Abstract
A primary mechanism of lentivirus persistence is the ability of these viruses to evolve in response to biological and immunological selective pressures with a remarkable array of genetic and antigenic variations that constitute a perpetual natural experiment in genetic engineering. A widely accepted paradigm of lentivirus evolution is that the rate of genetic variation is correlated directly with the levels of virus replication: the greater the viral replication, the more opportunities that exist for genetic modifications and selection of viral variants. To test this hypothesis directly, we examined the patterns of equine infectious anemia virus (EIAV) envelope variation during a 2.5-year period in experimentally infected ponies that differed markedly in clinical progression and in steady-state levels of viral replication as indicated by plasma virus genomic RNA assays. The results of these comprehensive studies revealed for the first time similar extents of envelope gp90 variation in persistently infected ponies regardless of the number of disease cycles (one to six) and viremia during chronic disease. The extent of envelope variation was also independent of the apparent steady-state levels of virus replication during long-term asymptomatic infection, varying from undetectable to 105 genomic RNA copies per ml of plasma. In addition, the data confirmed the evolution of distinct virus populations (genomic quasispecies) associated with sequential febrile episodes during acute and chronic EIA and demonstrated for the first time ongoing envelope variation during long-term asymptomatic infections. Finally, comparison of the rates of evolution of the previously defined EIAV gp90 variable domains demonstrated distinct differences in the rates of nucleotide and amino acid sequence variation, presumably reflecting differences in the ability of different envelope domains to respond to immune or other biological selection pressures. Thus, these data suggest that EIAV variation can be associated predominantly with ongoing low levels of virus replication and selection in target tissues, even in the absence of substantial levels of plasma viremia, and that envelope variation continues during all stages of persistent infection as the virus successfully avoids clearance by host defense mechanisms.
Experimental infection of horses with equine infectious anemia virus (EIAV) results in a uniquely rapid and dynamic disease process that is characterized by three defined stages: acute, chronic, and long-term asymptomatic (19). The initial acute disease is usually observed within 3 to 4 weeks postinfection and is associated with high levels of viremia and clinical symptoms including fever, diarrhea, lethargy, edema, thrombocytopenia, and anemia. While some EIAV-infected horses may experience only a single episode of EIA, most infections typically progress to chronic EIA, characterized by repeated cycles of disease and associated waves of viremia. EIA disease cycles occur at irregular intervals, separated by weeks or months, with an average of six to eight disease episodes within the first year postinfection. At this time, most infected horses become asymptomatic for EIA indefinitely, presumably due to the development of enduring protective host immunity. These inapparent carriers, however, remain infected for life, with the maintenance of markedly different subclinical levels of steady-state virus replication (11, 12). Thus, EIAV offers a unique model for characterizing natural immunologic control of lentivirus replication and disease and for elucidating the nature and role of viral variation in persistence and pathogenesis.
Studies from our laboratory have previously demonstrated that the cyclic disease episodes observed during chronic EIA are associated with distinct viral populations that can be distinguished as genomic and antigenic variants (16, 22). Detailed molecular characterization of EIAV envelope variation during sequential disease cycles in experimentally infected ponies has revealed the presence of distinct EIAV envelope variants with each wave of viremia. These observations suggest that the cyclic nature of chronic EIA is due to the sequential production and selection of viral envelope variants that are able to temporarily escape established host immune responses. The predominant site of EIAV variation during persistent infection is the gp90 surface envelope glycoprotein, and the pattern of gp90 nucleotide and amino acid variation has been analyzed to define distinct conserved and variable protein domains (16) as observed with other animal and human lentiviruses (8, 15, 28–30). These characterizations of EIAV envelope variation and definition of conserved and variable envelope domains have been confirmed by others (33). While these studies have examined the nature of EIAV variation during chronic disease and clinical levels of virus replication (i.e., >107 copies of genomic RNA per ml of plasma), there is no information on the evolution of viral quasispecies in inapparent carriers during long-term asymptomatic infections and relatively low levels of viral replication.
