Abstract
Proteostasis is vital for cellular health, with disruptions leading to pathologies including aging, neurodegeneration and metabolic disorders. Traditionally, proteotoxic stress responses were studied as acute reactions to various noxious factors; however, recent evidence reveals that many proteostasis stress-response genes exhibit ~12-hour ultradian rhythms under physiological conditions in mammals. These rhythms, driven by an XBP1s-dependent 12h oscillator, are crucial for managing proteostasis. By exploring the chromatin landscape of the murine 12h hepatic oscillator, we identified RBBP5, a key subunit of the COMPASS complex writing H3K4me3, as an essential epigenetic regulator of proteostasis. RBBP5 is indispensable for regulating both the hepatic 12h oscillator and transcriptional response to acute proteotoxic stress, acting as a co-activator for proteostasis transcription factor XBP1s. RBBP5 ablation leads to increased sensitivity to proteotoxic stress, chronic inflammation, and hepatic steatosis in mice, along with impaired autophagy and reduced cell survival in vitro. In humans, lower RBBP5 expression is associated with reduced adaptive stress-response gene expression and hepatic steatosis. Our findings establish RBBP5 as a central regulator of proteostasis, essential for maintaining mammalian organismal health.
Introduction:
Proteostasis, the maintenance of proper protein folding, trafficking, and turnover, is a major challenge for the cell (Labbadia & Morimoto, 2015). Much of the cellular energy expenditure is dedicated to maintaining a healthy proteome, as millions of molecules are produced every single minute (Buttgereit & Brand, 1995). Challenges to proteostasis, exemplified by the accumulation of unfolded or misfolded proteins, can lead to a range of cellular dysfunctions and pathological conditions, such as aging, neurodegenerative diseases, cancer, and metabolic disorders (Klaips et al, 2018; Labbadia & Morimoto, 2015; Martinez et al, 2017; Taylor & Hetz, 2020; Zhao & Ackerman, 2006). To cope with proteotoxic stress, organisms have developed a wide range of stress response mechanisms, including the heat shock response, the unfolded protein response (UPR), and the more generalized integrated stress response (ISR) (Costa-Mattioli & Walter, 2020; Hetz et al, 2020; Richter et al, 2010). Although the detailed signaling molecules vary, these pathways generally involve transmitting the information of cellular stress to the nucleus by activating various transcription factors (TF) containing the basic leucine zipper domain (bZIP), such as UPR TFs spliced form of XBP1 (XBP1s), ATF4 and ATF6 and heat shock factor 1 (HSF1). After binding to the stress response element in the promoters/enhancers, these TFs in turn upregulate a plethora of genes aimed to restore proteostasis (Balch et al, 2008; Ron & Walter, 2007).
While proteotoxic stress responses were traditionally viewed and studied as distinct acute responses to different “noxious” factors, recent evidence by our group indicates that at the physiological level (and in the absence of evident extrinsic stress), the expression of hundreds of proteotoxic stress response regulatory and output genes exhibit cell-autonomous ultradian rhythms cycling with a ~12h period in multiple mammalian cell-types and tissues, including humans (Asher & Zhu, 2022b; Ballance & Zhu, 2021; Dion et al, 2022a; Meng et al, 2022; Meng et al, 2020; Pan et al, 2020; Scott et al, 2023; Zhu et al, 2023; Zhu et al, 2024; Zhu et al, 2017b). These proteostasis genes encompass both output genes involved in protein processing in the ER/Golgi, redox regulation, protein folding, ER-associated protein degradation (ERAD), autophagy, and upstream regulatory molecules and TFs like Xbp1, Atf4, Atf6, Ddit3, Perk and Ire1α (Asher & Zhu, 2022a; Ballance & Zhu, 2021). Many of these ~12h ultradian rhythms are established by an XBP1s-dependent 12h oscillator separate from both the ~24h circadian clock and the cell cycle (Pan et al., 2020; Zhu & Liu, 2023a; Zhu et al, 2017a). It is hypothesized that the mammalian 12h oscillator may have evolved from the circatidal clock of marine animals, and later co-opted to adapt to the ~12h cycle of metabolic stress peaking at transition times at dawn and dusk in terrestrial animals (Asher & Zhu, 2022b; Pan et al., 2020; Zhu et al., 2024). The 12h oscillator is well-studied in the liver, and mice with liver-specific 12h oscillator ablation exhibited accelerated liver aging and steatosis (Meng et al., 2020), manifested with impaired proteostasis, lipid metabolism, and mitochondria dysfunction (Dion et al, 2022b; Meng et al., 2022; Meng et al., 2020). Since the 12h oscillator integrates multiple proteotoxic stress signaling, it provides us with a unique opportunity to study proteostasis using a holistic approach: rather than studying individual stress response separately, by investigating the 12h oscillator we could gain new mechanistic insights for universal proteostasis regulation.
While significant work has been done on upstream proteotoxic stress sensing and protein-folding mechanisms in the ER and cytosol, there remains a lack of knowledge regarding transcriptional regulation in proteostasis in general, particularly concerning the temporal epigenome dynamics, chromatin landscapes, and co-regulatory networks underlying global proteostasis control. In this study, we seek to address an important unanswered question: is there a designated epigenetic regulator that orchestrates global proteostasis? By examining the chromatin landscape of the murine 12h hepatic oscillator, we identified RBBP5—an essential subunit of the Complex Proteins Associated with Set1 (COMPASS) complex responsible for depositing H3K4me3 (Shilatifard, 2012a) —as a pivotal epigenetic regulator that governs global proteostasis dynamics. RBBP5 plays an indispensable role in regulating both the 12h oscillator and transcriptional response to acute proteostasis stress, functioning as a co-activator for key proteostasis master TFs, such as XBP1s. Ablation of RBBP5 leads to heightened sensitivity to proteotoxic stress, resulting in chronic inflammation and hepatic steatosis in mice, as well as defective autophagy and impaired cell survival in vitro. In humans, reduced RBBP5 expression is linked to a diminished adaptive stress response and the onset of hepatic steatosis. Our findings establish RBBP5 as a crucial epigenetic regulator of proteostasis dynamics, essential for maintaining organismal health.
Results:
Global RBBP5 binding to chromatin exhibits a ~12h ultradian rhythm, correlated with the promoter-proximal ~12h H3K4me3 rhythms in mouse liver.
The 12h oscillator is transcriptionally regulated by the UPR TF XBP1s in mice (Meng et al., 2020; Pan et al., 2020; Zhu et al., 2017b), and likely under control by additional bZIP TFs, such as ATF4 and ATF6 (Zhu, 2020). However, decades of research of eukaryotic gene regulation demonstrated that TFs themselves are not sufficient to initiate transcription, and additional so-called “co-regulators” are required for full gene activation by shaping the epigenetic landscape (Dasgupta et al, 2014; Roeder, 2005; Spiegelman & Heinrich, 2004). Currently, the epigenetic landscape of both the 12h oscillator and proteotoxic stress responses are poorly characterized. To identify epigenetic regulators of global proteostasis control, we first examined a published temporal hepatic epigenome dataset for histone modifications associated with the active promoters of ~12h genes (Koike et al, 2012). We found ~12h rhythms of histone H3 trimethylated at lysine 4 (H3K4me3), but not two histone acetylation markers, histone H3 acetylated at lysine 9 (H3K9Ac) and lysine 27 (H3K27Ac), at the promoters of ~12h genes (Fig. 1A, B). This is particularly prominent for those ~12h genes with rhythmic expressions peaking at ZT0 and ZT12 that are enriched for proteostasis pathways (Pan et al., 2020), with H3K4me3 level also peaking at these two time points (Fig. 1A, B).
Fig. 1. Global ~12h RBBP5 cistrome is associated with promoter-proximal ~12h H3K4me3 epigenome in mouse liver.
(A) Heatmap of ~12h hepatic transcriptome aligned with H3K4me3, H3K9Ac and H3K27Ac cistromes. (B) Quantification of temporal H3K4me3, H3K9ac and H3K27Ac. (C) Heatmap showing RBBP5 and H3K4me3 chromatin occupancy 1kb ± of TSS of 6,451 genes. (D) RBBP5 chromatin occupancy aligned with nucleosome positioning. (E) Quantification of temporal average RBBP5 binding intensity on different nucleosomes. (F) RBBP5 chromatin occupancy aligned with H3K4me3 occupancy.
In eukaryotes, H3K4 is tri-methylated by the COMPASS complex, which includes regulatory subunits RBBP5, ASH2L, WDR5, DPY30 and catalytic subunits SETD1A/B/MLL1-4 (Fig. S1A) (Qu et al, 2018; Shilatifard, 2012b; Takahashi et al, 2011). Via writing H3K4me3, COMPASS is essential for the eviction of paused RNA polymerase II (RNAPII) and promotes transcriptional elongation (Hu et al, 2023; Wang et al, 2023). Landscape In Silico deletion Analysis (LISA) (Qin et al, 2020) further inferred COMPASS subunits WDR5, ASH2L, DYP30 and RBBP5 as putative epigenetic regulators of hepatic ~12h, but not circadian genes (Fig. S1B). Being the commonly shared regulatory subunits for all MLL family H3K4 methyl transferase complex (including SETD1A/B and MLL1-4), RBBP5/ASH2L forms the minimal heterodimer sufficient to activate all MLL histone methyl transferase (Li et al, 2016; Ruthenburg et al, 2007) and are essential for the assembly of the whole COMPASS complex (Han et al, 2019; Qu et al., 2018). Between the two essential subunits, we initially focused on RBBP5. Eigenvalue/pencil method (Antoulas et al, 2018b) revealed that the levels of both hepatic Rbbp5 mRNA and nuclear RBBP5 protein exhibit ~12h oscillations, with the nuclear protein level peaking at ~CT0 and ~CT12 (Fig. S1C, D), in line with the acrophases of H3K4me3 and gene expression oscillations (Fig. 1A, B).
To further evaluate the potential role of RBBP5 in establishing the epigenetic landscape of hepatic ~12h rhythms, we performed temporal RBBP5 ChIP-seq at a 4-h interval for a total of 48 hours in the liver of male C57BL/6 mice housed under constant darkness condition fed ad libitum. Chromatin occupancies of RBBP5 align well with nucleosome positioning downstream of the transcription start sites (TSS) and the H3K4me3 epigenome, with ‘ridges’ corresponding to 130bp, 310bp, 490bp, 670bp downstream of TSS at all time points, thus demonstrating the high quality of our ChIP-seq dataset (Fig. 1C-F). In total, we identified 7,963 hepatic RBBP5 binding sites that are within 2kb (up and downstream) of TSS of 6,451 genes (Fig. 1C and Table S1). The average binding intensities of RBBP5 on all post-TSS nucleosomes displayed robust 12h rhythms, with peaks occurring at CT0, CT12, CT24, and CT36 (Fig. 1C-E). These rhythms aligned with the 12h oscillations of promoter-proximal H3K4me3 (Fig. 1C–E), but not with those of H3K9Ac or H3K27Ac (Fig. S1E). This pattern is exemplified by Xbp1, one of the major UPR TFs and the transcriptional regulator of the 12h oscillator, and Hsph1, a canonical 12h oscillator output gene encoding a heat shock protein (Fig. S1F). Using the eigenvalue/pencil and RAIN (Thaben & Westermark, 2014) algorithms, we identified 3,028 and 1,864 (p<0.05) RBBP5 binding sites that cycle with a ~12h period, respectively (Fig. S1G, H and Table S1). Collectively, these results establish the epigenetic landscape of hepatic 12h oscillator is marked by RBBP5-H3K4me3.
Hepatic RBBP5 cistrome coincides with that of XBP1s and hepatic ~12h transcriptome.
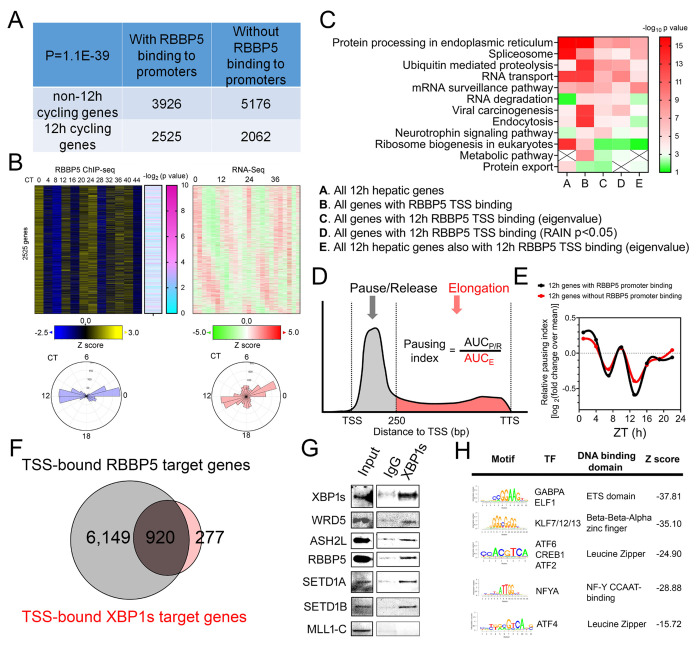
To investigate if RBBP5 regulates hepatic ~12h rhythms of gene expression, we first examined the correlation between RBBP5 proximal promoter binding status and ~12h rhythms of gene expression. As illustrated in Fig. 2A, genes with proximal promoter RBBP5 binding are enriched for ~12h rhythms of gene expression (P=1.1e-39 by Chi-squared test). For those 2,525 ~12h genes with RBBP5 binding, the phases of RBBP5 chromatin binding center around CT0 and CT12, slightly preceding the phases of gene expression observed at ~CT1 and ~CT13 (Fig. 2B). This temporal relationship suggests that RBBP5 acts as a driver, rather than a consequence of ~12h rhythms of gene expression.
