Abstract
The rotavirus nonstructural protein NSP2 self-assembles into homomultimers, binds single-stranded RNA nonspecifically, possesses a Mg2+-dependent nucleoside triphosphatase (NTPase) activity, and is a component of replication intermediates. Because these properties are characteristics of known viral helicases, we examined the possibility that this was also an activity of NSP2 by using a strand displacement assay and purified bacterially expressed protein. The results revealed that, under saturating concentrations, NSP2 disrupted both DNA-RNA and RNA-RNA duplexes; hence, the protein possesses helix-destabilizing activity. However, unlike typical helicases, NSP2 required neither a divalent cation nor a nucleotide energy source for helix destabilization. Further characterization showed that NSP2 displayed no polarity in destabilizing a partial duplex. In addition, helix destabilization by NSP2 was found to proceed cooperatively and rapidly. The presence of Mg2+ and other divalent cations inhibited by approximately one-half the activity of NSP2, probably due to the increased stability of the duplex substrate brought on by the cations. In contrast, under conditions where NSP2 functions as an NTPase, its helix-destabilizing activity was less sensitive to the presence of Mg2+, suggesting that in the cellular environment the two activities associated with the protein, helix destabilization and NTPase, may function together. Although distinct from typical helicases, the helix-destabilizing activity of NSP2 is quite similar to that of the ςNS protein of reovirus and to the single-stranded DNA-binding proteins (SSBs) involved in double-stranded DNA replication. The presence of SSB-like nonstructural proteins in two members of the family Reoviridae suggests a common mechanism of unwinding viral mRNA prior to packaging and subsequent minus-strand RNA synthesis.
Rotaviruses, members of the family Reoviridae, are the major cause of severe gastroenteritis in infants and young children (16). The viruses are icosahedrons made up of three concentric layers of protein and contain a genome composed of eleven segments of double-stranded RNA (dsRNA) (8). The outermost capsid layer consists of the spike protein, VP4, and the glycoprotein, VP7, and the intermediate layer is formed by VP6 trimers (34). The innermost layer consists of 60 dimers of VP2, arranged as a T=1 icosahedron (21). Positioned at the vertices of the VP2 icosahedron are one copy each of the RNA-dependent RNA polymerase (RdRP), VP1, and the capping enzyme, VP3 (21). Together, VP1, VP2, VP3, and the dsRNA genome make up the core of the virion.
Double-layered particles, representing cores surrounded by VP6, have an associated transcriptase activity that catalyzes the synthesis of viral mRNAs (6, 22). The mRNAs not only direct protein synthesis but also serve as templates for the synthesis of minus-strand RNA to form dsRNA (5). Minus-strand synthesis occurs soon after or as viral mRNAs are packaged into core-like replication intermediates (RIs) (9, 32). In addition to the core proteins, several lines of evidence suggest that the nonstructural protein NSP2 is involved in mRNA packaging and/or minus-strand synthesis: (i) a mutant rotavirus with a temperature-sensitive lesion in the NSP2 gene produces empty virus particles at nonpermissive temperatures (4, 35); (ii) NSP2 is a major component of RIs that support packaging and replication (9, 31); (iii) cross-linking of infected cell lysates has revealed that NSP2 is associated with the viral RdRP (17) and with partially replicated RNA (1); and (iv) NSP2 accumulates in cytoplasmic inclusions that form in infected cells (33), the sites where packaging, dsRNA synthesis, and the assembly of double-layered particles occurs.
NSP2 is a highly conserved, basic 35-kDa protein that binds single-stranded RNA (ssRNA) nonspecifically and with high affinity (18, 40). The protein self-assembles into stable homomultimers consisting of 8 subunits (37), and these NSP2 octamers bind ssRNA cooperatively to form large RNA-protein complexes (40). NSP2 also possesses a Mg2+-dependent nucleoside triphosphatase (NTPase) activity (40). These features of NSP2, viz., NTPase activity, affinity for ssRNA, lack of template specificity, association with RIs, and oligomeric nature, are characteristic of known viral helicases (15). In this study we report that NSP2 possesses helix-destabilizing activity, but unlike typical helicases the activity is dependent on neither Mg2+ nor ATP. The destabilization activity of the protein is nondirectional and causes the disruption of both DNA-RNA and RNA-RNA duplexes. The helix-destabilizing activity of NSP2 is similar to those of ssDNA binding proteins (SSBs) and the reovirus nonstructural protein ςNS and may promote rotavirus RNA replication by removing secondary structures in the mRNA templates that impede packaging and minus-strand synthesis.
MATERIALS AND METHODS
Expression and purification of NSP2.
NSP2 was expressed in Escherichia coli M15 cells as described previously (40). Recombinant NSP2 was purified using Ni-nitrilotriacetic acid (NTA) affinity chromatography following the protocol provided by the manufacturer (Qiagen). The protein in the final eluate was dialyzed against low-salt buffer (LSB) (2 mM Tris-HCl [pH 7.2], 0.5 mM EDTA, 0.5 mM dithiothreitol [DTT]) for 48 h. The concentration of the purified NSP2 was determined by Bradford assay using bovine serum albumin as the protein standard and by comparison with known amounts of bovine serum albumin coelectrophoresed on sodium dodecyl sulfate (SDS)–12% polyacrylamide gels (Novex) and stained with Coomassie blue. Purified NSP2 was adjusted to a concentration of 0.5 to 1 mg per ml and stored at 4°C.
In vitro synthesis of RNAs.
The T7 transcription vector, SP65g8 5′-3′SacII, was generated using PCR to delete residues 88 to 1010 of the 1,059-nucleotide (nt) gene 8 cDNA contained within the vector SP65g8R (29). To produce the A11-StyI and A11-SacII RNAs, SP65g8 5′-3′SacII was linearized with StyI and SacII, respectively, blunt-ended by treatment with T4 DNA polymerase, and transcribed with T7 RNA polymerase using an Ambion MAXIscript kit (30). After DNase treatment, the RNA products were purified by phenol-chloroform extraction and isopropanol precipitation. The A11-StyI and A11-SacII RNAs were purified by electrophoresis and elution from 8% polyacrylamide gels containing 7 M urea (28). RNA concentrations were calculated from optical densities at 260 nm.
Preparation of duplexes for strand displacement assays.
The sequences of the DNA (Life Technologies) and RNA (Dharmacon) oligonucleotides used to prepare DNA-RNA and RNA-RNA duplexes are given in Table 1. To prepare 5′-end-labeled oligonucleotides, 50-μl reaction mixtures containing 100 pmol of an oligonucleotide, 10 μCi of [γ-32P]ATP (3,000 Ci/mmol; NEN), and 20 U of T4 polynucleotide kinase were incubated at 37°C for 1 h (Life Technologies). The reactions were terminated by incubation at 65°C for 20 min. The unincorporated nucleotides were removed from the reaction mixtures by centrifugation through Sephadex G-25 spin columns (Roche). Following electrophoresis on 20% polyacrylamide gels containing 7 M urea, the concentrations of the end-labeled oligonucleotides were estimated using a Molecular Dynamics PhosphorImager 445SI.