Genomic and antigenic variation, as observed in EIAV, is a distinguishing characteristic of animal and human lentiviruses and is believed to result from errors made by the viral reverse transcriptase (RT) in copying genomic RNA to proviral DNA (1, 23, 26). Moreover, it is widely accepted that the potential for genomic variation is directly related to the levels of virus replication and associated rounds of reverse transcription and to the presence of a selective pressure, e.g., antibodies or antiviral drugs (3, 14, 32). According to this paradigm, one would predict a greater extent of envelope variation in EIAV-infected ponies that experience multiple disease cycles compared to infected ponies that become inapparent carriers after the initial acute episode. To test this hypothesis directly, we have in this study characterized and compared the patterns of envelope antigenic variation in four ponies experimentally infected in parallel with EIAV, also utilizing previous studies of the viral replication dynamics and host immune responses during the progression from chronic EIA to a long-term asymtomatic state (11). As a basis for this study the four experimentally infected ponies were fortuitously divided into two distinct groups, based on clinical course of disease and steady-state virus replication levels (11). Two of the ponies (561 and 562), designated nonprogressor ponies, experienced only a single acute disease episode and then remained asymptomatic for EIA for the entire observation period. Measurements of EIAV plasma genomic RNA levels in these ponies indicate a rapid suppression of viral replication after the acute disease episode and a maintenance of low levels of plasma viral genomic RNA (<102 RNA copies per ml) (11). In contrast, the other two experimentally infected ponies (564 and 567), designated progressor ponies, experienced six disease cycles characteristic of chronic EIA and maintained relatively high levels of plasma viral RNA (>104 copies per ml) during prolonged asymptomatic stages of infection during the 3-year observation period (11). In the present study, we have examined the patterns of viral envelope variation in these progressor and nonprogressor ponies to determine the influence of viral replication levels on the evolution of EIAV envelope proteins. This study also provided an opportunity to compare the evolution of EIAV envelope quasispecies during sequential disease cycles in two ponies infected in parallel with the same reference viral strain.
MATERIALS AND METHODS
Experimental infections, clinical evaluation, and sample collection.
Four outbred, mixed-breed ponies (animals 561, 562, 564, and 567) were experimentally inoculated intravenously with 103 50% tissue culture infective doses (TCID50) of the pathogenic strain EIAVPV. The clinical and immune responses in these experimentally infected ponies during persistent infection have been extensively described (10, 11). Rectal temperatures and clinical status were recorded daily. Clinical EIA episodes were determined on the basis of rectal temperature and platelet count in combination with the presence of infectious plasma virus (10, 11, 31). Whole-blood samples were fractionated for enumeration of platelets (Unopette microcollection system; Becton Dickinson, Rutherford, N.J.). Plasma samples were collected during each disease cycle (defined as rectal temperature above 39°C and platelet number below <100,000/μl of whole blood) and stored at −80°C until RNA extraction was performed.
Isolation of EIAV from MDM.
EIAV genomic RNA was isolated directly from the plasma of experimentally infected ponies during cycles of disease as previously described (16). However, previous data from our lab and others indicate that the infectious EIAV titer in plasma declines to undetectable levels during afebrile periods in infected ponies and that the level of plasma viral RNA may also be undetectable in asymptomatic carriers (11–13, 20). Thus, while the levels of EIAV circulating in the plasma during asymptomatic infections were detectable, viral RNA levels were too low to be amplified by our protocol of long-range RT-PCR. To overcome this limitation and obtain representative virus isolates during long-term asymptomatic infection, we isolated virus from the experimentally infected ponies by in vitro culture of monocyte-derived macrophages (MDM) obtained at 800 days postinfection (dpi) as described by Raabe et al. (24). The production of EIAV from the cultured macrophages was monitored by measurement of the RT activity in the supernatants (17). EIAV was successfully isolated from macrophages cultured from ponies 562, 564, and 567, but repeated attempts to isolate virus from pony 561 were uniformly unsuccessful.
RNA purification and RT-PCR.
