Abstract
Mitochondria are hubs of metabolism and signaling and play an important role in tumorigenesis, therapeutic resistance, and metastasis in many cancer types. Various laboratory models of cancer demonstrate the extraordinary dynamics of mitochondrial structure, but little is known about the role of mitochondrial structure in resistance to anticancer therapy. We previously demonstrated the importance of mitochondrial structure and oxidative phosphorylation in the survival of chemotherapy-refractory triple negative breast cancer (TNBC) cells. As TNBC is a highly aggressive breast cancer subtype with few targeted therapy options, conventional chemotherapies remain the backbone of early TNBC treatment. Unfortunately, approximately 45% of TNBC patients retain substantial residual tumor burden following chemotherapy, associated with abysmal prognoses. Using an orthotopic patient-derived xenograft mouse model of human TNBC, we compared mitochondrial structures between treatment-naïve tumors and residual tumors after conventional chemotherapeutics were administered singly or in combination. We reconstructed 1,750 mitochondria in three dimensions from serial block-face scanning electron micrographs, providing unprecedented insights into the complexity and intra-tumoral heterogeneity of mitochondria in TNBC. Following exposure to carboplatin or docetaxel given individually, residual tumor mitochondria exhibited significant increases in mitochondrial complexity index, area, volume, perimeter, width, and length relative to treatment-naïve tumor mitochondria. In contrast, residual tumors exposed to those chemotherapies given in combination exhibited diminished mitochondrial structure changes. Further, we document extensive intra-tumoral heterogeneity of mitochondrial structure, especially prior to chemotherapeutic exposure. These results highlight the potential for structure-based monitoring of chemotherapeutic responses and reveal potential molecular mechanisms that underlie chemotherapeutic resistance in TNBC.
INTRODUCTION
Triple negative breast cancer (TNBC), an aggressive subtype of breast cancer, comprises 10–15% of breast cancer cases1. Because TNBCs do not express estrogen or progesterone receptors or overexpress the human epidermal growth factor receptor 2, there are limited targeted therapies for this subtype1,2. Most patients with TNBC receive pre-surgical neoadjuvant chemotherapy (NACT) that includes several conventional chemotherapeutic agents, usually given in simultaneous and/or sequential combinations. NACT now also includes the immunotherapy agent Programmed Cell Death 1 (PD-1) inhibitor with a marginal but statistically significant improvement in response rate3. Conventional chemotherapies typically include DNA-damaging agents (e.g., Adriamycin, cyclophosphamide, carboplatin) and taxanes (e.g., docetaxel, paclitaxel). As there is no single accepted standard NACT regimen for TNBC, various regimens are used throughout the United States4. Residual cancer burden (RCB) following NACT is assessed by a pathology evaluation of the resected surgical specimen. Unfortunately, approximately 45% of TNBC patients retain substantial RCB after NACT, which is strongly associated with high rates of metastatic relapse and poor overall survival5,6. Thus, understanding the molecular features of residual tumor cells that survive chemotherapeutic regimens will be key to developing novel strategies to combat residual disease.
Mitochondria are adaptive organelles that allow cells to meet energetic and signaling demands in response to exogenous stressors, in part through mitochondrial structural rearrangements7. In the context of cancer, metabolic adaptations can encompass oxidative phosphorylation (oxphos), glycolysis, reactive oxygen species (ROS), nucleotide synthesis, and fatty acid oxidation as we recently reviewed8. The interplay of mitochondrial fission and fusion that regulates mitochondrial structure has been studied in cancer therapy resistance and metastasis. For TNBC, mitochondrial structure dynamics play a role in metastatic phenotypes, although the findings seem to be context-dependent9,10. We showed that mitochondrial fusion drove to chemotherapy-induced oxphos that promoted the survival of residual tumor cells that were refractory to conventional chemotherapies11. Importantly, we demonstrated that chemotherapy-induced mitochondrial structure and function adaptations resulted in a new therapeutic dependency for residual TNBC, wherein we could delay tumor regrowth by inhibiting mitochondrial fusion or by direct inhibition of Complex I11–13.
Serial block-face scanning electron microscopy (SBF-SEM) was first developed in 2004 by Denk and Horstmann to study mitochondrial structure and location within the neuron14,15. SBF-SEM produces high-resolution micrographs that capture subcellular ultrastructural features through three-dimensional (3D) rendering16,17 that, unlike other volumetric electron microscopy techniques, provide larger x- and y-axis ranges16,17. These visualizations of changes in mitochondrial morphology, orientation, configuration, size, and shape provide insights into mitochondrial physiology and function important for physiology and disease18–21. We and others have shown that using 2D transmission electron microscopy (TEM) to characterize mitochondrial structures in breast cancer provides useful, albeit limited, insights into the extent of the mitochondrial network10,11,22,23. Previous SBF-SEM studies of mitochondria demonstrate dynamic 3D structures and networks associated with the physiology of skeletal muscle24, aging25–27, pancreatic ductal adenocarcinoma28, and non-small cell lung cancer (NSCLC)29. Of note, the recent NSCLC study employed genetically engineered mouse models to demonstrate that 3D mitochondrial structure was spatially heterogeneous within tumor cells and functionally affected mitochondrial respiration29.
In TNBC, the comprehensive nature of 3D mitochondrial networks remains unexplored. Further, SBF-SEM has not been applied to patient-derived xenograft (PDX) models nor to understand therapeutic resistance in cancer. To address these knowledge gaps and based on our prior work demonstrating the importance of mitochondrial structure and oxphos for TNBC chemoresistance, we used SBF-SEM to characterize 3D mitochondrial structures in our deeply characterized orthotopic PDX model of TNBC. PDXs provide valuable models for longitudinal studies to evaluate chemotherapy response as they effectively recapitulate the molecular features and the heterogeneity observed in human TNBC30–34. Our longitudinal analyses enabled us to compare residual tumors surviving exposure to a variety of conventional chemotherapies to the treatment-naïve state. We found significant changes in mitochondrial shape, size, and complexity in residual tumors compared to the treatment-naïve tumor. Of note, these changes were more pronounced in tumors that had been exposed to single, rather than combination, chemotherapies. Further, we found striking intratumoral heterogeneity (ITH) of mitochondria within individual PDX tumors, and this was most pronounced in the treatment-naïve tumor. This finding highlights the incredible complexity within individual TNBC tumors and reaffirms the extensive ITH that has been thoroughly documented at the genomic, transcriptomic, epigenetic, and proteomic levels35,36.
METHODS
Patient-derived xenograft mouse studies
Animal studies were conducted in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals with approval of the BCM IACUC under protocol AN-8243. Mice were ethically euthanized as recommended by the Association for Assessment and Accreditation of Laboratory Animal Care. The PIM001-P PDX model was originally obtained from the University of Texas MD Anderson Cancer Center, where it was generated and characterized and was then propagated as described previously13,32. Briefly, cryo-preserved PDX cells were suspended as a 1:1 volume mixture in a total of 20 μl of complete cell growth medium (Dulbecco’s modified Eagle’s medium:F12, Cytiva HyClone, SH30023.01) supplemented with 5% fetal bovine serum and Matrigel (Corning, 354234). 5.0 × 105 viable tumor cells were injected unilaterally into the fourth mammary fat pads of 5- to 8-week-old female Nod-Rag-Gamma mice (NRG, Nod.Cg-Rag1tm1Mom/ IL2rgtm1wjl/SzJ, The Jackson Laboratory).
To validate the PDX model’s identity, the Cytogenetics and Cell Authentication Core at MD Anderson Cancer Center performed short-tandem repeat (STR) DNA fingerprinting on DNA extracted from PDX tumors. The Promega 16 High Sensitivity STR Kit (Catalog # DC2100) was used for fingerprinting analysis, and profiles were compared to online databases (DSMZ/ ATCC/ JCRB/ RIKEN).