TABLE 1.
Oligonucleotides used in the preparation of DNA-RNA and RNA-RNA duplexes
| Name | Type | Sequence (5′ to 3′) |
|---|---|---|
| 18AD | DNA | GCCGCTCAAACGGCGACTGAGGAT |
| 5′18AD | DNA | TGAGACGCTTTAAAAGCC |
| 3′22AD | DNA | GGTCACATAAGCGCTTTCTATT |
| 14AR | RNA | UGAAAACGGCGACUGAGGAUACC |
| 10AR | RNA | UGAAAACGGCGACGAUACC |
The following procedure was used to prepare the DNA-RNA duplexes A11-StyI–18AD, A11-StyI–5′18AD, and A11-StyI–3′22AD and the RNA-RNA duplexes A11-StyI–14AR and A11-StyI–10AR (the numbers refer to the number of annealed nucleotides present in DNA-RNA [AD] and RNA-RNA [AR] duplexes). Twenty picomoles of 5′-end-labeled DNA or RNA oligonucleotide was mixed with 40 to 200 pmol of unlabeled A11-StyI or A11-SacII RNA in buffer containing 10 mM Tris, pH 8, and 200 mM NaCl. After being heated to 95°C for 5 min, the mixture was cooled gradually to 25°C over a 5-h period. The quality of the duplexes was assessed by electrophoresis on nondenaturing 20% polyacrylamide gels in Tris-glycine buffer (28). The concentrations of the duplexes were determined by comparison with known amounts of radiolabeled oligonucleotides using a PhosphorImager.
Strand displacement assay.
In a standard strand displacement assay, between 1 and 200 pmol of NSP2 was added to 0.1 pmol of a radiolabeled DNA-RNA or RNA-RNA duplex in buffer containing 25 mM HEPES-KOH (pH 7.5), 20 mM NaCl, and 1 mM DTT. In some cases, the reaction mixtures also included 1 to 10 mM MgCl2, MnCl2, or CaCl2, 50 to 250 mM NaCl, and/or a 1 or 5 mM concentration of a nucleoside triphosphate (NTP). After incubation at 37°C for 30 min, the products of the reactions were treated with 40 μg of proteinase K for 15 min at 37°C. The digestion reactions were terminated by the addition of an equal volume of sample buffer (50 mM Tris, 50 mM glycine, 20 mM EDTA, 0.2% SDS, 0.04% Triton X-100, 25% glycerol and bromophenol blue [pH 8.8]). The products of the assay were analyzed by electrophoresis on nondenaturing 20% polyacrylamide gels, visualized by autoradiography, and quantitated with a PhosphorImager. The helix-destabilizing activity of NSP2 was calculated as the amount of single-stranded (unannealed) 32P-labeled oligonucleotide detected in assays performed with NSP2 minus the amount of single-stranded 32P-labeled oligonucleotide detected in control assays performed in the absence of NSP2. The calculated helix-destabilizing values were then reported as the percentage of the total amount of radiolabeled oligonucleotide (annealed plus unannealed) in the assay.
RESULTS
Detection of helix-destabilizing activity.
NSP2 containing a C-terminal His tag was expressed in E. coli and purified to homogeneity as described previously (40) (Fig. 1). NSP2 prepared in this manner consists nearly exclusively of 12 S homomultimers, each consisting of 8 protein subunits, which possess ssRNA binding activity (37, 40). NSP2 was examined for the ability to destabilize a short partial DNA-RNA duplex constructed by annealing the 5′-end-labeled 27-mer DNA oligonucleotide, 18AD, to the unlabeled 48-mer RNA, A11-StyI (Fig. 2A). The resulting duplex, A11-StyI–18AD, had single-stranded 5′ and 3′ overhangs on both the RNA and DNA strands and was stabilized by 18 complementary nucleotides. The strand displacement assay was performed by incubating 0.1 pmol of the duplex with increasing amounts of purified recombinant NSP2 (1 to 200 pmol) for 30 min at 37°C. Based on previous experiments showing that NSP2 binds to ssRNA as an octamer and protects a region of 10 to 25 nt of an RNA oligonucleotide from RNase digestion (37, 1), NSP2 should saturate the 14- and 16-nt ssRNA tails of the duplex when the amount of NSP2 in the assay is ≥10 pmol. After incubation of the duplex and NSP2, the reaction mixtures were treated with proteinase K and then diluted into sample buffer containing 0.1% SDS. The reaction mixtures were analyzed by electrophoresis on a nondenaturing 20% polyacrylamide gel and by autoradiography. As shown in Fig. 2B, the partial duplex substrate was stable when NSP2 was not added to the reaction mixture (lane 2). However, with the addition of increasing amounts of NSP2 to the reaction mixtures, a proportional decrease of the duplex substrate and an increase in the amount of the 5′-end-labeled DNA oligonucleotide, 18AD, was observed (lanes 4 to 9). From this we concluded that NSP2 catalyzed the displacement of the radiolabeled DNA oligonucleotide from the partial DNA-RNA duplex and, therefore, that NSP2 possessed a helix-destabilizing activity. Since the reaction buffer for the assay lacked Mg2+ and ATP, the helix-destabilizing activity of NSP2 is dependent on neither of these.
FIG. 1.

Expression and purification of recombinant NSP2. NSP2 expressed in E. coli with a C-terminal His tag was purified by NTA affinity chromatography, and the final eluate was dialyzed against LSB. The eluate (lane 2) and protein dialyzed in LSB (lane 1) were resolved by SDS-polyacrylamide gel electrophoresis and stained with Coomassie blue. M, molecular size marker.
FIG. 2.
NSP2 possesses helix-destabilizing activity. (A) A schematic representation of the 32P-labeled DNA-RNA partial duplex A11-StyI–18AD. (B) From 1 to 200 pmol of NSP2 (lanes 4 to 9) was incubated with 0.1 pmol of A11-StyI–18AD for 30 min at 37°C. Afterwards, the reaction mixtures were analyzed by nondenaturing gel electrophoresis and autoradiography. Reaction mixtures containing 0.1 pmol of the 32P-labeled 18AD DNA oligonucleotide instead of the duplex (lane 1), 0.1 pmol of the duplex and no NSP2 (lane 2), and 0.1 pmol of the duplex denatured by heating at 95°C for 2 min and containing no NSP2 (lane 3) were also analyzed.
Directionality the of helix-destabilizing activity.
