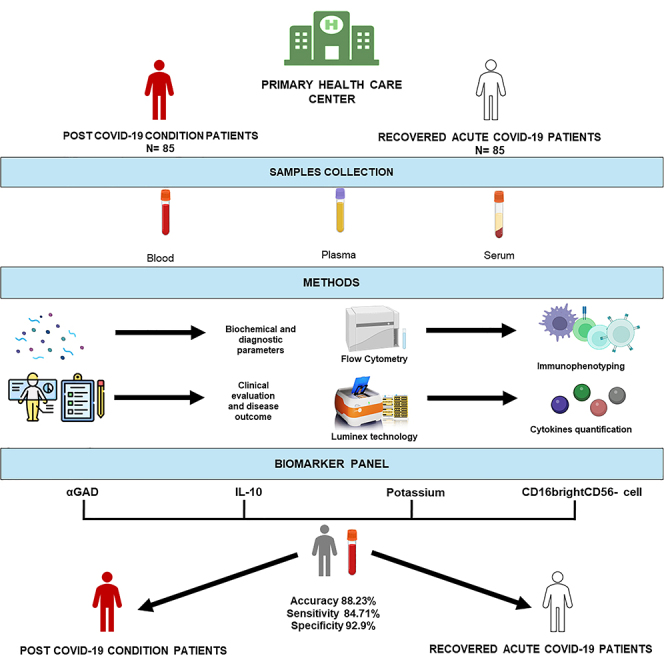
Summary
Post-coronavirus disease condition (PCC) continues to affect many people globally, yet there remains a lack of diagnostic biomarkers to distinguish PCC from those recovered from acute COVID-19. This study compared biomarkers between two age- and gender-matched groups: PCC individuals and those recovered within three months of acute COVID-19 in 2020 (n = 85 each). Biomarkers were assessed 12–24 months after initial diagnosis, examining biochemical profiles, blood cell counts, coagulation status, antibody serology, lymphocyte populations, and cytokine levels. PCC individuals exhibited significant alterations in 49 of 167 markers, including K+ levels, αGAD antibodies, antithrombin III, insulin-like growth factor-binding protein 3 (IGFBP3), and interleukin-10 (IL-10). A panel of αGAD, IL-10, potassium levels, and CD16brightCD56− cell presence distinguished PCC individuals from recovered patients with >88% accuracy and <92% precision.
Subject areas: Immunology, Virology, Biochemistry
Graphical abstract
Highlights
-
•
10% of COVID-19 patients have prolonged symptoms, but we lack proper diagnostic panel
-
•
αGAD, IL-10, K+, and CD56-CD16bright were significantly altered in PCC patients
-
•
Biomarkers identify PCC from recovered individuals with 88% accuracy and 92% precision
-
•
This model aids clinicians guiding PCC diagnosis and treatment
Immunology; Virology; Biochemistry
Introduction
Since 2020, coronavirus disease 2019 (COVID-19) caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has spread rapidly throughout the world, seriously threatening global public health.1 Globally, as of January 2024, there have been 774,291,287 confirmed cases of COVID-19 including 7,019,704 deaths.2 Most people with an acute SARS-CoV-2 infection recover within 1 month after diagnosis, depending on the severity of the symptoms.3,4 However, it is estimated that regardless of the severity of the symptomatology, approximately 20% of people maintain and/or develop multisystem symptoms after 5 weeks onward, whereas more than 10% of people do so after 12 weeks onward since the acute infection.5,6 The World Health Organization (WHO) registered in December 2022 the official definition as “post-COVID-19 condition” (PCC)7 that is defined as the persistence or development of new symptoms 3 months after the initial SARS-CoV-2 infection, which may last for at least 2 months with no other explanation.7
Apart from being confusing and not very specific, characteristic symptoms of PCC may be persistent or fluctuate over time8,9 Profound fatigue, shortness of breath, fever, cough, headache, chest and/or throat pain, and muscle and body aches are symptoms that frequently appear in the acute phase of COVID-19 and persist beyond the month following the infection and may last more than 3 months. In addition, PCC patients over time develop cognitive deficits, myalgias, neurological symptoms, and depressive and anxiety symptoms.10,11,12
Acute COVID-19 develops after upper airway infection with SARS-CoV-2, and it is characterized by a hyperinflammatory cascade response resulting in a so-called “cytokine storm” in severe cases.13,14,15,16 As a result, severe disease is further characterized by endothelial damage and activation, as well as intravascular coagulation, followed by lymphopenia and thrombocytopenia.17,18 In most cases, the individual’s biological immune response can control the infection, keeping symptoms mild or absent.9 Whether and how the alterations in the immune, inflammatory, and coagulation responses in acute COVID-19 are mechanistically linked to the development of PCC is not very well known.9,19,20 Initial studies comparing COVID-19 recovery cohorts with PCC individuals revealed a statistically significant correlation between PCC symptoms and the increased levels of circulating interleukin-6 (IL-6), IL-10, interferon gamma (IFN-γ), and tumor necrosis factor alpha (TNF-α).21,22 It is also already known that excess activation of the immune response directly correlates with cognitive-behavioral changes,23 as potentially ensues in PCC.
Although much PCC research is being performed, PCC cohorts are rarely compared with matched controls that were infected by the same strain of virus at the same time and managed in the same outpatient setting. In this study, we analyzed more than 150 biomarkers to survey the immunological, biochemical, and coagulation characteristics of a cohort of individuals who suffered an episode of acute COVID-19 during 2020 and early 2021, comparing individuals who developed PCC versus individuals who overcame COVID-19 without persistent or new symptoms. Using this large set of markers on a well-defined cohort of participants, this study aims to describe a panel of biomarkers to aid in diagnosis of PCC.
Results
Description of study population
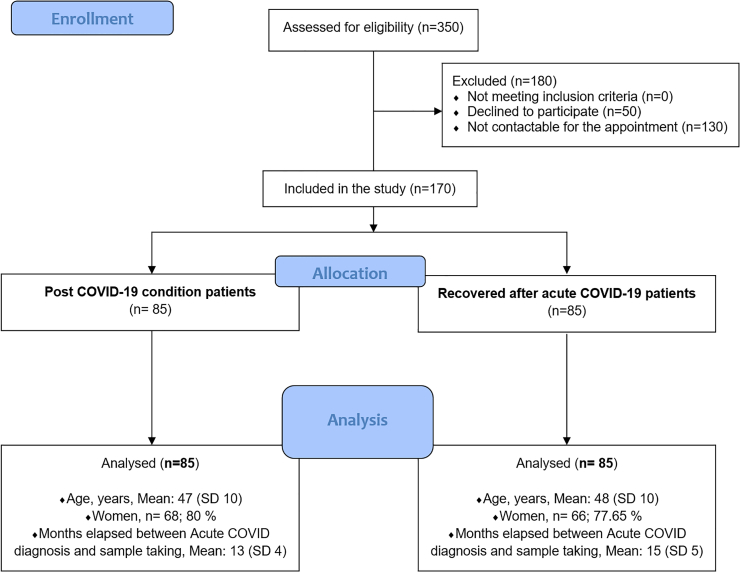
A total of 170 individuals, 85 with PCC and 85 recovered individuals, were recruited. The inclusion of the participants is detailed in the flowchart for parallel observational studies for sample selection (Figure 1). Individuals were recruited among those attending the Primary Health Care Centers (PHCCs) with PCC, which included many more women than men (68 versus 17). The small differences between the two groups for age, gender, and months elapsed between acute COVID-19 diagnosis and sample taking were not statistically significant.
Figure 1.
The flowchart of participant’s selection
Out of 350 participants (with a COVID-19 diagnosis) who were assessed for eligibility, 170 participants (85 PCC patients and 85 recovered patients) were included in the study. The two parallel groups were matched for age, sex, and date of acute COVID-19 diagnosis. The two groups are compared for mean age (years), gender composition (Nº and percentage), and time elapsed between COVID-19 diagnosis and sample taking (months).