Mouse body weights and tumor volumes, measured using digital calipers, were recorded twice weekly for the duration of the experiment. When mammary tumors reached an average size of 150 mm3, mice were randomized into five treatment groups (n = 6 mice per group) to monitor tumor responses and collect samples for molecular analyses. For the treatment-naïve group, the tumors were harvested at 150 mm3 without any treatment. Mice in the Adriamycin plus cyclophosphamide (AC) treatment group received one dose of Adriamycin and cyclophosphamide every three weeks for a total of three doses. Adriamycin (doxorubicin, ChemieTek, CT-DOXO) was solubilized in sterile water immediately before administration and protected from light prior to intraperitoneal (i.p.) injection at a dose of 0.5 mg/kg. Cyclophosphamide (Sandoz, 0781-3244-94) was solubilized in sterile water immediately prior to administration via i.p. injection at 50 mg/kg. Mice in the docetaxel (DTX), the carboplatin (CRB), and the docetaxel plus carboplatin combination (DTX+CRB) treatment groups received weekly doses for three weeks. Docetaxel (Hospira, 0409-0201-10) was supplied in a solution of polysorbate 80NF, 4 mg anhydrous citric acid USP, 23% v/v dehydrated alcohol USP, q.s. with polyethylene glycol 300 NF at 20 mg/kg. DTX was administrated by i.p. injection at a dose volume of 2 ml/kg. Carboplatin (TEVA Pharmaceuticals, 0703-4248-01) was supplied at 10 mg/ml in water for i.p. injection at 50 mg/kg at a dose volume of 5 ml/kg. For the DTX+CRB treatment group, each compound was administered consecutively by i.p. injection. For each group, residual tumors were harvested one full cycle after the third dose of chemotherapy (i.e., after a total of 28 days for DTX, CRB, DTX+CRB, and after 63 days for AC.
Formalin-fixed paraffin-embedded (FFPE) tissue staining
After harvesting the tumors, we fixed tumor fragments in formalin (Sigma HT501128–4L) for 48 hours at room temperature on a rocker, washed them with 1× phosphate-buffered saline, and stored them in 70% ethanol at 4 °C until they were submitted to the BCM Pathology Core and Lab to be processed into paraffin blocks, cut into 3um sections, and stained with hematoxylin and eosin (H&E). Immunohistochemical staining with antibodies against Ku80 (CST 2180, 1:100) and human mitochondria (Abcam 92824, 1:1,000) was conducted as described previously13.
SBF-SEM sample processing
Tumors were harvested from the mammary gland immediately after mice were euthanized. A tumor slice ~1 mm thick was placed into 1 ml of fixative (2% paraformaldehyde + 2% glutaraldehyde in 0.15 M cacodylate buffer) and stored at 4°C until they were sent to the Mayo Microscopy and Cell Analysis Core for processing and imaging. Tissue samples for SBF-SEM were prepared using a modified procedure20,37. Briefly, fixed tissue was rinsed in 0.1 M cacodylate buffer and placed in 2% osmium tetroxide + 1.5% potassium ferracyanide in 0.1 M cacodylate, washed with nH2O (MilliQ water), incubated at 50°C in 1% thiocarbohydrazide, incubated again in 2% osmium tetroxide in nH2O, rinsed in nH2O and placed in 2% uranyl acetate overnight. The next day, the tissue was rinsed again in nH2O, incubated with Walton’s lead aspartate, dehydrated through an ethanol series, and embedded in Embed 812 resin. To prepare embedded tissue for placement into the SBF-SEM, a ~1.0 mm3 piece was trimmed of excess resin and mounted on an 8 mm aluminum stub using silver epoxy Epo-Tek (EMS, Hatfield, PA). The mounted sample was then carefully trimmed into a smaller ~0.5 mm3 tower using a Diatome diamond trimming tool (EMS, Hatfield, PA) and vacuum sputter-coated with gold-palladium to help dissipate any charge. Sectioning and imaging of the sample were performed using a VolumeScope 2 SEM™ (Thermo Fisher Scientific, Waltham, MA). Imaging was performed under low vacuum/water vapor conditions with a starting energy of 3.0 keV and a beam current of 0.10 nA. Sections of 50 nm thickness were cut, providing a final image spatial resolution of either 15 nm × 15 nm × 50 nm or 10 nm × 10 nm × 50 nm.
SBF-SEM data analysis
Image analyses including registration, volume rendering, and visualization, were performed using Amira (Thermo Fisher) software packages. The selection criteria for a region of interest (ROI) included tumor regions and excluded necrotic and extracellular matrix regions. A minimum of three ROIs were selected per sample. For each sample, images were collected from at least 300 serial sections that were then stacked, aligned, and visualized using Amira to make videos and quantify volumetric structures. A random number generator was used to select 350 segmented mitochondria for each tumor for the final analysis. See Supplementary Table 1 for the specific number of mitochondria segmented per ROI and those selected for final analysis. Previously defined protocols using Amira software provide a workflow for 3D reconstructed organelles by manual or semi-automated segmentation17. Amira provides parameters for quantifying 3D structures, many of which are included in the software, such as volume and surface area. We have added sphericity, which is measured through the Feret’s Diameter. Feret’s diameter refers to the distance between any two most distant points in a 3D object. Sphericity refers to the adherence of the object to that of a sphere and is calculated by the equation (π1/3(6V)2/3)/SA, where V is the volume of the object, and SA is the surface area of the object. We also quantified the mitochondrial complexity index, calculated by the equation SA3/16π2V2, and the mitochondrial branching index, which calculates the relative branching between the transverse and longitudinal mitochondrial orientations24.
Statistical analyses
We evaluated the means for each measurement between treatment groups by the Mann–Whitney U test followed by the Holm method of p-value adjustment38–40. We performed Bartlett’s test followed by pairwise F-tests to determine whether the variance of each measurement differed between treatment groups using R 4.1.0 (R Core Team, 2023) and the stats package (base statistical package in R)41. Bartlett’s test assesses whether all the treatment groups have the same variance. A p-value less than 0.05 for a given measurement by Bartlett’s test indicates that some of the treatment groups have different variances for that measurement. To identify which treatment groups differed, we carried out a two-tailed F test on each possible pairwise comparison between treatment groups. In a two-tailed F test, the F value is defined as the quotient of the variance of one group divided by the variance of the other. This F value is used to test whether the variance in the numerator equals that in the denominator. For a given F test, a p-value was determined by taking the area under the F distribution beyond that test’s F value and then subtracting that area from 1. Once all possible comparisons had been made for a measurement, the p-values for that measurement were adjusted by the Holm method to account for multiple comparisons.
RESULTS
Residual tumors persisted in a PDX model following various chemotherapy treatments.
We previously deeply an orthotopic PDX model of primary treatment naïve TNBC, PIM001-P13,32. Our previous work with the conventional front-line chemotherapy combination Adriamycin + cyclophosphamide (AC) revealed partial tumor regression followed by regrowth upon discontinuation of treatment. The regressed, aka ‘residual’, tumors had heightened oxphos compared to treatment-naïve tumors, resulting in a novel therapeutic window for inhibition of electron transport chain complex I13. We went on to show that following chemotherapy exposure, residual tumor cells had altered mitochondrial structure, which elicited heightened vulnerability to inhibition of mitochondrial fusion protein optic atrophy 1 (OPA1) with the small molecule inhibitor MYLS2211,12.
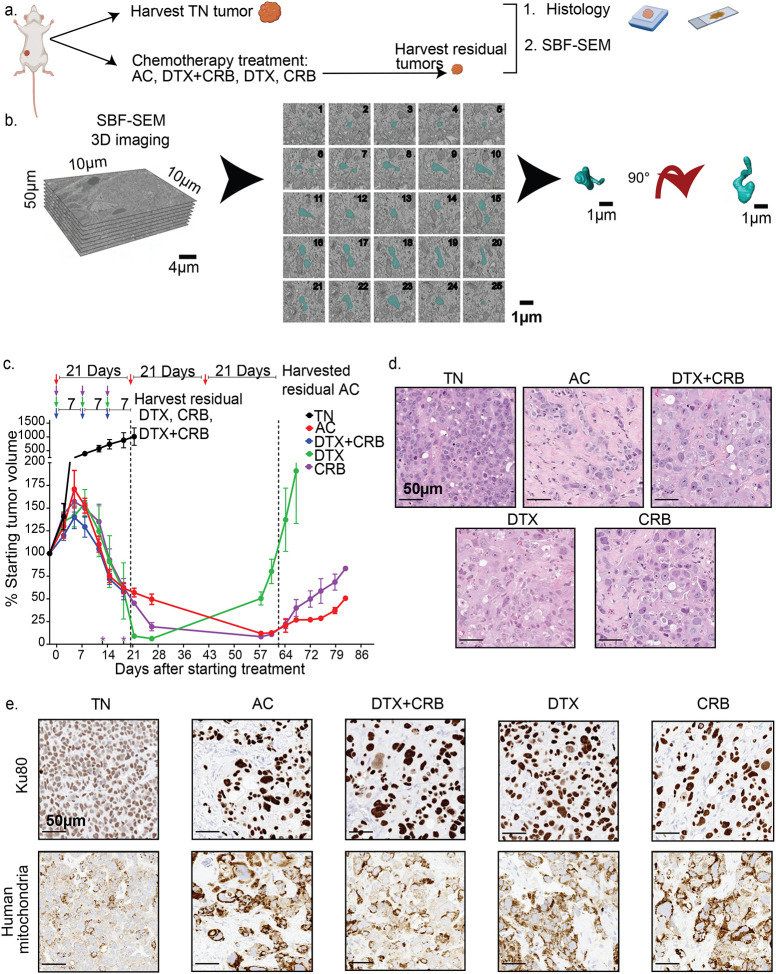
Here, using the PIM001-P PDX model, we compared AC with docetaxel (DTX), carboplatin (CRB), or combined DTX+CRB (Fig. 1a). As we observed previously with AC treatment of PIM001-P13, tumors in all treatment groups exhibited partial regression. After three cycles of chemotherapy, residual tumors that were harvested (on the day that the fourth cycle would have began) showed an average decrease in tumor volume across all groups of 53.6% (DTX, 52.9%; CRB, 61.0%; DTX+CRB, 61.5%; and AC, 34.0%) (Fig. 1c). Animals maintained body weight with all chemotherapy regimens as we observed previously13,42. For feasibility, we randomly selected one mouse per group to be analyzed by SBF-SEM. Two additional mice per group were euthanized at the same endpoint and conventional IHC was conducted. In the three remaining mice, the tumor was allowed to re-grow until it reached 100% of the tumor volume at the start of treatment, demonstrating these residual tumors had the capacity to regrow back to their original size.
