Abstract
Cytotherapy is a strategy to deliver modified cells to a diseased tissue, but targeting solid tumours remains challenging. Here we design macrophages, harbouring a surface glypican-3-targeting peptide and carrying a cargo to combat solid tumours. The anchored targeting peptide facilitates tumour cell recognition by the engineered macrophages, thus enhancing specific targeting and phagocytosis of tumour cells expressing glypican-3. These macrophages carry a cargo of the TLR7/TLR8 agonist R848 and INCB024360, a selective indoleamine 2,3-dioxygenase 1 (IDO1) inhibitor, wrapped in C16-ceramide-fused outer membrane vesicles (OMV) of Escherichia coli origin (RILO). The OMVs facilitate internalization through caveolin-mediated endocytosis, and to maintain a suitable nanostructure, C16-ceramide induces membrane invagination and exosome generation, leading to the release of cargo-packed RILOs through exosomes. RILO-loaded macrophages exert therapeutic efficacy in mice bearing H22 hepatocellular carcinomas, which express high levels of glypican-3. Overall, we lay down the proof of principle for a cytotherapeutic strategy to target solid tumours and could complement conventional treatment.
Subject terms: Drug delivery, Nanoparticles, Cancer immunotherapy, Monocytes and macrophages
Macrophages are considered a good candidate for cancer cytotherapy because of their phagocytotic capacity, enabling them to deliver cargo to tissues. Here authors engineer macrophages that are targeted to glypican-3-expressing tumour cells and equipped with drug-loaded exosomes and show therapeutic efficiency in a mouse model of hepatocellular cancer.
Introduction
Cytotherapy has made major breakthroughs in current cancer therapy, and it will continue to be developed in the future1. As a popular marketed therapy, chimeric antigen receptor (CAR) T-cell therapy has benefited many patients with haematologic malignancies. However, its clinical efficacy in solid tumours has been unfavorable2. Previous attempts have demonstrated that solving the problems of restricted tumour trafficking, poor infiltration and an unfavourable immunosuppressive tumour microenvironment (TME) is necessary to increase the curative effect of cytotherapy on solid tumours3.
To enhance the poor efficacy of cytotherapy in the treatment of solid tumours, alternative cell types that could be used for cytotherapy have received increasing attention. As a result, genetically engineered chimeric antigen receptor macrophages (CAR-M) were developed. CAR-Ms demonstrated encouraging early results in the treatment of solid tumours and entered clinical trials (NCT04660929)4. This macrophage-based cytotherapeutic has inherited the phagocytosis ability and deep penetration capacity of macrophages5, which effectively solves the dilemma of T-cell therapy in solid tumour treatment and exhibits broad prospects for clinical application2,6,7. Moreover, macrophages lead to lower off-target toxicity due to their limited circulation time, and a large number of clinical trials have demonstrated the feasibility and safety of adoptively transferred macrophages8,9. Therefore, macrophages are a promising alternative cell type for cytotherapy against solid tumours. However, the intrinsic immunosuppressive TME highly restricted the effect of injected macrophages by re-polarizing macrophages to the anti-inflammatory and pro-tumoural (M2) phenotype instead of the proinflammatory and anti-tumoural (M1) phenotype, resulting in poor transplantation feasibility and weakened antitumour potency10. Therefore, there is an ever-growing demand to develop therapeutic macrophages with the ideal phenotype and high immunological activity.
Extensive efforts have demonstrated that phagocytosis plays a crucial role in macrophage-based cytotherapy11,12. In the process, recognition and adhesion are the primary processes necessary for macrophages to initiate phagocytosis13,14. Compared with M2-type macrophages, M1-type macrophages can downregulate the expression of SIRPα15,16 and have a stronger ability to directly phagocytose tumour cells, release cytokines and continuously activate downstream adaptive immune responses17–20. However, the nonspecific tumour phagocytosis of M1-type macrophages limits the antitumour effect. Therefore, increasing the chance of specific recognition and adhesion between M1-type macrophages and tumour cells may be an effective way to achieve tumour-specific phagocytosis and improve the therapeutic effect. Glypican-3 (GPC3), a critical molecular target in hepatocellular carcinoma (HCC) and melanoma, is a heparan sulfate proteoglycan that is attached to the cell membrane surface via a glycosylphosphatidylinositol anchor21. Our previous studies have proven that the GPC3 targeting strategy has the specific targeting ability for HCC cells, which exhibits promising applications for early diagnosis, targeted delivery, and circulating tumour cell capture effects22,23. Therefore, we hypothesized that anchoring the GPC3 peptide on the membrane surface could further promote macrophage–tumour cell recognition, thus enhancing tumour-specific targeting, phagocytosis and the killing of tumour cells with high GPC3 expression.
The TME has a fatal influence on the phenotype and biological functions of injected macrophages and is difficult to comprehensively regulate by currently marketed cytotherapy formulations24–26. As an important class of immune cells, tumour-associated macrophages (TAM) are polarized towards the M2 phenotype, which is one of the main drivers responsible for the immunosuppressive TME27–29. TAMs secrete immunosuppressive cytokines while accelerating the catabolism of tryptophan (Trp) by elevating the expression level of indoleamine 2,3-dioxygenase 1 (IDO1). Its metabolite, kynurenine (Kyn), can aggravate the phenotypic polarization of TAMs to M2, promote regulatory T-cell (Treg) expansion and increase immune tolerance30,31. Toll-like receptor (TLR) 7/TLR8 is the main receptor of pathogen-related molecular patterns that can monitor and recognize various pathogen molecules and initiate the immune response, and its agonists can polarize TAMs towards the M1 phenotype32. IDO1 is mainly overexpressed by TAMs and tumour cells at the tumour site, and its inhibitors competitively bind to the IDO1 catalytic site with Trp, block Kyn production and alleviate immune tolerance33. Accumulating studies have shown that combining TLR7/TLR8 agonists with IDO1 inhibitors is expected to regulate the TAM phenotype, enhance T-cell viability, and then remodel the immunosuppressive TME to provide a beneficial environment34. Furthermore, exosomes produced by M1-type macrophages have a phospholipid bilayer structure, showing great potential as drug carriers, and can stimulate TAM polarization, inducing a proinflammatory TME35. Therefore, we proposed that M1-type macrophages packed TLR7/TLR8 agonists and IDO1 inhibitors delivered by generated drug exosomes to the tumour site could simultaneously regulate the TAM phenotype, enhance T-cell viability and reshape the immunosuppressive TME to improve the effectiveness of macrophage-based cytotherapy.
Efficient drug loading and the controllable drug release of macrophages are the main requirements for inner drug packing36,37. The current inner packing strategies mainly include free-drug loading or drug-nanoparticle loading. Compared with free drug loading, drug-nanoparticle loading has the advantages of maintaining a highly effective drug loading and a fixed synergistic drug ratio while avoiding drug-induced damage to macrophages38,39. Our group previously developed a packed macrophage with sorafenib-nanoparticle loading strategy, which showed a negligible effect on the functions of macrophages themselves, including targeting, deep penetration and cytokine secretion abilities, and improved their therapeutic effect through a combined cyto- with chemo-therapeutic mechanism40. However, the current drug-nanoparticle loading process requires passage through lysosomes, which results in nanostructural degradation and rapid drug release, ultimately affecting drug-loading capacity41. Therefore, an efficient drug-loading process requires both high drug loading and a stable nanostructure. Outer membrane vesicles (OMV) are nanoscale spherical bilayer-membrane vesicles naturally secreted during the growth of Escherichia coli (E. coli) that retain outer membrane and periplasmic components similar to those of the source bacteria42. The FimH-positive E. coli can enter macrophages through caveolin-mediated endocytosis, which enables them to bypass lysosome processing and to maintain a stable nanostructure43,44. After efficient drug loading, regulating drug release processes is another critical challenge. Ceramides are sphingolipids composed of sphingosine and a fatty acid. Ceramide containing fatty acyl chains of 16 carbon atoms (C16-ceramide) is a conical structure lipid that can induce membrane invagination and promote exosome biogenesis45,46. Altogether, we hypothesized that drug nanoparticles wrapped by C16-ceramide fused E. coli-originated OMVs could enter macrophages to form drug-packed macrophages by caveolin-mediated endocytosis in vitro and furtherly generate drug exosomes at the tumour site. Perhaps these drug nanoparticles would possess higher internalization efficiency than drug nanoparticles without OMV wrapping and these generated drug exosomes would trigger the antitumour immune response.
Herein, we report macrophages obtained by surface GPC3-targeting peptide (GTP) harbouring and inner R848/INCB024360-lipid OMV (RILO) carrying to promote specific phagocytosis of tumour cells expressing GPC3 and R848/INCB024360-exosome (RI-exosome) generation for combating solid tumours (Fig. 1). After intravenous (i.v.) injection, GPC3-targeted RILO-loaded macrophage (RILO@MG) actively targets and accumulates at the tumour site based on innate tumour chemotaxis and GPC3-mediated targeting. RILO@MG exerts therapeutic efficacy in mice bearing H22 hepatocellular carcinomas and B16F10 malignant melanomas. This study suggests that producing modified macrophages with standardization and personalization may be a solution to the great clinical demand for cytotherapy for the treatment of solid tumours.
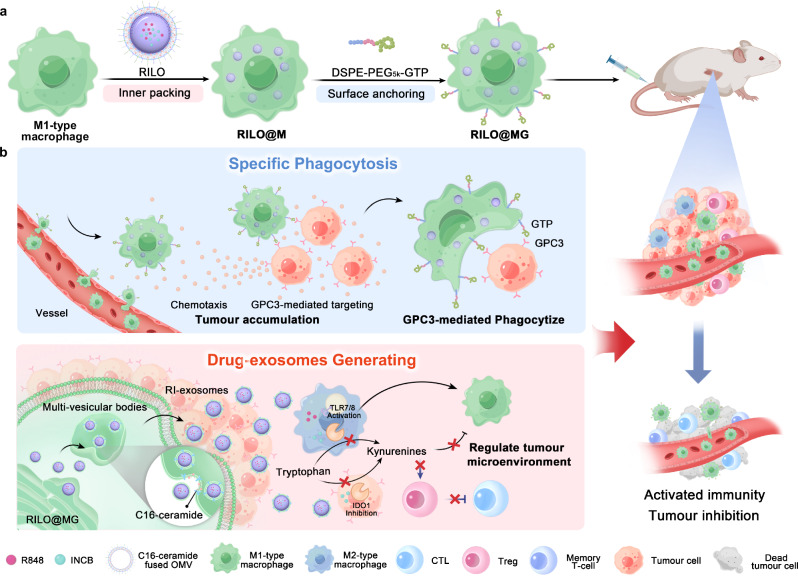
Fig. 1. Schematic illustration of RILO@MG promoting specific tumour phagocytosis and generating drug exosomes that play a role in triggering an antitumour immune response to combat solid tumour.
a Preparation of RILO@MG. First, RILO@M was prepared by the inner packing of M1-type macrophages and RILO. Then, DSPE-PEG5k-GTP was anchored on the surface of RILO@M to prepare RILO@MG. b RILO@MG accumulated in the tumour site through chemotaxis and GPC3-mediated targeting after i.v. administration directly killed tumour cells by GPC3-mediated phagocytosis and generated RI-exosomes containing R848 and INCB to regulate the TAM phenotype and enhance T-cell viability. Therefore, RILO@MG exerted antitumour efficacy by directly killing tumour cells and reversing the suppressive TME. CTL cytotoxic T-cell.
Results
RILO was stable in macrophages, and RILO@MG maintained the M1 phenotype in different culture environments
As illustrated in Fig. 2a, the TLR7/TLR8 agonist R848 and IDO1 inhibitor INCB024360 (INCB) lipid nanoparticle (RIL) was first prepared, and RIL was wrapped by C16-ceramide-fused FimH-positive E. coli MG1655-originated OMVs to further prepare RILO. Followed by the preparation of RILO-loaded macrophage (RILO@M), the GTP was anchored to the membrane surface via DSPE-PEG5k, and the RILO@MG was finally prepared. In short, the preparation of RILO@MG was mainly divided into two parts, the process of inner packing by coincubating M1-type macrophages with RILO and the implementation of surface anchoring by inserting GTP into the lipid bilayers of the cell membrane via hydrophobic interactions,. We determined that the mass ratio of R848 to INCB was between 6:5 and 6:6 according to the changes in M1/M2 ratio (Supplementary Fig. 1a, b), and the optimal concentration of C16-ceramide incubated with M1-type macrophages was 100 µM (Supplementary Fig. 1c). Inspired by macrophages in naturally fighting bacteria and other pathogens47,48, we selected OMVs obtained from the culture medium of nonpathogenic E. coli MG1655 by multiple centrifugation and ultrafiltration steps as a component of RILO. The RIL was coextruded with C16-ceramide fused OMVs to prepare RILO. Transmission electron microscopy (TEM) images and dynamic light scattering analysis showed that RILO was ~50.54 nm in size with a core–shell-structured spherical morphology (Fig. 2b–d), indicating the successful assembly of RILO. R848 and INCB were efficiently loaded into RILO with drug loadings (DL, %) of 6.17 ± 0.02% and 5.13 ± 0.15% for R848 and INCB, respectively (Supplementary Table 1). Protein profiles in RILO were determined by SDS–polyacrylamide gel electrophoresis (SDS–PAGE), which confirmed that the proteins from the OMVs were retained in RILO (Fig. 2e). Moreover, RILO remained stable under storage conditions without substantial changes in particle size, zeta potential and DL of R848 and INCB for at least 14 days, respectively (Supplementary Fig. 1d, e). RILO could also remain stable under physiological conditions for at least 48 h (Supplementary Fig. 1f). To mimic clinical settings, bone marrow-derived macrophages (BMDM) were used in RILO@MG preparation unless marked RAW superscript. BMDMs were identified by morphology and flow cytometry (Supplementary Fig. 2a, b).