Fig. 2. Hepatic RBBP5 cistrome coincides with that of XBP1s and hepatic ~12h transcriptome.
(A) The table showing the number of genes with or without ~12h rhythms and with or without promoter RBBP5 binding. (B) Heatmap showing RBBP5 binding intensity along with -log10 transformed P values for having 1~2h rhythm by RAIN and ~12h rhythms of gene expression on 2,525 genes. (C) Heat map summary of GO analysis demonstrating the -log10 transformed P value of different enriched pathways for different genes. (D) A representative diagram depicting a typical Gro-Seq signal from TSS to transcription stop site (TTS) of a gene and using AUC to calculate the pausing index. (E) Log2 mean-normalized temporal pausing index calculated from the GRO-Seq data for ~12h genes with or without promoter RBBP5 binding. (F) Venn diagram demonstrating distinct and common cistromes for RBBP5 and XBP1s. (G) Western blot showing co-IP in liver nuclear extracts from CT0 using anti-XBP1s and normal IgG control antibody. (H) Motif analysis of RBBP5 binding sites of 6,149 genes.
We next performed Gene Ontology (GO) analysis on those hepatic genes with proximal promoter RBBP5 binding and revealed strong enrichment of proteostasis pathways including protein processing in the ER and ubiquitin-mediated proteolysis (Fig. 2C). Additionally, pathways involved in mRNA processing, such as spliceosome, RNA transport, and mRNA surveillance, were also enriched (Fig. 2C). These findings align with previous observations that both mRNA and protein metabolism pathways are enriched in genes exhibiting ~12h rhythms in both mice and humans (Dion et al., 2022a; Zhu & Liu, 2023b; Zhu et al., 2024). The coupling of mRNA and protein metabolism is orchestrated by a SON-XBP1s axis and involves the liquid-liquid phase separation (LLPS) dynamics of nuclear speckles (Dion et al., 2022a). Similar pathways are also enriched in genes exhibiting both ~12h rhythms of RBBP5 promoter binding and gene expression (Fig. 2C).
Recent studies indicated that by depositing H3K4me3, RBBP5 (as a part of COMPASS) is essential for the eviction of paused RNAPII and promotes transcriptional elongation (Hu et al., 2023; Wang et al., 2023). To investigate whether the ~12h rhythms of RBBP5 promoter binding may translate into ~12h rhythms of RNAPII pause release, we performed a post-hoc analysis of a published Global run-on sequencing (GRO-Seq) dataset in mouse liver (Fang et al, 2014) that measured nascent hepatic RNA transcription at a 3h interval for a total of 24 hours. While the sampling frequency and duration of this dataset are not optimal for a comprehensive and rigorous detection of ~12h rhythms, we believe it may still offer valuable mechanistic insights into the transcriptional regulation of the hepatic 12-hour oscillator. By calculating the temporal pausing index of each ~12h hepatic gene (Fig. 2D), we observed that, on average, genes with RBBP5 promoter binding exhibited a greater amplitude in pausing index oscillations, peaking at ZT10 and ZT22, compared to those without RBBP5 binding (Fig. 2E).
XBP1s is the major transcriptional factor regulating the hepatic 12h oscillator and also exhibits a global 12h rhythm of chromatin binding to the promoter regions peaking at CT0, 12, 24 and 36 (Meng et al., 2020; Pan et al., 2020). Comparing the cistromes of RBBP5 and XBP1s revealed a significant overlap between the two, with 920 genes sharing RBBP5 and XBP1s proximal promoter binding (Fig. 2F). Co-immunoprecipitation in the hepatic nuclear extracts at CT0 confirmed the physical interaction between XBP1s and subunits of COMPASS, including RBBP5, ASH2L, WDR5, and two histone methyltransferase SETD1A and SETD1B (Fig. 2G). Motif analysis of the DNA sequences around the 6,149 RBBP5 binding sites that do not overlap with XBP1s cistrome identified binding motifs for GABPA, KLF, ATF6, NFYA and ATF4 (Fig. 2H). These findings suggest that, in addition to acting as a co-activator for XBP1s, RBBP5 may also work with other transcription factors to shape the hepatic 12-hour epigenome.
RBBP5 is an epigenetic regulator of the ~12h oscillator, but not the canonical ~24h circadian clock.
To establish the causality between RBBP5 and hepatic 12h rhythms of gene expression, we generated RBBP5 liver hepatocyte-specific knockout (RBBP5 LKO) mice using the CRE-loxP system. Exon 10 of mouse Rbbp5, which encodes the SET/ASH2L binding domain, was flanked by loxP sites (Fig. 3A). CRE-mediated deletion of Exon 10 is expected to lead to a frameshift and nonsense-mediated RNA decay of truncated Rbbp5 transcript (Fig. S2A). Crossing homozygous floxed Rbbp5 mice with Albumin-CRE mice resulted in liver-specific deletion of RBBP5, which was confirmed by DNA genotyping, immunofluorescence against RBBP5 and western blot analysis (Figs. 3B and S2B–D). The resulting RBBP5 LKO mice weighed slightly less than wild-type counterparts (Fig. S3A) but maintained normal rhythmic locomotor activity and fasting-feeding cycles under both 12h:12h L/D and constant darkness conditions (Fig. S3B–I).
Fig. 3. RBBP5 is an epigenetic regulator of the hepatic ~12h oscillator, but not the canonical ~24h circadian clock.
(A) Schematic of the design of Rbbp5Flox mice. (B) Immunofluorescence of RBBP5 in the liver of Rbbp5 Flox and Rbbp5 LKO mice. (C) Histograms showing the period distributions of all rhythmic genes uncovered from Rbbp5 Flox and Rbbp5 LKO mice (with adjP values or FDR <0.05) (n=2 at each time point for either genotype). (D, E) Cumulative distribution of 10~12h (D) and 22~28h (E) genes in Rbbp5 Flox and Rbbp5 LKO mice with different FDR cut-offs. (F) Heat map of 15 core circadian clock (top) and canonical ~12h gene expression (bottom) in the liver of Rbbp5 Flox and Rbbp5 LKO mice. Log2 normalized fold change of average gene expression across 48 hours between Rbbp5 Flox and Rbbp5 LKO mice was also shown. (G) Heat map of 6,025 ~12h gene expression (FDR<0.05) in the liver of Rbbp5 Flox and Rbbp5 LKO mice, with 5,601 of them lost in Rbbp5 LKO mice, along with -log10 transformed adjP values for having 10-12h rhythm by RAIN. Log2 normalized fold change of average gene expression across 48 hours between Rbbp5 Flox and Rbbp5 LKO mice was also shown. (H) RNA-Seq data for representative proteostasis and mRNA processing genes in the liver of Rbbp5 Flox and Rbbp5 LKO mice. (I) Heat map summary of GO analysis demonstrating the -log10 transformed P values of different enriched pathways for different genes. Data: Mean ± S.E.M.
To identify the RBBP5-dependent oscillating hepatic transcriptome, we performed bulk RNA sequencing (RNA-Seq) analysis in the liver of male Rbbp5 Flox and Rbbp5LKO mice at 2-h intervals for a total of 48 hour under constant darkness in duplicates (Table S2). To identify genes cycling with a period ranging from 6 to 36 hours, we initially applied the RAIN algorithm (Thaben & Westermark, 2014) to each genotype’s temporal transcriptome (Thaben & Westermark, 2014) (Table S3). Compared to alternative methods like JTK_CYCLE, RAIN detects rhythms with arbitrary waveforms and therefore more robustly uncovers ultradian rhythms (Antoulas et al, 2018a; Pan et al., 2020; Zhu et al., 2017a). Consistent with past studies (Hughes et al, 2009; Meng et al., 2022; Pan et al., 2020; van der Veen & Gerkema, 2017; Zhu et al., 2017b), two populations of hepatic transcripts cycling with periods of ~12h and ~24h were identified in Rbbp5 Flox mice liver (Fig. 3C, D). By contrast, in Rbbp5 LKO mice, the ~12h rhythms are all but abolished and the circadian oscillations remain largely intact, which includes all core circadian clock genes (such as Bmal1, Per1/2/3, and Nr1d1/2) displaying robust circadian rhythms with the same phases and amplitudes as those observed in Rbbp5 Flox mice (Figs. 3C-F and S4A). The maintenance of ~24h circadian rhythms and abolishment of ~12h ultradian rhythms in Rbbp5 LKO mice is further confirmed by the principal component analysis (PCA) (Fig. S4B, C). Specifically, using RAIN with an FDR cutoff of 0.01 and 0.05, we identified a total of 3,549 and 6,025 hepatic transcripts cycling at periods between 10-12h in Rbbp5 Flox mice, respectively. By contrast, only 61 and 813 10-12h genes were observed in Rbbp5 LKO mice with the same FDR thresholds, indicating over 93% ~12h hepatic transcriptome was abolished with liver RBBP5 ablation (Figs. 3G, H, S4D and Table S3). GO analysis confirmed that these RBBP5-depenent ~12h hepatic transcriptome is enriched in various mRNA processing and proteostasis pathways, distinct from those of RBBP5-independent circadian genes enriched in lipid metabolism and phosphorylation (Fig. 3I).
To determine the robustness of our results to different analytic methods, we also performed spectrum analysis with the eigenvalue/pencil method (Table S4) (Antoulas et al., 2018a; Dion et al., 2022b; Meng et al., 2022; Meng et al., 2020; Pan et al., 2020; Zhu et al., 2017a), which unlike statistical methods such as JTK_CYCLE and RAIN does not require pre-assignment of period range, enabling unbiased identification of multiple superimposed oscillations for any given gene (Antoulas et al., 2018a; Dion et al., 2022b; Meng et al., 2022; Meng et al., 2020; Pan et al., 2020; Zhu et al., 2017a). Revealing up to three oscillations from each gene, eigenvalue/pencil analyses also revealed prevalent ~24h and ~12h oscillations in Rbbp5 Flox mice, along with a third population cycling at a period around 8h (Fig. S5A–E). Hepatic ~8h oscillations were also previously reported but their regulation and function remained elusive (Asher & Zhu, 2022a; Hughes et al., 2009). Rbbp5 LKO mice, on the other hand, exhibited a drastically different spectrum, characterized by the near complete loss of the ~12h rhythms, de novo gain of many faster ultradian rhythms with periods between 6 and 7h, and the maintenance of most circadian rhythms (Fig. S5A–E). Similar findings were also observed for dominant oscillations (which is the one with the largest amplitude among the three superimposed oscillations for each gene) for each gene (Fig. S5A–E). For the ~12h genes originally identified in Rbbp5 Flox mice but lost in Rbbp5 LKO mice, the period spectrum in the Rbbp5 LKO mice displayed a diverse distribution. This ranged from a complete lack of rhythmicity (~39%) to shorter ultradian periods of 6-7 hours, and even to longer periods exceeding 24 hours (Fig. S5F). RBBP5-dependent ~12h transcriptome uncovered by the eigenvalue/pencil showed substantial overlap with the results obtained through the RAIN method.
This convergence was evident both in the specific genes identified (Fig. S5C) and in the enriched biological pathways (Fig. S5G, H), thereby confirming the robustness of our findings. These results thus establish RBBP5 as a central epigenetic regulator of the hepatic 12h oscillator, with more than 90% ~12h hepatic transcriptome abolished in its absence.
In addition to liver, cell-autonomous ~12h rhythms of proteostasis gene expression including Xbp1 can also be observed in mouse embryonic fibroblasts (MEFs) synchronized by low concentration of ER stress inducer tunicamycin (Tu) (Pan et al., 2020; Zhu et al., 2017a). We observed a ~12h rhythm of Rbbp5 expression in Tu-synchronized MEFs, which is independent of BMAL1 but require XBP1s (Fig. S6A), indicating RBBP5 itself is also under 12h oscillator control. To test the functional role of COMPASS in regulating the cell-autonomous ~12h rhythms, we knocked down either Rbbp5 or Ash2l via siRNA in a previously described 12h oscillator reporter MEFs that stably express Manf promoter-driven destabilized luciferase (dluc) (Pan et al., 2020), and found that both siRNAs significantly dampen the ~12h oscillation of luciferase activity as well as shorten the period (Fig. S6B). By contrast, neither RBBP5 nor ASH2L knocking down affects the circadian oscillation of Bmal1 promoter activity in human U2OS cells in a previous genome-wide siRNA screen study (Zhang et al, 2009) (Fig. S6C). Collectively, our results demonstrate that RBBP5 is an integral component of the 12h oscillator, but dispensable for the canonical circadian clock both in vivo and in vitro.
RBBP5 modulates the hepatic transcriptional response to acute proteotoxic stress in vivo.
