Abstract
The matrix (M) protein of vesicular stomatitis virus (VSV) is a potent inhibitor in vivo of transcription by all three host RNA polymerases (RNAP). In the case of host RNA polymerase II (RNAPII), the inhibition is due to lack of activity of the TATA-binding protein (TBP), which is a subunit of the basal transcription factor TFIID. Despite the potency of M protein-induced inhibition in vivo, experiments presented here show that M protein cannot directly inactivate TFIID in vitro. Addition of M protein to nuclear extracts from uninfected cells did not inhibit transcription activity, indicating that the inhibition is indirect and is mediated through host factors. The host factors that are known to regulate TBP activity include phosphorylation by host kinases and association with different TBP-associated factor (TAF) subunits. However, TBP in VSV-infected cells was found to be assembled normally with its TAF subunits, as shown by ion exchange high-pressure liquid chromatography and sedimentation velocity analysis. A normal pattern of phosphorylation of TBP in VSV-infected cells was also observed by pH gradient gel electrophoresis. Collectively, these data indicate that M protein inactivates TBP activity in RNAPII-dependent transcription by a novel mechanism, since the known mechanisms for regulating TBP activity cannot account for the inhibition.
Many viruses inhibit the expression of host genes. In most cases, the role of this inhibition of host gene expression in the viral replicative cycle is to inhibit the expression of gene products that are involved in the antiviral response of the host (17). In the case of vesicular stomatitis virus (VSV), the prototype member of the rhabdovirus family, much of the inhibition of host gene expression has been attributed to the activity of the viral matrix (M) protein. M protein plays a role in two very different aspects of VSV replication. Most of the M protein is present in the cytoplasm, where it functions in virus assembly by binding the nucleoprotein core to the cytoplasmic surface of the host plasma membrane and inducing the budding process that generates the viral envelope (15). M protein is also present in the nuclei of infected cells, which is consistent with its major role in the inhibition of host gene expression (21). This role includes inhibition of transcription by all three host RNA polymerases (RNAP) (1, 3) and inhibition of nuclear-cytoplasmic transport of host RNAs and proteins (13, 25, 29). The role of M protein in the inhibition of host gene expression is genetically separable from its function in virus assembly, as shown by M protein mutants that are defective in the inhibition of host gene expression but function as well as wild-type M protein in virus assembly. Conversely, other mutants are defective in virus assembly but are as potent as wild-type M protein in the inhibition of host gene expression (4, 8, 10, 18).
The molecular mechanisms involved in inhibiting host gene expression are of considerable interest, both because of their implications for viral pathogenesis and because they may reveal new features of the regulation of gene expression in host cells. In the case of host RNA polymerase II (RNAPII), the inhibition by VSV M protein is due to inactivation of the basal transcription initiation factor TFIID (31). This inhibition was shown by reconstituting transcription initiation in vitro using partially purified transcription initiation factors, in which the TFIID fraction from VSV-infected cells was not able to reconstitute transcription initiation in the presence of the other basal transcription factors from uninfected cells. Conversely, TFIID from uninfected cells was able to fully restore transcription initiation in nuclear extracts from infected cells. Thus, TFIID was the only basal transcription factor whose inhibition could be detected (31).
TFIID is a multisubunit complex consisting of a DNA-binding subunit, the TATA-binding protein (TBP), and a set of TBP-associated factors (TAFs) (5). TFIID is the first basal transcription factor assembled onto RNAPII-dependent promoters through binding of TBP to the TATA box DNA sequence located upstream of most promoters. TBP is the only subunit of TFIID required for basal transcription in vitro. However, activation of transcription by proteins that bind DNA sequence-specific enhancer elements requires interaction with one or more TAF subunits, either directly or indirectly through so-called adapter proteins (32).
M protein does not directly inactivate host TFIID in vitro.
We have shown previously that purified recombinant TBP is able to fully restore basal transcription initiation in nuclear extracts from VSV-infected cells, consistent with the inhibition occurring at the level of basal transcription initiation (31). This finding raises the question of how the activity of TBP is inhibited in infected cells. The inhibition is not due to a reduction in the amount of TBP, which is present in nuclear extracts from both infected and uninfected HeLa cells at a level of 0.13 ng of TBP per μg of protein (31). One possibility is that M protein inhibits TBP activity by binding directly to TFIID. We examined this possibility by testing the ability of M protein to inhibit the activity of TFIID from uninfected cells in vitro.
