Abstract
Mantle cell lymphoma (MCL) is genetically characterized by the IG::CCND1 translocation mediated by an aberrant V(D)J rearrangement. CCND1 translocations and overexpression have been identified in occasional aggressive B-cell lymphomas with unusual features for MCL. The mechanism generating CCND1 rearrangements in these tumors and their genomic profile are not known. We have reconstructed the IG::CCND1 translocations and the genomic profile of 13 SOX11-negative aggressive B-cell lymphomas using whole genome/exome and target sequencing. The mechanism behind the translocation was an aberrant V(D)J rearrangement in three tumors and by an anomalous IGH class-switch recombination (CSR) or somatic hypermutation (SHM) mechanism in ten. The tumors with a V(D)J-mediated translocation were two blastoid MCL and one high-grade B-cell lymphoma. None of them had a mutational profile suggestive of DLBCL. The ten tumors with CSR/SHM-mediated IGH::CCND1 were mainly large B-cell lymphomas, with mutated genes commonly seen in DLBCL and BCL6 rearrangements in 6. Two cases, which transformed from marginal zone lymphomas, carried mutations in KLF2, TNFAIP3 and KMT2D. These findings expand the spectrum of tumors carrying CCND1 rearrangement that may occur as a secondary event in DLBCL mediated by aberrant CSR/SHM and associated with a mutational profile different from that of MCL.
Subject terms: Oncogenesis, B-cell lymphoma
Introduction
Mantle cell lymphoma (MCL) is a mature B-cell neoplasm genetically characterized by the rearrangement of CCND1 with immunoglobulin (IG) genes leading to cyclin D1 overexpression. Two molecular subtypes have been identified with different clinical and biological behavior [1]. Conventional MCL (cMCL) expresses the transcription factor SOX11 and usually presents with disseminated disease involving nodal and extranodal tissues and frequently aggressive behavior. The leukemic non-nodal MCL (nnMCL) subtype is SOX11-negative, with common splenomegaly without adenopathies, and usually indolent evolution for long periods. The morphological spectrum of MCL includes tumors with small sized to blastoid or large pleomorphic cells, that may be difficult to distinguish from lymphoblastic lymphoma or diffuse large B-cell lymphoma (DLBCL). The detection of CCND1 rearrangement (CCND1-R) and protein expression are of paramount relevance in the recognition of MCL. In addition to MCL, cyclin D1 overexpression has been identified in ~1–2% of DLBCL [2, 3]. However, most such cases do not carry a translocated CCND1 and cyclin D1 is usually expressed only in a proportion of the tumor cells. Intriguingly, CCND1-R has been identified as a secondary alteration acquired in the progression of chronic lymphocytic leukemia (CLL) [4, 5] and follicular lymphoma (FL) [6]. CCND1-R has been also described in occasional DLBCL [7–10], primary central nervous system (CNS) LBCL [11] and several aggressive B-cell lymphomas associated with translocations of other oncogenes such as BCL2, BCL6 or MYC, suggesting that it may correspond to a secondary event in these tumors [12]. The distinction between blastoid/pleomorphic MCL and DLBCL is relevant since these tumors are managed with different therapeutic strategies [13, 14]. CD5 and SOX11 transcription factors are usually expressed in cMCL but not in large B-cell lymphomas with cyclin D1 expression, and therefore, these markers may aid in the differential diagnosis. However, SOX11 is not expressed in nnMCL, and CD5 may be negative in 19% of these tumors [15, 16].
CCND1-R in both molecular subtypes of MCL, cMCL and nnMCL, occurs in immature pro/pre-B cells of the bone marrow as an error in the V(D)J IG gene rearrangement mediated by recombination-activating gene (RAG) activity in virtually all cases [17]. The mechanism of CCND1-R in non-MCL lymphomas is not known but the recent identification of an acquired IGH::CCND3 in a Richter transformation from CLL mediated by somatic hypermutation (SHM) suggests that a similar mechanism may be responsible in these tumors [18]. Therefore, if this hypothesis holds true, the identification of the IG breakpoint in CCND1-R in aggressive B-cell lymphomas might be relevant for the differential diagnosis. In addition, recent studies have extensively defined the mutational profile of MCL and different subtypes of DLBCL [17, 19–24]. Although individual genes may be found mutated in both types of tumors, the genomic profile is remarkably different and, therefore, it may also assist in the differential diagnosis of aggressive lymphomas carrying CCND1-R [25].
The aims of this study were to identify the mechanism of CCND1-R in aggressive non-MCL B-cell lymphomas and to characterize their genomic mutational profile to determine the potential value of these parameters in the differential diagnosis of these tumors with blastoid and pleomorphic MCL.
Material and methods
Patients and samples
We searched our files for aggressive mature B-cell lymphomas SOX11-negative expressing cyclin D1 with CCND1-R identified by conventional cytogenetics or fluorescence in situ hybridization (FISH). We identified 13 tumors conforming to different subtypes that had been previously diagnosed as blastoid or pleomorphic MCL or large B-cell lymphoma with CCND1-R and cyclin D1 overexpression. The morphology of these cases was reviewed before performing the genomic studies. Nine cases were considered DLBCL, one with focal nodular areas, 3 tumors had blastoid morphology, and one was diagnosed as high-grade B-cell lymphoma, NOS, with intermediate features between DLBCL and Burkitt lymphoma. Two large B-cell lymphomas were transformed tumors from a previous splenic marginal zone lymphoma (SMZL) or a nodal marginal zone lymphoma (NMZL). The tissue samples available were six lymph nodes, one tonsil, three testicular masses, two gastric biopsies, one mediastinal biopsy, one duodenal biopsy and one cervical mass. Two patients had more than one sample. One patient had a splenectomy specimen, and a subsequent lymph node obtained 14 years later (Table 1 and Supplementary Table 1). The initial biopsy of the NMZL was not available. All samples had been fixed in formalin and embedded in paraffin (FFPE). Frozen tumor tissue was also available in three patients. Non-involved peripheral blood for DNA extraction could be obtained in these three patients with frozen tissue and in one additional patient. All cases were studied by a routine panel of lymphoid markers by immunohistochemistry (Table 1). None of these cases had been previously reported.
Table 1.
Clinical and pathological features of patients.
