Abstract
A novel species of Mucor was identified as the causal agent of a brown rot of Prunus domestica (European plum), widely grown in the south of Xinjiang, China. This disease first appears as red spots after the onset of the fruits. With favorable environmental conditions, fruit with infected spots turn brown, sag, expand, wrinkle, and harden, resulting in fruit falling. Fungal species were isolated from infected fruits. A phylogenetic analysis based on internal transcribed spacer (ITS) regions and the large subunit (LSU) of the nuclear ribosomal RNA (rRNA) gene regions strongly supported that these isolates made a distinct evolutionary lineage in Mucor (Mucoromycetes, Mucoraceae) that represents a new taxonomic species, herein named as Mucor xinjiangensis. Microscopic characters confirmed that these strains were morphologically distinct from known Mucor species. The pathogenicity of M. xinjiangensis was confirmed by attaching an agar disk containing mycelium on fruits and re-isolation of the pathogen from symptomatic tissues. Later, fourteen fungicides were selected to determine the inhibitory effect on the pathogen. Further, results showed that difenoconazole had the best effect on the pathogen and the strongest toxicity with the smallest half maximal effective concentration (EC50) value, followed by a compound fungicide composed of difenoconazole with azoxystrobin, mancozeb, prochloraz with iprodione, pyraclostrobin with tebuconazole, and trifloxystrobin with tebuconazole and ethhylicin. Present study provides the basis for the prevention and control of the novel plum disease and its pathogen.
Keywords: chemical control, new taxon, plant pathogen, Prunus domestica (European plum), taxonomy, Xinjiang (China)
Introduction
Prunus domestica is a flowering plant species belonging to the family Rosaceae. It is commonly known as European plum, common plum, or prune and produces stone fruits, which are typically called plums (Zhebentyayeva et al., 2019). Prusnusdomestica is a rich source of vitamins, minerals, organic acids, and fiber, including anthocyanins, flavonol derivatives, and phenolic acids (Soares et al., 2023). A number of studies have shown that the consumption of plums promotes a wide range of health benefits; prevents a wide variety of diseases, such as cancer, diabetes, and obesity; has anti-inflammatory properties; improves digestive function; and has significant applications in the fields of medicine and food (Silvan et al., 2020; Abraao et al., 2022; Bahrin et al., 2022; Rybak and Wojdyło, 2023).
Over the past few years, plum-growing areas have increased in Xinjiang, China (Wang et al., 2021). The total plum-growing area (in Chinese pinyin: Xinmei) in Kashgar Jiashi County is more than 28,000 ha, yielding an estimated production volume of 85,000 tons. It accounts for 40% of China’s total area of plum production and 60% of China’s total plum output (Fan and Zhang, 2023). Due to the gradual expansion of the plum growth area, coupled with mismanagement and inadequate technology, plums are prone to diseases and pests such as spot disease, brown rot, and food worms, which negatively influence their growth.
Mucor rot is a common postharvest disease of pome and stone fruits such as apples, cherries, nectarines, pears, peaches, plums, and prunes, as well as other commercial berries and citrus fruits. This disease is mainly caused by Mucor piriformis and a few other Mucor species, such as Mucor circinelloides (Saito et al., 2016), Mucor fragilis (Abbas et al., 2018; Khan and Javaid, 2022), Mucor hiemalis (Saito et al., 2016), Mucor mucedo (Eseigbe and Bankole, 1996; Saito et al., 2016), Mucor racemosus (Kwon and Hong, 2005; López et al., 2016), and Mucor strictus (Suh et al., 2018). However, in cold storage, only M. piriformis has been frequently found to cause significant losses (López et al., 2016; Saito et al., 2016). The causal agent of Mucor rot belongs to the genus Mucor which is the type of subkingdom Mucoromyceta (Mucoromycota, Mucoromycetes, Mucorales, and Mucoraceae), with conserved type M. mucedo (Tedersoo et al., 2018; Turland et al., 2018). Typically, it has fast growth, aerial and luxuriant hyphae, sporangiophores with no branching, and zygospores with opposed suspensors (Schipper, 1978). All over the world, various species of Mucor are widely collected from soil and dung (Walther et al., 2013).
In the realm of agricultural practices, combating fungal diseases remains a critical concern, particularly in safeguarding the health and yield of essential agricultural and fruit crops. Despite advancements in various agricultural techniques, chemical control continues to stand out as a fundamental tool in the fight against fungal pathogens (Kettles and Luna, 2019). Currently, Mucor species associated with plums were mainly reported from Norway (Børve and Vangdal, 2007), Nigeria (Eseigbe and Bankole, 1996), Pakistan (Hassan et al., 2022), Poland (Tuszyński and Satora, 2003), Saudia Arabia (Gherbawy and Hussein, 2010), South Africa (Kwinda et al., 2015), Turkey (Ghimire et al., 2022), and USA (Hong et al., 2000). Despite the advancements in agricultural practices, the control of Mucor species remains a significant concern for farmers and researchers alike. Plant diseases caused by Mucor species can be prevented and reduced by various management strategies. There have been some reports to control these diseases caused by fungi, such as Mucor species, with fungicides (Koka et al., 2021; Saito et al., 2023), plant extracts (Kinge and Besong, 2021; Jangid and Begum, 2022), and biological control agents (Wallace et al., 2018; Oufensou et al., 2023). The Mucor species associated with plums in China have not been reported. In our investigation of plum diseases and their management in China, a total of 37 Mucor strains were isolated. Based on the morphological and phylogenetic analysis, these isolates were identified as Mucor xinjiangensis sp. nov. A detailed description and illustration are provided for the new species and compared with other closely related taxa. Pathogenicity test confirmed that M. xinjiangensis causative agent of brown rot of plum. Further, 14 fungicides were analyzed to check the inhibitory effect on isolated pathogens.
Materials and methods
Sample collection and isolation
Samples were collected during plum disease surveys conducted between 2019 and 2020, from affected orchards in Yingmaili Township (39° 31′ 11″ N 76° 55′ 17″ E), Jiashi County, Kashgar Prefecture, Xinjiang, China. We screened out 15 orchards with disease symptoms, and a total of 28,972 fruits were investigated. A total of 10 trees were observed in each orchard, and a 5-point sampling method was used for the observation (Ni et al., 2008). Disease incidence for each orchard was calculated by observing diseased fruits/total number of fruits observed. A total of 200 diseased fruits were collected and brought to the laboratory. Among them, 70 diseased fruits with typical diseased symptoms were separated for fungal isolation. When conidia were visible, single-spore isolation was performed (Zhang et al., 2013; Brahmanage et al., 2020). Alternatively, diseased tissues were grown on potato dextrose agar (PDA) for tissue isolation, as described by Raza et al. (2019). A total of 37 strains of Mucor species were isolated, and mycelial plugs were stored in 2 mL tubes for long-term storage at 4°C under sterile water. Dry and living cultures were deposited in the Herbarium of Microbiology, Academia Sinica (HMAS), and China General Microbiological Culture Collection Center (CGMCC), respectively. Taxonomic novelty description and nomenclature were deposited in MycoBank.
Culture description
Observations of morphological features were made on 4- to 7-day-old fungal colonies incubated at room temperature (28°C) under near-ultraviolet (near-UV) light with 12-h photoperiod and 12-h darkness. A color guide by Kornerup and Wanscher (1967) was used to describe colony color on PDA. The morphological characters were photographed using a Nikon Eclipse Ci-L light microscope (Yokohama, Japan) and an Oplenic D2000 digital camera (USA). Columellae, chlamydospore, sporangia, sporangiophore, and sporangiospores were also observed on slides mounted in 100% lactic acid.
Genomic DNA extraction, polymerase chain reaction amplification, and sequencing
The genomic deoxyribonucleic acid (gDNA) was extracted using the cetyltrimethylammonium bromide (CTAB) method (Doyle, 1990). Amplification of the internal transcribed spacer (ITS) of isolates was conducted using primer pairs ITS 1 forward (5 TCCGTAGGTGAACCTGCGG-3) and ITS 4 reverse (5 TCCTCCGCTTATTGATATGC-3) (Khan and Javaid, 2022). An initial basic local alignment search tool (BLASTn) analysis was conducted to screen Mucor species based on their ITS sequences. Mucor strains were also amplified and sequenced for a fragment of 28S rRNA gene with primer pairs NL1 and NL4 (Raza et al., 2019). Each locus was amplified using the polymerase chain reaction (PCR) protocol described by Hurdeal et al. (2021) and Zhao et al. (2023). The PCR reaction was performed in a 25-μL reaction volume using a 15-μL rapid Taq master mix (Vazyme, Nanjing, China), 0.1-mM primers, and 10-ng gDNA. PCR was performed in the following conditions: predenaturation at 95°C for 5 min; denaturation at 95°C for 30 s; annealing at 55°C (for ITS) or 56°C (for large subunit [LSU]) for 40 s; extension at 72°C for 45 s, 35 cycles; and elongation at 72°C for 10 min. The PCR products were detected by 1% agarose gel electrophoresis, and sequencing was done by Sangong Bioengineering (Shanghai) Co., Ltd.
