Abstract
Detecting severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) reinfections is challenging with current serology assays and is further complicated by the marked decrease in routine viral testing practices as viral transmission increased during Omicron. Here, we provide proof-of-principle that high-avidity anti-nucleocapsid (N) antibodies detects reinfections after a single infection with higher specificity (85%; 95% confidence interval [95% CI], 80%–90%) compared to anti-N antibody levels (72%; 95% CI, 66%–79%) in a vaccinated cohort. This method could be used to retroactively investigate the epidemiology and incremental long-term health consequences of SARS-CoV-2 reinfections.
Keywords: SARS-CoV-2, COVID-19, avidity, serology, nucleocapsid, reinfection
Measuring the high-avidity anti-nucleocapsid (N) IgG detected SARS-CoV-2 reinfections after de novo infection with improved specificity compared to anti-N IgG levels. This method allows retroactive study of the epidemiology and health consequences of asymptomatic or unreported reinfections.
Antibody testing is the gold standard for diagnosing prior severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infections. Current methods are based on measuring anti-nucleocapsid (N) antibody titers, which increase following infection but not after vaccination [1]. In the context of limited viral testing and frequent asymptomatic infections, it becomes difficult to detect reinfections [2]. Solely measuring N antibody levels poorly identifies reinfections once someone has already been infected [1]. Siddiqui et al showed distinct antibody features after reinfections, including changes in anti-N immunoglobulin G (IgG) levels and Fc binding [3]. However, this approach relies on longitudinal paired sera and fold-change differences in antibody responses to detect reinfections. Therefore, a gap remains in methods for identifying reinfections in cross-sectional cohorts without paired sampling. In contrast to antibody levels, which wane rapidly after infection, avidity increases after reinfections [4–7] due to antigen-specific B-cell affinity maturation [8]. Here, we report a novel method to accurately detect SARS-CoV-2 reinfections based on antibody avidity.
METHODS
Cohorts, Sample Collection, and Grouping
Blood samples were selected from a longitudinal seroprevalence study (parent study) of 4900 school workers in Vancouver, British Columbia (BC), Canada followed from February 2021 to March 2023 [9, 10]. In the original study, participants were asked to complete serial health, and SARS-CoV-2 exposure and vaccination questionnaires, and to provide blood samples annually for diagnostic serology testing (see “diagnostic serology testing” paragraph below). Data on vaccine doses (dates and types) and infections confirmed by viral nucleic acid amplification testing (NAAT) or rapid antigen testing (RAT) on nasal swabs were obtained. Sera were collected by venipuncture in gold-top serum separator tubes (catalog No. 367989; BD Biosciences). After 30–60 minutes at room temperature, tubes were centrifuged at 1400g for 10 minutes and frozen at −80°C within 4 hours. Written consent was obtained from all participants. The study was approved by the University of British Columbia Children's and Women's Research Ethics Board (H20–03593).
To optimize the chaotrope concentration for detection of changes in SARS-CoV-2 N antibody avidity profiles following reinfection, we used sera from 8 uninfected, 27 infected once, and 12 infected twice individuals (optimization cohort). For further determination of the accuracy of the anti-N antibody avidity assay for detection of 1 versus 2 infections, a separate set of sera were retrieved for a validation cohort of 183 individuals infected once and 46 individuals infected at least twice.
Blood samples for the optimization and validation cohorts were selected based on number of infections confirmed by diagnostic viral testing and serology results. To minimize misclassification, uninfected samples were also selected from the earliest longitudinal serum collections when community seroprevalence was very low, and therefore least likely to contain a false negative. Sera from uninfected individuals were thus selected based on having no history of positive viral NAAT/RAT, a negative diagnostic serology, and before April 2021, when SARS-CoV-2 seroprevalence in BC and in the parent cohort was less than 3% [10, 11]. Sera from individuals infected once (referred to as infected) were selected based on having a positive diagnostic serology or a history of only 1 infection confirmed by diagnostic viral NAAT/RAT collected after April 2022, after the first Omicron wave in BC. Sera from reinfected individuals were selected based on having a history of 2 or more positive diagnostic viral tests at least 30 days apart, confirmed by a positive diagnostic serology, from sera collected between February 2022 and January 2023 as the seroprevalence in BC rose from approximately 15% to 75% [11]. The study used a convenience sample size, as a proof-of-concept study.
Diagnostic serology testing on samples was performed using the Food and Drug Administration-licensed Ortho T VITROS Anti-SARS-CoV-2 Total Antibody Assay (Ortho Clinical Diagnostics) on a Vitros 5600 analyzer for unvaccinated individuals, and using the total antibody Roche Elecsys Anti-SARS-CoV-2 anti-N assay (Roche) on a Cobas e601 analyzer for vaccinated individuals. For both assays, a positivity cutoff ≥1.00 was used.