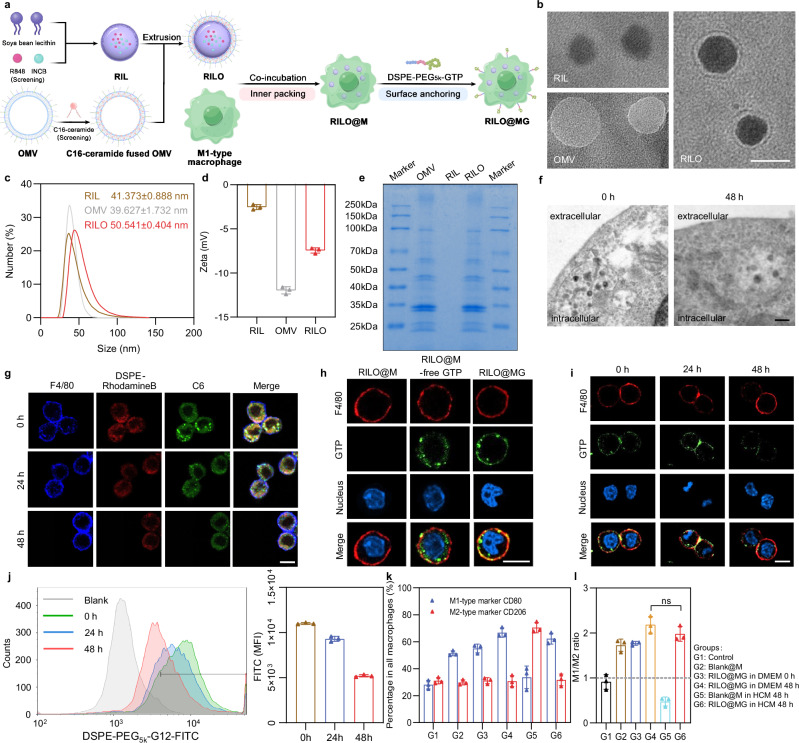
Fig. 2. RILO maintains a stable nanostructure in macrophages and RILO@MG maintains the M1 phenotype in an immunosuppressed environment.
a Schematic illustration of the preparation process of RILO@MG. b Micromorphological characterizations of RIL, OMV and RILO (n = 3 biologically independent experiments). Scale bar, 50 nm. c, d Dynamic light scattering (c) and zeta potential (d) analyses of RIL, OMV and RILO (n = 3 biologically independent experiments). e SDS–PAGE protein analysis in OMV, RIL and RILO (n = 3 biologically independent experiments). f TEM images of RILO@M at 0 and 48 h after preparation (n = 3 biologically independent experiments). Scale bar, 200 nm. g Representative confocal images showing the stability of the nanostructure of fluorescently labelled RILO in M1-type macrophages from RAW264.7 cells (M1-type macrophagesRAW) (n = 3 biologically independent experiments). The cell membrane was stained with anti-F4/80 antibody (blue). DSPE-Rhodamine B and C6 were selected to label lipids (red) and replace the drug (green), respectively. Scale bar, 10 µm. h Representative confocal images of RILO@M, RILO@M-free GTP and RILO@MG (n = 3 biologically independent experiments). Scale bar, 10 µm. RAW264.7 cells were used in (h). i, j Representative confocal images (i) and flow cytometric analysis (j) of RILO@MGRAW after preparation for 24 and 48 h (n = 3 biologically independent experiments). Scale bar, 10 µm. FITC was selected to label GTP in (h–j). All cells were stained with anti-F4/80 antibody (red) and Hoechst 33342 (blue) in (h and i). k, l Phenotype analysis of RILO@MG including the percentage of macrophages with different phenotypes (k) and M1/M2 ratio (l) in different culture environments (n = 3 biologically independent experiments). Data are expressed as the mean ± SD and were processed by one-way analysis of variance (ANOVA) with Tukey’s multiple comparisons test (l). ns no significance. BMDMs were used in all experiments involving macrophages unless marked RAW superscript.
Next, we determined the optimal formulations of inner RILO packing by investigating the effects of incubation concentration and time on cell viability and DL. We found that 200 μg/mL (quantified by R848) and 2 h were the optimal conditions for the preparation of RILO@M with greater biocompatibility and higher DL, respectively (Supplementary Fig. 2c–e). We also studied the endocytic pathways and the subsequent intracellular trafficking in terms of Coumarin-6 (C6) intensity (Supplementary Fig. 3a). Flow cytometry showed that the uptake of C6-L and C6-LO was completely blocked at 4 °C, which suggested energy-dependent internalization. Moreover, the cellular uptake of C6-LO was significantly inhibited by caveolin-mediated inhibitors (genistrin and methyl-β-cyclodextrin) but not by clathrin-mediated inhibitors (chlorpromazine) or macropinocytosis inhibitors (cytochalasin D), which suggested that C6-LO was mainly internalized into M1-type macrophages through caveolin-mediated endocytosis, unlike the uptake of C6-L (Supplementary Fig. 3b). As displayed in Supplementary Fig. 3c and d, C6-LO gradually colocalized with the Golgi tracker, and the highest Pearson correlation coefficient reached 0.78 after 6 h of incubation, which indicated that most of the internalized C6-LO was transported to the Golgi. In contrast, the Pearson correlation coefficients for colocalization with lysosomes or macropinosomes were 0.55 (P < 0.01) and 0.44 (P < 0.001), respectively. Surprisingly, the DL (µg/106) of RILO@M was increased to 29.75 ± 4.76 and 25.13 ± 2.75 µg/106 for R848 and INCB, respectively (Supplementary Fig. 3e). Collectively, the encapsulation of OMVs changed the endocytic pathway of C6-LO and increased the DL of M1-type macrophages. To further reveal the mechanism of endocytic pathway changes, we performed proteomic analyses on the OMVs. The FimH protein was detected (Supplementary Table 2), which can bind to CD48 on the cell surface, causing E. coli to infect macrophages through caveolin-mediated endocytosis43,44. Therefore, the FimH protein contained in OMVs may contribute to the caveolin-mediated endocytosis of RILOs. Furthermore, the stability of RILO in RILO@M was evaluated and visualized by TEM at 0 and 48 h. As shown in Fig. 2f, spherical nanostructures were observed, which represented RILO stabilized in RILO@M without degradation. The representative confocal images also showed the same results (Fig. 2g). DSPE-Rhodamine B and C6 were selected to label lipids (red) and replace the drug (green), respectively, and stronger colocalization (yellow) was observed in confocal images at 0, 24 and 48 h. These results indicated that RILO could maintain a stable nanostructure in M1-type macrophages.
We used GTP based on our previous work22, which was synthesized and verified by proton nuclear magnetic resonance (1H-NMR, Supplementary Fig. 4). We also determined optimal formulations of surface GPC3 peptide anchoring by investigating the effects of incubation concentration and time on cell viability and GTP carrying. 100 μg/mL (quantified by DSPE-PEG5k-GTP) and 20 min were set as the optimal incubation concentration and time for the preparation, respectively (Supplementary Fig. 5a–c). To characterize the resulting RILO@MG, GTP was tagged by FITC. Free GTP without DSPE-PEG5k linked groups (RILO@M-free GTP) was also prepared as a contrast. As displayed in Fig. 2h, the RILO@MG group, but not the RILO@M-free GTP group, showed that GTP colocalized with the macrophage surface marker F4/80, suggesting that GTP was successfully retained on the cell surface by a lipid anchoring strategy. We then tested the stability and retention of GTP on the cell surface by confocal microscopy imaging and flow cytometry. GTP cells stayed attached to the surface of cells for up to 48 h at 37 °C in DMEM containing 10% foetal bovine serum (FBS) (Fig. 2i, j). Finally, we analysed the phenotype of RILO@MG in different culture environments by flow cytometry. We noted that compared to the Blank@M group, the RILO@MG group could maintain high expression of CD80 with low expression of CD206, even in HCC conditioned medium (HCM) (Fig. 2k, l), indicating that RILO@MG could resist the immunosuppressed TME to maintain the M1 phenotype.
RILO@MG generated RI-exosomes achieved the release of RILOs
We evaluated the release of RILOs in RILO@MG by the biogenetic mechanism of exosomes. As shown in the schematic illustration, the conical structure of C16-ceramide could spontaneously induce negative curvature in the phospholipid bilayer of the membrane, causing membrane invagination and promoting the formation of vesicles. The released drugs mainly included free and nonfree forms, among which the nonfree drug form could be divided into RI-exosomes and RI-microvesicles (RI-MV) (Fig. 3a). First, the release profiles of all forms of R848 and INCB from R@MG or I@MG, RILO@MG- (RILO@MG without C16-ceramide) and RILO@MG were measured over 72 h (Supplementary Fig. 6). We further clarified the release profiles of free and nonfree drugs from RILO@MG, which could be separated by ultrafiltration centrifugation (100 kDa) and quantified by high-performance liquid chromatography (HPLC) (Fig. 3b, c). The proportion of cumulative released nonfree R848 to all forms from RILO@MG at 72 h was 75.38 ± 1.16% (Fig. 3d), which was significantly higher than the proportion of released free R848 from RILO@MG (24.62 ± 1.16%, P < 0.001) and the proportion of released nonfree R848 from RILO@MG- (57.03 ± 0.48%, P < 0.001). The analysis based on INCB quantitative data showed the same conclusion (Fig. 3e). In stark contrast, the released medium from RILO@MG at 24 h after preparation rather than Blank@M or RILO@MG- contained a large number of spherical nanoparticles (Fig. 3f). These results demonstrated that RILO@MG released more nonfree drugs with the help of C16-ceramide.
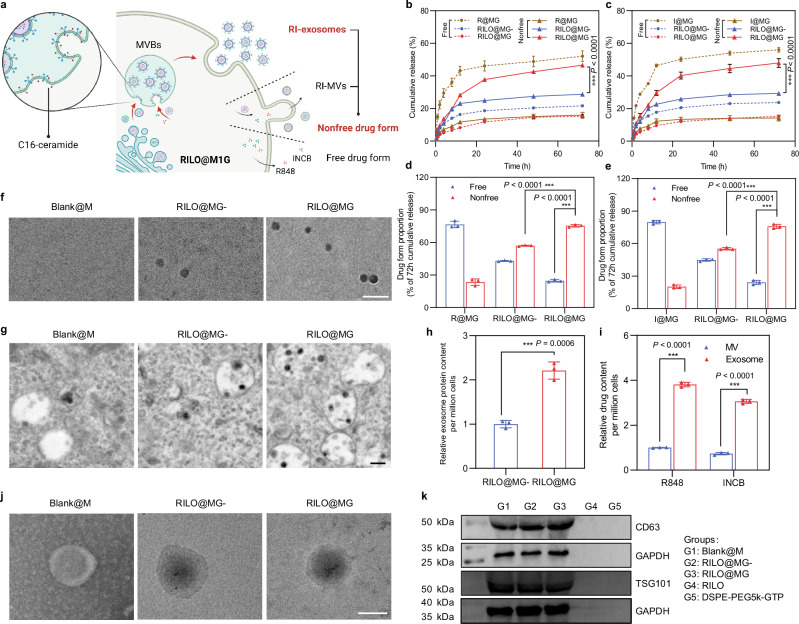
Fig. 3. RILOs are released from RILO@MG in the form of RI-exosomes.
a Schematic illustration of regulating the release form of the RILOs packed in RILO@MG. b–e Release profiles during 72 h for R848 (b) or INCB (c) and the proportion of cumulative release at 72 h for R848 (d) or INCB (e) of free drug form or nonfree drug form from different groups, respectively (n = 3 biologically independent experiments). f Representative TEM analyses about released media from Blank@M, RILO@MG- and RILO@MG groups at 24 h after preparation (n = 3 biologically independent experiments). Scale bar, 200 nm. g Representative TEM images showed increasing MVBs and ILVs formation containing RILO in RILO@MG with the help of C16-ceramide (n = 3 biologically independent experiments). Scale bar, 200 nm. h The relative released exosome protein content 24 h after RILO@MG- and RILO@MG were prepared (n = 3 biologically independent experiments). i R848 and INCB content encapsulated in exosomes or MVs by RILO@MG as measured by HPLC, indicating that the drugs were mostly contained in exosomes, not MVs (n = 3 biologically independent experiments). j TEM images of exosomes from Blank@M and RI-exosomes from RILO@MG- and RILO@MG (n = 3 biologically independent experiments). Scale bar, 50 nm. k Detection of exosome markers (CD63 and TSG101) of released exosomes from different groups by western blotting (n = 3 biologically independent experiments). Data are expressed as the mean ± SD. Two-tailed Student’s t-test (d, e, h, i) or two-way ANOVA with Tukey’s multiple comparisons test (b, c) was carried out for statistical analysis. *P < 0.05; ***P < 0.001. BMDMs were used in all experiments involving macrophages.
Since extracellular vesicles released by living cells are mainly divided into exosomes and MVs, we focused on RI-exosomes and RI-MVs in the nonfree drug form subgroups. TEM images depicted an increased amount of multivesicular bodies (MVB) and intralumenal vesicles (ILV) and embryonic exosomes existing within MVBs in RILO@MG group compared with the RILO@MG- group (Fig. 3g). Total protein quantification of released exosomes using a bicinchoninic acid assay further confirmed that C16-ceramide increased the generation of exosomes in RILO@MG (P < 0.001, Fig. 3h). Subsequently, we separated exosomes and MVs released from RILO@MG by standard multistep ultracentrifugation. Importantly, the R848 and INCB content encapsulated in exosomes was 3.81-fold higher than that in MVs (Fig. 3i), indicating that the drugs were mostly contained in exosomes, not MVs. TEM images of exosomes from the Blank@M, RILO@MG- and RILO@MG groups showed no differences in the appearance of exosomes from these groups, but exosomes from the RILO@MG- and RILO@MG groups contained nanoparticles dyed black by phosphotungstic acid (Fig. 3j). Exosome markers (CD63 and TSG101) were detected by western blot in RI-exosomes released by RILO@MG with a size of 87.57 ± 4.21 nm (Fig. 3k and Supplementary Table 3). This finding supports RILO@MG generating RI-exosomes to achieve the release of RILOs.
RILO@MG displayed tumour accumulation and deep penetration capabilities
The effect of RILO@MG on cellular uptake was evaluated in H22 cells and TAMRAW. TAMRAW was generated by culturing RAW264.7 cells in HCM for 24 h. Fluorescent-labelled C6 formulations were prepared. To elucidate drug uptake at the tumour site, C6 released cumulatively at 48 h from LO@M or LO@MG was collected for these experiments. For H22 cells, at 1 and 4 h, the fluorescence intensity of C6 released from the LO@MG group was higher than that of C6 released from LO@M (Fig. 4a), and similar results were confirmed based on the data of flow cytometry (P < 0.01, P < 0.01, Fig. 4b). For TAMRAW, the internalization of C6 released from LO@MG was time-dependent. Compared with C6-LO and C6 released from LO@M groups, the fluorescence intensity of C6 released from LO@MG group showed no significant difference (Fig. 4c, d), which suggested that C6 released from LO@MG could be ingested by TAM as expected. Furthermore, we found that the RI-exosomes accumulated at 48 h from RILO@MG contained GTP by examining FITC-tagged GTP (Supplementary Fig. 7a). In the uptake experiments of these RI-exosomes by H22 cells, the fluorescence intensity of the C6-exosomes released from the LO@MG group was stronger than that of the C6-exosomes released from the LO@M group. The experimental results of the competitive inhibition assay showed that GPC3 mediated C6-exosome release from LO@MG into H22 cells rather than TAMRAW (Supplementary Fig. 7b–e). These results demonstrated that C6 released from LO@MG could be efficiently internalized into H22 cells and TAMs.