Phylogenetic analysis
Phylogenetic relationship and taxonomic distinction for novel species were determined using genetic markers recommended in a recent bibliography of the genus Mucor (Hurdeal et al., 2021; Zhao et al., 2023). A sequence assembly was performed, and necessary corrections were made manually wherever necessary using BioEdit 7.2.5 (Hall, 1999). Bayesian inference (BI) and maximum likelihood (ML) analyses were employed to reconstruct the phylogeny, respectively, with MrBayes 3.2.7 (Ronquist et al., 2012) and RAxML 8.2.10 (Stamatakis, 2014). Based on the Akaike information criterion, MrModeltest 2.3 (Nylander, 2004) was used to estimate the best-fit evolutionary models for the two-locus dataset. The posterior probability (PP) distribution convergence was ensured by running 6,000,000 generations of Markov chain Monte Carlo (MCMC) with a random seed and a stopval = 0.01 MCMC algorithm of four chains. Based on the 50% majority rule and removing the first 25% of the trees sampled, we calculated consensus trees based on the 50% majority rule and PP. It was considered significant if the PP value was greater than 0.95. Selected bootstrap replicates were 1,000, and bootstrap support (BS) ≥70 was considered significant (Li et al., 2023). Sequences generated in this study were deposited in GenBank, and their accession numbers can be found in Table 1.
Table 1.
Reference specimens and their GenBank accession numbers were used for phylogenetic analysis in this study.
T, ET, NT, and IsoT denote type, ex-epitype, ex-neotype, and ex-isotype cultures, respectively. ITS, internal transcribed spacer; LSU, large subunit. Novel taxon is indicated in bold.
Pathogenicity test
Two representative strains of Mucor species (CGMCC 3.27539 and 39Z29) were tested for pathogenicity (Hussain et al., 2016). Selected fresh and healthy new plum fruits were surface sterilized with 75% ethanol, and then a 4-mm diameter wound was made in the middle of the fruit using a wound retractor. Mycelial plugs were taken from the margin of the growing colonies of isolates using a 4-mm diameter cork borer. Fresh wounds were inoculated by placing mycelial plugs into the wounds. A plug of PDA with no fungal growth was included as a control. The inoculated fruits were arranged in a sterile humidity (65–70%) chamber for 7 days, and fruits were observed every day. Twenty fruits were inoculated with two representative strains and repeated 3 times. After the incubation period, disease lesions were measured, and pathogens were re-isolated (Song et al., 2020). The morphological characteristics and ITS sequences of the re-isolated fungus were compared to those of the original strains. The inoculation experiment was conducted 2-times to ensure reliability.
Fungicides and adjuvants against pathogen
A total of 14 fungicides, including 10 fungicides and 4 adjuvants (Table 2), were tested in vitro against the pathogen (type strain) (Hussain et al., 2014). The fungicides selected in this study were registered under the Pesticide Inspection Institute, Ministry of Agriculture and Rural Affairs, China.1 The fungicide solutions were prepared according to the label instructions provided by their manufacturers (Table 2). The recommended concentration was diluted, and 1 mL was added to 49 mL PDA medium and poured into a 9 cm Petri dish. An equal volume of sterile water was added to the control Petri dish. A plug of mycelium (5 mm) was inoculated into a petri dish containing fungicides and kept at 28°C for 7 days with daily checks (Wagner et al., 2019). Each treatment was replicated 4 times. The colony diameter was measured, and the inhibition rate was calculated as follows:
Table 2.
Fungicides and fungicides/adjuvants mixture tested against Mucor xinjiangensis in this study.
| Commercial name (Chinese pinyin) | Active ingredient (AC) | AC rate (g/L) | Dosage form | Recommended dosage by company | Production company |
|---|---|---|---|---|---|
| Lóngdēng tǒng wàng | Carbendazim | 500 | Suspension concentrate | 120–150 mL/acre | Jiangsu Longdeng Chemical Co., Ltd. |
| Bǎi jūn qīng | Chlorothalonil | 750 | Wettable powder | 150–200 g/acre | Shaanxi Hengtian Biological Agriculture Co., Ltd. |
| Shì jié | Difenoconazole | 200 | Emulsion in water | 30–40 mL/acre | Shaanxi Thompson Biotechnology Co. Ltd. |
| Yǐ mì fēn | Ethirimol | 250 | Suspension concentrate | 65–95 mL/acre | Jiangxi Heyi Chemical Co., Ltd. |
| Yībiàn jìng | Ethylicin | 800 | Emulsifiable concentrate | 25–30 g/acre | Henan Kebang Chemical Co., Ltd. |
| Fú guī zuò | Flusilazole | 400 | Emulsifiable concentrate | 10–20 mL/acre | Beijing Agonon Biopharmaceutical Co. Ltd. |
| Ruì pǔ shēng | Mancozeb | 800 | Wettable powder | 150–240 g/acre | Shandong Baishiwei Crop Protection Co., Ltd. |
| Bǎo fēng | Pyraclostrobin | 250 | Suspending agent | 60–120 mL/acre | Anyang City Ruipu Agrochemical Co., Ltd. |
| Wù zuò chún | Tebuconazole | 430 | Suspension concentrate | 20–30 mL/acre | Xi ‘an Dingsheng Bio-Chemical Co., Ltd. |
| Dà fēng tuō | Thiophanatemethyl | 500 | Suspension concentrate | 100–150 mL/acre | Shaanxi Hengtian Biological Agriculture Co., Ltd. |
| Měi shí lè | Difenoconazole + azoxystrobin | 150; 250 | Suspension concentrate | 30–40 mL/acre | Qingdao Hansheng Biotechnology Co., Ltd. |
| Xīng líng | Prochloraz + aprodione | 100; 100 | Suspension concentrate | 200–240 mL/acre | Shaanxi Hengrun Chemical Industry Co., Ltd. |
| Yōu zé shí | Pyraclostrobin + aebuconazole | 250; 430 | Microemulsion | 60–120 mL/acre | Qingdao Hansheng Biotechnology Co., Ltd. |
| Shè xǐ | Trifloxystrobin + aebuconazole | 250; 500 | Water dispersible granule | 12–15 g/acre | Qingdao Odis Biotechnology Co., Ltd. |
In addition, the average growth rate of mycelium in each treatment was used to screen out fungicides that were effective, and the half maximal effective concentration (EC50) value was calculated to measure the toxicity of the agent, using the recommended concentration as the center (used as control). A total of five concentrations were set with four replicates for each concentration.
Statistical analysis
Each measurement was repeated at least 3 times. Dunnett tests were used to compare mean values based on univariate analysis of variance (ANOVA) with Statistical Package for the Social Sciences (SPSS) version 19.0 (IBM) software. Different letters above the bars indicate statistical differences (p < 0.05).
Results
Disease symptoms and incidence
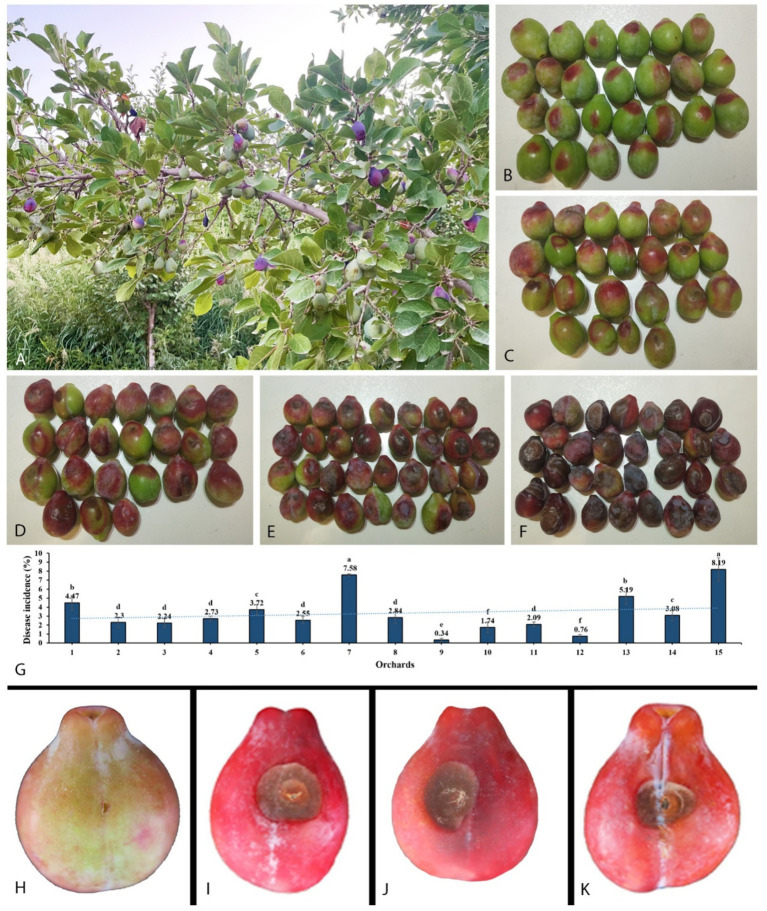
The initial symptom of the newly observed disease was small, scattered red spots appearing after the onset of the fruit (Figures 1A–F). It was found that the disease spreads swiftly when the temperature rises, resulting in brown spots that sag and expand around, wrinkle, and harden, and eventually lead to fruit falling from the tree. At high humidity, especially early in the morning, white mycelia were observed on infected fruits. In our investigation, we found that a total of 1,032 fruits were diseased out of 28,972. The disease incidence caused by Mucor species for each orchard was between 0.31 and 7.63% (Figure 1G).
Figure 1.
Disease development in the field on plum tree and pathogenicity test. (A) Infected plant. (B−F) Gradually disease expansion on fruit collected from different orchards. (G) Disease incidence in 15 orchards. (H) Control. (I) Inoculated with Mucor xinjiangensis, strain CGMCC 3.27539. (J) Inoculated with M. xinjiangensis, strain 39Z29. (K) Inoculated directly with diseased plum tissue collected from the field.