| Total (n = 13) | V(D)J-mediated (n = 3) | CSR/SHM-mediated (n = 10) | |
|---|---|---|---|
| Demographics of patients | |||
| Median age (years) | 62.5 (45–86) | 64 (59–84) | 61 (45–86) |
| Male | 10 (77%) | 3 (100%) | 7 (70%) |
| Localization | |||
| Lymphadenopathy | 6 (50%) | 2 (67%) | 7 (70%) |
| Tonsil | 1 (8%) | 1 (33%) | 0 |
| Testis | 3 (25%) | 0 | 3 (30%) |
| Gastrointestinal tract | 3 (23%) | 1 (33%) | 2 (20%) |
| Bone marrow | 3/9 (33%) | 2 (67%) | 1/6 (17%) |
| Peripheral blood | 2/9 (22%) | 2 (67%) | 0/6 |
| Morphology | |||
| Blastoid | 3 (23%) | 2 (676.%) | 1 (10%) |
| Large cell | 9 (69%) | 0 | 9 (90%) |
| High-grade B-cell | 1 (8%) | 1 (33%) | 0 |
| FISH/cytogenetics | |||
| MYC-R | 1 (8%) | 1 (33%) | 0 |
| BCL2-R | 0 | 0 | 0 |
| BCL6 -R | 6 (46%) | 0 | 6 (60%) |
| IRF4-R | 1/3 | 1/3 | |
| Immunophenotype | |||
| CD5 | 3/13 (23%) | 2/3 (67%) | 1/10 (10%) |
| CD10 | 3/13 (23%) | 1/3 (33%) | 2/10 (22%) |
| BCL6 | 7/13 (54%) | 1/3 (33%) | 6/10 (60%) |
| MUM1 | 9/13 (69%) | 0/3 | 9/10 (90%) |
| BCL2 | 9/11 (82%) | 1/2 | 8/9 (89%) |
| TDT | 0/6 (14%) | 0/2 | 0/4 (20%) |
| MYC | 3/10 (30%) | 2/3 (67%) | 1/7 (14%) |
| TP53 | 4/7 (57%) | 3/3 (100%) | 1/4 (25%) |
| Follow-up | |||
| Alive no evidence of disease | 5/10 (50%) | 0 | 5/7 (3–72 m)* |
| Died of disease | 5/10 (50%) | 3 (2–21 m) | 2/7 (11–12 m) |
R: rearranged
*One additional patient in this group had a complete remission after treatment, and in another one, the treatment was discontinued because of comorbidities. No additional follow-up could be obtained in these two patients (Supplementary Table 18).
FISH
FISH studies were performed using dual color dual fusion IGH::CCND1 and/or CCND1 break-apart probes. BCL6, MYC, BCL2 and IRF4 rearrangements were also investigated by FISH using break-apart commercial probes (CCND1 break-apart: Metasystems D-5071-100-OG; IGH::CCND1 dual color dual fusion: Metasystems D-5140-100-OG; BCL2 break-apart: Metasystems D-6018-100-OG; Abbott Molecular 07J75-001; BCL6 break-apart: Metasystems D-6016-100-OG; Abbott Molecular 01N23-020; MYC break-apart Metasystems D-6023-100-OG; Abbott Molecular 05J91-001; IRF4 break-apart Metasystems D-6040-100-OG and the non-commercial custom probes detailed in supplementary method). In case #8, a combination of IRF4 break-apart probes labeled in aqua and commercial dual fusion dual color IGH::CCND1 was used (Supplementary Methods). Digital image acquisition, processing, and evaluation were performed using ISIS digital image analysis version 5.0 (MetaSystems). The FISH results are summarized in Supplementary table 2.
Whole genome and exome sequencing
DNA was extracted from the three tumors with frozen tissue available and from FFPE tissues. Whole-genome sequencing (WGS) was performed in the 3 tumors in which DNA was obtained from frozen tissues and their respective paired germline DNA. Whole exome sequencing (WES) was performed in four tumors, one of them with germline DNA obtained from non-involved peripheral blood (Supplementary Table 3).
Target panels for next-generation sequencing
Different custom target panels interrogating 120–440 genes were used for the next-generation sequencing (NGS) analysis of the tumors in 11 patients. These panels were routine diagnostic NGS assays used in the respective hospitals of origin as well as our custom DLBCL-oriented panel previously published (Supplementary Table 4) [26]. WES and WGS data were also available from 4 and 2 cases respectively (Supplementary Table 3).
Custom IG and MCL target panel
To characterize the IG gene rearrangements and identify the IG breakpoint regions and CCND1 rearrangements, we designed a customized capture-based NGS panel (IG-MCL panel) covering the full-length IGH V(D)J regions (from FR1 to FR4), IGK and IGL VJ, and all breakpoint positions in IGH, IGK and IGL loci. The IGHV identity and gene usage of the tumors in this cohort were compared to those of MCL, MZL and DLBCL previously published [17, 27–29]. This panel also included 10 MCL driver genes (TP53, NOTCH1, NOTCH2, SP140, NSD2, HNRNPH1, CCND1, SMARCA4, MYC, CDKN2A/CDKN2B) and other genes potentially associated with treatment resistance (Supplementary Table 5).
Bioinformatic analyses
The bioinformatic analyses of the WGS, WES and target NGS data were performed using updated versions of our recently described pipelines (Supplementary Methods) [18, 30, 31].
Immunoglobulin gene rearrangements
Immunoglobulin gene rearrangements, identity, and IG::CCND1 rearrangement breakpoints were initially analyzed from WGS using IgCaller (version 1.4) [27]. The IGH breakpoints were mapped at base pair resolution and searched for evidence of aberrant V(D)J recombination generated by the activity of RAG enzymes or mediated by activation-induced cytidine deaminase (AID) through aberrant class-switch recombination (CSR) or SHM as previously described [17]. We confirmed the results using the IG-MCL target panel with the DNA extracted from the FFPE tissues of the same cases (Supplementary Table S5). The IG gene rearrangements and translocations breakpoints were then examined in the remaining cases with FFPE DNA only applying the same IG-MCL panel.
Results
Genomic characterization of the CCND1 rearrangements
CCND1 was rearranged with IG genes in all tumors, 12 cases with IGH and 1 with IGL. In 3 cases, the IG breaks had evidence of being mediated by RAG enzymes during V(D)J recombination (cases #1–3), whereas the IG breaks in the remaining 10 tumors involved AID-related mechanisms through aberrant CSR in 9 cases and SHM in 1, therefore, generated in a mature B-cell probably during a follicular germinal center reaction (Fig. 1A, Supplementary Tables 6, 7).
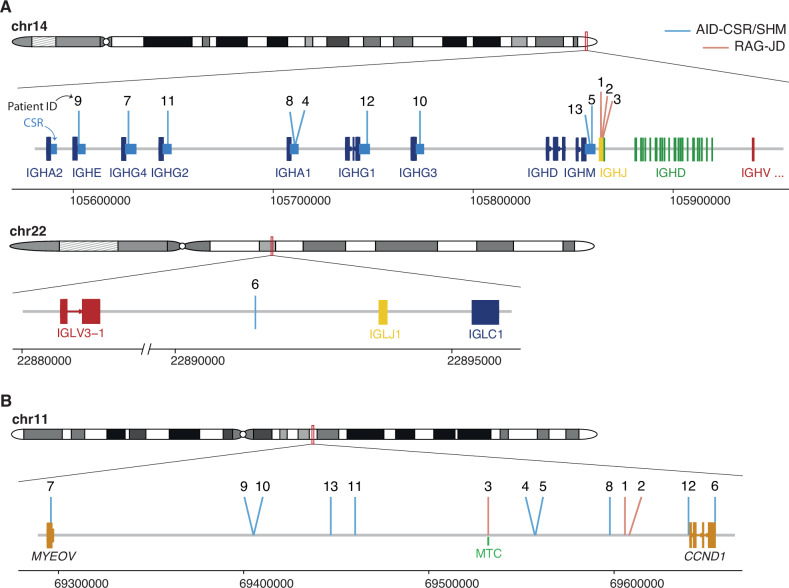
Fig. 1. IG and CCND1 breakpoints.
A Schematic representation of the location of the breakpoints found in the IG loci (IGH, top, IGL, bottom). CSR, class-switch recombination region. IGHJ genes (1–6) are shown as a single gene due to their small size and compactness for illustrative purposes. Genomic locations are shown on the x axis. Breakpoints are colored based on their underlying mechanism as AID-mediated through aberrant CSR/SHM or RAG-mediated during the initial J-D recombination. B Representation of the breakpoints found in chromosome 11. MTC major translocation cluster.