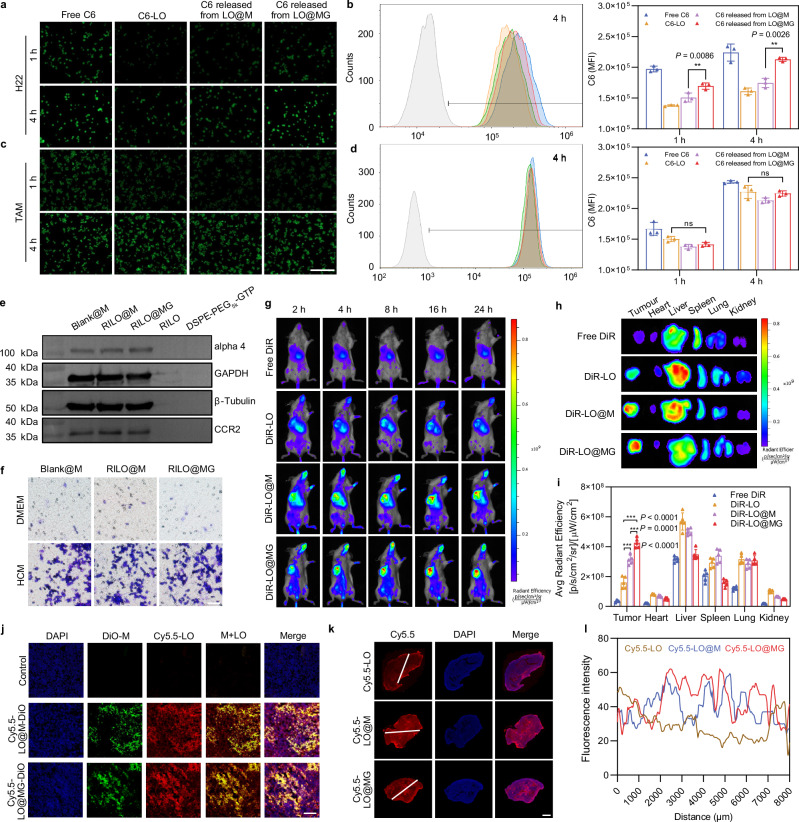
Fig. 4. The tumour accumulation and deep penetration capabilities of RILO@MG.
a, b Fluorescence microscopy images (a) and flow cytometric analysis (b) of the uptake by H22 cells at the same concentration (200 ng/mL) of C6 (n = 3 biologically independent experiments). c, d Fluorescence microscopy images (c) and flow cytometric analysis (d) of the uptake by TAMRAW at the same concentration (200 ng/mL) of C6 (n = 3 biologically independent experiments). The drug was replaced by C6 (green) in (a–d). Scale bar = 200 μm in (a–d). e Detection of chemotaxis-associated proteins (alpha 4 and CCR2) by western blotting (n = 3 biologically independent experiments). f Tumour-migrating capability was evaluated by Transwell assay in vitro (n = 3 biologically independent experiments). Representative microscopy images of migrating Blank@M, RILO@M and RILO@MG after incubation for 16 h. Scale bar = 200 μm. g–i In vivo images (g) of biodistribution in the H22 tumour-bearing mouse model after treatment with different formulations (i.v.). Ex vivo images (h) and average radiant efficiency (i) of tumours and main organs at 24 h (n = 5 biologically independent animals per group). DiR was a near-infra-red tracer in (g–i). j Representative confocal images of the H22 tumour tissue sections after i.v. administration at 24 h, which indicated that injected M1-type macrophages could release drugs at the tumour site (n = 3 biologically independent experiments). DiO and Cy5.5 were selected to label injected M1-type macrophages (green) and replace the drug (red), respectively. Scale bar: 100 μm. k, l Penetration overviews (k) of the whole H22 tumour tissue sections after i.v. administration at 24 h (n = 3 biologically independent experiments). The fluorescence intensity of the white line marked region was quantified with ImageJ (l). Cy5.5 was selected to replace the drug (red). Scale bar, 2 mm. Data are expressed as the mean ± SD and were processed by one-way ANOVA with Tukey’s multiple comparisons test (b, d, i). **P < 0.01; ***P < 0.001; ns, no significance. BMDMs were used in all experiments involving macrophages unless marked RAW superscript.
Next, chemotaxis-associated proteins (alpha 4 and CCR2) from different formulations were determined by western blotting, which confirmed that the protein expression of M1-type macrophages was not changed by RILO packing and GTP anchoring (Fig. 4e). Subsequently, we observed no reduction in the migration capacity of RILO@M and RILO@MG compared with unmodified M1-type macrophages by Transwell assay (Fig. 4f). These results suggested that RILO@MG maintained an intact chemokine-mediated tumour accumulation capacity. We chose H22 tumour-bearing mice to evaluate the tumour accumulation ability of RILO@MG. Fluorescently labelled RILO@MG and other formulation groups were prepared by the near-infra-red tracer DiR. In vivo biodistribution images after treatment with DiR-LO@MG (i.v.) showed enduring fluorescent accumulation in the tumour site at corresponding time intervals (Fig. 4g). Compared with the free DiR, DiR-LO and DiR-LO@M groups, the DiR-LO@MG group exhibited the highest fluorescence intensity, suggesting that M1-type macrophages, especially M1-type macrophages with GTP modification, could enhance accumulation at the tumour site. Ex vivo images (Fig. 4h) and average radiant efficiency at 24 h (Fig. 4i) were performed to accurately assess the distribution of different formulations in tumours and main organs. Compared with the DiR-LO and DiR-LO@M groups, the DiR-LO@MG group had a significantly increased tumour fluorescence signal (P < 0.001, P < 0.001). These results indicated that RILO@MG has strong tumour-directing abilities through chemotaxis and GPC3-mediated targeting action.
As expected, RILO@MG could release the packed RILOs in tumour tissues according to confocal images of the H22 tumour tissue sections (Fig. 4j). DiO and Cy5.5 were selected to label injected M1-type macrophages (green) and replace the drug (red), respectively. There was strong red and green fluorescence colocalization (yellow) in Fig. 4j, which indicated that RILO@MG could successfully deliver drugs to tumour tissues. At the same time, red fluorescence was observed outside the colocalization region, indicating that drugs could be released from injected RILO@MG at the tumour tissues. We also used Cy5.5 (red) to replace the drug and investigate the tumour penetration capability. Penetration overviews of tumour sections (Fig. 4k) and fluorescence intensity of the white line marked region (Fig. 4l) showed that there was a much stronger fluorescence intensity farther away from the tumour boundary in the Cy5.5-LO@M and Cy5.5-LO@MG groups than in the Cy5.5-LO group. Collectively, these results suggested that drugs loaded in RILO@M or RILO@MG tended to penetrate deeper than those loaded in RILO, which could be attributed to the chemotactic mobility of M1-type macrophages within tumours36.
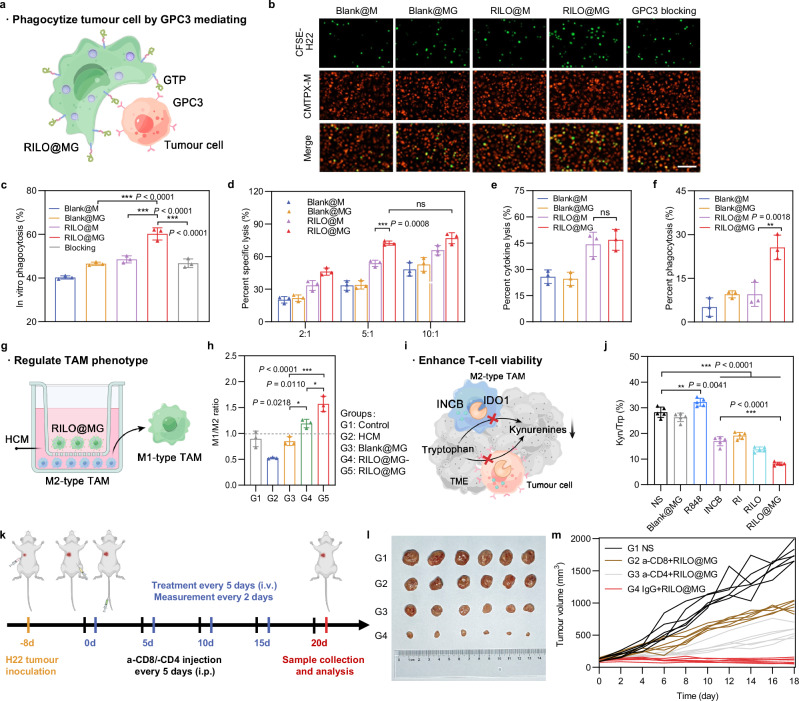
RILO@MG played an immunotherapeutic role by specifically phagocytizing tumour cells, regulating the TAM phenotype and reducing the percentage of Kyn/Trp
In our study, RILO@MG exerted immunotherapeutic effects primarily through three pathways, including tumour cell phagocytosis (Fig. 5a), TAM phenotype regulation (Fig. 5g) and Kyn/Trp percentage reduction (Fig. 5i), which jointly promoted the antitumour immune response.
Fig. 5. RILO@MG promotes antitumour immunity by specifically phagocytizing tumour cells, regulating the TAM phenotype and enhancing T-cell viability.
a Schematic illustration of RILO@MG phagocytizing H22 cells mediated by GPC3 and GTP. b, Fluorescence microscopy images of different formulations after coculture with H22 cells for 4 h (E:T = 2:1). Scale bar = 100 μm. E:T, effector cell (different formulations prepared using M1-type macrophage) to target cell (H22 cell) ratio. c Anchoring of GTP on RILO@MG promoted the phagocytosis of H22 cells (n = 3 biologically independent experiments). Different formulations were cocultured with H22 cells for 4 h (E:T = 1:1) and evaluated by flow cytometric analysis. d Specific lysis of H22 cells after coculture for 12 h at different E:T ratios measured by CCK-8 assay (n = 3 biologically independent experiments). e, f Percent cytokine lysis (e) and phagocytosis lysis (f) of H22 cells after coculture with different formulations for 12 h (E:T = 5:1) by using Transwell plates (pore size 0.4 μm) (n = 3 biologically independent experiments). g Experimental process for promoting the polarization from M2 phenotype to M1 phenotype. h Phenotype analysis of TAMs after coculture with different formulations in HCM for 24 h (n = 3 biologically independent experiments). i Schematic illustration of RILO@MG enhancing T cells by inhibiting the production of Kyn. j IDO1 activity was evaluated according to the percentage of Kyn/Trp within tumours (n = 5 biologically independent animals per group). k Regimen of the antitumour experiment after removal of CD4+ or CD8+ T cells. l, m Photographs of tumours (l) and individual tumour growth curves (m) showed the critical role of T cells in antitumour immunity mediated by RILO@MG (n = 6 biologically independent animals per group). Data are expressed as the mean ± SD and were processed by one-way ANOVA with Tukey’s multiple comparisons test (c, e, f, h, j) or two-way ANOVA with Tukey’s multiple comparisons test (d). *P < 0.05; **P < 0.01; ***P < 0.001; ns no significance. BMDMs were used in all experiments involving macrophages.
To further substantiate the surface anchoring strategy in promoting the phagocytosis of M1-type macrophages through receptor–ligand interactions. The in vitro coculture assay of RILO@MG and cancer cells was employed for validation from multiple angles. We employed fluorescence microscopy to observe the phagocytosis of CFSE-labelled H22 cells (green) by different CMTPX-labelled formulations (red), including Blank@M, Blank@MG, RILO@M and RILO@MG (Fig. 5b). Considering that macrophages were adherent cells and H22 cells were suspended cells, we added CFSE-labelled H22 cells to different CMTPX-labelled preparations that were already adherent. After coincubation for an additional 4 h, the CFSE-labelled H22 cells were washed to remove the unattached cells. As indicated in Fig. 5b, more CFSE-labelled H22 cells were present in the RILO@MG group than in the other groups. The strongest colocalization (yellow) of CFSE-labelled H22 cells and CMTPX-labelled RILO@MG was observed in the RILO@MG group, suggesting that RILO@MG could effectively recognize, adhere to and phagocytize H22 cells. Similar results were obtained from flow cytometric data (Fig. 5c and Supplementary Fig. 8). The percentage of phagocytosis was the ratio of dual-positive M1-type macrophages (CFSE+DiD+) to total M1-type macrophages (CFSE+). The Blank@MG and RILO@M groups only slightly enhanced the percentage of phagocytosis. In contrast, RILO@MG increased the percentage of phagocytized H22 cells by M1-type macrophages more than 2-fold compared with that of Blank@MG and RILO@M (P < 0.001, P < 0.001). Moreover, the competitive inhibition experiment significantly abrogated phagocytosis enhanced by GTP anchoring (P < 0.001). This result demonstrated that combining surface anchoring with inner packing could significantly improve the tumour-specific and pro-phagocytic function of macrophages. Additionally, we measured the specific lysis capacity of Blank@M, Blank@MG, RILO@M or RILO@MG in H22 cells for 12 h at different E:T ratios by cell counting kit-8 (CCK-8) assay and determined that the optimal E:T was 5:1 (Fig. 5d). The direct killing effect of macrophages on tumour cells is reflected in two aspects: cytokine killing and phagocytosis killing49. To illuminate the specific mechanism by which RILO@MG directly kills H22 cells, we cocultured different formulations (Blank@M, Blank@MG, RILO@M or RILO@MG) and H22 cells for 12 h using 0.4 μm-sized Transwell plates that could allow free passage of bioactive molecules rather than cells to evaluate specific and cytokine lysis. The phagocytosis-killing ability could be obtained from the percentage difference between specific lysis and cytokine lysis. The resultant data showed that compared with RILO@M, RILO@MG further enhanced the direct killing of tumours through phagocytosis rather than cytokine killing (P < 0.01, Fig. 5e, f). All of these results illustrated that RILO@MG could effectively recognize, adhere, phagocytize and kill H22 cells.
Theoretically, highly activated M1-type macrophages could reverse the phenotype of adjacent M2-type macrophages by secreting cytokines, exosomes and other mechanisms. In our study, RI-exosomes generated from RILO@MG could further enhance M2-to-M1 reversion. TAMs were provided by culturing BMDMs in HCM for 24 h. We explored the influence of RILO@MG on TAMs by continuing to coincubate the two kinds of cells in HCM in Transwell plates (pore size 0.4 μm). The TAMs incubated in the lower chamber were collected for flow cytometry. Based on the calculation of the M1/M2 ratio (Fig. 5h), it was found that the ratio of freshly harvested TAMs dropped from 0.89 ± 0.15 to 0.52 ± 0.02 after continuing culture in HCM for 24 h when the upper chamber was free of formulations. In contrast, the calculation of the M1/M2 ratio was elevated sharply to 1.57 ± 0.15 when RILO@MG was introduced in coculture for 24 h, which showed a higher ratio than that of the RILO@MG- group (P < 0.05), indicating that releasing drugs in exosome form exerted an excellent effect on the M2-to-M1 reversion of TAM in the lower chamber. Adoptively transferred macrophage and TAM in the tumour could be distinguished by fluorescent labelling technology12. Based on this, we used IVISense 680 fluorescent dye labelling injected macrophage of Blank@M, RILO@M and RILO@MG groups to evaluate the phenotypes of adoptively transferred macrophage and TAM in an H22 tumour-bearing mouse model (Supplementary Fig. 9a, b). We found that RILO@M and RILO@MG could maintain the M1 phenotype in vivo. And the experiment results further showed that endogenous TAM of mice treated with RILO@MG was polarized to M1 phenotype, as evidenced by significantly increased expressions of CD80 compared with TAM of mice treated with Blank@M (P < 0.001), which indicated the action of RILO@MG could convert TAM to play anti-tumour role.
Moreover, the activity of IDO1 was studied in vivo. Compared with the NS group, the INCB, RI, RILO and RILO@MG groups showed lower percentages of Kyn/Trp (P < 0.001, Fig. 5j), which was related to the inhibitory effect of INCB on IDO1. Compared with the INCB group, the RILO@MG group effectively decreased the percentage of Kyn/Trp, which may be related to the tumour accumulation and deep penetration capabilities of RIL@MG (P < 0.001). To further confirm the efficacy of enhanced T cells in tumour cell killing, the therapeutic effect of RILO@MG was evaluated after removing either CD8+ or CD4+ T cells (Fig. 5k). Antitumour effects were significantly attenuated in the RILO@MG group after depleting CD8+ or CD4+ T cells (P < 0.001, P < 0.001, Fig. 5l, m and Supplementary Fig. 10), presenting the critical role of CD8+ or CD4+ T cells in immunotherapy achieved by RILO@MG. Overall, RILO@MG exerted strong antitumour effects by specifically phagocytizing tumour cells, regulating the TAM phenotype and enhancing T-cell viability due to the combined contributions from both the surface anchoring and inner packing strategies. These results motivated our next comprehensive assessment of immune cells and cytokine levels in the TME.