Koch’s postulates for pathogenicity test
A total of 70 diseased fruit samples with typical disease symptoms were collected, from which fungi were isolated in 21 samples. This yielded 60 fungal isolates, of which 37 were morphologically identified as Mucor species, representing 61.67% of the total isolates. Plum fruits inoculated with two representative strains showed brown rot symptoms of 1.5–2 cm after 7 days. The symptoms first consisted of brown spots that expanded. As brown spots develop, the infected area becomes wrinkled and hard, then fades to dark brown. Infected fruits were found to have white mycelia. Fruits infected with the representative strains developed similar symptoms to those observed in the field. There were no visible symptoms of brown rot on the non-inoculated control fruit (Figures 1H–K). Strains were recovered from the inoculated diseased fruit symptoms, which were conspecific to the original isolates from the natural diseased fruits, based on microscopic characteristics and 100% similarity in ITS and LSU sequences.
Phylogenetic analyses
Among the total Mucor isolates, five strains were randomly selected basis of different collection sites (fields) and successfully amplified with single ITS fragments. A preliminary comparison of ITS sequences via BLASTn search showed that each strain belongs to Mucor but representative strains (CGMCC 3.27539 and 39Z29) were most closely related to Mucor pseudolusitanicus (MF495059; 96% query cover and 98% identity and MF495059; 98% query cover and 99% identity, respectively). By analyzing the combined dataset of ITS and LSU loci, strains collected from diseased plums were further identified. A total of 93 reference sequences were used, including 91 Mucor species, and two species of the Backusella were used as outgroups, retrieved from GenBank (Table 1). In the concatenated alignment of five Mucor strains and 93 reference taxa, 957 distinct alignment patterns and 29.22% proportion of gaps and completely undetermined characters. Based on the combined ITS and LSU phylogenetic analyses, our strains formed an exclusive and well-supported clade (93/0.98 for ML/PP) (Figure 2). There was a strong correlation between the topologies of the individual gene trees and the concatenated tree, indicating that both strains recovered from brown spots on plum fruit were distinct species of Mucor.
Figure 2.
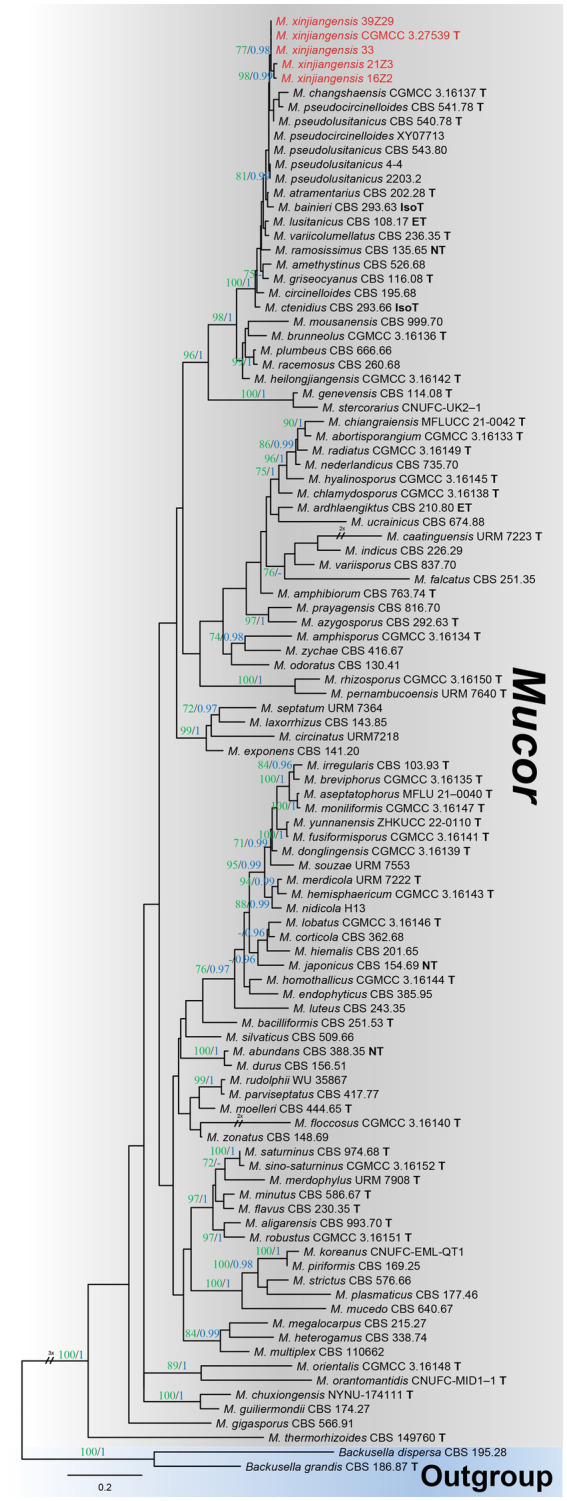
Maximum likelihood (ML) phylogenetic tree inferred from a two-locus concatenated alignment (ITS and LSU). Bootstrap values >70% for ML in green and posterior probability (PP) >0.95 in blue were added on the above and below the branch length (ML/PP). The type, epitype, neotype, and isotype strains were indicated in bold with T, ET, NT, and IsoT, respectively. The strains introduced in this study are represented in red. The tree is rooted using Backusella dispersa (CBS 195.28) and Begonia grandis (CBS 186.87).
Taxonomy
Mucor xinjiangensis B. Song & M. Raza, sp. nov. (Figures 3A–O).
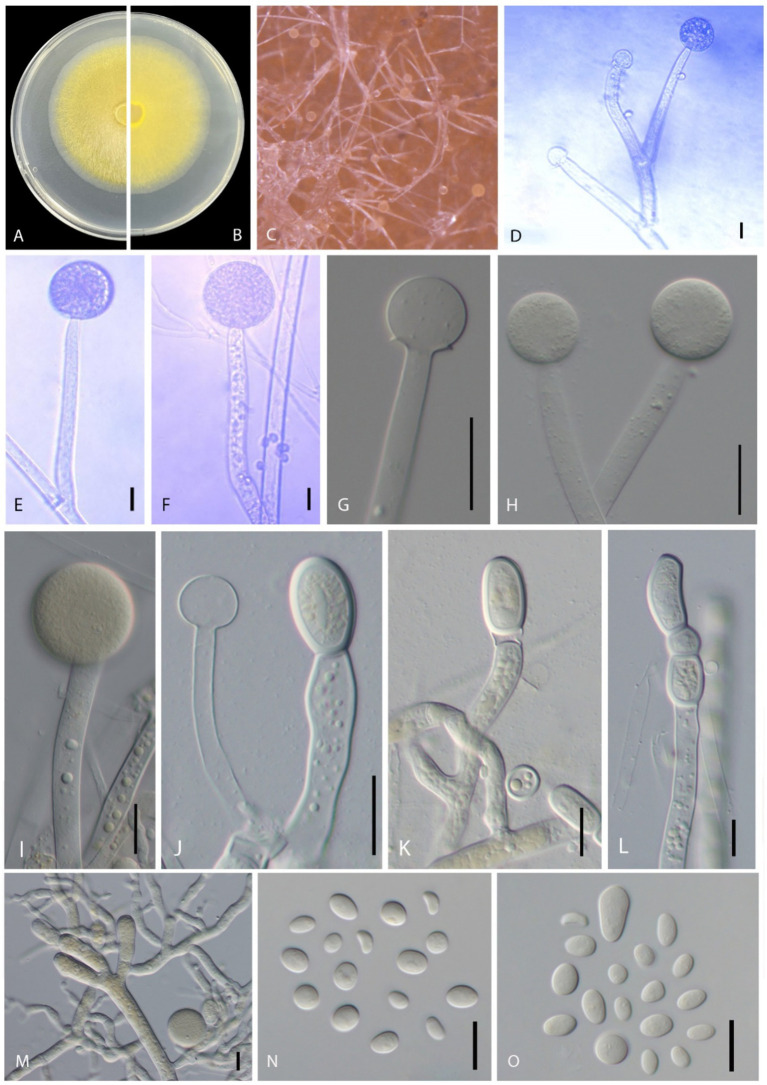
Figure 3.
Disease symptoms and morphological characteristics of Mucor xinjiangensis (CGMCC 3.27539). (A,B) Colony on PDA—(A) from above and (B) from below. (C) Sporulation on PDA. (D,E) Columellae. (F–I) Sporangia. (J) Columella and chlamydospores. (K–M) Chlamydospores. (N–O) Sporangiospores. Scale bars: (D–F,N) = 10 μm; (G–M) = 20 μm.
MycoBank: MB853833.
Etymology: refers to Xinjiang Uyghur Autonomous Region in China from which the holotype was isolated.
Typification: China, Xinjiang Uyghur Autonomous Region, Kashgar prefecture, Jiashi county, on P. domestica (European plum), July 2019, B. Song (HMAS 352969, ex-type living culture 19Z3 = CGMCC 3.27539).
Morphology: Hyphae smooth, branched, aseptate, hyaline to yellowish, 5.5–13 μm diameter. Sporangiophore erects directly from aerial hyphae, small and tall, colorless, simple or 1–2 times sympodially branched, 35–160 μm in length (average = 93.04 ± 40.82 μm), 6–12 μm in diameter (average = 8.70 ± 1.67 μm), branches often subterminal and longer than the main stems, all terminating with a sporangium, non-apophysate below the sporangium. Sporangia non-apophysate, globose to slightly depressed globose, 16–42.5 μm in width (average = 26.88 ± 9.73 μm), the wall is slowly dissolving or broken, grayish brown. Columellae globose or subglobose, 11–19.5 μm width (average = 16.35 ± 2.42 μm), hyaline or pale orange–brown, no collar. Sporangiospores variable in shape, ellipsoidal to obovoid, 4–11 × 3–7 μm (average = 6.89 ± 1.13 × 4.83 ± 0.63 μm) wide, colorless. Chlamydospore occurring in vegetative hyphae, smooth, thin walled, intercalary, single, in pairs or chains, globose, subglobose, 15–35 × 12–19 μm width (average = 21.45 ± 6.09 × 15.37 ± 1.93 μm). Rhizoids present. Zygospores not observed.