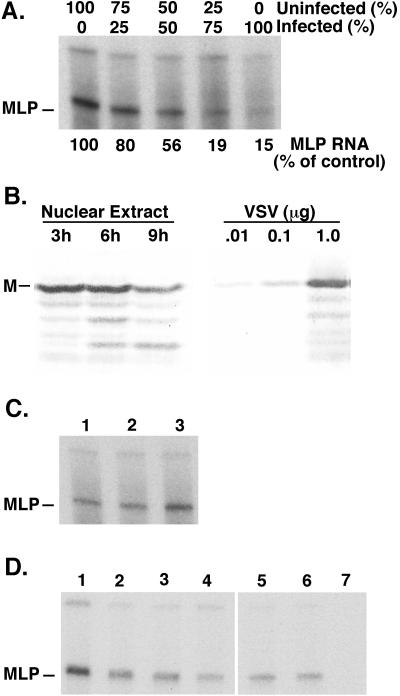
Figure 1A shows that M protein in nuclear extracts from VSV-infected cells was not able to inhibit TFIID in nuclear extracts from uninfected cells in mixing experiments. Nuclear extracts from infected or uninfected cells were mixed in the proportions shown in Fig. 1A, and RNAPII-dependent transcription was assayed using a plasmid DNA template that contained the adenovirus major late promoter (MLP). The plasmid lacked upstream activating sequences so that only basal transcription was assayed. Transcripts were measured by an RNase protection assay with a radiolabeled riboprobe, as described previously (31). The upper band and the diffuse background in Fig. 1A result from random initiation on the plasmid DNA template, whereas the lower band results from initiation at the MLP (indicated in Fig. 1A). Only the MLP-initiated product is an assay for the activity of RNAPII transcription initiation factors. The activity of these factors was readily apparent in nuclear extract from uninfected cells (lane 1), while activity in nuclear extract from infected cells (lane 5) was markedly reduced. Mixing nuclear extracts (lanes 2 to 4) resulted in transcription activities that were proportional to the amounts of uninfected cell nuclear extract present in the mixture. This result indicated that nuclear extract from infected cells did not inhibit MLP-initiated transcription by nuclear extract from uninfected cells. Thus, the inhibitory activity in infected cell nuclear extracts was either not present in excess or was not freely diffusible between TFIID complexes from infected and uninfected cells.
FIG. 1.
Effect of M protein on transcription by nuclear extracts in vitro. (A) Nuclear extracts from uninfected or VSV-infected cells prepared at 6 h postinfection were mixed in the indicated amounts and incubated in an in vitro transcription reaction with a plasmid DNA template containing the adenovirus MLP. The transcription products were analyzed by an RNase protection assay using a radiolabeled riboprobe, an autoradiograph of which is shown, and quantitated by densitometry as indicated. The position of the MLP-initiated product is indicated. (B) Nuclear extracts were prepared from VSV-infected HeLa cells at 3, 6, and 9 h postinfection. The amount of M protein in the nuclear extracts was determined by Western blot, using the indicated amounts of purified VSV as a standard. (C) Nuclear extract from uninfected cells was mixed with M protein purified from VSV virions by ion exchange chromatography (lane 3). Nuclear extract with no additions (lane 1) or with column buffer used to elute M protein (lane 2) were used as controls. Transcription in vitro was assayed as in panel A. (D) Nuclear extract from uninfected cells was mixed with in vitro translation reactions containing M protein (lanes 2 to 4) or N protein (lanes 5 to 7) or with no additions (lane 1). In vitro transcription reactions contained 8 μl of nuclear extract and 1 μl (lanes 2 and 5), 2.5 μl (lanes 3 and 6), or 5.5 μl (lanes 4 and 7) of translation mixture in a total volume of 25 μl.
Western blot analysis was used to determine whether M protein was present in excess over host TFIID in nuclear extracts from VSV-infected cells (Fig. 1B). Nuclear extracts were prepared as described previously (31) from VSV-infected HeLa cells at 3, 6, and 9 h postinfection and analyzed in a Western blot using a monoclonal M protein antibody, 23H12 (21). Serial dilutions of purified virions were analyzed in parallel as a standard. The amount of M protein in nuclear extracts was nearly constant from 3 to 9 h postinfection, although in this experiment the amount of M protein declined slightly by 9 h postinfection. The amount of M protein in the nuclear extracts was calculated to be 2.0 ng of M protein per μg of extract protein. The concentration of TBP is 0.13 ng of TBP per μg of protein (31). Thus, there is a 15-fold excess of M protein compared to TBP. Since TFIID contains only a fraction of cellular TBP (32), the excess of M protein compared to TFIID in nuclear extracts was greater than 15-fold.
