Abstract
Background
Although 7 human adenovirus (HAdV) species are known to exist, only F (types 40 and 41) and G are identified as diarrheal disease agents. The role of other HAdV species in diarrheal disease remains unclear, and data on their prevalence are limited. We describe HAdV species and types in hospitalized children with diarrhea in coastal Kenya.
Methods
Three hundred twenty-nine stool samples collected between June 2022 and August 2023 from children aged <13 years were screened for HAdV using quantitative polymerase chain reaction (qPCR). Positive HAdV cases were genotyped by adenovirus primers from the RespiCoV panel by amplification, next-generation sequencing, and phylogenetic analysis.
Results
Sixty-five samples (20%) tested HadV positive, of which 5 HAdV species were identified. Other than HAdV F, other species included A, B, C, and D; these were detected as either mono-detections or coinfections. Six HAdV F identified by NGS had been missed by our qPCR typing method. This appeared to be as a result of a 133-nucleotide deletion in the long fiber protein, which abrogated a primer and probe binding site. Based on grading of diarrheal disease severity using VESIKARI scores, 93% of the HAdV cases presented with severe disease. One child with an HAdV F infection died.
Conclusions
Our study shows the enormous diversity and clinical characteristics of HAdV species in children with diarrhea in coastal Kenya. These data offer an opportunity to improve current diagnostic assays and increase knowledge of HAdV in Africa for control of outbreaks in the future.
Keywords: enteric, human adenovirus, Kenya, ONT, RespiCoV
Human adenoviruses (HAdVs) are nonenveloped, double-stranded DNA viruses that belong to the Mastadenovirus genus [1]. To date, 7 different species, A to G, and 114 different types of HAdV have been described by the Human Adenovirus Working Group (http://hadvwg.gmu.edu/). These HAdVs are associated with a variety of disease presentations including gastroenteritis (F and G) [2, 3], respiratory tract infections (A, B, C and E) [4], and keratoconjunctivitis (D) [5].
HAdV types F40 and F41 are a common etiology of mild to severe diarrhea among children under the age of 5 years [3, 6–11]. However, other HAdV species such as A, B, C, D, and E have been detected among diarrhea cases, although their contribution to diarrhea disease is unclear [3, 7, 10]. In China, HAdV B3 has been associated with diarrhea (adjusted odds ratio, 9.205; P < .001) [3]. In Canada, HAdV F40/41 detections had the highest fraction attributable (96%; 95% CI, 92.3%–97.7%) to diarrhea symptoms among HAdV F40/41 cases, but HAdV C1, C2, C5, and C6 were attributed to 52% (95% CI, 12%–73%) of the symptoms among HAdV C cases [10]. A previous study in Kenya reported a predominance of HAdV species D and F in urban and rural settings, respectively, among cases with diarrhea, but other types including B3, B21, C2, C5, and C6 were also detected among diarrheal cases [12].
The KEMRI-Wellcome Trust Research Programme (KWTRP) has been conducting a prospective hospital-based rotavirus surveillance study at Kilifi County Hospital pediatric ward in coastal Kenya [6, 13]. The prevalence of adenoviruses of any species among pediatric diarrhea cases in coastal Kenya has been reported to be 15.9% (95% CI, 12.8%–19.5%). However, HAdV F only accounts for approximately half of the HAdV detections (7.3%; 95% CI, 5.2%–10.1%), with the rest of the HAdVs untyped [13].
This study aimed to genotype HAdV-positive samples detected between June 2022 and August 2023 to determine the circulating non-F HAdVs and any HAdV Fs that may have been missed by real-time polymerase chain reaction (PCR) screening. HAdV genotyping is typically done by amplifying a region of the hexon gene, followed by Sanger sequencing [14]. Here we used adenovirus primers from the RespiCoV panel [15], applying these primers to stool-derived nucleic acid extracts for the first time and sequencing the amplicons on the Oxford Nanopore Technologies (ONT) platform.
METHODS
Study Site and Population
The target population was children below the age of 13 years admitted to Kilifi County Hospital (KCH) who presented with diarrhea as one of their illness symptoms, that is, 3 or more loose stools in a 24-hour period [16].