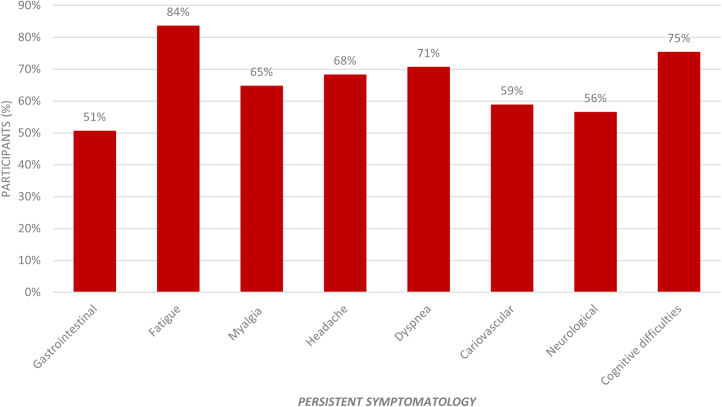
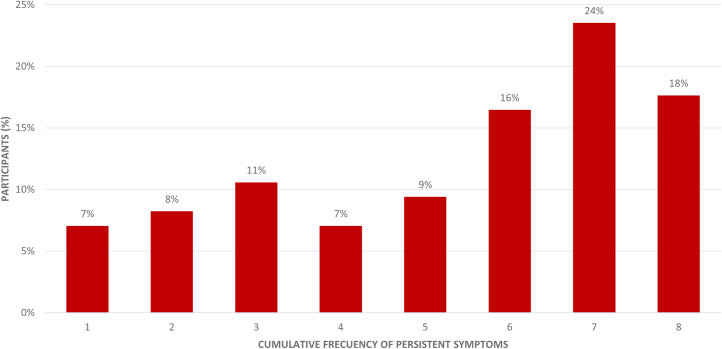
Delving into the persistent symptoms, as can be observed in Figure 2, the most frequent symptoms are tiredness or fatigue (84%), memory loss or mental confusion (75%), dyspnea (71%), headache (68%), and myalgia (65%). Furthermore, in Figure 3, as can be observed, 42% of the participants with PCC currently have seven or more persistent symptoms, with a mean of 5.35 symptoms (SD 2.23).
Figure 2.
Persistent symptomatology in the PCC group
The figure shows the percentage of individuals with PCC (85 participants) who reported the persistent symptoms listed on the x axis.
Figure 3.
Presence of total persistent symptoms in the PCC group
The figure shows the percentage of individuals with PCC (85 participants) who reported the number of persistent symptoms listed on the x axis.
We assessed a total of 167 biomarkers previously related to the severity or outcome of acute COVID-19 disease or those routinely used in patient care at the Hospital Universitario Miguel Servet. The tables below show the statistically significant results of the biochemical profile, blood cell counts, coagulation test, serology, counts of specialized lymphocyte populations, and levels of pro-inflammatory cytokines, comparing both groups of patients. Biomarkers whose levels were not statistically significant between the two groups can be viewed in Tables S1–S4. The values of most of the markers with significant differences between the two cohorts lie within the normal ranges of those parameters for known diseases. However, such values may still be clinically relevant in the new and unknown context of PCC. Small differences may discriminate between patient groups, even when values lie within normal boundaries.
Differences across biochemical, antibodies, and blood count profiles between PCC and recovered individuals
PCC individuals showed significantly lower levels of creatine kinase, glycohemoglobin, sodium, potassium, and glomerular filtration rate compared to recovered individuals (Table 1). By contrast, PCC individuals showed higher creatinine, cholesterol, and albumin levels than recovered individuals.
Table 1.
Comparison of the biochemical profiles, liver and thyroid function tests, and measurements of electrolyte statistics between recovered and PCC individuals
| Variables | Reference range | Recovered (n = 85) | PCC (n = 85) | p value |
|---|---|---|---|---|
| Biochemical profile | ||||
| Creatine kinase, ng/mL, median (IQR) | 0.6–6.3 | 1.60 (1.80) | 1.20 (1.30) | 0.001 |
| A1C, %, median (IQR) | <5.6 | 5.40 (0.40) | 5.30 (0.50) | 0.016 |
| Creatinine, mg/dL, median (IQR) | 0.67–117 | 0.69 (0.19) | 0.78 (0.18) | 0.001 |
| Cholesterol total, mg/dL, mean ± SD | 120–220 | 209.68 ± 46.74 | 223.41 ± 37.98 | 0.037 |
| Cholesterol no-HDL, mg/dL, mean ± SD | <130 | 146.23 ± 38.51 | 159.11 ± 35.98 | 0.026 |
| Albumin, g/dL, median (IQR) | 3.5–5.2 | 4.40 (0.40) | 4.50 (0.40) | 0.008 |
| Na +, mEq/L, median (IQR) | 136–146 | 140 (2) | 139 (3) | 0.003 |
| K +, mEq/L, median (IQR) | 3.5–5.1 | 4.40 (0.40) | 4.20 (0.40) | <0.001 |
| GFR, mL/min∗1.73mˆ2, median (IQR) | >90 | 102.61 (15.52) | 94.23 (18.77) | 0.002 |
| Antibody levels | ||||
| IgG total, pg/mL, median (IQR) | 650–1600 | 1150 (210) | 1007 (334) | 0.009 |
| IgG1, pg/mL, median (IQR) | 382.4–928.6 | 521.80 (232.90) | 453.85 (231.70) | 0.004 |
| IgE UI/mL, median (IQR) | 0–115 | 21.80 (793.61) | 41.150 (58.47) | 0.009 |
| Complement C3, mg/dL, median (IQR) | 79–152 | 94.80 (23.0) | 99.70 (26.2) | 0.014 |
| Complement C4, mg/dL, median (IQR) | 16–38 | 21.40 (6.50) | 25.00 (9.00) | 0.004 |
| αGAD, U/mL, median (IQR) | <17 | 5.33 (0.93) | 6.03 (1.09) | <0.001 |
| IGFBP3, μg/mL, mean ± SD | 3.2–6.6 | 5.64 ± 1.10 | 6.33 ± 1.24 | <0.001 |
| pANCA, u, mean ± SD | <20 | 3.31 ± 1.32 | 2.95 ± 0.52 | 0.020 |
| Blood count and coagulation test | ||||
| MCV, fL, median (IQR) | 80–100 | 88.90 (5.4) | 90.25 (5.0) | 0.029 |
| MCHC, g/dL, median (IQR) | 31–37 | 29.90 (2.2) | 30.2 (2.1) | 0.006 |
| ESR, mm/h, mean ± SD | <15 | 11.10 ± 9.05 | 14.41 ± 12.00 | 0.044 |
| INR, mean ± SD | 0.8–1.2 | 0.92 ± 0.05 | 0.94 ± 0.07 | 0.019 |
| Partial thromboplastin time, seg, mean ± SD | 24.8–37.2 | 29.74 ± 2.06 | 30.77 ± 2.72 | 0.006 |
| APTT ratio, mean ± SD | 0.8–1.2 | 0.95 ± 0.06 | 0.99 ± 0.08 | 0.008 |
| Prothrombin time, seg, median (IQR) | 9.5–14.3 | 11.10 (0.80) | 11.00 (1.40) | 0.047 |
| Fibrinogen, g/L, mean ± SD | 2–4 | 4.21 ± 0.86 | 4.57 ± 1.04 | 0.017 |
| Antithrombin III, %, mean ± SD | 80–120 | 97.03 ± 10.17 | 104.83 ± 15.86 | <0.001 |
| COVID-19 serology antibodies | ||||
| SARS-CoV-2 NC IgG positive, n (%) | 80 (94.12) | 68 (80) | 0.011 | |
| SARS-CoV-2 S1 IgG levels, pg/mL, mean ± SD | 1794.16 ± 645.09 | 1525.98 ± 828.61 | 0.020 | |
p value from χ2, Mann-Whitney’s U-test, or Student’s t test. Recovered: individuals who completely recovered within 3 months after acute COVID-19; PCC: individuals diagnosed with post-COVID-19 condition; A1c: glycohemoglobin; Na+: sodium; K+: potassium; GFR: glomerular filtration rate; Ig: immunoglobulin; αGAD: GAD antibody; IGFBP3: insulin-like growth factor-binding protein 3; pANCA: perinuclear anti-neutrophil cytoplasmic antibody; MCV: mean corpuscular volume; MCHC: mean corpuscular hemoglobin concentration; ESR: erythrocyte sedimentation rate; INR: international normalized ratio; APTT: activated partial thromboplastin time; NC, nucleocapsid; S1, Spike; SD, standard deviation; IQR, interquartile range. The reference range includes each biomarker’s upper and lower limits based on a group of healthy people, indicated by the Miguel Servet University Hospital (Zaragoza, Spain).
PCC individuals showed significantly lower levels of total immunoglobulin G (IgG), IgG1, and perinuclear anti-neutrophil cytoplasmic antibodies (pANCAs) compared to recovered individuals (Table 1). By contrast, PCC individuals showed higher levels of IgE, complement C3 and C4, αGAD, and insulin growth factor-binding protein 3 (IGFBP3) compared to recovered individuals (Table 1).
PCC individuals showed significantly lower prothrombin time levels than recovered individuals (Table 1). By contrast, PCC individuals showed higher levels of mean corpuscular volume (MCV), mean corpuscular hemoglobin concentration (MCHC), erythrocyte sedimentation rate (ESR), internation normalized ratio (INR), activated partial thromboplastin time (APTT) ratio, fibrinogen, and antithrombin III, compared to recovered individuals (Table 1).
