Abstract
Background:
Analyzing continuous glucose monitoring (CGM) data is a mandatory step for multiple purposes spanning from reporting clinical trial outcomes to developing new algorithms for diabetes management. This task is repetitive, and scientists struggle in computing literature glucose control metrics and waste time in reproducing possibly complex plots and reports. For this reason, to provide the diabetes technology community a unified tool, here we present Automated Glucose dATa Analysis (AGATA), an automated glucose data analysis toolbox developed in MATLAB/Octave.
Methods:
Automated Glucose dATa Analysis is an open-source software program to visualize and preprocess CGM data, compute glucose control metrics, detect adverse events, evaluate the effectiveness of users’ prediction algorithms, and compare study arms. Automated Glucose dATa Analysis can be used as a standalone computer application accessible through a dedicated graphical user interface, particularly suitable for clinicians, or by integrating its functionalities in user-defined MATLAB/Octave scripts, which fits the need of researchers and developers. To demonstrate its features, we used AGATA to analyze CGM data of two subjects extracted from a publicly available data set of individuals with type one diabetes. Finally, AGATA’s features are compared against those of 12 noncommercial software programs for CGM data analysis.
Results:
Using AGATA, we easily preprocessed, analyzed, and visualized CGM data in a handy way, in compliance with the requirements and the standards defined in the literature. Compared to the other considered software programs, AGATA offers more functionalities and capabilities.
Conclusion:
Automated Glucose dATa Analysis is easy to use and reduces the burden of CGM data analysis. It is freely available in GitHub at https://github.com/gcappon/agata.
Keywords: continuous glucose monitoring, data analysis, software, toolbox
Introduction
In diabetes, analysis of glucose data collected via continuous glucose monitoring (CGM) sensors represents a fundamental step for a multitude of purposes, for example, evaluating the efficacy of a certain treatment, assessing glucose control during clinical trials, and adjusting the insulin therapy according to specific glucose patterns just to mention a few.
This task usually involves computation of several well-established metrics to quantify the overall glycemic control (eg, percentage of time spent in specific glucose ranges, glucose variability, estimated A1c), the occurrence of adverse events (eg, number of hyperglycemic events per week, mean duration of hypoglycemia), and the users’ adherence to CGM use (eg, percentage of available glucose measurements over the monitoring period). However, every time these metrics are computed, original, raw, CGM data must be first preprocessed to remove outliers, then homogenized on a uniform time grid, and eventually imputed to fill the missing portion of data. This is clearly a delicate, repetitive, and time-consuming task that needs to be standardized to make results comparable to those of other works. Furthermore, even if well defined in the literature, several of those metrics are not trivial to calculate, and coded implementations are not always officially available, forcing data scientists to “reinvent the wheel” and possibly introduce multiple errors.
Another necessary task when dealing with CGM data analysis regards their effective visualization. Indeed, multiple plots (eg, the plot of CGM trace over the period of monitoring and the plot of distribution of daily CGM profiles displayed as a single day) and reports (eg, the ambulatory glucose profile [AGP] report), which are well established both in the literature1,2 and in common clinical practice, must be produced, forcing researchers and analysts to reproduce them at their best with a clear waste of time and resources.
In this context, current commercially available analysis and visualization tools that CGM devices are sold with (eg, Dexcom Clarity System 3 [Dexcom Inc, San Diego, CA, USA], Eversense Diabetes Management System 4 (Ascensia Diabetes Care Holdings AG, NJ, USA), and Abbott LibreView 5 (Abbott, IL, USA) compute just a limited subset of CGM-derived metrics, and more importantly, they often visualize CGM data without following the latest guidelines for the scope.1,2
To solve these issues and provide the diabetes technology community with a unified comprehensive tool for these purposes, we developed Automated Glucose dATa Analysis (AGATA), a freely available open-source toolbox, written in MATLAB/Octave, that allows investigators to ease, standardize, and speed up both the processing and the analysis of CGM data.
Methods
Software Description
Automated Glucose dATa Analysis is developed in MATLAB/Octave and aims to facilitate the analysis of glucose data sets acquired through CGM. We decided to develop AGATA in such a framework since it represents one of the most popular and widely spread research/prototyping environment adopted by the diabetes technology community.
Automated Glucose dATa Analysis is hosted in GitHub, and it is freely available as an open-source software program for download from https://github.com/gcappon/agata and documented at https://gcappon.github.io/agata. The toolbox is distributed under the GNU General Public License v3. 6 Such setup allows users to identify and notify code issues, ask for new functionalities, and contribute actively to its development, making AGATA maintainable and robust. To maximize the former, the toolbox supports continuous integration via ad-hoc test units to automatically detect major bugs introduced by code variations.
Automated Glucose dATa Analysis can be used directly in MATLAB/Octave, via either the usual command line or integrated in dedicated user-defined MATLAB/Octave scripts, to fit the needs of researchers/developers who want to incorporate its functionalities within their code. As an alternative, AGATA is also available as a standalone application (compatible with Windows, MacOS, and Linux systems) that can be used via a dedicated graphical user interface (GUI) for a more user-friendly and immediate experience, which is particularly suitable for practitioners. Installers of AGATA’s standalone version can be found in the remote folder https://github.com/gcappon/agata/tree/master/standalone.
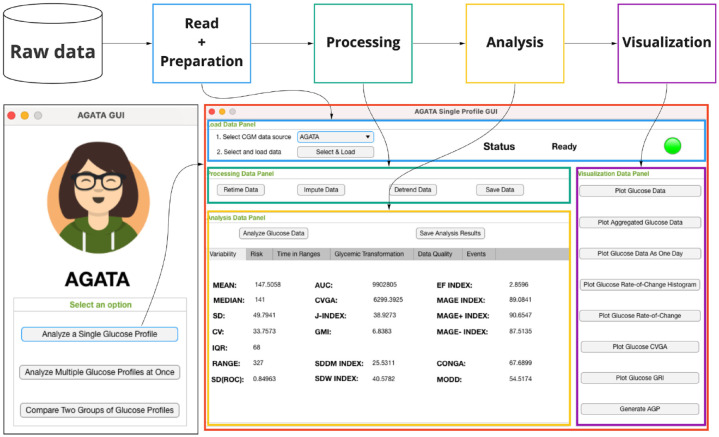
Leveraging the set of utilities described below in the “AGATA’s features” section and depending on the specific user’s needs, AGATA can be used to implement different preprocessing and analysis pipelines including data reading and preparation, data processing, data analysis, and data visualization. This is exemplified in Figure 1, where it is possible to observe how each functionality matches a dedicated panel in one of the window panels of the standalone version of AGATA.