Although possessing strong affinity for ssRNA, NSP2 has little or no affinity for dsRNA (40). This difference in binding activities provides an explanation as to why NSP2 can destabilize duplexes that contain single-stranded overhangs (i.e., partial duplexes) but not duplexes that lack single-stranded overhangs (i.e., complete duplexes) (data not shown). When NSP2 binds to the ssRNA region of a partial DNA-RNA duplex such as A11-StyI–18AD (Fig. 2A), displacement of the annealed DNA oligonucleotide may occur in a 5′-to-3′ or 3′-to-5′ direction or in both directions. In order to assess the directionality of the strand displacement activity of NSP2, the two 5′-end-labeled DNA oligonucleotides, 5′18AD (18-mer) and 3′22AD (22-mer), were simultaneously annealed to the 144-mer A11-SacII RNA. The DNA-RNA duplex formed with 5′18AD lacks a 5′-ssRNA overhang, while the DNA-RNA duplex formed with 3′22AD lacks a 3′-ssRNA overhang (Fig. 3A). The product of the annealing reaction, containing A11-SacII RNA and 5′18AD and 3′22AD, migrated as multiple poorly resolved bands upon electrophoresis (Fig. 3B, labeled duplex mixture), suggesting either that they represented a mixture of three different partial duplexes or that the RNA component of the duplexes had alternative secondary structures. Strand displacement assays showed that the addition of increasing amounts of NSP2 to the duplexes resulted in a corresponding release of both the 5′18AD and 3′22AD DNA oligonucleotides from the DNA-RNA duplexes (Fig. 3B). Thus, NSP2 was able to bind to the ssRNA component of the DNA-RNA duplexes and displace DNA oligonucleotides in both 5′-to-3′ and 3′-to-5′ directions. From this, the helix-destabilizing activity of NSP2 was inferred to lack directionality.
FIG. 3.
Directionality of the unwinding activity of NSP2. (A) A schematic representation of the possible duplexes formed by annealing the 32P-labeled DNA oligonucleotides 5′18AD and 3′22AD to the A11-SacII RNA. (B) A standard strand displacement assay was performed by incubating 0.1 pmol of the duplex mixture as shown in panel A with 1 to 100 pmol of recombinant NSP2 (rNSP2) (lanes 4 to 7). The reaction mixtures were analyzed by nondenaturing gel electrophoresis and autoradiography. Reaction mixtures containing 32P-labeled 5′18AD (lane 1) or 5′22AD (lane 2) and no duplexes were also analyzed.
Kinetics of strand separation.
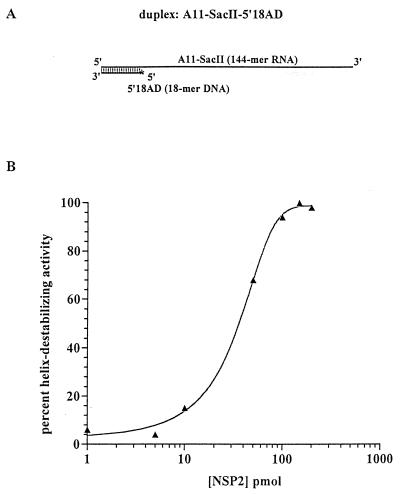
NSP2 binds ssRNA cooperatively (40). To determine if NSP2 could function cooperatively to destabilize a helix, the A11-SacII–5′18AD duplex was prepared by annealing the 5′18AD DNA oligonucleotide to the A11-SacII RNA (Fig. 4A). The duplex has a 126-nt ssRNA tail to which more than one NSP2 octamer can bind. After incubation of 0.1 pmol of the duplex with 1 to 200 pmol of NSP2, the reaction mixtures were analyzed by nondenaturing gel electrophoresis and the amount of annealed versus released DNA oligonucleotide was determined with a PhosphorImager. The values were used to calculate the percent helix-destabilizing activity of the protein (as described in Materials and Methods) and were plotted as functions of the NSP2 concentration in the reaction mixtures (Fig. 4B). Consistent with the results presented in Fig. 2, strand displacement was readily observed when the ratio of NSP2 to duplex was 100:1 (10 pmol of NSP2; 0.1 pmol of A11-SacII–5′18AD) (Fig. 4B). As the NSP2 concentration was increased from 10 to 100 pmol in the reaction mixtures, a steep rise in the level of helix-destabilizing activity was observed. An additional increase in the NSP2 concentration from 100 to 200 pmol did not result in a greater level of helix destabilization, indicating that the reaction had reached equilibrium (Fig. 4B). Similar results were obtained when the DNA oligonucleotide used in the assay (5′18AD) was annealed to the 48-mer A11-StyI RNA instead of the 144-mer A11-SacII RNA (data not shown). The sigmoidal (nonlinear) curve generated by the results of this experiment (Fig. 4B) indicated that NSP2 operated cooperatively to destabilize the duplex.
FIG. 4.
Helix destabilization by NSP2 is cooperative. (A) A schematic representation of the 32P-labeled DNA-RNA duplex A11-SacII–5′18AD, formed by incubating the DNA oligonucleotide, 5′18AD, with the A11-SacII RNA. (B) A set of standard strand displacement assays was performed in parallel by incubating 0.1 pmol of the duplex with 1 to 200 pmol of NSP2. The reaction mixtures were analyzed by nondenaturing gel electrophoresis, and the percent helix-destabilizing activity was determined using a PhosphorImager. The percent helix-destabilizing activity of NSP2 was plotted as a function of the amount of NSP2 in the reaction mixtures.
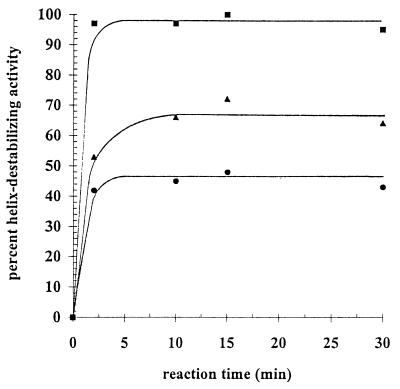
The rate at which NSP2 was able to destabilize a helix was evaluated by combining 50, 100, or 200 pmol of the protein with the DNA-RNA duplex A11-SacII–5′18AD. During incubation, aliquots were taken from the reaction mixtures and were analyzed by gel electrophoresis for the level of 5′18AD DNA oligonucleotide displaced from the duplex. Similar to the rate of interaction of NSP2 with ssRNA (unpublished results), the results showed that the helix destabilization activity by NSP2 was also rapid (Fig. 5), reaching equilibrium or nearly so within 2 min of incubation of the protein with the duplex. The percent helix-destabilizing activity was approximately 45, 65, or 98% when 50, 100, or 200 pmol, respectively, of NSP2 was present in the reaction mixture, and in each case little or no additional change in the level of helix destabilization was observed after 2 min of incubation. The net amount of helix destabilization was, therefore, limited not by the reaction time but rather by how much NSP2 was incubated with the duplex substrate. These results suggest that NSP2 destabilizes a helix not by an enzymatic process but by a passive process that is driven by the affinity of the protein for single-stranded regions of a partial duplex.