Anti-N Avidity Antibody Profiling
The current method was adapted from a previous one for anti-pertussis toxin vaccine responses [12]. In the current study, a chaotrope solution of ammonium thiocyanate (98.9% NH4SCN 4.0 M; catalog No. A1479; Millipore Sigma) was prepared in 1 × phosphate-buffered solution (PBS), serially diluted to working solutions of 2.0, 1.0, 0.75, and 0.5 M. These concentrations were selected to resolve a broad avidity profile consisting of very low (0–0.5 M; weakest binding), low (0.5–0.75 M), medium (0.75–1.0 M), high (1.0–2.0 M), and very high avidity (>2.0 M; strongest binding). To obtain the relative fractional anti-N avidity, IgG lost at each step of thiocyanate increasing from 0 to 2.0 M was quantified relative to the total anti-N IgG level in an untreated well. For example, if 1 of 10 of all anti-N IgG were stripped between 0 and 0.5 M (very low binding), the very low avidity fraction would represent 10% of the avidity profile. The fraction of high-avidity IgG antibodies was determined by the amount of residual antibody removed between 1.0 and 2.0 M divided by the total anti-N IgG level (termed the high-avidity fraction). Antibodies remaining after 2.0 M constitute the highest avidity antibodies (ie, very high).
Anti-N IgG levels and avidity were measured using a modified V-PLEX SARS-CoV-2 Panel 31 (IgG) Kit (catalog No. K15642U; Meso Scale Diagnostics). Briefly, after blocking, prediluted sera in assay diluent (Diluent 100) were added in quintuplicate (5 times) along with standards and internal controls in duplicate. After 2 hours of incubation, plates were washed and either 1 × PBS or ammonium thiocyanate (0 to 2.0 M) was added to each plate, followed by incubation for 30 minutes at 37°C. After incubation, the manufacturer's protocol was resumed by washing and adding 100 µL of anti-human SULFO-TAG–conjugated IgG detection antibody. The levels of anti-N IgG antibody with and without the chaotrope (ie, PBS only) were extrapolated by a 4-parameter logistic regression standard curve (7-point + blank). Uninfected sera were tested at a 1:500 dilution. Infected and reinfected sera were tested at 1:5000 and 1:10 000 dilutions. For a complete experimental protocol see the Supplementary Materials.
Statistical Analyses
For optimization of the chaotrope concentration, very low, low, medium, high, and very high anti-N fractional antibody avidity, as a percentage of total anti-N antibody, were compared between infected and reinfected individuals of the optimization cohort using a 2-sample Wilcoxon signed rank test. To determine the performance of the avidity anti-N IgG method, an empirical receiver operating characteristic (ROC) curve was constructed using true-positive and false-positive rates at various cutoffs specified by the observed data. The area under the curve (AUC) and 95% confidence intervals (CIs) were calculated using the normal approximation and were reported as a metric of the overall assay accuracy. The cutoff value for anti-N high-avidity IgG antibodies that best discriminated 1 versus 2 infections was determined by maximizing the Youden index, according to the following formula: sensitivity + specificity − 1. The sensitivity and 1 − specificity at the optimized cutoff were reported with 95% CIs based on a normal distribution. Effects of time on IgG levels and high-avidity fraction were determined in linear regression models. Statistical analyses were performed in R version 4.22.