Before the discovery of the mammalian 12h oscillator, the dynamics of proteostasis were primarily studied as transient responses to various proteotoxic stresses, with a classic example being the UPR triggered by ER stress. In response to the accumulation of misfolded proteins in the ER, the UPR initiates a signaling cascade from the ER to the nucleus, ultimately activating three key transcription factors: XBP1s, ATF4, and ATF6. This activation leads to a beneficial adaptive response, upregulating proteostasis genes to restore ER homeostasis (Fig. 4A) (Metcalf et al, 2020; Walter & Ron, 2011; Wiseman et al, 2022). However, ER stress can also trigger maladaptive and deleterious responses, such as the activation of pro-inflammatory and immune gene expression through the TRAF2-mediated activation of AP1 (comprised of c-Fos and c-Jun), NF-κB or STAT3 TFs (Fig. 4A) (So, 2018). Our findings that RBBP5 can co-activate XBP1s to regulate the 12h oscillator suggest that RBBP5 is essential for the adaptive UPR TFs-mediated activation of proteostasis genes but is not required for the maladaptive activation of immune genes by immune-regulatory TFs, whose binding motifs are absent from RBBP5 binding sites (Fig. 2H).
Fig. 4. RBBP5 regulates the hepatic transcriptional response to proteotoxic stress.
(A) The diagram of UPR. (B) PCA showing the hepatic transcriptional response to Tu in Rbbp5 Flox and Rbbp5 LKO mice. (C) Representative RNA-seq data for proteostasis (top) and immune genes (bottom) with RBBP5 promoter-binding status also shown below. n=3 for Rbbp5 Flox DMSO and Rbbp5 Flox Tu, and n=4 for Rbbp5 LKO DMSO and Rbbp5 LKO Tu. (D) Heat map summary of GO analysis demonstrating the -log10 transformed P values of different enriched pathways for different genes. (E) Log 2 normalized fold induction by Tu in Rbbp5 Flox and Rbbp5 LKO mice for genes with or without RBBP5 promoter binding. (F) Western blot analysis of different proteins in Rbbp5 Flox and Rbbp5 LKO mice treated with or without Tu. (G, H) Mice tail-vein injected with AAV8-TGB-GFP or AAV8-TGB-RBBP5 were treated with or without Tu. Western blot analysis (G) and qPCR (H). n=3 for AAV8-TBG-EGFP DMSO, n=4 for AAV8-TBG-EGFP Tu, n=4 for AAV8-TGB-mRBBP5 DMSO and n=4 for AAV8-TGB-mRBBP5 Tu. Data: Mean ± S.E.M.
To test this hypothesis, we intraperitoneally injected male Rbbp5 Flox and Rbbp5 LKO mice with vehicle control or 0.05mg/kg Tu to induce acute ER stress and performed RNA-seq on hepatic transcriptome isolated 8 hours post injection (Table S5). This lower dose of Tu was selected for its ability to induce the UPR, but that was significantly less toxic than the more common experimental dose of 1 mg/kg—which is known to elicit hepatocellular death and severe toxicity (Rutkowski et al, 2008). PCA demonstrated the separation of the four groups on the transcriptome space (Fig. 4B). Using an adjusted p value (FDR) of 0.10 as cut-off, we identified 139 and 45 genes significantly induced by Tu in the liver of Rbbp5 Flox and Rbbp5 LKO mice, respectively, with 21 commonly shared between the two (Figs. 4C, S7A–G and Table S6). In Rbbp5 Flox mice, these 139 genes are enriched in pathways of proteostasis and immune functions as expected, with pathways of ‘response to ER stress’ and ‘ERAD’ among the top two GO terms (Fig. 4C, D). By contrast, in Rbbp5 LKO mice, these 45 genes are only enriched in immune functions, with proteostasis genes not being significantly induced by Tu (Fig. 4C, D). In general, proteostasis genes were induced by Tu to a much greater fold in Rbbp5 Flox mice (such as Creld2, Xbp1, Hsp90b1), while the opposite was found for blood coagulation and immune genes in Rbbp5 LKO mice (such as S100a9 and Fgb) (Figs. 4C, D and S7E, F, H). Further integrating RBBP5 ChIP-seq results, we found that, on average, the ablation of RBBP5 markedly diminished the fold induction by Tu for genes with RBBP5 promoter binding, while it had no effects for those without RBBP5 chromatin recruitment (Fig. 4C, E). To rule out the possibility that RBBP5 regulates UPR via influencing the upstream ER stressing in the ER, we performed western blot analysis of p-IRE1α and p-PERK in the liver of Rbbp5 Flox and Rbbp5 LKO mice and did not observe any notable difference in the increase of their phosphorylated level in response to Tu between the two genotypes (Fig. 4F).
To determine if RBBP5 overexpression is also sufficient to amplify UPR, we tail-vein injected AAV8-TBG-GFP and AAV8-TBG-RBBP5 viruses into wild-type male mice followed by intraperitoneal injection with vehicle control or 0.025mg/kg Tu 30 days later. Thyroxine binding globulin (TBG) promoter is liver-specific and directs RBBP5 transgene expression in hepatocytes (Yan et al, 2012). qPCR and western blot analysis confirmed an increase of total hepatic expression of RBBP5 of ~1.4 fold in AAV8-TBG-RBBP5 group under vehicle control condition (Fig. 4G, H). Consequently, RBBP5 hepatic overexpression significantly amplified the UPR of proteostasis genes including Manf, Hspa5 (BiP), Pdia4, Hyou1, and Syvn1 without affecting the upstream ER stress sensing (Fig. 4G, H). In addition, we found that RBBP5 overexpression attenuated the expression of several immune and blood coagulation genes such as C3, Tifa, Fgl1 and Fgb under ER stress conditions (Fig. 4H). Taken together, these results demonstrate that RBBP5 is both necessary and sufficient for the transcriptional regulation of hepatic response to acute proteotoxic stress in vivo. RBBP5 activates the expression of proteostasis genes while repressing those involved in immune functions.
RBBP5 protects mice from chronic ER stress-induced hepatic inflammation and steatosis.
Contrary to acute ER stress, chronic ER stress, experimentally reconstituted by repeated injection of Tu, paradoxically leads to feedback-mediated suppression of the UPR signaling, to even below basal levels (Rutkowski et al., 2008). Proposed mechanisms of this suppression include silencing of the ATF6a branch of UPR, enhancement of mRNA degradation via IRE1-dependent decay (RIDD), and the direct inhibition of UPR stress sensors by the ER chaperone HSPA5 (BiP) (Rutkowski et al., 2008). To investigate whether RBBP5 also modulates the feedback suppression of UPR under chronic ER stress conditions, we repeatedly intraperitoneally injected male Rbbp5 Flox and Rbbp5 LKO mice with vehicle control or 0.025mg/kg Tu for six consecutive days and harvested liver tissues 24 hours after the last injection for analysis (Fig. 5A). Western blot analysis revealed attenuation of p-IRE1α and ATF4 level in both Rbbp5 Flox and Rbbp5 LKO mice with repeated Tu injection (Fig. 5B). In agreement with the Western blot result, qPCR analysis also confirmed the downregulation of proteostasis genes Atf4, Hyou1, and Manf in both mice under chronic ER stress condition (Fig. 5C). By contrast, compared to Rbbp5 Flox mice, the expression of pro-inflammatory cytokine genes Il1α and Il1b were higher in Tu-injected Rbbp5 LKO mice, consistent with higher levels of p-IRE1α and p-c-Jun (Ser73) observed in Rbbp5 LKO mice (Fig. 5B, C). The persistent elevated levels of p-IRE1α/p-c-JUN and pro-inflammatory cytokine gene expression in Rbbp5 LKO mice suggest that these mice exhibit heightened sensitivity to Tu-induced ER stress and inflammation. In summary, our results indicate that while wild-type mice can sustain persistent cycles of UPR activation and deactivation in response to ER stress, as previously described (Rutkowski et al., 2008), the loss of RBBP5 disrupts this balance. Although RBBP5-deficient mice retain the ability to attenuate UPR, they are unable to properly activate it. Consequently, Rbbp5 LKO mice exhibit consistently lower expression of adaptive proteostasis genes under ER stress condition, leading to heightened proteotoxic stress and chronic inflammation likely via activating the p-IRE1α-TRAF2-AP1 signaling cascade.
Fig. 5. RBBP5 protects mice from chronic ER stress-induced hepatic inflammation and steatosis.
(A) A schematic of experimental design. (B) Western blot and quantification of the expression of different proteins in Rbbp5 Flox and Rbbp5 LKO mice with or without repeated Tu injection. (C) qPCR analysis of different proteostasis and immune gene expressions in the same cohort of mice. (D) Representative images of Oil Red O staining and quantification of lipid droplet size, number and total lipid content (total lipid droplet area). n=4 for Rbbp5 Flox DMSO, n=4 for Rbbp5 Flox Tu, n=2 for Rbbp5 LKO DMSO, n=3 for Rbbp5 LKO Tu. Data: Mean ± S.E.M.
Both chronic ER stress and inflammation can result in hepatic steatosis (Ajoolabady et al, 2023; Gehrke & Schattenberg, 2020; Gong et al, 2024; Guo & Li, 2014). We therefore performed Oil Red O staining, which stained all neutral lipids, on liver tissues from Rbbp5 Flox and Rbbp5 LKO mice after repeated vehicle or Tu injection. Rbbp5 LKO mice liver displayed an increased number of lipid droplets and total lipid content, compared to wild-type counterparts under both basal and chronic ER stress conditions (Fig. 5D). Our data thus revealed that RBBP5 protects mice from chronic ER stress-induced hepatic inflammation and steatosis.
Ablation of RBBP5 in MEFs compromises the transcriptional response to proteotoxic stress and nutrient-deprivation, resulting in impaired autophagy and reduced cell survival.
To determine whether RBBP5 regulates transcriptional responses to proteotoxic stress in a cell-autonomous manner beyond the liver, we examined MEFs, which exhibit both a robust cell-autonomous XBP1s-dependent 12h oscillator and strong transcriptional responses to proteotoxic stress (Dion et al., 2022a; Dion et al, 2024; Pan et al., 2020; Zhu et al., 2017b). Rbbp5 mRNA expression was induced by Tu (Fig. 6A), and transient knockdown of RBBP5 using siRNA (Fig. S8A) significantly dampened the UPR of proteostasis gene expressions in response to Tu, mirroring the effects observed with XBP1s knockdown (Fig.6A). Similarly, knocking down ASH2L also markedly dampened the UPR, whereas knocking down SETD1A, one of the H3K4me3 methyltransferases in the COMPASS complex, had only modest effects—likely due to the redundancy among the many H3K4me3 methyltransferases that exist (Fig. S8B). As a negative control, we further knocked down both GCN5 and PCAF, proteins that function to catalyze H3K9Ac as subunits in two distinct histone acetyltransferase (HAT) complexes, SAGA (Spt-Ada-Gcn5 acetyltransferase) and ATAC (Ada2a-containing) (Gates et al, 2017a; Gates et al, 2017b; Jin et al, 2011) and found it had no effects on the induction of Xbp1 and Manf expression in response to ER stress (Fig. S8C). The lack of effects on UPR for HAT subunits is consistent with the absence of ~12h rhythms of both H3K9Ac and H3K27Ac levels around the promoter of ~12h proteostasis genes in mouse liver (Fig. 1A, B). ChIP-qPCR analysis confirmed a gradual, temporal increase in the recruitment of RBBP5 to the proximal promoters of select proteostasis genes in response to ER stress, preceding the rise in H3K4me3 levels (Fig. 6B).
Fig. 6. RBBP5 is required for transcriptional responses to diverse proteotoxic stresses.
(A) qPCR of different genes in MEFs transfected with scrambled, Rbbp5 or Xbp1 siRNAs and treated with DMSO or Tu (100ng/ml) for 8 hours. (B) Selected genes aligned for RBBP5 and H3K4me3 ChIP-seq signal from CT0 mouse liver and ChIP-qPCR of RBBP5 and H3K4me3 in MEFs treated with Tu for different hours. (C, D) qPCR of different genes in Rbbp5 (fl/fl) ROSA26-CreERT2 (+/+) MEFs treated with vehicle (WT) or tamoxifen (Rbbp5 KO) followed by treatment with vehicle control or 1mM DTT for 5 hours (C) or treatment with full DMEM media, leucine-free media or EBSS for 16 hours (D). (E) Diagram of the mCherry-GFP-LC3 autophagy reporter. (F) Quantification of autophagic flux in mCherry-GFP-LC3-expressing Rbbp5 (fl/fl) ROSA26-CreERT2 (+/+) MEFs treated with vehicle (WT) or tamoxifen (Rbbp5 KO) followed by treatment with full DMEM media or leucine-free media for 16 hours. (G) Diagram of the ER-phagy reporter using the mCherry cleavage from ER assay. (H, I) Western blot of different proteins (H) and quantification (I) of mCherry cleavage in RAMP4-mCherry-expressing Rbbp5 (fl/fl) ROSA26-CreERT2 (+/+) MEFs treated with vehicle (WT) or tamoxifen (Rbbp5 KO) followed by treatment with full DMEM media or EBSS for 16 hours. Data: Mean ± S.E.M.