Laboratory Methods
Total Nucleic Acid Extraction and Screening
Total nucleic acid (TNA) was extracted from a 0.2-g (or 200 µL if liquid) stool sample using the QIAamp Fast DNA Stool Mini kit (Qiagen, Manchester, UK), as previously described. Pan-HAdV (forward primer: 5′-GCCCCAGTGGTCTTACATGCACATC-3′; probe: “FAM-TCGGAGTACCTGAGCCCGGGTCTGGTGCA-MGBNFQ”; and reverse primer: 5′-GCCACGGTGGGGTTTCTAAACTT-3′) and HAdV-F (forward primer: 5′-CACTTAATGCTGACACGGGC-3′; probe: “FAM-TGCACCTCTTGGACTAGT-MGBNFQ’; and reverse primer: 5′-ACTGGATAGAGCTAGCGGGC-3′) primers and probes and TaqMan Fast Virus 1-Step Master Mix were used for screening, as previously described [17]. The thermocycling conditions were 95°C for 20 seconds and 35 cycles of 94°C for 15 seconds and 60°C for 30 seconds. A positive case was determined using a cycle threshold value (Ct value; ie, the number of quantitative PCR [qPCR] cycles required to replicate detectable HAdV DNA) cutoff of 35.
Other enteric viruses including rotavirus group A, norovirus genogroup II, astrovirus, and sapovirus were also screened using the extracted TNA and TaqMan Fast Virus 1-Step Master Mix, as described in our previous work [6].
Partial Hexon Sequencing
DNA Amplification. The primers used in amplification were adopted from the RespiCov panel [15]. Briefly, 14 adenovirus primers were pooled into 1 tube and resuspended in nuclease-free water to generate a 10-μM working concentration. TNA from HAdV-positive samples were amplified using the Q5 Hot Start HighFidelity 2X Master Mix (NEB, Oxford, UK) kit. The master mix was prepared as follows: Q5 Hot Start High-Fidelity 2X Master Mix (6.25 μL), H2O (3 μL), HAdV Primer pool (2 μL), and DNA (1.25 μL). The reaction was then incubated on a thermocycler using the following conditions: 98°C for 30 seconds followed by 35 cycles of 98°C for 15 seconds, 65°C for 30 seconds, and 72°C for 20 seconds, and a final extension of 72°C for 5 minutes.
Library Preparation and Oxford Nanopore Technologies Sequencing. Library preparation was performed using the SQK-LSK114 ligation kit with the SQK-NBD114.96 barcoding kit. Briefly, the amplicons were end-repaired, barcoded, and pooled into 1 tube, and adapters were ligated to the library and the final library sequenced using the FLOW-MIN106D R9.4.1 flow cell on the GridION platform (Oxford Nanopore Technologies) for 1 hour.
Long Fiber Amplification and Illumina Sequencing
Primers that could amplify the long fiber protein were obtained by picking forward (HAdV-F41_1kb_jh_85_LEFT:ACACTACAMTCCCCTTGACATCC) and reverse primers (HAdV-F41_1kb_jh_87_RIGHT:AAGAAAATGAGCAGCAGGGGATG) from whole-genome sequencing (WGS) HAdV primers designed elsewhere (Quick F41 WGS primers). The mastermix reaction was prepared as described in the above section and incubated on a thermocycler using the following conditions: 98°C for 30 seconds followed by 35 cycles of 98°C for 15 seconds and 65°C for 5 minutes. The amplicons generated were used for library preparation using an Illumina library preparation kit as recommended by the manufacturer. Briefly, the amplicons were tagmented, indexed, and amplified. The libraries were then normalized, pooled, and sequenced as paired-end reads (2*150 bp).
Whole-Genome Sequencing
For samples that were classified as HAdV A, C, and F, their DNA was amplified using the Q5 Hot Start High-Fidelity 2X Master Mix (NEB, Oxford, UK) kit and HAdV A, C, and F WGS primers (https://github.com/quick-lab/HAdV/tree/main), as previously described [17]. The amplicons were then used for library preparation and sequenced on the Illumina Miseq platform as described in the above section.
For samples that were classified as HAdV B and D, metagenomics sequencing was performed as WGS primers were not available. The DNA was directly used to perform library preparation and sequenced on the Illumina Miseq platform as described in the above section.
Data Analysis
Consensus Genome Generation From WGS Data
Short-read paired fastq data obtained from the Illumina MiSeq platform were trimmed using fastp with a phred scrore of q30. The cleaned reads were also assembled using a de novo approach using MetaSPAdes, version 3.13 (https://github.com/ablab/spades). The assembled contigs were then mapped to the HAdV references (NC_001460.1, NC_001454.1, NC_001405.1, AC_000018.1, and AC_000008.1) to check for completeness and generate consensus genomes.