We next assessed the antibody titers against SARS-CoV-2 (specifically S1 and N) in the plasma of all study subjects. PCC individuals showed lower αSpike IgG levels, as well as α-nucleocapsid IgG positivity, compared to individuals who recovered normally after acute COVID-19 (Table 1).
Differences across flow cytometry and cytokine analysis between PCC and recovered individuals
PCC individuals had higher levels of naive CD8+ T cells and lower levels of effector CD8+ T cells (% among CD8+ cells) than recovered individuals (Table 2). PCC individuals also showed higher levels of cells with natural killer (NK) phenotype CD56+CD16+, as well as CD8+CD4low cells, than recovered participants. On the contrary, the unconventional cell population with CD56−CD16bright phenotype was significantly reduced in PCC individuals.
Table 2.
Comparison of flow cytometry statistics between recovered and PCC individuals
| Variables | Recovered (n = 85) | PCC (n = 85) | p value |
|---|---|---|---|
| CD56+CD16+ cells among NK + NKT, % | 63.20 (25.30) | 73.45 (28.72) | 0.038 |
| CD56+CD16− cells among NK + NKT, % | 35.70 (27.00) | 23.30 (26.50) | 0.032 |
| CD56−CD16bright among CD56− cells, % | 2.21 (2.22) | 1.60 (1.17) | <0.001 |
| Naive CD8+ cells among CD8+ cells, % | 22.10 (20.73) | 29.40 (25.93) | 0.023 |
| Effector CD8+ among CD8+ cells, % | 16.60 (14.80) | 13.15 (12.18) | 0.015 |
| CD8+CD4low cells among T lymphocytes % | 0.37 (0.39) | 0.50 (0.44) | 0.013 |
Subsets of cells in human peripheral blood based on the relative expression of either CD16 and CD56 or CD8 and either CD45RA or CD62L. Frequencies (in %) among CD3−CD56+ NK cells, CD56− lymphocytes cells, and CD3+CD8+ cells are indicated for each patient group. Recovered: individuals who completely recovered within 3 months after acute COVID-19; PCC: individuals diagnosed with post-COVID-19 condition; values are presented as median (interquartile range). Statistical differences were calculated using Mann-Whitney’s U-test.
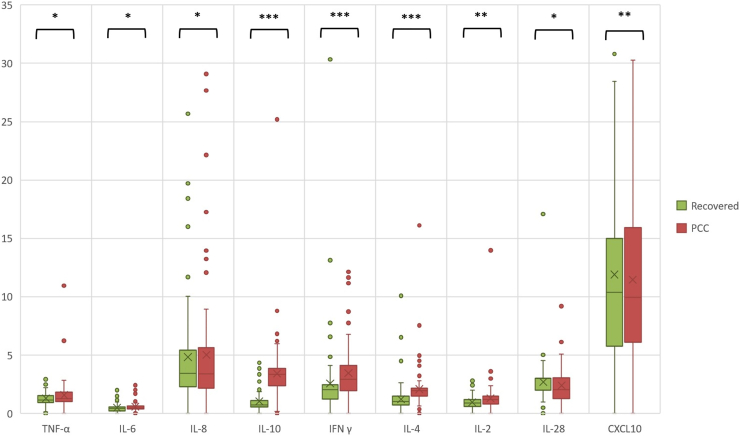
We also studied levels of several circulating cytokines. We selected markers previously identified as relevant for the severity of acute COVID-19 disease (CXCL10 and IL-6), additional markers relevant for diagnosis and disease severity in acute and PCC (TNF-α, IFN-γ, IL-2, IL-4, IL-8, IL-10),24 and cytokines involved in antiviral defense (IL-10, IFN-α, IFN-β, IL-28B).25 Due to low signal-to-noise ratios (SNRs), data for IFN-α and IFN-β were not included in further analysis. Peripheral blood from PCC individuals showed higher levels of TNF-α, IFN-γ, IL-6, IL-8, IL-10, IL-4, IL-2, and CXCL10 compared to recovered individuals. However, PCC individuals showed lower levels of IL-28 (Figure 4). Pro-inflammatory cytokines analysis (values of median and interquartile range) for the analytic sample can be viewed in Table S5.
Figure 4.
Comparison of concentrations of circulating cytokines between recovered and PCC individuals
Cytokine levels have been analyzed by Luminex. The boxplot shows the levels of the cytokines indicated (horizontal axis) and their concentrations (y axis: pg/mL). The median values and interquartile ranges in each group (recovered controls in green; patients with PCC in red) can be viewed in Table S5. The statistical significance of differences from Mann-Whitney’s U-test between the two groups is indicated as: ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
Predictors of PCC revealed by multivariate logistic regression analysis
Table 3 shows two multivariate logistic regression analyses, including model A with 15 biomarkers and model B with 4 biomarkers. Both multivariate logistic regression (MLR) models explain around 80% of the possibilities of PCC characterization, the highest in the prediction model being with four biomarkers (αGAD, IL-10, potassium, and CD56−CD16bright). This prediction MLR model showed that the values of αGAD and IL-10 were an independent predictor of PCC. In addition, this analysis showed that the values of CD56−CD16bright cells and K+ were protective factors against persistent COVID-19. This model explains 81.8% of the possibilities of PCC characterization.
Table 3.
Multivariate logistic regression analysis of PCC predictors
| Model | Biomarker | OR | 95% CI |
|---|---|---|---|
| A) | |||
| αGAD | 1.626 | [1.084, 2.440] | |
| IL-10 | 3.412 | [2.252, 5.170] | |
| Antithrombin III | 1.116 | [1.030, 1.208] | |
| Potassium | 0.003 | [0.000, 0.129] | |
| CD56−CD16bright cells | 0.349 | [0.189, 0.643] | |
| pANCA | 0.000 | 0.000 | |
| Prothrombin activity | 1.034 | [0.974, 1.099] | |
| Aldolase | 0.879 | [0.614, 1.260] | |
| Effector CD8+ T cells | 0.996 | [0.923, 1.074] | |
| MCHC | 0.627 | [0.251, 1.570] | |
| Sodium | 0.418 | [0.233, 0.752] | |
| IGFBP3 | 0.523 | [0.252, 1.085] | |
| CD16+ NKT-like cells | 1.012 | [0.851, 1.202] | |
| CD16+ NK cells | 1.002 | [0.849, 1.183] | |
| NT-proBNP | 0.984 | [0.964, 1.005] | |
| R2 Nagelkerke = 0.801 | |||
| B) | |||
| αGAD | 1.956 | [1.177, 3.249] | |
| IL-10 | 5.750 | [2.829, 11.684] | |
| Potassium | 0.025 | [0.003, 0.236] | |
| CD56−CD16bright cells | 0.546 | [0.379, 0.786] | |
| R2 Nagelkerke = 0.818 | |||
Multivariate logistic regression analysis using the stepwise method. Odds ratios (ORs) were calculated with a 95% confidence interval. αGAD: GAD antibody; IGFBP3: insulin-like growth factor-binding protein 3; CD16: CD16 natural killer cells; CD56: CD56 natural killer cells; NT-proBNP: N-terminal pro b-type natriuretic peptide; CD8: CD8 T lymphocytes or cytotoxic T cells; pANCA: perinuclear anti-neutrophil cytoplasmic antibody.
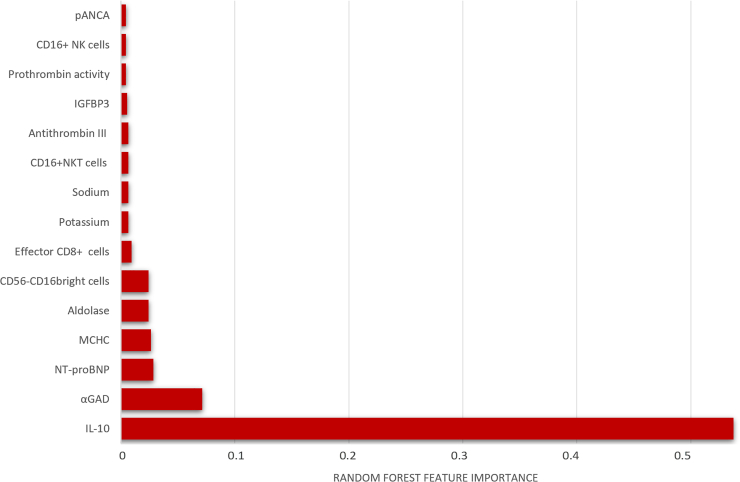
Predictors of PCC revealed by random forest analysis
In terms of significant variables, Figure 5 shows the feature importance of the variables included for the random forest model to observe which values of these biomarkers achieve a better characterization of PCC.
Figure 5.
The feature importance plot of Random Forest model for PCC prediction
Each plot is a bar graph with the feature importance on the x axis and the names of each feature on the y axis. The x axis represents the feature importance, with the values indicating the contribution of the feature to the model. A higher value on the x axis indicates a higher importance of the feature in the model.