To broadly assess the function of RBBP5 in the transcription regulation of proteotoxic stress response, we utilized orthogonal approaches to both deplete RBBP5 expression and trigger cellular stress. We took advantage of our homozygous Rbbp5 Flox mice by crossing them with homozygous ROSA26 Cre-ERT2 mice, then isolated and immortalized primary MEFs from homozygous F2 embryos (Fig. S8D). Treating these Rbbp5 (flox/flox); ROSA26 Cre-ERT2 (+/+) MEFs with tamoxifen for 7 days resulted in the complete deletion of both floxed Rbbp5 alleles (Fig. S8E) and a subsequent ~90% reduction in RBBP5 protein level (Fig. S8F). The remaining RBBP5 protein is likely extremely long-lived and remains stable even after the deletion of underlying coding genes. To trigger ER stress, we used a different stimulus: Dithiothreitol (DTT), which disrupts protein disulfide formation in the ER (G & Singh, 2022; Higa et al, 2014). Similar to Tu, RBBP5 depletion significantly dampened the induction of proteostasis gene expression in response to DTT (Fig. 6C).
Finally, we tested two additional proteotoxic stressors that are more physiologically relevant: leucine deprivation to simulate amino acid restriction, and a more severe nutrient deprivation by culturing cells in Earle’s Balanced Salt Solution (EBSS) for 16 hours. Both treatments not only trigger the UPR but also activate the broader integrated stress response (ISR), resulting in the induction of autophagy (Pakos-Zebrucka et al, 2016). RBBP5 depletion impaired the transcriptional response to both leucine deprivation and EBSS, leading to a reduced induction of autophagy-promoting genes (Fig. 6D). To quantify the autophagic flux, we utilized a tandem LC3 reporter mCherry-GFP-LC3 where an increase in the number of red-fluorescent cytosolic puncta indicates increased autolysosome formation and autophagic flux (Figs. 6E and S8G) (Pakos-Zebrucka et al., 2016). Consistent with the qPCR results, RBBP5 depleted MEFs failed to induce a robust general autophagy in response to leucine deprivation, and further exhibited lower basal level of autophagic flux (Fig. 6F). Western blot analysis of p62/SQSTM1, a marker of autophagic flux due to its degradation during autophagy (Bjørkøy et al, 2009), showed a significant reduction in wild-type, but not RBBP5 depleted cells following leucine deprivation, thus confirming the defective autophagy phenotype associated with RBBP5 depletion (Fig. S8H, I). Since prolonged nutrient deprivation, such as 16h of EBSS treatment, can robustly induce both general autophagy and autophagy of the ER (ER-phagy) (Liang et al, 2020), we further measured ER-phagy by utilizing a previously published ER-phagy reporter system where a stably expressed ER-targeting mCherry-RAMP4 fusion is cleaved to free mCherry during starvation-induced ER-phagy (Liang et al, 2018) (Fig. 6G). As shown in Fig. 6H, I, RBBP5 depletion prevented the cleavage of mCherry in response to EBSS, indicating an impaired ER-phagy. Western blot analysis of p62/SQSTM1 further showed undetectable levels in wild-type cells following EBSS treatment, whereas a significant level remained in RBBP5 depleted cells under the same conditions, thus confirming a reduction of the general autophagy in RBBP5 depleted cells in response to EBSS (Fig. 6H). Consistent with weakened stress response and impaired autophagy, RBBP5 depleted MEFs displayed reduced cell survival under nutritional stress (Fig. S8J). In sum, our results demonstrate RBBP5 is indispensable for regulating the transcriptional responses to a plethora of proteotoxic stresses in a cell-autonomous manner.
Reduced RBBP5 expression is associated with aging in mice and hepatic steatosis in humans.
The roles of RBBP5 in diseases are incompletely understood (Cenik & Shilatifard, 2021; Shilatifard, 2012b). Since the decline of proteostasis and stress response is causally associated with both aging and metabolic diseases (Chen et al, 2020; Ghemrawi et al, 2018; Kourtis & Tavernarakis, 2011; Mohan et al, 2019), we aim to uncover potential implications of RBBP5 in these two processes. Intriguingly, in a recent study, RBBP5 (and DPY30) is predicted to be a positive regulator of maximum lifespan across 26 different species, with its major effect predicted to be related to liver (Lu et al, 2022). Consistent with this prediction, an overall reduction of Rbbp5 mRNA level is observed in the liver of aging mice (Almanzar et al, 2020), largely concordant with the expression dynamics of 97 stress response genes (those genes exhibiting 12h expression, directly bound by RBBP5 and can be induced by ER stress) (Fig. 7A).
Fig. 7. Reduced RBBP5 expression is associated with aging in mice and hepatic steatosis in humans.
(A) Z score-normalized expression of mouse Rbbp5 and average expression of stress response genes across mouse life span according to Tabua Muris dataset (Almanzar et al., 2020). (B-F) Data in humans with NAFLD/MAFLD was from (Ahrens et al., 2013). Expression of RBBP5 (B) and the eigengene of 97 stress genes (C). (D) Scatter plot showing a positive correlation between RBBP5 and eigengene expression. (E) Heatmap of 97 stress genes expression in different cohorts of human subjects. (F) GSEA analysis showing stress gene expression are downregulated in steatosis subjects.
Leveraging human gene-derived correlations across tissues (GD-CAT) dataset (Zhou et al, 2024), we identified in human liver, RBBP5 expression is positively correlated with those involved in proteostasis (such as ribosome biogenesis, translation regulation) and mRNA metabolism (mRNA splicing and processing) and negatively associated with those implicated in immune response (acute phase response, complement activity, and immunoglobin activity) (Fig. S9).
These results are in line with our RNA-seq and ChIP-seq data obtained from Rbbp5 Flox and Rbbp5 LKO mice (Figs. 2C, 3I, 4D). In humans with NAFLD/MAFLD (Ahrens et al, 2013), compared to healthy obese, RBBP5 mRNA level is significantly reduced in steatosis subjects (Fig. 7B), concordant with reduced stress gene expression in the same cohorts (Fig. 7C-F). Overall, a positive correlation between Rbbp5 and the eigengene expression of stress response genes [eigengene (Langfelder & Horvath, 2007) is the principle component of 97 stress response genes expression in each subject] is observed across all subjects (Fig. 7D). Collectively, these data suggests that downregulation of Rbbp5 and proteostatic stress response genes expression is associated with aging and MAFLD in mice and humans, respectively.
Discussion:
While the epigenetic landscape of the mammalian ~24h circadian clock was well-studied (Aguilar-Arnal & Sassone-Corsi, 2015; Koike et al., 2012; Masri et al, 2015; Nakahata et al, 2008; Takahashi, 2017), the epigenome of the mammalian ~12h ultradian oscillator remains completely unexplored. In this study, we uncovered that unlike the circadian clock, which was predominantly regulated by histone acetyltransferase (HAT) (Doi et al, 2006; Stashi et al, 2014; Zhu et al, 2015a) and histone deacetylase (HDAC) (Alenghat et al, 2008; Feng et al, 2011; Nakahata et al., 2008), the epigenome of the 12h oscillator is marked by COMPASS-mediated histone methylation, specifically H3K4me3 at the proximal promoters. Loss of one of the essential subunits of COMPASS, RBBP5, abolished more than 90% of the hepatic ~12h transcriptome, while having no effects on the core circadian clock. Knocking down RBBP5 in MEFs further dampened the cell-autonomous 12h oscillations, but not the circadian rhythms. We further expanded our findings to the epigenetic regulation of response to acute and chronic proteotoxic stress and found both increased RBBP5 promoter recruitment and H3K4me3 level are associated with the transcription activation of adaptive stress response genes. Subsequently, depletion of RBBP5 significantly dampened the adaptive transcriptional responses to various proteotoxic stresses.
In our study, we uncovered distinct roles for H3K4me3 and H3K9Ac/H3K27Ac in regulating the 12h oscillator and transcriptional responses to acute proteotoxic stress. While all three histone modifications—H3K4me3, H3K9Ac, and H3K27Ac—are traditionally linked to transcriptional activation (Chen et al, 2022; Gates et al., 2017a; Gates et al., 2017b), our findings suggest that H3K9Ac and H3K27Ac are dispensable for the transcriptional regulation of proteostasis under both basal (the 12h oscillator) and stress conditions. Interestingly, recent research by (Policarpi et al, 2024) demonstrated that among various epigenetic marks associated with transcriptional activation, the installation of H3K4me3 at promoters resulted in the most potent transcriptional activation. This not only underscores the causal role of H3K4me3 in gene activation but also highlights the differential impacts of specific histone modifications on transcription. We speculate that among the many histone modifications, H3K4me3 has been evolutionarily selected as the primary epigenetic modification for stress response due to its ability to trigger the most robust gene activation, thereby providing a rapid adaptive advantage under adverse environmental conditions. Future investigations are needed to further test this hypothesis.
The physiological and pathological roles of mammalian COMPASS subunits, particularly RBBP5, remain incompletely understood (Cenik & Shilatifard, 2021). While extensive research has primarily focused on their contributions to organogenesis, early development, and neurodevelopmental disorders, our study significantly broadens the scope of RBBP5’s physiological roles by uncovering its involvement in proteostasis and hepatic metabolism. This expansion not only advances our understanding of RBBP5’s function but also opens new avenues for exploring its broader impact on cellular and systemic processes.
Of particular interest is a recent study that identified heterozygous loss-of-function mutations in RBBP5 in humans, which were primarily linked to neurodevelopmental disorders (Huang et al). This finding raises intriguing questions about the full spectrum of RBBP5’s physiological roles. Given our discovery of RBBP5’s involvement in metabolic regulation, it is tempting to hypothesize that individuals with RBBP5 loss-of-function mutations may also exhibit metabolic defects. Such a connection would have significant implications, suggesting that RBBP5 plays a multifaceted role in maintaining both neural and metabolic homeostasis. Future research should investigate whether these mutations influence metabolic pathways, potentially contributing to a broader spectrum of clinical symptoms in affected individuals. This line of inquiry could ultimately lead to a deeper understanding of how COMPASS subunits like RBBP5 integrate signals across different tissues to maintain overall physiological balance.
Finally, we propose that the 12h oscillator serves as an effective ‘discovery tool’ for uncovering previously unknown mechanisms of proteostasis regulation. By leveraging this approach, we recently identified a novel XBP1s-SON axis, which links nuclear speckle LLPS dynamics with the UPR (Dion et al., 2022b). Herein, using a similar approach, we identified RBBP5 as an epigenetic regulator of proteostasis. We believe that continued exploration of the 12h oscillator will unveil further hidden principles governing proteostasis.
Methods:
Plasmids
pCDH-EF1a-mCherry-EGFP-LC3B was a gift from Sang-Hun Lee (Addgene plasmid # 170446; http://n2t.net/addgene:170446; RRID: Addgene_170446) (Wulansari et al, 2021). pLenti-X1-hygro-mCherry-RAMP4 was a gift from Jacob Corn (Addgene plasmid # 118391; http://n2t.net/addgene: 118391; RRID:Addgene_118391) (Liang et al., 2018).
The generation of Rbbp5 (flox/flox) mice
The Rbbp5-flox allele was generated by the Mouse Embryo Services Core (University of Pittsburgh, Department of Immunology). The targeting strategy is based on the Easi-CRISPR method (Quadros et al, 2017). In brief, fertilized embryos from C57BL/6J background, produced by natural mating, were microinjected in the pronuclei with a mixture of 0.67 μM EnGen Cas9 protein (New England Biolabs, Cat.No. M0646T), two Cas9 guides RNA: Rbbp5-695rev and Rbbp5-951forw (21.23 ng/μl each) and a long single stranded oligonucleotides Rbbp5-flox-ssODN (5 ng/μl). The long single stranded oligonucleotide encoding the donor sequence was synthesized by Integrated DNA Technologies (IDT).
To produce the sgRNA, a double strand linear DNA template is created by annealing of the following target specific oligonucleotides with a common primer (gRNA-Scaffold-R: 5’-AAAAAAGCACCGACTCGGTGCCACTTTTTCAAGTTGATAACGGACTAGCCTTATTTTAACTTGCTATTTCTAGCTCTAAAAC-3’) containing the full tracrRNA sequence and then PCR amplified as previously described (Pelletier et al, 2015). The sgRNA was synthesized using HiScribe T7 Quick High Yield RNA Synthesis Kit (New England Biolabs, Cat. No. E2050S).
T7-Rbbp5-695rev sequence:
TAATACGACTCACTATAGGAAGCATTCTAAGATTAAACCGTTTTAGAGCTAGAAATAGCA
T7-Rbbp5-951forw sequence:
TAATACGACTCACTATAGGAGTAACAGATAGTATTCCGAGTTTTAGAGCTAGAAATAGCA
Rbbp5-flox-ssODN sequence:
gcagttaataaacttgaagtgtttatatagaaaatacatcaggatggcaaccaaaccctttagatacctgagattttattctcttatcccaggtATAACTTCGTATAGCATACATTATACGAAGTTATgaattcttaatcttagaatgcttagtctgtttcatgctgaagataagttgtcagaactcacattgttactgatttttcgttcagtgactcgtctgttttgtcataggaaaattggagtgcatttgcaccagacttcaaagagttggatgaaaatgtagaatatgaggaaagagaatcagaatttgatattgaggatgaagataagagtgagcctgagcaaacaggtgatgcctctcagtaacagatagtattgaattcATAACTTCGTATAGCATACATTATACGAAGTTATccgaagggaaggactgtcttactcaatgctctttgaattagtgaatgcatttatttactttgtgttgtaactgtgctcaaagcttcatctgccacgtttccctcacttgtacatagttttcacagtgcttgaagagtcttactg
The injected zygotes were cultured overnight, the next day the embryos that developed to the 2-cell stage were transferred to the oviducts of pseudo pregnant CD1 female surrogates. Following optimization of the cycling conditions, the potential founder mice were genotype with the following primers pair, using Q5 High-Fidelity DNA Polymerase (NEW ENGLAND BIOLABS, Cat. no. M0491L). The body weight of male adult mice was measured weekly.