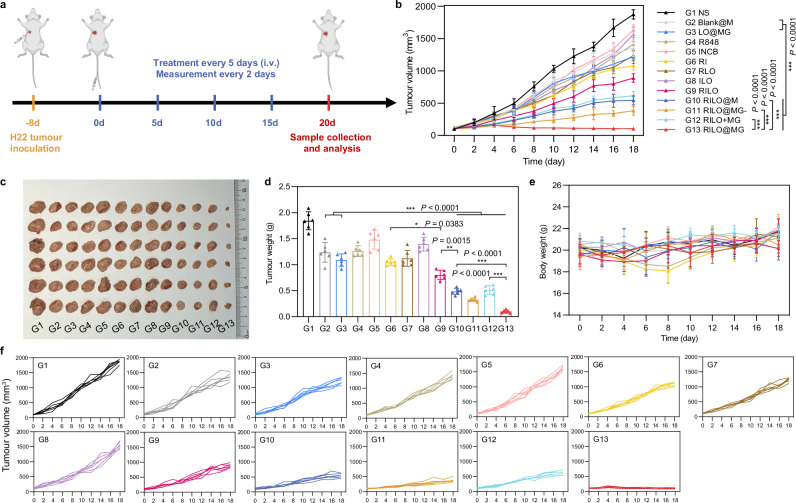
RILO@MG significantly improved antitumour efficacy in an H22 tumour-bearing mouse model
We next evaluated the antitumour efficacy of RILO@MG in H22 tumour-bearing mice, which expressed GPC3. In order to investigate the antitumour effects of different preparations, the mice were randomly divided into 13 groups for a single-factor test and given four i.v. injections with one of the following formulations as in Fig. 6a. NS group was set as the blank control group. First, we set up R848, INCB, RI, RLO, ILO and RILO groups to prove the advantages of combining TLR7/TLR8 agonists with IDO1 inhibitors (compared R848 and INCB groups with RI group), nanomedicine delivery (compared R848 and INCB groups with RLO and ILO groups) and co-loading drugs into one nanoparticle (compared RI group with RILO group). Second, we set up Blank@M, LO@MG, RILO, combination treatment of RILO and MG (RILO + MG) and RILO@MG groups to prove the necessity for an inner packing strategy. Third, RILO@M and RILO@MG- groups were used to demonstrate the role of surface anchoring strategy and releasing drugs in exosome form, respectively. Tumour volume was monitored after the first treatment. The RILO@MG group had the slowest trend of tumour growth, with almost no growth in tumour volume over 20 days (Fig. 6b, f). The combination of TLR7/TLR8 agonists and IDO1 inhibitors could simultaneously regulate the TAM phenotype and enhance T-cell viability to produce anti-tumour effects. The RILO group, with a 56.46 ± 4.90% tumour inhibition rate, had a lower tumour inhibition rate than that of the RLO (P < 0.001), ILO (P < 0.001) and RI group (P < 0.05), indicating the effectiveness of combination R848 and INCB, especially when co-loaded into the one particle (Supplementary Fig. 11). Compared with Blank@M and LO@MG groups, RILO@M, RILO@MG- or RILO@MG group resulted in smaller tumour volumes at the study endpoint (P < 0.001, P < 0.001, P < 0.001, Fig. 6b), indicating the necessity of modifying M1-type macrophagesfor drug delivery. Compared with RILO@MG, the RILO + MG group exhibited only moderate antitumour effects, which proved that merely a combination of RILO and MG did not trigger the antitumour effects, and more strongly indicated that only inner packing strategy could effectively inhibit tumour growth. Compared with RILO@MG, RILO@M only inhibited tumour growth by 73.72 ± 3.24% under the same dose regimen (P < 0.001, Supplementary Fig. 11), which showed the antitumour role of GPC3-mediated targeting. The tumour volume in the RILO@MG group was 102.49 ± 16.78 mm3, which was significantly smaller than that in the RILO@MG- group (P < 0.001, Fig. 6b), suggesting that the release form of RI-exosomes contributed to the promotion of antitumour efficacy. These data fully demonstrated the contribution of surface anchoring and inner packing strategies to antitumour efficacy.
Fig. 6. The antitumour activity of RILO@MG in an H22 tumour-bearing mouse model.
a Regimen of i.v. administration in H22 tumour-bearing mice (at a dosage of 3.0 × 106 cells per mouse per injection, equal to 4 mg/kg R848 and 3.4 mg/kg INCB). Mice requiring injected formulations made by M1-type macrophages each received the equivalent number of cells (3.0 × 106 cells per mouse). Mice requiring injection of other formulations each received the equivalent dose of medicine (4 mg/kg R848 and 3.4 mg/kg INCB). When the tumour volumes reached ~2000 mm3, the mice were euthanized by CO2. b–f Average tumour growth curves (b), tumour photographs (c), tumour weights (d), body weight changes (e) and individual tumour growth curves (f) of H22 tumour-bearing mice receiving the indicated treatments (n = 6 biologically independent animals per group). Data are expressed as the mean ± SD. One-way ANOVA with Tukey’s multiple comparisons test (d) or two-way ANOVA with repeated measures (b) was used for statistical analysis. *P < 0.05; **P < 0.005; ***P < 0.001. BMDMs were used in all experiments involving macrophages.
Mice in each group were euthanized by CO2 per institutional policy and dissected at 20 days post-first treatment, and the major organs and tumours were collected for analysis. Consistent with the tumour growth profiles of the analysed tumours, the tumours in the RILO@MG group were the smallest in all the groups, as shown in the tumour weights and tumour images (Fig. 6c, d). Based on the results of Ki-67 immunohistochemistry, haematoxylin and eosin (H&E) staining and TUNEL immunofluorescence, we analysed the proliferation and apoptosis of tumours. The sectioning results showed that the RILO@MG group had more tumour cell apoptosis and less tumour cell proliferation (Supplementary Fig. 12). In addition, none of the groups showed significant loss of body weight at the study endpoint (Fig. 6e) or tissue damage (Supplementary Fig. 13). Furthermore, RILO@MG did not cause the appearance of a cytokine storm after initial administration, which would be evident by elevated serum IL-6 and TNF-α levels, a significant change in the organ/body weight ratio at the study endpoint and an increase in the levels of liver or kidney injury markers at the study endpoint (BUN, LDH, ALT, AST and ALP, Supplementary Fig. 14). These results confirmed that RILO@MG had preliminary safety and efficacy in H22 tumour-bearing mice.
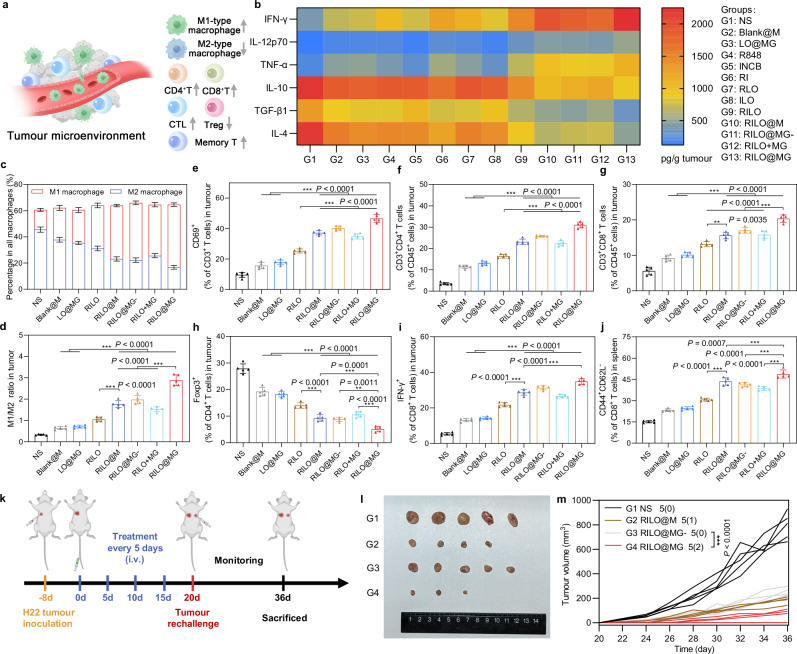
RILO@MG reversed the suppressive TME by regulating the TAM phenotype and enhancing T-cell viability, and produced long-term immune memory in an H22 tumour-bearing mouse model
The frequencies of immune cells (M1-type macrophage, M2-type macrophage, CD69+ T cell, CD4+ T cell, CD8+ T cell, CTL and Treg) and the concentrations of cytokines (including IFN-γ, IL-12p70, TNF-α, IL-10, TGF-β1 and IL-4) in tumour tissues collected on Day 20 and the frequencies of effector memory T-cell in spleens collected on Day 20 were used to monitor the changes in immune status of the TME (Fig. 7a).
Fig. 7. The RILO@MG regulates TAM phenotype and enhances T-cell viability to remodel the suppressive TME in an H22 tumour-bearing mouse model.
a Schematic diagram of the regulatory effect of RILO@MG on the immunosuppressive TME, indicating an increase about immune-active cells and a decrease in the proportion of immune-suppressing cells within the TME. b H22 tumour-bearing mice were treated as in Fig. 6a. The intratumoural levels of cytokines were quantified using ELISA analysis at the study endpoint. c–i H22 tumour-bearing mice were treated as in Fig. 6a. Flow cytometric analysis of M1-type macrophage and M2-type macrophage (c, d), CD69+ T cells (e), CD4+ T cells (f), CD8+ T cells (g), CD4+Foxp3+ T cells (h), and CD8+IFN-γ+ T cells (i) within the TME. j Flow cytometric analysis quantification of effector memory (CD44+CD62L-) CD8+ T cells in spleens. k Experimental timeline of rechallenged tumour model establishment. l Secondary tumour photographs of the sacrificed mice at the study endpoint. m Rechallenged tumour growth curves of tumour-bearing mice were monitored over time. The dosage regimen of all data in Fig. 7 was the same as that shown in Fig. 6a. Data are expressed as the mean ± SD with five biologically independent animals per group and were processed by one-way ANOVA with Tukey’s multiple comparisons test (c–j) or two-way ANOVA with repeated measures (m). **P < 0.01; ***P < 0.001. BMDMs were used in all experiments involving macrophages.
To evaluate the overall immune status of the tumour, we first measured the levels of intratumoural cytokines. The heatmap showed that mice given RILO@MG had increased intratumoural levels of immunostimulatory cytokines, including IFN-γ, IL-12p70 and TNF-α, along with reduced intratumoural levels of immunosuppressed cytokines, including IL-10, TGF-β1 and IL-4 (Fig. 7b), indicating that RILO@MG had an immunomodulatory effect. Encouraged by these expected results, we next examined the tumour infiltration of immune cells.
Recent preclinical and clinical insights indicated that macrophages were the most abundant nonneoplastic critical effector cells of cancer immunotherapy in the TME50,51. The population of M1-type macrophages within the TME of mice in the RILO@MG group was 3.19-fold higher than that in the NS group, and the opposite trend was observed for M2-type macrophages (Fig. 7c and Supplementary Fig. 16a, b). The M1/M2 ratio in the RILO@M group was higher than that in the RILO group (P < 0.001, Fig. 7d), implying that inner RILO packing of M1-type macrophages could repolarize M2-type macrophages to M1-type macrophages in vivo. Compared with the Blank@M, LO@MG and RILO@MG- groups, RILO@MG had the highest M1/M2 ratio (P < 0.001, P < 0.001, P < 0.001), which implied that the released drugs, especially in exosome form, played a key role in M2-to-M1 reversion of macrophage in the tumour. This conclusion was in agreement with the experimental results verified in vitro (Fig. 5g).
Based on the fact that T cells played a key role in the immunotherapy achieved by RILO@MG (Fig. 5k, l), we further evaluated the frequencies of multiple intratumoural T cell subtypes. CD69 was one of the earliest markers of upregulation after T-cell activation, and CTL-infiltrating tumour tissues are generally considered the primary mediator of tumour killing. The RILO@MG and RILO@M groups revealed a higher percentage than the RILO group of CD69+ T cells (P < 0.001, P < 0.001, Fig. 7e and Supplementary Fig. 16c), CD4+ T cells (P < 0.001, P < 0.001, Fig. 7f and Supplementary Fig. 16d), CD8+ T cells (P < 0.001, P < 0.01, Fig. 7g) and CTLs (IFN-γ+CD8+ T cells) (P < 0.001, P < 0.001, Fig. 7i and Supplementary Fig. 16f), indicating that packing RILOs in M1-type macrophages was required to further enhance the activation of multiple T cells in tumour tissue. Next, the RILO@MG group greatly decreased the proportion of Tregs compared with the other groups (Fig. 7h and Supplementary Fig. 16e). Considering the antitumour role of GPC3-mediated targeting, we focused on comparing the immune cell changes between RILO@M group and RILO@MG group. Significantly, the RILO@MG group had more CD69+ T cells, CD4+ T cells, CD8+ T cells and CTLs (IFN-γ+CD8+ T cells) and fewer Tregs than the RILO@M group, showing better immune cell regulation in tumour tissues (Fig. 7e–i). These results collectively suggested that RILO@MG could change the immune status of the TME from immune suppressive to immune active, thus promoting the antitumour immune response.
Additionally, RILO@MG treatment resulted in the highest percentage of effector memory (CD44+CD62L-) CD8+ T cells compared with that after treatment with the other macrophage-based formulations (Fig. 7j and Supplementary Fig. 17a). We were thus interested in determining whether RILO@MG could induce memory immune responses. To establish a rechallenged tumour model, H22 tumour-bearing mice that had been treated four times were reinoculated with H22 cells (Fig. 7k). Rechallenged tumours in mice administered RILO@MG were significantly delayed, while rapid growth was observed in the NS group (Fig. 7m and Supplementary Fig. 17b). Remarkably, two out of five mice were still tumour free on Day 36 in the RILO@MG group (Fig. 7l). These data indicated that RILO@MG could effectively activate the long-term antitumour response and effectively inhibit tumour growth in mice.