Figure 1.
On the top, the processing pipeline that can be implemented with AGATA thanks to its functionalities. On the bottom left, the home screen of AGATA’s standalone version, which allows to easily access specific interfaces to either analyze a single/multiple glucose profiles or compare two groups of glucose profiles. On the bottom right, as representative use case, the dedicated GUI that allows to analyze a single glucose profile.
Abbreviations: AGATA, Automated Glucose dATa Analysis; GUI, graphical user interface.
AGATA’s Features
As introduced, the toolbox implements a set of functions to preprocess glucose data, compute glucose control metrics, detect glucose adverse events, and visualize results. Moreover, AGATA includes several functions to calculate common prediction error metrics between the original data and predicted glucose profiles, run automatic analysis of a given data set, and compare two study arms. Processed data and the computed metrics resulting from their analysis can be easily saved for further future analyses.
Below, we list the functionalities of AGATA, grouping them by category. For additional details on AGATA’s functions, as well as details on the required inputs and resulting outputs, we refer the reader to the online documentation available at https://gcappon.github.io/agata.
Data Reading and Preparation Utilities
All functions of AGATA require input CGM data to be in the timetable format. The timetable format is a proprietary The MATLAB/Octave format is used to represent timeseries data in a tabular form (see https://it.mathworks.com/help/matlab/timetables.html). Timetables store column-oriented data variables and provide a set of dedicated robust time-specific functions to align, combine, and perform calculations with time-stamped data. As such, we selected timetable as an input format of choice of AGATA as it represents the natural handy solution to manage CGM timeseries. Therefore, as a preliminary step, users are required to convert raw CGM data (commonly stored in parallel timestamp-double vectors in .mat MATLAB/Octave files or in .csv Excel files) to timetable before using AGATA’s functionalities. To facilitate this process, AGATA implements four utility functions to help user comply with this constraint by converting to timetable CGM data that are stored as parallel timestamp-double vectors or raw CGM data in a .csv format downloaded from three major diabetes management applications, including Dexcom Clarity System, 3 Eversense Diabetes Management System, 4 and Abbott LibreView. 5
Preprocessing Utilities
Automated Glucose dATa Analysis provides two utilities to perform common preprocessing tasks: retiming CGM data on a uniform time grid, and imputing missing values using linear interpolation, which consists of replacing a specific data gap with data obtained by interpolating the values between the last and the next available glucose values that delimit such portion of data. As a technical detail, to guarantee that the imputation procedure will work correctly, data must have been preliminarily retimed on a uniform time grid with the dedicated preprocessing utility of AGATA.
Moreover, three functions are available to locate missing data sequences, obtain the number of days the CGM was worn, and to calculate the percentage of missing data that are presented in the recorded trace. Finally, an additional function is available to detrend CGM data. It is important to remark that detrending data affects the computation of CGM-derived metrics, and therefore, such preprocessing must not be applied to CGM data by default. Indeed, the use of the detrending functionality has been implemented and integrated into AGATA to provide to the final users with another “tool in their belt” to process CGM data. To exemplify a possible application of this utility, when data are affected by a systematic error due to CGM faults rather than an actual trend in patients’ glycemia, users of AGATA might want to apply the detrending utility to test and explore how CGM metrics are affected. This allows the users of AGATA to evaluate if detrending CGM data is or is not worthy to be included in their custom processing pipeline.
Data Analysis Utilities
Glucose control metrics
The toolbox provides six functions to calculate glucose control metrics related to the percentage of time spent in a specific glycemic zone, that is, hypoglycemia, hyperglycemia (%THYPER), target range (%TTARGET), severe hypoglycemia, severe hyperglycemia, and tight target range, as defined in the study by Battelino et al, 1 plus a seventh function to compute the percentage of time spent in a user-defined range.
Automated Glucose dATa Analysis also implements 34 functions to compute literature metrics used to quantify glucose variability, that is, mean, median, standard deviation (SD), interquartile range (IQR), coefficient of variation, glucose range, J-index, 7 SD of glucose rate of change, area between the glucose curve and zero, area between the glucose trace and a user-defined value, continuous overall net glycemic action index, 8 mean of daily difference, 9 glucose management indicator, 10 glycemic risk index (GRI), 11 SD of within-day means index, 12 mean of within-day SD index, 12 mean amplitude of glycemic excursion index (MAGE), 13 positive MAGE, 13 negative MAGE, 13 excursion frequency index, 13 control variability glucose analysis (CVGA), 14 blood glucose risk index, 15 low blood glucose index, 15 high blood glucose index, 15 dynamic risk, 16 average daily risk range, 17 hypoglycemic index, 18 hyperglycemic index, 18 index of glucose control, 18 M-value, 19 glycemic risk assessment diabetes equation score (GRADE), 20 GRADE in euglycemia, 20 GRADE in hypoglycemia, 20 and GRADE in hyperglycemia. 20
Adverse event detection
Automated Glucose dATa Analysis provides three utilities to detect and compute statistics of hypoglycemic, hyperglycemic, and prolonged hypoglycemic event occurrence as defined in the study by Battelino et al. 1
Automatic data set analysis utilities
Two functions are implemented to automatically analyze a single CGM profile and a set of multiple CGM profiles (ie, a study arm). A third utility is also available to compare two study arms and identify statistically meaningful differences across metric distributions.
Visualization utilities
The toolbox incorporates nine utilities to generate literature-standard visual reports, including (1) a standard plot of a CGM trace over multiple days, (2) a plot of CGM trace over multiple days with superimposed aggregated glucose values to facilitate the identification of meaningful hyperglycemic and hypoglycemic events, (3) plot of a CGM trace as a distribution of daily CGM profiles (median and percentiles) displayed as a single day, (4) plot of CVGA, 14 (5) plot of GRI, 10 (6) histogram of CGM rate of change, (7) plot of CGM rate of change over the observation period, (8) AGP report, 1 and (9) plot of Clarke Error Grid. 21
Moreover, five additional functions are available to generate plots to visually compare two CGM profiles or, in general, two study arms, by aligning the data under comparison on the same plot in a convenient and visually immediate fashion.