We constructed two types of models (Random Forest [RF] and MLR) to evaluate the effects of the independent variables on PCC. To assess the importance of the included biomarkers, two types of classifications were used (type A, 15 biomarkers; type B, 4 biomarkers classified among the main ones included in both models; see in Table 4).
Table 4.
Performance of different models and predictors
| Model | Accuracy (95% CI) | Precision (95% CI) | Recall (95% CI) | F1 (95% CI) | Sensitivity | Specificity | PPV | NPV |
|---|---|---|---|---|---|---|---|---|
| A) | ||||||||
| MLR | 86.47 (84.35–87.65) | 88.45 (85.59–90.41) | 84.33 (81.75–86.25) | 85.36 (83.06–86.95) | 85.88 | 91.76 | 91.25 | 86.67 |
| RF | 81.69 (79.50–82.50) | 84.44 (81.44–86.56) | 83.00 (80.44–85.56) | 81.09 (79.35–82.65) | 84.71 | 87.06 | 86.75 | 85.06 |
| B) | ||||||||
| MLR | 88.23 (87.10–88.90) | 92.43 (90.50–93.50) | 84.29 (82.05–85.95) | 87.19 (85.95–88.05) | 84.71 | 92.94 | 92.31 | 85.87 |
| RF | 85.25 (83.50–86.50) | 84.97 (81.75–86.25) | 86.76 (84.05–87.95) | 84.59 (82.50–85.50) | 85.88 | 89.41 | 89.02 | 86.36 |
Describe different models for separation of PCC patients and controls etc. Variables included Model A: αGAD + IL-10 + Antithrombin III + Potassium + CD56−CD16bright + pANCA + Prothrombin activity + Aldolase + Effector CD8+ T cells MCHC + Sodium + IGFBP3 + CD16+NKT cells + CD16+ NK cells + NT-proBNP. Variables included Model B: αGAD + IL-10+ Potassium + CD56−CD16bright.
The results indicated that, by including 15 biomarkers (Model A), the MLR model showed better accuracy, precision, recall, and F1 than the RF model (Table 4). Meanwhile, by including only four biomarkers (Model B), both models showed better results compared to those obtained by including 15 biomarkers. In this case, the MLR model achieved better accuracy, precision, and F1 results; however, the RF model had the highest recall rate (Table 4). The prediction MLR model with four biomarkers (αGAD, IL-10, potassium, and CD56−CD16bright) showed an accuracy of 88.23%, a high precision of 92.43, and an F1 of 87.19%.
In all the results of the predictive models (Table 4), a high specificity and sensitivity could be observed, the highest in the prediction model being the MLR with four biomarkers (αGAD, IL-10, potassium, and CD56−CD16bright) with a specificity of 92.94%, sensitivity of 84.71%, and positive and negative predictive values of 92.31% and 85.87%, respectively. In this model, 13 participants out of the 85 PCC individuals were classified by the algorithm as recovered patients (false negatives), whereas 6 participants out of the 85 recovered individuals were classified as PCC patients (false positives) in this same model. The levels of biomarkers were compared between individuals with false-negative PCC compared to the average of individuals with PCC or between cases of false positives and the average of recovered individuals. These biomarkers and persistent symptoms of those cases of false positives or false negatives obtained in each of the different models are compiled in Table S6. Specifically, the levels of several biomarkers (IL-10, αGAD, antithrombin III) are lower in patients with false-negative PCC compared to the average of individuals with PCC and loser to the values observed in recovered patients (such as IL-10, αGAD, antithrombin III).
Correlation between biomarkers obtained in the prediction models and persistent symptoms
Table 5 shows the bivariant analysis relating the 15 biomarkers obtained in the prediction models and the presence of persistent symptoms. As Table 5 shows, the presence of IL-10 and IGFBP3 was moderate correlated with all persistent symptoms analyzed, whereas higher counts of CD56−CD16bright cell counts were inversely correlated with the persistent symptoms. On the other hand, alterations in the levels of biomarkers such as αGAD, antithrombin III, and CD16+ NKT-like cell counts were more specifically associated with symptoms such as muscle pain, headache, dyspnea, and fatigue.
Table 5.
Correlation between biomarkers and persistent symptoms
| Model | Gastrointestinal | Fatigue | Muscular pain | Headache | Dyspnea | Cardiovascular | Neurological | Cognitive difficulties | Total of persistent symptoms |
|---|---|---|---|---|---|---|---|---|---|
| αGAD | 0.102 | 0.129 | 0.154∗ | 0.069 | 0.231∗∗ | 0.045 | 0.098 | 0.115 | 0.145 |
| IL-10 | 0.300∗∗∗ | 0.433∗∗∗ | 0.281∗∗∗ | 0.264∗∗ | 0.363∗∗∗ | 0.337∗∗∗ | 0.258∗∗ | 0.426∗∗∗ | 0.412∗∗∗ |
| Antithrombin III | 0.111 | 0.234∗∗ | 0.242∗∗ | 0.193∗ | 0.242∗∗ | 0.085 | 0.164∗ | 0.127 | 0.219∗∗ |
| Potassium | −0.142 | −0.141 | −0.103 | −0.183∗ | −0.196∗ | −0.164∗ | −0.147 | −0.151∗ | −0.198∗∗ |
| CD56−CD16bright cells | −0.214∗∗ | −0.296∗∗∗ | −0.254∗∗ | −0.192∗ | −0.252∗∗ | −0.181∗ | −0.220∗∗ | −0.220∗∗ | −0.283∗∗∗ |
| pANCA | −0.122 | −0.152∗ | −0.133 | −0.153∗ | −0.154∗ | −0.135 | −0.107 | −0.124 | −0.167∗ |
| Prothrombin activity | 0.005 | −0.031 | 0.083 | −0.000 | 0.063 | 0.025 | −0.030 | 0.026 | 0.023 |
| Aldolase | −0.013 | 0.063 | 0.111 | 0.091 | 0.050 | 0.083 | 0.110 | 0.075 | 0.086 |
| Effector CD8+ T cells | −0.075 | −0.061 | −0.016 | −0.107 | −0.068 | −0.111 | −0.145 | −0.079 | −0.105 |
| MCHC | −0.099 | −0.166∗ | −0.125 | −0.112 | −0.162∗ | −0.202∗∗ | −0.105 | −0.0201∗∗ | −0.181∗ |
| Sodium | −0.053 | −0.097 | −0.065 | −0.098 | −0.114 | −0.183∗ | −0.056 | −0.105 | −0.128 |
| IGFBP3 | 0.222∗∗ | 0.184∗ | 0.255∗∗ | 0.211∗∗ | 0.200∗∗ | 0.256∗∗ | 0.262∗∗ | 0.168∗ | 0.264∗∗ |
| CD16+ NKT-like cells | 0.198∗ | 0.102 | −0.010 | 0.086 | 0.035 | 0.027 | 0.043 | 0.0281 | 0.071 |
| CD16+ NK cells | −0.124 | −0.113 | −0.097 | −0.000 | −0.123 | −0.076 | −0.113 | −0.091 | −0.071 |
| NT-proBNP. | 0.083 | 0.098 | 0.003 | −0.014 | 0.149 | 0.071 | 0.024 | 0.007 | 0.065 |
Relations between the levels of the 15 biomarkers obtained in the prediction models (indicate Figure/Table) and the presence of persistent symptoms were analyzed through bivariate analysis using Point-Biserial rpb correlation coefficients. Point-Biserial’s rpb correlations are listed., Their statistical significance of differences from Point-Biserial’s rpb correlation is indicated as follows: ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
Discussion
This study compares the immunological and biochemical characteristics of samples taken from two groups of individuals matched by sex and age, who developed acute COVID-19 during the years 2020–2021 but experienced a different evolution. One group recovered within 3 months and has been asymptomatic since. The other group developed PCC, defined as the continuation of symptoms or the appearance of novel symptoms more than 3 months after SARS-CoV-2 diagnosis. We have investigated differences between the two groups at the immunological level (serology, cell populations, and cytokine levels) and in terms of commonly analyzed biochemical markers.
Our objective was to create a panel of biomarkers that differentiates between PCC individuals and recovered controls. Such a panel may suggest underlying mechanisms, but relating the panel to the pathogenesis is a different task. Therefore, clinical interpretation was not the direct goal of our analysis. We have been trying to understand the pathogenesis in separate studies, partially based on the biomarkers identified here.
Among the 167 markers analyzed, several showed statistically different values in PCC patients compared to controls.
Several of these markers have been previously associated with alterations in PCC: the immunosuppressive cytokine IL-1026 and the natriuretic NT-ProBNP.27 These earlier studies examined individuals with specific symptoms of PCC, such as somatic and affective symptoms,26 autonomic nervous system impairment,27 and a much more limited set of biomarkers. In our study, we confirmed reported results, using a larger sample and with a control group of asymptomatic post-COVID-19 patients.