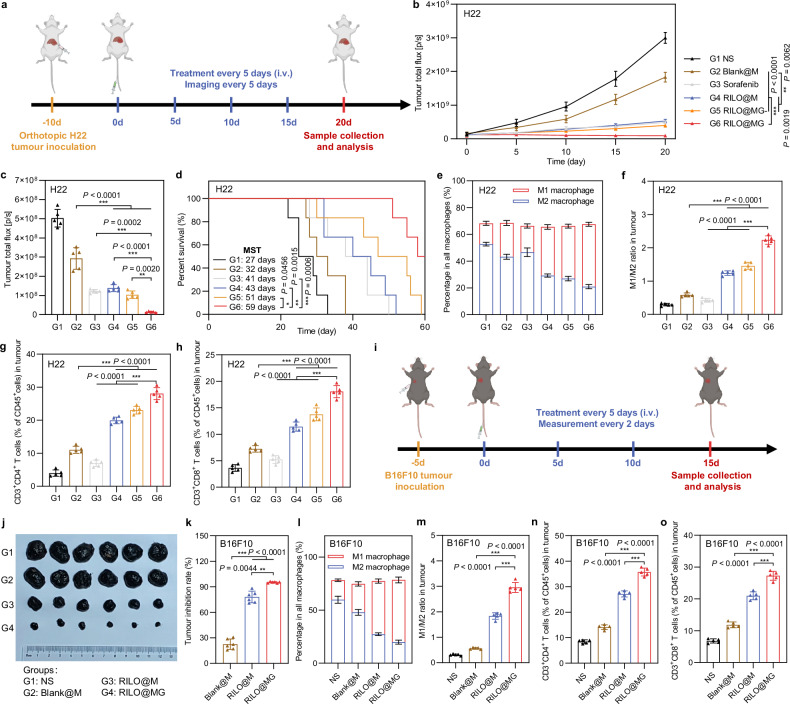
RILO@MG showed antitumour efficacy in an orthotopic HCC mouse model and B16F10 tumour-bearing mouse model
The antitumour efficacy of RILO@MG was further evaluated in orthotopic HCC model mice. Anaesthetised mice were injected with H22-luc cells in the left lobe of the liver, and the development of tumours was monitored via the bioluminescence of H22-luc cells. The treatment regimen was identical to that for the subcutaneous tumour model (Fig. 8a). Furthermore, we selected sorafenib, a first-line systemic therapeutic drug for patients with HCC, as a positive control to evaluate the clinical therapeutic potential of RILO@MG. Based on the results of in vivo bioluminescence intensity (Fig. 8b and Supplementary Fig. 18a) and ex vivo bioluminescence quantification of livers (Fig. 8c and Supplementary Fig. 18b), we found that RILO@MG showed therapeutic effects. Compared with Blank@M group, four doses of RILO@M, RILO@MG- or RILO@MG resulted in lower ex vivo bioluminescence quantification of livers at the study endpoint (P < 0.001, P < 0.001, P < 0.001, Fig. 8c), again indicating the necessity for inner packing strategy. When comparing RILO@M and RILO@MG groups, we found the antitumour role mediated by GPC3 (P < 0.01, Fig. 8c). As shown in Fig. 8d, the median survival time (MST) of orthotopic HCC model mice was increased from 27 days (NS group) to 59 days after treatment with RILO@MG, which was significantly longer than the MST of the sorafenib group (41 days, P < 0.001). Furthermore, three of six mice receiving RILO@MG treatment survived at least 60 days. In contrast, no mice from all other groups survived after 60 days. Consistent with the results observed in the subcutaneous tumour model, following treatment with RILO@MG, the proportions of M1-type macrophages (Fig. 8e, f and Supplementary Fig. 19a), CD4+ T cells (Fig. 8g), and CD8+ T cells (Fig. 8h) were significantly activated in tumours. In contrast, the proportion of M2-type macrophages (Fig. 8e, f and Supplementary Fig. 19b) was significantly decreased in tumours. Altogether, these results suggested that RILO@MG exhibited antitumour efficacy through immune action in orthotopic HCC model mice.
Fig. 8. Efficacy validation of RILO@MG in the orthotopic H22 tumour model and B16F10 tumour-bearing mouse model.
a Schematic of the orthotopic H22 tumour model experiment (dose of 3.0 × 106 cells per mouse per injection, equal to 4 mg/kg R848 and 3.4 mg/kg INCB; sorafenib: 10 mg/kg). b–d In vivo bioluminescence intensity curves (b), ex vivo livers on Day 20 of bioluminescence quantification (c) and animal survival (d) of the orthotopic H22 tumour model receiving the indicated treatments (n = 5 biologically independent animals for b and c and n = 6 biologically independent animals for survival). e-h, Flow cytometry quantitative data of M1-type macrophage and M2-type macrophage (e, f) and CD4+ and CD8+ T cells (g, h) in tumours of the orthotopic H22 tumour model sacrificed on Day 20 (n = 5 biologically independent animals). i Schematic of the B16F10 tumour-bearing mouse model experiment (dose of 3.0 × 106 cells per mouse per injection, equal to 4 mg/kg R848 and 3.4 mg/kg INCB). j, k Tumour photographs (j) and tumour inhibition rate (k) of the sacrificed B16F10 tumour-bearing mice at the study endpoint (n = 6 biologically independent animals). l–o Flow cytometry quantitative data of M1-type macrophage and M2-type macrophage (l, m) and CD4+ and CD8+ T cells (n, o) in tumours of the sacrificed B16F10 tumour-bearing mice (n = 5 biologically independent animals). Data are expressed as the mean ± SD. One-way ANOVA with Tukey’s multiple comparisons test (c, f–h, m–o), the Welch ANOVA with Dunnett’s T3 multiple comparisons test (k), two-way ANOVA with repeated measures (b) and log-rank tests for survival data (d) were used for statistical analysis. **P < 0.01; ***P < 0.001. BMDMs were used in all experiments involving macrophages.
To investigate the generalizability of RILO@MG, we further verified the anti-tumour effects on B16F10 tumour-bearing mouse model with GPC3 expression (Fig. 8i)52,53. The tumour photographs and tumour growth curves among different groups are shown in Fig. 8j and Supplementary Fig. 20a. After treatment, the tumour inhibition rate of RILO@MG group was 95.22 ± 0.81%, which was significantly higher than that in RILO@M and Blank@M group (P < 0.01, P < 0.001), demonstrating surface anchoring and inner packing strategies could better inhibit the tumour progress (Fig. 8k). During the treatment, no significant difference was observed in the body weight changes, exhibiting the preliminary safety (Supplementary Fig. 20b). Similar to the H22 tumour-bearing mouse model, the RILO@MG group significantly enhanced intratumoural M1-type macrophages (Fig. 8l, m), CD4+ T cells (Fig. 8n), and CD8+ T cells (Fig. 8o) infiltration, again indicating that the immunomodulatory capacity of RILO@MG in the B16F10 tumour-bearing mouse model.
Discussion
Haematologic malignancies rapidly develop, and the efficacy of cytotherapy for the treatment of solid tumours has been difficult to enhance, which hinders the progress of this therapy in cancer treatment2. Considering the phagocytosis, specific targeting and deep penetration capacity of macrophages, macrophage-based cytotherapy has emerged. Clinical trials have provided evidence for the feasibility and safety of adoptively transferred macrophages8. Nevertheless, injected macrophages tend to change into the M2 phenotype in the immunosuppressive TME and lose immunological activity, resulting in poor transplantation efficacy, which is the key challenge to overcome to improve cytotherapy for the treatment of solid tumours. Here, we developed a GPC3-targeted macrophage, termed RILO@MG, obtained by surface GPC3 peptide anchoring and inner RILO packing to increase the immunological activity of injected macrophages.
According to our previous studies, the GTP with DHLASLWWGTEL amino acid sequence was selected, which showed the highest specific recognition ability for multiple tumour cells expressing GPC322. We experimentally confirmed that RILO@MG released RI-exosomes and displayed tumour accumulation and deep penetration capabilities by maintaining intact tumour chemotactic mobility and GPC3-mediated recognition. RILO@MG exerted immunotherapeutic effects primarily through three pathways, including the phagocytosis of tumour cells, regulation of the TAM phenotype and enhancement of T-cell activity, which jointly play the antitumour immune role. Notably, administration of RILO@MG exerted remarkable antitumour efficacy in the rechallenged tumour model and tumour-bearing mouse model expressing GPC3 compared with that of all other experimental treatments. Compared with our previously reported nanoparticle-loaded macrophages, our current study focused on using surface anchoring and inner packing strategies to enhance the inherent function of M1-type macrophages in specifically killing tumours and remodelling the TME and emphasized the promotion of specific phagocytosis and drug exosome generation.
For the engineering of macrophages, the components of RILOs and DSPE-PEG5k-GTP could be industrially prepared and stably stored according to a standardized process and only need to be easily incubated with isolated and purified autologous or allogeneic macrophages in vitro before patient use. In conclusion, our proposed GPC3-targeted macrophage strategy bridges the gap between standardized and personalized treatment, shortens the waiting period for patients, and helps accelerate the commercialization of macrophage-based cytotherapy. In addition, the strategies presented here could be applied to other types of cells, such as dendritic cells and natural killer cells. As macrophages play crucial roles in various immune processes, the engineered macrophages presented here may help in the treatment of other immune-related diseases.
Methods
This research complies with all relevant ethical regulations approved by the Laboratory Animal Ethical and Welfare Committee of Shandong University Cheeloo College of Medicine.
Materials
Soya lecithin was purchased from AVT Pharmaceutical Technology (Shanghai, China). INCB, methylthiazolyldiphenyl-tetrazolium bromide (MTT) and CCK-8 were obtained from TargetMol (MA, USA). R848 was purchased from Aladdin Biochemical Technology (Shanghai, China). ELISA kits for IFN-γ, IL-6, IL-10, IL-12, TGF-β and TNF-α were purchased from MultiSciences Biotech (Hangzhou, China). Percoll cell isolation solution was purchased from Solarbio Life Sciences (Beijing, China). Cell stimulation cocktail plus protein transport inhibitors (500X) was purchased from eBioscience (CA, USA). Cell lysis buffer for IP and western blot was purchased from Sperikon Life Science & Biotechnology (Sichuan, China). FBS was purchased from Inner Mongolia Opcel Biotechnology (Inner Mongolia, China). DSPE-PEG5k-Mal was obtained from Ruixi Biological Technology (Xi’an, China). GTP (amino acid sequence: D-Cys-DHLASLWWGTEL) and FITC labelled GTP (GTP-FITC) were synthesized by Leon Biotechnology (Nanjing, China). Recombinant Mouse IFN-γ was purchased from Novoprotein (Shanghai, China). Murine monocyte-colony stimulating factor (M-CSF) was purchased from PeproTech (NJ, USA). Cell proliferation tracer fluorescent probe CFSE was purchased from MeilunBio (Dalian, China). D-luciferin potassium salt was purchased from Shanghai Life-iLab Biotech Co., Ltd. (Shanghai, China). Analytical grade reagents were used unless otherwise indicated.
Antibodies
PerCP/Cyanine5.5 anti-mouse/human CD11b antibody (BioLegend; Catalogue number: 101228; Clone name: M1/70; 1:200 dilution), Brilliant Violet 421™ anti-mouse F4/80 antibody (BioLegend; Catalogue number: 123132; Clone name: BM8; 1:200 dilution), Alexa Fluor® 647 anti-mouse F4/80 antibody (BioLegend; Catalogue number: 123122; Clone name: BM8; 1:400 dilution), PerCP/Cyanine5.5 anti-mouse F4/80 antibody (BioLegend; Catalogue number: 123128; Clone name: BM8; 1:50 dilution), PE anti-mouse CD80 antibody (BioLegend; Catalogue number: 104708; Clone name: 16-10A1; 1:100 dilution), APC anti-mouse CD206 (MMR) antibody (BioLegend; Catalogue number: 141708; Clone name: C068C2; 1:100 dilution), Pacific Blue™ anti-mouse CD45 antibody (BioLegend; Catalogue number: 103126; Clone name: 30-F11; 1:400 dilution), APC anti-mouse CD3 antibody (BioLegend; Catalogue number: 100236; Clone name: 17A2; 1:100 dilution), PE/Cyanine7 anti-mouse CD69 antibody (BioLegend; Catalogue number: 104512; Clone name: H1.2F; 1:50 dilution), FITC anti-mouse CD4 antibody (BioLegend; Catalogue number: 100405; Clone name: GK1.5; 1:400 dilution), PE anti-mouse CD4 antibody (BioLegend; Catalogue number: 100408; Clone name: GK1.5; 1:200 dilution), PE anti-mouse CD8a antibody (BioLegend; Catalogue number: 100708; Clone name: 53-6.7; 1:200 dilution), APC anti-mouse CD8a antibody (BioLegend; Catalogue number: 100712; Clone name: 53-6.7; 1:200 dilution), PE anti-mouse IFN-γ antibody (BioLegend; Catalogue number: 505808; Clone name: XMG1.2; 1:200 dilution), Alexa Fluor® 647 anti-mouse FOXP3 antibody (BioLegend; Catalogue number: 126408; Clone name: MF-14; 1:100 dilution), PerCP/Cyanine5.5 anti-mouse/human CD44 antibody (BioLegend; Catalogue number: 103032; Clone name: IM7; 1:200 dilution), FITC anti-mouse CD62L antibody (BioLegend; Catalogue number: 104406; Clone name: MEL-14; 1:400 dilution), FITC anti-mouse CD206 (MMR) antibody (BioLegend; Catalogue number: 141703; Clone name: C068C2; 1:400 dilution), anti-GAPDH (Affinity; Catalogue: AF7021; Source: Rabbit; 1:10,000 dilution), anti-CD63 (Affinity; Catalogue: AF5117; Source: Rabbit; 1:1000 dilution), anti-TSG101 (Affinity; Catalogue: DF8427; Source: Rabbit; 1:2000 dilution), anti-alpha 4 (Affinity; Catalogue: DF6135; Source: Rabbit; 1:1000 dilution), anti-β-Tubulin (Affinity; Catalogue: DF7967; Source: Rabbit; 1:2000 dilution), anti-CCR2 (Affinity; Catalogue: DF7507; Source: Rabbit; 1:2000 dilution).
Cell culture
BMDMs were obtained as previously reported54. Briefly, female BALB/c mice were selected. Obtained bone marrow cells were cultured in BMDM growth medium (DMEM + 10% FBS + 20 ng/mL M-CSF) under standard conditions (37 °C incubator with 5% CO2). The medium was renewed, and nonadherent cells were removed on Day 3. On Day 7, the formation of BMDMs was evaluated using immunofluorescence double staining with PerCP/Cyanine5.5 anti-mouse/human CD11b antibody (clone: M1/70, BioLegend) and Brilliant Violet 421™ anti-mouse F4/80 antibody (clone: BM8, BioLegend). In cases where polarized BMDMs were desired, the culture was changed to a fresh stimulation medium on Day 7. In detail, for M1-type macrophages, DMEM containing 10% FBS, 20 ng/mL IFN-γ and 100 ng/mL LPS was used for 24 h; for M2-type macrophages, HCM (50% DMEM containing 10% FBS + 50% filtered H22 cell culture supernatant collected at 48 h) was used for 24 h. BMDMs were used in all experiments involving macrophages unless marked RAW superscript.
The mouse hepatocarcinoma cell strains H22 (catalogue number: FH1029) and H22-luc (catalogue number: FH0962), mouse malignant melanoma cell strains B16F10 (catalogue number: FH0361) and the macrophage cell strain RAW264.7 (catalogue number: FH0328) were purchased from Fuheng Biotechnology (Shanghai, China). H22, H22-luc and B16F10 cells were cultured in 10% FBS-containing RPMI 1640 medium with streptomycin and penicillin (1%). RAW264.7 cells were cultured in 10% FBS-containing DMEM. All cells were cultured under standard conditions.