Despite the excess of M protein over TFIID in nuclear extracts (Fig. 1B), the M protein in nuclear extracts from VSV-infected cells was not able to inhibit TFIID in nuclear extracts from uninfected cells (Fig. 1A). This was confirmed by the addition of purified M protein (Fig. 1C) or M protein translated in vitro (Fig. 1D) to nuclear extracts from uninfected cells. M protein was purified from virions by a phosphocellulose column as described previously (23) and was added to nuclear extracts from uninfected cells at concentrations similar to that in nuclear extracts from infected cells. As a control for purified M protein, an equal amount of column buffer was added to nuclear extracts (Fig. 1C, lane 2). There was no inhibition of MLP-initiated transcription when purified M protein from virions was added to nuclear extracts prepared from uninfected cells (Fig. 1C, lane 3). In fact, the reaction containing M protein actually yielded more MLP-initiated RNA in the experiment shown in Fig. 1C, although this was not consistently observed in all experiments.
In the experiment shown in Fig. 1D, M protein synthesized by in vitro translation was tested for its ability to inhibit RNAPII-dependent transcription. RNA transcribed from a plasmid containing the M gene driven by the bacteriophage T7 promoter (4) was translated in a reticulocyte lysate (Promega Corp.). The in vitro translation mixture containing M protein was added to nuclear extract from uninfected cells in various amounts (Fig. 1D, lanes 2 to 4). Equal volumes of in vitro-translated N protein of VSV were added as a negative control (lanes 5 to 7), since N protein does not inhibit host transcription (2, 10). The addition of in vitro-translated M protein did inhibit MLP-initiated transcription slightly (Fig. 1D, lanes 2 to 4), but this was attributable to other components in the in vitro translation mixture, since a similar inhibition was observed with the N protein control (lanes 5 to 7).
The data in Fig. 1 indicate that M protein cannot inactivate TFIID by a direct interaction in vitro. The data leave open the possibility that M protein assumes a conformation in vivo that differs from the M protein tested in vitro. However, such a conformation would have to be distinct from that found in the M protein that is present in nuclear extracts from infected cells, as well as that produced by reticulocyte lysates or purified from virions. The M protein in all three of these preparations was active in other in vitro assays for M protein activity, such as the ability to assemble into viral nucleocapsid-M protein complexes involved in virus assembly (19; unpublished data). Thus, although this possibility remains, the fact that we tested three different types of M protein preparations made it unlikely that M protein was in a nonnative conformation in vitro. It is also possible that M protein binds to TFIID in vivo by a mechanism that is not recreated in vitro. However, M protein and TBP did not coimmunoprecipitate with antibodies to either protein (data not shown). This suggests that M protein acts indirectly in vivo through host mechanisms that are capable of regulating TFIID activity, in particular the activity of TBP, which is the only subunit required for basal transcription.
Association of TBP with TAF subunits in VSV-infected cells.
There are two ways that the activity of TBP is known to be regulated: (i) through its subunit interactions and (ii) by phosphorylation. TBP is a subunit of three different transcription factors that function in the activation of all three host RNAPs through association with different TAF subunits (32). The TAF subunits of TFIID are referred to as TAFIIs. Association of TBP with a set of TAFIs forms the transcription factor SL1, which functions in the activation of RNAPI, and association with a set of TAFIIIs forms the transcription factor TFIIB, which functions in the activation of RNAPIII. The activity of all three of these transcription factors can be either enhanced or inhibited by phosphorylation, depending on which site is phosphorylated (6, 11, 12, 14, 16, 22).
