Abstract
Recent analyses have identified a number of binding partners for E6, including E6AP, ERC55, paxillin, hDlg, p300, interferon regulatory factor 3, hMCM7, Bak, and E6TP1. Notably, association with E6 targets p53, E6TP1, myc, hMCM7, and Bak for degradation. However, the relative importance of the various E6 targets in cellular transformation remains unclear. E6 alone can dominantly immortalize normal human mammary epithelial cells (MECs), permitting an assessment of the importance of various E6 targets in cellular transformation. Studies in this system indicate that E6-induced degradation of p53 and E6 binding to ERC55 or hDlg do not correlate with efficient immortalization. Here, we have examined the role of E6TP1, a Rap GTPase-activating protein, in E6-induced immortalization of MECs. We tested a large set of human papillomavirus type 16 E6 mutants for their ability to bind and target E6TP1 for degradation in vitro and in vivo. We observed a strict correlation between the ability of E6 protein to target E6TP1 for degradation and its ability to immortalize MECs. Recent studies have identified telomerase as a target of E6 protein. Previous analyses of E6 mutants have revealed this trait to closely correlate with MEC immortalization. We examined our entire panel of E6 mutants for rapid induction of telomerase activity and found in general a strong correlation with immortalizing ability. The tight correlation between E6TP1 degradation and MEC immortalization strongly supports a critical role of functional inactivation of E6TP1 in E6-induced cellular immortalization.
The human papillomaviruses (HPVs) have been strongly implicated in the pathogenesis of tumors as well as benign warts of oral and urogenital epithelium (36, 37). Those associated with carcinomas, such as HPV type 16 (HPV16) and HPV18, are classified as high-risk HPVs, whereas those associated with benign lesions, such as HPV11 and HPV6, are referred to as low-risk HPVs (36, 37). Two early genes of the high-risk HPVs, E6 and E7, are essential and sufficient for oncogenic transformation of cells in vitro (10, 22), and their expression is an invariant feature of HPV-associated human epithelial malignancies (2, 27). Both E6 and E7 genes are required for efficient immortalization of cervical keratinocytes, imposing limitations on elucidation of the biochemical pathways selectively targeted by these two oncogenes. However, as we demonstrated earlier, E6 alone can efficiently immortalize normal human mammary epithelial cells (MECs) (3, 4, 31). This single-gene immortalization model provides a valuable system to dissect the biochemical pathways involved in E6-induced transformation (3, 4, 7, 21, 31).
E6 protein is a small polypeptide of 151 amino acids, with no known intrinsic enzymatic activities. It is generally accepted that E6 functions as a dominant oncogene by interacting with and altering the function of critical cellular proteins. In the last few years, a large number of cellular proteins have been reported to interact with high-risk HPV E6 proteins. These include p53 (11), E6BP (E6-binding protein or ERC55, a putative calcium-binding protein) (5), paxillin (a focal adhesion protein involved in transducing signals from the plasma membrane to the actin cytoskeleton) (29), hDlg (the human homologue of the Drosophila melanogaster discs large tumor suppressor protein) (16, 20), interferon regulatory factor 3 (a component of virus-activated transcription factor complex) (24), multicopy maintenance protein 7 (a subunit of the replication licensing factor M) (18), Bak (bcl-2 homologous antagonist/killer, a protein that promotes apoptosis) (28), p300 (a transcriptional coactivator) (23, 35), E6TP1 (E6-targeted protein, a Rap GTPase-activating protein [GAP] homologue) (9), and PKN (a Rho-regulated serine threonine kinase) (9a). The relative contribution of the various E6 targets in cellular transformation is an area of intense investigation, but a clear consensus is lacking at present. The interaction of HPV E6 protein with the p53 tumor suppressor protein has been most widely analyzed thus far (7, 8, 15, 21). Interaction of E6 with p53 is indirect, mediated by the ubiquitin ligase E6AP, which binds to both E6 and p53 (11–13, 26). Bound E6 targets the p53 tumor suppressor protein for E6AP-mediated ubiquitination, followed by proteasome-mediated degradation. Our previous mutational analysis of HPV16 E6 demonstrated a strong correlation between p53 degradation and immortalization of MECs by E6 (7). Similarly, other investigators have shown that the HPV E6-induced in vivo p53 degradation closely correlates with abrogation of actinomycin D-induced growth arrest in human keratinocytes (8). However, we and others have recently isolated three distinct E6 mutants (8S9A10T, F2V, and Y54H) that are unable to target p53 for degradation but retain their ability to immortalize MECs (15, 21). Thus, it is clear that E6-induced immortalization of MECs can proceed in the absence of p53 inactivation. These results suggest that other E6 targets may play a critical role in E6-induced cellular transformation.
Recent studies have implicated telomerase as one such target (15, 17). It has been shown previously that introduction of E6 into MECs leads to increased telomerase activity (17). Mutational analysis demonstrated that E6 mutants that increased the telomerase activity were able to induce MEC immortalization, whereas E6 mutants that did not induce telomerase activity were defective in immortalization (15, 17). Notably, certain immortalization-competent E6 mutants that did not target p53 for degradation were capable of increasing telomerase activity (15). Based on these studies, it was concluded that activation of telomerase, but not p53 degradation, is required for E6-induced immortalization of MECs. However, one mutant, Δ118-122, reportedly failed to induce telomerase activity but still retained its ability to immortalize MECs (7, 15, 17). Recent studies of E6 mutants suggest that binding of E6 to hDlg, E6BP, and interferon regulatory factor 3 is not essential for E6-induced immortalization (15, 21, 24). These studies underscore the need for in-depth studies of various E6 targets to critically delineate their roles in cellular transformation.