Animals
Female BALB/c mice (6–8 weeks, Stock number: B201) and female C57BL/6 mice (6 ~ 8 weeks, Stock number: B204) were obtained from SPF Biotechnology (Beijing, China). Standard feeding environment (light cycle 12:12, temperature 25 ± 2 °C and humidity 60 ± 10%) for mice. Mice had free access to food and water. All experiments were approved by the Laboratory Animal Ethical and Welfare Committee of Shandong University Cheeloo College of Medicine (No.19031). All animal experiments were performed under the Guide for Care and the Animal Management Rules of the Ministry of Health of the People’s Republic of China. The maximum tumour size in mice permitted by the Laboratory Animal Ethical and Welfare Committee of Shandong University Cheeloo College of Medicine was 2000 mm3. When the tumour size reached this limit, the mice were immediately euthanized by CO2 per institutional policy. The sex was not considered in the study design because there was no direct correlation between the selected tumour model and sex.
Preparation of RIL, OMV and RILO
RIL was prepared by nanoprecipitation methods. Briefly, R848 (9 mg/mL) and INCB (9 mg/mL) were dissolved in ethanol to obtain the organic phase. Soya lecithin (12 mg/mL) was dissolved in 0.5% (w/v) Tween-80 buffer solution was used to obtain the water phase. The volume ratio of the water phase to the organic phase was 10:1. With the help of a microsyringe pump (KDS100, KD Scientific, MA, USA), the organic phase was introduced into the water phase under ice bath stirring conditions at a drop acceleration of 10 mL/h. The resulting liquid after ethanol volatilization was filtered with a 0.22 μm filter. Then, RIL was obtained. For the preparation of C6-L, DiR-L, Cy5.5-L, RL and IL, drugs were replaced with C6 (10 μg/mL or 2 mg/mL for further preparing C6 released from LO@M and LO@MG), DiR (8 mg/mL), Cy5.5 (5 mg/mL), R848 (9 mg/mL) and INCB (9 mg/mL).
OMVs were prepared from the E. coli MG1655 strain following a previously reported protocol55. Briefly, E. coli MG1655 was added to 250 mL LB medium and incubated at 37 °C with shaking at 200 rpm. When the optical density at 600 nm (OD600) measured by spectrophotometer reached 1.2, the supernatant was collected by centrifugation at 4000×g for 10 min at 4 °C and filtered. Next, the filtrate was made into 50 mL using a 100 kDa (MWCO) ultrafiltration tube by centrifugation at 4000×g for 10 min at 4 °C. OMVs were collected and washed with PBS by ultracentrifugation (XPN-100, Beckman Coulter Life Sciences, CA, USA) at 150,000×g for 3 h at 4 °C. Finally, OMVs were resuspended in 1 mL PBS, filtered with a 0.22 μm filter and stored at −80 °C. The bicinchoninic acid (BCA) assay was used to evaluate the total protein.
C16-Ceramide (450 μM) was added to OMVs (400 μg/mL quantified by total protein) followed by sonication for 2 min at 4 °C to obtain C16-ceramide-fused OMVs. Then, the prepared C16-ceramide-fused OMV was mixed with freshly prepared C6-L, DiR-L, Cy5.5-L, RL, IL or RIL at a volume ratio of 1:1 under vortex conditions and extruded 12 times through a LiposoFast-Basic extruder (LF-1, Avestin, Ottawa, Canada) with a 50 nm membrane to obtain C6-LO, DiR-LO, Cy5.5-LO, RLO, ILO or RILO.
Characterization of RILO
The zeta potential, polydispersity index (PDI) and size of RIL, OMV or RILO were measured using a dynamic light scattering analyser (Nano-ZS90, Malvern, UK). The morphology of RIL, OMV or RILO was observed by TEM (HT7700, Hitachi, Tokyo, Japan) after staining with 1% phosphorus tungstate.
The protein expression profiles of OMVs were identified by label-free quantitative proteomics technology. All nanoLC‒MS/MS data of OMVs were analysed by Proteome Discover software. Positive results were obtained for proteins with unique peptide numbers greater than or equal to 2. Finally, the proteins were sequenced according to emPAI scores for further analysis. We also manually checked the subcellular localizations of the detected proteins in the UniPort database. The whole proteins of RIL, OMV and RILO were compared by SDS–PAGE. ~20 μg of whole proteins were loaded, fractionated by SDS–PAGE, and stained with Coomassie blue G250 staining solution.
The loading of R848 and INCB in RIL and RILO was quantified by HPLC after methanol demulsification. The encapsulation efficiency (EE, %) was calculated by the following equations: EE % = Wthe weight of drug in RILs or RILOs/Wthe weight of total drug input × 100%. The DL was calculated by the following formula: DL % = Wthe weight of drug in RILs or RILOs/Wthe weight of all added into RILs or RILOs × 100%.
The stability of RILO under physiological conditions was evaluated. Briefly, RILO was diluted 10 times with PBS, DMEM containing 10% FBS, 10% FBS or normal saline, and then we measured the size and zeta potential of RILO at each preset time point for 48 h with incubation on a shaker at 37 °C. In addition, the storage stability was evaluated by recording the size, zeta potential and DL of RILO for 14 days.
Preparation of RILO@M
RILO@M was obtained by incubating M1-type macrophages with RILO. A 150 mm diameter dish with a 30 mL system, a 12-well plate with a 1 mL system, a 24-well plate with a 600 μL system or a 96-well plate with a 100 μL system was used depending on the experimental purpose. The medium was exchanged with serum-free DMEM. RILO (200 μg/mL quantified by R848) was added to M1-type macrophages at a density of ~1 × 105 cells per cm2 for 2 h and placed in a cell culture incubator. After washing, RILO@M was obtained and used immediately for the subsequent study.
To determine the optimal formulations of RILO@M, cell viability and DL were used as indices to investigate the incubation concentration (400, 200, 100, 50 or 25 μg/mL quantified by R848) and time (1, 2, 4 h) by a single-factor study.
To calculate the DL (µg/106) of RILO@M, R848 and INCB, their content in the medium before and after incubation with M1-type macrophages was quantitatively measured using HPLC (1200 Series, Agilent Technologies, CA, USA). The medium was mixed with 5 times methanol and diluted with mobile phase, and then the sample was injected into the HPLC system after filtration. The DL (µg/106) of RILO@M was calculated as follows: (Wthe weight of drug before incubating−Wthe weight of drug after incubating)/Nthe amount of cells being incubated × 106.
The cell viability of RILO@M was tested by MTT assay. M1-type macrophages or M1-type macrophagesRAW were polarized in 96-well plates. Medium was used to dilute RILO so that the concentrations of R848 in each well were 400, 200, 100, 50 or 25 μg/mL in a 100 μL system. Plates were further incubated for 1, 2 or 4 h. Then, excess RILO was washed away using DMEM supplemented with 10% FBS. Then, 20 μL of MTT (5 mg/mL) was added after 48 h for another 4 h of incubation. A microplate reader (Cytation 5, BioTek, VT, USA) was used to measure cell viability in 200 μL of DMSO at 570 nm.
Characterization of RILO@M
To evaluate the endocytic pathway of RILO in M1-type macrophage, fluorescently labelled nanoparticles were used. Genistrin (200 μM), methyl-β-cyclodextrin (800 μM), chlorpromazine (50 μM) and cytochalasin D (5 μM) were added to M1-type macrophages in 12-well plates (4.5 × 105 cells per well) for 30 min to achieve preincubation. Subsequently, C6-L or C6-LO (200 ng/mL) was added to the wells while maintaining the inhibitor concentration. After 2 h, the cells were harvested and examined by a flow cytometer (Accuri C6 Plus, BD, NJ, USA).
To evaluate the subcellular localization of C6-LO, M1-type macrophagesRAW were polarized in a pretreated glass-bottom dish. The medium was renewed containing C6-LO (200 ng/mL) followed by incubation for 2 h. After another 4 h, the cells whose nucleus and Golgi apparatus, macropinosomes or lysosomes had been stained by Hoechst 33342 and Golgi tracker red, macropinosome tracker (70 kDa RhoB-Dextran) or lysosome tracker red were imaged using a laser confocal imaging system (LSM 900 with Airyscan 2, Zeiss, Germany). Pearson correlation coefficients of C6-LO and subcellular structure tracker fluorescence were calculated using ImageJ software by pixel intensity.
To evaluate the stability of the nanostructure in RILO@M, RILO@M was prepared and cultured for 0 and 48 h in fresh DMEM supplemented with 10% FBS. The cells were collected, fixed with 2.5% glutaraldehyde aqueous solution and imaged by TEM (HT7700, Hitachi, Tokyo, Japan) at predetermined time points. Rhodamine B-labelled C6-LO was obtained by dissolving DSPE-Rhodamine B and soya lecithin in 0.5% (w/v) Tween-80 buffer solution with subsequent steps similar to those for C6-LO. M1-type macrophagesRAW were polarized in a pretreated glass-bottom dish. Rhodamine B-labelled C6-LO (200 ng/mL quantified by C6) was added for 2 h incubation. The cells were incubated with fresh DMEM containing 10% FBS for 0, 24 or 48 h after washing with PBS. At the corresponding point in time, the cells were stained with Alexa Fluor® 647 anti-mouse F4/80 antibody (clone: BM8, BioLegend) and observed using a laser confocal imaging system.
Synthesis of DSPE-PEG5k-GTP
DSPE-PEG5k-Mal (1.5 mM) and GTP-SH (6 mM) were dissolved in 4 and 2 mL of buffer solution (pH 7.4 with 1 mM EDTA), respectively. GTP-SH reaction liquid was added dropwise into the DSPE-PEG5k-Mal reaction liquid under nitrogen protection, stirred at 25 °C for 12 h, and purified via 3.5 kDa (MWCO) dialysis for 48 h, followed by lyophilization. DSPE-PEG5k-GTP was verified by 1H-NMR (Avance DPX-300, Bruker, Rheinstetten, Germany), and the grafting rate of GTP in DSPE-PEG5k-GTP was determined by HPLC. Whenever necessary, DSPE-PEG5k-GTP-FITC was synthesized through the reaction of DSPE-PEG5k-Mal with FITC-GTP-SH.
Preparation and characterization of RILO@MG
RILO@MG was obtained by incubating RILO@M with DSPE-PEG5k-GTP. In brief, freshly prepared RILO@M was treated with DSPE-PEG5k-GTP at a concentration of 100 μg/mL for 20 min with shaking. Then, the cells were washed with PBS to remove residual DSPE-PEG5k-GTP and collected for subsequent in vitro or in vivo studies.
To determine the optimal formulations for anchoring GTP to the cell surface, the cell viability and GTP carrying capacity were used as indices to investigate the incubation concentration (400, 200, 100, 50 or 25 μg/mL quantified by DSPE-PEG5k-GTP) and time (10, 20, 30 min) by a single-factor study.
To calculate the GTP carrying (µg/106) of RILO@MG, the DSPE-PEG5k-GTP content in the medium before and after incubation with RILO@M was quantitatively measured using HPLC. The GTP carrying (µg/106) of RILO@MG was calculated as follows: (Wthe weight of DSPE-PEG5k-GTP before incubating−Wthe weight of DSPE-PEG5k-GTP after incubating)/Nthe amount of cells being incubated × 106 × 19.78%. Here, 19.78% represented the grafting rate of GTP in DSPE-PEG5k-GTP.
To confirm that GTP was anchored to the cell surface by lipids, GTP-FITC and DSPE-PEG5k-GTP-FITC were synthesized. RILO@MG-FITCRAW and RILO@M-free GTP-FITCRAW cells were stained with Alexa Fluor® 647 anti-mouse F4/80 antibody (clone: BM8, BioLegend) and Hoechst 33342 and imaged by confocal microscopy. Furthermore, RILO@MG-FITCRAW cells were cultured with 10% FBS-containing DMEM for different periods (0, 24 and 48 h) to investigate the stability of GTP on the surface of cells by confocal imaging and flow cytometry analysis.
The phenotype of RILO@MG was evaluated in different culture environments, including 10% FBS-containing DMEM and HCM. Briefly, freshly prepared Blank@M and RILO@MG were incubated with 10% FBS-containing DMEM or HCM for 0 or 48 h. BMDMs incubated in 10% FBS-containing DMEM were used to prepare the control group. Then, the cells were analysed by flow cytometry by staining with PE anti-mouse CD80 antibody (clone: 16-10A1, BioLegend), Brilliant Violet 421™ anti-mouse F4/80 antibody (clone: BM8, BioLegend) and APC anti-mouse CD206 (MMR) antibody (clone: C068C2, BioLegend). The same experiment was performed with Blank@MRAW and RILO@MGRAW.
Release of drug from RILO@MG
In order to obtain the total drug release profiles from different formulations prepared using macrophages, R@MG, I@MG, RILO@MG- or RILO@MG was incubated with serum-free DMEM for 0.5, 1, 2, 4, 8, 12, 24, 48 or 72 h. The supernatant sample was collected, and fresh medium was added quickly at predetermined time points until 72 h. The amount of total R848 or INCB released into the supernatant was determined using HPLC. To further obtain the release profiles of free and nonfree drug forms from different formulations prepared using macrophages, the supernatants collected at various time points mentioned above were centrifuged (5000×g, 15 min) by ultrafiltration centrifugation (100 kDa). The amount of free R848 or INCB form was obtained by determining the drug in the ultrafiltrate. The amount of nonfree R848 or INCB form (Qcumulative total drug release−Qcumulative free drug form release) was calculated. In addition, media released from Blank@M, RILO@MG- and RILO@MG were taken and analysed by TEM at 24 h after preparation.
Formation and characterization of RI-exosomes
To observe the formation process of exosomes, Blank@M, RILO@MG- and RILO@MG were prepared. Six hours after preparation, Blank@M, RILO@MG- or RILO@MG were fixed with 2.5% glutaraldehyde aqueous solution, and the MVBs and ILVs were carefully imaged by TEM (HT7700, Hitachi, Tokyo, Japan).
To collect and purify the MVs and exosomes secreted by Blank@M, RILO@MG- and RILO@MG with serial centrifugation and ultracentrifugation following a previously reported protocol56. Briefly, freshly prepared Blank@M, RILO@MG- and RILO@MG were cultured in serum-free DMEM for 24 h. MVs were obtained by serial centrifugation (300×g for 10 min; 2000×g for 20 min and 16,500×g for 30 min at 4 °C) of the cell culture medium. Exosomes were obtained by serial centrifugation (300×g for 10 min; 2000×g for 20 min; 16,500×g for 30 min and 120,000×g for 90 min at 4 °C) of the cell culture medium. Exosomes were quantified by total protein concentration by BCA.
The size and zeta potential of exosomes were measured using a dynamic light scattering analyser. The morphology of exosomes stained with 1% phosphorus tungstate was observed by TEM. Exosome markers (CD63 and TSG101) were detected by western blotting. In brief, exosome protein extracts were isolated using cell lysis buffer. Approximately 20 μg of exosome protein was loaded, subjected to fractionation by SDS–PAGE and transmembrane, and probed with anti-GAPDH, anti-CD63 or anti-TSG101 (Affinity Biosciences). Then, the signal was detected using an automatic chemiluminescence imaging analysis system (5200, Tanon, Shanghai, China).
To determine the optimal concentration of exosomes induced by C16-ceramide in M1-type macrophages, M1-type macrophagesRAW were treated with various concentrations of C16-ceramide (200, 100, 50, 25 or 0 μM) before the study began released exosomes in 24 h were quantified by protein concentration (μg/106 cells).