FIG. 5.
Kinetics of helix destabilization by NSP2. The 32P-labeled DNA-RNA duplex A11-StyI–5′18AD (0.1 pmol) was incubated with 50 (●), 100 (▴), or 200 (■) pmol of NSP2 for 2, 10, 15, or 30 min at 37°C. Afterwards, the reaction mixtures were analyzed by nondenaturing gel electrophoresis, and the percent helix-destabilizing activity was determined using a PhosphorImager. The percent helix-destabilizing activity of NSP2 was plotted as a function of reaction time.
Effect of ions and nucleotides on the helix-destabilizing activity of NSP2.
Although the helix-destabilizing activity of NSP2 requires neither Mg2+ nor NTP, the NTPase activity of the protein requires the cation to carry out the hydrolysis of NTP to NDP (40). To determine whether conditions that allow the NTPase of NSP2 to function have any effect on its helix-destabilizing activity, we analyzed the impact of Mg2+ and NTP separately and in combination on the ability of NSP2 to disrupt the helix of the DNA-RNA duplex A11-StyI–18AD. The results showed that when increasing amounts (0 to 10 mM) of only Mg2+ were included in the strand displacement assay, the helix-destabilizing activity of the protein progressively decreased to a level that, at 10 mM Mg2+, was only 25% of that in a Mg2+-free control reaction (Fig. 6A). Mg2+ is known to neutralize the strong repulsions between closely packed phosphates in highly folded RNA (26) and dsDNA (38, 44) and as a result may increase the stability of the DNA-RNA duplex used in the strand separation assay, making it more difficult for NSP2 to disrupt the helix. If this is the mechanism by which Mg2+ reduces the helix-destabilizing activity of NSP2 in the strand separation assay, then other divalent cations such as Mn2+ and Ca2+ should have similar effects. Indeed, experiments performed with Mn2+ and Ca2+ showed that they reduced the helix-destabilizing activity of NSP2 in a manner similar to that of Mg2+ (Fig. 6A), suggesting that these cations affected this activity by increasing the stability of the DNA-RNA duplex. In comparison to the divalent cations, the monovalent cation Na+ had little effect on strand displacement by NSP2 (Fig. 6B). For example, even at 250 mM NaCl, the helix-destabilizing activity of NSP2 was reduced by <20%. Thus, divalent cations inhibit strand displacement by NSP2 more effectively than monovalent cations such as NaCl. It is unlikely that the inhibitory effect of the divalent cations was due to a reduction of the affinity of NSP2 for ssRNA, since we observed no significant effect of Mg2+, Ca2+, or Mn2+ on the ability of NSP2 to bind ssRNA in gel shift assays (data not shown). It is possible that concentrations of a monovalent cation that are much higher than those considered physiological (150 mM) could inhibit helix destabilization by NSP2 to the same extent as a 10 mM concentration of a divalent cation. However, these concentrations (>250 mM) could not be tested because of their interfering effects on gel electrophoresis. Notably, at physiological concentrations of divalent (1 mM) and monovalent (150 mM) cations, the extent of strand displacement by NSP2 was reduced by less than 20% (Fig. 6).
FIG. 6.
Effect of cations on helix destabilization by NSP2. The DNA-RNA duplex, A11-StyI–18AD (0.1 pmol), was incubated with NSP2 (200 pmol) in the presence of 0 to 10 mM MgCl2 (○), CaCl2 (□), or MnCl2 (◊) (panel A) or 0 to 250 mM NaCl (●) (panel B). The reaction mixtures were analyzed by nondenaturing gel electrophoresis, and the percent helix-destabilizing activity was determined using a PhosphorImager. The percent helix-destabilizing activity was plotted as a function of salt concentration, with the value obtained for the reaction mixture lacking salt normalized to 100%.
Incubation of 200 pmol of NSP2, 0.1 pmol of the DNA-RNA duplex A11-StyI–18AD, and 5 mM ATP, GTP, CTP, or UTP showed that the NTPs, in the absence of Mg2+, did not significantly affect the helix-destabilizing activity of the protein (Fig. 7A). Consistent with the results shown in Fig. 6A, the strand displacement activity of NSP2 was reduced by approximately 50% when Mg2+ and not NTPs was added to reaction mixtures (Fig. 7B). However, when ATP or CTP was added to the reaction mixture along with Mg2+, the strand displacement activity of NSP2 was restored to levels (80 or 95%, respectively) approaching those achieved when Mg2+ was left out of the reaction mixtures. In contrast, the addition of UTP or GTP to reaction mixtures containing Mg2+ only minimally increased the extent of strand displacement activity (65 or 60%, respectively). The mechanism by which any of the NTPs act to overcome the Mg2+-mediated inhibition of the helix-destabilizing activity of NSP2 is not known. But given that NSP2 is a nonspecific NTPase, the finding that ATP and CTP were more effective than UTP and GTP in restoring the activity of NSP2 is particularly perplexing. However, this effect of NTPs and Mg2+ on the helix destabilization activity of NSP2 was repeatedly observed even when the concentration in the reaction mixtures of either the NTPs or Mg2+ was decreased to 1 mM or when the amount of NSP2 was decreased to 100 pmol (data not shown).
FIG. 7.
Combined effect of Mg2+ and NTP on helix destabilization by NSP2. (A) NSP2 (200 pmol) was incubated with 0.1 pmol of the DNA-RNA duplex, A11-StyI–18AD, in the absence or presence of 5 mM ATP, GTP, CTP, or UTP for 30 min at 37°C. (B) Reaction mixtures containing the same components as those of the reaction mixtures in panel A, except that these contained 5 mM MgCl2, were also incubated. (C) NSP2 (100 pmol) was incubated with 0.1 pmol of the DNA-RNA duplex, A11-StyI–18AD, in the absence or presence of 1 mM MgCl2 or in the presence of 1 mM MgCl2 and 1 mM ATP, ADP, or ATP-γ-S. The reaction mixtures were analyzed by nondenaturing gel electrophoresis, and the percent helix-destabilizing activity was determined using a PhosphorImager. The values were normalized to that of 100% for the assays performed in the absence of MgCl2.