Rbbp5 genotyping forward primer: AGTTCAGAGCTGGTTTCCAAC
Rbbp5 genotyping reverse primer: AGGTAATGCCCATGTGAGCC
Biological rhythm study in mice
All mice used for biological rhythm study are in C57BL/6J background, male and between 3 and 4 months of age. Wild-type C57BL/6J mice (n=12), Rbbp5 (flox/flox); Alb-CRE (−/−) (n=48) and Rbbp5 (flox/flox); Alb-CRE (+/−) mice (n=48) [Rbbp5 (flox/flox); Alb-CRE (−/−) and Rbbp5 (flox/flox); Alb-CRE (+/−) mice were littermates] were first entrained under LD12:12 conditions for 2 weeks before transferred to constant darkness for 24hrs. Mice were then sacrificed via cervical dislocation at a 2h interval [for Rbbp5 (flox/flox); Alb-CRE (−/−) and Rbbp5 (flox/flox); Alb-CRE (+/−) mice] or 4h interval (for wild-type C57BL/6J mice) for a total of 48 hours under constant darkness. Mice were fed ad libitum during the entire experiment. The animal studies were carried out in accordance with the National Institutes of Health guidelines and were granted formal approval by the University of Pittsburgh’s Institutional Animal Care and Use Committee (approval number IS00013119 and IS00013119).
Food intake and locomotor activity monitoring
Promethion Multi-plexed Metabolic Cage System was used for real-time measuring of food intake and locomotor activity. Male Rbbp5 Flox and Rbbp5 LKO mice between 3 and 4 months of age were acclimated to the chambers and ad libitum food intake was monitored for 48 hours under LD12:12 followed by 48 hours of constant darkness. n=6 for Rbbp5 Flox and n=8 for Rbbp5 LKO mice for LD12:12 study. n=5 for Rbbp5 Flox and n=3 for Rbbp5 LKO mice for DD study.
ER stress induction study in mice
For acute ER stress induction experiment, male littermates Rbbp5 (flox/flox); Alb-CRE (−/−) and Rbbp5 (flox/flox); Alb-CRE (+/−) mice between 5 and 7 months of age were randomly divided into two groups, and intraperitoneally injected with 0.05mg/kg body weight of tunicamycin dissolved in 500ul 3% DMSO in PBS or vehicle control (3% DMSO in PBS), respectively. Mice were injected with Tu between 9~10am in the morning and 8 hours later dissected for transcriptomic and western blot analysis for different tissues. The sample size is n=3 for Rbbp5 Flox DMSO, n=3 for Rbbp5 Flox Tu, n=4 for Rbbp5 LKO DMSO, n=4 for Rbbp5 LKO Tu.
For experiments involving RBBP5 liver-overexpressing mice, AAV8-TBG-EGFP and AAV8-TGB-mRBBP5 viruses were customarily designed and produced from Vector Biolab (Malvern, PA). Male wildtype C57BL/6J mice between 2 and 3 months of age were tail-vein injected with 0.5x10^11 genome copies of AAV8-TBG-EGFP or AAV8-TGB-mRBBP5 virus diluted in 100 μl of PBS. 30 days later, mice injected with either viruses were randomly divided into two cohorts and intraperitoneally injected with 0.05mg/kg body weight of tunicamycin dissolved in 500ul 3% DMSO in PBS or vehicle control (3% DMSO in PBS). Mice were injected with Tu between 9~10am in the morning and 8 hours later dissected for transcriptomic and western blot analysis for different tissues. The sample size is n=3 for AAV8-TBG-EGFP DMSO, n=4 for AAV8-TBG-EGFP Tu, n=4 for AAV8-TGB-mRBBP5 DMSO and n=4 for AAV8-TGB-mRBBP5 Tu.
For chronic ER stress induction experiment, male littermates Rbbp5 (flox/flox); Alb-CRE (−/−) and Rbbp5 (flox/flox); Alb-CRE (+/−) mice between 5 and 7 months of age were intraperitoneally injected with 0.025mg/kg body weight of tunicamycin dissolved in 500ul 3% DMSO in PBS or vehicle control (3% DMSO in PBS) daily for six consecutive days. Mice were injected with Tu between 9~10am in the morning and 24 hours after the last injection dissected for transcriptomic, western blot analysis, histology for liver tissues. The sample size is n=4 for Rbbp5 Flox DMSO, n=4 for Rbbp5 Flox Tu, n=2 for Rbbp5 LKO DMSO, n=3 for Rbbp5 LKO Tu.
Oil Red O Staining
Frozen sections were rinsed with PBS and then fixed with 10% neutral buffered formalin for 30 min at RT. After washing twice with deionized water, 60% isopropanol was applied to the fixed cells for 5 min, followed by a freshly prepared working solution of 1.5 mg/ml Oil Red O in isopropanol for 5 min at RT. The stained, fixed tissues were then rinsed with tap water until clear, covered with tap water and viewed on a phase contrast microscope. Size and areas of lipid droplet were quantified by customarily written pipelines in CellProfiler (v4.1.3).
Generation of stable autophagy reporter and ER-phagy reporter cell line
Primary MEFs were isolated from Rbbp5 (flox/flox); Rosa26 CRE (+/+) mice and immortalized with SV40 lentivirus as previously described (Xu, 2005). For mCherry-EGFP-LC3B-expressing autophagy reporter MEFs, lentiviruses packaged in HEK293T cells with co-transfection of pCDH-EF1a-mCherry-EGFP-LC3B, pMD2.G and psPAX2 plasmids were used to infect Rbbp5 (flox/flox); Rosa26 CRE (+/+) MEFs with a multiplicity of infection (MOI) of 3 for three times to achieve near complete infection. For mCherry-RAMP4-expressing ER-phagy reporter MEFs, lentiviruses packaged in HEK293T cells with co-transfection of pLenti-X1-hygro-mCherry-RAMP4, pMD2.G and psPAX2 plasmids were used to infect Rbbp5 (flox/flox); Rosa26 CRE (+/+) MEFs with a multiplicity of infection (MOI) of 3. Stable MEFs were selected in the presence of 200μg/ml hygromycin.
siRNA Transient Transfections
Immortalized MEFs isolated from wild-type C57BL/6J mice were transfected with 10μM of different siRNAs for 24~48 hours with Lipofectamine RNAiMAX reagents (Life technologies) per the manufacturer’s instructions. Source of siRNA are as follows: siGENOME non-targeting siRNA pool (Dharmacon, D-001206-1305), siGENOME SMARTpool mouse Rbbp5 siRNA (Dharmacon, L-054560-01-0005), siGENOME SMARTpool mouse Ash2l siRNA (Dharmacon, L-048754-01-0005), siGENOME SMARTpool mouse Setd1a siRNA (Dharmacon, L-051358-01-0005), siGENOME SMARTpool mouse Kat2a/Gcn5 siRNA (Dharmacon, L-040665-01-0005), siGENOME SMARTpool mouse Kat2b/Pcaf siRNA (Dharmacon, L-049885-01-0005) and siGENOME SMARTpool mouse Xbp1 siRNA (Dharmacon, L-040825-00-0005). MEFs were transfected with 10μM of different siRNAs for 48 hours and treated with DMSO or 100ng/ml of tunicamycin for 8h.
Real-time Luminescence Assay
Stable Manf-dluc MEFs were transfected with non-targeting, Rbbp5, or Ash2l siRNA for 48 hours in DMEM (4.5g/L glucose) supplemented with 10% FBS and treated with 25ng/ml tunicamycin in DMEM for 2h before subjected to real-time luminescence assay using a Lumicycle (Actimetrics) as previously described (Zhu et al, 2015b). Briefly, after tunicamycin treatment, MEFs were washed with 1x PBS and cultured with DMEM (4.5g/L glucose) supplemented with 0.1 mM luciferin and 10mM HEPES buffer in 35 mm tissue culture dishes in the absence of serum and transferred immediately to Lumicycle for real-time luminescence analysis. Periods of oscillation were identified by embedded Periodogram function.
Immunofluorescence (IF)
Immunofluorescence was performed as previously described (Dion et al., 2022a). Briefly, liver OCT sections were fixed in cold acetone for 10 mins at −20 °C. The sections were then air dried, rehydrated with PBS and permeabilized with PBS+ 0.1% Triton X-100. The sections were then blocked with 10% goat serum at room temperature for 1 hour. Primary antibody against RBBP5 (Bethyl, A300-109A) was added to the OCT section at 1:1000 dilution overnight at 4 °C. Next day, sections were washed three times with PBS and stained with Alexa-555 anti-rabbit secondary antibody at room temperature for 2 hours. After that, the sections were washed with PBS three times and counterstained with DAPI before mounting (with ProlongGold Glass) and imaging using Leica SP8 lightening confocal microscope (Leica Microsystems).
Time-lapse microscopy for autophagy reporter MEFs
Time-lapse imaging was performed using SP8 lightening confocal microscope (Leica Microsystems) with Okolab stage top incubator to maintain constant CO2 (5%), temperature (37 °C) and humidity (90%). mCherry-EGFP-LC3B-expressing Rbbp5 (flox/flox); Rosa26 CRE (+/+) MEFs were plated into 8-well chamber slides in full DMEM media and treated with (to deplete RBBP5) or without tamoxifen (3μM/ml) for seven days before replacing with full DMEM or leucine-free media counterstained with DAPI and images were taken every 2 hours for a total of 16 hours. The number of cytosolic autophagosomes (GFP and mCherry double positive foci) and autolysosomes (GFP negative and mCherry positive foci) per cell were quantified manually.
Immunoblot
Nuclear extracts were made from liver according to previously published protocol (Malovannaya et al, 2011). Whole cell lysates were lysed in RIPA buffer as previously described (Dion et al., 2022a). Protein concentrations were determined by Bradford assays (Bio-Rad), and aliquots were snap-frozen in liquid nitrogen and stored at −80°C until usage. Immunoblot analyses were performed as described previously (Zhu et al., 2015a). Briefly, 25~50μg proteins separated by 4~20% gradient SDS-PAGE gels (Biorad) were transferred to nitrocellulose membranes, blocked in TBST buffer supplemented with 5% bovine serum albumin (BSA) or 5% fat-free milk and incubated overnight with primary antibody anti-PERK (Cell signaling, #3192), anti-phospho-PERK (Thr908) (Thermo Fisher, MA5-15033), anti-ATF4 (Cell signaling, #11815), anti-IRE1α (Cell signaling, #3294), anti-phospho-IRE1α (Ser724) (ABclonal, AP0878), anti-XBP1s (Biolegend, 658802), anti-RBBP5 (Bethyl, A300-109A), anti-RBBP5 (D3I6P) (Cell signaling, #13171), anti-WDR5 (Cell signaling, #13105), anti-ASH2L (Bethyl, A300-489A), anti-ASH2L (Bethyl, A300-112A), anti-SETD1A (Diagenode, CS-117-100), anti-SETD1B (Diagenode, CS-118-100), anti-MLL1-c (D6G8N) (Cell signaling, #14197), anti-mCherry (E5D8F) (Cell signaling, #43590), anti-p62/SQSTM1 (Santa Cruz, sc-28359) and anti-β-ACTIN (Cell signaling, #12620) at 4°C. Blots were incubated with an appropriate secondary antibody coupled to horseradish peroxidase at room temperature for 1 hour, and reacted with ECL reagents per the manufacturer’s (Thermo) suggestion and detected by Biorad ChemiDoc MP Imaging System.