E6TP1 is a novel cellular target of high-risk HPV E6 proteins which was recently isolated in our laboratory using a yeast two-hybrid approach (9). E6TP1 shows high sequence homology with Rap GAPs, including SPA-1, Rap1GAP, and tuberin (9). The cellular targets of these GAPs are the Rap family of Ras-related small G proteins (19, 25, 32). Although earlier studies suggested that Rap may function as an antagonist of Ras (14), a large number of recent studies have clearly shown that Rap participates in a signaling cascade that promotes cell proliferation, and overexpression of Rap1 was shown to oncogenically transform cells (1, 30, 34). Thus, Rap-specific GAPs, such as E6TP1, are likely to negatively regulate the mitogenic signaling pathways mediated by Rap. Targeting of such cell growth suppressor proteins by E6 could represent an important element in cellular transformation.
In this study, we have examined the ability of a large panel of E6 mutants to bind and target E6TP1 for degradation and have related these traits to E6-induced telomerase activity and immortalization of human MECs. The panel of mutants included two immortalizing mutants that are defective for p53 degradation, as well as a mutant that was reported to be defective in inducing telomerase activity but was still capable of immortalizing MECs. In this panel of mutants, we observed a perfect correlation between the ability of HPV16 E6 mutants to target E6TP1 for degradation and their ability to immortalize MECs, suggesting that loss of E6TP1 function is crucial for cellular immortalization.
In vitro binding of HPV16 E6 mutants to E6TP1.
We and others have previously characterized a large panel of HPV16 E6 mutants, and the abilities of these mutants to bind to E6AP, E6BP, and p53 have been well defined (6–8, 15, 21). Here, we have utilized this panel of E6 mutants to analyze the requirement for E6TP1 degradation in E6-induced immortalization of MECs.
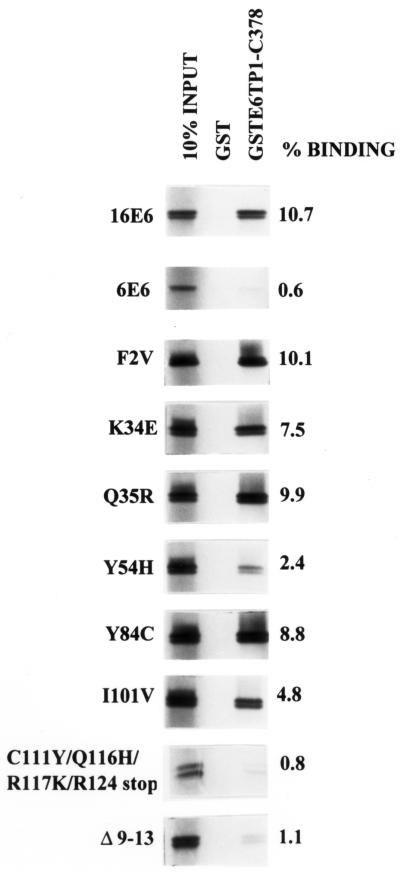
As a first step, we examined the ability of various E6 mutants to bind to E6TP1, an interaction mediated by the C-terminal 194 amino acids of E6TP1 (9). Binding between E6 proteins and E6TP1 was assessed in vitro using a glutathione S-transferase (GST) fusion protein incorporating the C-terminal 378 amino acids of E6TP1 (GST-E6TP1-C378) and proteins generated by in vitro translation of the wild type or mutants of HPV16 E6 in a wheat germ lysate system in the presence of [35S]cysteine, as described previously (9). For each E6 protein, the extent of binding was quantified relative to its input control, which was resolved side by side with the binding reaction. The data are presented as percent binding relative to input of labeled E6 protein as well as relative to binding of wild-type E6 (within parentheses in Table 1). Binding of representative E6 mutants to GST-E6TP1-C378 is shown in Fig. 1, and the entire data are summarized in Table 1. Only a small fraction of E6 mutants (C111Y/Q116H/R117K/R124stop, Q90R/C111R/E113 stop, K94 stop, Δ9-13, and C63R/Y70C/K72R/T86S) show low binding to E6TP1 (binding comparable to that of low-risk HPV6 E6, <20% compared to wild-type HPV16 E6). The vast majority of remaining mutants showed substantial binding to E6TP1, providing a large panel of mutants for analysis of E6TP1 degradation and its correlation with immortalization of MECs.
TABLE 1.