To investigate the possible connection between the NTPase activity of NSP2 and the ability of ATP to overcome the inhibitory effect of Mg2+ on helix destabilization, NSP2 and the DNA-RNA duplex, A11-StyI–18AD, were incubated in reaction mixtures containing Mg2+ and either ATP, ADP, or the nonhydrolyzable ATP analog, ATP-γ-S. As shown in Fig. 7C, neither ADP nor ATP-γ-S increased the helix-destabilizing activity of NSP2, although ATP restored the activity to a level approximating that seen in reaction mixtures lacking Mg2+. The results indicate that under conditions where NSP2 can hydrolyze ATP the helix-destabilizing activity is less sensitive to Mg2+, and thus these two activities of the protein may be related.
NSP2 destabilizes an RNA-RNA duplex.
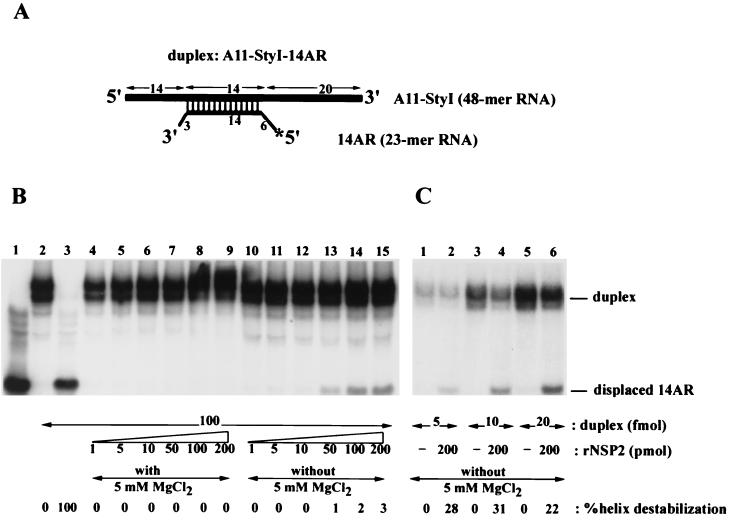
The substrates for the helix-destabilizing activity of NSP2 in the infected cell are likely to be the helical regions present in the secondary structures of viral mRNAs. As a consequence, we evaluated whether the helix-destabilizing activity of NSP2 could disrupt an RNA-RNA duplex in addition to the DNA-RNA duplexes that were used in our previous experiments. The radiolabeled RNA-RNA duplex, A11-StyI–14AR, was prepared by annealing the radiolabeled RNA, 14AR, to the unlabeled RNA, A11-StyI. The duplex contained a 14-nt annealed region and 5′ and 3′ single-stranded overhangs (Fig. 8A). The assay was performed by incubating increasing amounts of NSP2 (1 to 200 pmol) with 0.1 pmol of the RNA-RNA duplex in the absence (Fig. 8B, lanes 10 to 15) or presence (lanes 4 to 9) of 5 mM Mg2+. The results showed that in the absence of Mg2+, the duplex was disrupted when 50 or more pmol of NSP2 was present in the reaction mixture. Thus, the helix-destabilizing activity of NSP2 functions on both DNA-RNA and RNA-RNA duplexes and does not require the cofactors, i.e., NTPs or Mg2+, normally required by helicases. Consistent with data obtained using DNA-RNA duplexes as substrates, Mg2+ in the absence of NTPs inhibited the destabilizing activity of NSP2 (lanes 4 to 9). Although both types of duplexes were used as substrates, the destabilizing activity observed with NSP2 was much greater with DNA-RNA than RNA-RNA duplexes under identical reaction conditions. For example, in assays where the ratio of NSP2 to DNA-RNA duplex was 500:1, approximately 75% of the duplex was disrupted (Fig. 2, lane 7). In contrast, in assays where the ratio of NSP2 to RNA-RNA duplex was also 500:1, only 1% of the duplex was disrupted (Fig. 8B, lane 13). Further increases in the ratio of NSP2 to RNA-RNA duplex to 1:1,000 and 1:2,000 marginally increased the percentage of helix-destabilization by NSP2 (Fig. 8B, lanes 14 and 15).
FIG. 8.
Destabilization of an RNA-RNA duplex by NPS2. (A) A schematic representation of the 32P-labeled RNA-RNA duplex A11-StyI–14AR produced by annealing the RNAs A11-StyI and 14AR. (B) Strand displacement assays were performed by incubating 0.1 pmol of the RNA-RNA duplex with 1 to 200 pmol of NSP2 in the presence (lanes 4 to 9) or absence (lanes 10 to 15) of 5 mM MgCl2. As controls, reaction mixtures were also prepared that contained 0.1 pmol of the 14AR RNA instead of the duplex (lane 1), 0.1 pmol of A11-StyI–14AR duplex and no NSP2 (lane 2), or 0.1 pmol of A11-StyI–14AR and no NSP2, denatured by heating at 95°C for 2 min (lane 3). (C) Strand displacement assays were performed by incubating 5 to 20 fmol of the RNA-RNA duplex in the presence or absence of recombinant NSP2 (rNSP2). The reaction mixtures were analyzed by nondenaturing gel electrophoresis and autoradiography. A PhosphorImager was used to determine the percent helix-destabilizing activity for each reaction. The values were normalized to that of 100% for the assay reaction wherein the substrate duplex was denatured by heating.
To test if even higher ratios of NSP2 to RNA-RNA duplex further increased the extent of destabilization, assays were performed in which the amount of substrate included was reduced from 0.1 pmol to 5, 10, or 20 fmol and the amount of NSP2 added was 200 pmol. As shown in Fig. 8C, when protein was present in molar excess over substrate by 10,000- to 40,000-fold, the percentage of helix destabilization increased to 20 to 30%.
The limited destabilizing activity of NSP2 on the RNA-RNA duplex, compared to that of a DNA-RNA duplex, may be due to the increased stability of the helix of the RNA-RNA duplex. If so, then NSP2 should be able to destabilize RNA-RNA duplexes which have shorter annealed regions more efficiently than RNA-RNA duplexes with longer annealed regions. To test this hypothesis, the partial duplex, A11-StyI–10AR, was constructed by annealing the radiolabeled 10AR RNA to the unlabeled A11-StyI RNA. The A11-StyI–10AR duplex contained a 10-nt annealed region instead of the 14-nt annealed region of A11-StyI–14AR (Fig. 9A). When increasing amounts of NSP2 (1 to 200 pmol) were incubated with A11-StyI–10AR, 30 and 38% of the duplex was destabilized in reactions with 100 and 200 pmol of NSP2, respectively (Fig. 9B, lanes 9 and 10). Thus, under conditions where the ratio of NSP2 octamer to duplex was 12.5:1 to 25:1 (8 pmol of NSP2 equals 1 pmol of NSP2 octamer), significant amounts of the duplex were destabilized. The fact that NSP2 more effectively destabilized an RNA-RNA duplex with a 10-nt annealed region than an RNA-RNA duplex with a 14-nt annealed region indicates that the stability of the duplex has an impact on the destabilizing activity of NSP2. Based on the computed predictions of the secondary structures of rotavirus mRNAs, nearly all of the RNA-RNA helices formed by folding of the RNAs are shorter than 10 nt (data not shown). As a consequence, given the high level of NSP2 produced in the infected cell and associated with rotavirus replication intermediates (9), the ability of NSP2 to efficiently destabilize the 10-mer duplex of A11-StyI–10AR suggests that NSP2 can interact with mRNA templates to destabilize secondary structures during dsRNA synthesis.