Figure 1. PDX TNBC tumors persist following conventional chemotherapy treatments.
(a) The experimental approach used in the study. Images were adapted from BioRender. (b) SBF-SEM 3D rendering approach. (c) Tumor volumes were monitored biweekly following administration of chemotherapies to mice bearing orthotopic PIM001-P tumors (n = 6 mice/group). Arrows at the top indicate when chemotherapy was administered. Asterisks (*) above the x-axis indicate early euthanasia due to animal health concerns. Residual tumors were harvested, as noted by arrows. Error bars represent the standard error of the mean. (d) Residual tumors were harvested, processed for FFPE, and analyzed by H&E staining, (e) IHC for Ku80 and human-specific mitochondria. Scale bars are 50 μm.
H&E staining of FFPE tissue sections revealed poorly differentiated cellular architecture with abundant mitotic figures in the treatment-naïve tumors. All residual tumors in chemotherapy treatment groups showed extensive desmoplastic response with significant tumor cell pleomorphism, including atypical and enlarged nuclear and cytoplasmic sizes and shapes, pronounced nucleoli, and areas of necrosis, consistent with chemotherapy-induced cytological changes observed previously in PDXs and in patients13,43 (Fig 1e). IHC staining with Ku80 to distinguish human tumor cells from mouse stromal cells revealed abundant stromal infiltration in all residual tumors (Fig. 1e), as shown previously with AC treatment13. IHC, using an antibody specific to human mitochondria (excluding mouse mitochondria), revealed increased staining in residual tumor cells compared to treatment-naïve cells, consistent with our prior findings for AC-treated residual tumors (Fig. 1e)13. Therefore, various conventional chemotherapy agents induced partial tumor regression in PIM001-P, and residual tumors persisting after these four treatments had aberrant histology architecture and increased mitochondrial content in the tumor cell compartment.
3D rendering of mitochondria by SBF-SEM was used to quantify mitochondrial features.
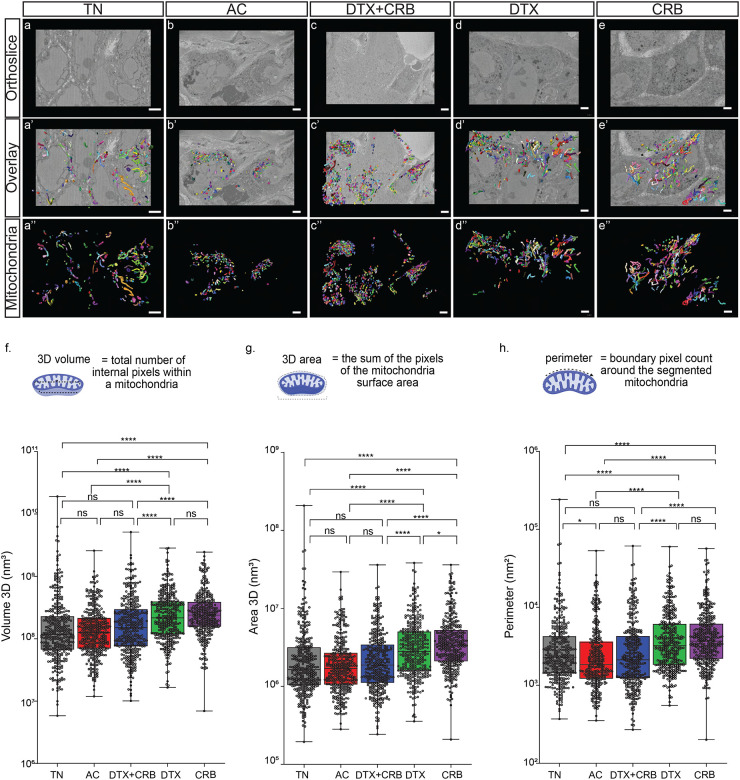
From each treatment group, we selected one representative tumor with a volume near each group’s mean tumor volume to generate blocks for SBF-SEM. Electron micrographs were taken at either 15 nm × 15 nm × 50 nm or 10 nm × 10 nm × 50 nm resolution with a minimum of three ROIs, each with 300 slices per sample. We inspected ROIs prior to imaging to avoid necrotic and stromal tissue. Mitochondria segmentation was conducted on different areas of the orthogonal (ortho) slice (Fig. 2a–e) to include any spatial heterogeneity within each tumor. Segmented mitochondria used for final analyses were each analyzed manually to ensure they were completely segmented (i.e., not missing a slice) (Fig. 2a’–e’). From each tumor, we randomly selected 350 mitochondria for further analysis (Fig. 2a’–e”). For each mitochondrion, we computed area, volume, perimeter, width, length, mitochondria complex index, and mitochondria branching index as described previously17,44.
Figure 2. Several mitochondrial features were significantly altered by single-agent chemotherapy treatments.
(a-e) Representative SBF-SM orthoslices. (a’-e’) Representative 3D SBF-SEM orthoslices with overlays of mitochondria segmentations. (a”-e”) Representative 3D SBF-SEM mitochondria segmentations for each treatment group: treatment-naïve (TN), Adriamycin + cyclophosphamide (AC), docetaxel + carboplatin (DTX+CRB), docetaxel (DTX), and carboplatin (CRB). The scale bar is 3 μm. Mitochondrial measurements were calculated using Amira software for 350 segmented mitochondria (represented as dots) from each tumor for (f) volume, (g) 3D area, (h), and perimeter. Adjusted p-values (*p < 0.05, **p < 0.01, ***p < 0.001, ****p<0.0001) are shown on box and whisker plots as determined using the Mann–Whitney U test followed by the Holm method.
We compared the mean values of each mitochondrial feature between tumors. To test for differences in the mean of a measured value across treatment groups, we performed a Mann–Whitney U test because q–q plots revealed that mitochondrial features were non-normally distributed for all treatment groups, precluding the use of ANOVA (see Supplementary Fig. 1). The single-agent treatment groups, DTX and CRB, had significantly increased mitochondrial area, volume, and perimeter compared to treatment-naïve tumors (Fig 2f–h), which was not observed for residual tumors after either of the combination therapies. These trends were also apparent in density plots (see Supplementary Fig. 2 & 3), which further illustrate the distinct distribution of mitochondrial features observed between each tumor. Thus, residual tumors that persisted after single-agent chemotherapies had significantly increased mitochondrial length, area, volume, and perimeter compared to the treatment-naïve mitochondria. In contrast, most of these measurements did not increase significantly in residual tumors after treatment with combination therapies. Of note, several significant structural differences were observed between residual mitochondria following single-agent versus combination chemotherapy treatment.
Residual tumors exhibited increased mitochondrial complexity.