In the 3 cases with RAG-mediated rearrangements, the IG breaks occurred in IGHD and IGHJ genes, likely during the initial IGHD–IGHJ recombination. We identified the RAG recombination signal sequence (RSS) at the IGHD and IGHJ breakpoints with the addition of non-templated nucleotides (N-nucleotides) at both derivative junctions. The nine tumors with AID-mediated rearrangement had breakpoints in the IGHA1 and IGHM defined CSR region in two cases each, and IGHG3, IGHG1, IGHG2, IGHG4, and IGHE in one case, respectively. The remaining case had the IG breakpoint downstream of IGLJ1 generated by SHM mechanism (Fig. 1A).
Breakpoints on chromosome 11 (chr11) were found upstream of the 5’ of CCND1 in 12 tumors and in the 3’ region in one (case #6) (Fig. 1B). Only one tumor with RAG-mediated breakpoint had the 11q13 breakpoint in the previously recognized major translocation cluster (MTC) region in MCL whereas the breakpoints in the other 11 tumors were located upstream or downstream of this region (Fig. 1B). As previously described for MCL [17], most breaks on chr11 occurred near CpG sites and AID motifs (Supplementary Table 7). The G allele of SNP rs9344 has been associated with t(11;14) in Multiple Myeloma but not MCL [32]. We did not observe differences in the G and A allele in DLBCL with CCND1-R (Supplementary Table 8).
Immunogenetic profile
The three tumors with RAG-mediated translocations had IG rearrangements with 99.65, 97.89, and 95.14% identity (cases #1–3, respectively) with the germline IGHV sequences (Supplementary Fig. 1, Supplementary Table 9). The IGHV genes used were IGHV4-34 in 2 cases and IGHV3-23 in one, both commonly used in different types of lymphomas, including MCL [17]. The SMZL and the subsequent DLBCL (case #4) had the same IG rearrangement IGHV1-2*04–IGHD3-10*01–IGHJ6*02, frequently used in SMZL [28], with 97.22% identity. The transformed NMZL (case #9) had an IG rearrangement using IGHV4-34. In three DLBCL we observed the usage of IGHV4-34, frequently used in DLBCL. The uncommon rearranged genes in DLBCL IGHV1-3, IGHV1-46, IGHV3-48, and IGHV3-7 were observed in one case each (Supplementary Fig. 1) [27]. In one DLBCL (case #6) the IGHV gene rearrangement could not be properly identified, probably due to the high number of SHM. The identity with the germline in these tumors was very variable from 90.28 to 98.25% (Supplementary Fig. 1, Supplementary Table 9).
Genomic profile
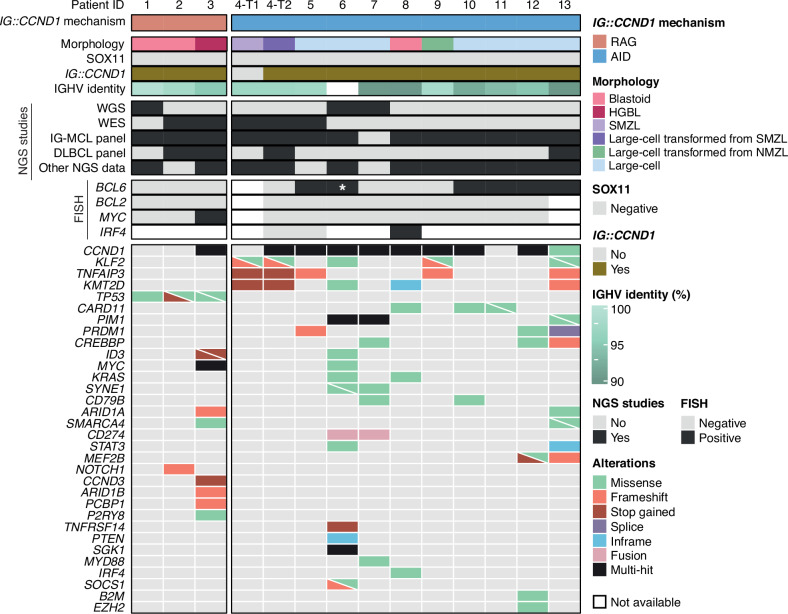
The results of the sequencing studies are summarized in Fig. 2 and Supplementary Tables 10–19. Tumor and paired germline DNA from cases #1 and #2 with CCND1-R mediated by RAG were studied by WGS and WES, respectively. Among all coding mutations identified, only TP53 mutations in both cases, and NOTCH1 truncating mutation in case #2 were previously described as recurrent in lymphoid neoplasms. Case #3 had an unusual genomic profile with MYC translocation and mutations in TP53, ID3, ARID1A, ARID1B, CCND3, SMARCA4, PCBP1 and P2RY8, among others (Fig. 2, Supplementary Fig. 2). The two cases (cases #4 and #9) diagnosed as transformed SMZL and NMZL had an AID-meditated CCND1-R and mutations in KLF2 and TNFAIP3, frequently described in these tumors (Fig. 2). The SMZL and its transformation shared the mutations in these genes and in KMT2D. The transformed tumor also acquired mutations in BCL10 and the translocated CCND1 allele (Fig. 2, and Supplementary Fig. 2). Of the other eight tumors with AID-mediated CCND1-R, 6 had mutations in CCND1, 3 in CARD11, and mutations of KMT2D, PIM1, KRAS, DDX3X, SYNE1, PRMD1, CD79B, and CREBBP were observed in two cases each. Mutated genes in one single case were MYD88, CD70, ATM, SMARCA4, and IRF4, among others (Fig. 2, Supplementary Fig. 2).
Fig. 2. Profile of putative driver mutations in CCND1-R neoplasms.
Oncoprint showing the morphology, SOX11 status, presence/absence of IG::CCND1, IGHV identity, NGS and FISH studies performed, and putative driver gene mutations found in the studied samples. Cases are grouped based on the mechanism underlying the IG::CCND1 rearrangement. *, Cryptic BCL6 rearrangement only identified by WGS.
In addition to coding driver mutations, genome-wide mutations also differ between RAG-mediated and AID-mediated CCND1-R cases. Case #1 (RAG-mediated) carried 7,074 single nucleotide variants, while cases #6 and #7 harbored 64,415 and 12,086 mutations, respectively. In terms of mutational signatures previously identified in MCL and DLBCL [17, 33], case #1 carried mutations explained by signatures 1 (spontaneous or enzymatic deamination of 5-methylcytosine to thymine; clock-like), 5 (unknown; clock-like) and 8 (unknown), cases #6 and #7 also had signatures 2 (APOBEC activity), 9 (polymerase eta SHM activity), 17b (unknown), and/or 18 (damage by reactive oxygen species) (Supplementary Fig. 3).
In terms of structural alterations, BCL2 and BCL6 rearrangements were not observed in RAG-mediated CCND1-R cases, while 6/10 cases with an AID-mediated CCND1-R carried a BCL6 rearrangement demonstrated by FISH/WGS. Of note, one of the latter cases carried a cryptic BCL6 rearrangement due to an inversion detected only by WGS (Supplementary Fig. 4). MYC rearrangement was only observed in case #3 (RAG-mediated), while no MYC rearrangements were observed in AID-mediated cases (Fig. 2). Case #8 showed an IRF4::IGH that was concordant with the mutations observed in the 5’ region of the gene (Fig. 2). Two IGH rearrangements, IGH::CCND1 and IGH::IRF4, were detected by FISH in the same cells, confirming the co-occurrence of the two alterations (Supplementary Fig. 5).