By contrast, other markers we identify in this study have not been previously linked to PCC. Not surprisingly, symptoms associated with several of the markers we found significantly modified in the present study do partially overlap with PCC symptoms. This holds for debilitating symptoms, including post-exertional malaise, fatigue, neurological, and muscular pain associated with high IL-10 levels28; headache, tiredness, myalgia, cardiac arrhythmias, and fatigue associated with altered Na+/K+ levels29,30; and neurological symptoms, cognitive difficulties, and fatigue associated with impaired GABA neurotransmission by high levels of αGAD antibodies31; dysautonomia has been associated with the persistence of elevated levels of NT-ProBNP.27 Moreover, altered antithrombin levels are associated with several pathophysiological processes associated with COVID-19 disease and possibly PCC such as prolonged activation of coagulation, microvascular injury, and thrombosis.32 IGFBP3 is the main carrier of insulin growth factor 1 (IGF-1) in the body. The production of IGFBP3 in the liver as well as circulating levels are reportedly altered in chronic inflammatory diseases,33 although we are unaware of previous reports linking IGFBP3 to PCC.
Immunological alterations and virus-specific T cell responses have been studied previously in acute COVID-19 disease. The extent to which dysregulated immune responses contribute to PCC remains poorly understood. The immunological profile of the individuals with PCC from our cohort appears to indicate a general tendency toward an immunological disorder, exemplified by a decrease in effector CD8+ T cells along with an increase in naive CD8+T cells. These combined data may point toward an impaired transition to CTLs (cytotoxic T lymphocytes) or memory cells. A decrease in the activation of CD8+ memory T cells in PCC patients has been reported previously.34 SARS-CoV-2 evolution appears driven by CD8 T cell escape, providing indirect evidence for a potential role of effector CD8+ T cell responses in PCC,34 consistent with our own observations. The overexpression of cytokine IL-10 associated with decreased numbers of CD8+ CTLs in our PCC group is reminiscent of the long-term evasion of pathogens from immune response detected in persistent infections.35 In addition, we observed increased levels of highly cytotoxic NK cells with CD56+CD16+ phenotype in individuals with PCC, in accordance with previous reports.36 These cells may compensate for the reduced levels of CTLs and would likely contribute to the proinflammatory environment along with the increased plasma levels of IL-6, IL-8, IL-2, IFN-γ, and TNF-α we observed in our PCC. On the other hand, lower levels of cells with CD56−CD16bright phenotype were also found in the PCC group, as reported previously for acute COVID-19 patients.36 These cells are unconventional cytotoxic mediators for chronic diseases,19 and in fact, they are present in high levels in people living with HIV.20
Statistical analysis of the combined data identified two panels of markers whose levels permit the separation of PCC patients from patients who recovered within 3 months from acute COVID-19 disease with a high accuracy and precision, similar in both statistical methods. One of this panel had 15 biomarkers, with most significances were the values of both CD8+ effector cells and CD56−CD16bright cells; lower sodium and potassium ion concentrations; higher levels of IL-10, NT-proBNP, antithrombin III, prothrombin activity, IGFBP3, pANCA, aldolase, MCHC, and αGAD antibodies; and higher numbers of CD16+NKT cells and CD16+ NK cells, with an accuracy of 0.864 and a precision of 0.884. The second panel found produced a highly accurate diagnostic model based on a combination of four markers (potassium decreased, αGAD antibodies levels, the frequency of CD56−CD16bright cells, and higher values of IL-10) with an accuracy of 0.882 and a precision of 0.924. Because of the lower number of markers included in this second panel, it may be more easily adapted into clinical practice. Distinct statistical methods used in our research give similar results when establishing the profile characterization of the PCC patients, which bolsters our findings.
The main strength of our research is that we have compared specifically between PCC patients and individuals recovered after 3 months after a confirmed episode of COVID-19 disease, since we have not found in the literature similar studies, as PCC is usually compared with a general population cohort; in addition, we compared paired cases, reducing the possibility of age, gender and the dates of infection being confounding factors. We also analyzed many markers in a reasonable number of PCC outpatients and the controls.
All participants were recruited among those who passed COVID-19 on an outpatient basis, so we surmised that our findings were more related to the SARS-CoV-2 infection itself, eliminating the potential effects of severe disease and hospitalization. Fulfilling a widely reported necessity, we here present an accurate panel of biomarkers for the diagnosis of PCC.
The addition of CD56−CD16bright cell counts to the diagnostic panel improved the accuracy and precision of the model. As this marker is not easily available in a primary care/hospital setting, we continue the search for substitute markers. Nevertheless, our results may prove to be of widespread use to the primary care services, setting where these patients are routinely cared for. Future implementation of this panel in a care setting may help separate PCC individuals from other individuals with similar symptoms but different causes and disease courses. This is essential to find therapeutic solutions for PCC individuals who suffer from a series of highly disabling symptoms in many cases.37
The classification models presented may be hard to implement in clinical practice in their present configuration, because of the inability to measure specific cell counts in a hospital/primary care setting and insufficient computational literacy among primary care physicians. In addition to the recruitment and analysis of a validation cohort, efforts are on the way to evaluate an array of markers either related or not to the specific cell populations identified. It is our hope that we can substitute cell counts for single-molecule biochemical assays and/or immunoassays. Using the present models as a starting point, we aim to incorporate additional markers for the development of a next generation of models based on more readily available biomarkers. As examples, proteomic studies found that several markers of vascular health related to ANGPT1 and vascular endothelial growth factor (VEGF) signaling, and the sustainability of the vascular bed (including HIF1, PDGFA, PDGFRA) may be associated with PCC and/or progression from acute COVID-19 disease to PCC.38 A separate study has indicated specifically high ANG-1/P-SEL levels in PCC.39 Addition of these and related markers may improve future versions of these panels and facilitate its integration into clinical practice. The integration of validated classification models into clinical practice may improve through the contribution of specialized units (at a hospital or regional scale) or the implementation of handheld devices.
We present data on markers that may differentiate PCC individuals from recovered individuals, independent of symptoms. The profile representing our individuals with PCC was characterized by lower levels of potassium and CD56−CD16bright cells and higher levels of IL-10 and αGAD. This model will help clinicians in decision-making and reduce the professionals’ uncertainty about this new disease. We hope to proceed toward a diagnostic panel of markers that underlie post COVID-19 syndrome and work on a predictive algorithm for PCC and patient stratification.
Limitations of the study
Several limitations must be considered in this report. Sample taking, and subsequent data were collected once per participant, rather than longitudinally. This cross-sectional analysis limits the ability to draw causal inferences from the data obtained, which provides no predictive value regarding the evolution or progress of the disease. Our sample size in both cohorts (n = 170) was small from the statistical and machine learning perspective. Future prospective studies with a larger sample size and lower SNR thresholds are warranted to validate our findings. This is especially true for the levels of TNF-α, IL-6, and IL-10 that we reported here, as the signal-to-noise ratio we calculated for these data was close to a minimum threshold of 14 dB for acceptance.
Future corroboration on an independent group of participants (preferably representing either a more diverse set of individuals or distinct geographical localizations) and/or set of external data obtained under a similar experimental protocol is pending. Moreover, similarities and reproducibility among other PCC-biomarkers’ conducted studies have not been consistent, so there is a need for standardization and validation of the specificity/sensibility of the biomarkers, to be able to draw more rigorous conclusions and valuable comparisons.40 Even considering these limitations, we hope that our study may contribute to a better definition of diagnostic markers specific for PCC.
Although the diagnosis of PCC was established 3 months after acute infection and disease (Centers of Disease Control and Prevention [CDC] and WHO definitions), samples for the analysis of biomarkers were taken at 1–2 years after the acute infection. It would have been preferable to take and analyze timed samples, in order to describe the alterations of markers over time. The results of such an analysis may elucidate changes and markers that predict PCC condition and help understand the pathophysiology of PCC. As only 10%–20% of individuals would develop PCC,5,6 this approach would have required an enormous amount of samples to do. Unfortunately, neither the personnel nor funding was available during the pandemic. As carried out, the biomarkers identified in our study lack predictive value. We do provide biomarkers that are hopefully applicable to clinical practice for diagnosing PCC in a next step.
Likewise, the influence of sex and/or gender on these results cannot be studied, given that the population diagnosed with CCP has been predominantly female and therefore the percentage of male population in question is very low. That is why, in order to generalize our results comparing CCP with recovered individuals, we made a selection of the two parallel groups that were matched by age, sex/gender, and date of acute COVID-19 diagnosis.
Resource availability
Lead contact
Further information and requests should be directed to and will be fulfilled by the lead contact, Fátima Méndez-López (fmendez@iisaragon.es).
Materials availability
This study did not generate new materials.