Prediction error metrics
Finally, to facilitate the evaluation of glucose prediction algorithms, six functions can be used to compute the common prediction error metrics between the original glucose profile and a given inferred glucose trace, that is, root mean square error, coefficient of deviation, mean absolute relative difference, Clarke’s Error Grid Analysis, 21 delay between traces, and the glucose root mean square error. 22 We would like to point out that commercially available CGM systems do not commonly provide inferred glucose data profiles together with the actual recorded glucose time-course. As such, there is no need to use these utilities to analyze data collected by CGM systems per se. Nevertheless, as glucose prediction techniques represent one on the most popular categories of algorithms in the field of diabetes technology, we decided to still add these utilities to AGATA. In particular, because the computation of these metrics is usually needed only by researchers and developers at the end of the prediction algorithm development process, we decided to provide these functions as MATLAB/Octave scripts only, as it represents the easiest solution to let scientists integrate AGATA in their custom scripts.
Results
Application of AGATA to a Representative Use-Case
As a representative use-case, here we used AGATA to process, analyze, and compare glucose data of two subjects included in the Ohio Type 1 Diabetes Mellitus (T1DM) data set, 23 a publicly available data set, which can be downloaded upon request at http://smarthealth.cs.ohio.edu/OhioT1DM-dataset.html, involving subjects with T1DM that was released in 2020 to foster research on glucose prediction algorithms. The study included 12 adult subjects monitored for eight weeks collecting CGM measurements in parallel to meal intakes, insulin infusions, physical activity recordings, and other additional features of potential impact for the glucose concentration, for example, galvanic skin response and sleep reports. CGM data were collected.
Specifically, raw CGM data of subject Ohio544 and Ohio596 have been extracted from the data set and converted to the timetable format using the data reading and preparation utilities presented in the “Data reading and preparation utilities” section. Then, we used AGATA to process raw data by making CGM samples uniform on a homogeneous five-minute sampling grid and imputing missing data gaps smaller than 30 minutes. In particular, we chose to impute only data gaps that are smaller than 30 minutes as this represents a common imputation strategy performed in the literature.24,25 Furthermore, this choice allows to mitigate the introduction of artifacts within data, which impacts the computation of CGM-derived metrics. Indeed, performing interpolation on a long portion of missing data hides not only the possible presence of both glucose peaks and nadirs but also critical adverse events, inevitably affecting data analysis.
Processed data have been analyzed by computing the abovementioned metrics of glucose control and adverse event occurrence via the dedicated AGATA’s automatic analysis utility defined in the “Automatic data set analysis utilities” section.
The obtained results, in terms of median and IQR, are reported in Table 1 for both subjects. Focusing on the analysis of hypoglycemia/hyperglycemia occurrence, it can be observed that Ohio544 is worse controlled than Ohio596 showing a lower %TTARGET (65.14% vs 74.08%) due to a higher %THYPER. In Ohio544, the occurrence of hypoglycemic events is lower than that in Ohio596 (2.07 events/week vs 4.30 events/week), further indicating that the latter is more prone to lower glycemic concentrations. Glucose excursions are more pronounced in Ohio544 with SD and MAGE being higher than 8.51 mg/dL and 16.45 mg/dL, respectively. Finally, both subjects present more than 15% of missing data (15.55% in Ohio544 and 18.21% in Ohio596). Similar considerations can be made by analyzing the other computed glucose control metrics reported in Table 1.
Table 1.
Analysis Results of Subjects Ohio544 (in Blue) and Ohio596 (in Red).
| %THYPO (%) 1.33 2.06 |
%THYPER (%) 33.52 23.85 |
%TTARGET (%) 65.14 74.08 |
%TSHYPO (%) 0.12 0.19 |
%TSHYPER (%) 8.50 3.57 |
%TTTARGET (%) 31.67 38.96 |
| Mean (mg/dL) 162.72 147.51 |
Median (mg/dL) 155.00 141.00 |
SD (mg/dL) 58.30 49.79 |
IQR (mg/dL) 79.00 68.00 |
CV (%) 35.83 33.76 |
RANGE (mg/dL) 352.00 327.00 |
| J-Index (−) 48.85 38.93 |
SDROC (mg/dL/min) 0.90 0.85 |
AUC (mg2/dL2) 10686080.00 9902805.00 |
GMI (−) 7.20 6.83 |
CONGA (mg/dL) 77.80 67.69 |
MODD (mg/dL) 52.62 54.52 |
| SDDM (mg/dL) 25.62 25.53 |
SDW (mg/dL) 50.05 40.58 |
MAGE (mg/dL) 105.53 89.08 |
MAGE+ (mg/dL) 116.35 90.65 |
MAGE− (mg/dL) 94.72 87.51 |
EF (−) 3.87 2.86 |
| CVGA (−) 7200.00 6299.39 |
BGRI (−) 7.85 5.68 |
LBGI (−) 0.51 0.70 |
HBGI (−) 7.34 4.98 |
ADRR (−) 35.09 29.84 |
GRI (%) 36.89 27.01 |
| HYPO (−) 0.03 0.07 |
HYPER (−) 0.84 0.45 |
IGC (−) 0.87 0.51 |
M-value (−) 19.61 12.62 |
GRADE (−) 8.76 6.91 |
GRADEEU (−) 32.93 43.15 |
| GRADEHYPO (−) 0.81 1.85 |
GRADEHYPER (−) 66.26 55.00 |
Hypoglycemic events per week (event/week) 2.07 4.30 |
Hyperglycemic events per week (event/week) 16.85 12.65 |
Prolonged hypoglycemic events per week (event/week) 0.00 0.00 |
Mean hypoglycemic event duration (minutes) 51.56 35.71 |
| Mean hyperglycemic event duration (minutes) 169.23 155.49 |
Mean prolonged hypoglycemic event duration (minutes) 0.00 0.00 |
Percentage of missing data (%) 15.55 18.21 |
Number of monitoring days (days) 54.00 57.00 |
Abbreviations: %THYPO, percentage of time spent in hypoglycemia; %THYPER, percentage of time spent in hyperglycemia; %TTARGET, percentage of time spent in the target range; %TSHYPO, percentage of time spent in severe hypoglycemia; %TSHYPER, percentage of time spent in severe hyperglycemia; %TTTARGET, percentage of time spent in the tight target range; ADRR, average daily risk range; AUC, area between the glucose curve and zero; BGRI, blood glucose risk index; CONGA, continuous overall net glycemic action; CVGA, control variability glucose analysis; EF, excursion frequency index; GMI, glucose management indicator; GRADE, glycemic risk assessment diabetes equation score; GRADEEU, GRADE in euglycemia; GRADEHYPER, GRADE in hyperglycemia; GRADEHYPO, GRADE in hypoglycemia; HBGI, high blood glucose index; HYPER, hyperglycemic index; HYPO, hypoglycemic index; IGC, index of glucose control; IQR, interquartile range; LBGI, low blood glucose index; MAGE, mean amplitude of glycemic excursion index; MAGE−, negative MAGE; MAGE+, positive MAGE; MODD, Mean of Daily Difference; RANGE, glucose range; SD, standard deviation; SDW, mean of within-day standard deviation index; SDROC, SD of glucose rate of change.