Regarding copy number alterations (CNA) identified by WGS/WES analyses, cases with a CCND1-R mediated by RAG carried alterations frequently observed in MCL [17], such as gains in 3q, 4q, 8q (MYC) and 18q, as well as deletions in 6q, 9p (CDKN2A/B), 9q, 10q, 13q and 17p (TP53). In contrast, cases with an AID-mediated CCND1-R harbored CNA recurrently detected in DLBCL [34], including gains in 1q, focal 2p affecting REL and BCL11A, 11q, 18q, and trisomies of chromosomes 3, 7, and X (Supplementary Fig. 6). Additionally, WGS analyses reveal in case #1 (RAG-mediated) the presence of a complex genomic profile including chromothripsis-like patterns in chromosome 15 and breakage-fusion bridge cycles frequently seen in MCL [17], as well as CD274 (PD-L1) alterations truncating the 3’UTR region in two cases with a CCND1-R mediated by AID (cases #6 and #7). These two cases had strong PD-L1 protein expression, whereas it was negative in the other 5 cases studied (Supplementary Fig. 7). CNA was also studied in case #8 using OncoScan assay (Thermo Fisher Scientific) detecting gains in 10p15.3-q26.3, 17p11.2-q25.3 and 21q11.1-q22.3 and losses in 8p22-p21.3, 17p13.3-p11.2/TP53 and 19p13.3-p13.2/CD70/TNFSF9. Additionally, this case had a 1p36.33-p11.2 CN neutral loss of heterozygosity.
Pathological characteristics
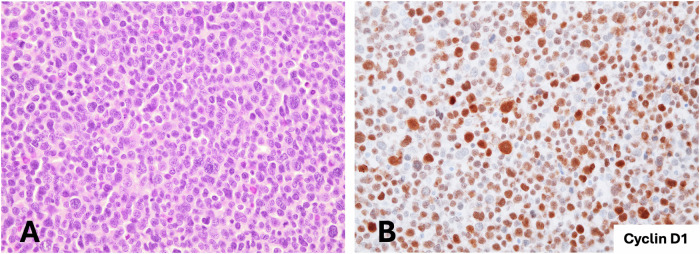
Tumors carrying RAG-mediated CCND1-R had blastoid (case #1 and #2) or high-grade morphology (case #3) (Table 1 and Supplementary Table 1). Cases #1 and #2 were composed of medium to large-sized cells, with round nuclei, fine disperse chromatin and high number of mitosis. In addition, smaller atypical cells with irregular nuclei, dense chromatin, and scarce cytoplasm were also seen (Fig. 3). These cases were positive for CD20, CD5 and TP53 (Table 1). One case expressed MYC and both were negative for SOX11, CD10, BCL6 and IRF4. Cyclin D1 was positive in virtually all cells, including small cells. These cases were diagnosed as SOX11-negative blastoid MCL based on the morphology of the cells, and the presence of small cells with strong expression of cyclin D1 and CD5-positivity. The RAG-mediated CCND1-R and the genomic alterations were consistent with this diagnosis.
Fig. 3. Blastoid MCL lymphoma with RAG-mediated CCND1 rearrangement.
A The tumor cells are predominantly medium to large size with round nuclei and blastoid chromatin. Smaller cells with irregular nuclei are also present (H&E stain, original magnification ×400). B Cyclin D1 immunohistochemical staining shows expression in all range of tumor cells (immunoperoxidase, original magnification ×400).
Case 3 had two biopsies, one from a cervical mass and a subsequent gastric sample, both with similar features. The tumor was diagnosed as HGBL, NOS. The tumor cells were positive for CD20, CD10, BCL6, MYC and TP53 and negative for CD5 and IRF4/MUM1. Ki67 was positive in virtually all cells.
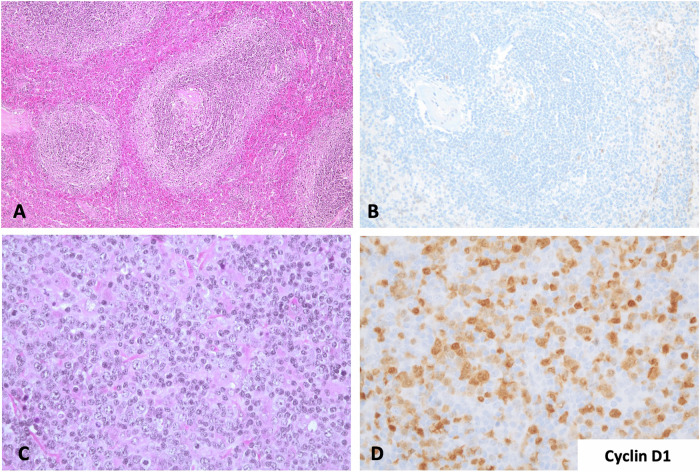
From the 10 tumors carrying an AID-mediated CCND1-R, nine had large cell morphology, whereas one was blastoid (case #8). Two tumors (cases #4 and #9) were transformed from a SMZL and a NMZL, respectively (Table 1 and Supplementary Table 1). The initial SMZL of case #4 was diagnosed in the splenectomy specimen and had typical features of this tumor with negative cyclin D1 expression and CCND1-R by FISH. The cytogenetic study found a subclonal del(7)(q31q35) and del(6)(q12q16). The subsequent transformed DLBCL was diagnosed in a cervical lymph node 14 years later with the same IGH clonal rearrangement. A subpopulation of the tumor cells was positive for cyclin D1 (Fig. 4) and CCND1-R was detected by FISH in around 20% of the cells. The transformed NMZL (case #9) was diagnosed in a core needle biopsy showing areas with small cells with clear cytoplasm and areas where large cells were predominant. The tumor cells in both MZL were positive for CD20 and BCL2 and negative for CD5, CD10, BCL6, IRF4/MUM1 and MYC.
Fig. 4. SMZL and transformed LBCL with CSR-mediated CCND1 rearrangement.
A Splenic marginal zone lymphoma with expansion of the marginal areas of the white pulp (H&E stain, original magnification ×100). B Cyclin D1 staining was negative (immunoperoxidase, original magnification ×200). C Lymph node with diffuse large B-cell lymphoma obtained 14 years later. Both tumors had the same clonal IGHV1-2*04 - IGHD3-10*01 - IGHJ6*02 rearrangement (H&E stain, original magnification ×200). D The transformed tumor was positive for cyclin D1 in the large cells but also in occasional atypical smaller cells (immunoperoxidase, original magnification ×200).
The remaining eight tumors had a diffuse growth pattern with only one (case #6) having focal nodular areas with a meshwork of follicular dendritic cells suggestive of a transformed follicular lymphoma (Table 1 and Supplementary Table 1). Two cases (cases #6 and #8) had a GC phenotype with expression of CD10 and BCL6, but case #8 had a blastoid morphology and strong expression of IRF4/MUM1 (Fig. 5). Six cases had a non-GCB phenotype by Hans algorithm including the three with a testicular mass, and two nodal and one duodenal tumors. CD5 was only positive in one testicular tumor. Ki67 was very high in all these tumors with >80% positive cells.