Cellular uptake analysis
To visualize and quantify the cellular uptake process, C6 was selected to prepare fluorescently labelled formulations. Free C6, C6-LO, C6-LO@M and C6-LO@MG were prepared, and C6 released cumulatively at 48 h from C6-LO@M (C6 released from LO@M) or C6-LO@MG (C6 released from LO@MG) was collected for experiments. Next, 1 × 106 H22 cells were incubated with C6 released from LO@MG, C6 released from LO@M, C6-LO or free C6 (200 ng/mL) for 1 or 4 h. For imaging, the H22 cells were washed with PBS three times, fixed with 4% paraformaldehyde, stained with Hoechst 33342 for 15 min and then imaged using a fluorescence microscope (Cytation 5, BioTek, VT, USA). For quantification, H22 cells were collected, washed and fixed. The fluorescence of C6 was analysed by flow cytometry (CytoFLEX S, Beckman Coulter, CA, USA), and the data were processed using FlowJo (V10). To prepare TAMRAW, RAW264.7 cells were seeded in 12-well plates (5 × 105 cells per well) and treated with HCM for 24 h. The cellular uptake evaluated on TAMRAW was performed in a similar way to that on H22 cells.
Evaluation of RILO@MG chemotaxis
Chemotaxis-associated proteins were detected by western blotting. Whole-cell protein extracts of Blank@M, RILO@M, RILO@MG, RILO and DSPE-PEG5k-GTP were isolated using cell lysis buffer. Approximately 20 μg of total protein was loaded, and anti-GAPDH, anti-alpha 4, anti-β-Tubulin and anti-CCR2 (Affinity Biosciences) antibodies were used. The in vitro tumour-migrating capability was evaluated by using 24-well Transwell plates (pore size 8 μm, Corning), Blank@M, RILO@M or RILO@MG were seeded into the upper chambers (3 × 104 cells per well) in serum-free DMEM with the lower chambers containing DMEM supplemented with 10% FBS or HCM. After 16 h of incubation under standard conditions, the migrated cells on the lower side of the Transwell membranes were stained with crystal violet and photographed by a fluorescence microscope (IX73-DP80, Olympus Corporation, Tokyo, Japan).
Biodistribution and tumour accumulation in vivo
H22 tumour-bearing BALB/c mice were used to evaluate biodistribution. DiR, a near-infra-red tracer, was selected to prepare different formulations. For this tumour model, 1 × 106 H22 cells were subcutaneously injected into the right axilla of BALB/c mice. The mice were given one i.v. injection with free DiR, DiR-LO, DiR-LO@M or DiR-LO@MG (at a dosage of 3.0 × 106 cells per mouse, equal to 2.5 mg/kg DiR) after 12 days. At 2, 4, 8, 12 and 24 h postadministration, the mice were imaged after anaesthesia induction with an IVIS spectrum imaging system (IVIS Kinetic, Calliper Life Sciences, MA, USA). After the in vivo imaging was completed, the mice were sacrificed. We carried out ex vivo imaging by harvesting the tumours and main organs.
To demonstrate that the drugs could be released from injected macrophages at the site of the tumour in vivo, Cy5.5 was selected as a tracer to replace the drugs. Cy5.5-LO@M and Cy5.5-LO@MG were prepared and stained with the cell membrane dye DiO to obtain Cy5.5-LO@M-DiO and Cy5.5-LO@MG-DiO. H22 tumour-bearing BALB/c mice were i.v. injected with Blank@M, Cy5.5-LO@M-DiO or Cy5.5-LO@MG-DiO (at a dosage of 3.0 × 106 cells per mouse, equal to 1.5 mg/kg Cy5.5) 24 h before harvesting the tumours. The tumours were fixed, embedded in paraffin, sectioned and stained with DAPI. Subsequently, the sections were imaged. For the deep penetration evaluation in vivo, Cy5.5-LO@MG, Cy5.5-LO@M and Cy5.5-LO were prepared, and H22 tumour-bearing BALB/c mice were i.v. injected with Cy5.5-LO@MG, Cy5.5-LO@M or Cy5.5-LO (at a dosage of 3.0 × 106 cells per mouse, equal to 1.5 mg/kg Cy5.5) 24 h before harvesting the tumours. The tumours were fixed, embedded in paraffin, sectioned and stained with DAPI. The fluorescence intensity was quantified in the selected region with ImageJ.
Specific tumour cell phagocytosis
For the microscopy-based assay, Blank@M, Blank@MG, RILO@M and RILO@MG were prepared in 12-well plates (5 × 105 cells per well) and then labelled with CMTPX. A suspension of CFSE-labelled H22 cells (2.5 × 105 cells per well) was added to the wells. Notably, CFSE-labelled H22 cells incubated with free GTP for 30 min in advance to saturate GPC3 were used in the GPC3 blocking group. After coincubation for 4 h, the unbound CFSE-labelled H22 cells were subsequently removed, and images were obtained with a fluorescence microscope (Cytation 5, BioTek, VT, USA).
For the flow cytometry-based assay, CFSE-labelled Blank@M, Blank@MG, RILO@M or RILO@MG and DiD-labelled H22 cells were incubated together at a ratio of 1:1 for 4 h under standard conditions. Then, flow cytometry was performed to assess the phagocytosis of H22 cells by macrophage-based formulations.
To assess specific lysis capacity, H22 cells were added to Blank@M, Blank@MG, RILO@M or RILO@MG prepared in 24-well plates (2 × 105 cells per well with 600 μL system) at E:T ratios of 2:1, 5:1 and 10:1. Following 12 h of incubation, CCK-8 reagent (60 μL) was added to each well. After 2 h of incubation, the absorbance at 450 nm was measured.
To evaluate cytokine lysis ability, a Transwell-mediated assay was used. In brief, H22 cells were seeded into each lower chamber of 0.4 μm-sized 24-well Transwell plates (2 × 105 cells per lower chamber with a 600 μL system). Then, freshly prepared Blank@M, Blank@MG, RILO@M or RILO@MG were added to the upper chambers at an E:T of 5:1. Following 12 h of incubation, CCK-8 reagent (60 μL) was added to each well. After 2 h of incubation, the absorbance at 450 nm was measured. Then, the phagocytosis-killing ability could be calculated from the percentage difference between specific lysis and cytokine lysis.
TAM phenotype regulation
Macrophage-based formulations were cocultured with TAMs using a Transwell system in HCM. TAMs were prepared in the lower chamber of 0.4 μm-sized 12-well Transwell plates with 1 × 106 cells per chamber. Then, freshly prepared Blank@MG, RILO@MG- or RILO@MG was added to the upper chambers with 5 × 105 cells per chamber. The control group consisted of freshly obtained BMDMs cultured in DMEM containing 10% FBS, and the HCM group consisted of TAMs cultured in HCM without any formulations. Following 24 h of incubation, the cells in the lower chamber were stained with APC anti-mouse CD206 (MMR) antibody (clone: C068C2, BioLegend), PE anti-mouse CD80 antibody (clone: 16-10A1, BioLegend) and Brilliant Violet 421™ anti-mouse F4/80 antibody (clone: BM8, BioLegend) for flow cytometry analysis.
For the conditional screening of the mass ratio of R848 to INCB, the M1/M2 ratio was used as an index to investigate the mass ratio of R848 to INCB (6:8, 6:7, 6:6, 6:5, 6:4) in 12-well plates by a single-factor study before the study began.
H22 tumour-bearing BALB/c mice were used to evaluate TAM phenotype regulation in vivo. IVISense 680, a fluorescent cell labelling dye, was selected to labelling injected macrophage of Blank@M, RILO@M and RILO@MG groups. The mice were given two i.v. injection 5 days apart with Blank@M, RILO@M or RILO@MG (at a dosage of 3.0 × 106 cells per mouse). 48 h after the second injection, mice were euthanized by CO2 per institutional policy, and the tumours were isolated to prepare single-cell suspensions for flow cytometry. Cells were stained with Pacific Blue™ anti-mouse CD45 antibody (clone: 30-F11, BioLegend), PerCP/Cyanine5.5 anti-mouse F4/80 antibody (clone: BM8, BioLegend), PE anti-mouse CD80 antibody (clone: 16-10A1, BioLegend) and FITC anti-mouse CD206 (MMR) antibody (clone: C068C2, BioLegend). The injected macrophages were distinguished by flow cytometry (CytoFLEX S, Beckman Coulter, CA, USA), and the data were processed using FlowJo (V10).
T-cell viability enhancement
Mice bearing H22 tumours were treated with different formulations for four i.v. injections, as in Fig. 6a. The tumours were collected 20 days after the first injection. Furthermore, the obtained tumours were made into a tissue homogenate. Then, the resulting homogenate was subjected to albumin precipitation, followed by centrifugation and filtration. The concentrations of Trp and Kyn were determined using HPLC.
In vivo immunization study
The tumours and spleens were obtained from H22 tumour-bearing mice after four doses of treatment. The obtained tumours were weighed, cut into small pieces and digested using hyaluronidase (2.5 U/mL), collagenase type IV (1 mg/mL) and DNase I (0.1 mg/mL) at 37 °C under gentle shaking and filtered. The obtained single-cell suspension was treated with Percoll solution to obtain tumour-infiltrating lymphocytes and subsequently blocked. The collected cells were stained with the corresponding antibodies. For T-cell analysis, Pacific Blue™ anti-mouse CD45 antibody (clone: 30-F11, BioLegend), APC anti-mouse CD3 antibody (clone: 17A2, BioLegend), PE/Cyanine7 anti-mouse CD69 antibody (clone: H1.2F3, BioLegend), FITC anti-mouse CD4 antibody (clone: GK1.5, BioLegend) and PE anti-mouse CD8a antibody (clone: 53-6.7, BioLegend) were used. For CTL analysis, a cell stimulation cocktail (plus protein transport inhibitors) (1X) was used for pretreatment at 37 °C for 4 h followed by blocking and staining with Pacific Blue™ anti-mouse CD45 antibody (clone: 30-F11, BioLegend), APC anti-mouse CD8a antibody (clone: 53-6.7, BioLegend) and PE anti-mouse IFN-γ antibody (clone: XMG1.2, BioLegend). For Treg analysis, cells were stained with Pacific Blue™ anti-mouse CD45 antibody (clone: 30-F11, BioLegend), PE anti-mouse CD4 antibody (clone: GK1.5, BioLegend) and Alexa Fluor® 647 anti-mouse FOXP3 antibody (clone: MF-14, BioLegend). For TAM analysis, cells were stained with Pacific Blue™ anti-mouse CD45 antibody (clone: 30-F11, BioLegend), PerCP/Cyanine5.5 anti-mouse F4/80 antibody (clone: BM8, BioLegend), PE anti-mouse CD80 antibody (clone: 16-10A1, BioLegend) and APC anti-mouse CD206 (MMR) antibody (clone: C068C2, BioLegend). Splenic lymphocytes were separated from the single-cell suspension and stained with Pacific Blue™ anti-mouse CD45 antibody (clone: 30-F11, BioLegend), APC anti-mouse CD3 antibody (clone: 17A2, BioLegend), PE anti-mouse CD8a antibody (clone: 53-6.7, BioLegend), PerCP/Cyanine5.5 anti-mouse/human CD44 antibody (clone: IM7, BioLegend) and FITC anti-mouse CD62L antibody (clone: MEL-14, BioLegend) to assay effector memory T cells. The changes in the immune cells were measured by flow cytometry (CytoFLEX S, Beckman Coulter, CA, USA), and the data were processed using FlowJo (V10). Gating strategies are provided in Supplementary Figs. 15 and 17.
In addition, the levels of immunostimulatory cytokines (IFN-γ, IL-12p70 and TNF-α) and immunosuppressed cytokines (IL-10, TGF-β1 and IL-4) in tumours were measured using ELISA kits according to the manufacturer’s protocol and analysed by logistic curve fitting 2 (four parameters).
In vivo antitumour activity
For the antitumour experiment in vivo, mice were subcutaneously inoculated with 1 × 106 H22 cells in the right axilla. After 8 days, the mice were randomly assigned to 13 groups, which were given four i.v. injections (at Days 0, 5, 10 and 15) as in Fig. 6a with one of the following formulations: (G1) NS, (G2) Blank@M, (G3) LO@MG, (G4) R848, (G5) INCB, (G6) RI, (G7) RLO, (G8) ILO, (G9) RILO, (G10) RILO@M, (G11) RILO@MG-, (G12) RILO + MG and (G13) RILO@MG. Mice that were injected with formulations made with M1-type macrophages each received the equivalent number of cells (at a dosage of 3.0 × 106 cells per mouse per injection, equal to 4 mg/kg R848 and 3.4 mg/kg INCB). Mice requiring injection of other formulations each received the equivalent dose of medicine (4 mg/kg R848 and 3.4 mg/kg INCB). The body weight, tumour length (L) and tumour width (W) were monitored every 2 days after the first treatment, and the tumour volume (V) was calculated (V = L × W2 × 0.5). The mice were euthanized by CO2 per institutional policy when the tumour volume reached ~2000 mm3. Mice were sacrificed on Day 20, and the resulting tumours were photographed and weighed. The tumour inhibition rate was calculated according to tumour weight. The heart, liver, spleen, lung, and kidney were obtained and weighed to calculate the organ/body weight ratio. Furthermore, the tumour sections were stained with H&E, Ki-67 and TUNEL for the assessment of tumour proliferation and apoptosis. The major organs were also analysed with H&E staining.
For the immune memory experiment, H22 tumour-bearing mice were given four i.v. injections with RILO@M, RILO@MG- or RILO@MG as indicated above. The administration regimen was consistent with the antitumour experiment in vivo. To establish a rechallenged tumour model, 1 × 106 H22 cells were reinoculated into the left axilla of those mice on Day 20. Tumour size was measured every 2 days, and the rechallenged tumours were obtained on Day 36.
To investigate the role of T cells in RILO@MG-mediated antitumour activity, one day before the first injection, CD4+ or CD8+ T cells in mice were depleted by intraperitoneal injection of antibodies (200 μg/mouse). The mice were treated every 5 Days 5 times. The antibodies used for depletion were Ultra-LEAF™ Purified Rat IgG2a, κ Isotype Ctrl Antibody (clone: RTK2758, BioLegend), Ultra-LEAF™ Purified Rat IgG2b, κ Isotype Ctrl Antibody (clone: RTK4530, BioLegend), Ultra-LEAF™ Purified anti-mouse CD8a antibody (clone: 53-6.7, BioLegend) and Ultra-LEAF™ Purified anti-mouse CD4 antibody (clone: GK1.5, BioLegend). The body weight and tumour size were measured every 2 days, and the tumours were obtained on Day 20.
Evaluation of serum cytokines and liver and kidney function
Serum samples from mice at 48 h after the first treatment according to the dosage given in Fig. 6a were collected and analysed for concentrations of IL-6 and TNF-α using the ELISA Kit. Serum samples from mice treated according to the administration regimen given in Fig. 6a were collected at Day 20 and analysed for levels of ALP, ALT, AST, LDH and BUN.