Figures 2 to 5 demonstrate how CGM data can be visualized with AGATA.
Figure 2.
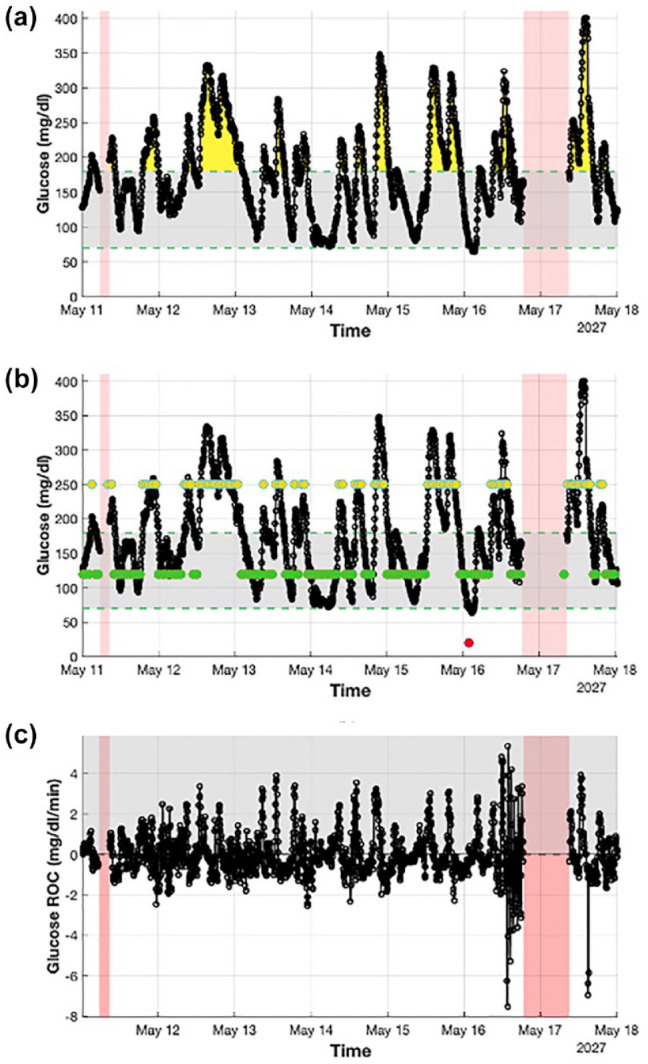
Detailed visualization of a week of CGM data extracted from a representative subject (Ohio544) obtained using AGATA’s utilities. Panel (a) (plot I) shows with a black solid line the CGM trace and highlights the hyperglycemic events with yellow areas, the hypoglycemic events with red areas, and the missing portion of data with shaded red areas. It also marks the target glycemic range with a shaded gray shaded area delimited by two green dashed lines. Panel (b) (plot II) shows with a black solid line the CGM trace and highlights, with superimposed dots, aggregated glucose values in hyperglycemia (in yellow), hypoglycemia (in red), and target (in green). It also highlights missing portion of data with shaded red areas and marks the target glycemic range with a shaded gray area delimited by two green dashed lines. Panel (c) (plot VII) shows with a black solid line the CGM rate of change. It also highlights positive values with a gray shaded area and the missing portion of data with red shaded areas.
Abbreviations: CGM, continuous glucose monitoring; AGATA, Automated Glucose dATa Analysis.
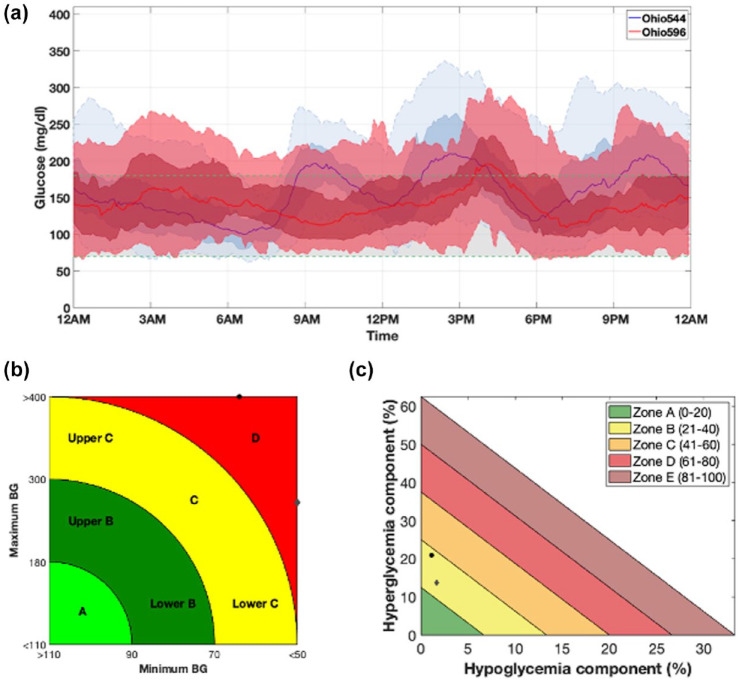
Figure 5.
Comparison between CGM data of two representative subjects (Ohio544 and Ohio596) obtained using AGATA’s utilities. Panel (a) (plot III) compares the daily glucose profile distributions highlighted in blue for Ohio544 and in red for Ohio596, in terms of median, 25th to 75th percentile, and 5th to 95th percentile. Panels (b and c) compare the CVGA and GRI of the two subjects where data of Ohio544 and Ohio596 are reported as black circles and gray diamonds, respectively.
Abbreviations: CGM, continuous glucose monitoring; AGATA, Automated Glucose dATa Analysis; CVGA, control variability glucose analysis; GRI, glycemic risk index; BG, blood glucose.
Figure 2 is composed of three plots (ie, plots I, II, and VII defined in the “Visualization utilities” section) that offer a detailed visualization of a week of data of subject Ohio544. Figure 2a (plot I) shows CGM data in solid black and highlights notable features in the recorded trace, that is, hyperglycemic events in yellow, hypoglycemic events in red, and missing data in shaded red. Similarly, Figure 2b (plot II) shows the same CGM data but with superimposed aggregated glucose values in hypoglycemia (in red), hyperglycemia (in yellow), and euglycemia (in green) to facilitate their visualization. Finally, Figure 2c (plot VII) reports in solid black the CGM rate of change for the considered portion of data discriminating positive and negative values.