Co-Immunoprecipitation (Co-IP)
Nuclear extracts (NE) were made from liver according to our published protocol (Malovannaya et al., 2011). Protein concentrations were determined by Bradford assays (Bio-Rad), and aliquots were snap-frozen in liquid nitrogen and stored at −80°C until usage. 200 μg of NE was used for per IP. 5 μg of different antibodies or control IgG were coupled to 1.5 mg of magnetic Dynabeads (Life Technologies) for every IP using Dynabeads Antibody Coupling Kit (Life Technologies) per manufactures’ protocol. Co-IP was essentially carried out as previously described (Zhu et al, 2015c) except that coupled antibody Dynabeads was added to the NEs for incubation. The antibodies used for Co-IP are as follows: anti-XBP1s (Biolegend, 658802).
qRT-PCR
Total mRNA was isolated from murine embryonic fibroblasts (MEFs) or mouse liver with PureLink RNA mini kit (Life Technologies) with additional on-column DNase digestion step to remove genomic DNA per the manufacturer’s instructions. Reverse transcription was carried out using 5μg of RNA using Superscript III (Life Technologies) per the manufacturer’s instructions. For gene expression analyses, cDNA samples were diluted 1/30-fold (for all other genes except for 18sRNA) and 1/900-fold (for 18sRNA). qPCR was performed using the SYBR green system with sequence-specific primers. All data were analyzed with 18S or β-actin as the endogenous control. qPCR primer sequences are as follows and all primers span introns, except for primers for quantifying pre-mRNAs:
Mouse Rbbp5 forward primer: CAGAGCCTATGCAGAAGCTGCA
Mouse Rbbp5 reverse primer: CTTCACCAGGTTGCCAATGCTC
Mouse Ddit3 forward primer: GGAGGTCCTGTCCTCAGATGAA
Mouse Ddit3 reverse primer: GCTCCTCTGTCAGCCAAGCTAG
Mouse Creld2 forward primer: CTACACCAAGGAGAGTGGACAG
Mouse Creld2 reverse primer: TCTGTCTCCTCAAAGCCTTCCG
Mouse Hspa5 forward primer: TGTCTTCTCAGCATCAAGCAAGG
Mouse Hspa5 reverse primer: CCAACACTTCCTGGACAGGCTT
Mouse Dnajb11 forward primer: TGTGACCGTCTCACTGGTTGAG
Mouse Dnajb11 reverse primer: CCCTTTCTTCCACAGCTTGGCT
Mouse Pdia4 forward primer: TGGGCTCTTTCAGGGAGATGGT
Mouse Pdia4 reverse primer: GGGAGACTTTCAGGAACTTGGC
Mouse Hsp90b1 forward primer: GTTTCCCGTGAGACTCTTCAGC
Mouse Hsp90b1 reverse primer: ATTCGTGCCGAACTCCTTCCAG
Mouse Syvn1 forward primer: CCAACATCTCCTGGCTCTTCCA
Mouse Syvn1 reverse primer: CAGGATGCTGTGATAAGCGTGG
Mouse Gdf15 forward primer: ACTCAGTCCAGAGGTGAGATTG
Mouse Gdf15 reverse primer: GGGGCCTAGTGATGTCCCAG
Mouse Il1a forward primer: ACGGCTGAGTTTCAGTGAGACC
Mouse Il1a reverse primer: CACTCTGGTAGGTGTAAGGTGC
Mouse Il1b forward primer: TGGACCTTCCAGGATGAGGACA
Mouse Il1b reverse primer: GTTCATCTCGGAGCCTGTAGTG
Mouse Fga forward primer: GGATTCTAACTCACTGACCAGGA
Mouse Fga reverse primer: CCTCAGGATCTCAATTCTGCGC
Mouse C3 forward primer: CGCAACGAACAGGTGGAGATCA
Mouse C3 reverse primer: CTGGAAGTAGCGATTCTTGGCG
Mouse Tifa forward primer: AGCAAGAAAACCAGTTTGATGGTAG
Mouse Tifa reverse primer: GCAACAGGAACTGATACTCTCCG
Mouse Fgl1 forward primer: GGAAACTGTGCTGAGGAAGAGC
Mouse Fgl1 reverse primer: TCCGTTTCTGCCCTGTAGGAAC
Mouse Fgb forward primer: TGACACCTCCATCAAGCCGTAC
Mouse Fgb reverse primer: GGTCCCATTTCCTGCCAAAGTC
Mouse Il6 forward primer: CTTCCATCCAGTTGCCTTCT
Mouse Il6 reverse primer: CTCCGACTTGTGAAGTGGTATAG
Mouse Herpud1 forward primer: acctgagccgagtctaccc
Mouse Herpud1 reverse primer: aacagcagcttcccagaataaa
Mouse Nbr1 forward primer: GGATTTAAAGCACCTCCTGATTCC
Mouse Nbr1 reverse primer: ATTGGGTCCCACTCAGGTCT
Mouse Sesn2 forward primer: CTGGCCGAACTCATCCAAG
Mouse Sesn2 reverse primer: CTGCCTCATGCGTTCCATC
Mouse Sqstm1 forward primer: AACAGATGGAGTCGGGAAAC
Mouse Sqstm1 reverse primer: AGACTGGAGTTCACCTGTAGA
Mouse Stbd1 forward primer: CTGGGAAGTAGATGGGAAAGTG
Mouse Stbd1 reverse primer: TCGGTGTTTGTGGTGTAGTG
Mouse Sirt1 forward primer: TGACAGAACGTCACACGCCA
Mouse Sirt1 reverse primer: ATTGTTCGAGGATCGGTGCCA
Mouse Gcn5 forward primer: TTGATTGAGCGCAAACAGGC
Mouse Gcn5 reverse primer: CAGCCTGTCTCTCGAATGCC
Mouse Pcaf forward primer: CGTGAAGAAGGCGCAGTTG
Mouse Pcaf reverse primer: CAGGACTCCTCTGCCTTGC
Mouse Ash2l forward primer: gctgtgtctgctagtgggaac
Mouse Ash2l reverse primer: catcttgctgcttacgcttg
Mouse Setd1a forward primer: gggtagcaccccctactctc
Mouse Setd1a reverse primer: gggtttgaaggaggttgaagt
Mouse Eif2ak3 forward primer: ccttggtttcatctagcctca
Mouse Eif3ak3 reverse primer: atccagggaggggatgat
Mouse total Xbp1 forward primer: gggtctgctgagtcc
Mouse total Xbp1 reverse primer: cagactcagaatctgaagagg
Mouse Sec23b forward primer: tgaccaaactggacttctgga
Mouse Sec23b reverse primer: aaagaatctcccatcaccatgt
Mouse Manf forward primer: gacagccagatctgtgaactaaaa
Mouse Manf reverse primer: tttcacccggagcttcttc
Mouse Hyou1 forward primer: GAGGCGAAACCCATTTTAGA
Mouse Hyou1 reverse primer: GCTCTTCCTGTTCAGGTCCA
Mouse Atf4 forward primer: CCACTCCAGAGCATTCCTTTAG
Mouse Atf4 reverse primer: CTCCTTTACACATGGAGGGATTAG
Mouse Atf6 forward primer: CATGAAGTGGAAAGGACCAAATC
Mouse Atf6 reverse primer: CAGCCATCAGCTGAGAATTAGA
Mouse 18s RNA forward primer: ctcaacacgggaaacctcac
Mouse 18s RNA reverse primer: cgctccaccaactaagaacg
Mouse β-actin forward primer: aaggccaaccgtgaaaagat
Mouse β-actin reverse primer: gtggtacgaccagaggcatac
RNA-Seq to identify RBBP5-dependent 12h hepatic transcriptome
Mouse liver tissues were collected from Rbbp5 Flox (n=2) and Rbbp5 LKO (n=2) mice at 2h intervals for a total of 48 hours under constant darkness condition. Total RNA was isolated from mouse liver with miRNeasy Mini Kit (Qiagen) per the manufacturer’s instructions. RNA was assessed for quality using an Agilent TapeStation 4150/Fragment Analyzer 5300 and RNA concentration was quantified on a Qubit FLEX fluorometer. Libraries were generated with the Illumina Stranded mRNA Library Prep kit (Illumina: 20040534) according to the manufacturer’s instructions. Briefly, 200 ng of input RNA was used for each sample. Following adapter ligation, 12 cycles of indexing PCR were completed, using IDT for Illumina RNA UD Indexes (Illumina: 20040553-6). Library quantification and assessment was done using a Qubit FLEX fluorometer and an Agilent TapeStation 4150/Fragment Analyzer 5300. The prepared libraries were pooled and sequenced using NoveSeq 6000 (Illumina), generating an average of 40 million 2×100 bp paired end reads per sample. RNA-Seq library preparation and sequencing were performed at UPMC Genome Center. Raw RNA-seq FASTQ files were analyzed by FastQC for quality control. Adaptors and low-quality reads were filtered by Trimmomatic (Bolger et al, 2014b). Then the processed reads were aligned by HISAT2 (Kim et al, 2015) against mouse reference mm10. Gene and isoform FKPM values were calculated by Cufflinks. Since the background gene expression level is FPKM=0.1 in mouse liver (Pan et al., 2020), only those genes with average gene expression across 48 hours in Rbbp5 Flox mice larger than 0.1 were included for rhythm-identification analysis. Averaged FPKM values at each time were used for cycling transcripts identification by the eigenvalue/pencil method. Raw temporal data was subject to polynomial detrend (n=2) first, and superimposed oscillations were identified using previously described eigenvalue/pencil method on the detrended dataset (Antoulas et al., 2018a; Zhu et al., 2017a). Specifically, three oscillations were identified from each gene. Criterion for circadian gene is period between 21h to 27h, decay rate between 0.9 and 1.1; for ~12hr genes: period between 10h to 13h, decay rate between 0.9 and 1.1. The relative amplitude is calculated via dividing the amplitude by the mean expression value of each gene. All analysis were performed in MatlabR2017A. Heat maps were generated by Gene Cluster 3.0 and TreeView 3.0 alpha 3.0 using log2 mean-normalized values. RAIN analysis was performed as previously described in Bioconductor (3.4) (http://www.bioconductor.org/packages/release/bioc/html/rain.html) (Thaben & Westermark, 2014) using the full detrended dataset with duplicated at each time point.
For the eigenvalue method, every gene consists of multiple superimposed oscillations. Therefore, we define a circadian gene as any gene that exhibits a superimposed circadian rhythm, regardless of its relative amplitude compared to other superimposed oscillations. Similar definitions apply to 12hr genes. Under this definition, a gene can be both a circadian and 12hr-cycling gene. By comparison, we define a dominant circadian gene as any gene whose superimposed circadian rhythm has the largest amplitude among all oscillations. Similar definitions also apply to 12hr genes. Under this definition, dominant circadian and dominant 12hr genes are mutually exclusive.
RNA-seq for identifying RBBP5-dependent acute transcriptional response to ER stress
Mouse liver tissues were collected from Rbbp5 Flox and Rbbp5 LKO mice intraperitoneally treated with or without Tu for 8 hours. Total mRNA was isolated from mouse liver with PureLink RNA mini kit (Life Technologies) with additional on-column DNase digestion step to remove genomic DNA per the manufacturer’s instructions. Detailed procedures for transcriptome sequencing were described previously(Liu et al, 2024) and were supported by the Genomics and Systems Biology Core of the Pittsburgh Liver Research Center. Briefly, the mRNA samples per mouse were processed into short-read libraries using the Bio-Rad SEQuoia Dual Indexed Primers. Next, the circularization was performed using the Element Biosciences Adept Library Compatibility Kit v1.1 based on the manufacturer’s instructions, followed by the quantification using qPCR with the provided standard. Finally, the libraries were measured by the Element Biosciences AVITI system to sequence paired end reads with 75 bp each. The raw FASTQ files from RNA-seq were trimmed by Trimmomatic (Bolger et al, 2014a) to filter out low-quality reads. The surviving reads were then aligned to the mouse mm10 reference genome by STAR aligner(Dobin et al, 2013). Fragments Per Kilobase of transcript per Million mapped reads (FPKM) values per library were quantified by the tool Cufflinks(Trapnell et al, 2010). Log 2 normalized fold induction and adjusted p values between Rbbp5 Flox DMSO and Rbbp5 LKO DMSO, Rbbp5 Flox Tu and Rbbp5 LKO Tu, Rbbp5 Flox DMSO and Rbbp5 Flox Tu, and Rbbp5 LKO DMSO and Rbbp5 LKO Tu were calculated by using the iDEP (integrated Differential Expression and Pathway analysis) online application (Ge et al, 2018).
Chromatin Immunoprecipitation (ChIP)-Seq and ChIP-qPCR
ChIP for RBBP5 was performed using anti-RBBP5 antibody (Bethyl, A300-109A) as previously described (Pan et al., 2020). Briefly, mouse liver samples were submerged in PBS + 1% formaldehyde, cut into small (~1 mm3) pieces with a razor blade and incubated at room temperature for 15 minutes. Fixation was stopped by the addition of 0.125 M glycine (final concentration). The tissue pieces were then treated with a TissueTearer and finally spun down and washed twice in PBS. Chromatin was isolated by the addition of lysis buffer, followed by disruption with a Dounce homogenizer. The chromatin was enzymatically digested with MNase. Genomic DNA (Input) was prepared by treating aliquots of chromatin with RNase, Proteinase K and heated for reverse-crosslinking, followed by ethanol precipitation. Pellets were resuspended and the resulting DNA was quantified on a NanoDrop spectrophotometer. An aliquot of chromatin (30 μg) was precleared with protein A agarose beads (Invitrogen). Genomic DNA regions of interest were isolated using 4 μg of antibody. Complexes were washed, eluted from the beads with SDS buffer, and subjected to RNase and proteinase K treatment. Crosslinking was reversed by incubation overnight at 65 °C, and ChIP DNA was purified by phenol-chloroform extraction and ethanol precipitation. The DNA libraries were prepared at the University of Pittsburgh and sequenced at Gene by Gene, Ltd per standard protocols. DNA libraries were prepared with Ovation® Ultralow V2 DNA-Seq library preparation kit (NuGen) using 1ng input DNA. The size selection for libraries was performed using SPRIselect beads (Beckman Coulter) and purity of the libraries were analyzed using the High Sensitivity DNA chip on Bioanalyzer 2100 (Agilent). The prepared libraries pooled and sequenced using Nova-Seq 6000 (Illumina), generating ~30 million 75 bp single-end reads per sample. ChIP-qPCR for MEFs were essentially performed the same way as previously described with anti-RBBP5 (Bethyl, A300-109A) and anti-H3K4me3 (Active Motif, 61379) antibodies, except that the MEFs were directly fixed with 1% formaldehyde before subject to nuclei isolation and chromatin immunoprecipitation. The primers used for ChIP-qPCR are as follows:
Negative control region 1 forward primer: GCAACAACAACAGCAACAATAAC
Negative control region 1 reverse primer: CATGGCACCTAGAGTTGGATAA
Manf promoter forward primer: CCCTTAAATGGGTCAACGTCTC
Manf promoter reverse primer: GGCGCTAAACCCAAGGAAA
Xbp1 promoter forward primer: TCCGTACGGTGGGTTAGAT
Xbp1 promoter reverse primer: ACCTTCTTCTGTGCCTGTG
Hyou1 promoter forward primer: CCGTGTGGGTACGTCCT
Hyou1 promoter reverse primer: GACGGCTGCTCCATCCT
Sesn2 promoter forward primer: ACAGGAGGCCGGGACTA
Sesn2 promoter reverse primer: CTGGGCTGAAAGGAGTGTCTAT
ChIP-Seq analysis
The sequences identified were mapped to the mouse genome (UCSC mm10) using BOWTIE function in Galaxy Deeptool (https://usegalaxy.org/) (Ramirez et al, 2016). Only the sequences uniquely mapped with no more than 2 mismatches were kept and used as valid reads. PCR duplicates were also removed. Peak calling was carried out by MACS2 (version 2.1.1.20160309) in Galaxy (options --mfold 5, 50 --pvalue 1e-4), on each ChIP-seq file against input. To account for the different sequencing depths between samples, the BAM files generated from MACS2 were RPKM normalized to sequencing depth using the bamCoverage function in Galaxy Deeptool and the bigwig files were generated accordingly. The relative intensity of each RBBP5 binding site is further calculated via the computeMatrix function with the RPKM normalized bigwig files and bed files from the peak calling as inputs by calculating the area under the curve.