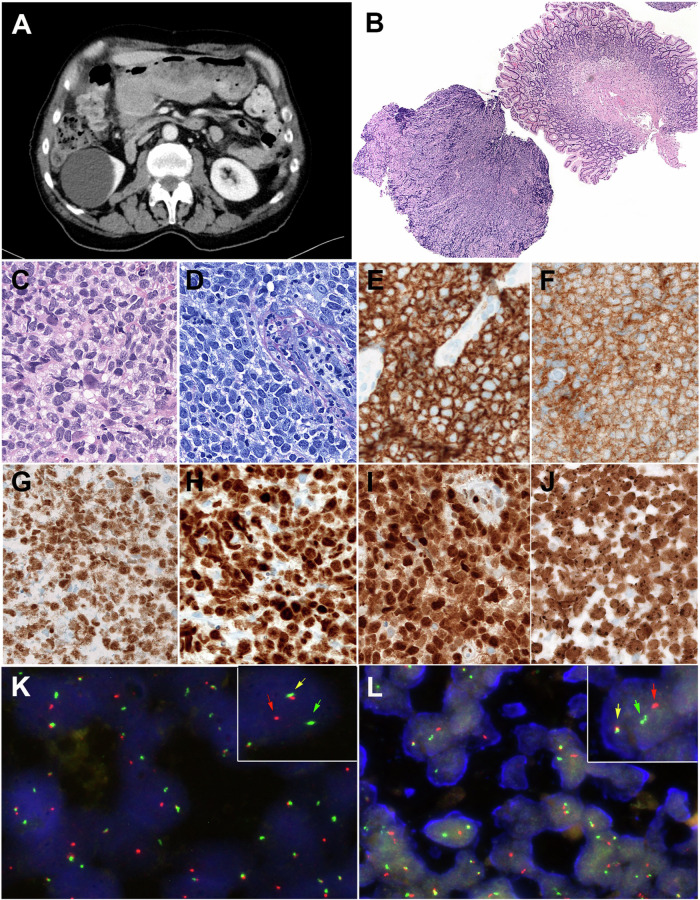
Fig. 5. Radiological, Morphological and FISH findings in a large B-cell lymphoma with IRF4 and CCND1 rearrangement (Patient #8).
A Computed tomography (CT)-scan demonstrates a large mass in the stomach that was biopsied. B Panoramic view of the gastric biopsy obtained at relapse with a diffuse lymphoid infiltration in one of the fragments. (hematoxylin and eosin (H&E) stain, original magnification, ×50). C Higher magnification shows that the tumor is composed or medium to large-sized lymphoid cells with irregular nuclei, blastic chromatin, inconspicuous nucleoli and clear cytoplasm. (H&E stain, original magnification ×400). D Giemsa stain highlights the blastic cytology of the lymphoid cells (Giemsa stain, original magnification, ×400). E The tumor cells are positive for CD20, CD10 (F), BCL6 (G), MUM1/IRF4 (H), and cyclin D1 (I). (immunoperoxidase, original magnification ×400). J The MIB1/Ki67 stain shows a proliferation index of 100%. K FISH analysis using an IRF4 break-apart assay demonstrates a signal constellation of one colocalized signal (inset, yellow arrow) and one split signal (inset, red and green arrows) consistent with gene rearrangement. L FISH analysis using an CCND1 break-apart assay shows a signal constellation of one colocalized signal (inset, yellow arrow) and one split signal (inset, red and green arrows) consistent with gene rearrangement.
Clinical features
The clinical features, treatment, and follow-up of the patients are summarized in Table 1 and Supplementary Table 20. The three patients carrying tumors with RAG-mediated CCND1-R were males with a median age of 64 years (59–84 years). The initial manifestation of the disease was generalized lymphadenopathy (case #1), isolated enlarged tonsil (case #2), or an explosive disseminated disease (case #3). Two of the patients (cases #1 and #3) had bone marrow and peripheral blood involvement. The disease did not respond to therapy in these two patients, and they died 2 and 6 months after diagnosis. The disease in patient #2 had an initial complete response but relapsed 5 months after diagnosis and the patient died 16 months later.
Patients with tumors carrying an AID-mediated CCND1-R were 7 males and 3 females with a median age of 61 years (45–86 years) (Supplementary Table 20). The two patients with transformed MZL had been initially diagnosed with a SMZL and NMZL 14 and one year before, respectively. The transformed SMZL LBCL was localized in a cervical lymph node and responded to Ibrutinib, being in complete remission 26 months later. The other eight patients had a presentation with lymphadenopathy in 3, testicular mass in 4, duodenal lesion in a patient with anemia, one patient had a mediastinal mass with also gastric involvement, and one patient had bone lesions in addition to generalized lymphadenopathy and a testicular mass. Patient 8 with an additional IGH::IRF4-translocation was a 76-year-old man with a bulky mediastinal mass and infiltration of the right bronchus. PET-CT scan demonstrated, in addition, small lymph nodes in the inguinal region (Fig. 5). The patient received 6 cycles of R-CHOP-14 achieving complete remission. One year later, the tumor recurred with orbital and gastric involvement. The patient received local radiotherapy to the orbital mass and R-bendamustine with poor response and the patient died 3 months after the recurrence. The clinical presentation and evolution of this patient suggest that this tumor does not correspond to the large B-cell lymphoma with IRF4 rearrangement observed in children and young adults.
Discussion
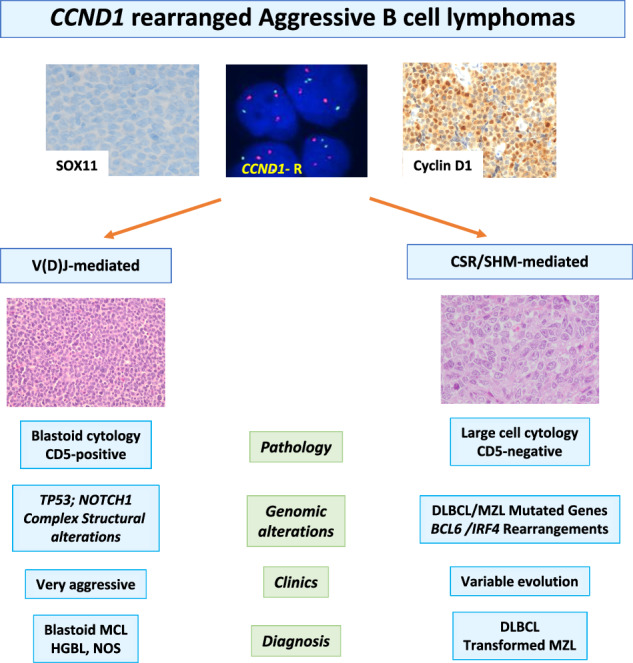
Cyclin D1 expression and CCND1-R are considered the molecular hallmarks of MCL. However, in the last years this translocation has been identified in occasional CLL, FL and large B-cell lymphomas with unusual features for MCL [4–10]. Regarding the major differences of therapeutic choices between MCL and aggressive LBCL, especially in younger patients, it is crucial to determine if CCND1-R can occur outside MCL. In this study, we have characterized the breakpoints of the CCND1-R and the genomic profile of SOX11-negative aggressive B-cell lymphomas to determine whether these alterations were different from those observed in MCL. We identified two subsets of tumors based on the mechanism of the rearrangements. In most tumors (10 out of 13) the CCND1-R occurred in a mature B-cell generated by aberrant CSR or SMH of IG genes. Three additional tumors carried CCND1-R mediated by RAG activity during an anomalous V(D)J rearrangement in immature B cells. These two subsets of tumors had different pathological, genomic, and clinical features suggesting that they correspond to different entities.