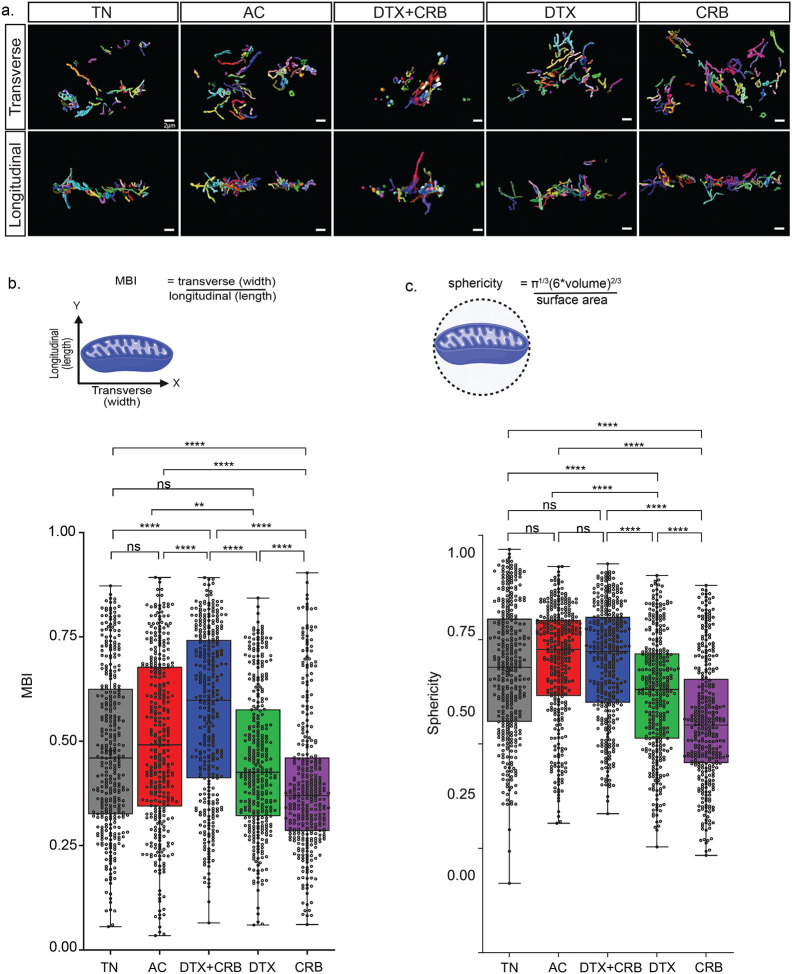
We then analyzed mitochondria for complex measurements, including sphericity and mitochondrial branching index (MBI). Transverse and longitudinal views of the segmented mitochondria were analyzed to determine mitochondrial complexity (Fig. 3a). Sphericity scores range from 0–1, with 1 representing a perfect sphere (Fig. 3c). MBI compares transverse (length) to longitudinal (width) measurements to determine the directionality of any branching (Fig. 3b). For example, if MBI is greater than 1, then the width measurement is greater than the length measurement, and the branching is along the x-axis. Conversely, if the MBI is less than 1, the branching is along the y-axis. If MBI is around 1, then the branching along the x-axis and y-axis is similar. Single-agent chemotherapy-treated mitochondria had significantly decreased sphericity and MBI compared to treatment-naïve tumors, consistent with increased narrow, branched mitochondria. The combination treatment groups had an increased MBI and sphericity compared to the treatment-naïve group, consistent with less branched mitochondria and more spherical mitochondria (Fig. 3b–c).
Figure 3. Single chemotherapeutic agent treated residual tumors have the greatest mitochondrial complexity.
Mitochondrial networks revealed through 3D mitochondria rendered using SBF-SEM. (a) Representative longitudinal and transverse views of the segmented mitochondria for each treatment group: treatment-naïve (TN), Adriamycin + cyclophosphamide (AC), docetaxel + carboplatin (DTX+CRB), docetaxel (DTX), and carboplatin (CRB). The scale bar is 3 μm. Mitochondrial measurements were calculated using Amira software for 350 segmented mitochondria (represented as dots) from each tumor for (b) mitochondrial branching index (MBI) and (c) sphericity, along with their respective method of measurements below. Adjusted p-values (*p < 0.05, **p < 0.01, ***p < 0.001, ****p<0.0001) are shown on box and whisker plots as determined using the Mann–Whitney U test followed by the Holm method.
The extensive intra-tumoral structural heterogeneity of treatment-naïve mitochondria was reduced in residual tumors.
ITH is a well-documented characteristic of TNBC and other cancers35,45, yet the specific heterogeneity within mitochondrial structures remains unexplored in breast cancer. To address this, we quantified mitochondrial ITH by calculating the variance for each mitochondrial feature between all 350 mitochondria within each individual tumor, then compared these variances between tumors using an F-test (see Supplementary Fig. 4a). This revealed that the treatment-naïve tumor exhibited significantly higher variance in all mitochondrial features compared to residual tumors post-chemotherapy (see Supplementary Fig. 4b–i). Specifically, the variance in mitochondrial area, volume, perimeter, width, length, and MCI was significantly greater in treatment-naïve tumors (see Supplementary Fig. 4b–i). Furthermore, combination chemotherapy treatments resulted in a greater reduction in the variance of area, perimeter, width, and MCI compared to single-agent treatments (see Supplementary Fig. 4c&d, 4g&h). These findings indicate that there is significant 3D mitochondrial ITH in this PDX model of TNBC, and that chemotherapy, particularly combination regimens, drastically reduced this heterogeneity.
Mito-otyping revealed heterogeneous and complex mitochondrial structures in treatment-naïve and residual tumors.
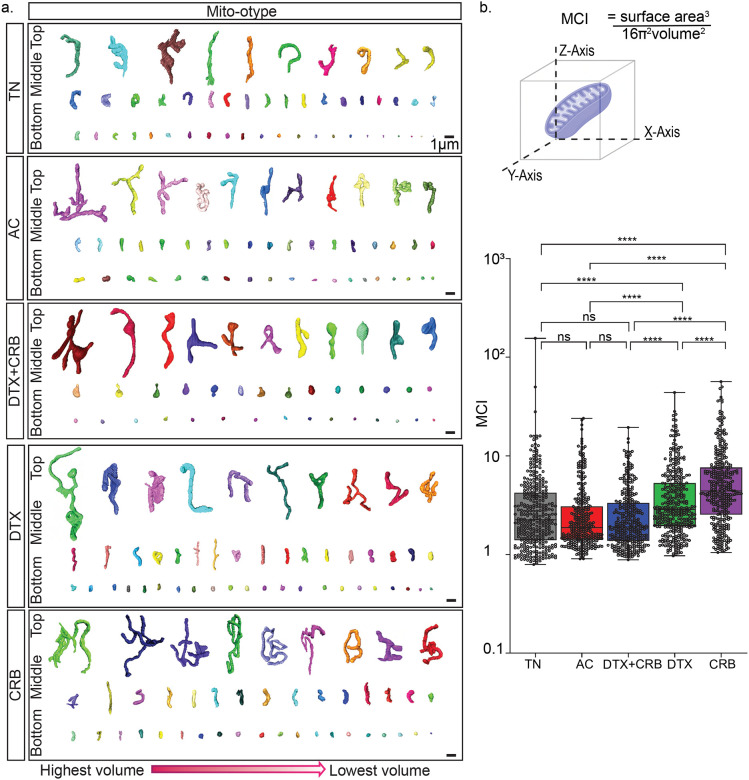
To visualize the complexity of mitochondrial structures, we performed mito-otyping24, or karo-type-like arrangement of mitochondria to visualize complexity relative to volume (Fig. 4a). As a result of the extensive ITH of mitochondria, we displayed a representative subset of the top, middle, and bottom 10% of mitochondria, based on volume, within each tumor. Consistent with the quantitative data described above, we found extensive ITH in each specimen with a shift in the size and complexity of mitochondrial structures after chemotherapy. We computed the mitochondrial complexity index (MCI), measuring mitochondria as a 3D object by treating surface area as a function of volume17,24 (Fig. 4b). In agreement with the aforementioned mitochondrial alterations, single-agent chemotherapy-treated mitochondria had significantly greater MCI than treatment-naïve mitochondria (Fig. 4b). Combination-treated mitochondria showed a non-significant trend towards decreased MCI compared to treatment-naïve mitochondria.
Figure 4. Mito-otyping of the diverse mitochondria structures observed within each treatment group.
