1126/science.2173140. [DOI] [PubMed] [Google Scholar]
- 11.Faisst S, Faisst S R, Dupressoir T, Plaza S, Pujol A, Jauniaux J-C, Rhode S L, Rommelaere J. Isolation of a fully infectious variant of parvovirus H-1 supplanting the standard strain in human cells. J Virol. 1995;69:4538–4543. doi: 10.1128/jvi.69.7.4538-4543.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fotedar A, Cannella D, Fitzgerald P, Rousselle T, Gupta S, Doree M, Fotedar R. Role for cyclin A-dependent kinase in DNA replication in human S phase cell extracts. J Biol Chem. 1996;271:31627–31637. doi: 10.1074/jbc.271.49.31627. [DOI] [PubMed] [Google Scholar]
- 13.Fotedar R, Roberts J M. Association of p34cdc with replicating DNA. Cold Spring Harbor Symp Quant Biol. 1991;56:325–333. doi: 10.1101/sqb.1991.056.01.039. [DOI] [PubMed] [Google Scholar]
- 14.Jackson P K, Chevalier S, Philippe M, Kirschner M W. Early events in DNA replication require cyclin E and are blocked by p21CIP1. J Cell Biol. 1995;130:755–769. doi: 10.1083/jcb.130.4.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jonsson Z O, Hubscher U. Proliferating cell nuclear antigen: more than a clamp for DNA polymerases. Bioessays. 1997;19:967–975. doi: 10.1002/bies.950191106. [DOI] [PubMed] [Google Scholar]
- 16.Krude T, Jackman M, Pines J, Laskey R A. Cyclin/Cdk-dependent initiation of DNA replication in a human cell-free system. Cell. 1997;88:109–119. doi: 10.1016/s0092-8674(00)81863-2. [DOI] [PubMed] [Google Scholar]
- 17.Maga G, Hubscher U. DNA replication machinery: functional characterization of a complex containing DNA polymerase alpha, DNA polymerase delta, and replication factor C suggests an asymmetric DNA polymerase dimer. Biochemistry. 1996;35:5764–5777. doi: 10.1021/bi952455k. [DOI] [PubMed] [Google Scholar]
- 18.Maul G G. Nuclear domain 10, the site of DNA virus transcription and replication. Bioessays. 1998;20:660–667. doi: 10.1002/(SICI)1521-1878(199808)20:8<660::AID-BIES9>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 19.Ni T-H, McDonald W F, Zolotukhin I, Melendy T, Waga S, Stillman B, Muzyczka N. Cellular proteins required for adeno-associated virus DNA replication in the absence of adenovirus coinfection. J Virol. 1998;72:2777–2787. doi: 10.1128/jvi.72.4.2777-2787.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nüesch J P F, Dettwiler S, Corbau R, Rommelaere J. Replicative functions of minute virus of mice NS1 protein are regulated in vitro by phosphorylation through protein kinase C. J Virol. 1998;72:9966–9977. doi: 10.1128/jvi.72.12.9966-9977.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Oleksiewicz M B, Costello F, Huhtanen M, Wolfinbarger J B, Alexandersen S, Bloom M E. Subcellular localization of Aleutian mink disease parvovirus proteins and DNA during permissive infection of Crandell feline kidney cells. J Virol. 1996;70:3242–3247. doi: 10.1128/jvi.70.5.3242-3247.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oleksiewicz M B, Wolfinbarger J B, Bloom M E. A comparison between permissive and restricted infections with Aleutian mink disease parvovirus (ADV): characterization of the viral protein composition at nuclear sites of virus replication. Virus Res. 1998;56:41–51. doi: 10.1016/s0168-1702(98)00053-7. [DOI] [PubMed] [Google Scholar]
- 23.Op De Beeck A, Anouja F, Mousset S, Rommelaere J, Caillet-Fauquet P. The nonstructural proteins of the autonomous parvovirus minute virus of mice interfere with the cell cycle, inducing accumulation in G2. Cell Growth Differ. 1995;6:781–787. [PubMed] [Google Scholar]
- 24.Op De Beeck A, Caillet-Fauquet P. The NS1 protein of the autonomous parvovirus minute virus of mice blocks cellular DNA replication: a consequence of lesions to the chromatin? J Virol. 1997;71:5323–5329. doi: 10.1128/jvi.71.7.5323-5329.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pagano M, Pepperkok R, Verde F, Ansorge W, Draetta G. Cyclin A is required at two points in the human cell cycle. EMBO J. 1992;11:961–971. doi: 10.1002/j.1460-2075.1992.tb05135.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanaka S, Hu S Z, Wang T S, Korn D. Preparation and preliminary characterization of monoclonal antibodies against human DNA polymerase alpha. J Biol Chem. 1982;257:8386–8390. [PubMed] [Google Scholar]
- 27.Tsurimoto T, Stillman B. Replication factors required for SV40 DNA replication in vitro. I. DNA structure-specific recognition of a primer-template junction by eukaryotic DNA polymerases and their accessory proteins. J Biol Chem. 1991;266:1950–1960. [PubMed] [Google Scholar]