Lymphomas with CSR/SMH mediated IG::CCND1 translocations had predominantly large cell morphology with heterogeneous phenotype. Virtually all tumors were CD5 negative and, according to Hans algorithm, eight of the 10 cases had a non-germinal center (non-GC) phenotype. One tumor with blastoid morphology was triple positive for CD10, BCL6 and IRF4/MUM1 and this case carried an IRF4 rearrangement, as previously described in LBCL with this triple phenotype [35, 36]. One additional tumor had a GC phenotype with nodular areas suggestive of transformation from a FL. Two additional tumors were transformed large B-cell lymphomas from NMZL and SMZL suggesting that the CCND1-R in LBCL may be a secondary event mediated by an anomalous CSR/SHM. These observations and the finding of the CCND1-R only in a subpopulation of tumor cells in cases 5 and 12 (Supplementary Table 2) suggest that this rearrangement may be a secondary event in DLBCL.
The genomic alterations observed in tumors with CSR/SHM mediated IG::CCND1 translocation had been previously described in different subtypes of DLBCL but were uncommon in MCL. The two LBCL transformed from MZL had mutations in KLF2 and TNFAIP3 frequently seen in MZL [25, 37]. The DLBCL with GC phenotype had mutations in TNFRSF14, KMT2D, PTEN, SGK1, STAT3 and DDX3X frequently described in this DLBCL subtype but also in ID3, CD70 or KLF2 not common in GCB-DLBCL [22, 38]. The LBCL with IRF4 rearrangement had the expected mutations in IRF4 and CARD11 but also mutations in KMT2D and KRAS, uncommon in pediatric LBCL with this translocation [37]. Other tumors carried mutations seen in extranodal DLBCL such as CARD11, MYD88, PIM and CD79B [25].
BCL6 rearrangement seems particularly frequent in this group of tumors since it was detected in 6 of the 10 DLBCLs, but BCL2 and MYC rearrangement were absent. The BCL6 rearrangement in case #6 was an inversion detected by WGS but not by FISH. This alteration has been described in a transformed FL [39], and the tumor in which we detected this alteration had nodular areas suggestive of an FL transformation. BCL6 translocations have been described in occasional MCL, but they are very rare [40, 41]. The association of BCL6 and CCND1 rearrangements with other oncogenic translocations such as MYC and BCL2 seen in DLBCL, together with the absence of expression of SOX11 and CD5, have questioned the diagnosis of MCL in these tumors [12]. Interestingly, two tumors carried CD274 (PD-L1) rearrangements leading to the truncation of the 3’ UTR region and strong expression of the protein, as described in multiple cancers including 8% of DLBCL [42]. The truncation of this region stabilizes the mRNA leading to protein overexpression. The relatively high frequency of this translocation in this group of tumors is intriguing and suggests that target immunotherapy could be of benefit.
Most tumors with CSR/SMH mediated IG::CCND1 rearrangements were disseminated at the time of diagnosis with extranodal presentation in 7 of them, including 4 testicular. Only one of the 5 cases studied had bone marrow involvement, and no leukemic expression was documented. The follow-up information is limited and with a median follow-up of 11.5 months, two patients died of disease (11 and 12 months) and 5 were alive with no evidence of lymphoma (3–72 months). One additional patient of this group had a complete remission after treatment and in one additional patient the treatment was discontinued because of comorbidities. No additional follow-up could be obtained in these two patients (Supplementary Table 20).
These pathological, genomic, and clinical features contrast with those of the tumors with a RAG-mediated IG::CCND1 translocation. Two of them expressed CD5 and had blastoid morphology with a component of small, atypical cells intermingled with the large cells suggestive of SOX11-negative blastoid MCL. One of them presented with disseminated disease, bone marrow and peripheral blood involvement. Both patients died of progressive disease 2 and 21 months after diagnosis. The identification of a RAG-mediated breakpoint in these two cases and the profile of structural genomic alterations are consistent with the diagnosis of MCL [17]. However, the mutational profile, examined by WGS and WES, is more difficult to interpret. Although the tumors had a high number of mutated genes, only TP53 in the 3 tumors and NOTCH1 in one, were recognized as recurrently mutated in previous genomic studies of MCL, DLBCL or other lymphoid neoplasms. This may be due in part to the limited number of genomic studies in SOX11-negative MCL and particularly its blastoid variant [17, 20, 21, 41, 43]. However, TP53 is the most frequent mutated gene in SOX11-negative MCL and NOTCH1 mutations have been described also in these tumors [17, 44].
The third lymphoma with RAG-mediated IG::CCND1 translocation had unusual pathological and genomic features with MYC rearrangement and mutations frequently seen in BL (ID3, CCND3, SMARCA4, ARID1A, PCBP1 and P2RY8) but unusual in MCL [45]. We have not identified the breakpoint of the IGH::MYC rearrangement with our target panel which captures all the IG V(D)J and CSR regions. This negative observation suggests that the breakpoint in the IGH locus may be located in the intronic region between the V and IGHM genes, not fully covered by our panel, and possibly mediated by SHM. This situation is similar to the IGH::MYC in BL, which is generated by an erroneous CSR/SHM mechanism [45]. This configuration of the IGH::MYC rearrangement, the mutational profile and the phenotype of this tumor are closer to those of BL than MCL, but the presence of a RAG-mediated IG::CCND1 is intriguing and challenges the precise taxonomy of this tumor. Independently of their classification, the clinical course of the three tumors with the RAG-mediated IG::CCND1 translocation was very aggressive without response to different lines of treatment.
In conclusion, this study expands the spectrum of mature B-cell lymphomas carrying CCND1-R including different subtypes of aggressive B-cell lymphomas. The mutational profile and identification of a CSR/SHM mechanism underlying the translocation may support the diagnosis of DLBCL rather than MCL. This diagnosis may be also suggested by the large cell morphology, negative expression of CD5 and SOX11 in most cases, and additional translocations such as BCL6 or IRF4 in a subset of them. However, tumors with blastoid morphology and CD5 expression with a RAG-mediated CCND1-R may correspond to aggressive variants of SOX11-negative MCL. The precise diagnosis of aggressive B-cell lymphomas with CCND1-R requires an integrated analysis of clinical, phenotypic, and molecular information. Additional studies of similar tumors are needed to confirm these observations.
Supplementary information
Acknowledgements
This study was supported by the PMP21/00015 project from the Instituto de Salud Carlos III (ISCIII) and the “NextGenerationEU/Mechanism for Recovery and Resilience (MRR)/PRTR, by PID2021-123054OB-I00 project from Ministry of Science and Innovation (MCIN) and FEDER “Una manera de hacer Europa” to E.C, by PI22/00203 project from ISCIII and FEDER (to EG and SB); and the Generalitat de Catalunya Suport Grups de Recerca AGAUR (2021-SGR-01172 to E.C. and 2021-SGR-01293 to S.B.). M.P. was supported by the 2022 Estela Matutes Fellowship. F.N. acknowledges research support European Hematology Association (EHA Junior Research Grant 2021, RG-202012-00245). E.C. is an Academia Researcher of the “Institució Catalana de Recerca i Estudis Avançats” (ICREA) of the Generalitat de Catalunya.