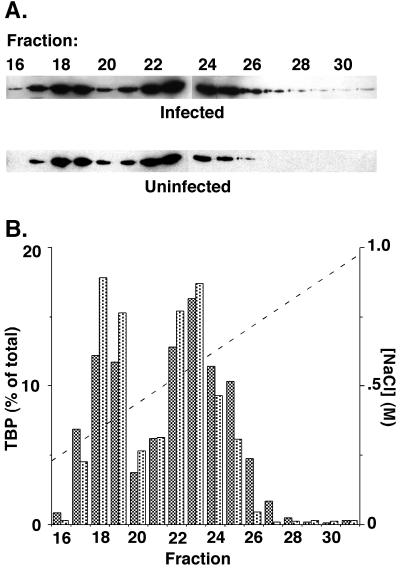
TBP-containing transcription factors in nuclear extracts from VSV-infected or uninfected cells were analyzed by ion exchange chromatography to determine whether there was an alteration in the association of TBP with TAF subunits in infected cells. The nature of the TAF subunits dictates the behavior of these transcription factors in ion exchange chromatography, and this technique has been used previously to assay association of TBP with TAF subunits (33). Nuclear extracts were prepared from uninfected or VSV-infected HeLa cells at 6 h postinfection and were chromatographed on a 1.7-ml preparative cation exchange high-pressure liquid chromatography (HPLC) column (POROS HS; Perseptive Biosystems) eluted with a 17-ml gradient of 50 mM to 1.0 M NaCl in buffer D, as described by Dignam et al. (9), but lacking KCl. TBP in column fractions was determined by Western blot analysis (Fig. 2A) as described previously (31), and the results of densitometry analysis of the Western blots are shown in Fig. 2B as the percentage of the total TBP eluted from the column. TBP in nuclear extracts from both the infected cells (Fig. 2B, dark bars) and the uninfected cells (light bars) eluted in two peaks. The first peak eluting at lower salt concentrations was centered around fraction 18 and contained TFIIIB (9). The second peak was centered around fraction 23 and primarily contained TFIID and the SL1. The second peak was broader because TFIID is heterogeneous in its composition of TAF subunits (5). The elution profiles for TBP-containing transcription factors from infected cells were remarkably similar to those from uninfected cells. In both cases, approximately 40% of TBP eluted in the first peak and 60% of TBP eluted in the second. These data indicate that there was no difference in the association of TBP with TAF subunits in infected cells versus uninfected cells that could be detected by ion exchange HPLC. The same conclusion was reached when the extracts were analyzed by conventional chromatography on a phosphocellulose column, as described previously (33), rather than by HPLC (not shown). In addition, the TFIID from infected cells was inactive following chromatography (31). Thus, the mechanism of M protein inactivation of TFIID does not involve direct, but labile, protein-protein or protein-inhibitor interactions that are dissociated during chromatography.
FIG. 2.
Ion exchange HPLC of TBP-containing transcription factors. Nuclear extracts were prepared from VSV-infected or uninfected HeLa cells at 6 h postinfection and chromatographed on an ion exchange HPLC column eluted with a NaCl concentration gradient. (A) Column fractions were assayed for TBP content by Western blot. (B) Densitometry of the Western blots in panel A was used to determine the percent of total TBP in each fraction for nuclear extracts from infected (dark bars) or uninfected cells (light bars). The concentration gradient of NaCl used to elute the column is also shown.
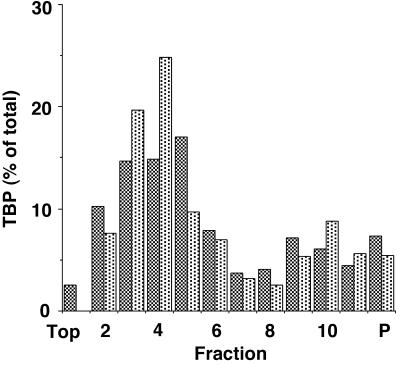
The conclusion that there was no difference in the association of TBP with TAF subunits in infected cells versus uninfected cells was supported by sedimentation velocity analysis of TBP-containing transcription factors. TFIID, which has an s20,w of 17S (26, 34) is easily resolved from TFIIIB and SL1, which have s20,w of about 5 to 7S (28). Nuclear extracts were prepared from uninfected or VSV-infected HeLa cells at 6 h postinfection and loaded onto 10 to 30% sucrose gradients. After centrifugation at 40,000 rpm in a TLS-55 rotor (Beckman Instruments) for 8 h at 4°C, fractions were collected and analyzed by Western blotting using antibody against TBP. Figure 3 shows the amount of TBP in each fraction quantitated by densitometry and represented as the percentage of the total TBP recovered from the gradient. Two peaks of TBP were evident. The peak in the upper half of the gradient (fractions 2 through 6) corresponds to SL1 and TFIIIB, for which the sedimentation velocity is about 5S. The TBP in fractions 2 through 6 together amounted to 69% of the total for nuclear extracts from uninfected cells and 65% for nuclear extracts from infected cells. The peak in the lower half of the gradient (fractions 8 through 11) is TFIID, for which the sedimentation velocity is about 17S. The TBP in fractions 8 through 11 amounted to 22% of the total for nuclear extracts from both uninfected cells and infected cells. In repeated experiments, there was no significant difference in the level of TFIID (18% for nuclear extracts from uninfected cells versus 22% for nuclear extracts from infected cells in three experiments). Also, there was no significant difference in sedimentation velocity of TFIID, which sedimented 9.33 ± 0.57 fractions for nuclear extracts from uninfected cells and 9.66 ± 0.57 fractions for nuclear extracts from infected cells. These data support the conclusion that TBP associates normally with TAF subunits in transcription factors from VSV-infected cells.