Viral RNA was extracted from plasma samples or supernatants of RT-positive macrophage cultures by Trizol (Gibco BRL, Rockville, Md) treatment of virus pellets obtained by centrifugation at 120,000 × g at 4°C for 45 min. All primers were derived from the pEIAV19-2 proviral sequence (GenBank accession number U01866). In the following oligonucleotides, the EIAVPV specific sequences are indicated by uppercase letters, and the added restriction sites (EcoRI or KpnI) are indicated by lowercase letters. Reverse transcription of 2 to 5 μl of purified viral RNA was performed with the Superscript PreAmplification system (Gibco BRL) as specified by the manufacturer, using the EIAV-specific primer PV12AS (5′-cggggtaccccgTGAGTAGAGAATTATATTTATTAC-3′, nucleotides [nt] 8293 to 8270). Amplifications of the specific 3,901-bp fragment from plasma RNA samples obtained during febrile episodes and from viral RNA extracted from the MDM culture supernatant were performed as previously described (16), using the Elongase mix (Taq/Pyrococcus species GB-D DNA polymerase mixture; Gibco BRL) with a mixture of 60 mM Tris-SO4 (pH 9.1), 18 mM (NH4)2SO4, 1.5 mM MgSO4, 0.01 mM each deoxynucleoside triphosphate, 0.4 μM each primer, 2 μl of Elongase mix, and 3 μl of cDNA in a final volume of 50 μl. All the primers used for RT reaction and PCR were described in a previous publication (16). An initial PCR was performed with primers PV2S (5′-cggaattcCTCAGAGAGGGGATAAAGG-3′; nt 4276 to 4294) and PV12AS. A second seminested PCR was then carried out with primers PV11S (5′-ccggaattccggGTACAGGAGTATTCTGGGTAG-3′; nt 4303 to 4323) and PV12AS and 3 μl of the first PCR product. The following conditions were used: 1 min at 95°C for the initial denaturation step; 20 s at 95°C, 20 s at 50°C, and 6 min at 68°C for 35 cycles; and 10 min at 68°C for one cycle. For plasma RNA from episode V of pony 564 and episode III of pony 567, a smaller fragment corresponding to the variable regions V2 through V8 of gp90 was amplified with primers Var1 (5′-GTTCCTTCCCGGGGTGTAGACC-3′; nt 5692 to 5713) and Var2 (5′-GAGGAGTTATATTGGTTAAAGCTTTGG-3′; nt 6544 to 6518) as previously described (17).
Cloning of RT-PCR products.
Several independent RT-PCR products (at least two independent RT reactions and three or four independent PCRs) were generated from plasma samples taken during each febrile episode or viral RNA from the supernatants of MDM, purified using the Wizard PCR Preps DNA purification system (Promega, Madison, Wis.), and cloned. Due to the highly unstable nature of EIAV env sequences when associated with high-copy-number plasmids in transformed Escherichia coli (7), the 3,901-bp fragments of the EIAV genome were cloned into the polylinker of the low-copy-number vector pLG338 using the EcoRI and KpnI sites created in the sense (PV11S) and antisense (PV12AS) primers, respectively. The short RT-PCR products generated from episode V of pony 564 and III of pony 567 were cloned into the pGEM5Zf (+) T-A vector (Promega). The ligation products were used to transform competent E. coli DH5α. The clones were screened by a standard colony hybridization technique (27) using α 32P-labeled EIAV probe. The positive clones were then checked by restriction enzyme digestion for the proper-size insert.
Sequencing of RT-PCR Clones.
Plasmid DNA was extracted and purified with a midiprep kit (Qiagen, Valencia, Calif.). The clones were automatically sequenced with a Taq Dye Deoxy Terminator Cycle Sequencer kit (Applied Biosystems, Foster City, Calif.), using internal EIAV primers as previously described (16). DNA sequences were resolved with an ABI Prism 373 DNA sequencer (Applied Biosystems). Error rate associated with amplification with the Elongase DNA polymerase was previously determined to be 0.015% (3 substitutions per 19,510 bp sequenced) (16).
Sequence analysis.
The sequences were analyzed using the Genetics Computer Group package analyses software (9), Clustal W multiple sequence alignment program, and the Phylip package. Phylogenetic trees were generated with the Clustal W software after an alignment using the default parameters (complete sequence alignment, slow/accurate; gap open penalty, 15.00; gap extension penalty, 6.66; delay divergent, 40%). A Phylip file was generated to calculate the distance matrix with Prodist and DNAdist.
Nucleotide sequence accession numbers.
The sequences analyzed were submitted to GenBank and have been assigned accession numbers AF29866 through AF29870 for pony 561, AF298671 through AF298684 for pony 562, AF298685 through AF298702 for pony 564, and AF298703 through AF298762 for pony 567.
RESULTS
Clinical and virologic profiles of EIAVPV-infected ponies.