Consensus Partial Hexon Sequence Generation From ONT Data
The FASTQ reads from the GridION were trimmed using porechop, version 0.2.4, and mapped to the HAdV reference genomes (DQ923122.2, NC_001460.1, NC_001454.1, NC_001405.1, AC_000006.1, AC_000018.1, AC_000008.1, NC_012959.1) using minimap2, version 2.24-r1122 (https://github.com/lh3/minimap2). Variant calling and consensus sequence generation were done using ivar, version 1.3.1 (https://github.com/andersen-lab/ivar), with a minimum read depth of 20. Taxonomic classification was done using BLASTN (https://blast.ncbi.nlm.nih.gov/).
Genotyping
Genotyping was done initially using the partial hexon sequence from the RespiCoV panel. This was then compared using recovered penton, hexon, fiber, and whole-genome sequences from WGS data. The generated partial- and full-consensus genomes were aligned with contemporaneous global HAdV sequences on GenBank using MAFFT (https://mafft.cbrc.jp/alignment/software/) and maximum likelihood trees generated using iqtree 2 (http://www.iqtree.org/). The trees were then annotated and visualized using ggtree (https://guangchuangyu.github.io/software/ggtree).
Long Fiber Primer Check Validation
Short-read paired fastq data obtained from the Illumina MiSeq platform were trimmed using fastp with a phred scrore of q30. The cleaned reads were then mapped to the HAdV-F reference genome using bwa (https://github.com/lh3/bwa). The primers were then trimmed from the BAM files and consensus genomes generated using ivar.
For quality check, the cleaned reads were also assembled using a de novo approach using MetaSPAdes, version 3.13 (https://github.com/ablab/spades). The generated contigs were compared with consensus genomes from the reference guide approach.
Real-time PCR primer and probe sequences were then aligned to the generated consensus long fiber sequences to check for differences in binding sites using Geneious Prime 2023.2.1 (https://www.geneious.com).
Disease Severity
The Vesikari Clinical Severity Scoring System Manual was used to estimate disease severity using data obtained at enrollment as previously described [18]. The following parameters were used: maximum number of stools and vomiting per day, duration of diarrhea and vomiting episodes in days, temperature, dehydration status, and treatment. The Vesikari grading categories were mild, moderate, and severe for scores of <7, 7–10, and ≥11, respectively.
RESULTS
HAdV Epidemiology
Between June 3, 2022, and August 28, 2023, 329 children with diarrhea as one of their illness symptoms were consented and gave stool samples for enteric virus screening. A total of 65 (20%) cases had an HAdV detection in their sample when using a pan-adenovirus real-time PCR assay. Forty-three samples were successfully sequenced and genotyped using adenovirus RespiCoV primers to determine the circulating HAdV species and types in stool. Twenty-five samples yielded partial to near complete genomes and were also used for genotyping using the penton, hexon, and fiber sequences. This approach was used to confirm the species and types detected based on the adenovirus RespiCoV primers.
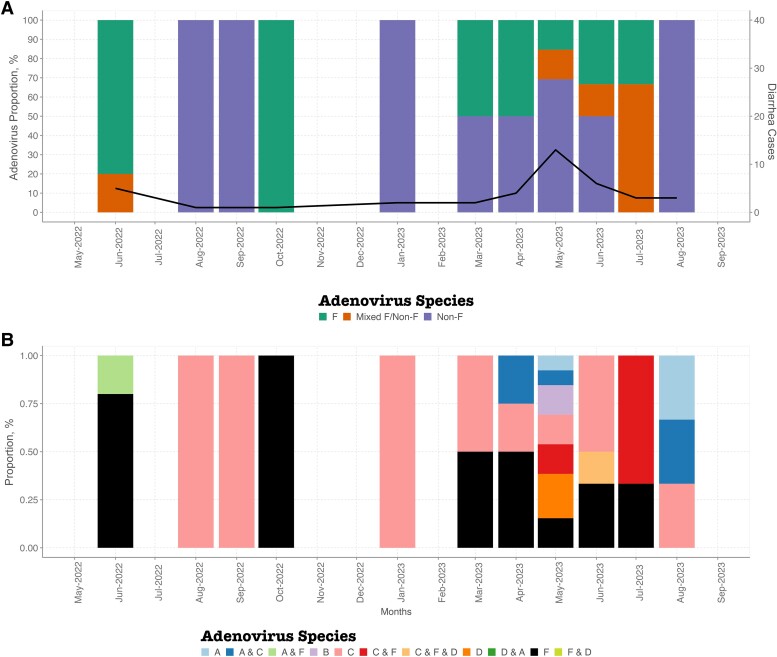
Single or multiple HAdV types were detected in the samples (Figure 1). Single HAdV type detections were as follows: F (n = 13), C (n = 12), D (n = 3), B (n = 2), and A (n = 2). Codetection of HAdV samples was also observed in 10 samples: C and F (n = 4), A and C (n = 3), A and D (n = 1), F and D (n = 1), and C, F, and D (n = 1) (Figure 1B).