Other specimens examined: China, Xinjiang Uyghur Autonomous Region, Kashgar prefecture, Jiashi county, on P. domestica (European plum), July 2019, B. Song, living culture 39Z29.
Cultural characteristics: Colonies on PDA are fast growing, reaching 6.8 cm in diameter in 2 days after incubation at 28 ± 1°C, colony medium, slightly raised with an erose edge, rough surface, effuse, well-defined margin; colony from above; dull, medium, whitish to pale yellow, later blackish; from below, pale yellow; not producing pigment in PDA media. Sporulate on PDA.
Notes: Five strains of Mucor xinjiangensis clustered together and closely related to Mucor changshaensis, Mucor pseudocircinelloides, and Mucor pseudolusitanicus, but type isolate (CGMCC 3.27539) differs in producing smaller sized sporangia (23.5–52 μm in M. changshaensis, up to 90-μm in diameter in M. pseudocircinelloides, up to 75-μm in diameter in M. pseudolusitanicus, and up to 16–42.5 μm in M. xinjiangensis), columellae (10–28.5 × 10.5–28 μm in M. changshaensis, 27–46 × 34–58 μm in M. pseudocircinelloides, 35–52 μm in M. pseudolusitanicus, and 11–19.5 μm in M. xinjiangensis), and larger chlamydospores (8.5–20 × 7–16.5 μm in M. changshaensis, 2.3–26.7 × 9.8–17.4 μm in M. pseudocircinelloides, 10.4–19.7 × 6.7–15.4 μm in M. pseudolusitanicus, and 15–35 × 12–19 μm in M. xinjiangensis). M. pseudocircinelloides and M. pseudolusitanicus produce hyaline to pale brown sporangia, while M. xinjiangensis produces hyaline to grayish brown sporangia (Wagner et al., 2019). In the case of M. changshaensis, these sporangia are light brown to black (Zhao et al., 2023). Our collection (CGMCC 3.27539) ITS loci are 3.2% (18 out of 560 bp) different from both M. changshaensis and M. pseudocircinelloides, while 0.8% (5 out of 560 bp) different from those of M. pseudolusitanicus. For LSU, this difference is 1.32% (9 out of 679 bp), with both M. pseudocircinelloides and M. pseudolusitanicus and LSU of M. changshaensis is not available. Furthermore, M. xinjiangensis produces a white to pale yellow color on PDA compared to those M. changshaensis (light to strontian yellow), M. pseudocircinelloides (white to pale brown, reverse uncolored), and M. pseudolusitanicus (white to light gray, reverse uncolored).
Evaluation of fungicides and fungicide adjuvants
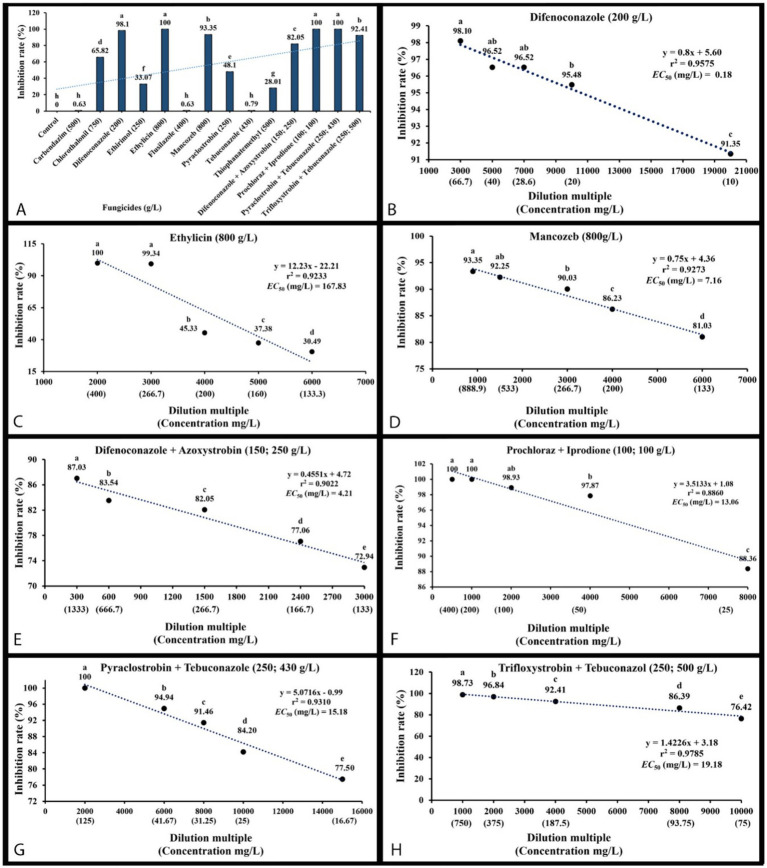
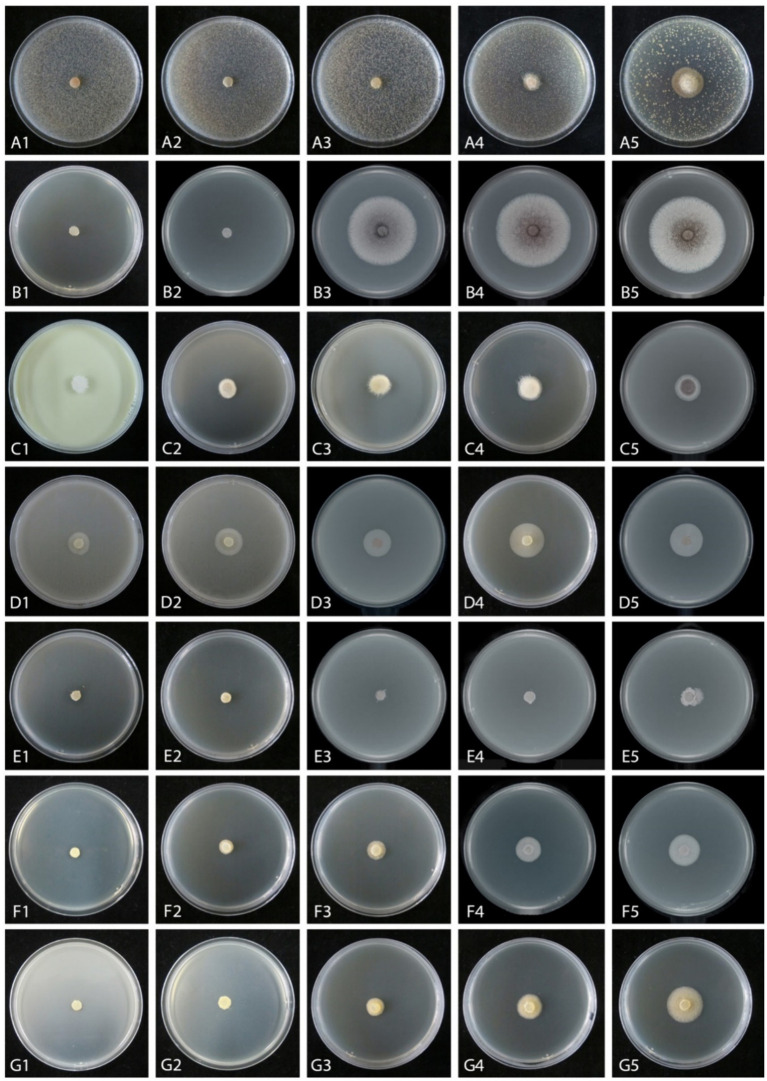
An in vitro sensitivity test (Bras and Deloron, 1983) of total 14 fungicides, including 10 fungicides named carbendazim, chlorothalonil, difenoconazole, ethirimol, ethylicin, flusilazole, mancozeb, pyraclostrobin, tebuconazole, and thiophanatemethyl, and 4 fungicide adjuvants including difenoconazole + azoxystrobin, prochloraz + iprodione, pyraclostrobin + tebuconazole, and trifloxystrobin + tebuconazole showed the different results. Our isolated strain (CGMCC 3.27539) was more sensitive to all fungicide adjuvants, and three fungicides, such as difenoconazole, ethylicin, and mancozeb, compared to other fungicides (Figures 4–6). First, ethylicin, prochloraz + iprodione, pyraclostrobin + tebuconazole showed the best inhibitory effect on the growth of the pathogen, and this inhibitory rate was as high as 100% at the recommended concentration. Difenoconazole, mancozeb, and trifloxystrobin + tebuconazole showed growth inhibitory effects of 98.10, 93.35, and 92.41%, respectively. Difenoconazole + azoxystrobin inhibitory effect was more than 80% (82.05%), and chlorothalonil showed general inhibition of 65.82%. Ethirimol, pyraclostrobin, and thiophanatemethyl have small inhibitory effects with inhibition rates of 33.07, 48.1, and 28.01%, respectively (Figures 4A–O). Carbendazim, flusilazole, and tebuconazole have very little effect on the growth of the isolated pathogen.