Figure 3 is composed of four panels (ie, plots III, IV, V, and VI defined in the “Visualization utilities” section) that show a summary visualization of Ohio544 CGM data. They can be used in practice to visually analyze the overall glucose control. Figure 3a (plot III) represents the glucose frequency distribution by time regardless of date, where the black solid line is the median glucose profile, the shaded blue zone is the area between the 25th and the 75th glucose percentiles, and the shaded light blue zone is the area enclosed between the 5th and the 95th glucose percentiles. Figure 3b (plot VI) summarizes via a histogram the distribution of CGM rate of change. Figure 3c (plot IV) and 3d (plot V) are the CVGA and GRI plots as defined in the studies by Magni et al 14 and Bergenstal et al, 10 respectively.
Figure 4 visualizes an AGP report (ie, plot VIII defined in the “Visualization utilities” section) of the last two weeks of data of Ohio544. In details, key CGM statistics and the respective target values are reported in the upper left panel. A breakdown analysis of the percentage of time spent in the different glycemic zones is shown in the upper right panel. The central panel shows glucose frequency distribution (in terms of median glucose profile and 5th, 25th, 75th, and 95th glucose percentiles) by time regardless of date. The bottom panel reports each daily profile, from midnight to midnight, for the observation period, where adverse events are highlighted in yellow (hyperglycemia) and red (hypoglycemia).
Figure 5 visually compares, through three panels (ie, plots III, IV, and V defined in the “Visualization utilities” section), the CGM data recorded in the two subjects. In particular, Figure 5a (plot III) compares the glucose frequency distribution by time regardless of date, where data of Ohio544 are in blue, and those of Ohio596 are in red. Figure 5b (plot IV) and 3d (plot V) are the CVGA and GRI plots as defined in the studies by Magni et al 14 and Bergenstal et al, 10 respectively, where the black circles represent Ohio544, and the gray diamonds are Ohio596.
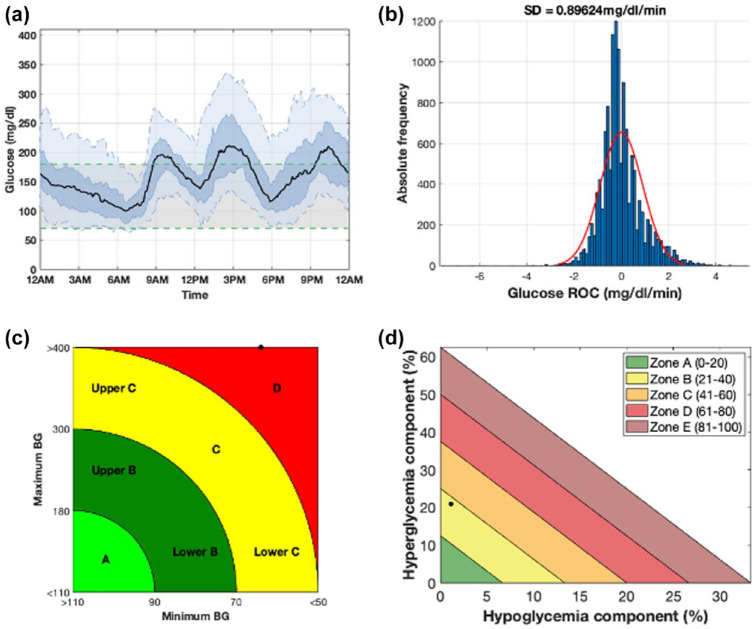
Figure 3.
Summary visualization of a week of CGM data extracted from a retrospective subject (Ohio544) obtained using AGATA’s utilities. Panel (a) (plot III) shows the daily glucose profile distribution resulting from the combination of every single daily profile. The median daily profile is represented with a black bold line, while the shaded dark blue and the shaded light blue areas highlight the 25th to 75th and the 5th to 95th percentiles, respectively. Panel (b) (plot IV) shows the distribution of CGM rate of change as an histogram (in blue) and the respective fitted Gaussian distribution (in red). Panel (c) (plot V) shows the CVGA plot. Panel (d) (plot VI) shows the GRI plot.
Abbreviations: CGM, continuous glucose monitoring; AGATA, Automated Glucose dATa Analysis; CVGA, control variability glucose analysis; GRI, glycemic risk index; BG, blood glucose.
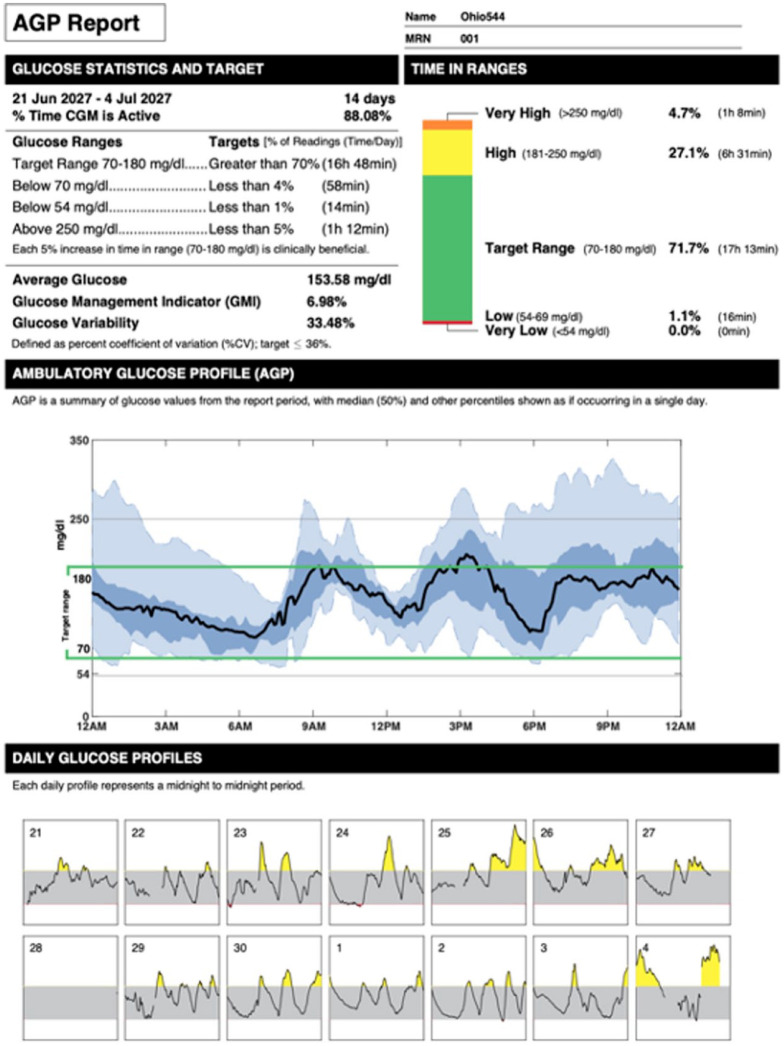
Figure 4.