Summary of E6TP1 binding, degradation in vitro and in vivo, p53 degradation in vivo, telomerase activation, and MEC immortalization by HPV16 E6 mutants
| Construct | E6TP1 binding in vitroa | E6TP1 degradationb
|
p53 degradation in vivo in MECsc | Telomerase activationd | MEC immortalizationc | E6 proteine | |
|---|---|---|---|---|---|---|---|
| In vitro | In vivo | ||||||
| HPV16 E6 | 10.7 (100) | + | + | + | + | + | + |
| HPV6 E6 | 0.6 (5.6) | − | −f | − | − | − | +f |
| F2V | 10.1 (94) | + | + | − | + | + | + |
| K34E | 7.5 (70) | +f | +f | + | + | + | +f |
| Q35R | 9.9 (92) | +f | + | + | + | + | + |
| Y54H | 2.4 (22.4) | + | + | − | + | + | + |
| Y84C | 8.8 (82.2) | +f | + | + | + | + | + |
| I101V | 4.8 (44.8) | +f | +f | + | + | + | +f |
| R124G/H126R | 4.1 (38.3) | − | − | − | − | − | + |
| W132R | 6.8 (63.5) | − | − | − | − | − | + |
| C111Y/Q116H/R117K/R124stop | 0.8 (7.4) | − | − | − | − | − | + |
| Q90R/C111R/E113stop | 2.1 (19.6) | −f | −f | − | − | − | +f |
| K94stop | 2 (18.6) | − | − | − | − | − | − |
| C63S | 3.5 (32.7) | −f | −f | − | − | − | +f |
| C63R/Y70C/K72R/T86S | 0.6 (5.6) | −f | −f | − | − | − | +f |
| C103R/D120G/I128M/R131P | 6.1 (57) | −f | −f | − | − | − | +f |
| C136G | 10.5 (98) | − | − | −g | − | −g | + |
| Δ9-13 | 1.1 (10.2) | − | − | − | − | − | + |
| Δ73-77 | 10.8 (101) | − | − | − | − | − | + |
| Δ101-105/N127D | 5.8 (54.2) | − | − | −g | − | −g | + |
| Δ106-110/E113G | 8.6 (80.3) | − | − | −g | − | −g | + |
| Δ111-115/N127K | 7.2 (67.3) | − | − | − | − | − | + |
| Δ118-122 | 12.8 (119) | − | + | + | +h | + | + |
| Δ123-127 | 6.7 (62.9) | − | − | − | − | − | + |
| Δ128-132 | 6.4 (59.8) | − | − | − | − | − | + |
| Δ143-147 | 11.3 (105) | + | + | +g | + | +g | + |
Binding of GST-E6TP1-C378 to HPV16 E6 or its mutants (Fig. 1). Percent binding was calculated by densitometry in comparison with input signals. Values in parentheses are percent binding compared to wild-type HPV16 E6 (considered 100%).
Data for in vitro and in vivo degradation of E6TP1 are from three independent experiments (Fig. 2 and 3).
Induction of telomerase activity from three independent transient-transfection experiments (Fig. 5).
E6 expression data from Fig. 3B and reference 9. + and −, ability and inability, respectively, of a mutant E6 protein to function in the assay indicated at the top of the column.
Data from reference 9.
Data from experiments done in this study.
FIG. 1.
In vitro binding of wild-type and mutant HPV E6 proteins to E6TP1. The HPV16 E6, HPV6 E6, and HPV16 E6 mutant proteins were generated by in vitro translation in the presence of [35S]cysteine using a wheat germ lysate-based coupled transcription-translation system. The 35S-labeled in vitro-translated proteins were incubated with 1 μg of GST or GST-E6TP1-C378 fusion proteins in 300 μl of lysis buffer for 2 h at 4°C. Bound 35S-labeled proteins were resolved by SDS–17% PAGE and visualized by fluorography. The first lane in each panel represents 10% of the in vitro-translated protein used for the binding assay. The films were scanned with an Epson Expression 800 scanner (Torrance, Calif.), and the density of the bands was quantified with NIH Image. Percent binding (shown on the right) was calculated in comparison with signals of 10% input lane.
In vitro and in vivo degradation of E6TP1 protein by HPV16 E6 mutants.
Earlier analysis of a small set (four immortalizing and four nonimmortalizing) of HPV16 E6 mutants suggested a correlation between degradation of E6TP1 and immortalization of MECs by E6. To more critically establish this correlation, we examined the entire panel of E6 mutants used above (8 immortalizing and 16 nonimmortalizing) for their ability to induce E6TP1 degradation.
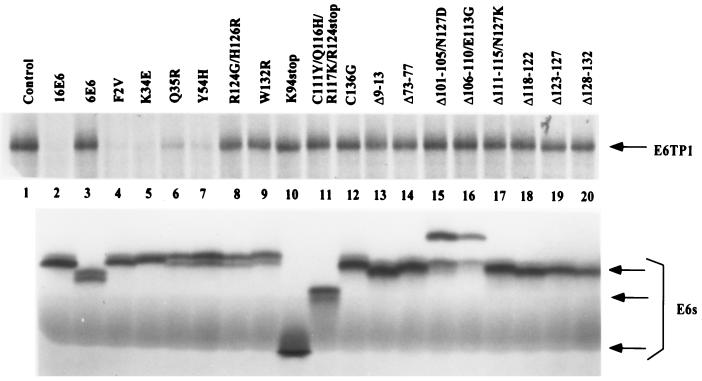
To assess the ability of E6 and its mutants to induce E6TP1 degradation in vitro, both the E6 proteins and E6TP1 were in vitro translated in the presence of [35S]cysteine. The labeled proteins were incubated together overnight at 30°C, and the level of labeled E6TP1 remaining at the end of incubation with E6 protein was determined in comparison with incubation in the absence of E6, as described earlier (9). As shown in Fig. 2, upper panel, and summarized in Table 1, incubation of E6TP1 with wild-type HPV16 E6 led to its complete degradation within the incubation period utilized (Fig. 2, lane 2, upper panel). In contrast, and as anticipated (9), no degradation of E6TP1 was observed upon incubation with HPV6 E6 (Fig. 2, lane 3, upper panel). Out of 12 amino acid substitution mutants, six were able to induce degradation of E6TP1, whereas the other six were defective (Fig. 2, upper panel, and Table 1). The three truncation mutants of E6 (C111Y/Q116H/R117K/R124stop, K94stop, and Q90R/C111R/E113stop) were defective in inducing degradation of E6TP1. Only one of nine small deletion mutants of E6, Δ143-147, was able to induce E6TP1 degradation in vitro (Fig. 2 and Table 1). All of the E6 mutants were expressed at comparable levels, as shown in Fig. 2, lower panel. Thus, representative panels of E6 mutants that were either capable or incapable of inducing E6TP1 degradation in vitro were defined.
FIG. 2.