RESULTS
Optimization of Chaotrope Concentrations for Detection of Reinfections
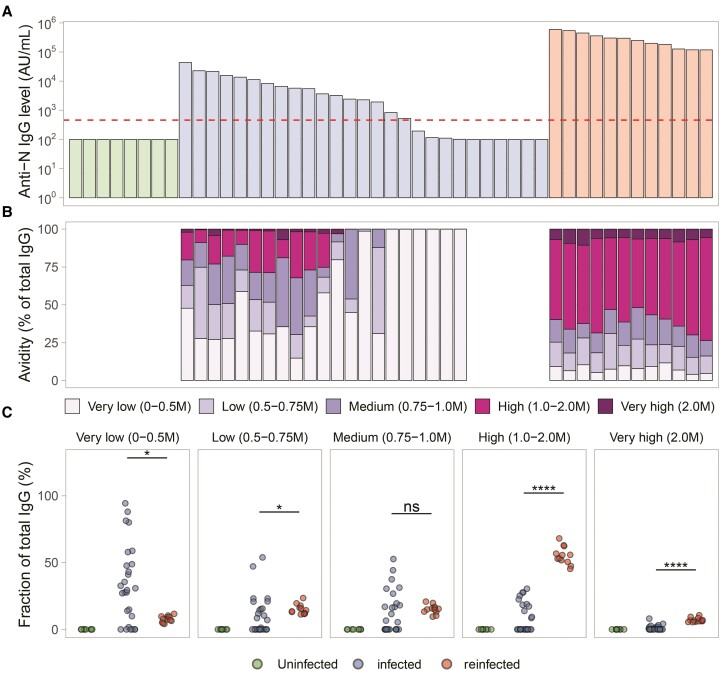
Baseline characteristics between the uninfected, infected (once), and reinfected groups in the optimization cohort are reported in Supplementary Table 1. Serum anti-N IgG levels from these individuals in the optimization cohort are shown in Figure 1A. As expected, bound anti-N IgG levels decreased with each increase in concentration of ammonium thiocyanate (Supplementary Figure 1). From these curves, the corresponding fraction of very low, low, medium, high, and very high avidity antibody were quantified as a percentage of N IgG antibody binding lost following increasing ammonium thiocyanate from 0 to 2.0 M for very low to high-avidity fractions, and residual N IgG binding at 2.0 M for quantification of the very high-avidity fraction (Figure 1B). Proportions of avidity fractions between groups are compared in Figure 1C. Statistically significant differences between the infection and reinfection groups were only observed in the high and very high-avidity antibody fractions (>1.0 M, P < .001; Figure 1C). The largest separation was observed in the high-avidity fraction (1.0 to 2.0 M). Based on this, we used the high (1.0 to 2.0 M) avidity fraction for further validation.
Figure 1.
Optimization of the antibody avidity method. A, Levels of anti-N IgG antibodies (AU/mL) between uninfected (n = 9), infected (n = 27), and reinfected (n = 12) individuals from the optimization cohort. The limit of detection of the assay is shown as a dashed red line. B, Anti-N IgG avidity profiles normalized to total IgG in corresponding individuals (vertically aligned with A). C, Fractions of anti-N IgG antibody detectable after increasing concentration of chaotrope agent, with averaged levels for uninfected, infected, and reinfected groups. Samples below the limit of detection of the assay (100 AU/mL) are not shown. Statistical comparisons were performed using 2-sample Wilcoxon signed rank tests. **** P < .0001, * P < .05, P > .05 ns. Abbreviations: IgG, immunoglobulin G; N, nucleocapsid; ns, not significant.
Performance of the Antibody Avidity Method
The age, sex, and number of vaccine doses were similar between infected and reinfected groups in the validation cohort, whereas the average time since the most recent infection was longer in the reinfected than infected group due to how samples were selected in relation to population seroprevalence, timing of SARS-CoV-2 waves, and blood sampling in the original cohort (Supplementary Table 2).
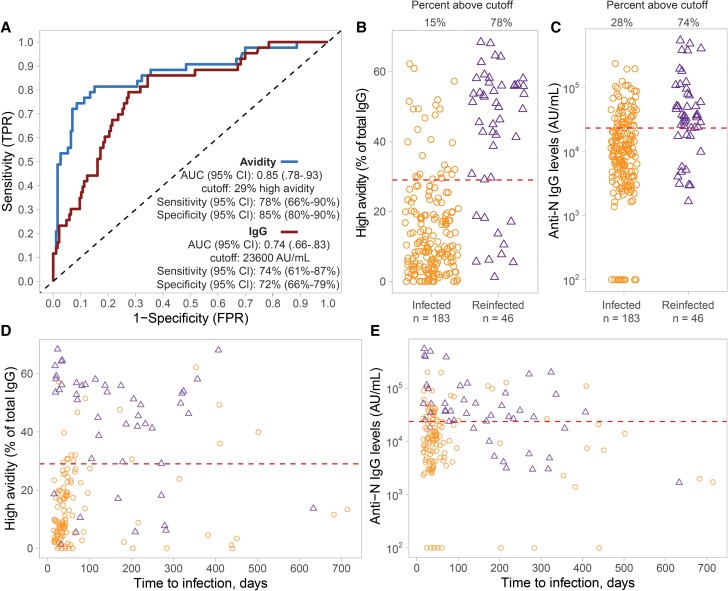
Based on ROC curves, the fraction of high-avidity anti-N IgG antibody over the total concentration of bound anti-N IgG (in the absence of ammonium thiocyanate) accurately classified infection and reinfections (AUC, 0.85; 95% CI, .78–.93). Moreover, this high-avidity fraction performed better in predicting reinfection than anti-N IgG levels (AUC, 0.74; 95% CI, .66–.83; Figure 2A). The cutoff thresholds for detecting reinfection for both avidity and IgG levels was determined, and the sensitivity and specificity of this optimized cutoff was calculated. The optimized cutoff for high avidity was 29% of total IgG. Using this cutoff, the sensitivity and specificity were 78% (95% CI, 66%–90%) and 85% (95% CI, 80%–90%), respectively. The optimized cutoff for anti-N IgG levels was 23 600 AU/mL, yielding a sensitivity of 74% (95% CI, 61%–87%) and specificity of 72% (95% CI, 66%–79%). The avidity method demonstrated improved specificity (ability to classify infections) compared to IgG levels (Figure 2B and 2C).