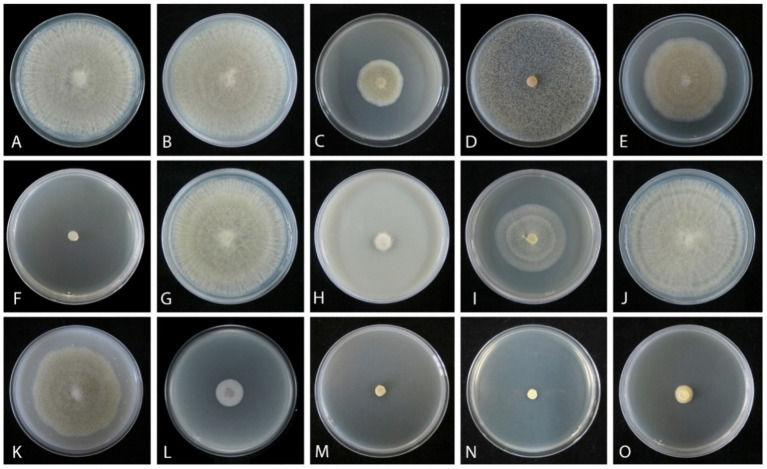
Figure 4.
Mycelial growth inhibition of Mucor xinjiangensis (CGMCC 3.27539) from different fungicides and fungicide adjuvants with recommended dosage, after 5 days. (A) Control. (B) Carbendazim (500 g/L). (C) Chlorothalonil (750 g/L). (D) Difenoconazole (PDA texture differentiation was caused by fungicide addition) (200 g/L). (E) Ethirimol (250 g/L). (F) Ethylicin (800 g/L). (G) Flusilazole (400 g/L). (H) Mancozeb (800 g/L). (I) Pyraclostrobin (250 g/L). (J) Tebuconazole (430 g/L). (K) Thiophanatemethyl (500 g/L). (L) Difenoconazole + Azoxystrobin (150, 250 g/L). (M) Prochloraz + Iprodione (100, 100 g/L). (N) Pyraclostrobin + tebuconazole (250, 430 g/L). (O) Trifloxystrobin + tebuconazole (250, 500 g/L).
Figure 6.
Mycelial growth inhibition percentage of Mucor xinjiangensis (CGMCC 3.27539) to fungicides and fungicide adjuvants and correlation of the sensitivity (EC50 value) to them. (A) Inhibition of M. xinjiangensis to a total of 14 fungicides and fungicide adjuvants. (B) EC50 value to difenoconazole. (C) EC50 value to ethylicin. (D) EC50 value to mancozeb. (E) EC50 value to difenoconazole + azoxystrobin. (F) EC50 value to prochloraz + iprodione. (G) EC50 value to pyraclostrobin + tebuconazole. (H) EC50 value to trifloxystrobin + tebuconazole.
The effective fungicides and fungicide adjuvants were further tested with different concentrations, including the recommended dosage for virulence determination with the EC50 value (Figure 5). There are some differences in the effective medium concentration of EC50 value for the seven agents against the M. xinjiangensis (Figures 6A–H). Among them, difenoconazole showed the best antifungal effects, the highest toxicity, and the lowest EC50 (0.18 mg/L). In the next two, difenoconazole + azoxystrobin and mancozeb were found to have EC50 values of 4.21 and 7.16 mg/L, respectively. The antifungal effect of ethylicin was relatively less, with the highest EC50 value of 167.83 mg/L. The EC50 value of other fungicide adjuvants ranged from about 13–20 mg/L including prochloraz + iprodione (13.06 mg/L), pyraclostrobin + tebuconazole (15.18 mg/L), and trifloxystrobin + tebuconazole (19.18 mg/L).
Figure 5.
Mycelial growth inhibition of Mucor xinjiangensis (CGMCC 3.27539) from different fungicides and fungicide adjuvants at different dilution concentrations after 5 days. (A1–A5) Difenoconazole diluted concentration (PDA texture differentiation was caused by fungicide addition) (A1. at 3,000; A2. at 5,000; A3. at 7,000; A4 at 10,000; A5. at 20,000). (B1–B5) Ethylicin diluted concentration (B1. at 2,000; B2. at 3,000; B3. at 4,000; B4 at 5,000; B5. at 6,000). (C1–C5) Mancozeb (C1. at 900; C2. at 1,500; C3. at 3,000; C4 at 4,000; C5. at 6,000). (D1–D5) Difenoconazole + azoxystrobin diluted concentration (D1. at 300; D2. at 600; D3. at 1,500; D4. at 2,400; D5. at 3,000). (E1–E5) Prochloraz + iprodione diluted concentration (E1. at 500; E2. at 1,000; E3. at 2,000; E4. at 4,000; E5. at 8,000). (F1–F5) Pyraclostrobin + tebuconazole diluted concentration (F1. at 2,000; F2. at 6,000; F3. at 8,000; F4 at 10,000; F5. at 15,000). (G1–G5) Trifloxystrobin + tebuconazole diluted concentration (G1. at 1,000; G2. at 2,000; G3. at 4,000; G4. at 8,000; G5. at 10,000).
Discussion
Prunus domestica is widely grown in high temperature and cool humid regions of northwest China (Cao et al., 2014). An unknown brown rot disease on plum fruit was found in several locations in Xinjiang, and its causal agent was unidentified. In the present study, we identified and described the pathogen as a new species, M. xinjiangensis. In the combined phylogenetic analysis of ITS and LSU, M. xinjiangensis formed a sister clade to M. pseudocircinelloides and M. pseudolusitanicus. Although M. xinjiangensis shares 99% ITS identity with M. pseudocircinelloides (XY07713) and M. pseudolusitanicus (CBS 543.8), LSU shares 100% ITS identity with M. pseudolusitanicus (CBS 540.78), but no LSU blast matches any M. pseudocircinelloides strain. By removing the ambiguous sequences of M. pseudolusitanicus in combined phylogenetic analysis, we found that M. pseudolusitanicus has a wide phylogenetic distribution even within its clade but is different in comparison with its type species (see above notes section). Furthermore, M. xinjiangensis is also genetically close to M. circinelloides and M. ctenidius, which are also pathogenic to plants (Nishijima et al., 2011; Sha and Meng, 2016). Nevertheless, these two species were differentiated from our collection based on morphological characteristics of sporangiophores, sporangia, columellae, and sporangiospores. M. xinjiangensis produces relatively larger sporangiophores (35–160 μm in M. xinjiangensis, 12–20 μm in M. circinelloides, 3–10 μm in M. ctenidius), sporangiospores (4–11 × 3–7 μm M. xinjiangensis, 4–7 × 3–6.2 μm in M. circinelloides, 4–8 × 3.2–6.4 μm in M. ctenidius), and smaller sporangia (40–53.5 × 39–53 μm in M. circinelloides, 50–70 μm in M. ctenidius, 16–42.5 μm in M. xinjiangensis), columellae (16–44 × 15–35 μm in M. circinelloides, 45–60 × 35–45 μm in M. ctenidius, 11–19.5 μm in M. xinjiangensis) compared to those of M. circinelloides and M. ctenidius (Walther et al., 2013).
Among the plum and prunes diseases, the most important postharvest diseases are brown rot, blue mold rot, gray rot, Mucor rot, Rhizopus rot, and bitter rot caused by Monilinia species (Monilinia laxa or Monilinia fructicola), Penicilliun expansum, Botrytis cinerea, M. piriformis, Rhizopus spp., Colletotrichum spp. (Colletotrichum gloeosporioides or Colletotrichum acutatum), respectively (Børve and Vangdal, 2007). Usually, these diseases begin with punctured wounds or insect bites. It is important to note, however, that Mucor rot and Rhizopus rot typically share the same symptoms. Due to Mucor rot or Rhizopus rot, the plums become soft, watery, and covered with black spore masses as the infection develops rapidly (Shahnaz et al., 2021; Seethapathy et al., 2022). In Mucor rot, it is often found that fungal structures are stiffer than those of Rhizopus rot, orientated at specific angles to the fruit surface at the time of maturation (Michalltdes, 1990). Rhizopus fruit rot is usually of less importance than the Mucor brown rot in the field, but both can cause important postharvest losses (Dennis and Mountford, 1975). However, the new brown rot disease reported here is the most severe in the field during June and July under high temperatures following continuous rainfall, when plum fruit faces continuous sunshine. The effects of sunburn are slow tree decline and browning of the skin on fruit exposed to too much heat (Racsko and Schrader, 2012; Lal and Sahu, 2017). It might be possible that continuous sunshine toward the premature plum promotes the initial infection (red spots), and then airborne Mucor pathogen attacks it and enhances the process of the premature fall of the fruit. Additionally, M. xinjiangensis was isolated from symptomatic plum fruit tissues, which showed different symptoms from Rhizopus rot and abiotic stress (sunburn). Due to these reasons, we propose a new name for the disease, “Mucor brown rot,” in order to distinguish it from two well-known biotic and abiotic diseases. Furthermore, healthy plum fruits inoculated with diseased fruit tissue collected from the field and with an isolated strain (M. xinjiangensis) also showed the same symptoms.
To date, M. xinjiangensis has only been isolated from P. domestica among stone fruits. The inoculation and re-isolation tests confirmed that M. xinjiangensis is pathogenic against P. domestica, and may be even more pathogenic. A range of hosts for M. xinjiangensis is unknown at the moment. Other stone fruits should be investigated for M. xinjiangensis infection.
It was found that different fungicides and fungicide adjuvants have different inhibition effects on pathogenic fungi (Nita et al., 2007). In the present study, 14 fungicides and their combinations were tested and compared with recommended dosage. These fungicides were commonly available in the Xinjiang market to control plant diseases. Among them, the antifungal rate of seven agents was more than 80%. As broad-spectrum low-toxicity, these fungicides are widely used to control fungal diseases on fruit trees, including apple, grape, peach, pear, and plum, in addition to combination formations. They are mainly classified into triazole (Toda et al., 2021), strobilurin (Balba, 2007), imidazole, benzimidazole, dicarboximidie, protection (chlorthalonil, mancozeb), and the plant bionic pesticide ethylicin (Zhang et al., 2020). There is an increasing tendency to combine two fungicides with different mechanisms of action to increase activity and efficacy (Rashid et al., 2014; Cohen et al., 2018) and to delay the emergence of fungicide resistance (Dooley et al., 2015). There are some research reports that explore different classes of fungicides to identify compounds capable of inhibiting Mucor species growth and spore germination. This includes triazoles, benzimidazoles, strobilurins, and other chemical groups commonly used against fungal pathogens (Suárez-García et al., 2021; Yamleshwar and Rai, 2023; Bai et al., 2024).
