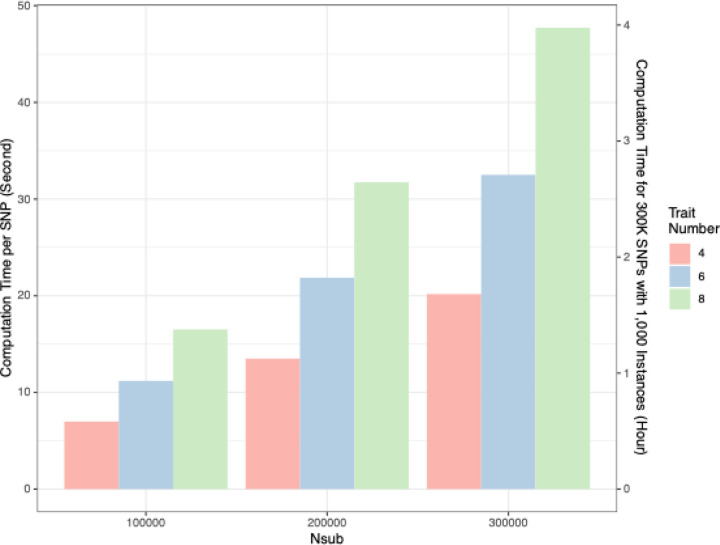
Figure 4. Computational performance of SCAMPI.
Computational run time of SCAMPI for different sample sizes and number of traits using High-Performance Computing (HPC) cluster hosted by Emory University Rollins School of Public Health (RSPH). Computational run time is based on average of 1,000 simulations for different scenarios with varying trait number and sample size. The first y-axis in Figure 4 displays the time in seconds to complete SCAMPI for one SNP at different configurations. The second y-axis shows the hours required to complete analyses assuming 1,000 job instances on a high-performance cluster.