Figure 1.
Temporal distribution of human adenoviruses in coastal Kenya between June 2022 and August 2023. A, Temporal plot showing distribution of F, mixed F (F and non-F coinfections), and non-F. The primary y-axis shows the proportions of adenovirus species, while the secondary y-axis shows the total number of diarrhea monthly cases. B, Temporal plot showing the distribution of HAdV species between June 2022 and August 2023. Abbreviation: HAdV, human adenovirus.
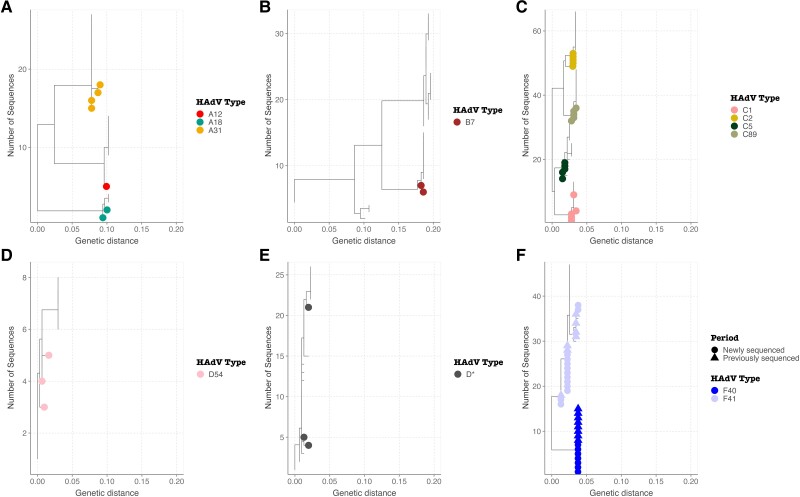
The HAdV species were further characterized into types (Figure 2). Both HAdV F40 (n = 7) and F41 (n = 13) were detected in HAdV species F-positive cases. Within HAdV species A, A31 (n = 4), A18 (n = 2), and A12 (n = 1) were identified. Only HAdV B7 was identified in species B. HAdV types C1 (n = 5), C2 (n = 5), C5 (n = 5), and C89 (n = 5) were identified within species HAdV C. Within HAdV D, D54 was identified in 2 samples while the rest of the sequences could not be genotyped further phylogenetically (Figure 2).
Figure 2.
Maximum likelihood trees showing the divergence of detected HAdV types A HAdV A, B HAdV B, C HAdV C, D HAdV D54, E HAdV D*, and F HAdV F using partial hexon sequence data. Abbreviation: HAdV, human adenovirus.
Among the samples that had the hexon, penton, and fiber genes sequenced, the species and type matched the genotyping results obtained from the adenovirus RespiCoV panel primers. The phylogenetic trees showing clustering of the newly sequenced data on the global phylogeny are shown in Supplementary Figure 1. Within species F, lineages 1 and 3 were observed in type F40, and lineages 1, 2A, and 3A were observed in type F41.
Adenovirus Outbreak in May 2023
In May 2023, there was a noticeable increase in adenovirus detections among children presenting with diarrhea over the study period. Of the 13 HAdV cases in May 2023, 69% of the cases were genotyped as non-HAdV F (D [n = 3], B [n = 2], C [n = 2], A and C [n = 1], and A [n = 1]), and only 4 samples were genotyped as HAdV F. Notably, 38% of the 13 HAdV cases had coinfections with norovirus GII (n = 4) and astrovirus (n = 1).
Demographic and Clinical Characteristics of HAdV Cases
The majority of the HAdV cases were male (60.5%) and between the ages of 12 and 59 months (65.1%). Coinfections with rotavirus A, norovirus GII, sapovirus, and astrovirus were identified across the different HAdV species. The majority of the cases (93%) presented with severe disease, including all the non-F cases (Table 1). Only 1 fatality was identified in the HAdV F cases and none in the non-F cases (Table 1).
Table 1.