Our results showed that difenoconazole has the strongest toxicity and the smallest EC50. The smaller the EC50, the stronger the toxicity of the agent and the better the antifungal effect (Halling-Sørensen, 2000; Hu et al., 2013). This fungicide belongs to the systemic fungicide, which can inhibit the formation of sporangium and prevent the infection of fungi. At the same time, it has a lasting protection and treatment effects, so as to improve crop yield and quality. Zeng et al. (2023) found that thiophanatemethyl, tebuconazole, and difenoconazole showed significant field control effects on pathogens of fruit brown rot on Prunus salicina var. taoxingli. The fungicide test results of our study broaden the control of brown rot disease on plum fruit and the selection of fungicides. Antifungal fungicides were tested for virulence in vitro only, with the EC50 value serving as a reference value during the in vitro test. Further verification is needed to screen out the effects in the field. Also, we need to examine the application method, the application time, the climate conditions, etc. The sensitivity of pathogenic fungi to fungicides may vary, so screening targeted fungicides is essential (Masiello et al., 2019). Mucor brown rot can be prevented and controlled with difenoconazole, followed by difenoconazole + azoxystrobin and mancozeb as alternatives or rotation agents, in order to prevent the development of pathogen resistance due to long-term or repeated use of the same fungicide.
Funding Statement
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was financially supported by the Project of Renovation Capacity Building for the Young Sci-Tech Talents sponsored by Xinjiang Academy of Agricultural Sciences (xjnkq-2020020), Forestry development subsidy funds of Xinjiang Uyghur Autonomous Region (XJLYKJ-2023-19), Project of Fund for Stable Support to Agricultural Sci-Tech Renovation (xjnkywdzc-2022004). M. Raza is grateful to the High-Level Talent Recruitment plan of Xinjiang Uyghur Autonomous Region (“Young Talents” Program) and the second phase of the Xinjiang Uyghur Autonomous Region “Tianchi Talents” introduction plan, 2023.
Footnotes
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author contributions
BS: Conceptualization, Data curation, Investigation, Writing – original draft, Writing – review & editing. MR: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing. L-JZ: Data curation, Investigation, Writing – review & editing. B-QX: Data curation, Methodology, Writing – review & editing. PZ: Data curation, Methodology, Writing – review & editing. X-FZ: Conceptualization, Investigation, Supervision, Writing – review & editing.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The reviewer DST declared a past co-authorship with the author MR to the handling editor.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
- Abbas M. F., Naz F., Batool S., Qamar M. I., Naeem M. (2018). First evidence of Mucor rot infecting loquat (Eriobotrya japonica L.) in Pakistan. Mycopathologia 16, 81–85. [Google Scholar]
- Abraao A. S., Fernandes N., Silva A. M., Dominguez-Perles R., Barros A. (2022). Prunus lusitanica L. fruits as a novel source of bioactive compounds with antioxidant potential: exploring the unknown. Antioxidants (Basel) 11:1738. doi: 10.3390/antiox11091738, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abramczyk B. M., Wiktorowicz D. G., Okrasińska A., Pawłowska J. Z. (2024). Mucor thermorhizoides—a new species from post-mining site in Sudety Mountains (Poland). Curr. Microbiol. 81:201. doi: 10.1007/s00284-024-03708-7, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez E., Cano J., Stchigel A. M., Sutton D. A., Fothergill A. W., Salas V., et al. (2011). Two new species of Mucor from clinical samples. Med. Mycol. 49, 62–72. doi: 10.3109/13693786.2010.499521 [DOI] [PubMed] [Google Scholar]
- Bahrin A. A., Moshawih S., Dhaliwal J. S., Kanakal M. M., Khan A., Lee K. S., et al. (2022). Cancer protective effects of plums: a systematic review. Biomed. Pharmacother. 146:112568. doi: 10.1016/j.biopha.2021.112568, PMID: [DOI] [PubMed] [Google Scholar]
- Bai S., Zhang M., Tang S., Li M., Wu R., Wan S., et al. (2024). Research progress on Benzimidazole fungicides: a review. Molecules 29:1218. doi: 10.3390/molecules29061218, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balba H. (2007). Review of strobilurin fungicide chemicals. J. Environ. Sci. Health B 42, 441–451. doi: 10.1080/03601230701316465 [DOI] [PubMed] [Google Scholar]
- Borkar S. G. (2021). Mucormycosis: a surge in Mucorales fungal infection in post-covid patients in Indian states and insight into known and unknown actors. J. Glob. Health 1, 26–60. doi: 10.14302/issn.2693-1176.ijgh-21-3907 [DOI] [Google Scholar]
- Børve J., Vangdal E. (2007). Fungal pathogens causing fruit decay on plum (Prunus domestica L.) in Norway. Acta Hortic. 734, 367–369. doi: 10.17660/ActaHortic.2007.734.53 [DOI] [Google Scholar]
- Brahmanage R. S., Dayarathne M. C., Wanasinghe D. N., Thambugala K. M., Jeewon R., Chethana K. T., et al. (2020). Taxonomic novelties of saprobic Pleosporales from selected dicotyledons and grasses. Mycosphere 11, 2481–2541. doi: 10.5943/mycosphere/11/1/15 [DOI] [Google Scholar]
- Bras J. L., Deloron P. (1983). In vitro study of drug sensitivity of plasmodium falciparum: evaluation of a new semi-micro test. Am. J. Trop. Med. Hyg. 32, 447–451. doi: 10.4269/ajtmh.1983.32.447 [DOI] [PubMed] [Google Scholar]
- Cao K., Zheng Z., Wang L., Liu X., Zhu G., Fang W., et al. (2014). Comparative population genomics reveals the domestication history of the peach, Prunus persica, and human influences on perennial fruit crops. Genome Biol. 15:415. doi: 10.1186/s13059-014-0415-1, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chai C., Liu W., Cheng H., Hui F. (2019). Mucor chuxiongensis sp. nov., a novel fungal species isolated from rotten wood. Int. J. Syst. Evol. Microbiol. 69, 1881–1889. doi: 10.1099/ijsem.0.003166, PMID: [DOI] [PubMed] [Google Scholar]
- Cohen Y., Rubin A. E., Galperin M. (2018). Oxathiapiprolin-based fungicides provide enhanced control of tomato late blight induced by mefenoxam-insensitive Phytophthora infestans. PLoS One 13:e0204523. doi: 10.1371/journal.pone.0204523, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crous P. W., Wingfield M. J., Burgess T. I., Hardy G. E. S. J., Gene J., Guarro J., et al. (2018). Fungal planet description sheets: 716-784. Persoonia 40, 240–393. doi: 10.3767/persoonia.2018.40.10, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Souza C. A. F., Voigt K., Gurgel L. S., Cordeiro T. R. L., Oliveira R. J. V., Lima D. X., et al. (2018). A new species of Mucor (Mucoromycotina, Mucorales) isolated from an enclave of upland Atlantic Forest in the semi-arid region of Brazil. Phytotaxa 351, 53–62. doi: 10.11646/phytotaxa.351.1.4 [DOI] [Google Scholar]
- Dennis C., Mountford J. (1975). The fungal flora of soft fruits in relation to storage and spoilage. Ann. Appl. Biol. 79, 141–147. doi: 10.1111/j.1744-7348.1975.tb01528.x [DOI] [PubMed] [Google Scholar]
- Dooley H., Shaw M. W., Spink J., Kildea S., Shaw M. (2015). Effect of azole fungicide mixtures, alternations and dose on azole sensitivity in the wheat pathogen Zymoseptoria tritici. Plant Path. 65, 124–136. doi: 10.1111/ppa.12395 [DOI] [Google Scholar]
- Doyle J. J. (1990). Isolation of plant DNA from fresh tissue. Focus 12, 13–15. [Google Scholar]
- Eseigbe D. A., Bankole S. A. (1996). Fungi associated with post-harvest rot of black plum (Vitex doniana) in Nigeria. Mycopathologia 136, 109–114. doi: 10.1007/bf00437504, PMID: [DOI] [PubMed] [Google Scholar]
- Fan X. K., Zhang A. P. (2023). Research on current situation and development strategy of Xinjiang Jiashi prune industry integration. J. Smart Agric. 3, 110–113. doi: 10.20028/j.zhnydk.2023.20.026 [DOI] [Google Scholar]
- Gajanayake A. J., Karunarathna S. C., Jayawardena R. S., Luangharn T., Balasuriya A. (2023). Fungicolous Mucor on mushrooms: one novel species and six host records from Southwest China and northern Thailand. Mycologia 115, 674–692. doi: 10.1080/00275514.2023.2220166, PMID: [DOI] [PubMed] [Google Scholar]
- Gherbawy Y. A., Hussein N. A. (2010). Molecular characterization of Mucor circinelloides and Rhizopus stolonifer strains isolated from some Saudi fruits. Foodborne Pathog. Dis. 7, 137–142. doi: 10.1089/fpd.2009.0359, PMID: [DOI] [PubMed] [Google Scholar]
- Ghimire D., ErdoĞAn A., Baran A., GÜRses M., Meral AktaŞ H. (2022). Determination of mold diversity of some fruits sold in eastern Turkey. Iğdır Univ. Fen Bilim. Enst. Derg. 12, 2199–2208. doi: 10.21597/jist.1084083 [DOI] [Google Scholar]
- Hall T. A. (1999). BioEdit: a user-friendly biological sequence alignment editor and analysis program for windows 95/98/NT. In Nucleic acids symposium series 41, 95–98.