Data and code availability
-
•
Data have been deposited at Zenodo and are publicly available as of the date of publication. The DOI is listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Acknowledgments
This study has been funded by Carlos III Health Institute (ISCIII), grant numbers PI22/01070, the Aragonese Primary Care Research Group (GAIAP, B21_23R), and ERDF Funds “A way of making Europe” and co-financed by the European Union. The work described in this manuscript received funding from the European Union “Horizon Europe” research and innovation program under the grant agreement No 101057302 (HERVCOV). Funders had no role in the study design, data collection and analysis, decision to publish, or manuscript preparation. The research will be audited once a year by the funding organization. We would like to give special thanks to the project PI17/02208 funded by Health Institute Carlos III (ISCIII) and “COVID funds” provided by IISA, both of which have contributed funding for the realization of this article. J.S. and D.L.-I. were supported by the HERVCOV project, funded by the HORIZONHLTH-2021-DISEASE project (Personalized medicine and infectious disease: understanding the individual host response to virus) of the European Commission under the Horizon Europe Framework Program (G.A.101057302). We would like to thank the Aragonese Primary Care Research Group (GAIAP, B21_23R) and the Placental pathophysiology and fetal programming group (B46_20R), both part of the Department of Innovation, Research and University at the Government of Aragón (Spain), the Research Network on Chronicity, Primary Care and Health Promotion (RICAPPS, RD21/0016/0005) that is part of the Results-Oriented Cooperative Research Networks in Health (RICORS) (Health Institute Carlos III), and the European Regional Development Fund (EDRF) “A way of making Europe”, for their support in the development of the study. In addition, we would like to thank other members of the Biocomputing Unit, IACS and the Biobank of the Aragon Health System (PT20/00112) integrated in the Platform ISCIII Biobanks and Biomodels, for their continued support and interest. We thank Julian Pardo for his suggestions regarding marker selection, Mark Strunk for additional data management, and Antonella Minutolo and Claudia Matteucci for sharing a template for the graphical abstract. We particularly want to acknowledge all the participants for their collaboration in this study. We also thank the Long Covid Aragon Patients Association for their contributions and collaboration in carrying out the study.
Author contributions
R.M.-B. and J.S. contributed to the coordination, design, and supervision of the study. M.R.-H. performed the sample extraction and data acquisition. J.G. and B.J.-B. carried out flow cytometry and Luminex analysis. M.E.-B. and I.A. did the maintenance and management of biobank samples. D.L.-I. and P.G.-I. checked, cleaned, and consolidated datasets and performed bibliographical searches. F.M.-L., M.B.-O., and B.O.-B. performed the data extraction and statistical analysis. A.M., J.I.-R., and C.T. performed the complex statistical analysis. M.E.-B., A.M.-M., I.A., J.G., B.J.-B., D.L.-I., V.C.-V., and P.G.-I. provided administrative, technical, or material support. F.M.-L., M.B.-O., and B.O.-B. participated in the preliminary report and assisted in the preparation of the manuscript draft. F.M.-L., M.C., C.T., and A.M.-M. provided essential knowledge for data interpretation. F.M.-L., R.M.-B., and J.S. wrote the manuscript. All authors contributed to the study and have reviewed, edited, and approved the final manuscript.
Declaration of interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| CD4-FITC/CD8-PE/CD3-PE Cy5.1 antibody Cocktail | Beckman Coulter | IM1650 |
| CD10 | Cytognos | CYT-10AP |
| CD56 | Cytognos | CYT-56F |
| CD45 | Immunostep | Cat# 45CFB1-100T, RRID:AB_11140404 |
| CD45RA | Immunostep | Cat# 45RAA2-100T, RRID:AB_11141726 |
| CD14 | Immunostep | Cat# 14A-100T, RRID:AB_11140663 |
| CD16 | Immunostep | Cat# 16PE2-100T, RRID:AB_11142639 |
| CD3 Antibody, anti-human, APC | Miltenyi Biotec | Cat# 130-113-687, RRID:AB_2726228 |
| CD62L (L-Selectin) Monoclonal Antibody (DREG-56 (DREG56)), APC-eFluor™ 780, eBioscience | Thermo Fisher Scientific | Cat# 47-0629-42, RRID:AB_1582224 |
| Critical commercial assays | ||
| Custom Luminex Assays | R&D Systems | https://www.bio-techne.com/luminex-assay-customization-tool |
| Deposited data | ||
| Data | Zenodo | Zenodo database: https://doi.org/10.5281/ZENODO.8211331 |
| Software and algorithms | ||
| IBM SPSS Statistics for Windows, Version 24.0 (Armonk, NY: IBM Corp). | IBM SPSS® | https://www.ibm.com/cn-zh/products/spss-statistics |
| xPONENT 3.1 software | R&D Systems | https://www.luminexcorp.com/xponent/ |
| Kaluza Analysis Software | Beckman Coulter | https://www.beckman.com/flow-cytometry/software/kaluza |
| Python 3.0 | Python Software Foundation | https://www.python.org/download/releases/3.0/ |
| Optuna optimization framework 3.1.1 |
Takuya Akiba, Shotaro Sano, Toshihiko Yanase, Takeru Ohta, and Masanori Koyama. 2019. Optuna: A Next-generation Hyperparameter Optimization Framework. In KDD. | https://optuna.org/ |
| Other | ||
| RBC lysis solution | Cytognos | CYT-QL-1 |
Experimental model and study participant details
A cross-sectional study was conducted comparing two parallel groups of human participants: individuals diagnosed with PCC (PCC individuals) versus individuals who completely recovered within 3 months after acute COVID-19 (recovered individuals).
For the sample size, we used the prevalence of PCC obtained in Global Burden of Disease Long COVID Collaborators study41 as the main variable. For a one-sided test, with a confidence level of 95% and a statistical power of 80%, we assumed empirically that up to 30% of analytical abnormalities can persist in the control group and up to 50% of them in the intervention group (considering only a 20% difference between them). Total sample size required was 156 (73 participants in each group to find that difference).
All participants were enrolled during 2022 in Primary Health Care Centers (PHCCs) of Zaragoza (Spain). Participants have been recruited through different methods: (1) during visits that were part of regular medical care, (2) advertising on posters inside PHCCs, and (3) telephone calls to individuals who were selected through review of electronic medical records. The two parallel groups were matched by age, sex/gender, and date of acute COVID-19 diagnosis. All human participants were interviewed to collect information on age, biological sex, and gender. The diagnosis of PCC was determined by a general practitioner, following the WHO criteria,7 before or at the time of inclusion in the study. Recovered individuals were required to have passed acute COVID-19, confirmed by RT-qPCR, antigen test, or SARS-CoV-2 serology. The exclusion criteria for participants were: (1) the presence of symptoms prior to acute SARS-CoV-2 infection; (2) the refusal or inability to consent or communicate; (3) be institutionalized at the time of appointment.
The project was approved by the Clinical Research Ethics Committee of Aragon (Spain) (protocol reference PI21/278). The study was developed in accordance with the Declaration of Helsinki. Since the project involves the collection and processing of personal data (including personal information) the collection, treatment, communication, and transfer of personal data of all participating subjects comply with the provisions of the General Data Protection Regulation (EU) (GDPR 2016/679) and the applicable national legislation, Organic Law 3/2018, of December 5, on the Protection of Personal Data. Samples and data from patients included in this study were provided by the Biobank of the Aragon Health System (National Registry of Biobanks B. B.0000873) (PT20/00112), integrated in the Platform ISCIII Biobanks and Biomodels and they were processed following standard operating procedures with the appropriate approval of the Ethics and Scientific Committees. Informed consent was obtained from all participants. This study has been reported according to Strengthening the Reporting of Observational studies in Epidemiology STROBE guidelines.
Method details
Peripheral blood samples were collected by intravenous puncture using different vacuum collection systems. For each analysis, samples of the same type were used from all individuals: serum for biochemical profiles, sodium-citrate plasma for coagulation status, and EDTA plasma for blood cell counts, flow cytometry and cytokine analysis.
The main outcome variable was the diagnosis of PCC, used as a dichotomous variable (Yes/No). The persistent symptoms of the participants were also recorded. Persistent symptoms included were: Gastrointestinal symptoms, tiredness or fatigue, dyspnea, headaches, cardiovascular symptoms (dizziness, tachycardia, orthostatic hypotension), muscular pain, neurological symptoms (tingling, spasms, etc.), and cognitive difficulties (memory loss, brain fog or confusion or poor attention and concentration capacity). Samples were analyzed to determine biomarkers routinely used in Patients’ Care at the Hospital Universitario Miguel Servet (Zaragoza, Spain): biochemical profile, complete blood cell counts, coagulation status, and serology (See more details in Table S7). In addition, we measured counts of specialized lymphocyte populations (T cell populations, Natural Killer cells, monocytes, and neutrophil maturation) and levels of pro-inflammatory cytokines. Flow cytometry and cytokine analysis using Luminex technology were carried out using standard procedures with modifications.