(a) Representative mitochondria structures from the bottom, middle, and top 10% of the 3D volume for tumors are displayed. Scale bars are 1μm. (b) Diagram of the mitochondrial measurements used to characterize mitochondria structure and mitochondria network through the mitochondrial complex index (MCI), as calculated by Amira software for 350 segmented mitochondria (represented as dots) from each tumor. The scale bar is 3 μm. Equations for these measurements are shown in Fig. 4b. Adjusted p-values (*p < 0.05, **p < 0.01, ***p < 0.001, ****p<0.0001) on the box and whisker plots were determined using the Mann–Whitney U test, followed by the Holm method.
DISCUSSION
Although chemotherapy is the preeminent treatment modality for neoadjuvant management of TNBC, molecular rationale for choice of specific chemotherapeutic regimens is lacking. Although there are efforts to identify underlying molecular features to inform targeted treatments, such as with PARP inhibitors2, immune checkpoint inhibitors46, and predictive biomarkers42,47, there is still little information on the chemotherapies that would be most effective for a patient based on the molecular features of the tumor. Thus, it is critically important to elucidate the biological characteristics of tumor cells that survive NACT. Improving chemotherapeutic responses has the potential to improve long-term prognoses for those ~45% of TNBC patients whose tumors persist despite enduring months of harsh chemotherapeutic exposure.
We previously established the importance of mitochondria structure dynamics in supporting oxphos in chemoresistant TNBC. We further demonstrated that inhibition of oxphos13 or mitochondrial fusion11 in residual tumors persisting after chemotherapy exposure was able to significantly prolong tumor control. Although we and others demonstrated that the cells that survive some conventional chemotherapies rely heavily on mitochondrial oxphos11, little is known about mitochondrial complexity in TNBC, and 3D mitochondrial structures remain unexplored in this disease. Our study provides evidence that the mitochondrial network is substantially altered in residual tumors, and that this is associated with the type of chemotherapeutic regimen administered.
We observed greater area, perimeter, length, and MCI in the mitochondria of residual tumors after single-agent chemotherapy than in the mitochondria of treatment-naïve tumors. These mitochondria also had greater volume and width compared to mitochondria from tumors after combination chemotherapy. Thus, the mitochondria in residual tumors after single-agent chemotherapies may support increased metabolic capacity, as evidenced by our previous study of TNBC showing that mitochondrial elongation led to enhanced oxphos in residual tumors13. Mitochondrial dysfunction, a complex condition with a wide range of health consequences, cannot be definitively determined from the 3D mitochondrial structure. However, given the importance of understanding mitochondrial dynamics in TNBC11, the unique mitochondrial shapes we observed in this study merit further functional investigations that may lead to therapeutic insights.
Mitochondria interact and communicate with the endoplasmic reticulum (ER), and mitochondrial fission occurs at mitochondria-ER contact sites. Mitochondria also interact with the cellular machinery, including lipid droplets, the nucleus, Golgi, and lysosomes19,48–50. However, the prevalence and importance of these mitochondria-organelle contact sites are poorly understood in cancer and remain drastically under-explored in TNBC51. We observed increased surface area in the mitochondria of residual tumors after single-agent chemotherapy, which could increase opportunity for interactions with other organelles. Future structural studies mapping the connections between mitochondria and key organelles, such as the ER or lipid droplets, will provide critical insights into these important subcellular mechanisms of communication.
We found that mitochondrial structural adaptations were more pronounced in tumors treated with a singleagent compared to those with a combination treatment. Combinations of chemotherapies or chemotherapies with other drugs can improve treatment efficacy, but there is evidence for antagonism or weak additive effects52–54. Therefore, it is crucial to understand mechanisms of resistance to individual therapies that make up the backbone of our standard cancer treatments. Differences in mitochondria in tumors treated with a single agent vs. a combination therapy may indicate a dose-dependency, i.e., mitochondria that adapt their shape in response to single agents cannot adapt when cells receive multiple agents simultaneously. This is important because multiple chemotherapies can be given to TNBC patients either simultaneously or sequentially as part of a standard neoadjuvant regimen. Our data, which suggest that the scheduling of chemotherapies affects the ability of mitochondria to adapt, will be important for the success of future mitochondria-targeting therapies developed for translation in TNBC patients.
We observe extensive ITH in mitochondrial structure, especially in the treatment-naïve tumors. Mitochondrial structural heterogeneity is tissue-specific and is determined by the need for mitochondrial activity; for example, calf muscles that are a mixture of mitochondria-rich and mitochondria-poor fibers have greater mitochondrial structural heterogeneity than cardiac muscle that is in constant motion and require continuous ATP24. The mitochondria of neuronal axons and dendrites have different morphologies that match the different energy demands of these neuronal compartments27. Thus, the ITH in the treatment-naïve tumor may reflect the spatial ITH of many proteins and other molecules that have been found in human TNBC specimens55–57. Heterogeneous mitochondria may provide energetic adaptability for cell survival in the face of stressors such as chemotherapy, which will need to be determined experimentally. All chemotherapy treatments we tested greatly reduced mitochondrial ITH, suggesting that mitochondria may undergo a ‘selective bottleneck’ to improve cell survival after the assault of chemotherapy. Indeed, many studies in breast cancer have provided evidence for enrichment of a cancer stem cell-like, or ‘tumor-initiating cell,’ subpopulation after a variety of therapeutic exposures58,59. Another potential mechanism underpinning these changes is dynamic adaptation of mitochondrial structure that arises within cells following chemotherapeutic exposure but is not necessarily restricted to a subpopulation or subclone. There is substantial evidence in breast cancer for this mechanism as well8,13,60,61. Indeed, mitochondria are incredibly dynamic, with fusion and fission coordinated with multiple cellular events such as cell cycle. It will be important that future investigations specifically investigate mitochondrial structure and function in this context in TNBC.
We showed previously in several TNBC cell lines that DNA-damaging chemotherapies such as CRB and Adriamycin induce mitochondrial fusion and that genetic or pharmacological inhibition of OPA1 increases chemosensitivity and reduces oxphos11. Conversely, taxane chemotherapies (including DTX) do not induce fusion or oxphos. Our present study revealed a greater increase in mitochondrial area, volume, perimeter, length, and MCI in mitochondria from CRB-treated tumors compared to DTX-treated tumors. It will be important to validate this finding in additional TNBC models, as this chemotherapy-specific effect on mitochondria could inform mitochondrial-targeting therapy development.
Mitochondria are not static organelles, with cycles of fission and fusion supporting their dynamic nature. Our imaging only captured a snapshot of mitochondrial structures in our tumor samples, and we suspect this may represent the ‘average’ mitochondrial state in each tumor specimen. As 3D methods with subcellular resolution evolve for longitudinal, or even real-time analyses, the potential may arise to evaluate rates of mitochondrial structure changes in experimental models. As we showed previously, both pharmacologic and genetic perturbations can induce these mitochondrial structures11. It is reasonable to speculate that these mitochondrial structures are plastic and can reverse, as reported prior in this PDX model, PIM001-P, treated with AC that there is a reversible drug-tolerant state13. In this previous study, barcode-mediated clonal tracking was used and did not provide evidence for clonal enrichment13.
Our study has several limitations. While SBF-SEM microscopy provides detailed information on the 3D landscape of subcellular organelles, it is exceptionally labor- and time-intensive, requiring specialized expertise16, which precludes its routine use in research labs and the clinic. For this reason, we herein conducted an in-depth analysis of mitochondria for one tumor per treatment group, providing spatially distinct ROIs for mitochondrial quantifications to cover the possible spatial heterogeneity within each tumor. It remains to be determined how reproducible mitochondrial features will be across large cohorts of mouse or human tumors. However, based on the extensive similarities between the two single-agent groups as well as between the two combination groups, we suspect the hundreds of mitochondria sampled within each tumor are a fair representation of the true disease. Although there are efforts to automate the analyses of the mitochondria and other organelles, algorithms often need to be retrained on new data sets62. It is also critically important to expand these studies to additional TNBC specimens from other patients because inter-patient heterogeneity within TNBC is well documented at the DNA, RNA, and protein levels56,63,64. There is interest in including 3D modeling to provide a more comprehensive understanding of cancer progression and drug responses65,66. Mitochondrial structural differences among TNBC subtypes are not yet characterized but will be important in future research studies. Finally, PDX models lack a fully intact immune system, so it will be important to follow up these studies on bona fide human tumor specimens. Nonetheless, publications from ours and many other groups provide evidence that PDX models accurately capture many features of human cancer, including ITH, metabolic phenotypes, and chemotherapeutic responses30,42,67.
Supplementary Material
Supplementary Table 1. Selected segmented mitochondria used for analyses. Number of segmented mitochondria characterized during this study and the process for selecting 350 mitochondria per treatment for analyses.