Demographic and Clinical Characteristics of HAdV Genotyped Cases in Coastal Kenya
| HAdV F (n = 13), No. (%) or Median (IQR) | HAdV Codetection F and Non-F (n = 7), No. (%) or Median (IQR) | HAdV Non-F (n = 23), No. (%) or Median (IQR) | Total (n = 43), No. (%) or Median (IQR) | |
|---|---|---|---|---|
| Sex | ||||
| Female | 7 (53.8) | 2 (28.6) | 6 (26.1) | 15 (34.9) |
| Male | 6 (46.2) | 4 (57.1) | 16 (69.6) | 26 (60.5) |
| No data | 0 (0.0) | 1 (14.3) | 1 (4.3) | 2 (4.7) |
| Age, mo | 13 (7–21) | 11.5 (11–28.5) | 15.5 (10.2–20) | 14 (10–21) |
| Age group, mo | ||||
| <12 | 1 (7.7) | 2 (28.6) | 3 (13.0) | 6 (14.0) |
| 12–23 | 5 (38.5) | 1 (14.3) | 9 (39.1) | 15 (34.9) |
| 24–59 | 5 (38.5) | 2 (28.6) | 6 (26.1) | 13 (30.2) |
| ≥60 | 2 (15.4) | 2 (28.6) | 5 (21.7) | 9 (20.9) |
| Coinfections | ||||
| Rotavirus A | 1 (7.7) | 2 (28.6) | 3 (13.0) | 6 (14.0) |
| Norovirus GII | 0 (0.0) | 1 (14.3) | 4 (17.4) | 5 (11.6) |
| Sapovirus | 1 (7.7) | 0 (0.0) | 2 (8.7) | 3 (7.0) |
| Astrovirus | 0 (0.0) | 1 (14.3) | 0 (0.0) | 1 (2.3) |
| Disease severity | ||||
| Moderate | 2 (15.4) | 1 (14.3) | 0 (0.0) | 3 (7.0) |
| Severe | 11 (84.6) | 6 (85.7) | 23 (100.0) | 40 (93.0) |
| Outcome | ||||
| Alive | 12 (92.3) | 6 (85.7) | 22 (95.7) | 40 (93.0) |
| Dead | 1 (7.7) | 0 (0.0) | 0 (0.0) | 1 (2.3) |
| No data | 0 (0.0) | 1 (14.3) | 1 (4.3) | 2 (4.7) |
Abbreviations: HAdV, human adenovirus; IQR, interquartile range.
Failure of HAdV-F Real-Time PCR Assay
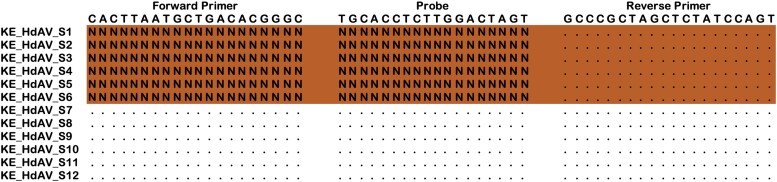
The genotyping results detected 6 additional HAdV F positives that were missed by real-time PCR. Successful sequencing of the long fiber protein where the real-time PCR primers bind showed that the sequences had a 133 base deletion that spanned the forward primer and probe binding regions, leading to failure in the HAdV F real-time PCR assay (Figure 3).
Figure 3.
An alignment of HAdV F sequences from missed and detected HAdV F samples mapped to the primers and probe sequences. Dots show consensus, and Ns show gaps in the primer and probe binding sites. Abbreviation: HAdV, human adenovirus.
Ct Value Distribution
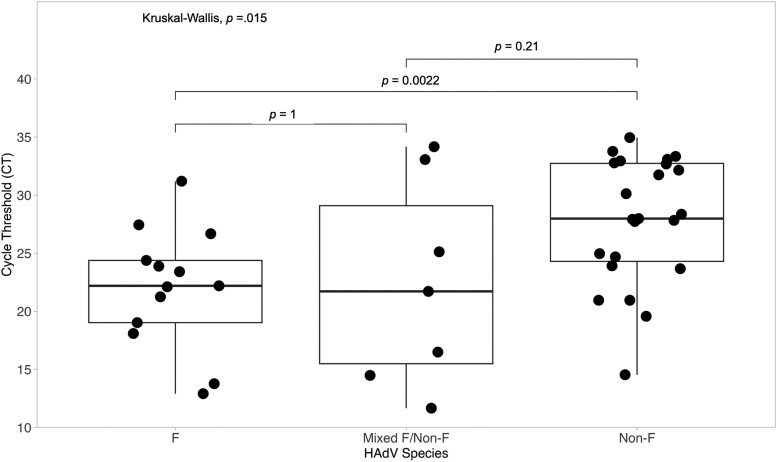
HAdV F cases had significantly lower Ct values compared with non-F cases (P = .002) (Figure 4). There was no significant difference in Ct values among HAdV F cases and cases that had codetections of HAdV-F and non-F. Eight non-F cases had a Ct value of <25 (high viral load), but 6 of these samples also had coinfections with norovirus GII, rotavirus A, and sapovirus.