Gene ontology analysis
DAVID (Version 2021) (Huang da et al, 2009) (https://david.ncifcrf.gov) was used to perform Gene Ontology analyses. Briefly, gene names were first converted to DAVID-recognizable IDs using Gene Accession Conversion Tool. The updated gene list was then subject to GO analysis using all Homo Sapiens as background and with Functional Annotation Chart function. GO_BP_DIRECT, KEGG_PATHWAY or UP_KW_BIOLOGICAL_PROCESS was used as GO categories. Only GO terms with a p value less than 0.05 were included for further analysis.
Motif analysis
Motif analysis was performed with the SeqPos motif tool (version 0.590) (He et al, 2010) embedded in Galaxy Cistrome using all motifs within the mouse reference genome mm10 as background. LISA analysis was performed using webtool (http://lisa.cistrome.org/).
Statistical Analysis
Data were analyzed and presented with GraphPad Prism software. Plots show individual data points and bars at the mean and ± the standard error of the mean (SEM). One-tailed t-tests were used to compare means between groups, with significance set at p < 0.05. In instances where the p value is not shown, *, **, ***, and **** represent p < 0.05, 0.01, 0.001, and 0.0001, respectively.
Supplementary Material
Acknowledgements:
We would like to thank Dr. Sebastien Gingras (University of Pittsburgh, Department of Immunology) for the design of the targeting strategy and identification of the founders carrying the properly targeted Rbbp5-flox allele. We would like to thank Chunming Bi and Zhaohui Kou of the Mouse Embryo Services Core (University of Pittsburgh, Department of Immunology) for microinjection of zygotes and production of Rbbp5-flox founder mice. We would also like to thank Dr. Heather Ballance, Dr. Naveen GV Kumar, Saad Irfan, C. Dupont, Dr. Bugra Zengin and Casey Edwards for assistance with mice colony maintenance, dissection, intraperitoneal injection and genotyping. For time course transcriptome study, the library generation was performed in the Health Sciences Sequencing Core at UPMC Children’s Hospital of Pittsburgh, Rangos Research Center. Services and instruments used in this project were graciously supported, in part, by the University of Pittsburgh, the Office of the Senior Vice Chancellor for Health Sciences, the Department of Pediatrics, the Institute for Precision Medicine, and the Richard K Mellon Foundation for Pediatric Research.
W.D. was supported by fellowship F31 AG080998 through NIA. B. Zhu was supported by grants 1DP2GM140924 and 1R21AG071893 through the NIH, and a grant from Richard King Mellon foundation. This research was supported in part by the University of Pittsburgh Center for Research Computing through the resources provided. Specifically, this work used the HTC cluster, which is supported by NIH award number S10OD028483. This research project was supported in part by the Pittsburgh Liver Research Centre supported by NIH/NIDDK Digestive Disease Research Core Center grant P30DK120531.
Footnotes
Disclosure and competing interests:
The authors have no competing interests to declare.
Data availability:
All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession numbers: GSE276155, GSE276157 and GSE276159. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
References:
- Aguilar-Arnal L, Sassone-Corsi P (2015) Chromatin landscape and circadian dynamics: Spatial and temporal organization of clock transcription. Proceedings of the National Academy of Sciences 112: 6863–6870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahrens M, Ammerpohl O, von Schönfels W, Kolarova J, Bens S, Itzel T, Teufel A, Herrmann A, Brosch M, Hinrichsen H et al. (2013) DNA Methylation Analysis in Nonalcoholic Fatty Liver Disease Suggests Distinct Disease-Specific and Remodeling Signatures after Bariatric Surgery. Cell Metabolism 18: 296–302 [DOI] [PubMed] [Google Scholar]
- Ajoolabady A, Kaplowitz N, Lebeaupin C, Kroemer G, Kaufman RJ, Malhi H, Ren J (2023) Endoplasmic reticulum stress in liver diseases. Hepatology 77: 619–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alenghat T, Meyers K, Mullican SE, Leitner K, Adeniji-Adele A, Avila J, Bućan M, Ahima RS, Kaestner KH, Lazar MA (2008) Nuclear receptor corepressor and histone deacetylase 3 govern circadian metabolic physiology. Nature 456: 997–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almanzar N, Antony J, Baghel AS, Bakerman I, Bansal I, Barres BA, Beachy PA, Berdnik D, Bilen B, Brownfield D et al. (2020) A single-cell transcriptomic atlas characterizes ageing tissues in the mouse. Nature 583: 590–595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoulas AC, Zhu B, Zhang Q, York B, O’Malley BW, Dacso CC (2018a) A novel mathematical method for disclosing oscillations in gene transcription: A comparative study. PLoS One 13: e0198503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoulas AC, Zhu B, Zhang Q, York B, O’Malley BW, Dacso CC (2018b) A novel mathematical method for disclosing oscillations in gene transcription: a comparative study. PLoS One 13: e0198503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher G, Zhu B (2022a) Beyond circadian rhythms: Emerging roles of ultradian rhythms in control of liver functions. Hepatology [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asher G, Zhu B (2022b) Beyond circadian rhythms: Emerging roles of ultradian rhythms in control of liver functions. Hepatology n/a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balch WE, Morimoto RI, Dillin A, Kelly JW (2008) Adapting proteostasis for disease intervention. Science 319: 916–919 [DOI] [PubMed] [Google Scholar]
- Ballance H, Zhu B (2021) Revealing the hidden reality of the mammalian 12-h ultradian rhythms. Cellular and Molecular Life Sciences [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjørkøy G, Lamark T, Pankiv S, Øvervatn A, Brech A, Johansen T (2009) Monitoring autophagic degradation of p62/SQSTM1. Methods Enzymol 452: 181–197 [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B (2014a) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B (2014b) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttgereit F, Brand MD (1995) A hierarchy of ATP-consuming processes in mammalian cells. Biochemical Journal 312: 163–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenik BK, Shilatifard A (2021) COMPASS and SWI/SNF complexes in development and disease. Nature Reviews Genetics 22: 38–58 [DOI] [PubMed] [Google Scholar]
- Chen K, Shen W, Zhang Z, Xiong F, Ouyang Q, Luo C (2020) Age-dependent decline in stress response capacity revealed by proteins dynamics analysis. Scientific Reports 10: 15211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YC, Koutelou E, Dent SYR (2022) Now open: Evolving insights to the roles of lysine acetylation in chromatin organization and function. Mol Cell 82: 716–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-Mattioli M, Walter P (2020) The integrated stress response: From mechanism to disease. Science 368: eaat5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta S, Lonard DM, O’Malley BW (2014) Nuclear receptor coactivators: master regulators of human health and disease. Annu Rev Med 65: 279–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion W, Ballance H, Lee J, Pan Y, Irfan S, Edwards C, Sun M, Zhang J, Zhang X, Liu S (2022a) Four-dimensional nuclear speckle phase separation dynamics regulate proteostasis. Science Advances 8: eabl4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion W, Ballance H, Lee J, Pan Y, Irfan S, Edwards C, Sun M, Zhang J, Zhang X, Liu S et al. (2022b) Four-dimensional nuclear speckle phase separation dynamics regulate proteostasis. Science Advances 8: eabl4150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion W, Tao Y, Chambers M, Zhao S, Arbuckle RK, Sun M, Kubra S, Nie Y, Ye M, Larsen MB et al. (2024) Nuclear speckle rejuvenation alleviates proteinopathies at the expense of YAP1. bioRxiv: 2024.2004.2018.590103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR (2013) STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29: 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doi M, Hirayama J, Sassone-Corsi P (2006) Circadian Regulator CLOCK Is a Histone Acetyltransferase. Cell 125: 497–508 [DOI] [PubMed] [Google Scholar]
- Fang B, Everett LJ, Jager J, Briggs E, Armour SM, Feng D, Roy A, Gerhart-Hines Z, Sun Z, Lazar MA (2014) Circadian enhancers coordinate multiple phases of rhythmic gene transcription in vivo. Cell 159: 1140–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng D, Liu T, Sun Z, Bugge A, Mullican SE, Alenghat T, Liu XS, Lazar MA (2011) A circadian rhythm orchestrated by histone deacetylase 3 controls hepatic lipid metabolism. Science 331: 1315–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- G G, Singh J (2022) Dithiothreitol causes toxicity in C. elegans by modulating the methionine–homocysteine cycle. eLife 11: e76021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gates LA, Foulds CE, O’Malley BW (2017a) Histone Marks in the ‘Driver’s Seat’: Functional Roles in Steering the Transcription Cycle. Trends Biochem Sci 42: 977–989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gates LA, Shi J, Rohira AD, Feng Q, Zhu B, Bedford MT, Sagum CA, Jung SY, Qin J, Tsai MJ et al. (2017b) Acetylation on histone H3 lysine 9 mediates a switch from transcription initiation to elongation. J Biol Chem 292: 14456–14472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge SX, Son EW, Yao R (2018) iDEP: an integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC bioinformatics 19: 1–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrke N, Schattenberg JM (2020) Metabolic Inflammation—A Role for Hepatic Inflammatory Pathways as Drivers of Comorbidities in Nonalcoholic Fatty Liver Disease? Gastroenterology 158: 1929–1947.e1926 [DOI] [PubMed] [Google Scholar]
- Ghemrawi R, Battaglia-Hsu SF, Arnold C (2018) Endoplasmic Reticulum Stress in Metabolic Disorders. Cells 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong H, He Q, Zhu L, Feng Z, Sun M, Jiang J, Yuan X, Shen Y, Di J (2024) Associations between systemic inflammation indicators and nonalcoholic fatty liver disease: evidence from a prospective study. Frontiers in Immunology 15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo B, Li Z (2014) Endoplasmic reticulum stress in hepatic steatosis and inflammatory bowel diseases. Frontiers in Genetics 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han J, Li T, Li Y, Li M, Wang X, Peng C, Su C, Li N, Li Y, Xu Y et al. (2019) The internal interaction in RBBP5 regulates assembly and activity of MLL1 methyltransferase complex. Nucleic Acids Research 47: 10426–10438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He HH, Meyer CA, Shin H, Bailey ST, Wei G, Wang Q, Zhang Y, Xu K, Ni M, Lupien M et al. (2010) Nucleosome dynamics define transcriptional enhancers. Nature Genetics 42: 343–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetz C, Zhang K, Kaufman RJ (2020) Mechanisms, regulation and functions of the unfolded protein response. Nature Reviews Molecular Cell Biology 21: 421–438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higa A, Taouji S, Lhomond S, Jensen D, Fernandez-Zapico ME, Simpson JC, Pasquet JM, Schekman R, Chevet E (2014) Endoplasmic reticulum stress-activated transcription factor ATF6α requires the disulfide isomerase PDIA5 to modulate chemoresistance. Mol Cell Biol 34: 1839–1849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Song A, Peng L, Tang N, Qiao Z, Wang Z, Lan F, Chen FX (2023) H3K4me2/3 modulate the stability of RNA polymerase II pausing. Cell Research 33: 403–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Huang Y, Jay KL, Huang AY-W, Wan J, Jangam SV, Chorin O, Rothschild A, Barel O, Mariani M, Iascone M et al. Loss-of-function in <em>RBBP5</em> results in a syndromic neurodevelopmental disorder associated with microcephaly. Genetics in Medicine [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes ME, DiTacchio L, Hayes KR, Vollmers C, Pulivarthy S, Baggs JE, Panda S, Hogenesch JB (2009) Harmonics of circadian gene transcription in mammals. PLoS Genet 5: e1000442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Q, Yu LR, Wang L, Zhang Z, Kasper LH, Lee JE, Wang C, Brindle PK, Dent SY, Ge K (2011) Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. Embo j 30: 249–262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12: 357–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaips CL, Jayaraj GG, Hartl FU (2018) Pathways of cellular proteostasis in aging and disease. J Cell Biol 217: 51–63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike N, Yoo SH, Huang HC, Kumar V, Lee C, Kim TK, Takahashi JS (2012) Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 338: 349–354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kourtis N, Tavernarakis N (2011) Cellular stress response pathways and ageing: intricate molecular relationships. The EMBO journal 