Abstract
Progressive multifocal leukoencephalopathy (PML) is a rare neurological condition associated with reactivation of dormant JC polyomavirus (JCPyV). In this study, we characterized gene expression and JCPyV rearrangements in PML brain tissue. Infection of white matter astrocytes and oligodendrocytes as well as occasional brain cortex neurons was shown. PML brain harbored exclusively rearranged JCPyV variants. Viral transcripts covered the whole genome on both strands. Strong differential expression of human genes associated with neuroinflammation, blood-brain barrier permeability, and neurodegenerative diseases was shown. Pathway analysis revealed wide immune activation in PML brain. The study provides novel insights into the pathogenesis of PML.
Keywords: brain, JC polyomavirus, JCPyV, neurodegeneration, NGS, PML, progressive multifocal leukoencephalopathy, sequencing, transcriptomics, viral transcripts
Brain tissue of patients with progressive multifocal leukoencephalopathy (PML) harbors rearranged variants of the causative agent JC polyomavirus. Strong regulation of genes associated with neuroinflammation, blood-brain barrier permeability, and neurodegenerative diseases provides new prospects for understanding PML pathogenesis.
JC polyomavirus (JCPyV) is a common childhood infection leading to lifetime asymptomatic persistence, for example in kidney, with occasional excretion into urine. Reactivation of dormant JCPyV may give rise to neurotropic rearrangements, typically in the noncoding control region (NCCR) and the major viral capsid protein (VP1) gene. Neurotropic JCPyV strains, or sequence variants, are strongly associated with the development of progressive multifocal leukoencephalopathy (PML), a severe neurological condition where lytic JCPyV infection in glial cells leads to destruction of brain white matter.
Risk of viral reactivation and developing PML is elevated in individuals with weakened immune defense due to human immunodeficiency virus (HIV), malignancy, especially lymphoma, use of immunomodulatory treatments for multiple sclerosis or rheumatoid arthritis, or due to transplant-related immunosuppression [1]. However, PML may also develop in the absence of immunosuppression and in seemingly immunocompetent individuals [2].
Here we present an analysis of JCPyV variants, viral gene expression, and the human transcriptome in brain tissue of live and deceased patients with confirmed PML. We show the predominance and diversity of rearranged viral variants in brain tissue, and the complexity of viral transcripts. We describe differential expression of genes implicated in neuroinflammation, blood-brain barrier (BBB) permeability, and neurodegenerative diseases, and wide immune activation in PML brain. Our study addresses in fundamental ways the pathophysiology of JCPyV infection in the brain.
METHODS
Study Population and Brain Samples
Our study included 9 PML cases and 2 non-PML controls. The cases have been described previously in more detail [3] and briefly in Supplementary Material. PML brain tissues were characterized by immunohistochemical staining described in detail in the Supplementary Material.
Study Design
Tissue from the brain lesions of 9 PML cases (1–9) and control brain tissue from 2 non-PML individuals was sectioned for DNA and RNA extraction. DNA from PML cases and controls was used for NCCR sequence analysis (Supplementary Material). RNA was used to prepare libraries for short-read RNA sequencing. Case 9 sample was inadequate for RNA sequencing. Data analysis is described in detail in Supplementary Material. The study was approved by the ethical board of the Helsinki and Uusimaa Hospital District.
Supplementary Material
Contributor Information
Anni Honkimaa, Department of Virology, Helsinki University Hospital and University of Helsinki, Helsinki, Finland.
Pia Laine, DNA Sequencing and Genomics Laboratory, Institute of Biotechnology, University of Helsinki, Helsinki, Finland.
Joni Suppula, DNA Sequencing and Genomics Laboratory, Institute of Biotechnology, University of Helsinki, Helsinki, Finland.
Olli Tynninen, Department of Pathology, Helsinki University Hospital and University of Helsinki, Helsinki, Finland.
Mika Saarela, Department of Neurology, Helsinki University Hospital and University of Helsinki, Helsinki, Finland.
Sini M Laakso, Department of Neurology, Helsinki University Hospital and University of Helsinki, Helsinki, Finland; Translational Immunology Research Program, University of Helsinki, Helsinki, Finland.