Total blood was stained with specific antibodies using 4 different panels. Panel 1 (T cell populations): CD3, CD4, CD8, CD45, CD45 RA and CD62L. Panel 2 NK Cells: CD3, CD45 and CD56. Panel 3 Monocytes: CD14, CD16 and CD45; and Panel 4 NKs and Neutrophil maturation (CD16, CD10, CD45 and CD56).
For the cytometry staining 50 μL of whole blood was stained with the different antibody cocktails with the volumes indicated for 15 min at 4°C in the dark. (Table S8). After incubation, RBCs were lysed for 30 min at room temperature with 950 μL of RBC lysis solution (Cytognos; CYT-QL-1) after vigorously stir. Data were acquired on a Gallios flow cytometer (Beckman Coulter) and analyzed using Kaluza Analysis Software (Beckman-Coulter, Miami, FL). The following antibodies were used for the immune phenotyping: (from Beckman Coulter) CD4-FITC/CD8-PE/CD3-PE Cy5.1 antibody Cocktail (IM1650); from Cytognos: CD10-APC (CYT-10AP) and CD56-FITC (CYT-56F); from IMMUNOSTEP: CD45-CF Blue (45CFB1-100T), CD45 RA-APC (45RAA2-100T), CD14-APC (14A-100T) and CD16-PE (16PE2-100T); from Miltenyi CD3-APC (130-113-687) and from eBiosciece CD62L-APC eFluor 780 (47-0629-42). See details about gating strategies in Figures S1–S4.
Luminex assay was run according to manufacturer’s instructions in 25 μL of plasma, samples were run in duplicate, a custom human cytokine panel (R&D Systems; LXSAHM) the next proteins were included: TNF α, IL2, IL4, IL6, IL8, IL10, IL28B, IFN α, IFN β, IFN γ and CXCL10 Supernatants (previously diluted 1:2 with the buffer provided in the kit) were mixed with magnetic beads coated with capture antibodies and incubated on a 96 well plate for 2 h after washing beads were incubated with biotin labeled antibodies for 1 h washed again and finally incubated with streptavidin-PE for 30 min. Assay plates were measured using a Lumimex 200 (luminex corporation) acquiring a minimum of 50 beads per analyte and analyzed with xPONENT 3.1 software.
Quantification and statistical analysis
A descriptive analysis (frequencies for categorical variables; means and standard deviation for parametric variables; median and interquartile range for non-parametric variables) was performed to determine the characteristics of the sample. Pearson’s χ2 test (qualitative variables), the Student’s t-test (parametric variables), and U Mann-Whitney (non-parametric variables) compared differences between independent variables. Different correlation analyses (Pearson’s r, Point-Biserial’s rpb, Kendall’s τ, and Spearman’s ρ correlation coefficients)42 were applied to the combined data (biochemical, cytokines, cell counts, and persistent symptoms) to identify the analytical parameters that had a greater resolution capacity in relation to the classification variables.
The statistical significance of differences from the different bivariante analysis is indicated as: ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001. The descriptive and correlation analyses were performed using IBM SPSS Statistics software (version 25.0). Also, we constructed two prediction models, an artificial intelligence (AI)-assisted regression and machine learning (Random Forest) (RF) and a Multivariate Logistic Regression (MLR) model to observe which values of these biomarkers achieve a better characterization of the PCC. In order to find optimal models to identify PCC patiens, we wrote our own code over Python3. Pandas library has been used for data preprocessing and manipulation, and scikit-learn provided us with a framework containing several classification algorithms and evaluation metrics, which have been boosted through the Optuna library to increase our model performance. To generate the classification model, we just included as input variables those considered biochemical markers, as these variables can provide an objective and generalizable characterization for any input subject. In this subset of data, outliers suspicious of being errors or artifacts were identified and removed through the expert knowledge of various professionals, including biologists, doctors, and nurses. Subsequently, the data were normalized using the Z score algorithm. At this point, the subset of data was cleaned and prepared for processing, but the number of selected dimensions was still too high. Therefore, we decided to proceed with a dimensional reduction using a set of algorithms that allow us to identify the most relevant variables for our purpose. Among the most important algorithms considered were the correlation between each independent variable and the dependent variable, as well as the implementation of random forest. This reduction allows us to get a simpler, easier computable and more reliable dataset. Finally, we used Optuna to search for the combination of biomarkers, along with a selection of classification models and their hyperparameters, aiming to increase the accuracy and recall of the resulting model. To achieve a more robust model, we used the cross-validation technique. Lastly, we aimed to obtain a good model using the MLR algorithm, as it is considered a 'white-box' model that allows us to easily understand why a patient has been classified into a specific category. The performance parameters taken while designing the classifiers RF, and MLR are presented in Table S9.
Additional resources
This study was registered with ISRCTN Registry during recruitment (ISRCTN27312680).
Published: August 30, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.110839.
Supplemental information
References
- 1.Cascella M., Rajnik M., Aleem A., Dulebohn S.C., Di Napoli R. StatPearls Publishing; 2023. Features, Evaluation, and Treatment of Coronavirus (COVID-19) [PubMed] [Google Scholar]
- 2.WHO WHO Coronavirus (COVID-19) Dashboard. 2023. https://covid19.who.int/
- 3.Sudre C.H., Murray B., Varsavsky T., Graham M.S., Penfold R.S., Bowyer R.C., Pujol J.C., Klaser K., Antonelli M., Canas L.S., et al. Attributes and predictors of long COVID. Nat. Med. 2021;27:626–631. doi: 10.1038/s41591-021-01292-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burn E., Tebé C., Fernandez-Bertolin S., Aragon M., Recalde M., Roel E., Prats-Uribe A., Prieto-Alhambra D., Duarte-Salles T. The natural history of symptomatic COVID-19 during the first wave in Catalonia. Nat. Commun. 2021;12:777. doi: 10.1038/s41467-021-21100-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mahase E. Covid-19: What do we know about “long covid”. BMJ. 2020;m2815:m2815. doi: 10.1136/bmj.m2815. [DOI] [PubMed] [Google Scholar]
- 6.Office for National Statistics (UK) The prevalence of long COVID symptoms and COVID-19 complications. 2020. https://www.ons.gov.uk/news/statementsandletters/theprevalenceoflongcovidsymptomsandcovid19complications
- 7.WHO Post COVID-19 condition (Long COVID) 2022. https://www.who.int/europe/news-room/fact-sheets/item/post-covid-19-condition
- 8.Greenhalgh T., Knight M., A’Court C., Buxton M., Husain L. Management of post-acute covid-19 in primary care. BMJ. 2020;370:m3026. doi: 10.1136/bmj.m3026. [DOI] [PubMed] [Google Scholar]
- 9.Davis H.E., McCorkell L., Vogel J.M., Topol E.J. Long COVID: major findings, mechanisms and recommendations. Nat. Rev. Microbiol. 2023;21:133–146. doi: 10.1038/s41579-022-00846-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carfì A., Bernabei R., Landi F., Gemelli Against COVID-19 Post-Acute Care Study Group Persistent Symptoms in Patients After Acute COVID-19. JAMA. 2020;324:603–605. doi: 10.1001/jama.2020.12603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.National Health Service in England (NHS) Long-term effects of coronavirus (long COVID) 2021. https://www.nhs.uk/conditions/coronavirus-covid-19/long-term-effects-of-coronavirus-long-covid/
- 12.Rojas-Bolivar D., Huaroto-Ramírez F., Curisinche-Rojas M., Gonzales Zurita D., Gutiérrez E. Prevalence, clinical manifestations, and associated factors of long COVID-19. Revista de la Facultad de Medicina Humana. 2022;22:572–583. doi: 10.25176/RFMH.v22i3.5009. [DOI] [Google Scholar]