Figure 4.
Comparison of HAdV viral load (inverse of cycle threshold value) among F, mixed F (coinfection of F and non-F), and non-F cases. Abbreviation: HAdV, human adenovirus.
DISCUSSION
The study findings show that multiple HAdV species and types were in circulation between June 2022 and August 2023 in coastal Kenya and that our current real-time PCR primers for detecting HAdV F may be missing positive cases due to a 133-nucleotide deletion in the long fiber protein, which abolishes a primer and probe binding site in some circulating variants. This is the first application of the adenovirus part of the RespiCov method to stool samples for genotyping enteric viral infections. Previous studies from China, Tunisia, Kenya, and Brazil have reported detection of non-F HAdVs similar to our findings [3, 7, 9, 10, 12, 19, 20]. Non-F types such as B3, C1, C2, C5, and C6 have been associated with increased risk of diarrhea, but direct causation is not yet clear [3, 10]. In this study, we did not detect HAdV B3 but detected the HAdV types C1 and C5, although we cannot conclude that they were the main cause of diarrhea in these individuals. The RespiCov method used could not clearly genotype all HAdV species D phylogenetically. This is likely due to the relatively short ∼300 base pair hexon region that is amplified and the highly recombinatorial nature of species D HAdVs within the hexon locus [21].
Within HAdV F, there is a high divergence in the strains that have been reported to be circulating in Kenya and other regions across the globe [17, 22–24]. If this divergence occurs in primer and probe binding sites, nucleic acid amplification methods used for detection may be affected. The genotyping results in this study showed that 6 HAdV F cases were missed by the F-specific real-time PCR assay. Amplification of the long fiber protein revealed a 133 base deletion that impacted the HAdV-F real-time PCR assay, and it is advisable that similar primers used in previous works [13, 17] may be missing some positive cases and need redesigning, a phenomenon previously seen in SARS-CoV-2 [25], HIV [26], and Chlamydia trachomatis [27].
HAdV F samples had a significantly higher viral load (lower Ct value) compared with non-F samples, similar to previous studies [10, 19]. HAdV species F is highly associated with diarrhea when the Ct value is <22.7, and cases and controls can be discriminated at Ct 30.5, suggesting that the HAdV F detected in this study may be clearly associated with diarrhea [9, 28]. Interestingly, the HAdV-F detections with a Ct value >30 had a rotavirus A coinfection, suggesting that they could be the secondary cause of diarrhea. The non-F HAdVs detected had a lower viral load and were detected with other enteric viruses including rotavirus A, norovirus GII, and sapovirus, suggesting that they may be associated with carriage due to prolonged shedding or gut contamination from respiratory infections rather than diarrhea [29, 30].
This study had some limitations. First, there were no clinical data on the respiratory symptoms of the patients to help interpret the nonenteric HAdVs detected in stool samples that are usually associated with respiratory illnesses. The enteric HAdV wave was preceded by a respiratory adenovirus wave that happened between January and April 2023, suggesting that this could be the cause of the nonenteric HAdVs (unpublished data). Secondly, only 4 common enteric viruses were screened for, and these were frequently detected as coinfections in non-F cases. Screening for additional causes such as bacteria, parasites, and helminths may have helped elucidate whether other enteric pathogens contribute to the detection of non-F cases.
In conclusion, there is a high diversity of HAdV types found among diarrhea cases in coastal Kenya, and this may reflect the situation in Africa where there are limited data. Timely and accurate genotyping of these HAdV cases are key for troubleshooting failure of molecular detection assays, estimation of diarrhea prevalence associated with HAdV-F, and implementation of interventions to reduce the burden of HAdV-associated diarrhea.
Supplementary Material
Acknowledgments
We are grateful to the study participants who provided samples and members of the pathogen epidemiology and omics group at KEMRI-Wellcome Trust Programme who did sample collection and laboratory processing. This manuscript was written with the permission of Director KEMRI CGMRC.