- Halling-Sørensen B. (2000). Algal toxicity of antibacterial agents used in intensive farming. Chemosphere 40, 731–739. doi: 10.1016/S0045-6535(99)00445-2, PMID: [DOI] [PubMed] [Google Scholar]
- Hassan M., Irshad G., Inam-ul-Haq Q., Khan M. A., Gull-elaala (2022). Characterization and evaluation of fungal flora associated with aflatoxins in dry plum by high performance liquid chromatography. J. Biomaterials and Bio-products Techn. 2, 1–24. [Google Scholar]
- Hong C. X., Michailides T. J., Holtz B. A. (2000). Mycoflora of stone fruit mummies in California orchards. Plant Dis. 84, 417–422. doi: 10.1094/pdis.2000.84.4.417, PMID: [DOI] [PubMed] [Google Scholar]
- Hu X. G., Liu L., Hu K., Yang X. L., Wang G. X. (2013). In vitro screening of fungicidal chemicals for antifungal activity against Saprolegnia. J. World Aquacult. Soc. 44, 528–535. doi: 10.1111/jwas.12052 [DOI] [Google Scholar]
- Hurdeal V. G., Gentekaki E., Hyde K. D., Nguyen T. T. T., Lee H. B. (2021). Novel Mucor species (Mucoromycetes, Mucoraceae) from northern Thailand. MycoKeys 84, 57–78. doi: 10.3897/mycokeys.84.71530, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain M., Ghazanfar M. U., Hamid M. I., Zahid M. A., Hussain T., Raza M., et al. (2014). In vitro evaluation of fungicides and plant extracts for suppressing mycelium growth of macrophomina phaseolinacauses charcoal rot of sunflower. Int. J. Appl. Agric. Sci. 6, 136–140. [Google Scholar]
- Hussain M., Hamid M. I., Ghazanfar M. U., Akhtar N., Raza M. (2016). First report of fruit rot of strawberry caused by Geotrichum candidum in Pakistan. Plant Dis. 100:1948. doi: 10.1094/pdis-03-16-0277-pdn [DOI] [Google Scholar]
- Jangid R., Begum T. (2022). Antimycotic activity of some medicinal plants against Mucor circinelloides. Biomed. Res. Int. 2022:3523920. doi: 10.1155/2022/3523920, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kettles G. J., Luna E. (2019). Food security in 2044: how do we control the fungal threat? Fungal Biol. 123, 558–564. doi: 10.1016/j.funbio.2019.04.006, PMID: [DOI] [PubMed] [Google Scholar]
- Khan I. H., Javaid A. (2022). Mucor fragilis causing rot of Seychelles pole bean in Pakistan. Australas. Plant Pathol. 51, 359–362. doi: 10.1007/s13313-022-00859-8 [DOI] [Google Scholar]
- Kinge T. R., Besong P. N. (2021). Fungi diversity on some fruits and biological control using two plants extracts. J. Adv. Biol. 24, 24–38. [Google Scholar]
- Koka J. A., Bhat M. Y., Wani A. H. (2021). In vitro efficacy of fungicides on mycelial growth and spore germination of Alternaria alternata and Mucor plumbeus. J. Drug Deliv. Ther. 11, 17–22. doi: 10.22270/jddt.v11i3.4692 [DOI] [Google Scholar]
- Kornerup A., Wanscher J. H. (1967). Methuen handbook of colour. London: Methuen & Co. Ltd. 1–243., PMID: [Google Scholar]
- Kwinda G. T., Jacobs A., Lebelo S. L., Rong I. H. (2015). Mucorales from selected spoilt fruit commodities in the Gauteng Province, South Africa. Afr. Plant Prot. 18, 1–5. [Google Scholar]
- Kwon J. H., Hong S. B. (2005). Soft rot of tomato caused by Mucor racemosus in Korea. Mycobiol. 33, 240–242. doi: 10.4489/myco.2005.33.4.240, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lal N., Sahu N. (2017). Management strategies of sun burn in fruit crops-a review. Int. J. Curr. Microbiol. Appl. Sci. 6, 1126–1138. doi: 10.20546/ijcmas.2017.606.131 [DOI] [Google Scholar]
- Li G. J., Hyde K. D., Zhao R. L., Hongsanan S., Abdel-Aziz F. A., Abdel-Wahab M. A., et al. (2016). Fungal diversity notes 253–366: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 78, 1–237. doi: 10.1007/s13225-016-0366-9 [DOI] [Google Scholar]
- Li M., Raza M., Song S., Hou L., Zhang Z. F., Gao M., et al. (2023). Application of culturomics in fungal isolation from mangrove sediments. Microbiome 11:272. doi: 10.1186/s40168-023-01708-6, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lima D. X., Barreto R. W., Lee H. B., Cordeiro T. R. L., Souza C. A. F., Oliveira R. J. V., et al. (2020). Novel mucoralean fungus from a repugnant substrate: Mucor merdophylus sp. nov., isolated from dog excrement. Curr. Microbiol. 77, 2642–2649. doi: 10.1007/s00284-020-02038-8, PMID: [DOI] [PubMed] [Google Scholar]
- Lima D. X., Souza-Motta C. M., Wagner L., Voigt K., De Souza C. A. F., De Oliveira R. J. V., et al. (2017). Circinella simplex—a misapplied name of Mucor circinatus sp. nov. Phytotaxa 329:269. doi: 10.11646/phytotaxa.329.3.8 [DOI] [Google Scholar]
- López S. N., Sangorrín M. P., Pildain M. B. (2016). Fruit rot of sweet cherries and raspberries caused by Penicillium crustosum and Mucor piriformis in South Patagonia, Argentina. Can. J. Plant Pathol. 38, 511–516. doi: 10.1080/07060661.2016.1243582 [DOI] [Google Scholar]
- Lu X. L., Najafzadeh M. J., Dolatabadi S., Ran Y. P., Gerrits van den Ende A. H., Shen Y. N., et al. (2013). Taxonomy and epidemiology of Mucor irregularis, agent of chronic cutaneous mucormycosis. Persoonia 30, 48–56. doi: 10.3767/003158513X665539, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masiello M., Somma S., Ghionna V., Logrieco A. F., Moretti A. (2019). In vitro and in field response of different fungicides against aspergillus flavus and fusarium species causing ear rot disease of maize. Toxins (Basel). 11:11. doi: 10.3390/toxins11010011, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalltdes T. J. (1990). Postharvest diseases of pome and stone fruits caused by Mucor piriformis in the Pacific northwest and California. Plant Dis. 74:537. doi: 10.1094/pd-74-0537 [DOI] [Google Scholar]
- Ni H. G., Lu F. H., Luo X. L., Tian H. Y., Wang J. Z., Guan Y. F., et al. (2008). Assessment of sampling designs to measure riverine fluxes from the Pearl River Delta, China to the South China Sea. Environ. Monit. Assess. 143, 291–301. doi: 10.1007/s10661-007-9982-x, PMID: [DOI] [PubMed] [Google Scholar]
- Nishijima K. A., Wall M. M., Chang L. C., Wei Y., Wong D. K. W. (2011). First report of association of Mucor circinelloides on noni (Morinda citrifolia) in Hawaii. Plant Dis. 95:360. doi: 10.1094/pdis-11-10-0815, PMID: [DOI] [PubMed] [Google Scholar]
- Nita M., Ellis M. A., Wilson L. L., Madden L. V. (2007). Evaluation of the curative and protectant activity of fungicides and fungicide–adjuvant mixtures on Phomopsis cane and leaf spot of grape: a controlled environment study. Crop Prot. 26, 1377–1384. doi: 10.1016/j.cropro.2006.11.010 [DOI] [Google Scholar]
- Nylander J. (2004). MrModeltest V2. Program distributed by the author. Bioinformatics 24, 581–583. doi: 10.1093/bioinformatics/btm388 [DOI] [Google Scholar]
- Oufensou S., Ul Hassan Z., Balmas V., Jaoua S., Migheli Q. (2023). Perfume guns: potential of yeast volatile organic compounds in the biological control of mycotoxin-producing fungi. Toxins 15:45. doi: 10.3390/toxins15010045, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phookamsak R., Hyde K. D., Jeewon R., Bhat D. J., Jones E. B. G., Maharachchikumbura S. S. N., et al. (2019). Fungal diversity notes 929–1035: taxonomic and phylogenetic contributions on genera and species of fungi. Fungal Divers. 95, 1–273. doi: 10.1007/s13225-019-00421-w [DOI] [Google Scholar]
- Racsko J., Schrader L. E. (2012). Sunburn of apple fruit: historical background, recent advances and future perspectives. Crit. Rev. Plant Sci. 31, 455–504. doi: 10.1080/07352689.2012.696453 [DOI] [Google Scholar]
- Rashid M. H., Hossain M. A., Kashem M. A., Kumar S., Rafii M. Y., Latif M. A. (2014). Efficacy of combined formulations of fungicides with different modes of action in controlling botrytis gray mold disease in chickpea. Sci. World J. 2014:639246. doi: 10.1155/2014/639246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raza M., Zhang Z. F., Hyde K. D., Diao Y. Z., Cai L. (2019). Culturable plant pathogenic fungi associated with sugarcane in southern China. Fungal Divers. 99, 1–104. doi: 10.1007/s13225-019-00434-5 [DOI] [Google Scholar]
- Ronquist F., Teslenko M., van der Mark P., Ayres D. L., Darling A., Hohna S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rybak M., Wojdyło A. (2023). Inhibition of α-Amylase, α-Glucosidase, Pancreatic Lipase, 15-Lipooxygenase and Acetylcholinesterase Modulated by Polyphenolic Compounds, Organic Acids, and Carbohydrates of Prunus domestica Fruit. Antioxidants. 12:1380. doi: 10.3390/antiox12071380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito S., Michailides T. J., Xiao C. L. (2016). Mucor rot-an emerging postharvest disease of mandarin fruit caused by Mucor piriformis and other Mucor spp. in California. Plant Dis. 100, 1054–1063. doi: 10.1094/PDIS-10-15-1173-RE, PMID: [DOI] [PubMed] [Google Scholar]