Antitumour activity in an orthotopic H22 tumour model
Orthotopic HCC model mice were established with female BALB/c mice to further evaluate the antitumour efficacy. Eight days after tumour incubation, the orthotopic HCC model mice were randomly assigned to 6 groups, which were given four i.v. injections (at Days 0, 5, 10 and 15) as in Fig. 8a with one of the following formulations: (G1) NS, (G2) Blank@M, (G3) sorafenib, (G4) RILO@M, (G5) RILO@MG- and (G6) RILO@MG. The dose of sorafenib was 10 mg/kg. Mice that were injected with formulations made with M1-type macrophages each received the equivalent number of cells (at a dosage of 3.0 × 106 cells per mouse per injection, equal to 4 mg/kg R848 and 3.4 mg/kg INCB). Every 3 days, the body weight was measured. The mice were monitored with a bioluminescence signal imaging system every 5 days. After the last imaging in vivo (Day 20), the mice were sacrificed. Simultaneously, the livers were immediately imaged. For survival period studies, the mice received the same treatment, and MST was calculated using GraphPad Prism 8. On Day 20, the tumours in the livers were isolated to prepare single-cell suspensions for flow cytometry.
Antitumour activity in B16F10 tumour model
Mice were subcutaneously inoculated with 1 × 106 B16F10 cells in the right axilla. After 5 days, the mice were randomly assigned to 4 groups, which were given three i.v. injections (at Days 0, 5 and 10) with one of the following formulations: NS, Blank@M, RILO@M and RILO@MG. Mice that were injected each received the equivalent number of cells (at a dosage of 3.0 × 106 cells per mouse per injection). The body weight, tumour length (L) and tumour width (W) were monitored every 2 days after the first treatment, and the tumour volume (V) was calculated (V = L × W2 × 0.5). The mice were euthanized by CO2 per institutional policy when the tumour volume reached ~2000 mm3. On Day 15, the tumours were isolated, photographed, weighed and prepared single-cell suspensions for flow cytometry.
Statistical analysis and schematic illustrations
GraphPad Prism 8 was used for statistical analyses. The data are expressed as the mean ± SD and the Shapiro–Wilk normality test was performed for evaluation of normal distribution. The data met the homogeneity test of variance, and the one-way ANOVA with Tukey’s multiple comparisons test among multiple groups or two-tailed Student’s t-test among two groups was used for statistical significance calculation. The data do not meet the homogeneity test of variance, the Welch ANOVA with Dunnett’s T3 multiple comparisons test among multiple groups or two-tailed Student’s t-test with Welch’s correction among two groups was used for statistical significance calculation. The two-way ANOVA with Tukey’s multiple comparisons test was used when two non-repeated measure parameters were considered. Further, the two-way ANOVA with repeated measures was used to analyse the effect of different formulations on tumour growth over time, the Geisser-Greenhouse correction was used when Mauchly’s test of sphericity was not satisfied, and Tukey’s post hoc test was used for inter-group comparison. *P < 0.05, **P < 0.01 and ***P < 0.001 were considered statistically significant. Schematic illustrations of Figs. 1, 2a, 5a, 5i and 7a were created by ourselves with Adobe Illustrator. Schematic illustrations of Figs. 3a, 5g, 5k, 6a, 7k, 8a, 8i and Supplementary Fig. 3a created with BioRender.com released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Source data
Acknowledgements
Our work was supported by the National Natural Science Foundation of China (No. 82173756 to Y.L., No. 82173757 to N.Z., No. 82204295 to W.M.), the Young Scholar Programme of Shandong University (YSPSDU, 2017WLJH40 to Y.L.), the Shandong Excellent Youth Fund (ZR2022YQ76 to Y.L.) and Shandong Provincial Natural Science Foundation (ZR2022QH224 to W.M.). We are grateful to the Translational Medicine Core Facility of Shandong University and the Pharmaceutical Biology Sharing Platform of Shandong University for supporting this work.
Author contributions
JH.L., N.Z., and Y.L. asked scientific questions and designed the experiments. JH.L. performed the experiments and analysed the data. H.Z. contributed to the extraction of BMDMs and immunological evaluation. Q.Y. and J.L. contributed to the preparation and characterization of RILO@MG. P.G., N.L., Q.M. and X.Z. helped with the evaluation of tumour accumulation and deep penetration capabilities. X.H., SJ.L., M.L. and S.Y. helped with in vivo antitumour evaluation. T.G. and S.F. helped with the assessment of immune cells and cytokine levels. W.M., S.L., and S.F. contributed to the design of GPC-3-targeted macrophages and wrote the instructions. JH.L., N.Z. and Y.L. wrote the manuscript. All authors analysed and interpreted the data, and the final draft was approved by all authors.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Data availability
The raw data generated in this study are provided in the Supplementary Information/Source Data file. The protein mass spectrometry raw data have been deposited to the ProteomeXchange Consortium via the iProX repository with the data set identifier Project ID IPX0009378000. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Na Zhang, Email: zhangnancy9@sdu.edu.cn.
Yongjun Liu, Email: liuyongjun@sdu.edu.cn.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-024-52500-5.
References
- 1.Yu, J. X., Upadhaya, S., Tatake, R., Barkalow, F. & Hubbard-Lucey, V. M. Cancer cell therapies: the clinical trial landscape. Nat. Rev. Drug Discov.19, 583–584 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Hou, A. J., Chen, L. C. & Chen, Y. Y. Navigating CAR-T cells through the solid-tumour microenvironment. Nat. Rev. Drug Discov.20, 531–550 (2021). [DOI] [PubMed] [Google Scholar]
- 3.Sorkhabi, A. D. et al. The current landscape of CAR T-cell therapy for solid tumors: Mechanisms, research progress, challenges, and counterstrategies. Front. Immunol.14, 1113882 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klichinsky, M. et al. Human chimeric antigen receptor macrophages for cancer immunotherapy. Nat. Biotechnol.38, 947–953 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Franken, L., Schiwon, M. & Kurts, C. Macrophages: sentinels and regulators of the immune system. Cell. Microbiol.18, 475–487 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Saez-Ibanez, A. R. et al. Landscape of cancer cell therapies: trends and real-world data. Nat. Rev. Drug Discov.21, 631–632 (2022). [DOI] [PubMed] [Google Scholar]
- 7.Maude, S. L. et al. Tisagenlecleucel in children and young adults with B-cell lymphoblastic leukemia. N. Engl. J. Med.378, 439–448 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen, J. et al. Activating innate immune responses repolarizes hPSC-derived CAR macrophages to improve anti-tumor activity. Cell Stem Cell31, 1–17 (2024). [DOI] [PubMed] [Google Scholar]
- 9.Subramanian, S. et al. Long-term culture-expanded alveolar macrophages restore their full epigenetic identity after transfer in vivo. Nat. Immunol.23, 458–468 (2022). [DOI] [PubMed] [Google Scholar]
- 10.Lee, S., Kivimae, S., Dolor, A. & Szoka, F. C. Macrophage-based cell therapies: The long and winding road. J. Control Release240, 527–540 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu, J. J. et al. Copper sulfide nanoparticle-redirected macrophages for adoptive transfer therapy of melanoma. Adv. Funct. Mater.31, 2008022 (2021). [Google Scholar]
- 12.Shields, C. W. et al. Cellular backpacks for macrophage immunotherapy. Sci. Adv.6, eaaz6579 (2020). [DOI] [PMC free article] [PubMed]
- 13.Gordon, S. Phagocytosis: an immunobiologic process. Immunity44, 463–475 (2016). [DOI] [PubMed] [Google Scholar]
- 14.Swanson, J. A. Shaping cups into phagosomes and macropinosomes. Nat. Rev. Mol. Cell Biol.9, 639–649 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei, Y. et al. Quantitatively lighting up the spatial organization of CD47/SIRPα immune checkpoints on the cellular membrane with single-molecule localization microscopy. ACS Nano17, 21626–21638 (2023). [DOI] [PubMed] [Google Scholar]
- 16.Hsu, S. P. C. et al. Rapamycin and hydroxychloroquine combination alters macrophage polarization and sensitizes glioblastoma to immune checkpoint inhibitors. J. Neuro-Oncol.146, 417–426 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xia, Y. Q. et al. Engineering macrophages for cancer immunotherapy and drug delivery. Adv. Mater.32, e2002054 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Zhao, C. Y. et al. X-ray-guided in situ genetic engineering of macrophages for sustained cancer immunotherapy. Adv. Mater.35, e2208059 (2023). [DOI] [PubMed] [Google Scholar]
- 19.Kamber, R. A. et al. Inter-cellular CRISPR screens reveal regulators of cancer cell phagocytosis. Nature597, 549–554 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tang, Z. H. et al. Inflammatory macrophages exploit unconventional pro-phagocytic integrins for phagocytosis and anti-tumor immunity. Cell Rep.37, 110111 (2021). [DOI] [PubMed] [Google Scholar]
- 21.Shih, T. C., Wang, L., Wang, H. C. & Wan, Y. Y. Glypican-3: a molecular marker for the detection and treatment of hepatocellular carcinoma. Liver Res.4, 168–172 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mu, W. W. et al. Promoting early diagnosis and precise therapy of hepatocellular carcinoma by glypican-3-targeted synergistic chemo-photothermal theranostics. ACS Appl. Mater. Int.11, 23591–23604 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Mu, W. W. et al. Multipoint costriking nanodevice eliminates primary tumor cells and associated-circulating tumor cells for enhancing metastasis inhibition and therapeutic effect on HCC. Adv. Sci.9, 2101472 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Seehawer, M. et al. Necroptosis microenvironment directs lineage commitment in liver cancer. Nature562, 69–75 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bruni, D., Angell, H. K. & Galon, J. The immune contexture and Immunoscore in cancer prognosis and therapeutic efficacy. Nat. Rev. Cancer20, 662–680 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Bayik, D. & Lathia, J. D. Cancer stem cell–immune cell crosstalk in tumour progression. Nat. Rev. Cancer21, 526–536 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Christofides, A. et al. The complex role of tumor-infiltrating macrophages. Nat. Immunol.23, 1148–1156 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Robinson, A., Han, C. Z., Glass, C. K. & Pollard, J. W. Monocyte regulation in homeostasis and malignancy. Trends Immunol.42, 104–119 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Su, S. C. et al. Immune checkpoint inhibition overcomes ADCP-induced immunosuppression by macrophages. Cell175, 442–457 (2018). [DOI] [PubMed] [Google Scholar]
- 30.Mantovani, A., Allavena, P., Marchesi, F. & Garlanda, C. Macrophages as tools and targets in cancer therapy. Nat. Rev. Drug Discov.21, 799–820 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takenaka, M. C. et al. Control of tumor-associated macrophages and T cells in glioblastoma via AHR and CD39. Nat. Neurosci.22, 729–CD740 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rodell, C. B. et al. TLR7/8-agonist-loaded nanoparticles promote the polarization of tumour-associated macrophages to enhance cancer immunotherapy. Nat. Biomed. Eng.2, 578–588 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin, S. M. et al. Overcoming chemoimmunotherapy-induced immunosuppression by assemblable and depot forming immune modulating nanosuspension. Adv. Sci.8, e2102043 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ito, H., Ando, T., Arioka, Y., Saito, K. & Seishima, M. Inhibition of indoleamine 2,3-dioxygenase activity enhances the anti-tumour effects of a Toll-like receptor 7 agonist in an established cancer model. Immunology144, 621–630 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen, Q. et al. Exosome-mediated crosstalk between tumor and tumor-associated macrophages. Front. Mol. Biosci.8, 764222 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Evans, M. A. et al. Macrophage-mediated delivery of hypoxia-activated prodrug nanoparticles. Adv. Ther.-Ger.3, 1900162 (2020). [Google Scholar]
- 37.Patel, S. et al. Brief update on endocytosis of nanomedicines. Adv. Drug Deliv. Rev.144, 90–111 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shen, S. et al. A nanotherapeutic strategy to overcome chemotherapeutic resistance of cancer stem-like cells. Nat. Nanotechnol.16, 104–113 (2021). [DOI] [PubMed] [Google Scholar]
- 39.Zhang, P. et al. Controlled interfacial polymer self-assembly coordinates ultrahigh drug loading and zero-order release in particles prepared under continuous flow. Adv. Mater.35, e2211254 (2023). [DOI] [PubMed] [Google Scholar]
- 40.Hou, T. et al. Nanoparticle-loaded polarized-macrophages for enhanced tumor targeting and cell-chemotherapy. Nano-Micro Lett.13, 6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sahay, G., Alakhova, D. Y. & Kabanov, A. V. Endocytosis of nanomedicines. J. Control. Release145, 182–195 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Toyofuku, M., Nomura, N. & Eberl, L. Types and origins of bacterial membrane vesicles. Nat. Rev. Microbiol.17, 13–24 (2019). [DOI] [PubMed] [Google Scholar]
- 43.Shin, J. S. & Abraham, S. N. Co-option of endocytic functions of cellular caveolae by pathogens. Immunology102, 2–7 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Baorto, D. M. et al. Survival of FimH-expressing enterobacteria in macrophages relies on glycolipid traffic. Nature389, 636–639 (1997). [DOI] [PubMed] [Google Scholar]
- 45.Trajkovic, K. et al. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science319, 1244–1247 (2008). [DOI] [PubMed] [Google Scholar]
- 46.Xu, Y. P. et al. Macrophages transfer antigens to dendritic cells by releasing exosomes containing dead-cell-associated antigens partially through a ceramide-dependent pathway to enhance CD4(+) T-cell responses. Immunology149, 157–171 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Long, Y. et al. Hedgehog artificial macrophage with atomic-catalytic centers to combat drug-resistant bacteria. Nat. Commun.12, 6143 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li, M. et al. Chemotaxis-driven delivery of nano-pathogenoids for complete eradication of tumors post-phototherapy. Nat. Commun.11, 1126 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.DeNardo, D. G. & Ruffell, B. Macrophages as regulators of tumour immunity and immunotherapy. Nat. Rev. Immunol.19, 369–382 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cassetta, L. & Pollard, J. W. Targeting macrophages: therapeutic approaches in cancer. Nat. Rev. Drug Discov.17, 887–904 (2018). [DOI] [PubMed] [Google Scholar]
- 51.Chen, Z. & Hambardzumyan, D. Immune microenvironment in glioblastoma subtypes. Front. Immunol.9, 1004 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kandil, D., Leiman, G., Allegretta, M. & Evans, M. Glypican-3 protein expression in primary and metastatic melanoma. Cancer Cytopathol.117, 271–278 (2009). [DOI] [PubMed] [Google Scholar]
- 53.Motomura, Y. et al. Embryonic stem cell-derived dendritic cells expressing glypican-3, a recently identified oncofetal antigen, induce protective immunity against highly metastatic mouse melanoma, B16-F10. Cancer Res.66, 2414–2422 (2006). [DOI] [PubMed] [Google Scholar]
- 54.Ying, W., Cheruku, P. S., Bazer, F. W., Safe, S. H. & Zhou, B. Investigation of macrophage polarization using bone marrow derived macrophages. J. Vis. Exp.76, 50323 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhao, X., Zhao, R. & Nie, G. Nanocarriers based on bacterial membrane materials for cancer vaccine delivery. Nat. Protoc.17, 2240–2274 (2022). [DOI] [PubMed] [Google Scholar]
- 56.Dechantsreiter, S. et al. Heterogeneity in extracellular vesicle secretion by single human macrophages revealed by super-resolution microscopy. J. Extracell. Vesicles11, e12215 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data generated in this study are provided in the Supplementary Information/Source Data file. The protein mass spectrometry raw data have been deposited to the ProteomeXchange Consortium via the iProX repository with the data set identifier Project ID IPX0009378000. Source data are provided with this paper.