Author contributions. C.A.N. and C.J.H. sourced the study funding. C.N.A., C.J.H., and A.W.L. designed the study laboratory assay. A.W.L. and M.M. did the laboratory experiments. E.K. and A.W.L. managed the study data and did the data analysis. A.W.L., C.A.N., and C.J.H. wrote the first manuscript draft. All authors read, revised, and approved the final manuscript.
Financial support. This study was funded in part by the Cambridge-Africa ALBORADA Research Fund to Drs. Agoti and Houldcroft. This research was funded in part by the Wellcome Trust (226002/Z/22/Z). For the purpose of Open Access, the author has applied a CC-BY public copyright license to any author accepted manuscript version arising from this submission.
Data availability. The data sets used and/or analyzed in the current study are available from the KWTRP Research repository via https://doi.org/10.7910/DVN/XCHBND. The HAdV sequences were deposited on GenBank and can be accessed using the accession numbers PP318651–PP318703.
Patient consent. The research protocol for the study was approved at Kenya Medical Research Institute (KEMRI) by the Scientific and Ethics Review Unit (SSC#2861) in Nairobi, Kenya.
Contributor Information
Arnold W Lambisia, Epidemiology and Demography Department, KEMRI-Wellcome Trust Research Programme, Kilifi, Kenya.
Martin Mutunga, Epidemiology and Demography Department, KEMRI-Wellcome Trust Research Programme, Kilifi, Kenya.
Esther N Katama, Epidemiology and Demography Department, KEMRI-Wellcome Trust Research Programme, Kilifi, Kenya.
Charles N Agoti, Epidemiology and Demography Department, KEMRI-Wellcome Trust Research Programme, Kilifi, Kenya; School of Health and Human Sciences, Pwani University, Kilifi, Kenya.
Charlotte J Houldcroft, School of Health and Human Sciences, University of Cambridge, Cambridge, UK.
Supplementary Data
Supplementary materials are available at Open Forum Infectious Diseases online. Consisting of data provided by the authors to benefit the reader, the posted materials are not copyedited and are the sole responsibility of the authors, so questions or comments should be addressed to the corresponding author.
References
- 1. Hulo C, de Castro E, Masson P, et al. ViralZone: a knowledge resource to understand virus diversity. Nucleic Acids Res 2011; 39(Suppl_1):D576–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bányai K, Martella V, Meleg E, et al. Searching for HAdV-52, the putative gastroenteritis-associated human adenovirus serotype in southern Hungary. New Microbiol 2009; 32:185–8. [PubMed] [Google Scholar]
- 3. Qiu FZ, Shen XX, Li GX, et al. Adenovirus associated with acute diarrhea: a case-control study. BMC Infect Dis 2018; 18:450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Lynch J, Kajon A. Adenovirus: epidemiology, global spread of novel serotypes, and advances in treatment and prevention. Semin Respir Crit Care Med 2016; 37:586–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Adhikary AK, Banik U. Human adenovirus type 8: the major agent of epidemic keratoconjunctivitis (EKC). J Clin Virol 2014; 61:477–86. [DOI] [PubMed] [Google Scholar]
- 6. Lambisia AW, Murunga N, Mutunga M, et al. Temporal changes in the positivity rate of common enteric viruses among paediatric admissions in coastal Kenya, during the COVID-19 pandemic, 2019–2022. Gut Pathog 2024; 16:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Afrad MH, Avzun T, Haque J, et al. Detection of enteric- and non-enteric adenoviruses in gastroenteritis patients, Bangladesh, 2012–2015. J Med Virol 2018; 90:677–84. [DOI] [PubMed] [Google Scholar]
- 8. Ghebremedhin B. Human adenovirus: viral pathogen with increasing importance. Microbiol Immunol 2014; 4:26–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Huang Z, He Z, Wei Z, et al. Correlation between prevalence of selected enteropathogens and diarrhea in children: a case-control study in China. Open Forum Infect Dis 2021; 8:XXX–XX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pabbaraju K, Tellier R, Pang XL, et al. A clinical epidemiology and molecular attribution evaluation of adenoviruses in pediatric acute gastroenteritis: a case-control study on behalf of the Alberta Provincial Pediatric EnTeric Infection TEam (APPETITE). 2020; 59:e02287-20. [DOI] [PMC free article] [PubMed]