FIG. 3.
Sedimentation analysis of TBP-containing transcription factors. Nuclear extracts were prepared from VSV-infected (dark bars) or uninfected (light bars) HeLa cells at 6 h postinfection and sedimented on 10 to 30% sucrose gradients. Gradient fractions and the pelleted material (P) were assayed for TBP content by Western blots as in Fig. 2, and the percentage of total TBP in each fraction was determined by densitometry.
Phosphorylation of TBP.
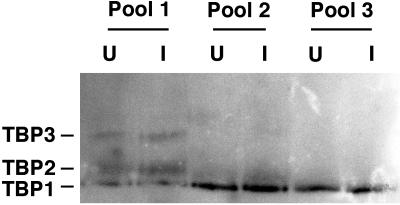
In addition to its association with different subunits, the activity of TBP in each of its three transcription factors can also be regulated by phosphorylation. Phosphorylation can either enhance or inhibit the activity of TBP, depending on which sites are phosphorylated. We considered that TBP could be inactivated in VSV-infected cells by phosphorylation at an inhibitory site. An alternative possibility involving dephosphorylation of an activating site cannot account for the inhibition of RNAPII-dependent transcription, since TBP is active in basal RNAPII-dependent transcription in its unphosphorylated form, while TBP in infected cells is almost completely inactive (31). HPLC column fractions containing TBP similar to those shown in Fig. 2 were combined into three pools, and the extent of phosphorylation of TBP was determined by electrophoresis in a pH gradient gel, followed by Western blotting with antibody against TBP (Fig. 4). This is a variant of the isoelectric focusing technique, in which proteins are separated according to their isoelectric points but are not electrophoresed to equilibrium as in the traditional isoelectric focusing technique (24, 27). This technique provides better resolution for basic proteins such as TBP and can detect single charge differences, such as those introduced by phosphorylation. There are three forms of TBP in nuclear extracts from HeLa cells (Fig. 4). The fastest migrating form (TBP1) corresponds to unphosphorylated TBP. TBP2 is probably a singly phosphorylated form, and TBP3 is a multiply phosphorylated form, as shown by conversion of TBP2 and TBP3 into the TBP1 form by treatment with alkaline phosphatase in vitro (not shown). Pool 1 consisted of HPLC fractions corresponding to fractions 17 through 19 in Fig. 2B, which contained TFIIIB. The TBP in pool 1 consisted primarily of the phosphorylated forms of TBP. This is consistent with recent data showing that phosphorylation of TBP is necessary for TFIIIB activity (11). Pool 2 was the first half of the TFIID peak, corresponding to fractions 21 through 23, and pool 3 was the second half of the TFIID peak (fractions 24 through 26). Both pools contained primarily unphosphorylated TBP and a small amount of the TBP2 form. This is consistent with activity of TBP in RNAPII-dependent transcription in its unphosphorylated form.
FIG. 4.
Analysis of TBP phosphorylation by pH gradient gel electrophoresis. Nuclear extracts were prepared from VSV-infected (I) or uninfected (U) HeLa cells at 6 h postinfection and chromatographed on an ion exchange HPLC column eluted with a NaCl concentration gradient as in Fig. 2. Column fractions were combined into pool 1 (fractions 17 through 19), pool 2 (fractions 21 through 23), and pool 3 (fractions 24 through 26) and electrophoresed on a pH gradient gel. TBP was detected by Western blot and migrated as three forms, with TBP1 being the fastest migrating form and TBP3 the slowest.
The most important result of the experiment in Fig. 4 was the finding that there was no difference in the state of phosphorylation of TBP between infected cells and uninfected cells. In particular, most of the TBP in TFIID was in the unphosphorylated form in the nuclear extracts from both infected and uninfected cells. The same results were obtained when the phosphatase inhibitor sodium vanadate (5 mM) was included in all the buffers used to prepare nuclear extracts and perform the HPLC, as well as when the vanadate was omitted, as it was in our previous study demonstrating that TFIID from infected cells is inactive (31). Since the unphosphorylated form of TBP is fully active in RNAPII-dependent transcription, this observation rules out phosphorylation of TBP as a mechanism of inactivation of TFIID in infected cells. Collectively, these data indicate that M protein inactivates TBP activity in RNAPII-dependent transcription by a novel mechanism, since the known mechanisms for regulating TBP activity cannot account for the inhibition.