In vitro degradation of E6TP1 by wild-type and mutant HPV E6 proteins. (Upper panel) HPV16 E6, HPV6 E6, HPV16 E6 mutants, and E6TP1 were translated in vitro in rabbit reticulocyte lysate in the presence of [35S]cysteine. Aliquots of 35S-labeled E6TP1 were incubated with water-primed lysate (control) or indicated wild-type or mutant E6 proteins overnight at 30°C in a 10-μl reaction mixture. The E6TP1 remaining at the end of the degradation assay was resolved by SDS–6% PAGE and visualized by fluorography. The arrow indicates the E6TP1 protein. (Lower panel) Aliquots of HPV16 E6, HPV6 E6, and HPV16 E6 mutant proteins were resolved by SDS–17% PAGE and visualized by fluorography. Arrows indicate different sizes of E6 proteins.
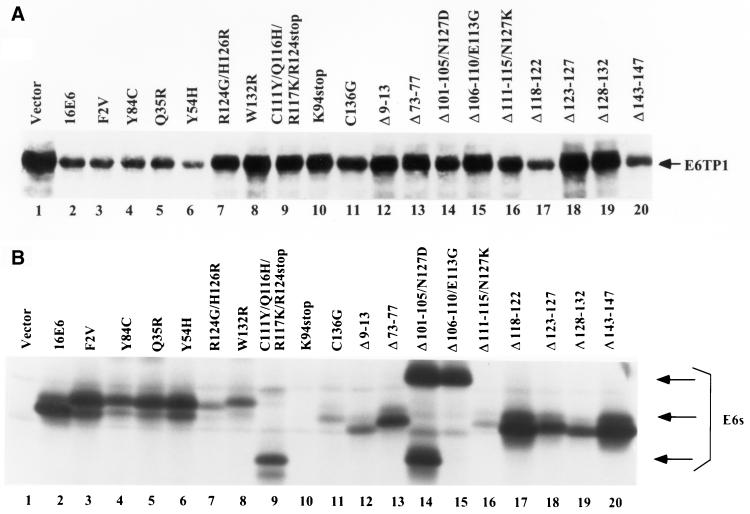
Next, we examined the ability of HPV16 E6 mutants to induce E6TP1 degradation in vivo, when expressed in mammalian cells. This was particularly important since previous studies have shown discrepancies between E6-induced p53 degradation in vitro and that in vivo (8, 21). pSG5 expression constructs encoding E6TP1 and various HPV16 E6 mutants were cotransfected into 293T cells by the calcium phosphate method (33), cells were lysed after 48 h, and the lysates were subjected to immunoblotting using a rabbit anti-E6TP1 antibody. Under the conditions utilized, immunoblotting with this antibody did not detect endogenous E6TP1 in 293T cells (data not shown), allowing an assessment of the effect of E6 coexpression on the levels of introduced E6TP1. As shown in Fig. 3A, all of the E6 mutants capable of inducing degradation of E6TP1 protein in vitro were capable of inducing degradation of E6TP1 in vivo as shown by a marked decrease in the steady-state E6TP1 levels. Notably, however, one E6TP1-binding E6 mutant, Δ118-122, which was unable to induce degradation of E6TP1 in vitro, reproducibly induced a marked decrease in E6TP1 levels in vivo. This result is reminiscent of previous results, which show discrepancies between in vitro and in vivo p53 degradation by certain E6 mutants (8, 21). Importantly, all the E6 mutants that could induce E6TP1 degradation either in vitro or in vivo were able to bind to E6TP1 to some extent. However, our analyses indicate that binding of E6 to E6TP1 is not sufficient for E6TP1 degradation. Six deletion mutants (Δ73-77, Δ101-105/N127D, Δ106-110/E113G, Δ111-115/N127K, Δ123-127, and Δ128-132) and three point mutants (C103R/D120G/I128M/R131P, C136G, and W132R) could bind to E6TP1 efficiently but were unable to target E6TP1 for degradation (Fig. 3A and Table 1). Similar results were obtained when these mutants were inoculated onto another cell line, C33A, a cervical carcinoma cell line (data not shown). These data suggest that binding of E6 to E6TP1 is required but not sufficient to target E6TP1 for degradation. All E6 mutants except K94stop used in this study are expressed in 293T cells (Fig. 3B). Although the expression levels of E6 protein varied among different E6 mutants, there was no correlation between the amount of E6 protein and the abilities of the mutants to degrade E6TP1 (Fig. 3 and Table 1).
FIG. 3.
(A) In vivo degradation of E6TP1 by wild-type and mutant HPV E6 proteins. 293T cells were plated overnight at 2 × 106 cells per 100-mm-diameter dish (two dishes for each construct) and cotransfected with 10 μg of pSG5-E6TP1 and 10 μg each of the indicated E6 mutants in pSG5 vector using the calcium phosphate coprecipitation method. The total DNA amount was kept constant at 20 μg. The cells in one dish each were harvested 48 h after transfection, and 100-μg aliquots of lysate were fractionated by SDS–6% PAGE and transferred to a polyvinylidene difluoride membrane. Membranes were immunoblotted with rabbit anti-E6TP1 antibody followed by enhanced chemiluminescence detection. The arrow indicates the E6TP1 protein. (B) Expression of mutant E6 protein in transfected 293T cells. Paired dishes of 293T transfectants shown above were labeled with [35S]cysteine, and lysates were immunoprecipitated with an anti-E6 antibody. Bound proteins were resolved by SDS–12% PAGE and visualized by fluorography. The nature of multiple-size polypeptide seen in certain lanes is undetermined.