- 11. Binder AM, Biggs HM, Haynes AK, et al. Morbidity and Mortality Weekly Report human adenovirus surveillance—United States, 2003–2016. MMWR Morb Mortal Wkly Rep 2017; 66:1039–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Magwalivha M, Wolfaardt M, Kiulia NM, van Zyl WB, Mwenda JM, Taylor MB. High prevalence of species D human adenoviruses in fecal specimens from urban Kenyan children with diarrhea. J Med Virol 2010; 82:77–84. [DOI] [PubMed] [Google Scholar]
- 13. Agoti CN, Curran MD, Murunga N, et al. Differences in epidemiology of enteropathogens in children pre- and post-rotavirus vaccine introduction in Kilifi, coastal Kenya. Gut Pathog 2022; 14:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Lu X, Erdman DD. Molecular typing of human adenoviruses by PCR and sequencing of a partial region of the hexon gene. Arch Virol 2006; 151:1587–602. [DOI] [PubMed] [Google Scholar]
- 15. Brinkmann A, Uddin S, Ulm SL, et al. RespiCoV: simultaneous identification of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and 46 respiratory tract viruses and bacteria by amplicon-based Oxford-nanopore MinION sequencing. PLoS One 2022; 17:e0264855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. World Health Organization . The Treatment of Diarrhoea: A Manual for Physicians and Other Senior Health Workers. Vol 17. 4th Rev. World Health Organization; 2005. [Google Scholar]
- 17. Lambisia AW, Makori TO, Mutunga M, et al. Genomic epidemiology of human adenovirus F40 and F41 in coastal Kenya: a retrospective hospital-based surveillance study (2013–2022). Virus Evol 2023; 9:vead023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lewis K. VAD_vesikari_scoring_manual. 2011. Accessed 10 January 2024.
- 19. do Nascimento LG, Fialho AM, de Andrade J da SR, de Assis RMS, Fumian TM. Human enteric adenovirus F40/41 as a major cause of acute gastroenteritis in children in Brazil, 2018 to 2020. Sci Rep 2022; 12:11220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bouazizi A, Ben Hadj Fredj M, Bennour H, et al. Molecular analysis of adenovirus strains responsible for gastroenteritis in children, under five, in Tunisia. Heliyon 2024; 10:e22969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Walsh MP, Chintakuntlawar A, Robinson CM, et al. Evidence of molecular evolution driven by recombination events influencing tropism in a novel human adenovirus that causes epidemic keratoconjunctivitis. PLoS One 2009; 4:e5635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Götting J, Cordes AK, Steinbrück L, Heim A. Molecular phylogeny of human adenovirus type 41 lineages. Virus Evol 2022; 20:veac098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chandra P, Lo M, Mitra S, et al. Genetic characterization and phylogenetic variations of human adenovirus-F strains circulating in eastern India during 2017–2020. J Med Virol 2021; 93:6180–90. [DOI] [PubMed] [Google Scholar]
- 24. Maes M, Khokhar F, Wilkinson SA, et al. Enteric adenovirus F41 genetic diversity comparable to pre-COVID-19 era: validation of a multiplex amplicon-MinION sequencing method. Microb Genom 2023; 9:mgen000920 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Volz E, Mishra S, Chand M, et al. Assessing transmissibility of SARS-CoV-2 lineage B.1.1.7 in England. Nature 2021; 593:266–9. [DOI] [PubMed] [Google Scholar]
- 26. Müller B, Nübling CM, Kress J, Roth WK, De Zolt S, Pichl L. How safe is safe: new human immunodeficiency virus type 1 variants missed by nucleic acid testing. Transfusion 2013; 53:2422–30. [DOI] [PubMed] [Google Scholar]
- 27. Ripa T, Nilsson PA. A Chlamydia trachomatis strain with a 377-bp deletion in the cryptic plasmid causing false-negative nucleic acid amplification tests. Sex Transm Dis 2007; 34:255–6. [DOI] [PubMed] [Google Scholar]
- 28. Liu J, Platts-Mills JA, Juma J, et al. Use of quantitative molecular diagnostic methods to identify causes of diarrhoea in children: a reanalysis of the GEMS case-control study. Lancet 2016; 388:1291–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bartholomeusz A, Locarnini S. Associated with antiviral therapy. Antivir Ther 2006; 55:52–5. [Google Scholar]
- 30. Kim JS, Lee SK, Ko DH, et al. Associations of adenovirus genotypes in Korean acute gastroenteritis patients with respiratory symptoms and intussusception. Biomed Res Int 2017; 2017:1602054. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.