AGP report (plot VIII) of two weeks of data of a representative subject (Ohio 544). The upper panels report several CGM-derived metrics together with their ideal target values. The middle panel combines the daily glucose profiles to make a 24-hour summary of glucose data. The bottom panel reports each single daily glucose profile and highlights the presence of hyperglycemia (in yellow) and hypoglycemia (in red).
Abbreviations: AGP, ambulatory glucose profile; CGM, continuous glucose monitoring; CV, coefficient of variation.
Comparison of AGATA With Available Software Packages and Tools for CGM Data Analysis
In the following paragraphs, we report the differences and advantages of AGATA when compared with other similar available software packages and tools for CGM data analysis. Specifically, we evaluated the features of AGATA against 12 similar noncommercial software programs identified in the literature by a recent review of Piersanti et al, 26 namely GlyCulator, 27 Easy Glycemic Variability, 28 Continuous Glucose Monitoring Graphical User Interface for Diabetes Evaluation, 29 Glycemic Variability Analyzer Program, 30 Tidepool, 31 CGManalyzer, 32 cgmanalysis, 33 GLU, 34 CGMStatsAnalyser, 35 iglu, 36 rGV, 37 and cgmquantify. 38
Table 2 summarizes the results of such comparison in terms of 12 main overall characteristics.
Table 2.
Comparison of AGATA Against Other Software Programs for CGM Data Analysis in Terms of Main Overall Features.
| Name | GlyCulator | EasyGV | CGM-GUIDE | GVAP | Tidepool | CGManalyzer | cgmanalysis | GLU | CGMStats analyzer | iglu | rGV | cgmquantify | AGATA |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Year of analyzed version | October 2021 | October 2020 | December 2021 | April 2015 | September 2015 | January 2018 | October 2019 | February 2020 | January 2021 | April 2021 | July 2021 | August 2021 | December 2022 |
| GUI | Yes | Yes | Yes | Yes | Yes | No | No | No | Yes | Yes | Yes | No | Yes |
| Open-source | No | No | No | Yes | Yes | Yes | Yes | Yes | No | Yes | No | Yes | Yes |
| Language | R | Excel | MATLAB | MATLAB | JS | R | R | R | App | R | R | R, Python | MATLAB |
| Supported devices | Any (after conversion) | Any (after conversion) | Any (after conversion) | Any (after conversion) | 11 | Abbott Freestyle Libre, Glutalor, Dexcom, Medtronic + Any (after conversion) | Diasens, Dexcom, Medtronic iPro 2, Carelink, Abbott Freestyle Libre + Any (after conversion) | Medtronic iPro 2, Abbott Freestyle Libre, Dexcom G6 + Any (after conversion) | Medtronic iPro 2 + Any (after conversion) | Dexcom, Abbott Freestlye Libre, Abbott Freestyle Libre Pro, Medtronic iPro + Any (after conversion) | Any (after conversion) | Dexcom, Abbott Freestyle Libre + Any (after conversion) | Abbott Freestyle Libre, Dexcom, Eversense + Any (after conversion) |
| Data format | csv, txt, xls, xlsx | xlsm | xls | xls | Device specific | Device specific | csv | csv | csv | csv | csv | csv | mat, csv, xls, xlsx |
| Input units | mg/dL | mmol/L | mg/dL | mg/dL | mg/dL, mmol/L | mg/dL, mmol/L | mg/dL | mmol/L | mmol/L | mg/dL | mg/dL | mg/dL | mg/dL, mmol/L |
| Reports/results download | Yes | Yes | No | No | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Units’ conversion | Yes | To mmol/L | No | No | Yes | No | No | No | No | No | No | Yes | Yes (automatic to mg/dL) |
| Docs | Yes | Yes | No | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Updating | Yes | Yes | No | No | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Toy example | Yes | Yes | No | Yes | Yes | Yes | Yes | Yes | No | Yes | No | Yes | Yes |
| Video tutorials | Yes | No | No | No | Yes | No | No | No | No | No | No | No | No |
Abbreviations: AGATA, Automated Glucose dATa Analysis; CGM, continuous glucose monitoring; EasyGV, Easy Glycemic Variability; GVAP, Glycemic Variability Analyzer Program; GUI, graphical user interface.
It can be appreciated that AGATA appears to be one of the most complete software programs among the noncommercial solutions. It provides a GUI (3/12 do not), being open source (5/12 are not), supporting CGM data collected using either mg/dL or mmol/L as the unit of measurement (9/12 do not), providing the possibility of downloading the results of the analysis and generating visual reports (2/12 do not), being able to convert the units of measurements between mg/dL and mmol/L (9/12 cannot), being constantly updated and maintained (2/12 do not), and providing a toy example to start with (3/12 do not). On the contrary, it can be observed that, compared to AGATA, Tidepool excels in terms of supported devices and documentation quality, which also includes a dedicated video tutorial to facilitate its usage among new users.
Finally, we evaluated AGATA and the other competing software programs in terms of implemented functionalities, that is, the preprocessing procedures, computable metrics, and visualization utilities. Results, summarized in Table 3, show that retiming on a homogeneous time grid and imputing missing glucose values are commonly possible also with the other available solutions. On the contrary, none of the other software programs implement a detrending utility as AGATA, while some of them allow removal of outliers observed in the recorded CGM trace. Considering the computable metrics, AGATA can calculate more metrics than the other packages, including all the key measures defined in the international consensus for CGM data analysis and reporting by Battelino et al. 1 Finally, focusing on the visualization utilities, AGATA can generate up to nine different reports versus an average of three using the other toolboxes. Specifically, it can be observed that AGATA and Tidepool are the only available software programs that integrate a routine to generate AGP reports, which is the current standard reporting tool for CGM data.
Table 3.