Iivo Hetemäki, Translational Immunology Research Program, University of Helsinki, Helsinki, Finland.
Hanna Liimatainen, Department of Virology, Helsinki University Hospital and University of Helsinki, Helsinki, Finland.
Petri Auvinen, DNA Sequencing and Genomics Laboratory, Institute of Biotechnology, University of Helsinki, Helsinki, Finland.
Eeva Auvinen, Department of Virology, Helsinki University Hospital and University of Helsinki, Helsinki, Finland.
RESULTS
Study Population
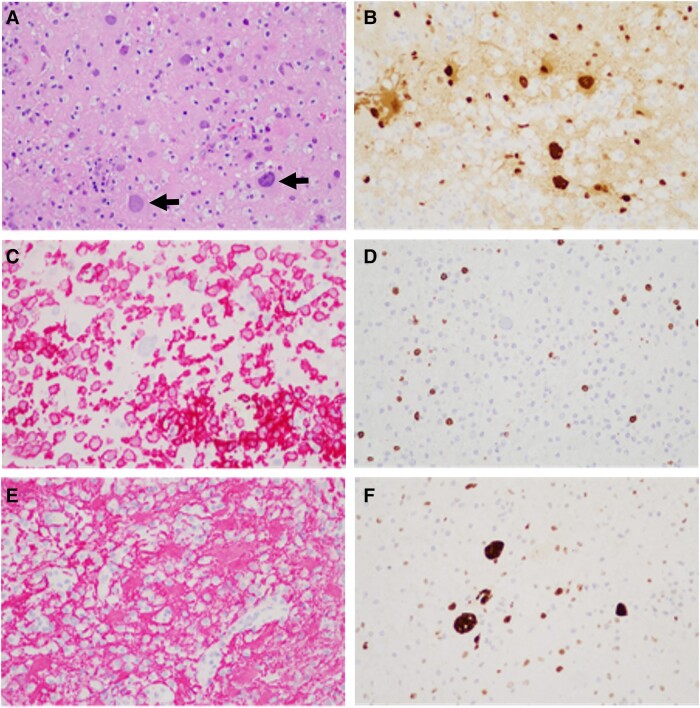
The study population included 9 PML patients and 2 control individuals (Supplementary Table 1) [3]. The medical history of cases 3, 4, 5, 7, and 9 revealed diseases and/or medications predisposing to PML. Intriguingly, no predisposing factor was identified for cases 1, 2, 6, and 8. Infected and transformed glial cells were visualized by hematoxylin-eosin staining (Figure 1A). PML diagnosis of each case was confirmed by immunohistochemical staining for viral large T antigen in brain tissue (Figure 1B). Inflammatory and glial cell reaction was characterized by staining for macrophages (Iba1; Figure 1C), T lymphocytes (CD3; Figure 1D), astrocyte marker (GFAP; Figure 1E), and tumor suppressor p53 (Figure 1F). Interestingly, accumulation of p53 protein was seen in infected glial cells (Figure 1F). Semiquantitative grading of expression levels revealed strong expression of each marker in majority of cases, except in case 6 (Supplementary Table 1). Besides infected astrocytes and oligodendrocytes, in at least 1 PML sample we also observed JCPyV infection in cortical neurons based on cellular morphology.
Figure 1.
Example of immunohistochemical staining in progressive multifocal leukoencephalopathy tissue. Hematoxylin-eosin staining (A) shows infected and transformed glial cells (arrows), further highlighted by an antibody for JC polyomavirus large T antigen (B). The inflammatory reaction is illustrated by staining for macrophages (C, Iba1 antibody), T lymphocytes (D, CD3 antibody, pan–T-cell marker), and reactive astrocytes (E, GFAP antibody). Infected glial cells show accumulation of p53 tumor suppressor protein (F). Magnification 200×.
JCPyV NCCR Variants
Short-read sequencing of the NCCR regions consisting of ABCDEF sequence blocks [4] revealed that PML brain tissue contained exclusively neurotropic JCPyV variants. Deletion of D block was most frequent (see more detailed analysis in Supplementary Material). Rearranged JCPyV variants were also found in 1 of the controls.