Supplementary Figure 1. Mitochondrial measurements across treatment groups do not have normal distributions. Q–q plots of the raw measurements across all tumors are shown. Each blue dot represents one of the segmented mitochondria from a total of 350 segmented mitochondria. The red line indicates a reference for where the normal distribution falls on the graph.
Supplementary Figure 2. Distribution of mitochondrial metrics within each treatment group. Density plots are of log10 transformed data showing the distribution of the respective metric for the segmented mitochondria. The line indicates the median for treatment-naïve tumors. Treatment groups: treatment-naïve (TN), Adriamycin + cyclophosphamide (AC), docetaxel + carboplatin (DTX+CRB), docetaxel (DTX), and carboplatin (CRB)
Supplementary Figure 3. Metrics width and length were used to calculate MBI for each treatment group. Mitochondrial measurements for width and length were calculated using Amira software for 350 segmented mitochondria (represented as dots) from each treatment group. (a) Width (b) Length. Adjusted p-values (*p < 0.05, **p < 0.01, ***p < 0.001, ****p<0.0001) are shown on box and whisker plots as determined using the Mann–Whitney U test followed by the Holm method. Density plots are of log10 transformed data showing the distribution of the respective metric for the segmented mitochondria. The line indicates the median for treatment-naïve tumors. (c) Density plot for width (d) Density plot for length. Treatment groups: treatment-naïve (TN), Adriamycin + cyclophosphamide (AC), docetaxel + carboplatin (DTX+CRB), docetaxel (DTX), and carboplatin (CRB).
Supplementary Figure 4. Significant ITH was observed in treatment-naïve mitochondria, while ITH is reduced upon chemotherapy treatment. (a) Schematic for comparing the variance between treatment groups across all mitochondrial features. (b-i) Variance bar graphs for all 350 segmented mitochondrial volume, area, perimeter, width, length, mitochondrial branching index (MBI), sphericity, and mitochondrial complex index (MCI). F-test and Holm method for adjusted p-values (*p < 0.05, **p < 0.01, ***p < 0.001, ****p<0.0001).
ACKNOWLEDGEMENTS
We thank the breast cancer patients who donated their biopsies for PDX models. The generation of the PDX model PIM001-P was supported by a generous gift from the Cazalot family and the MD Anderson Women’s Cancer Moonshot Program. Dr. Helen Piwnica-Worms provided the PIM001-P PDX model via a materials transfer agreement. The BCM Patient Derived Xenograft Core guided the treatment regimens for DTX, CRB, DTX+CRB funded by CPRIT Core Facility Award (RP220646) and P30 Cancer Center Support Grant (NCI-CA125123). Dr. Junegoo Lee and Ms. Emily Goff aided with animal experiments. Dr. Tao Wang provided guidance for statistical analyses. Ms. Janice Cowden provided patient research advocacy support. The Mayo Clinic Microscopy and Cell Analysis Core provided experimental technical support. H&E, Ku80, and human mitochondria staining of PDX tumor sections were conducted at the BCM Pathology Core and Lab, supported by the Breast Center and a variety of research grants awarded to its faculty, including one of nine Specialized Programs of Research Excellence (SPORE) in Breast Cancer granted by the National Institute of Health. S10 grant (1S10OD028671-01) for digital imaging of the IHC slides. Dr. George Miles aided with the histological review of IHC and H&E-stained slides.
FUNDING
GVE is a CPRIT Scholar in Cancer Research. Funding sources that supported this work include the Cancer Prevention and Research Institute of Texas RR200009 (to GVE); NIH 1K22CA241113-01 (to GVE); NIH T32 predoctoral training grant T32GM136560-02 (to MJB); BWF Ad-hoc Award, National Institutes of Health (NIH) Small Research Pilot Subaward to 5R25HL106365-12 from the NIH PRIDE Program, DK020593 (to AHJ); 1R37CA269783-01A1 (to GVE); a BCM Breast Program SPORE Career Enhancement Program Grant (to GVE); National Science Foundation Graduate Research Fellowship 2140736 (to MJB); American Cancer Society Research Scholar Grant RSG-22-093-01-CCB (to GVE), a Myra Branum Wilson Baylor Research Advocates for Student Scientists Scholarship (to MJB); and a Breast Cancer Alliance Young Investigator Grant (to GVE); UNCF/Bristol-Myers Squibb (UNCF/BMS) E.E. Just Postgraduate Fellowship in Life Sciences Fellowship and Burroughs Wellcome Fund/ PDEP #1022376 (to HKB) UNCF/BMS E.E. Just Faculty Fund, Career Award at the Scientific Interface (CASI Award) from Burroughs Welcome Fund ID # 1021868.01, Vanderbilt Diabetes and Research Training Center for Department of Medicine’s Diabetes Research and Training Center (DRTC) Alzheimer’s Disease Pilot & Feasibility Program, CZI Science Diversity Leadership grant number 2022-253529 from the Chan Zuckerberg Initiative DAF, and an advised fund of Silicon Valley Community Foundation (to AHJ). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, the Cancer Prevention and Research Institute of Texas, National Science Foundation, American Cancer Society, BRASS, UNCF/BMS, Burroughs Welcome Fund, DRTC, Chan Zuckerberg Initiative DAF, or Silicon Valley Community Foundation.
DISCLOSURES
MTL is a founder and limited partner in StemMed Ltd. and a manager in StemMed Holdings, its general partner. He is a founder and equity stakeholder in Tvardi Therapeutics Inc. Some PDX models, none of which are included in this study, are exclusively licensed to StemMed Ltd., resulting in royalty income to MTL. LED is a compensated employee of StemMed Ltd. Some PDX models, none of which are included in this study, are exclusively licensed to StemMed Ltd., resulting in royalty income to LED. GVE receives sponsored research funding from Chimerix, Inc and experimental compounds from the Lead Discovery Center of Germany. All other authors have nothing to disclose.
Funding Statement
GVE is a CPRIT Scholar in Cancer Research. Funding sources that supported this work include the Cancer Prevention and Research Institute of Texas RR200009 (to GVE); NIH 1K22CA241113-01 (to GVE); NIH T32 predoctoral training grant T32GM136560-02 (to MJB); BWF Ad-hoc Award, National Institutes of Health (NIH) Small Research Pilot Subaward to 5R25HL106365-12 from the NIH PRIDE Program, DK020593 (to AHJ); 1R37CA269783-01A1 (to GVE); a BCM Breast Program SPORE Career Enhancement Program Grant (to GVE); National Science Foundation Graduate Research Fellowship 2140736 (to MJB); American Cancer Society Research Scholar Grant RSG-22-093-01-CCB (to GVE), a Myra Branum Wilson Baylor Research Advocates for Student Scientists Scholarship (to MJB); and a Breast Cancer Alliance Young Investigator Grant (to GVE); UNCF/Bristol-Myers Squibb (UNCF/BMS) E.E. Just Postgraduate Fellowship in Life Sciences Fellowship and Burroughs Wellcome Fund/ PDEP #1022376 (to HKB) UNCF/BMS E.E. Just Faculty Fund, Career Award at the Scientific Interface (CASI Award) from Burroughs Welcome Fund ID # 1021868.01, Vanderbilt Diabetes and Research Training Center for Department of Medicine’s Diabetes Research and Training Center (DRTC) Alzheimer’s Disease Pilot & Feasibility Program, CZI Science Diversity Leadership grant number 2022-253529 from the Chan Zuckerberg Initiative DAF, and an advised fund of Silicon Valley Community Foundation (to AHJ). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, the Cancer Prevention and Research Institute of Texas, National Science Foundation, American Cancer Society, BRASS, UNCF/BMS, Burroughs Welcome Fund, DRTC, Chan Zuckerberg Initiative DAF, or Silicon Valley Community Foundation.
DATA AND CODE AVAILABILITY
Mitochondria images and raw data are available from the corresponding authors upon request. All computational codes used for statistical analyses are available https://github.com/audra-lane/berner-et-al-2024. The PDX model PIM001-P can be made available through a materials transfer agreement with The University of Texas MD Anderson Cancer Center.