Comparison of AGATA Against Other Software Programs for CGM Data Analysis in Terms of Implemented Functionalities.
| Name | GlyCulator | EasyGV | CGM-GUIDE | GVAP | Tidepool | CGManalyzer | cgmanalysis | GLU | CGMStatsAnalyser | iglu | rGV | cgmquantify | AGATA |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total computable metrics | 19 | 26 | 11 | 12 | 11 | 10 | 23 | 11 | 15 | 39 | 28 | 19 | 46 |
| Total computable standard metrics | 13 | 9 | 6 | 5 | 6 | 1 | 14 | 6 | 10 | 14 | 11 | 6 | 14 |
| Between group statistical analysis | No | No | No | No | No | No | No | Yes | No | No | No | No | Yes |
| Processing features | Imputation | Imputation | Imputation | Imputation | — | Retiming, imputation | Retiming, imputation, outlier removal | Retiming, imputation, outlier removal | — | — | Imputation | — | Retiming, imputation, detrending |
| # of Visualization utilities | 1 | 0 | 3 | 2 | 4 | 3 | 3 | 3 | 3 | 4 | 3 | 2 | 9 |
| AGP report | No | No | No | No | Yes | No | No | No | No | No | No | No | Yes |
Abbreviations: AGATA, Automated Glucose dATa Analysis; CGM, continuous glucose monitoring; Easy GV, Easy Glycemic Variability; GVAP, Glycemic Variability Analyzer Program; GUI, graphical user interface.
Conclusions
A lack of publicly available complete software packages often forces researchers of the diabetes technology community to “reinvent the wheel” even when standard analysis tasks and (apparently) easily computable metrics and indices must be calculated. In addition to wasting time and energies, development of codes from scratch can open the possibility to the introduction of errors and bugs. The proposed AGATA software provides a viable and reliable set of functionalities, to automate time-consuming and repetitive tasks involving CGM data, such as running standard preprocessing algorithms and computing literature glucose control metrics. Furthermore, it offers a set of visualization utilities that can be used to efficiently report and inspect CGM data through plots that are well established in both the clinical and technological communities. AGATA can be easily integrated in the custom script developed in MATLAB/Octave, one of the most widely used environment for data science and system development, especially in the diabetes technology community. Compared to 12 noncommercial packages for CGM data analysis, AGATA offers the possibility to compute more literature CGM metrics and to report data using a large set of visualization utilities. However, we saw that other software programs, for example, Tidepool, best our toolbox in terms of documentation quality and supported devices, which are key aspects to maximize its widespread use.
In conclusion, using AGATA, it is possible to sensitively reduce the burden of CGM data analysis and cut down the time needed to run it. The fact that AGATA is conceived as an open-source software program (in which anyone can examine and enhance the original source code) minimizes the risk of having results flawed because of programming errors and magnifies the possibility of providing the community with new/updated time-series analysis methodologies and technical functionalities much faster. Future work will focus on furtherly refining the utilities of AGATA to support a wider variety of devices, expanding the set of implemented features, and improving the available documentation with dedicated video tutorials.
Footnotes
Abbreviations: %THYPER, percentage of time spent in hyperglycemia; %TTARGET, percentage of time spent in the target range; AGATA, Automated Glucose dATa Analysis; AGP, Ambulatory Glucose Profile; AGP, ambulatory glucose profile; CGM, continuous glucose monitoring; CVGA, control variability glucose analysis; EasyGV, Easy Glycemic Variability; GRADE, glycemic risk assessment diabetes equation score; GRI, glycemic risk index; IQR, interquartile range; MAGE, mean amplitude of glycemic excursion index; SD, standard deviation; SDDM, standard deviation of within-day means index; T1DM, type one diabetes mellitus.
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was partially supported by MIUR (Italian Minister for Education, Universities and Research), under the initiatives of “Departments of Excellence” (Law 232/2016). This work was partially supported also by MIUR under the initiative “PRIN: Programmi di Ricerca Scientifica di Rilevante Interesse Nazionale (2020),” project ID: 2020X7XX2P, project title: “A noninvasive tattoo-based continuous GLUCOse Monitoring electronic system FOR Type-1 diabetes individuals (GLUCOMFORT).”
ORCID iDs: Giacomo Cappon  https://orcid.org/0000-0003-4358-9268
https://orcid.org/0000-0003-4358-9268
Giovanni Sparacino  https://orcid.org/0000-0002-3248-1393
https://orcid.org/0000-0002-3248-1393
Mode of Availability of the Software: The software is available for download at the following link: https://github.com/gcappon/agata
References
- 1. Battelino T, Danne T, Bergenstal RM, et al. Clinical targets for continuous glucose monitoring data interpretation: recommendations from the international consensus on time in range. Diabetes Care. 2019;42(8):1593-1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Clarke W, Kovatchev B. Statistical tools to analyze continuous glucose monitor data. Diabetes Technol Ther. 2009;11(suppl 1):S45-S54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dexcom Clarity. Date unknown. https://clarity.dexcom.eu/. Accessed November 7, 2022.
- 4. Eversense. Diabetes management. Ascensia Diabetes Care Holdings AG. Date unknown. https://www.ascensiadiabetes.com/eversense/eversense-cgm-system/data-management/. Accessed November 7, 2022. [Google Scholar]
- 5. FreeStyle Libre. Abbott LibreView. Date unknown. https://www.freestyle.abbott/ch-it/prodotti/libre-view.html. Accessed November 7, 2022.
- 6. GNU Operating System. GNU general public licence. Date unknown. https://www.gnu.org/licenses/gpl-3.0.html. Accessed November 7, 2022.