These results on the mechanism of inhibition of transcription by VSV provide an interesting comparison with the inhibition by poliovirus. Both viruses inhibit basal RNAPII-dependent transcription by inactivation of TBP. In the case of poliovirus, the viral 3C protease inactivates TBP directly by cleavage at one or more sites (7, 30). Unlike poliovirus 3C protease, M protein does not have any known catalytic activity that could be responsible for the inhibition of TBP activity. Also, unlike poliovirus 3C protease, M protein did not inhibit transcription in nuclear extracts from uninfected cells when added in vitro (Fig. 1). M protein is a very potent inhibitor of transcription in vivo. We have estimated that 50% inhibition of host RNAPII-dependent transcription in transfection experiments occurs when the amount of M protein is approximately 1,000-fold less than the amount of M protein expressed in VSV-infected cells (20). Thus, it is unlikely that the amounts of M protein added to nuclear extract from uninfected cells in the experiments shown in Fig. 1 were simply not sufficient, since they were similar to the amounts present in nuclear extract from VSV-infected cells. The conclusion that M protein does not directly inactivate TBP was supported by the inability to coimmunoprecipitate M protein and TBP with antibodies to either protein. The coimmunoprecipitation assays were sufficiently sensitive that if 10 to 25% of TFIID coprecipitated with M protein, the interaction would have been readily detected (data not shown). These results support the idea that M protein acts indirectly in vivo through host mechanisms that are capable of inhibiting TBP activity.
It has been proposed that the inhibition of transcription in VSV-infected cells is an indirect effect of the inhibition of nuclear-cytoplasmic transport by M protein, leading to a reduction in the levels of the critical transcription factors (13, 29). However, in the case of TFIID, we found that there was no reduction in the amount of TFIID in nuclear extracts from VSV-infected cells, but instead the TFIID was in an inactive form (31; Fig. 2 and 3). Our observation that the inactivation of TFIID was an indirect effect caused by M protein leaves open the possibility that the M protein-induced block in nuclear-cytoplasmic transport affects the activity of a host factor involved in regulating TFIID activity.
Phosphorylation of TBP by a cellular kinase was a likely candidate for the mechanism of inhibition of transcription initiation, since phosphorylation of TBP can either inhibit or enhance the activity of all three TBP-containing transcription initiation factors, depending on which sites are phosphorylated. For example, phosphorylation of TBP by cdc2/cyclin B kinase has been implicated in the silencing of transcription by all three host RNAPs during mitosis (12, 14, 16). Analysis of TBP phosphorylation in VSV-infected cells showed that there were dramatic differences among the different transcription factors, but there was little if any difference between infected and uninfected cells (Fig. 4). In particular, TFIIIB contained primarily phosphorylated TBP, while TFIID contained primarily unphosphorylated TBP. These results extend to HeLa cells the results obtained in the yeast Saccharomyces cerevisiae, in which phosphorylation of TBP in TFIIIB was originally demonstrated (11). In yeasts, phosphorylation of TBP by casein kinase II is required for TFIIIB activity. In contrast to TFIIIB, TFIID is active when TBP is in the unphosphorylated form, although its activity can be enhanced by phosphorylation by DNA-dependent protein kinase (6). Since most of the TBP in TFIID was in the unphosphorylated form in nuclear extracts from both infected and uninfected cells, differences in phosphorylation of TBP cannot account for the inactivation of TFIID.
The normal phosphorylation and assembly of TBP with TAF subunits in VSV-infected cells implies that there is a novel inhibitory factor responsible for the inactivation of TFIID. Such an inhibitory factor might act by associating with TFIID in a manner similar to the TAF subunits. If so, such a factor did not affect the behavior of TFIID in ion exchange HPLC nor affect its sedimentation velocity. Alternatively, TFIID could be inhibited by a posttranslational modification other than phosphorylation of TBP. If so, then this modification would not alter the charge on TBP, since there was no change in its isoelectric point. Both of these possibilities are currently being investigated.
Acknowledgments
We thank Barbara Yoza, Griffith Parks, and David Ornelles for helpful advice and comments on the manuscript.