REFERENCES CITED
- 1.Harbeck N. et al. Breast cancer. Nat Rev Dis Primers 5, 66 (2019). 10.1038/s41572-019-0111-2 [DOI] [PubMed] [Google Scholar]
- 2.Bianchini G., De Angelis C., Licata L. & Gianni L. Treatment landscape of triple-negative breast cancer - expanded options, evolving needs. Nat Rev Clin Oncol 19, 91–113 (2022). 10.1038/s41571-021-00565-2 [DOI] [PubMed] [Google Scholar]
- 3.Schmid P. et al. Pembrolizumab for Early Triple-Negative Breast Cancer. N Engl J Med 382, 810–821 (2020). 10.1056/NEJMoa1910549 [DOI] [PubMed] [Google Scholar]
- 4.Vagia E., Mahalingam D. & Cristofanilli M. The Landscape of Targeted Therapies in TNBC. Cancers (Basel) 12 (2020). 10.3390/cancers12040916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Symmans W. F. et al. Long-Term Prognostic Risk After Neoadjuvant Chemotherapy Associated With Residual Cancer Burden and Breast Cancer Subtype. J Clin Oncol 35, 1049–1060 (2017). 10.1200/JCO.2015.63.1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yau C. et al. Residual cancer burden after neoadjuvant chemotherapy and long-term survival outcomes in breast cancer: a multicentre pooled analysis of 5161 patients. Lancet Oncol 23, 149–160 (2022). 10.1016/S1470-2045(21)00589-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jenkins B. C. et al. Mitochondria in disease: changes in shapes and dynamics. Trends Biochem Sci 49, 346–360 (2024). 10.1016/j.tibs.2024.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pendleton K. E., Wang K. & Echeverria G. V. Rewiring of mitochondrial metabolism in therapy-resistant cancers: permanent and plastic adaptations. Front Cell Dev Biol 11, 1254313 (2023). 10.3389/fcell.2023.1254313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhao J. et al. Mitochondrial dynamics regulates migration and invasion of breast cancer cells. Oncogene 32, 4814–4824 (2013). 10.1038/onc.2012.494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Humphries B. A. et al. Enhanced mitochondrial fission suppresses signaling and metastasis in triple-negative breast cancer. Breast Cancer Res 22, 60 (2020). 10.1186/s13058-020-01301-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baek M. L. et al. Mitochondrial structure and function adaptation in residual triple negative breast cancer cells surviving chemotherapy treatment. Oncogene 42, 1117–1131 (2023). 10.1038/s41388-023-02596-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herkenne S. et al. Developmental and Tumor Angiogenesis Requires the Mitochondria-Shaping Protein Opa1. Cell Metab 31, 987–1003 e1008 (2020). 10.1016/j.cmet.2020.04.007 [DOI] [PubMed] [Google Scholar]
- 13.Echeverria G. V. et al. Resistance to neoadjuvant chemotherapy in triple-negative breast cancer mediated by a reversible drug-tolerant state. Sci Transl Med 11 (2019). 10.1126/scitranslmed.aav0936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Denk W. & Horstmann H. Serial block-face scanning electron microscopy to reconstruct threedimensional tissue nanostructure. PLoS Biol 2, e329 (2004). 10.1371/journal.pbio.0020329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiao W., Chatton J. Y. & Genoud C. Mitochondria morphometry in 3D datasets obtained from mouse brains with serial block-face scanning electron microscopy. Methods Cell Biol 177, 197–211 (2023). 10.1016/bs.mcb.2023.01.021 [DOI] [PubMed] [Google Scholar]
- 16.Marshall A. G. et al. Serial Block Face-Scanning Electron Microscopy as a Burgeoning Technology. Adv Biol (Weinh) 7, e2300139 (2023). 10.1002/adbi.202300139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garza-Lopez E. et al. Correction: Garza-Lopez et al. Protocols for Generating Surfaces and Measuring 3D Organelle Morphology Using Amira. Cells 2022, 11, 65. Cells 12 (2023). 10.3390/cells12101356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glancy B., Kim Y., Katti P. & Willingham T. B. The Functional Impact of Mitochondrial Structure Across Subcellular Scales. Front Physiol 11, 541040 (2020). 10.3389/fphys.2020.541040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beasley H. K., Rodman T. A., Collins G. V., Hinton A. Jr. & Exil V. TMEM135 is a Novel Regulator of Mitochondrial Dynamics and Physiology with Implications for Human Health Conditions. Cells 10 (2021). 10.3390/cells10071750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hinton A. Jr. et al. A Comprehensive Approach to Sample Preparation for Electron Microscopy and the Assessment of Mitochondrial Morphology in Tissue and Cultured Cells. Adv Biol (Weinh) 7, e2200202 (2023). 10.1002/adbi.202200202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neikirk K. et al. Systematic Transmission Electron Microscopy-Based Identification and 3D Reconstruction of Cellular Degradation Machinery. Adv Biol (Weinh) 7, e2200221 (2023). 10.1002/adbi.202200221 [DOI] [PubMed] [Google Scholar]
- 22.Chen L. et al. Positive feedback loop between mitochondrial fission and Notch signaling promotes survivin-mediated survival of TNBC cells. Cell Death Dis 9, 1050 (2018). 10.1038/s41419-018-1083-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Si L. et al. Silibinin inhibits migration and invasion of breast cancer MDA-MB-231 cells through induction of mitochondrial fusion. Mol Cell Biochem 463, 189–201 (2020). 10.1007/s11010-019-03640-6 [DOI] [PubMed] [Google Scholar]
- 24.Vincent A. E. et al. Quantitative 3D Mapping of the Human Skeletal Muscle Mitochondrial Network. Cell Rep 26, 996–1009 e1004 (2019). 10.1016/j.celrep.2019.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vue Z. et al. Three-dimensional mitochondria reconstructions of murine cardiac muscle changes in size across aging. Am J Physiol Heart Circ Physiol 325, H965–H982 (2023). 10.1152/ajpheart.00202.2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vue Z. et al. 3D reconstruction of murine mitochondria reveals changes in structure during aging linked to the MICOS complex. Aging Cell 22, e14009 (2023). 10.1111/acel.14009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Faitg J. et al. 3D neuronal mitochondrial morphology in axons, dendrites, and somata of the aging mouse hippocampus. Cell Rep 36, 109509 (2021). 10.1016/j.celrep.2021.109509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Novotny R. et al. Mitochondrial changes in adenocarcinoma of the pancreas. Ultrastruct Pathol 37, 227–231 (2013). 10.3109/01913123.2013.788306 [DOI] [PubMed] [Google Scholar]
- 29.Han M. et al. Spatial mapping of mitochondrial networks and bioenergetics in lung cancer. Nature 615, 712–719 (2023). 10.1038/s41586-023-05793-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bruna A. et al. A Biobank of Breast Cancer Explants with Preserved Intra-tumor Heterogeneity to Screen Anticancer Compounds. Cell 167, 260–274 e222 (2016). 10.1016/j.cell.2016.08.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dobrolecki L. E. et al. Patient-derived xenograft (PDX) models in basic and translational breast cancer research. Cancer Metastasis Rev 35, 547–573 (2016). 10.1007/s10555-016-9653-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Echeverria G. V. et al. High-resolution clonal mapping of multi-organ metastasis in triple negative breast cancer. Nat Commun 9, 5079 (2018). 10.1038/s41467-018-07406-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eirew P. et al. Dynamics of genomic clones in breast cancer patient xenografts at single-cell resolution. Nature 518, 422–426 (2015). 10.1038/nature13952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Savage P. et al. Chemogenomic profiling of breast cancer patient-derived xenografts reveals targetable vulnerabilities for difficult-to-treat tumors. Commun Biol 3, 310 (2020). 10.1038/s42003-020-1042-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baek M., Chang J. T. & Echeverria G. V. Methodological Advancements for Investigating Intra-tumoral Heterogeneity in Breast Cancer at the Bench and Bedside. J Mammary Gland Biol Neoplasia 25, 289–304 (2020). 10.1007/s10911-020-09470-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eccles S. A. et al. Critical research gaps and translational priorities for the successful prevention and treatment of breast cancer. Breast Cancer Res 15, R92 (2013). 10.1186/bcr3493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Deerinck T. J., Bushong E. A., Thor A. & Ellisman M. H. NCMIR methods for 3D EM: A new protocol for preparation of biological specimens for serial block face scanning electron microscopy. (2010).
- 38.Virtanen P. et al. SciPy 1.0: fundamental algorithms for scientific computing in Python. Nat Methods 17, 261–272 (2020). 10.1038/s41592-019-0686-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Seabold S. & Perktold J. Statsmodels: Econometric and Statistical Modeling with Python. Proceedings of the 9th Python in Science Conference 2010 (2010). [Google Scholar]
- 40.McKinney W. Data Structures for Statistical Computing in Python. (2010).