The panel of E6 mutants well characterized with respect to E6TP1 (above) and p53 (7–9, 15, 21; also Table 1) degradation allowed a direct correlation between the loss of E6TP1 versus the loss of p53 and E6-induced cellular immortalization. As summarized in Table 1, a direct one-to-one correlation between in vivo E6-induced loss of E6TP1 and E6-induced immortalization of MECs is observed. Interestingly, the one E6 mutant (Δ118-122) that did not induce detectable E6TP1 degradation in vitro but did so in vivo is capable of immortalizing MECs. None of those mutants that failed to induce E6TP1 degradation in vivo were capable of efficiently immortalizing MECs. Three E6 mutants, W132R, C63S, and R124G/H126R, which were unable to degrade E6TP1 either in vitro or in vivo, were able to inefficiently immortalize MECs. As shown earlier (7), W132R immortalized MECs in one out of four experiments, C63S immortalized MECs in one out of three experiments, and R124G/H126R immortalized MECs in one out of six experiments (7). Furthermore, immortalization with these mutants was preceded by a long crisis period, whereas wild-type E6 and other immortalizing mutants induced immortalization without any crisis period (7). Thus, E6TP1 degradation correlates with efficient E6-induced immortalization of MECs.
Notably, the correlation between E6-induced loss of E6TP1 and MEC immortalization emerged as stricter than even that between E6-induced p53 degradation and immortalization (Table 1). Interestingly, two of the E6 mutants used here, F2V and Y54H, are known to be defective for p53 degradation (21). As shown in Fig. 2 and 3, F2V and Y54H targeted E6TP1 for degradation to an extent similar to that with wild-type HPV16 E6 in both in vitro and in vivo assays.
Enhanced degradation of E6TP1 protein in the presence of HPV16 E6.
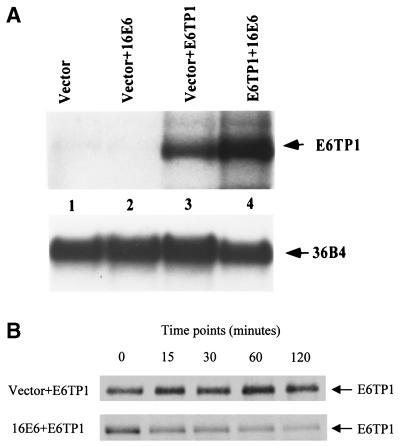
The experiments described above showed that E6 was able to induce a dramatic decrease in the levels of E6TP1 protein. To further evaluate if the lower levels of E6TP1 protein in the presence of E6 are due to protein degradation or to change at the mRNA level, the following experiments were done. 293T cells were transfected with E6TP1 with or without E6. As shown in Fig. 4A, similar levels of E6TP1 mRNA were observed in two sets of transfectants, suggesting that the E6-induced decrease in E6TP1 levels was not due to decreased transcription.
FIG. 4.
(A) E6TP1 mRNA expression in the presence or absence of HPV16 E6. 293T cells were cotransfected as described above with E6TP1 and vector or HPV16 E6. After 48 h, total RNA was prepared by the guanidinium-isothiocyanate method. Twenty micrograms of total RNA from these cells was probed with 32P-labeled full-length E6TP1 probe and visualized by autoradiography. 36B4 was used as a loading control. (B) Pulse-chase analysis of E6TP1. 293T cells were cotransfected with 2.5 μg of HA-tagged E6TP1 and 5 μg of vector or HPV16 E6. After 48 h, cells were metabolically pulse-labeled with 300 μCi of [35S]methionine plus [35S]cysteine for 30 min and chased for the indicated time periods (shown in minutes). Equal amounts of radiolabeled lysates (based on the amount of total protein) were immunoprecipitated with anti-HA antibody (12CA5) and analyzed by SDS-PAGE, followed by fluorography.
Despite our repeated efforts, we have thus far been unable to generate an anti-E6TP1 antibody of a sufficient titer to recognize the endogenous E6TP1 protein. We have so far made over 10 antipeptide antibodies and one GST-E6TP1 fusion protein antibody, but all of these antibodies were not suitable for detection of the endogenous E6TP1 protein. Therefore, to determine the E6TP1 protein stability, pSG5 expression constructs encoding hemagglutinin (HA)-tagged E6TP1 and HPV16 E6 were cotransfected into 293T cells. After 48 h, cells were metabolically labeled with [35S]methionine and [35S]cysteine for 30 min, followed by chase for various time periods. Equal amounts of radiolabeled lysates (based on the amount of total protein) were immunoprecipitated with anti-HA antibody (12CA5) and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), followed by fluorography. The results of a representative experiment are shown in Fig. 4B. E6TP1 in E6-cotransfected cells showed a dramatic decrease in the protein turnover (half-life of approximately 15 min) compared to that in vector-cotransfected cells (half-life of >2 h) (Fig. 4B). These data clearly demonstrate that E6 induces degradation of E6TP1 protein.
Effect of HPV16 E6 mutants on telomerase activity upon transient transfection in MECs.