JCPyV Transcription
We performed short-read RNA sequencing (RNAseq) of strand-specific libraries. Based on initial analysis of RNAseq data (Supplementary Material) we performed further analyses on cases 1–5 and controls. We first assessed the patterns of JCPyV transcription in brain tissue samples from cases 1–5. Considerably more reads (8.9-fold) represented transcripts from the plus strand (harboring late genes) as compared to the minus strand (harboring early genes) (Supplementary Figure 1). Read mapping covered the full length of the genome on both strands (Supplementary Figure 1), suggesting the presence of wraparound transcripts [5] although we cannot confirm this by short-read sequencing. The coverage of the reads representing transcripts from the plus strand was more or less equal throughout the genome, whereas the coverage of the reads representing transcripts from the minus strand was higher within protein coding regions. Although many sequencing reads mapped to the NCCR region, decreased coverage was seen due to deletions in the rearranged variants, particularly within block D (Supplementary Material). Complete transcripts or splicing patterns cannot be accurately described using short-read sequencing. However, the intron positions were assessed and they are shown in the Supplementary Material.
Expression of Human Genes in PML Brain
Differential expression (DE) of human genes was assessed by comparing RNAseq data from cases 1–5 to controls using iDEP version 1.1 with minimum fold-change 2 and false discovery rate < .1. The analysis revealed upregulation of 6316 genes and downregulation of 3938 genes. The DE genes are visualized by volcano plot (Supplementary Figure 2A) as well as a heatmap with hierarchical clustering (Supplementary Figure 2B). These analyses clearly differentiate the gene expression patterns of cases from controls.
When DE protein-coding genes were ordered according to adjusted P value (Padj), the top 20 upregulated genes (Supplementary Table 2) included genes implicated in cellular immune response, neuroinflammatory response, neurodegenerative diseases, and the BBB (Supplementary Table 2). Top 20 downregulated genes (Supplementary Table 1) include genes from the olfactory receptor family, but no associations with polyomaviruses, PML, or neurological disorders have been published. No relevant differences in DE between immunocompetent cases (n = 2) as compared to cases with known predisposing factor(s) (n = 3) were established (data not shown).
We explored the network involvement of DE genes using ingenuity pathway analysis (IPA). Most significant upstream regulators, both transcription factors and cytokines, with a significantly activated or inhibited status are listed (Table 1). These analyses confirmed a wide immune activation consistent with viral defense, as exemplified by activated cytokines type I, II, and III interferons (IFNG, IFNA2/1/13, and IFNB1) and interleukin 6 (IL-6) that trigger signaling through STAT1 and STAT3 [6]. Transcription factors IRF1 and IRF7 are also vital promoters of innate immune responses against viruses [7]. Among upregulated transcription factors is TP53 (Table 1). Interestingly, we showed accumulation of p53 protein in JCPyV infected cells of PML brain (Figure 1F and Supplementary Table 1).
Table 1.