This work was supported by Public Health Service grant AI 32983 from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Ahmed M, Lyles D S. Effect of vesicular stomatitis virus matrix protein on transcription directed by host RNA polymerases I, II, and III. J Virol. 1998;72:8413–8419. doi: 10.1128/jvi.72.10.8413-8419.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Black B L, Brewer G, Lyles D S. Effect of vesicular stomatitis virus matrix protein on host-directed translation in vivo. J Virol. 1994;68:555–560. doi: 10.1128/jvi.68.1.555-560.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Black B L, Lyles D S. Vesicular stomatitis virus matrix protein inhibits host cell-directed transcription of target genes in vivo. J Virol. 1992;66:4058–4064. doi: 10.1128/jvi.66.7.4058-4064.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Black B L, Rhodes R B, McKenzie M, Lyles D S. The role of vesicular stomatitis virus matrix protein in inhibition of host-directed gene expression is genetically separable from its function in virus assembly. J Virol. 1993;67:4814–4821. doi: 10.1128/jvi.67.8.4814-4821.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burley S K, Roeder R G. Biochemistry and structural biology of transcription factor IID (TFIID) Annu Rev Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- 6.Chibazakura T, Watanabe F, Kitajima S, Tsukada K, Yasukochi Y, Teraoka H. Phosphorylation of human general transcription factors TATA-binding protein and transcription factor IIB by DNA-dependent protein kinase—synergistic stimulation of RNA polymerase II basal transcription in vitro. Eur J Biochem. 1997;247:1166–1173. doi: 10.1111/j.1432-1033.1997.01166.x. [DOI] [PubMed] [Google Scholar]
- 7.Clark M E, Lieberman P M, Berk A J, Dasgupta A. Direct cleavage of human TATA-binding protein by poliovirus protease 3C in vivo and in vitro. Mol Cell Biol. 1993;13:1232–1237. doi: 10.1128/mcb.13.2.1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coulon P, Deutsch V, Lafay F, Martinet-Edelist C, Wyers F, Herman R C, Flamand A. Genetic evidence for multiple functions of the matrix protein of vesicular stomatitis virus. J Gen Virol. 1990;71:991–996. doi: 10.1099/0022-1317-71-4-991. [DOI] [PubMed] [Google Scholar]
- 9.Dignam J D, Martin P L, Shastry B S, Roeder R G. Eukaryotic gene transcription with purified components. Methods Enzymol. 1983;101:582–598. doi: 10.1016/0076-6879(83)01039-3. [DOI] [PubMed] [Google Scholar]
- 10.Ferran M C, Lucas-Lenard J M. The vesicular stomatitis virus matrix protein inhibits transcription from the human beta interferon promoter. J Virol. 1997;71:371–377. doi: 10.1128/jvi.71.1.371-377.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ghavidel A, Schultz M C. Casein kinase II regulation of yeast TFIIIB is mediated by the TATA-binding protein. Genes Dev. 1997;11:2780–2789. doi: 10.1101/gad.11.21.2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Heix J, Vente A, Voit R, Budde A, Michaelidis T M, Grummt I. Mitotic silencing of human rRNA synthesis: inactivation of the promoter selectivity factor SL1 by cdc2/cyclin B-mediated phosphorylation. EMBO J. 1998;17:7373–7381. doi: 10.1093/emboj/17.24.7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Her L S, Lund E, Dahlberg J E. Inhibition of Ran guanosine triphosphatase-dependent nuclear transport by the matrix protein of vesicular stomatitis virus. Science. 1997;276:1845–1848. doi: 10.1126/science.276.5320.1845. [DOI] [PubMed] [Google Scholar]
- 14.Kuhn A, Vente A, Doree M, Grummt I. Mitotic phosphorylation of the TBP-containing factor SL1 represses ribosomal gene transcription. J Mol Biol. 1998;284:1–5. doi: 10.1006/jmbi.1998.2164. [DOI] [PubMed] [Google Scholar]
- 15.Lenard J. Negative-strand virus M and retrovirus MA proteins: all in a family? Virology. 1996;216:289–298. doi: 10.1006/viro.1996.0064. [DOI] [PubMed] [Google Scholar]
- 16.Leresche A, Wolf V J, Gottesfeld J M. Repression of RNA polymerase II and III transcription during M phase of the cell cycle. Exp Cell Res. 1996;229:282–288. doi: 10.1006/excr.1996.0373. [DOI] [PubMed] [Google Scholar]
- 17.Lyles D S. Cytopathogenesis and the inhibition of host gene expression by RNA viruses. Microbiol Mol Biol Rev. 2000;64:709–724. doi: 10.1128/mmbr.64.4.709-724.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lyles D S, McKenzie M O. Activity of vesicular stomatitis virus M protein mutants in cell rounding is correlated with the ability to inhibit host gene expression and is not correlated with virus assembly function. Virology. 1997;229:77–89. doi: 10.1006/viro.1996.8415. [DOI] [PubMed] [Google Scholar]