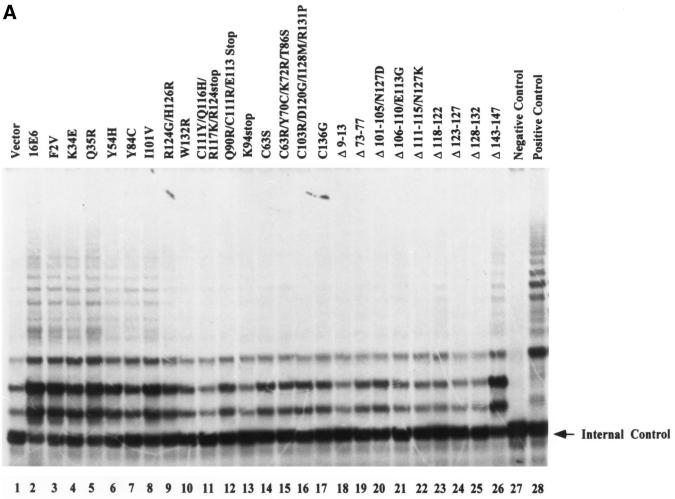
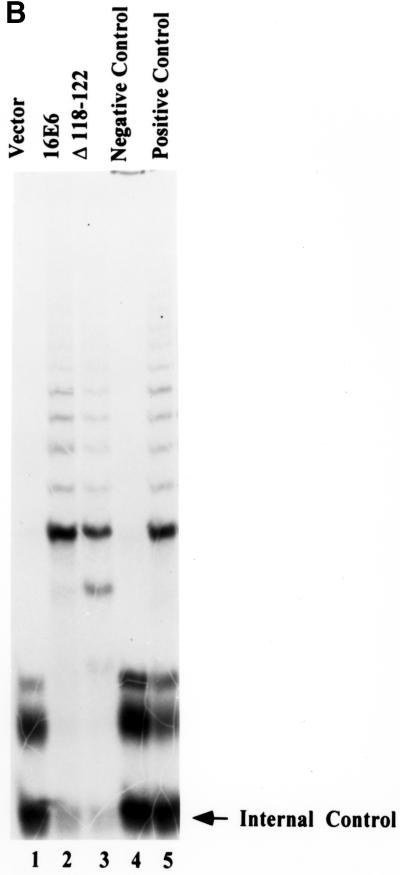
Recent reports indicate that, while p53 degradation and binding to hDlg or E6BP did not fully correlate with E6-induced immortalization of MECs, the ability of E6 to induce telomerase activity showed a stronger correlation (15, 21). Therefore, we wished to characterize the E6 mutants used in our analyses for their ability to induce telomerase activity so that the importance of E6TP1 degradation relative to the induction of telomerase could be compared in a single study. In order to reliably compare the abilities of E6 mutants to induce telomerase activity independent of their ability to induce immortalization, we established a transient-transfection protocol using Fugene-6 to transfect normal MECs. This method led to reproducible transfection of more than 20% of cells when cells were analyzed 48 h after transfection of a β-galactosidase reporter (data not shown). Using this transfection protocol, wild-type HPV16 E6 or its mutants were introduced into normal MEC strain 76N, and telomerase activity was assayed 48 h after transfection. As shown in Fig. 5 and summarized in Table 1, similar to wild-type HPV16 E6, all of the E6 mutants capable of immortalizing MECs (F2V, K34E, Q35R, Y54H, Y84C, I101V, and Δ143-147) induced telomerase activity. As expected, all of the nonimmortalizing mutants failed to induce telomerase activity. The mutants that were able to induce telomerase activity included F2V and Y54H, which did not induce p53 degradation but were able to induce E6TP1 degradation. One mutant, Δ118-122, which was previously reported not to activate telomerase in a transient expression system, also did not induce telomerase activity in our transient-transfection system (Fig. 5A). However, analysis of Δ118-122-immortalized MECs clearly showed telomerase activity comparable to that observed in wild-type E6-immortalized cells (Fig. 5B). These results indicate that transient-transfection experiments may not be sensitive enough to detect low-level induction of telomerase by certain E6 proteins. Importantly, Δ118-122 was able to induce E6TP1 degradation and to immortalize MECs. Overall, our data indicate strong concordance between the induction of telomerase activity and E6TP1 degradation on one hand and efficient E6-induced immortalization of MECs on the other.
FIG. 5.
Induction of telomerase activity in MECs upon introduction of the wild-type or mutant HPV E6 proteins. (A) Normal MECs (76N) were plated at 5 × 105 per 100-mm-diameter dish and transfected with 5 μg of pSG5 constructs encoding wild-type E6 or its mutants using Fugene-6 reagent. At 48 h after transfection, the cells were harvested in trypsin-EDTA, counted, and lysed. Telomerase activity was measured using the TRAP assay method (Intergen, Purchase, N.Y.). Lysates from 20,000 cells were used for each assay. The negative control was lysis buffer, and the positive control was lysates from 500 telomerase-positive cells, provided by the manufacturer. The arrow indicates internal control. (B) Comparison of telomerase activities of MECs immortalized by Δ118-122 at passage 10 (about 30 population doublings) and MECs immortalized by wild-type E6 at the same passage. The experimental conditions were the same as described for panel A.
Although the exact mechanism of E6-induced E6TP1 degradation and its role in MEC transformation are not yet understood, we speculate that E6-induced immortalization may involve deregulation of Rap signaling pathways, and by implication, E6TP1, a negative regulator of Rap GTPase, may function as a negative regulator of the mitogenic signaling pathway mediated by Rap. E6-targeted degradation of E6TP1 would then be expected to promote mitogenic signaling and to facilitate immortalization by E6. The tight linkage of E6TP1 degradation and telomerase induction during E6-induced immortalization of MECs is intriguing. At present, there is no direct biochemical connection between these pathways. Given that E6TP1 is likely a regulator of Rap, a Ras-like small G protein involved in regulating cell proliferation, a signaling pathway connecting Rap and regulation of telomerase activity may exist. Further analyses will be required to establish if such a connection indeed exists.
Acknowledgments
This work was supported by NIH grants CA64823, CA70195, and CA81076 to V.B. Ajay Kumar is a recipient of a fellowship from the Massachusetts Department of Public Health.
We thank Elliot Androphy and Karen Vousden for E6 mutants and Hamid Band for critical reading of the manuscript.