- 41.Team R. C. R: A Language and Environment for Statistical Computing, <<https://www.R-project.org/>> (2023).
- 42.Petrosyan V. et al. Identifying biomarkers of differential chemotherapy response in TNBC patient-derived xenografts with a CTD/WGCNA approach. iScience 26, 105799 (2023). 10.1016/j.isci.2022.105799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fisher E. R. et al. Pathobiology of preoperative chemotherapy: findings from the National Surgical Adjuvant Breast and Bowel (NSABP) protocol B-18. Cancer 95, 681–695 (2002). 10.1002/cncr.10741 [DOI] [PubMed] [Google Scholar]
- 44.Garza-Lopez E. et al. Protocols for Generating Surfaces and Measuring 3D Organelle Morphology Using Amira. Cells 11 (2021). 10.3390/cells11010065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gerlinger M. et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 366, 883–892 (2012). 10.1056/NEJMoa1113205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cortes J. et al. Pembrolizumab plus chemotherapy versus placebo plus chemotherapy for previously untreated locally recurrent inoperable or metastatic triple-negative breast cancer (KEYNOTE-355): a randomised, placebo-controlled, double-blind, phase 3 clinical trial. Lancet 396, 1817–1828 (2020). 10.1016/S0140-6736(20)32531-9 [DOI] [PubMed] [Google Scholar]
- 47.Anurag M. et al. Proteogenomic Markers of Chemotherapy Resistance and Response in Triple-Negative Breast Cancer. Cancer Discov 12, 2586–2605 (2022). 10.1158/2159-8290.CD-22-0200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Filadi R. et al. TOM70 Sustains Cell Bioenergetics by Promoting IP3R3-Mediated ER to Mitochondria Ca(2+) Transfer. Curr Biol 28, 369–382 e366 (2018). 10.1016/j.cub.2017.12.047 [DOI] [PubMed] [Google Scholar]
- 49.Giorgi C., Marchi S. & Pinton P. The machineries, regulation and cellular functions of mitochondrial calcium. Nat Rev Mol Cell Biol 19, 713–730 (2018). 10.1038/s41580-018-0052-8 [DOI] [PubMed] [Google Scholar]
- 50.Szymanski J. et al. Interaction of Mitochondria with the Endoplasmic Reticulum and Plasma Membrane in Calcium Homeostasis, Lipid Trafficking and Mitochondrial Structure. Int J Mol Sci 18 (2017). 10.3390/ijms18071576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu H., Sun C., Gong Q. & Feng D. Mitochondria-Associated Endoplasmic Reticulum Membranes in Breast Cancer. Front Cell Dev Biol 9, 629669 (2021). 10.3389/fcell.2021.629669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Osborne C. K., Kitten L. & Arteaga C. L. Antagonism of chemotherapy-induced cytotoxicity for human breast cancer cells by antiestrogens. J Clin Oncol 7, 710–717 (1989). 10.1200/JCO.1989.7.6.710 [DOI] [PubMed] [Google Scholar]
- 53.Palmer A. C., Chidley C. & Sorger P. K. A curative combination cancer therapy achieves high fractional cell killing through low cross-resistance and drug additivity. Elife 8 (2019). 10.7554/eLife.50036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Plana D., Palmer A. C. & Sorger P. K. Independent Drug Action in Combination Therapy: Implications for Precision Oncology. Cancer Discov 12, 606–624 (2022). 10.1158/2159-8290.CD-21-0212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ali H. R. et al. Imaging mass cytometry and multiplatform genomics define the phenogenomic landscape of breast cancer. Nature Cancer 1, 163–175 (2020). 10.1038/s43018-020-0026-6 [DOI] [PubMed] [Google Scholar]
- 56.Jackson H. W. et al. The single-cell pathology landscape of breast cancer. Nature 578, 615–620 (2020). 10.1038/s41586-019-1876-x [DOI] [PubMed] [Google Scholar]
- 57.Wagner J. et al. A Single-Cell Atlas of the Tumor and Immune Ecosystem of Human Breast Cancer. Cell 177, 1330–1345 e1318 (2019). 10.1016/j.cell.2019.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Creighton C. J. et al. Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci U S A 106, 13820–13825 (2009). 10.1073/pnas.0905718106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Balko J. M. et al. Profiling of residual breast cancers after neoadjuvant chemotherapy identifies DUSP4 deficiency as a mechanism of drug resistance. Nat Med 18, 1052–1059 (2012). 10.1038/nm.2795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Almendro V. et al. Inference of tumor evolution during chemotherapy by computational modeling and in situ analysis of genetic and phenotypic cellular diversity. Cell Rep 6, 514–527 (2014). 10.1016/j.celrep.2013.12.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kim C. et al. Chemoresistance Evolution in Triple-Negative Breast Cancer Delineated by Single-Cell Sequencing. Cell 173, 879–893 e813 (2018). 10.1016/j.cell.2018.03.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Heinrich L. et al. Whole-cell organelle segmentation in volume electron microscopy. Nature 599, 141–146 (2021). 10.1038/s41586-021-03977-3 [DOI] [PubMed] [Google Scholar]
- 63.Lehmann B. D. et al. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J Clin Invest 121, 2750–2767 (2011). 10.1172/JCI45014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shah S. P. et al. The clonal and mutational evolution spectrum of primary triple-negative breast cancers. Nature 486, 395–399 (2012). 10.1038/nature10933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Rozenblatt-Rosen O. et al. The Human Tumor Atlas Network: Charting Tumor Transitions across Space and Time at Single-Cell Resolution. Cell 181, 236–249 (2020). 10.1016/j.cell.2020.03.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Johnson B. E. et al. An omic and multidimensional spatial atlas from serial biopsies of an evolving metastatic breast cancer. Cell Rep Med 3, 100525 (2022). 10.1016/j.xcrm.2022.100525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guillen K. P. et al. A human breast cancer-derived xenograft and organoid platform for drug discovery and precision oncology. Nat Cancer 3, 232–250 (2022). 10.1038/s43018-022-00337-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1. Selected segmented mitochondria used for analyses. Number of segmented mitochondria characterized during this study and the process for selecting 350 mitochondria per treatment for analyses.
Supplementary Figure 1. Mitochondrial measurements across treatment groups do not have normal distributions. Q–q plots of the raw measurements across all tumors are shown. Each blue dot represents one of the segmented mitochondria from a total of 350 segmented mitochondria. The red line indicates a reference for where the normal distribution falls on the graph.
Supplementary Figure 2. Distribution of mitochondrial metrics within each treatment group. Density plots are of log10 transformed data showing the distribution of the respective metric for the segmented mitochondria. The line indicates the median for treatment-naïve tumors. Treatment groups: treatment-naïve (TN), Adriamycin + cyclophosphamide (AC), docetaxel + carboplatin (DTX+CRB), docetaxel (DTX), and carboplatin (CRB)
Supplementary Figure 3. Metrics width and length were used to calculate MBI for each treatment group. Mitochondrial measurements for width and length were calculated using Amira software for 350 segmented mitochondria (represented as dots) from each treatment group. (a) Width (b) Length. Adjusted p-values (*p < 0.05, **p < 0.01, ***p < 0.001, ****p<0.0001) are shown on box and whisker plots as determined using the Mann–Whitney U test followed by the Holm method. Density plots are of log10 transformed data showing the distribution of the respective metric for the segmented mitochondria. The line indicates the median for treatment-naïve tumors. (c) Density plot for width (d) Density plot for length. Treatment groups: treatment-naïve (TN), Adriamycin + cyclophosphamide (AC), docetaxel + carboplatin (DTX+CRB), docetaxel (DTX), and carboplatin (CRB).
Supplementary Figure 4. Significant ITH was observed in treatment-naïve mitochondria, while ITH is reduced upon chemotherapy treatment. (a) Schematic for comparing the variance between treatment groups across all mitochondrial features. (b-i) Variance bar graphs for all 350 segmented mitochondrial volume, area, perimeter, width, length, mitochondrial branching index (MBI), sphericity, and mitochondrial complex index (MCI). F-test and Holm method for adjusted p-values (*p < 0.05, **p < 0.01, ***p < 0.001, ****p<0.0001).
Data Availability Statement
Mitochondria images and raw data are available from the corresponding authors upon request. All computational codes used for statistical analyses are available https://github.com/audra-lane/berner-et-al-2024. The PDX model PIM001-P can be made available through a materials transfer agreement with The University of Texas MD Anderson Cancer Center.