FIG. 9.
NSP2 destabilizes a short RNA-RNA duplex with greater efficiency. (A) A schematic representation of the 32P-labeled RNA-RNA duplex A11-StyI–10AR produced by annealing the RNAs A11-StyI and 10AR. (B) Strand displacement assays were performed by incubating 0.1 pmol of the RNA-RNA duplex with 1 to 200 pmol of NSP2 (lanes 5 to 10). As controls, reaction mixtures were also prepared that contained 0.1 pmol of the 10AR RNA instead of the duplex (lane 1), 0.1 pmol of the A11-StyI–14AR duplex and no NSP2 (lane 2 and 4), or 0.1 pmol of A11-StyI–10AR and no NSP2, denatured by heating at 95°C for 2 min (lane 3). A PhosphorImager was used to determine the percent helix-destabilizing activity for each reaction. The values were normalized to that of 100% for the assay reaction wherein the substrate duplex was denatured by heating.
DISCUSSION
NSP2 is a helix-destabilizing protein but not a helicase.
In this study, we have shown that NSP2 possesses a helix-destabilizing activity which is independent of cofactor and energy requirements. Since the helix-destabilizing activities of known viral RNA helicases are characterized by a requirement for ATP and Mg2+ (15), we do not consider it appropriate to classify NSP2 as a helicase. Moreover, while the strand displacement activity of NSP2 was bidirectional, viral RNA helicases classically display a unidirectional strand displacement activity, mostly in the 3′-to-5′ direction of the ssRNA on which they are bound (15). The helix-destabilizing activity of NSP2 was detected only when the protein was added in saturating and stoichiometric amounts relative to the duplex substrate. The destabilization activity of NSP2 was also observed to rapidly reach equilibrium (within a few minutes). These characteristics are indicative not of an enzyme-based processive strand displacement activity (as is the case with helicases) but rather of a passive strand displacement activity that is the consequence of the saturative and high-affinity binding of NSP2 to a partial duplex.
Similarity of NSP2, ςNS of reovirus, and NS2 of BTV.
Other members of Reoviridae, orthoreovirus and bluetongue virus (BTV), encode proteins that, despite the lack of sequence identity, seem structurally and functionally related to NSP2. This includes the orthoreovirus nonstructural protein ςNS, which, like NSP2, self-assembles into homomultimers and binds ssRNA nonspecifically and with high affinity to form higher-order RNA-protein complexes (11). In infected cells, ςNS and NSP2 both localize to cytoplasmic inclusions (23) and have been implicated in viral RNA replication and packaging. In a recent report, Gillian et al. (12) showed that saturating concentrations of ςNS can unwind DNA-RNA duplexes in vitro in a manner that is ATP and Mg2+ independent. Unwinding was detected when the molar ratio of ςNS to the duplex was 100:1 and this activity was inhibited by the presence of Mg2+ in the reaction. These properties are reminiscent of those observed for the helix-destabilizing activity of NSP2. We have shown that NSP2 destabilizes both DNA-RNA and RNA-RNA duplexes, although the specific activity is reduced with RNA-RNA duplexes, possibly as a result of the increased stability of its helix. It was not possible to demonstrate the unwinding activity of ςNS on an RNA-RNA duplex (12).
Another putative homolog of NSP2 that may function as a helix-destabilizing protein is NS2 of BTV. Some of the features of NS2 are similar to those of NSP2 and ςNS, and these features are probably crucial for helix destabilization. These features include the ability of NS2 to bind nonspecifically to ssRNA (14, 41, 43), to form large 7S multimeric complexes (43) and to localize to cytoplasmic inclusions in the infected cell (3). However, there has been no report of a helix-destabilizing activity associated with NS2 protein.
Similarity of NSP2 and SSBs.
A number of SSBs involved in the replication of dsDNA have been reported to have a helix-destabilizing activity similar to that of NSP2. Examples of SSBs for eukaryotic dsDNA viruses include the ICP8 protein of herpes simplex virus type 1 (2), the adenovirus DNA binding protein (27), the ssDNA binding protein of Epstein-Barr virus (42), the I3 gene product of vaccinia virus (36), and the LEF-3 protein of baculovirus (24). SSBs with helix-destabilizing activity are also encoded by the bacteriophages T4 (13), φ29 (39), Nf, and GA-1 (10). In addition, SSBs are ubiquitous in bacterial and eukaryotic cells. Collectively, SSBs are noncatalytic proteins that bind ssDNA cooperatively, with high affinity and in a sequence-independent manner (19). Through these activities, SSBs destabilize the helix and prevent base pairing of complementary sequences (7). Generally, their function is to bind to ssDNA that has been unwound at the replication fork by helicases and to assist in the processivity of the replication complex. SSBs also stimulate the activity of helicases and origin binding proteins (19). Based on the similarities of the activities of SSBs and NSP2, NSP2 may be predicted to remove secondary structures in rotavirus mRNA templates that impede their packaging into cores and their replication to dsRNA. Indeed, using a cell-free system developed for the segmented dsRNA bacteriophage φ6, Mindich (25) has directly shown that secondary structures within mRNA templates can interfere with packaging and replication. The idea that NSP2 is an integral part of the rotavirus replication machinery is supported by the observation that the protein is a major component of RIs and is associated with the viral RdRP (1, 9, 17). Indeed, the amount of NSP2 associated with replication intermediates with replicase activity (i.e., the core and single-shelled intermediates) exceeds by severalfold the amount of the VP1 and VP3 that is present in these structures (9).
Mechanism of helix destabilization by NSP2.
The double-stranded regions of a partial helix are not necessarily stable and can occasionally “breathe,” producing regions that are transiently single stranded. Conditions that can stabilize a partial helix substrate, such as the presence of divalent cations (26), were found to interfere with the helix-destabilizing activity of NSP2. From this result, it can be inferred that binding of NSP2 to the transiently single-stranded regions prevents the reformation of the duplex and thereby destabilizes partial helices. The destabilizing activity of NSP2 is probably further amplified by the cooperativity that the protein displays in binding to ssRNA. The structural integrity of the NSP2 octamer also may be an important factor in the helix-destabilizing activity of the protein. This is supported by a recent study showing that 5 mM Mg2+ (but not 100 mM NaCl) causes the disassembly of NSP2 octamers into smaller units, most likely tetramers (37). Hence, the inhibitory effect of Mg2+ on helix destabilization could be the consequence of the effect of the cation not only on helix stability but also on the structural integrity of the functional unit of NSP2.