- 13.Odak I., Barros-Martins J., Bošnjak B., Stahl K., David S., Wiesner O., Busch M., Hoeper M.M., Pink I., Welte T., et al. Reappearance of effector T cells is associated with recovery from COVID-19. EBioMedicine. 2020;57:102885. doi: 10.1016/j.ebiom.2020.102885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Uranga-Murillo I., Morte E., Hidalgo S., Pesini C., García-Mulero S., Sierra J.L., Santiago L., Arias M., De Miguel D., Encabo-Berzosa M.D.M., et al. Integrated analysis of circulating immune cellular and soluble mediators reveals specific COVID19 signatures at hospital admission with utility for prediction of clinical outcomes. Theranostics. 2022;12:290–306. doi: 10.7150/thno.63463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frisoni P., Neri M., D’Errico S., Alfieri L., Bonuccelli D., Cingolani M., Di Paolo M., Gaudio R.M., Lestani M., Marti M., et al. Cytokine storm and histopathological findings in 60 cases of COVID-19-related death: from viral load research to immunohistochemical quantification of major players IL-1β, IL-6, IL-15 and TNF-α. Forensic Sci. Med. Pathol. 2022;18:4–19. doi: 10.1007/s12024-021-00414-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Müller S., Schultze J.L. Systems analysis of human innate immunity in COVID-19. Semin. Immunol. 2023;68:101778. doi: 10.1016/j.smim.2023.101778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tang N., Li D., Wang X., Sun Z. Abnormal coagulation parameters are associated with poor prognosis in patients with novel coronavirus pneumonia. J. Thromb. Haemost. 2020;18:844–847. doi: 10.1111/jth.14768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bikdeli B., Madhavan M.V., Jimenez D., Chuich T., Dreyfus I., Driggin E., Nigoghossian C.D., Ageno W., Madjid M., Guo Y., et al. COVID-19 and Thrombotic or Thromboembolic Disease: Implications for Prevention, Antithrombotic Therapy, and Follow-Up. J. Am. Coll. Cardiol. 2020;75:2950–2973. doi: 10.1016/j.jacc.2020.04.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Forconi C.S., Oduor C.I., Oluoch P.O., Ong’echa J.M., Münz C., Bailey J.A., Moormann A.M. A New Hope for CD56negCD16pos NK Cells as Unconventional Cytotoxic Mediators: An Adaptation to Chronic Diseases. Front. Cell. Infect. Microbiol. 2020;10:162. doi: 10.3389/fcimb.2020.00162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hu P.F., Hultin L.E., Hultin P., Hausner M.A., Hirji K., Jewett A., Bonavida B., Detels R., Giorgi J.V. Natural killer cell immunodeficiency in HIV disease is manifest by profoundly decreased numbers of CD16+CD56+ cells and expansion of a population of CD16dimCD56- cells with low lytic activity. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 1995;10:331–340. [PubMed] [Google Scholar]
- 21.Phetsouphanh C., Darley D.R., Wilson D.B., Howe A., Munier C.M.L., Patel S.K., Juno J.A., Burrell L.M., Kent S.J., Dore G.J., et al. Immunological dysfunction persists for 8 months following initial mild-to-moderate SARS-CoV-2 infection. Nat. Immunol. 2022;23:210–216. doi: 10.1038/s41590-021-01113-x. [DOI] [PubMed] [Google Scholar]
- 22.Peluso M.J., Lu S., Tang A.F., Durstenfeld M.S., Ho H.E., Goldberg S.A., Forman C.A., Munter S.E., Hoh R., Tai V., et al. Markers of Immune Activation and Inflammation in Individuals With Postacute Sequelae of Severe Acute Respiratory Syndrome Coronavirus 2 Infection. J. Infect. Dis. 2021;224:1839–1848. doi: 10.1093/infdis/jiab490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bortolato B., Carvalho A.F., Soczynska J.K., Perini G.I., McIntyre R.S. The Involvement of TNF-α in Cognitive Dysfunction Associated with Major Depressive Disorder: An Opportunity for Domain Specific Treatments. Curr. Neuropharmacol. 2015;13:558–576. doi: 10.2174/1570159X13666150630171433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patterson B.K., Guevara-Coto J., Yogendra R., Francisco E.B., Long E., Pise A., Rodrigues H., Parikh P., Mora J., Mora-Rodríguez R.A. Immune-Based Prediction of COVID-19 Severity and Chronicity Decoded Using Machine Learning. Front. Immunol. 2021;12:700782. doi: 10.3389/fimmu.2021.700782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Katze M.G., He Y., Gale M. Viruses and interferon: a fight for supremacy. Nat. Rev. Immunol. 2002;2:675–687. doi: 10.1038/nri888. [DOI] [PubMed] [Google Scholar]
- 26.Al-Hakeim H.K., Al-Rubaye H.T., Almulla A.F., Al-Hadrawi D.S., Maes M. Chronic Fatigue, Depression and Anxiety Symptoms in Long COVID Are Strongly Predicted by Neuroimmune and Neuro-Oxidative Pathways Which Are Caused by the Inflammation during Acute Infection. J. Clin. Med. 2023;12:511. doi: 10.3390/jcm12020511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Acanfora D., Nolano M., Acanfora C., Colella C., Provitera V., Caporaso G., Rodolico G.R., Bortone A.S., Galasso G., Casucci G. Impaired Vagal Activity in Long-COVID-19 Patients. Viruses. 2022;14:1035. doi: 10.3390/v14051035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Low R.N., Low R.J., Akrami A. A review of cytokine-based pathophysiology of Long COVID symptoms. Front. Med. 2023;10:1011936. doi: 10.3389/fmed.2023.1011936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Castro D., Sharma S. StatPearls Publishing; 2023. Hypokalemia. [PubMed] [Google Scholar]
- 30.Rondon H., Badireddy M. StatPearls Publishing; 2023. Hyponatremia. [PubMed] [Google Scholar]
- 31.Ortelli P., Ferrazzoli D., Sebastianelli L., Maestri R., Dezi S., Spampinato D., Saltuari L., Alibardi A., Engl M., Kofler M., et al. Altered motor cortex physiology and dysexecutive syndrome in patients with fatigue and cognitive difficulties after mild COVID-19. Eur. J. Neurol. 2022;29:1652–1662. doi: 10.1111/ene.15278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Turner S., Khan M.A., Putrino D., Woodcock A., Kell D.B., Pretorius E. Long COVID: pathophysiological factors and abnormalities of coagulation. Trends Endocrinol. Metab. 2023;34:321–344. doi: 10.1016/j.tem.2023.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martín A.I., Priego T., Moreno-Ruperez Á., González-Hedström D., Granado M., López-Calderón A. IGF-1 and IGFBP-3 in Inflammatory Cachexia. Int. J. Mol. Sci. 2021;22:9469. doi: 10.3390/ijms22179469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stanevich O.V., Alekseeva E.I., Sergeeva M., Fadeev A.V., Komissarova K.S., Ivanova A.A., Simakova T.S., Vasilyev K.A., Shurygina A.-P., Stukova M.A., et al. SARS-CoV-2 escape from cytotoxic T cells during long-term COVID-19. Nat. Commun. 2023;14:149. doi: 10.1038/s41467-022-34033-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Couper K.N., Blount D.G., Riley E.M. IL-10: the master regulator of immunity to infection. J. Immunol. 2008;180:5771–5777. doi: 10.4049/jimmunol.180.9.5771. [DOI] [PubMed] [Google Scholar]
- 36.Di Vito C., Calcaterra F., Coianiz N., Terzoli S., Voza A., Mikulak J., Della Bella S., Mavilio D. Natural Killer Cells in SARS-CoV-2 Infection: Pathophysiology and Therapeutic Implications. Front. Immunol. 2022;13:888248. doi: 10.3389/fimmu.2022.888248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levine R.L. Addressing the Long-term Effects of COVID-19. JAMA. 2022;328:823–824. doi: 10.1001/jama.2022.14089. [DOI] [PubMed] [Google Scholar]
- 38.Iosef C., Knauer M.J., Nicholson M., Van Nynatten L.R., Cepinskas G., Draghici S., Han V.K.M., Fraser D.D. Plasma proteome of Long-COVID patients indicates HIF-mediated vasculo-proliferative disease with impact on brain and heart function. J. Transl. Med. 2023;21:377. doi: 10.1186/s12967-023-04149-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patel M.A., Knauer M.J., Nicholson M., Daley M., Van Nynatten L.R., Martin C., Patterson E.K., Cepinskas G., Seney S.L., Dobretzberger V., et al. Elevated vascular transformation blood biomarkers in Long-COVID indicate angiogenesis as a key pathophysiological mechanism. Mol. Med. 2022;28:122. doi: 10.1186/s10020-022-00548-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamga E., Soulé A., Piché A., Emad A., Durand M., Rousseau S. Validation of ANG-1 and P-SEL as biomarkers of post-COVID-19 conditions using data from the Biobanque québécoise de la COVID-19 (BQC-19) Clin. Proteomics. 2023;20:44. doi: 10.1186/s12014-023-09436-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vos T., Hanson S.W., Abbafati C., Aerts J.G., Al-Aly Z., Ashbaugh C., Ballouz T., Blyuss O., Bobkova P., Bonsel G., et al. Estimated Global Proportions of Individuals With Persistent Fatigue, Cognitive, and Respiratory Symptom Clusters Following Symptomatic COVID-19 in 2020 and 2021. JAMA. 2022;328:1604–1615. doi: 10.1001/JAMA.2022.18931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sheskin D.J. Chapman and Hall/CRC; 2020. Handbook of Parametric and Nonparametric Statistical Procedures Fifth. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Data have been deposited at Zenodo and are publicly available as of the date of publication. The DOI is listed in the key resources table.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.