Figure 2.
ROC analysis of the performance of SARS-CoV-2 N antibody avidity method for predicting 1 or 2 SARS-CoV-2 infections. A, ROC curve for prediction of single versus 2 infections using the proportion of high-avidity anti-N IgG antibody fraction (blue line) versus anti-N IgG antibody levels (red line). B, Corresponding percentage of high-avidity N IgG antibody levels over the total N IgG antibody levels in infected (orange circles; n = 183) versus reinfected individuals (purple circles; n = 46). C, Corresponding anti-N IgG antibody levels in the same individuals. D, Proportion of high-avidity anti-N IgG antibody fraction and (E) anti-N IgG antibody levels between the same 2 groups over time to the most recent self-reported infection (days). The dashed lines in B to E represent the optimized cut-offs for detection of infection versus re-infections. Abbreviations: AUC, area under the curve; CI, confidence interval; FPR, false positive rate; IgG, immunoglobulin G; N, nucleocapsid; ROC, receiver operating characteristic; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2; TPR, true positive rate.
Using data from both infected (n = 117) and reinfected individuals (n = 46) who reported their infection date, there was no effect of time on avidity in the reinfection group, indicating a highly stable response (Supplementary Table 3). There was a significant decay in IgG levels in the reinfection group, but not in the infection group (Supplementary Table 4 and Figure 2D and 2E).
DISCUSSION
This proof-of-principle study demonstrated that quantifying high-avidity IgG levels against the SARS-CoV-2 N protein improved specificity for detecting reinfections compared to N IgG levels among individuals infected once. N IgG levels decreased over time in reinfected samples but remained stable in infected samples. Consequently, N IgG levels overlapped between the 2 groups with greater time since infection. In contrast, high-avidity IgG levels remained stable in the reinfected group and showed a statistically nonsignificant increasing trend in the infected group. Thus, high-avidity anti-N IgG predicted reinfection more accurately and remained more stable over time than anti-N IgG levels. To our knowledge, this is the first serology method to detect SARS-CoV-2 reinfections that does not require longitudinal samples.
Monitoring reinfections can be challenging in high-seroprevalence settings [2]. As public health authorities have now largely moved away from routine testing, drawing inferences solely based on viral testing data may carry a high risk of bias because it misses asymptomatic and/or unreported cases. The current method could be used to retrospectively track reinfections in existing cohorts. Even if most people have been infected multiple times, which may bolster their immunity, quantifying the trajectory of the health outcomes and incremental risk early on following the first few reinfections may be particularly important in understanding the resulting cumulative long-term health burden.
Our study has limitations. First, the sample size was limited for investigating long-term avidity changes more than 100 days postinfection. Similarly, this method, like other serology methods, may miss infections that occurred less than 14 days before sampling. Second, the empirical approach to validate the ammonium thiocyanate concentration used a predefined range of 1.0–2.0 M. In theory, the method could be optimized for detection of more than 1 reinfection, over longer time periods, but this still needs to be demonstrated. Third, it is possible that the infection status of some samples was misclassified, which could underestimate the accuracy of the method.
In conclusion, anti-N IgG antibody avidity can accurately detect SARS-CoV-2 reinfections after de novo infection. This method will be useful to quantify the incremental or cumulative risk, and the long-term health impact of reinfections, accounting for unreported and asymptomatic cases.
Supplementary Data
Supplementary materials are available at The Journal of Infectious Diseases online (http://jid.oxfordjournals.org/). Supplementary materials consist of data provided by the author that are published to benefit the reader. The posted materials are not copyedited. The contents of all supplementary data are the sole responsibility of the authors. Questions or messages regarding errors should be addressed to the author.
Supplementary Material
Contributor Information
Liam Golding, British Columbia Children’s Hospital Research Institute, University of British Columbia, Vancouver, British Columbia, Canada; Department of Pediatrics, Faculty of Medicine, University of British Columbia, Vancouver, British Columbia, Canada.