Network Involvement of Differentially Expressed Genes by Ingenuity Pathway Analysis
| z Score | P Valuea | |
|---|---|---|
| Transcription factor genes | ||
| STAT1 | 8.028 | 3.72E-25 |
| TP53 | 4.336 | 1.72E-20 |
| STAT3 | 4.777 | 3.56E-19 |
| IRF1 | 7.055 | 1.67E-18 |
| NFKBIA | 2.582 | 4.59E-18 |
| ETV3 | −6.083 | 5.53E-17 |
| PRDM1 | −4.538 | 3.02E-16 |
| MYC | 4.061 | 3.28E-16 |
| SPI1 | 3.887 | 3.83E-14 |
| KLF6 | 7.121 | 7.37E-14 |
| ETV6 | −5.642 | 8.35E-14 |
| CEBPB | 5.758 | 1.09E-13 |
| SMARCA4 | 6.382 | 2.58E-13 |
| NFKB1 | 3.805 | 3.1E-13 |
| IRF7 | 7.372 | 1.12E-12 |
| YAP1 | 5.259 | 1.45E-12 |
| ZBTB10 | 6.365 | 9.16E-12 |
| FOXO1 | 4.318 | 1.27E-11 |
| TP63 | 2.863 | 2.73E-11 |
| CTNNB1 | 3.861 | 6.19E-11 |
| Cytokine genes | ||
| IFNG | 10.669 | 5.29E-36 |
| CSF1 | 5.784 | 1.28E-29 |
| IL4 | 7.629 | 3.53E-29 |
| IL1B | 7.419 | 2.7E-28 |
| TNF | 9.327 | 1.67E-25 |
| IFNA2 | 7.993 | 2.34E-22 |
| IL2 | 7.522 | 3.27E-20 |
| CSF2 | 8.518 | 9.55E-18 |
| CD40LG | 6.449 | 8.85E-17 |
| IL33 | 6.21 | 6.7E-16 |
| IL6 | 7.237 | 4.14E-15 |
| IL27 | 6.116 | 3.15E-14 |
| TNFSF13B | 2.901 | 8.26E-14 |
| IL15 | 5.099 | 8.79E-14 |
| IFNL1 | 6.265 | 2.31E-13 |
| IL13 | 4.184 | 1.24E-12 |
| IFNA1/IFNA13 | 4.395 | 6.06E-12 |
| PRL | 6.084 | 1.47E-10 |
| IFNB1 | 3.559 | 1.58E-10 |
| CXCL12 | 4.223 | 4.85E-10 |
| Ingenuity canonical pathways | ||
| Pathogen-induced cytokine storm signaling pathway | 6.749 | 11 |
| Neuroinflammation signaling pathway | 6.915 | 9.88 |
| Multiple sclerosis signaling pathway | 5.735 | 9.17 |
| Th2 pathway | 4.429 | 7.73 |
| Th1 pathway | 5.397 | 7.72 |
| Role of osteoclasts in rheumatoid arthritis signaling pathway | 6.405 | 7.13 |
| Pyroptosis signaling pathway | 3.781 | 6.64 |
| Macrophage alternative activation signaling pathway | 4.737 | 6.47 |
| Hepatic fibrosis signaling pathway | 5.051 | 6.47 |
| PD-1, PD-L1 cancer immunotherapy pathway | −3.667 | 6.27 |
| Acute phase response signaling | 4.619 | 6.09 |
| Death receptor signaling | 3.667 | 5.75 |
| Tumor microenvironment pathway | 3.371 | 5.52 |
| Neutrophil extracellular trap signaling pathway | 3.22 | 5.5 |
| Crosstalk between dendritic cells and natural killer cells | 5.292 | 5.42 |
| Phagosome formation | 5.622 | 5.19 |
| Role of chondrocytes in rheumatoid arthritis signaling pathway | 4.718 | 5.19 |
| TREM1 signaling | 4.271 | 4.8 |
| NAD signaling pathway | 4.727 | 4.68 |
| Production of nitric oxide and reactive oxygen species in macrophages | 4.438 | 4.6 |
Upstream regulators of differentially expressed human genes in progressive multifocal leukoencephalopathy cases 1–5 were analyzed with ingenuity pathway analysis. Transcription factors and cytokines are shown separately and termed activated (not bold) or inhibited (bold) based on the z score. The 20 most significant transcription factors and cytokines are shown, all with absolute z score >2.0 and Padj ≤.05. The 20 most significant canonical pathways activated (not bold) and inhibited (bold) in cases 1–5 versus controls as identified by ingenuity pathway analysis are shown, all with absolute z score >2.0 and Padj ≤.05.
aFor transcription factor and cytokine genes the P value of overlap is given. For ingenuity canonical pathways the −log P value is given.
We performed further IPA analysis to explore the changes in canonical pathways (Table 1). Strong activation of both acquired and innate immune response pathways was observed. Among the 20 most significantly regulated pathways, the only inhibited pathway was the PD-1 immunotherapy pathway. Although the sample material was limited, we also applied IPA biomarker function to identify putative cerebrospinal fluid biomarker candidates for PML (Supplementary Table 3). The analysis yielded, for example, genes related to complement activation (C1QC, C1QB, C2, C3), chitinase −3-like 1-protein (CHI3L1), lysozyme, and type 1 inflammation (chemokines CXCL10 and CXCL9).