Relationship between the helix-destabilizing and NTPase activities of NSP2.
NSP2 is distinct from ςNS and most other SSBs, since in the presence of NTP and Mg2+ the protein also functions as an NTPase and undergoes autophosphorylation (40). Under conditions where the NTPase of NSP2 is active, the inhibitory effect of Mg2+ alone on the helix-destabilizing activity of NSP2 is lessened, particularly when the NTP present is ATP or GTP. This suggests that the NTPase and helix-destabilizing activities of the protein are linked. Perhaps the NTPase-induced phosphorylated form of NSP2 displays a higher affinity or cooperativity for binding ssRNA, and this overcomes the enhanced stability of the helix brought on by Mg2+. Such is the case for an SSB derived from ascites tumor cells, which binds ssDNA with higher affinity following phosphorylation (20). Alternatively, ligands such as NTP and Mg2+ may act in combination to affect the structure of the NSP2 multimer in a way that enhances its helix-destabilizing activity. Support for this possibility comes from the observation that in the presence of nucleosides, the NSP2 multimer changes to a significantly more compact conformation (37). The nucleoside-induced change in the structure of the NSP2 octamer is consistent with the hypothesis that the complex functions not only to destabilize secondary structures within viral mRNAs but also as a molecular motor helping to drive the packaging and replication of viral mRNAs.
ACKNOWLEDGMENT
We acknowledge Karen Kearney for critical review of the manuscript.
REFERENCES
- 1.Aponte C, Poncet D, Cohen J. Recovery and characterization of a replicase complex in rotavirus-infected cells using a monoclonal antibody against NSP2. J Virol. 1996;70:985–991. doi: 10.1128/jvi.70.2.985-991.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boehmer P E, Lehman I R. Herpes simplex virus type 1 ICP8: helix-destabilizing properties. J Virol. 1993;67:711–715. doi: 10.1128/jvi.67.2.711-715.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brookes S M, Hyatt A D, Eaton B T. Characterization of virus inclusion bodies in bluetongue virus-infected cells. J Gen Virol. 1993;74:525–530. doi: 10.1099/0022-1317-74-3-525. [DOI] [PubMed] [Google Scholar]
- 4.Chen D, Gombold J L, Ramig R F. Intracellular RNA synthesis directed by temperature-sensitive mutants of simian rotavirus SA11. Virology. 1990;178:143–151. doi: 10.1016/0042-6822(90)90387-7. [DOI] [PubMed] [Google Scholar]
- 5.Chen D, Zeng C Y, Wentz M J, Gorziglia M, Estes M K, Ramig R F. Template-dependent, in vitro replication of rotavirus RNA. J Virol. 1994;68:7030–7039. doi: 10.1128/jvi.68.11.7030-7039.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen J. Ribonucleic acid polymerase activity associated with purified calf rotavirus. J Gen Virol. 1977;36:395–402. doi: 10.1099/0022-1317-36-3-395. [DOI] [PubMed] [Google Scholar]
- 7.Dekker J, Kanellopoulos P N, Loonstra A K, van Oosterhout J A, Leonard K, Tucker P A, van der Vliet P C. Multimerization of the adenovirus DNA-binding protein is the driving force for ATP-independent DNA unwinding during strand displacement synthesis. EMBO J. 1997;17:1455–1463. doi: 10.1093/emboj/16.6.1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Desselberger U, McCrae M A. The rotavirus genome. Curr Top Microbiol Immunol. 1994;68:5945–5952. doi: 10.1007/978-3-642-78256-5_3. [DOI] [PubMed] [Google Scholar]
- 9.Gallegos C O, Patton J T. Characterization of rotavirus replication intermediates: a model for the assembly of single-shelled particles. Virology. 1989;172:616–627. doi: 10.1016/0042-6822(89)90204-3. [DOI] [PubMed] [Google Scholar]
- 10.Gascon I, Lazaro J M, Salas M. Differential functional behavior of viral phi29, Nf and GA-1 SSB proteins. Nucleic Acids Res. 2000;28:2034–2042. doi: 10.1093/nar/28.10.2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gillian A L, Nibert M L. Amino terminus of reovirus nonstructural protein ςNS is important for ssRNA binding and nucleoprotein complex formation. Virology. 1998;240:1–11. doi: 10.1006/viro.1997.8905. [DOI] [PubMed] [Google Scholar]
- 12.Gillian A L, Schmechel S C, Livny J, Schiff L A, Nibert M L. Reovirus protein sigma NS binds in multiple copies to single-stranded RNA and shares properties with single-stranded DNA binding proteins. J Virol. 2000;74:5939–5948. doi: 10.1128/jvi.74.13.5939-5948.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hosoda J, Takacs B, Brack C. Denaturation of T4 DNA by an in vitro processed gene 32 protein. FEBS Lett. 1974;47:338–342. doi: 10.1016/0014-5793(74)81043-4. [DOI] [PubMed] [Google Scholar]
- 14.Huismans H, Van Dijk A A, Bauskin A R. In vitro phosphorylation and purification of a nonstructural protein of bluetongue virus with affinity for their single-stranded RNA. J Virol. 1987;61:3589–3595. doi: 10.1128/jvi.61.11.3589-3595.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kadare G, Haenni A L. Virus-encoded RNA helicases. J Virol. 1997;71:2583–2590. doi: 10.1128/jvi.71.4.2583-2590.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kapikian A Z, Hoshino Y, Chanock R M, Perez-Schael I. Efficacy of a quadrivalent rhesus rotavirus-based human rotavirus vaccine aimed at preventing severe rotavirus diarrhea in infants and young children. J Infect Dis. 1996;1:S65–S72. doi: 10.1093/infdis/174.supplement_1.s65. [DOI] [PubMed] [Google Scholar]
- 17.Kattoura M D, Chen X, Patton J T. The rotavirus RNA binding protein NS35 (NSP2) forms 10S multimers and interacts with the viral RNA polymerase. Virology. 1994;202:803–813. doi: 10.1006/viro.1994.1402. [DOI] [PubMed] [Google Scholar]
- 18.Kattoura M, Clapp L L, Patton J T. The rotavirus non-structural protein, NS35, is a nonspecific RNA-binding protein. Virology. 1992;191:698–708. doi: 10.1016/0042-6822(92)90245-k. [DOI] [PubMed] [Google Scholar]
- 19.Kornberg A, Baker J. DNA replication. 2nd ed. New York, N.Y: W. H. Freeman & Co.; 1992. p. 280. [Google Scholar]