REFERENCES
- 1.Altschuler D L, Ribeiro-Neto F. Mitogenic and oncogenic properties of the small G protein Rap1b. Proc Natl Acad Sci USA. 1998;95:7475–7479. doi: 10.1073/pnas.95.13.7475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baker C C, Phelps W C, Lindgren V, Braun J M, Gonda A M, Howley P M. Structural and transcriptional analysis of human papillomavirus type 16 sequences in cervical carcinoma cell lines. J Virol. 1987;61:962–971. doi: 10.1128/jvi.61.4.962-971.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Band V, De Caprio J A, Delmolino L, Kulesa V, Sager R. Loss of p53 protein in human papillomavirus type 16 E6-immortalized human mammary epithelial cells. J Virol. 1991;65:6671–6676. doi: 10.1128/jvi.65.12.6671-6676.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Band V, Dalal S, Delmolino L, Androphy E J. Enhanced degradation of p53 protein in HPV-6 and BPV-1 E6-immortalized human mammary epithelial cells. EMBO J. 1993;12:1847–1852. doi: 10.1002/j.1460-2075.1993.tb05833.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen J J, Reid C E, Band V, Androphy E J. Interaction of papillomavirus E6 oncoproteins with a putative calcium-binding protein. Science. 1995;269:529–531. doi: 10.1126/science.7624774. [DOI] [PubMed] [Google Scholar]
- 6.Crook T, Tidy J A, Vousden K H. Degradation of p53 can be targeted by HPV E6 sequences distinct from those required for p53 binding and trans-activation. Cell. 1991;67:547–556. doi: 10.1016/0092-8674(91)90529-8. [DOI] [PubMed] [Google Scholar]
- 7.Dalal S, Gao Q, Androphy E J, Band V. Mutational analysis of human papillomavirus type 16 E6 demonstrates that p53 degradation is necessary for immortalization of mammary epithelial cells. J Virol. 1996;70:683–688. doi: 10.1128/jvi.70.2.683-688.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foster S A, Demers G W, Etscheid B G, Galloway D A. The ability of human papillomavirus E6 proteins to target p53 for degradation in vivo correlates with their ability to abrogate actinomycin D-induced growth arrest. J Virol. 1994;68:5698–5705. doi: 10.1128/jvi.68.9.5698-5705.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao Q, Srinivasan S, Boyer S, Wazer D, Band V. The E6 oncoproteins of high-risk papillomaviruses bind to a novel putative GAP protein, E6TP1, and target it for degradation. Mol Cell Biol. 1999;19:733–744. doi: 10.1128/mcb.19.1.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9a.Gao Q, Kumar A, Srinivasan S, Singh L, Mukai H, Ono Y, Wazer D E, Band V. PKN binds and phosphorylates human papillomavirus E6 oncoprotein. J Biol Chem. 2000;275:14824–14830. doi: 10.1074/jbc.275.20.14824. [DOI] [PubMed] [Google Scholar]
- 10.Hawley-Nelson P, Vousden K H, Hubbert N L, Lowy D R, Schiller J T. HPV16 E6 and E7 proteins cooperate to immortalize human foreskin keratinocytes. EMBO J. 1989;8:3905–3910. doi: 10.1002/j.1460-2075.1989.tb08570.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huibregtse J M, Scheffner M, Howley P M. A cellular protein mediates association of p53 with the E6 oncoprotein of human papillomavirus types 16 or 18. EMBO J. 1991;10:4129–4135. doi: 10.1002/j.1460-2075.1991.tb04990.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huibregtse J M, Scheffner M, Howley P M. Cloning and expression of the cDNA for E6-AP, a protein that mediates the interaction of the human papillomavirus E6 oncoprotein with p53. Mol Cell Biol. 1993;13:775–784. doi: 10.1128/mcb.13.2.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huibregtse J M, Scheffner M, Howley P M. Localization of the E6-AP regions that direct human papillomavirus E6 binding, association with p53, and ubiquitination of associated proteins. Mol Cell Biol. 1993;13:4918–4927. doi: 10.1128/mcb.13.8.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kitayama H, Sugimoto Y, Matsuzaki T, Ikawa Y, Noda M. A ras-related gene with transformation suppressor activity. Cell. 1989;56:77–84. doi: 10.1016/0092-8674(89)90985-9. [DOI] [PubMed] [Google Scholar]
- 15.Kiyono T, Foster S A, Koop J I, McDougall J K, Galloway D A, Klingelhutz A J. Both Rb/p16INK4a inactivation and telomerase activity are required to immortalize human epithelial cells. Nature. 1998;396:84–88. doi: 10.1038/23962. [DOI] [PubMed] [Google Scholar]
- 16.Kiyono T, Hiraiwa A, Fujita M, Hayashi Y, Akiyama T, Ishibashi M. Binding of high-risk human papillomavirus E6 oncoproteins to the human homologue of the drosophila discs large tumor suppressor protein. Proc Natl Acad Sci USA. 1997;94:11612–11616. doi: 10.1073/pnas.94.21.11612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Klingelhutz A J, Foster S A, McDougall J K. Telomerase activation by the E6 gene product of human papillomavirus type 16. Nature. 1996;380:79–82. doi: 10.1038/380079a0. [DOI] [PubMed] [Google Scholar]
- 18.Kuhne C, Banks L. E3-ubiquitin ligase/E6-AP links multicopy maintenance protein 7 to the ubiquitination pathway by a novel motif, the L2G box. J Biol Chem. 1998;273:34302–34309. doi: 10.1074/jbc.273.51.34302. [DOI] [PubMed] [Google Scholar]