DISCUSSION
In the current study we present the first comprehensive gene expression analysis in JCPyV-affected human brain tissue from PML patients. Our study population initially comprised 9 confirmed PML patients, including rare brain biopsies from living patients and post mortem tissue samples.
Traditionally, immunosuppression has been considered a requirement and the main cause of PML [1]. Among our PML patients 5 had predisposing treatment or disease, while as many as 4 patients had no disease or medication known to predispose to the development of PML. RNAseq data did not reveal any difference in the regulation of immune response-related genes among the immunocompetent individuals compared to those with known predisposing factors. Occasional PML cases in immunocompetent individuals have been reported previously [2]. More recent data on immune dysfunction may explain PML emergence without established predisposing factors. The surprisingly high proportion of PML patients without established predisposing factors in our patient cohort raises the question about the still unknown PML risk factor(s), such as past transient periods of immunosuppression.
Our short-read RNAseq analysis revealed complex patterns of viral transcription. Transcripts from both strands of the viral genome cover the complete genome length, including regions outside the established open reading frames. Similar findings have been reported from experimental polyomavirus infections. Approximately 40% of Merkel cell polyomavirus late strand transcripts have been reported to map to the antisense strand of the early region and the NCCR [8]. Analogous data have been presented for mouse polyomavirus [5]. Overall, the coverage within both strands varied heavily, seen in the spiky nature of the coverage curve, which may be due to noise when amplifying smaller RNA fragments, as suggested by Nomburg et al [5]. The coverage in the JCPyV microRNA (miRNA) gene region was low, in agreement with the low JC-5p and JC-3p miRNA read counts found in these samples [3]. Although our data suggest the existence of wraparound transcripts [5], due to our short-read sequencing approach we are not able to provide full characterization of viral transcripts.
Strong indications of abrupt and comprehensive immune activation and neuroinflammation were observed in RNAseq analysis, similar to our previous miRNA sequencing analysis [3]. A transmembrane protein, TYROBP, is a key regulator in immune systems [9], and its overexpression has been shown to lead to dramatic change in regulation of numerous downstream genes in brain microglia [10]. CHI3L1 is expressed by astrocytes, microglia, and monocytes in the brain, and it may have a significant role in modulating central nervous system (CNS) inflammation [11]. Increased CHI3L1 expression in cells of the white matter, seen also in our PML samples, is strongly correlated with an elevated number of Iba1-expressing microglia [12]. We observed elevated expression of Iba1 and CD3 proteins, indicating the enhanced presence of immune cells in PML brain. RNAseq analysis suggested putative viral effects or CNS entry through the BBB. We saw upregulation of M6PR, which is involved in BBB crossing by HIV-1 [13]. Of note, in the current PML brain samples we observed upregulation of miR-21–3p and miR-155, also implicated in BBB disruption, as we observed in a previous study [3], indicating that BBB permeability may indeed be affected in PML.
IPA analysis revealed downregulation of the PD-1 immunotherapy pathway. PD-1 inhibition using pembrolizumab has been reported to show clinical benefit [14] but also high failure rates thereafter in PML patients [15]. Future studies will show whether determination of the individual inhibitory status of the PD-1 immunotherapy pathway could help select PML patients for experimental pembrolizumab therapy. More detailed analysis of the immune cell composition in PML lesions, combined with transcriptomics, may unravel the failure of the immune system in the defense against neurotropic JCPyV.
In conclusion, PML as associated with enhanced replication of neurotropic JCPyV and lytic infection in brain white matter. Massive regulation of cellular gene expression including prominent immune activation was seen in PML brain tissue. Future studies will uncover how different rearranged variants, transcription of viral genes, and modifications in the cellular transcriptome contribute to PML pathogenesis and how these data could be employed in PML risk assessment and patient management.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Notes
Acknowledgments. We thank Ms Anu Kaitonen for valuable technical assistance. We thank the personnel of the DNA sequencing and genomics laboratory for running the NGS-related laboratory phases. We thank Dr Richard Frisque for providing us with the JCPyV antibody.