30: 2520–2531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbadia J, Morimoto RI (2015) The Biology of Proteostasis in Aging and Disease. Annual Review of Biochemistry 84: 435–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S (2007) Eigengene networks for studying the relationships between co-expression modules. BMC Systems Biology 1: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Han J, Zhang Y, Cao F, Liu Z, Li S, Wu J, Hu C, Wang Y, Shuai J et al. (2016) Structural basis for activity regulation of MLL family methyltransferases. Nature 530: 447–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang JR, Lingeman E, Ahmed S, Corn JE (2018) Atlastins remodel the endoplasmic reticulum for selective autophagy. J Cell Biol 217: 3354–3367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang JR, Lingeman E, Luong T, Ahmed S, Muhar M, Nguyen T, Olzmann JA, Corn JE (2020) A Genome-wide ER-phagy Screen Highlights Key Roles of Mitochondrial Metabolism and ER-Resident UFMylation. Cell 180: 1160–1177.e1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Obert C, Yu YP, Zhao J, Ren BG, Liu JJ, Wiseman K, Krajacich BJ, Wang W, Metcalfe K et al. (2024) Utility analyses of AVITI sequencing chemistry. BMC Genomics 25: 778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu JY, Simon M, Zhao Y, Ablaeva J, Corson N, Choi Y, Yamada KYH, Schork NJ, Hood WR, Hill GE et al. (2022) Comparative transcriptomics reveals circadian and pluripotency networks as two pillars of longevity regulation. Cell Metabolism 34: 836–856.e835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malovannaya A, Lanz RB, Jung SY, Bulynko Y, Le NT, Chan DW, Ding C, Shi Y, Yucer N, Krenciute G et al. (2011) Analysis of the human endogenous coregulator complexome. Cell 145: 787–799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez G, Duran-Aniotz C, Cabral-Miranda F, Vivar JP, Hetz C (2017) Endoplasmic reticulum proteostasis impairment in aging. Aging Cell 16: 615–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masri S, Kinouchi K, Sassone-Corsi P (2015) Circadian clocks, epigenetics, and cancer. Curr Opin Oncol 27: 50–56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng H, Gonzales NM, Jung SY, Lu Y, Putluri N, Zhu B, Dacso CC, Lonard DM, O’Malley BW (2022) Defining the mammalian coactivation of hepatic 12-h clock and lipid metabolism. Cell Reports 38: 110491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng H, Gonzales NM, Lonard DM, Putluri N, Zhu B, Dacso CC, York B, O’Malley BW (2020) XBP1 links the 12-hour clock to NAFLD and regulation of membrane fluidity and lipid homeostasis. Nature Communications 11: 6215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalf MG, Higuchi-Sanabria R, Garcia G, Tsui CK, Dillin A (2020) Beyond the cell factory: Homeostatic regulation of and by the UPR<sup>ER</>. Science Advances 6: eabb9614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan S, R PRM, Brown L, Ayyappan P, G RK (2019) Endoplasmic reticulum stress: A master regulator of metabolic syndrome. European Journal of Pharmacology 860: 172553. [DOI] [PubMed] [Google Scholar]
- Nakahata Y, Kaluzova M, Grimaldi B, Sahar S, Hirayama J, Chen D, Guarente LP, Sassone-Corsi P (2008) The NAD<sup>+</sup>-Dependent Deacetylase SIRT1 Modulates CLOCK-Mediated Chromatin Remodeling and Circadian Control. Cell 134: 329–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pakos-Zebrucka K, Koryga I, Mnich K, Ljujic M, Samali A, Gorman AM (2016) The integrated stress response. EMBO Rep 17: 1374–1395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Ballance H, Meng H, Gonzalez N, Kim S-M, Abdurehman L, York B, Chen X, Schnytzer Y, Levy O et al. (2020) 12-h clock regulation of genetic information flow by XBP1s. PLOS Biology 18: e3000580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelletier S, Gingras S, Green DR (2015) Mouse genome engineering via CRISPR-Cas9 for study of immune function. Immunity 42: 18–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Policarpi C, Munafò M, Tsagkris S, Carlini V, Hackett JA (2024) Systematic epigenome editing captures the context-dependent instructive function of chromatin modifications. Nature Genetics 56: 1168–1180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Q, Fan J, Zheng R, Wan C, Mei S, Wu Q, Sun H, Brown M, Zhang J, Meyer CA et al. (2020) Lisa: inferring transcriptional regulators through integrative modeling of public chromatin accessibility and ChIP-seq data. Genome Biology 21: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu Q, Takahashi YH, Yang Y, Hu H, Zhang Y, Brunzelle JS, Couture JF, Shilatifard A, Skiniotis G (2018) Structure and Conformational Dynamics of a COMPASS Histone H3K4 Methyltransferase Complex. Cell 174: 1117–1126.e1112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadros RM, Miura H, Harms DW, Akatsuka H, Sato T, Aida T, Redder R, Richardson GP, Inagaki Y, Sakai D et al. (2017) Easi-CRISPR: a robust method for one-step generation of mice carrying conditional and insertion alleles using long ssDNA donors and CRISPR ribonucleoproteins. Genome Biol 18: 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez F, Ryan DP, Gruning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dundar F, Manke T (2016) deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44: W160–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter K, Haslbeck M, Buchner J (2010) The heat shock response: life on the verge of death. Mol Cell 40: 253–266 [DOI] [PubMed] [Google Scholar]
- Roeder RG (2005) Transcriptional regulation and the role of diverse coactivators in animal cells. FEBS Lett 579: 909–915 [DOI] [PubMed] [Google Scholar]
- Ron D, Walter P (2007) Signal integration in the endoplasmic reticulum unfolded protein response. Nature Reviews Molecular Cell Biology 8: 519–529 [DOI] [PubMed] [Google Scholar]
- Ruthenburg AJ, Allis CD, Wysocka J (2007) Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell 25: 15–30 [DOI] [PubMed] [Google Scholar]
- Rutkowski DT, Wu J, Back SH, Callaghan MU, Ferris SP, Iqbal J, Clark R, Miao H, Hassler JR, Fornek J et al. (2008) UPR pathways combine to prevent hepatic steatosis caused by ER stress-mediated suppression of transcriptional master regulators. Dev Cell 15: 829–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott MR, Zong W, Ketchesin KD, Seney ML, Tseng GC, Zhu B, McClung CA (2023) Twelve-hour rhythms in transcript expression within the human dorsolateral prefrontal cortex are altered in schizophrenia. PLOS Biology 21: e3001688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A (2012a) The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annu Rev Biochem 81: 65–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilatifard A (2012b) The COMPASS family of histone H3K4 methylases: mechanisms of regulation in development and disease pathogenesis. Annual review of biochemistry 81: 65–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- So JS (2018) Roles of Endoplasmic Reticulum Stress in Immune Responses. Mol Cells 41: 705–716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegelman BM, Heinrich R (2004) Biological control through regulated transcriptional coactivators. Cell 119: 157–167 [DOI] [PubMed] [Google Scholar]
- Stashi E, Lanz RB, Mao J, Michailidis G, Zhu B, Kettner NM, Putluri N, Reineke EL, Reineke LC, Dasgupta S (2014) SRC-2 is an essential coactivator for orchestrating metabolism and circadian rhythm. Cell reports 6: 633–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi JS (2017) Transcriptional architecture of the mammalian circadian clock. Nat Rev Genet 18: 164–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi YH, Westfield GH, Oleskie AN, Trievel RC, Shilatifard A, Skiniotis G (2011) Structural analysis of the core COMPASS family of histone H3K4 methylases from yeast to human. Proc Natl Acad Sci U S A 108: 20526–20531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RC, Hetz C (2020) Mastering organismal aging through the endoplasmic reticulum proteostasis network. Aging Cell 19: e13265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaben PF, Westermark PO (2014) Detecting rhythms in time series with RAIN. J Biol Rhythms 29: 391–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnology 28: 511–U174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Veen DR, Gerkema MP (2017) Unmasking ultradian rhythms in gene expression. The FASEB Journal 31: 743–750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Ron D (2011) The unfolded protein response: from stress pathway to homeostatic regulation. Science 334: 1081–1086 [DOI] [PubMed] [Google Scholar]
- Wang H, Fan Z, Shliaha PV, Miele M, Hendrickson RC, Jiang X, Helin K (2023) H3K4me3 regulates RNA polymerase II promoter-proximal pause-release. Nature 615: 339–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiseman RL, Mesgarzadeh JS, Hendershot LM (2022) Reshaping endoplasmic reticulum quality control through the unfolded protein response. Molecular Cell 82: 1477–1491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wulansari N, Darsono WHW, Woo HJ, Chang MY, Kim J, Bae EJ, Sun W, Lee JH, Cho IJ, Shin H et al. (2021) Neurodevelopmental defects and neurodegenerative phenotypes in human brain organoids carrying Parkinson’s disease-linked DNAJC6 mutations. Sci Adv 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J (2005) Preparation, culture, and immortalization of mouse embryonic fibroblasts. Curr Protoc Mol Biol Chapter28: Unit 28.21 [DOI] [PubMed] [Google Scholar]
- Yan Z, Yan H, Ou H (2012) Human thyroxine binding globulin (TBG) promoter directs efficient and sustaining transgene expression in liver-specific pattern. Gene 506: 289–294 [DOI] [PubMed] [Google Scholar]
- Zhang EE, Liu AC, Hirota T, Miraglia LJ, Welch G, Pongsawakul PY, Liu X, Atwood A, Huss JW 3rd, Janes J et al. (2009) A genome-wide RNAi screen for modifiers of the circadian clock in human cells. Cell 139: 199–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Ackerman SL (2006) Endoplasmic reticulum stress in health and disease. Current Opinion in Cell Biology 18: 444–452 [DOI] [PubMed] [Google Scholar]
- Zhou M, Tamburini I, Van C, Molendijk J, Nguyen CM, Chang IY-Y, Johnson C, Velez LM, Cheon Y, Yeo R et al. (2024) Leveraging inter-individual transcriptional correlation structure to infer discrete signaling mechanisms across metabolic tissues. eLife 12: RP88863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B (2020) Decoding the function and regulation of the mammalian 12h-clock. Journal of Molecular Cell Biology [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Gates LA, Stashi E, Dasgupta S, Gonzales N, Dean A, Dacso CC, York B, O’Malley BW (2015a) Coactivator-Dependent Oscillation of Chromatin Accessibility Dictates Circadian Gene Amplitude via REV-ERB Loading. Mol Cell 60: 769–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Gates LA, Stashi E, Dasgupta S, Gonzales N, Dean A, Dacso CC, York B, O’Malley BW (2015b) Coactivator-Dependent Oscillation of Chromatin Accessibility Dictates Circadian Gene Amplitude via REV-ERB Loading. Mol Cell [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Gates LA, Stashi E, Dasgupta S, Gonzales N, Dean A, Dacso CC, York B, O’Malley BW (2015c) Coactivator-dependent oscillation of chromatin accessibility dictates circadian gene amplitude via REV-ERB loading. Molecular cell 60: 769–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Liu S (2023a) Preservation of ~12-hour ultradian rhythms of gene expression of mRNA and protein metabolism in the absence of canonical circadian clock. bioRxiv: 2023.2005.2001.538977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Liu S (2023b) Preservation of ~12-h ultradian rhythms of gene expression of mRNA and protein metabolism in the absence of canonical circadian clock. Frontiers in Physiology 14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Liu S, David NL, Dion W, Doshi NK, Siegel LB, Amorim T, Andrews RE, Kumar GN, Irfan S et al. (2023) Evidence for conservation of a primordial 12-hour ultradian gene program in humans. bioRxiv: 2023.2005.2002.539021 [Google Scholar]
- Zhu B, Liu S, David NL, Dion W, Doshi NK, Siegel LB, Amorim T, Andrews RE, Kumar GVN, Li H et al. (2024) Evidence for ~12-h ultradian gene programs in humans. npj Biological Timing and Sleep 1: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Zhang Q, Pan Y, Mace EM, York B, Antoulas AC, Dacso CC, O’Malley BW (2017a) A Cell-Autonomous Mammalian 12 hr Clock Coordinates Metabolic and Stress Rhythms. Cell Metab 25: 1305–1319 e1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu B, Zhang Q, Pan Y, Mace EM, York B, Antoulas AC, Dacso CC, O’Malley BW (2017b) A cell-autonomous mammalian 12 hr clock coordinates metabolic and stress rhythms. Cell metabolism 25: 1305–1319. e1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession numbers: GSE276155, GSE276157 and GSE276159. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.