Author contributions
E.O., M.P., G.F., O.B., E.C. collected the samples and reviewed the pathology. F.N., A.M., P.B.-N., X.S.P. designed the bioinformatics pipelines for WGS, WES and panel target sequences. A.M., G.F., P.B.-N., R.R., M.Ro., M.Ra, X.S.P., C.L., F.N. analyzed and interpreted genomic data. C.S., J.F., A.T., M.M.B., L.G., T.Z., S.C., S.V., L.d.L., J.B., F.C., S.R., M.B., E.G., L.Q., E.S.J. and P.B. provided samples and/or data, and interpreted data. D.C, S.B., J.S.-V., I.S. and C.L. performed FISH experiments. E.O., M.P., A.M., C.L., F.N. and E.C. contributed to manuscript preparation. E.C. reviewed and supervised the study, analyzed, and interpreted data, wrote the manuscript, and designed the study. All authors reviewed, commented, and approved the final version of the manuscript.
Data availability
The sequencing data has been deposited at European Genome–phenome Archive (http://www.ebi.ac.uk/ega/). The accession number is EGAS50000000564.
Competing interests
F.N. received honoraria from Janssen, AbbVie, AstraZeneca, and SOPHiA GENETICS for speaking in educational activities, and research funding from Gilead. E.C. has been a consultant for Takeda; has received honoraria from Janssen, EUSA Pharma and Roche for speaking at educational activities and research funding from AstraZeneca and is an inventor on 2 patents filed by the National Institutes of Health, National Cancer Institute: “Methods for selecting and treating lymphoma types,” licensed to NanoString Technologies, and “Evaluation of MCL and methods related thereof”, not related to this project. F.N. and E.C. licensed the use of the protected IgCaller algorithm for Diagnóstica Longwood. The remaining authors declare no competing financial interests.
Ethics declarations
The study was approved by the Ethic Committee of the Hospital Clinic of Barcelona (HCB/2021/1076). Patients had given the informed consent for large scale genomic studies and the use of their samples for research purposes. All methods were performed in accordance with the relevant guidelines and regulations.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Ece Özoğul, Anna Montaner, Melina Pol.
Supplementary information
The online version contains supplementary material available at 10.1038/s41408-024-01146-z.
References
- 1.Puente XA, Jares P, Campo E. Chronic lymphocytic leukemia and mantle cell lymphoma: crossroads of genetic and microenvironment interactions. Blood. 2018;131:2283–29. [DOI] [PubMed] [Google Scholar]
- 2.Hsiao SC, Cortada IR, Colomo L, Ye H, Liu H, Kuo SY, et al. SOX11 is useful in differentiating cyclin D1-positive diffuse large B-cell lymphoma from mantle cell lymphoma. Histopathology. 2012;61:685–93. [DOI] [PubMed] [Google Scholar]
- 3.Ok CY, Xu-Monette ZY, Tzankov A, O’Malley DP, Montes-Moreno S, Visco C, et al. Prevalence and clinical implications of cyclin D1 expression in diffuse large B-cell lymphoma (DLBCL) treated with immunochemotherapy: a report from the International DLBCL Rituximab-CHOP Consortium Program. Cancer. 2014;120:1818–29. [DOI] [PubMed] [Google Scholar]
- 4.Nishida Y, Takeuchi K, Tsuda K, Ugai T, Sugihara H, Yamakura M, et al. Acquisition of t(11;14) in a patient with chronic lymphocytic leukemia carrying both t(14;19)(q32;q13.1) and +12. Eur J Haematol. 2013;91:179–82. [DOI] [PubMed] [Google Scholar]
- 5.Schliemann I, Oschlies I, Nagel I, Murga Penas EM, Siebert R, Sander B. The t(11;14)(q13;q32)/CCND1-IGH translocation is a recurrent secondary genetic aberration in relapsed chronic lymphocytic leukemia. Leuk Lymphoma. 2016;57:2672–6. [DOI] [PubMed] [Google Scholar]
- 6.Koduru PR, Chen W, Garcia R, Fuda F. Acquisition of a t(11;14)(q13;q32) in clonal evolution in a follicular lymphoma with a t(14;18)(q32;q21) and t(3;22)(q27;q11.2). Cancer Genet 2015;208:303–9. [DOI] [PubMed] [Google Scholar]
- 7.Ehinger M, Linderoth J, Christensson B, Sander B, Cavallin-Stahl E. A subset of CD5- diffuse large B-cell lymphomas expresses nuclear cyclin D1 with aberrations at the CCND1 locus. Am J Clin Pathol. 2008;129:630–8. [DOI] [PubMed] [Google Scholar]
- 8.Juskevicius D, Ruiz C, Dirnhofer S, Tzankov A. Clinical, morphologic, phenotypic, and genetic evidence of cyclin D1-positive diffuse large B-cell lymphomas with CYCLIN D1 gene rearrangements. Am J Surg Pathol. 2014;38:719–27. [DOI] [PubMed] [Google Scholar]
- 9.Al-Kawaaz M, Mathew S, Liu Y, Gomez ML, Chaviano F, Knowles DM, et al. Cyclin D1-positive diffuse large B-cell lymphoma with IGH-CCND1 translocation and BCL6 rearrangement: a report of two cases. Am J Clin Pathol. 2015;143:288–99. [DOI] [PubMed] [Google Scholar]
- 10.Yoshida M, Ichikawa A, Miyoshi H, Kiyasu J, Kimura Y, Arakawa F, et al. Clinicopathological features of double-hit B-cell lymphomas with MYC and BCL2, BCL6 or CCND1 rearrangements. Pathol Int. 2015;65:519–27. [DOI] [PubMed] [Google Scholar]
- 11.Parrott AM, Haggiagi AM, Murty VV, Bhagat G, Alobeid B. Primary large B-cell lymphoma of the central nervous system with cyclin D1 expression and t(11;14) (IGH-CCND1): Diffuse large B-cell lymphoma with CCND1 rearrangement or mantle cell lymphoma? Hematol Oncol. 2020;38:817–22. [DOI] [PubMed] [Google Scholar]
- 12.Cheng J, Hashem MA, Barabe F, Cloutier S, Xi L, Raffeld M, et al. CCND1 genomic rearrangement as a secondary event in high grade B-cell lymphoma. Hemasphere. 2021;5:e505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilson MR, Barrett A, Cheah CY, Eyre TA. How I manage mantle cell lymphoma: indolent versus aggressive disease. Br J Haematol. 2023;201:185–98. [DOI] [PubMed] [Google Scholar]
- 14.Gerson JN, Handorf E, Villa D, Gerrie AS, Chapani P, Li S, et al. Outcomes of patients with blastoid and pleomorphic variant mantle cell lymphoma. Blood Adv. 2023;7:7393–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu J, Wang L, Li J, Saksena A, Wang SA, Shen J, et al. SOX11-negative mantle cell lymphoma: clinicopathologic and prognostic features of 75 patients. Am J Surg Pathol. 2019;43:710–16. [DOI] [PubMed] [Google Scholar]