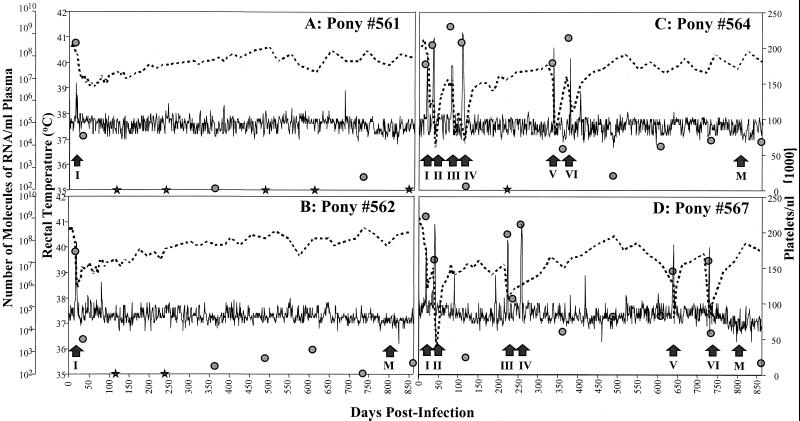
Four outbred ponies (561, 562, 564, and 567) were inoculated with 103 TCID50 of a reference stock of EIAVPV and monitored daily for EIA clinical symptoms as described by Hammond et al. (10). These experimental infections have been previously used to monitor the development of immune responses during the various stages of EIAV infection (10) and to characterize the evolution of viral quasispecies during early febrile episodes in pony 564 (16). Figure 1 summarizes the clinical profiles and virus replication levels over a 2.5-year observation period used as the basis for this study. Clinical EIA episodes were defined by rectal temperature above 39°C and by platelet count below 100,000 platelets per 1.0 μl of whole blood. All of the indicated EIA clinical episodes were associated with fever, thrombocytopenia, and plasma viral genomic RNA levels in excess of 107 copies per ml. After inoculation with EIAVPV, two markedly different profiles of clinical progression were observed. Ponies 561 and 562 experienced only a single febrile episode at 17 days postinfection (Fig. 1), corresponding to the initial acute disease episode, and these ponies remained asymptomatic for EIA for at least 4 years (unpublished data). Based on the lack of development of chronic EIA, these two ponies are designated for purposes of comparison as nonprogressors. In striking contrast to these nonprogressors, ponies 564 and 567 each experienced six febrile episodes at days 18 (I), 34 (II), 80 (III), 106 (IV), 337 (V), and 378 (VI) postinfection for 564 and at days 19 (I), 40 (II), 223 (III), 258 (IV), 640 (V), and 729 (VI) postinfection for pony 567 (Fig. 1). The development of recurring EIA disease episodes within the first year postinfection is characteristic of chronic EIA. Thus, these two ponies are referred to as progressors.
FIG. 1.
Clinical and virological profiles of ponies experimentally infected with EIAVPV. Ponies were experimentally infected with 103 TCID50 of the biological clone EIAVPV. Rectal temperature (——) and platelet count (⋯) were followed daily for up to 900 days (x-axis) after inoculation. Quantitation of the virus load ( ) was performed on viral RNA extracted from plasma at periodic time points during the fever episodes and asymptomatic stages. The symbol ✯ indicates a virus load below 100 copies. Febrile episodes I to VI were defined by a rectal temperature above 39°C in conjunction with a reduction in the number of platelets below 100,000/μl of whole blood and other clinical symptoms of EIA. M, virus isolation from macrophages. Data are adapted in part from reference 11.
) was performed on viral RNA extracted from plasma at periodic time points during the fever episodes and asymptomatic stages. The symbol ✯ indicates a virus load below 100 copies. Febrile episodes I to VI were defined by a rectal temperature above 39°C in conjunction with a reduction in the number of platelets below 100,000/μl of whole blood and other clinical symptoms of EIA. M, virus isolation from macrophages. Data are adapted in part from reference 11.
In addition to the differences observed in the clinical progression of disease in the progressor and nonprogressor ponies, there was a clear difference between the two groups with respect to the steady-state levels of virus replication during asymptomatic stages of infection. Steady state levels of plasma viral genomic RNA in the two progressor ponies ranged from 104 to 105 copies per ml, while the nonprogressor ponies typically maintained plasma virus levels ranging from undetectable to 102 copies per ml (Fig. 1). Thus, the apparent levels of virus replication during asymptomatic infection in the progressor ponies was at least 104-fold greater than that observed in the nonprogressor ponies. In addition, the multiple febrile episodes experienced by the two progressor ponies were associated with waves of viremia that averaged 109 copies of EIAV genomic RNA per ml. Based on these distinguishing differences in virus replication in the progressor and nonprogressor ponies, the following studies were performed to compare the patterns of envelope variation in parallel persistent infections with a common virus inoculum and to evaluate the correlation between EIAV envelope variation and the steady-state levels of virus replication during long-term persistent infection.