- 19.Kurachi H, Wada Y, Tsukamoto N, Maeda M, Kubota H, Hattori M, Iwai K, Minato N. Human SPA-1 gene product selectively expressed in lymphoid tissues is a specific GTPase-activating protein for Rap1 and Rap2. Segregate expression profiles from a rap1GAP gene product. J Biol Chem. 1997;272:28081–28088. doi: 10.1074/jbc.272.44.28081. [DOI] [PubMed] [Google Scholar]
- 20.Lee S S, Weiss R S, Javier R T. Binding of human virus oncoproteins to hDlg/SAP97, a mammalian homolog of the Drosophila discs large tumor suppressor protein. Proc Natl Acad Sci USA. 1997;94:6670–6675. doi: 10.1073/pnas.94.13.6670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y, Chen J J, Gao Q, Dalal S, Hong Y, Mansur C P, Band V, Androphy E J. Multiple functions of human papillomavirus type 16 E6 contribute to the immortalization of mammary epithelial cells. J Virol. 1999;73:7297–7307. doi: 10.1128/jvi.73.9.7297-7307.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Munger K, Phelps W C, Bubb V, Howley P M, Schlegel R. The E6 and E7 genes of the human papillomavirus type 16 together are necessary and sufficient for transformation of primary human keratinocytes. J Virol. 1989;63:4417–4421. doi: 10.1128/jvi.63.10.4417-4421.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patel D, Huang S M, Baglia L A, McCance D J. The E6 protein of human papillomavirus type 16 binds to and inhibits co-activation by CBP and p300. EMBO J. 1999;18:5061–5072. doi: 10.1093/emboj/18.18.5061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ronco L, Karpova A, Vidal M, Howley P M. Human papillomavirus 16 E6 oncoprotein binds to interferon regulatory factor-3 and inhibits its transcriptional activity. Genes Dev. 1998;12:2061–2072. doi: 10.1101/gad.12.13.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rubinfeld B, Munemitsu S, Clark R, Conroy L, Watt K, Crosier W J, McCormick F, Polakis P. Molecular cloning of a GTPase activating protein specific for the Krev-1 protein p21rap1. Cell. 1991;65:1033–1042. doi: 10.1016/0092-8674(91)90555-d. [DOI] [PubMed] [Google Scholar]
- 26.Scheffner M, Huibregtse J M, Vierstra R D, Howley P M. The HPV-16 E6 and E6-AP complex functions as a ubiquitin-protein ligase in the ubiquitination of p53. Cell. 1993;75:495–505. doi: 10.1016/0092-8674(93)90384-3. [DOI] [PubMed] [Google Scholar]
- 27.Schwarz E, Freese U K, Gissmann L, Mayer W, Roggenbuck B, Stremlau A, zur Hausen H. Structure and transcription of human papillomavirus sequences in cervical carcinoma cells. Nature. 1985;314:111–114. doi: 10.1038/314111a0. [DOI] [PubMed] [Google Scholar]
- 28.Thomas M, Banks L. Inhibition of Bak-induced apoptosis by HPV-18 E6. Oncogene. 1998;17:2943–2954. doi: 10.1038/sj.onc.1202223. [DOI] [PubMed] [Google Scholar]
- 29.Tong X, Howley P M. The bovine papillomavirus E6 oncoprotein interacts with paxillin and disrupts the actin cytoskeleton. Proc Natl Acad Sci USA. 1997;94:4412–4417. doi: 10.1073/pnas.94.9.4412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vossler R M, Yao H, York R D, Pan G M, Rim C S, Stork P J. cAMP activates MAP kinase and Elk-1 through a B-Raf- and Rap1-dependent pathway. Cell. 1997;89:73–82. doi: 10.1016/s0092-8674(00)80184-1. [DOI] [PubMed] [Google Scholar]
- 31.Wazer D E, Liu X L, Chu Q, Gao G, Band V. Immortalization of distinct human mammary epithelial cell types by human papilloma virus 16 E6 or E7. Proc Natl Acad Sci USA. 1995;92:3687–3691. doi: 10.1073/pnas.92.9.3687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wienecke R, Konig A, DeClue J E. Identification of tuberin, the tuberous sclerosis-2 product. Tuberin possesses specific Rap1GAP activity. J Biol Chem. 1995;270:16409–16414. doi: 10.1074/jbc.270.27.16409. [DOI] [PubMed] [Google Scholar]
- 33.Wigler M, Pellicer A, Silverstein S, Axel R, Urlaub G, Chasin L. DNA-mediated transfer of the adenine phosphoribosyltransferase. Proc Natl Acad Sci USA. 1979;76:1373–1376. doi: 10.1073/pnas.76.3.1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshida Y, Kawata M, Miura Y, Musha T, Sasaki T, Kikuchi A, Takai Y. Microinjection of smg/rap1/Krev-1 p21 into Swiss 3T3 cells induces DNA synthesis and morphological changes. Mol Cell Biol. 1992;12:3407–3414. doi: 10.1128/mcb.12.8.3407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zimmermann H, Degenkolbe R, Bernard H U, O'Connor M J. The human papillomavirus type 16 E6 oncoprotein can down-regulate p53 activity by targeting the transcriptional coactivator CBP/p300. J Virol. 1999;73:6209–6219. doi: 10.1128/jvi.73.8.6209-6219.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zur Hausen H. Papillomaviruses in human cancer. Appl Pathol. 1987;5:19–24. [PubMed] [Google Scholar]
- 37.Zur Hausen H, Salzman N P, editors. The Papovaviridae. 2. The Papillomaviruses. New York, N.Y: Plenum Press; 1987. pp. 245–263. [Google Scholar]