Allison W Watts, British Columbia Children’s Hospital Research Institute, University of British Columbia, Vancouver, British Columbia, Canada.
Jacob Shew, British Columbia Children’s Hospital Research Institute, University of British Columbia, Vancouver, British Columbia, Canada; Department of Pediatrics, Faculty of Medicine, University of British Columbia, Vancouver, British Columbia, Canada.
Marina Viñeta Paramo, British Columbia Children’s Hospital Research Institute, University of British Columbia, Vancouver, British Columbia, Canada; Department of Pediatrics, Faculty of Medicine, University of British Columbia, Vancouver, British Columbia, Canada; Women+ and Children's Health, Department of Obstetrics and Gynecology, University of British Columbia, Vancouver, British Columbia, Canada.
Louise C Mâsse, British Columbia Children’s Hospital Research Institute, University of British Columbia, Vancouver, British Columbia, Canada; School of Population and Public Health, University of British Columbia, Vancouver, British Columbia, Canada.
David M Goldfarb, British Columbia Children’s Hospital Research Institute, University of British Columbia, Vancouver, British Columbia, Canada; Department of Pathology and Laboratory Medicine, University of British Columbia, Vancouver, British Columbia, Canada.
Bahaa Abu-Raya, British Columbia Children’s Hospital Research Institute, University of British Columbia, Vancouver, British Columbia, Canada; Department of Pediatrics, Faculty of Medicine, University of British Columbia, Vancouver, British Columbia, Canada.
Pascal M Lavoie, British Columbia Children’s Hospital Research Institute, University of British Columbia, Vancouver, British Columbia, Canada; Department of Pediatrics, Faculty of Medicine, University of British Columbia, Vancouver, British Columbia, Canada; Women+ and Children's Health, Department of Obstetrics and Gynecology, University of British Columbia, Vancouver, British Columbia, Canada.
Notes
Acknowledgments . We thank the school staff who participated.
Financial support . This work was supported by the Government of Canada via its COVID-19 Immunity Task Force (2021-HQ-000130; to P. M. L. and L. C. M.). P. M. L. and L. C. M. were supported by salary awards from the British Columbia Children’s Hospital Research Institute. B. A. R. was support by a postdoctoral fellowship from Michael Smith Health Research (RT-2021-1868).
References
- 1. Fox T, Geppert J, Dinnes J, et al. Antibody tests for identification of current and past infection with SARS-CoV-2. Cochrane Database Syst Rev 2022; (11):CD013652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Team C-F. Past SARS-CoV-2 infection protection against re-infection: a systematic review and meta-analysis. Lancet 2023; 401:833–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Siddiqui SM, Bowman KA, Zhu AL, et al. Serological markers of SARS-CoV-2 reinfection. mBio 2022; 13:e0214121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Benner SE, Patel EU, Laeyendecker O, et al. SARS-CoV-2 antibody avidity responses in COVID-19 patients and convalescent plasma donors. J Infect Dis 2020; 222:1974–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Lofstrom E, Eringfalt A, Kotz A, et al. Dynamics of IgG-avidity and antibody levels after COVID-19. J Clin Virol 2021; 144:104986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Luo YR, Chakraborty I, Yun C, Wu AHB, Lynch KL. Kinetics of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) antibody avidity maturation and association with disease severity. Clin Infect Dis 2021; 73:e3095–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dapporto F, Marchi S, Leonardi M, et al. Antibody avidity and neutralizing response against SARS-CoV-2 Omicron variant after infection or vaccination. J Immunol Res 2022; 2022:4813199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Laidlaw BJ, Ellebedy AH. The germinal centre B cell response to SARS-CoV-2. Nat Rev Immunol 2022; 22:7–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Watts AW, Masse LC, Goldfarb DM, et al. SARS-CoV-2 cross-sectional seroprevalence study among public school staff in Metro Vancouver after the first Omicron wave in British Columbia, Canada. BMJ Open 2023; 13:e071228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Goldfarb DM, Masse LC, Watts AW, et al. SARS-CoV-2 seroprevalence among Vancouver public school staff in British Columbia, Canada: a cross-sectional study. BMJ Open 2022; 12:e057846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Skowronski DM, Kaweski SE, Irvine MA, et al. Serial cross-sectional estimation of vaccine-and infection-induced SARS-CoV-2 seroprevalence in British Columbia, Canada. CMAJ 2022; 194:E1599–E609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Abu-Raya B, Giles ML, Kollmann TR, Sadarangani M. Profiling avidity of antibodies elicited by vaccination using enzyme-linked immunosorbent assay-based elution—insights into a novel experimental and analytical approach. Vaccine 2020; 38:5389–92. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.