- Saito S., Wang F., Xiao C. L. (2023). Sensitivity of Mucor piriformis to natamycin and efficacy of natamycin alone and with salt and heat treatments against Mucor rot of stored mandarin fruit. Plant Dis. 107, 3602–3607. doi: 10.1094/PDIS-04-23-0796-RE, PMID: [DOI] [PubMed] [Google Scholar]
- Schoch C. L., Robbertse B., Robert V., Vu D., Cardinali G., Irinyi L., et al. (2014). Finding needles in haystacks: linking scientific names, reference specimens and molecular data for fungi. Database (Oxford) 2014, 1–21. doi: 10.1093/database/bau061, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schipper M. A. A. (1978). On certain species of Mucor with a key to all accepted species. Stud. Mycol. 17, 1–52., PMID: 24451623 [Google Scholar]
- Seethapathy P., Gothandaraman R., Gurudevan T., Malik I. (2022). “Diseases, pests, and disorders in plum: diagnosis and management” in Handbook of plum fruit. eds. Gull A., Nayik G. A., Wani S. M., Nanda V. (Taylor & Francis; ), 133–176. [Google Scholar]
- Sha R., Meng Q. (2016). Antifungal activity of rhamnolipids against dimorphic fungi. J. Gen. Appl. Microbiol. 62, 233–239. doi: 10.2323/jgam.2016.04.004, PMID: [DOI] [PubMed] [Google Scholar]
- Shahnaz E., Anwar A., Banday S. (2021). “Postharvest fungal diseases in stone fruits and their management” in Postharvest handling and diseases of horticultural produce. eds. Singh D., Sharma R. R., Devappa V., Kamil D. (Taylor & Francis; ). 259–266. [Google Scholar]
- Silvan J. M., Michalska-Ciechanowska A., Martinez-Rodriguez A. J. (2020). Modulation of antibacterial, antioxidant, and anti-inflammatory properties by drying of Prunus domestica L. plum juice extracts. Microorganisms 8:119. doi: 10.3390/microorganisms8010119, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares M. A. R., Pena A., Sendón R., Almeida C., Nieto G. A., Khwaldia K., et al. (2023). By-products of dates, cherries, plums and artichokes: a source of valuable bioactive compounds. Trends. Food Sci. 131, 220–243. doi: 10.1016/j.tifs.2022.12.004 [DOI] [Google Scholar]
- Song S. Q., Lü Z., Wang J., Zhu J., Gu M. Y., Tang Q. Y., et al. (2020). First report of a new potato disease caused by Galactomyces candidum F12 in China. J. Integr. Agric. 19, 2470–2476. doi: 10.1016/s2095-3119(20)63257-9 [DOI] [Google Scholar]
- Stamatakis A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics (Oxford, England) 30, 1312–1313. doi: 10.1093/bioinformatics/btu033, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suárez-García J., Cano-Herrera M. A., Ramírez-Villalva A., Fuentes-Benites A., Zavala-Segovia N., García-Eleno M. A., et al. (2021). Synthesis and antifungal activity evaluation of 1-(2-benzyloxy-2-phenylethyl)-1, 2, 3-triazole miconazole analogs. Pharm. Chem. J. 55, 436–440. doi: 10.1007/s11094-021-02440-x [DOI] [Google Scholar]
- Suh S. J., Yu J., Jang I. B., Moon J. W., Lee S. W., Jang I. B. (2018). Effects of storage temperature and seed treatment on emergence and growth properties of Panax ginseng at spring-sowing. Korean J. Crop Sci. 26, 401–407. doi: 10.7783/kjmcs.2018.26.5.401 [DOI] [Google Scholar]
- Tedersoo L., Sánchez-Ramírez S., Kõljalg U., Bahram M., Döring M., Schigel D., et al. (2018). High-level classification of the Fungi and a tool for evolutionary ecological analyses. Fungal Divers. 90, 135–159. doi: 10.1007/s13225-018-0401-0 [DOI] [Google Scholar]
- Tibpromma S., Hyde K. D., Jeewon R., Maharachchikumbura S. S. N., Liu J. K., Bhat D. J., et al. (2017). Fungal diversity notes 491–602: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers. 83, 1–261. doi: 10.1007/s13225-017-0378-0 [DOI] [Google Scholar]
- Toda M., Beer K. D., Kuivila K. M., Chiller T. M., Jackson B. R. (2021). Trends in agricultural Triazole fungicide use in the United States, 1992-2016 and possible implications for antifungal-resistant fungi in human disease. Environ. Health Persp. 129:55001. doi: 10.1289/EHP7484, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turland N. J., Wiersema J. H., Barrie F. R., Greuter W., Hawksworth D. L., Herendeen P. S., et al. (2018). International code of nomenclature for algae, fungi, and plants (Shenzhen code) adopted by the nineteenth international botanical congress Shenzhen, China, July 2017. Koeltz Botanical Books.
- Tuszyński T., Satora P. (2003). Microbiological characteristics of the węgierka zwykła plum orchard in submontane region. Pol. J. Food Nutr. Sci. 12, 43–48. [Google Scholar]
- Urquhart A. S., Douch J. K., Heafield T., Buddie A. G., Idnurm A. (2020). Diversity of Backusella (Mucoromycotina) in South-Eastern Australia revealed through polyphasic taxonomy. Persoonia 46, 1–25. doi: 10.3767/persoonia.2021.46.01 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale R. G., de Hoog G. S., Schwarz P., Dannaoui E., Deng S., Machouart M., et al. (2012). Antifungal susceptibility and phylogeny of opportunistic members of the order mucorales. J. Clin. Microbiol. 50, 66–75. doi: 10.1128/JCM.06133-11, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voglmayr H., Clémençon H. (2016). Identification and taxonomic position of two mucoralean endoparasites of Hysterangium (Basidiomycota) based on molecular and morphological data. Mycol. Progress. 15, 1–17. doi: 10.1007/s11557-015-1150-z [DOI] [Google Scholar]
- Vu D., Groenewald M., de Vries M., Gehrmann T., Stielow B., Eberhardt U., et al. (2019). Large-scale generation and analysis of filamentous fungal DNA barcodes boosts coverage for kingdom fungi and reveals thresholds for fungal species and higher taxon delimitation. Stud. Mycol. 92, 135–154. doi: 10.1016/j.simyco.2018.05.001, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner L., de Hoog S., Alastruey-Izquierdo A., Voigt K., Kurzai O., Walther G. (2019). A revised species concept for opportunistic Mucor species reveals species-specific antifungal susceptibility profiles. Antimicrob. Agents Chemother. 63, 10–1128. doi: 10.1128/aac.00653-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace R. L., Hirkala D. L., Nelson L. M. (2018). Efficacy of Pseudomonas fluorescens for control of Mucor rot of apple during commercial storage and potential modes of action. Can. J. Microbiol. 64, 420–431. doi: 10.1139/cjm-2017-0776, PMID: [DOI] [PubMed] [Google Scholar]
- Walther G., Pawlowska J., Alastruey-Izquierdo A., Wrzosek M., Rodriguez-Tudela J. L., Dolatabadi S., et al. (2013). DNA barcoding in Mucorales: an inventory of biodiversity. Persoonia 30, 11–47. doi: 10.3767/003158513X665070, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. H., Wang Y. G., Wang J. R., Cui D. F. (2021). Origin and conservation of European plum (Prunus domestica). For. Xinjiang 4, 29–31. doi: 10.3969/j.issn.1005-3522.2021.04.012 [DOI] [Google Scholar]
- Yamleshwar R. R. K., Rai D. (2023). Isolation of mycoflora associated with wheat seed and their management by chemicals. Environ. Ecol. 41, 266–272. [Google Scholar]
- Zeng Q., Ye S. S., Xie S. Y., Chen Z. Y., Ye B. H., Yang G., et al. (2023). Identification of the brown rot pathogen from Shengzhou nane fruit and screening of highly effective fungicide. J. Hunan Univ. Nat. Sci. (Nat. Sci.). 49, 321–328. [Google Scholar]
- Zhang K., Yuan-Ying S., Cai L. (2013). An optimized protocol of single spore isolation for fungi. Cryptogam. Mycol. 34, 349–356. doi: 10.7872/crym.v34.iss4.2013.349 [DOI] [Google Scholar]
- Zhang S., Zhang M., Khalid A. R., Li L., Chen Y., Dong P., et al. (2020). Ethylicin prevents potato late blight by disrupting protein biosynthesis of Phytophthora infestans. Pathogens 9:299. doi: 10.3390/pathogens9040299, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H., Dai Y. C., Liu X. Y. (2023). Outline and divergence time of subkingdom Mucoromyceta. Fungal Divers. 123, 49–157. doi: 10.1007/s13225-023-00525-4 [DOI] [Google Scholar]
- Zhebentyayeva T., Shankar V., Scorza R., Callahan A., Ravelonandro M., Castro S., et al. (2019). Genetic characterization of worldwide Prunus domestica (plum) germplasm using sequence-based genotyping. Hortic. Res. 6:12. doi: 10.1038/s41438-018-0090-6, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.