- 20.Koterov A N, Novoradovskaia N A, Pushkareva N B, Vorotnikov A V, Nikol'skii A V, Risnik V V. Phosphorylation and other properties of proteins binding single-stranded DNA (SSB-proteins) from chromatin and extrachromatin fractions of Ehrlich ascites carcinoma. Biokhimiya. 1991;56:666–673. [PubMed] [Google Scholar]
- 21.Lawton J A, Zeng C Q-Y, Mukherjee S K, Cohen J, Estes M K, Prasad B V V. Three-dimensional structural analysis of recombinant rotavirus-like particles with intact and amino-terminal deleted VP2: implications for the architecture of the VP2 capsid layer. J Virol. 1997;71:7353–7360. doi: 10.1128/jvi.71.10.7353-7360.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lawton J A, Estes M K, Prasad B V V. Three-dimensional visualization of mRNA release from actively transcribing rotavirus particles. Nat Struct Biol. 1997;2:118–121. doi: 10.1038/nsb0297-118. [DOI] [PubMed] [Google Scholar]
- 23.Mbisa J L, Becker M M, Zhou S, Dermody T S, Brown E G. Reovirus μ2 protein determines strain-specific differences in the rate of viral inclusion formation in L929 cells. Virology. 2000;272:16–26. doi: 10.1006/viro.2000.0362. [DOI] [PubMed] [Google Scholar]
- 24.Mikhailov V S. Helix-destabilizing properties of the baculovirus single-stranded DNA-binding protein (LEF-3) Virology. 2000;270:180–189. doi: 10.1006/viro.2000.0270. [DOI] [PubMed] [Google Scholar]
- 25.Mindich L. Precise packaging of the three genomic segments of the double-stranded-RNA bacteriophage phi6. Microbiol Mol Biol Rev. 1999;63:149–160. doi: 10.1128/mmbr.63.1.149-160.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Misra V K, Draper D E. On the role of magnesium ions on RNA stability. Biopolymers. 1999;48:113–135. doi: 10.1002/(SICI)1097-0282(1998)48:2<113::AID-BIP3>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 27.Monaghan A, Webster A, Ronald T H. Adenovirus DNA binding protein: helix-destabilizing activity. Nucleic Acids Res. 1994;22:742–748. doi: 10.1093/nar/22.5.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Patton J T. Rotavirus VP1 alone specifically binds to the 3′ end of viral mRNA but the interaction is not sufficient to initiate minus-strand synthesis. J Virol. 1996;70:7940–7947. doi: 10.1128/jvi.70.11.7940-7947.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patton J T, Chen D. RNA-binding and capping activities of proteins in rotavirus open cores. J Virol. 1999;73:1382–1391. doi: 10.1128/jvi.73.2.1382-1391.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Patton J T, Wentz M, Xiaobo J, Ramig R F. cis-acting signals that promote genome replication in rotavirus mRNA. J Virol. 1996;70:3961–3971. doi: 10.1128/jvi.70.6.3961-3971.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Patton J T, Gallegos C O. Structure and protein composition of the rotavirus replicase particle. Virology. 1988;166:358–365. doi: 10.1016/0042-6822(88)90506-5. [DOI] [PubMed] [Google Scholar]
- 32.Patton J T, Gallegos C O. Rotavirus RNA replication: single-stranded RNA extends from the replicase particle. J Gen Virol. 1990;71:1087–1094. doi: 10.1099/0022-1317-71-5-1087. [DOI] [PubMed] [Google Scholar]
- 33.Petrie B L, Greenberg H B, Graham D Y, Estes M K. Ultrastructural localization of rotavirus antigens using colloidal gold. Virus Res. 1984;1:133–152. doi: 10.1016/0168-1702(84)90069-8. [DOI] [PubMed] [Google Scholar]
- 34.Prasad B V V, Wang G J, Clerx J P M, Chiu W. Three-dimensional structure of rotavirus. J Mol Biol. 1988;199:269–275. doi: 10.1016/0022-2836(88)90313-0. [DOI] [PubMed] [Google Scholar]
- 35.Ramig R F, Petrie B L. Characterization of temperature sensitive mutants of simian rotavirus SA11: protein synthesis and morphogenesis. J Virol. 1984;49:665–673. doi: 10.1128/jvi.49.3.665-673.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rochester S C, Traktman P. Characterization of the single-stranded DNA binding protein encoded by the vaccinia virus 13 gene. J Virol. 1998;72:2917–2926. doi: 10.1128/jvi.72.4.2917-2926.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schuck, P., Z. Taraporewala, P. McPhie, and J. T. Patton. 2000. Rotavirus nonstructural protein NSP2 self-assembles into octamers that undergo ligand-induced conformational changes. J. Biol. Chem. (published online ahead of print December 19, 2000). [DOI] [PubMed]
- 38.Shaw S Y, Wang J C. Knotting of a DNA chain during ring closure. Science. 1993;260:533–536. doi: 10.1126/science.8475384. [DOI] [PubMed] [Google Scholar]
- 39.Soengas M S, Gutierrez C, Salas M. Helix-destabilizing activity of phi 29 single-stranded DNA binding protein: effect on the elongation rate during strand displacement DNA replication. J Mol Biol. 1995;253:517–529. doi: 10.1006/jmbi.1995.0570. [DOI] [PubMed] [Google Scholar]
- 40.Taraporewala Z, Chen D, Patton J T. Multimers formed by the rotavirus nonstructural protein NSP2 bind to RNA and have nucleoside triphosphatase activity. J Virol. 1999;73:9934–9943. doi: 10.1128/jvi.73.12.9934-9943.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thomas C P, Booth T F, Roy P. Synthesis of bluetongue virus-encoded phosphoprotein and formation of inclusion bodies by recombinant baculovirus in insect cells: it binds the single-stranded RNA species. J Gen Virol. 1990;71:2073–2083. doi: 10.1099/0022-1317-71-9-2073. [DOI] [PubMed] [Google Scholar]
- 42.Tsurumi T, Kishore J, Yokoyama N, Fujita M, Daikoku T, Yamada H, Yamashita Y, Nishiyama Y. Overexpression, purification and helix-destabilizing properties of Epstein-Barr virus ssDNA-binding protein. J Gen Virol. 1998;79:1275–1264. doi: 10.1099/0022-1317-79-5-1257. [DOI] [PubMed] [Google Scholar]
- 43.Uitenweerde J M, Theron J, Stoltz M A, Huismans H. The multimeric nonstructural NS2 proteins of bluetongue virus, African horsesickness virus and epizootic hemorrhagic disease virus differ in their single-stranded RNA-binding ability. Virology. 1995;209:624–632. doi: 10.1006/viro.1995.1294. [DOI] [PubMed] [Google Scholar]
- 44.Xu Y C, Bremer H. Winding of DNA helix by divalent cations. Nucleic Acids Res. 1997;25:4067–4071. doi: 10.1093/nar/25.20.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]