- 19.Lyles D S, McKenzie M O. Reversible and irreversible steps in assembly and disassembly of vesicular stomatitis virus: equilibria and kinetics of dissociation of nucleocapsid-M protein complexes assembled in vivo. Biochemistry. 1998;37:439–450. doi: 10.1021/bi971812j. [DOI] [PubMed] [Google Scholar]
- 20.Lyles D S, McKenzie M O, Ahmed M, Woolwine S C. Potency of wild-type and temperature-sensitive vesicular stomatitis virus matrix protein in the inhibition of host-directed gene expression. Virology. 1996;225:172–180. doi: 10.1006/viro.1996.0585. [DOI] [PubMed] [Google Scholar]
- 21.Lyles D S, Puddington L, McCreedy B J., Jr Vesicular stomatitis virus M protein in the nuclei of infected cells. J Virol. 1988;62:4387–4392. doi: 10.1128/jvi.62.11.4387-4392.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maldonado E, Allende J E. Phosphorylation of yeast TBP by protein kinase CK2 reduces its specific binding to DNA. FEBS Lett. 1999;443:256–260. doi: 10.1016/s0014-5793(98)01734-7. [DOI] [PubMed] [Google Scholar]
- 23.McCreedy B J, Jr, McKinnon K P, Lyles D S. Solubility of vesicular stomatitis virus M protein in the cytosol of infected cells or isolated from virions. J Virol. 1990;64:902–906. doi: 10.1128/jvi.64.2.902-906.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.O'Farrell P Z, Goodman H M, O'Farrell P H. High resolution two-dimensional electrophoresis of basic as well as acidic proteins. Cell. 1977;12:1133–1141. doi: 10.1016/0092-8674(77)90176-3. [DOI] [PubMed] [Google Scholar]
- 25.Petersen J M, Her L S, Varvel V, Lund E, Dahlberg J E. The matrix protein of vesicular stomatitis virus inhibits nucleocytoplasmic transport when it is in the nucleus and associated with nuclear pore complexes. Mol Cell Biol. 2000;20:8590–8601. doi: 10.1128/mcb.20.22.8590-8601.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Samuels M, Fire A, Sharp P A. Separation and characterization of factors mediating accurate transcription by RNA polymerase II. J Biol Chem. 1982;257:14419–14427. [PubMed] [Google Scholar]
- 27.Sanders M M, Groppi V E, Jr, Browning E T. Resolution of basic cellular proteins including histone variants by two-dimensional gel electrophoresis: evaluation of lysine to arginine ratios and phosphorylation. Anal Biochem. 1980;103:157–165. doi: 10.1016/0003-2697(80)90250-x. [DOI] [PubMed] [Google Scholar]
- 28.Tanese N. Small-scale density gradient sedimentation to separate and analyze multiprotein complexes. Methods. 1997;12:224–234. doi: 10.1006/meth.1997.0475. [DOI] [PubMed] [Google Scholar]
- 29.von Kobbe C, van Deursen J M A, Rodrigues J P, Sitterlin D, Bachi A, Wu X, Wilm M, Carmo-Fonseca M, Izaurralde E. Vesicular stomatitis virus matrix protein inhibits host cell gene expression by targeting the nucleoporin Nup98. Mol Cell. 2000;6:1243–1252. doi: 10.1016/s1097-2765(00)00120-9. [DOI] [PubMed] [Google Scholar]
- 30.Yalamanchili P, Harris K, Wimmer E, Dasgupta A. Inhibition of basal transcription by poliovirus: a virus-encoded protease (3Cpro) inhibits formation of TBP-TATA box complex in vitro. J Virol. 1996;70:2922–2929. doi: 10.1128/jvi.70.5.2922-2929.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yuan H, Yoza B K, Lyles D S. Inhibition of host RNA polymerase II-dependent transcription by vesicular stomatitis virus results from inactivation of TFIID. Virology. 1998;251:383–392. doi: 10.1006/viro.1998.9413. [DOI] [PubMed] [Google Scholar]
- 32.Zawel L, Reinberg D. Common themes in assembly and function of eukaryotic transcription complexes. Annu Rev Biochem. 1995;64:533–561. doi: 10.1146/annurev.bi.64.070195.002533. [DOI] [PubMed] [Google Scholar]
- 33.Zhou Q, Berk A J. The yeast TATA-binding protein (TBP) core domain assembles with human TBP-associated factors into a functional TFIID complex. Mol Cell Biol. 1995;15:534–539. doi: 10.1128/mcb.15.1.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou Q, Lieberman P M, Boyer T G, Berk A J. Holo-TFIID supports transcriptional stimulation by diverse activators and from a TATA-less promoter. Genes Dev. 1992;6:1964–1974. doi: 10.1101/gad.6.10.1964. [DOI] [PubMed] [Google Scholar]