- 7. Wójcicki JM. “J”-index. A new proposition of the assessment of current glucose control in diabetic patients. Horm Metab Res. 1995;27(1):41-42. [DOI] [PubMed] [Google Scholar]
- 8. McDonnell CM, Donath SM, Vidmar SI, Werther GA, Cameron FJ. A novel approach to continuous glucose analysis utilizing glycemic variation. Diabetes Technol Ther. 2005;7(2):253-263. [DOI] [PubMed] [Google Scholar]
- 9. Molnar GD, Taylor WF, Ho MM. Day-to-day variation of continuously monitored glycaemia: a further measure of diabetic instability. Diabetologia. 1972;8(5):342-348. [DOI] [PubMed] [Google Scholar]
- 10. Bergenstal RM, Beck RW, Close KL, et al. Glucose management indicator (GMI): a new term for estimating a1c from continuous glucose monitoring. Diabetes Care. 2018;41(11):2275-2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Klonoff DC, Wang J, Rodbard D, et al. A glycemia risk index (GRI) of hypoglycemia and hyperglycemia for continuous glucose monitoring validated by clinician ratings. J Diabetes Sci Technol. 2023;17(5):1226-1242. doi:10.1177/19322968221085273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Rodbard D. New and improved methods to characterize glycemic variability using continuous glucose monitoring. Diabetes Technol Ther. 2009;11(9):551-565. [DOI] [PubMed] [Google Scholar]
- 13. Service FJ, Molnar GD, Rosevear JW, Ackerman E, Gatewood LC, Taylor WF. Mean amplitude of glycemic excursions, a measure of diabetic instability. Diabetes. 1970;19(9):644-655. [DOI] [PubMed] [Google Scholar]
- 14. Magni L, Raimondo DM, Man CD, et al. Evaluating the efficacy of closed-loop glucose regulation via control-variability grid analysis. J Diabetes Sci Technol. 2008;2(4):630-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kovatchev BP, Cox DJ, Gonder-Frederick LA, Clarke W. Symmetrization of the blood glucose measurement scale and its applications. Diabetes Care. 1997;20(11):1655-1658. [DOI] [PubMed] [Google Scholar]
- 16. Guerra S, Sparacino G, Facchinetti A, Schiavon M, Man CD, Cobelli C. A dynamic risk measure from continuous glucose monitoring data. Diabetes Technol Ther. 2011;13(8):843-852. [DOI] [PubMed] [Google Scholar]
- 17. Kovatchev BP, Otto E, Cox D, Gonder-Frederick L, Clarke W. Evaluation of a new measure of blood glucose variability in diabetes. Diabetes Care. 2006;29(11):2433-2438. [DOI] [PubMed] [Google Scholar]
- 18. Rodbard D. Interpretation of continuous glucose monitoring data: glycemic variability and quality of glycemic control. Diabetes Technol Ther. 2009;11(suppl 1):S55-S67. [DOI] [PubMed] [Google Scholar]
- 19. Schlichtkrull J, Munck O, Jersild M. The M-value, an index of blood-sugar control in diabetes. Acta Medica Scandinavica. 1965;177:95-102. [DOI] [PubMed] [Google Scholar]
- 20. Hill NR, Hindmarsh PC, Stevens RJ, Stratton IM, Levy JC, Matthews DR. A method for assessing quality of control from glucose profiles. Diabet Med. 2007;24(7):753-758. [DOI] [PubMed] [Google Scholar]
- 21. Clarke WL, Cox DJ, Gonder-Frederick LA, Carter W, Pohl SL. Evaluating clinical accuracy of systems for self-monitoring of blood glucose. Diabetes Care Sep. 1987;10(5):622-628. [DOI] [PubMed] [Google Scholar]
- 22. Del Favero S, Facchinetti A, Cobelli C. A glucose-specific metric to assess predictors and identify models. IEEE Trans Biomed Eng. 2012;59(5):1281-1290. [DOI] [PubMed] [Google Scholar]
- 23. Marling C, Bunescu R. The OhioT1DM dataset for blood glucose level prediction: update 2020. CEUR Workshop Proc. 2020;2675:71-74. [PMC free article] [PubMed] [Google Scholar]
- 24. Prendin F, Del Favero S, Vettoretti M, Sparacino G, Facchinetti A. Forecasting of glucose levels and hypoglycemic events: head-to-head comparison of linear and nonlinear data-driven algorithms based on continuous glucose monitoring data only. Sensors (Basel). 2021;21(5):1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Faccioli S, Facchinetti A, Sparacino G, Pillonetto G, Del Favero S. Linear model identification for personalized prediction and control in diabetes. IEEE Trans Biomed Eng. 2022;69(2):558-568. [DOI] [PubMed] [Google Scholar]
- 26. Piersanti A, Giurato F, Göbl C, Burattini L, Tura A, Morettini M. Software packages and tools for the analysis of continuous glucose monitoring data. Diabetes Technol Ther. 2022. [DOI] [PubMed] [Google Scholar]
- 27. Pagacz K, Stawiski K, Szadkowska A, Mlynarski W, Fendler W. GlyCulator2: an update on a web application for calculation of glycemic variability indices. Acta Diabetol. 2018;55(8):877-880. [DOI] [PubMed] [Google Scholar]
- 28. Moscardó V, Giménez M, Oliver N, Hill NR. Updated software for automated assessment of glucose variability and quality of glycemic control in diabetes. Diabetes Technol Ther. 2020;22(10):701-708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rawlings RA, Shi H, Yuan LH, Brehm W, Pop-Busui R, Nelson PW. Translating glucose variability metrics into the clinic via continuous glucose monitoring: a graphical user interface for diabetes evaluation (CGM-GUIDE©). Diabetes Technol Ther. 2011;13(12):1241-1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Marics G, Lendvai Z, Lódi C, et al. Evaluation of an open access software for calculating glucose variability parameters of a continuous glucose monitoring system applied at pediatric intensive care unit. Biomed Eng Online. 2015;14:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Neinstein A, Wong J, Look H, et al. A case study in open source innovation: developing the Tidepool Platform for interoperability in type 1 diabetes management. J Am Med Inform Assoc. 2016;23(2):324-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang XD, Zhang Z, Wang D. CGManalyzer: an R package for analyzing continuous glucose monitoring studies. Bioinformatics. 2018;34(9):1609-1611. [DOI] [PubMed] [Google Scholar]
- 33. Vigers T, Chan CL, Snell-Bergeon J, et al. CGManalysis: an R package for descriptive analysis of continuous glucose monitor data. PLoS ONE. 2019;14(10):e0216851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Millard LAC, Patel N, Tilling K, Lewcock M, Flach PA, Lawlor DA. GLU: a software package for analysing continuously measured glucose levels in epidemiology. Int J Epidemiol. 2020;49(3):744-757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. De Livera AM, Shaw JE, Cohen N, Reutens A, Salim A. An interactive web application for the statistical analysis of continuous glucose monitoring data in epidemiological studies. J Diabetes Sci Technol. 2022;16(2):397-400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Broll S, Urbanek J, Buchanan D, et al. Interpreting blood GLUcose data with R package iglu. PLoS ONE. 2021;16(4):e0248560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Olawsky E, Zhang Y, Eberly LE, Helgeson ES, Chow LS. A new analysis tool for continuous glucose monitor data. J Diabetes Sci Technol. 2022;16(6):1496-1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Bent B, Henriquez M, Dunn J. CGMquantify: python and R software packages for comprehensive analysis of interstitial glucose and glycemic variability from continuous glucose monitor data. IEEE Open J Eng Med Biol. 2021;2:263-266. [DOI] [PMC free article] [PubMed] [Google Scholar]