- 16.Clot G, Jares P, Gine E, Navarro A, Royo C, Pinyol M, et al. A gene signature that distinguishes conventional and leukemic nonnodal mantle cell lymphoma helps predict outcome. Blood. 2018;132:413–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nadeu F, Martin-Garcia D, Clot G, Díaz-Navarro A, Duran-Ferrer M, Navarro A, et al. Genomic and epigenomic insights into the origin, pathogenesis, and clinical behavior of mantle cell lymphoma subtypes. Blood. 2020;136:1419–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nadeu F, Royo R, Massoni-Badosa R, Playa-Albinyana H, Garcia-Torre B, Duran-Ferrer M, et al. Detection of early seeding of Richter transformation in chronic lymphocytic leukemia. Nat Med. 2022;28:1662–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bea S, Valdes-Mas R, Navarro A, Salaverria I, Martín-Garcia D, Jares P, et al. Landscape of somatic mutations and clonal evolution in mantle cell lymphoma. Proc Natl Acad Sci USA. 2013;110:18250–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pararajalingam P, Coyle KM, Arthur SE, Thomas N, Alcaide M, Meissner B, et al. Coding and noncoding drivers of mantle cell lymphoma identified through exome and genome sequencing. Blood. 2020;136:572–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yi S, Yan Y, Jin M, Bhattacharya S, Wang Y, Wu Y, et al. Genomic and transcriptomic profiling reveals distinct molecular subsets associated with outcomes in mantle cell lymphoma. J Clin Invest. 2022;132:e153283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wright GW, Huang DW, Phelan JD, Coulibaly ZA, Roulland S, Young RM, et al. A probabilistic classification tool for genetic subtypes of diffuse large B cell lymphoma with therapeutic implications. Cancer Cell. 2020;37:551–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chapuy B, Stewart C, Dunford AJ, Kim J, Kamburov A, Redd RA, et al. Molecular subtypes of diffuse large B cell lymphoma are associated with distinct pathogenic mechanisms and outcomes. Nat Med. 2018;24:679–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lacy SE, Barrans SL, Beer PA, Painter D, Smith AG, Roman E, et al. Targeted sequencing in DLBCL, molecular subtypes, and outcomes: a Haematological Malignancy Research Network report. Blood. 2020;135:1759–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Leval L, Alizadeh AA, Bergsagel PL, Campo E, Davies A, Dogan A, et al. Genomic profiling for clinical decision making in lymphoid neoplasms. Blood. 2022;140:2193–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mozas P, Lopez C, Grau M, Nadeu F, Clot G, Valle S, et al. Genomic landscape of follicular lymphoma across a wide spectrum of clinical behaviors. Hematol Oncol. 2023;41:631–43. [DOI] [PubMed] [Google Scholar]
- 27.Nadeu F, Mas-de-Les-Valls R, Navarro A, Royo R, Martín S, Villamor N, et al. IgCaller for reconstructing immunoglobulin gene rearrangements and oncogenic translocations from whole-genome sequencing in lymphoid neoplasms. Nat Commun. 2020;11:3390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bikos V, Karypidou M, Stalika E, Baliakas P, Xochelli A, Sutton LA, et al. An immunogenetic signature of ongoing antigen interactions in splenic marginal zone lymphoma expressing IGHV1-2*04 receptors. Clin Cancer Res. 2016;22:2032–40. [DOI] [PubMed] [Google Scholar]
- 29.Traverse-Glehen A, Davi F, Ben Simon E, Callet-Bauchu E, Felman P, et al. Analysis of VH genes in marginal zone lymphoma reveals marked heterogeneity between splenic and nodal tumors and suggests the existence of clonal selection. Haematologica 2005;90:470–8. [PubMed] [Google Scholar]
- 30.Rivas-Delgado A, Nadeu F, Enjuanes A, Casanueva-Eliceiry S, Mozas P, Magnano L, et al. Mutational landscape and tumor burden assessed by cell-free DNA in diffuse large B-cell lymphoma in a population-based study. Clin Cancer Res. 2021;27:513–21. [DOI] [PubMed] [Google Scholar]
- 31.Diaz-Navarro A, Bousquets-Munoz P, Nadeu F, López-Tamargo S, Beà S, Campo E, et al. RFcaller: a machine learning approach combined with read-level features to detect somatic mutations. NAR Genom Bioinform. 2023;5:lqad056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weinhold N, Johnson DC, Chubb D, Chen B, Försti A, Hosking FJ, et al. The CCND1 c.870G>A polymorphism is a risk factor for t(11;14)(q13;q32) multiple myeloma. Nat Genet. 2013;45:522–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hubschmann D, Kleinheinz K, Wagener R, Bernhart SH, López C, Toprak UH, et al. Mutational mechanisms shaping the coding and noncoding genome of germinal center derived B-cell lymphomas. Leukemia. 2021;35:2002–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Grau M, Lopez C, Martin-Subero JI, Bea S. Cytogenomics of B-cell non-Hodgkin lymphomas: the “old” meets the “new. Best Pract Res Clin Haematol. 2023;36:101513. [DOI] [PubMed] [Google Scholar]
- 35.Frauenfeld L, Castrejon-de-Anta N, Ramis-Zaldivar JE, Streich S, Salmerón-Villalobos J, Otto F, et al. Diffuse large B-cell lymphomas in adults with aberrant coexpression of CD10, BCL6, and MUM1 are enriched in IRF4 rearrangements. Blood Adv. 2022;6:2361–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ramis-Zaldivar JE, Gonzalez-Farre B, Balague O, Celis V, Nadeu F, Salmerón-Villalobos J, et al. Distinct molecular profile of IRF4-rearranged large B-cell lymphoma. Blood. 2020;135:274–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bonfiglio F, Bruscaggin A, Guidetti F, Terzi di Bergamo L, Faderl M, Spina V, et al. Genetic and phenotypic attributes of splenic marginal zone lymphoma. Blood. 2022;139:732–47. [DOI] [PubMed] [Google Scholar]
- 38.Karube K, Enjuanes A, Dlouhy I, Jares P, Martin-Garcia D, Nadeu F, et al. Integrating genomic alterations in diffuse large B-cell lymphoma identifies new relevant pathways and potential therapeutic targets. Leukemia. 2018;32:675–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carlsen ED, Aarabi M, Swerdlow SH. Unexpected MYC::DMD translocation after transformation of follicular lymphoma with IGH::BCL2 and IGH::MYC. Br J Haematol. 2023;203:e74–e77. [DOI] [PubMed] [Google Scholar]
- 40.Camacho FI, Garcia JF, Cigudosa JC, Mollejo M, Algara P, Ruíz-Ballesteros E, et al. Aberrant Bcl6 protein expression in mantle cell lymphoma. Am J Surg Pathol. 2004;28:1051–6. [DOI] [PubMed] [Google Scholar]
- 41.Lopez C, Silkenstedt E, Dreyling M, Bea S. Biological and clinical determinants shaping heterogeneity in mantle cell lymphoma. Blood Adv. 2024;8:3652–3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kataoka K, Shiraishi Y, Takeda Y, Sakata S, Matsumoto M, Nagano S, et al. Aberrant PD-L1 expression through 3’-UTR disruption in multiple cancers. Nature. 2016;534:402–6. [DOI] [PubMed] [Google Scholar]
- 43.Hill HA, Qi X, Jain P, Nomie K, Wang Y, Zhou S, et al. Genetic mutations and features of mantle cell lymphoma: a systematic review and meta-analysis. Blood Adv. 2020;4:2927–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Federmann B, Frauenfeld L, Pertsch H, Borgmann V, Steinhilber J, Bonzheim I, et al. Highly sensitive and specific in situ hybridization assay for quantification of SOX11 mRNA in mantle cell lymphoma reveals association of TP53 mutations with negative and low SOX11 expression. Haematologica. 2020;105:754–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lopez C, Kleinheinz K, Aukema SM, Rohde M, Bernhart SH, Hübschmann D, et al. Genomic and transcriptomic changes complement each other in the pathogenesis of sporadic Burkitt lymphoma. Nat Commun. 2019;10:1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data has been deposited at European Genome–phenome Archive (http://www.ebi.ac.uk/ega/). The accession number is EGAS50000000564.