Author contributions. E. A. contributed conceptualization. A. H., P. L., J. S., O. T., S. M. L., I. H., P. A., and E. A. performed investigations and data analysis. M. S. performed sample acquisition. A. H., P. L., J. S., O. T., S. M. L., I. H., H. L., P. A., and E. A. contributed methodology. P. L., J. S., and O. T. contributed experimentation. A. H., P. L., S. M. L., I. H., and E. A. wrote the manuscript. All authors reviewed and commented on the manuscript.
Data availability . The data for this study have been deposited in the European Nucleotide Archive at EMBL-EBI under accession number PRJEB64568 (https://www.ebi.ac.uk/ena/browser/view/PRJEB64568).
Financial support. This work was supported by the Emil Aaltonen Foundation; the Finnish Society for the Study of Infectious Diseases; and the Maire Taponen Foundation.
References
- 1. Kartau M, Sipilä JO, Auvinen E, Palomäki M, Verkkoniemi-Ahola A. Progressive multifocal leukoencephalopathy: current insights. Degener Neurol Neuromuscul Dis 2019; 9:109–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zucker BE, Stacpoole SRL. Progressive multifocal leukoencephalopathy in the absence of immunosuppression. J Neurovirol 2018; 24:119–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Honkimaa A, Suppula J, Tynninen O, et al. JC polyomavirus modifies the expression of human microRNAs in PML brain. J Infect Dis 2023; 228:829–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wilczek MP, Pike AMC, Craig SE, Maginnis MS, King BL. Rearrangement in the hypervariable region of JC polyomavirus genomes isolated from patient samples and impact on transcription factor-binding sites and disease outcomes. Int J Mol Sci 2022; 23:5699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nomburg J, Zou W, Frost TC, et al. Long-read sequencing reveals complex patterns of wraparound transcription in polyomaviruses. PLoS Pathog 2022; 18:e1010401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Harrison AR, Moseley GW. The dynamic interface of viruses with STATs. J Virol 2020; 94:e00856-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jiang H, Li Y, Shen M, et al. Interferon-α promotes MHC I antigen presentation of islet β cells through STAT1-IRF7 pathway in type 1 diabetes. Immunology 2022; 166:210–21. [DOI] [PubMed] [Google Scholar]
- 8. Theiss JM, Günther T, Alawi M, et al. A comprehensive analysis of replicating Merkel cell polyomavirus genomes delineates the viral transcription program and suggests a role for mcv-miR-M1 in episomal persistence. PLoS Pathog 2015; 11:e1004974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ma J, Jiang T, Tan L, Yu JT. TYROBP in Alzheimer's disease. Mol Neurobiol 2015; 51:820–6. [DOI] [PubMed] [Google Scholar]
- 10. Zhang B, Gaiteri C, Bodea LG, et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer's disease. Cell 2013; 153:707–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Connolly K, Lehoux M, O’Rourke R, et al. Potential role of chitinase-3-like protein 1 (CHI3L1/YKL-40) in neurodegeneration and Alzheimer's disease. Alzheimers Dement 2023; 19:9–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Moreno-Rodriguez M, Perez SE, Nadeem M, Malek-Ahmadi M, Mufson EJ. Frontal cortex chitinase and pentraxin neuroinflammatory alterations during the progression of Alzheimer's disease. J Neuroinflammation 2020; 17:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dohgu S, Ryerse JS, Robinson SM, Banks WA. Human immunodeficiency virus-1 uses the mannose-6-phosphate receptor to cross the blood-brain barrier. PLoS One 2012; 7:e39565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cortese I, Muranski P, Enose-Akahata Y, et al. Pembrolizumab treatment for progressive multifocal leukoencephalopathy. N Engl J Med 2019; 380:1597–605. [DOI] [PubMed] [Google Scholar]
- 15. Boumaza X, Bonneau B, Roos-Weil D, et al. Progressive multifocal leukoencephalopathy treated by immune checkpoint inhibitors. Ann Neurol 2023; 93:257–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.