Envelope variation during disease and viremia cycles associated with acute and chronic EIA.
Previous studies from our lab (16, 17, 21) and others (33, 34) indicate that EIAV envelope variation during persistent infection is localized to specific segments of the gp90 surface glycoprotein, with negligible variation observed in the gp45 transmembrane protein. Therefore, for this study we focused on the 1.3-kb segment of the env gene encoding the gp90 envelope protein of EIAV. The EIAV gp90 sequences were determined using viral RNA recovered from the four experimentally infected ponies during a combined total of 14 febrile episodes and from in vitro supernatants of MDM obtained during asymptomatic infection in each pony at 800 dpi. A total of 140 clones were subjected to sequence analysis. We previously described the envelope variation observed in pony 564 during the first five disease cycles (16). In the present study, we added a sequence analysis of viral isolates associated with the sixth fever episode in pony 564 with the six episodes observed in pony 567 and the single episode in ponies 561 and 562.
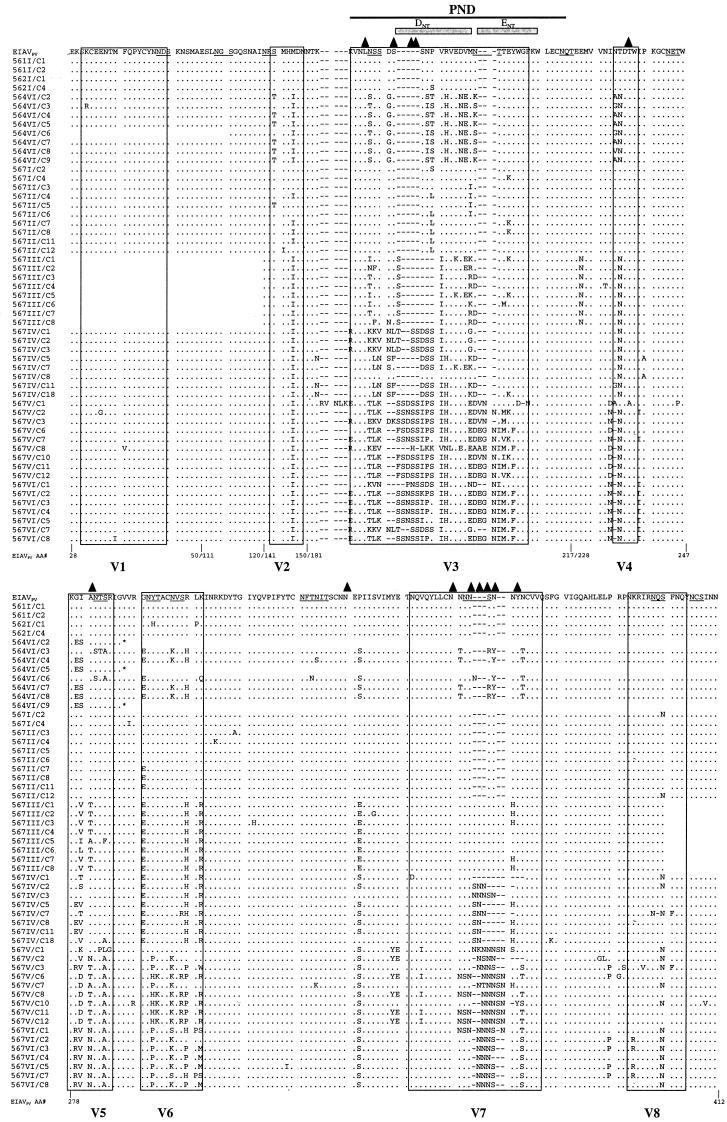
We compared the gp90 nucleotide and deduced amino acid sequences of the different clones derived from the virus recovered during the different febrile episodes to the EIAVPV inoculum nucleotide and deduced amino acid sequences (Fig. 2). After inoculation of EIAVPV into the ponies, the virus rapidly accumulated nucleotide changes in gp90. The overall percentages of divergence from the parental infectious virus observed in nucleotide and deduced amino acid sequences are summarized in Table 1. These data indicate that the virus species recovered in all cases, even the first febrile episodes, were different at the nucleotide and amino acid levels from the inoculated strain at rates higher than the PCR error rate of 0.015% and thus evidently reflective of in vivo changes occurring during the first several weeks post infection. The mutations observed among virus isolates from the first clinical episodes in all four ponies (between 17 and 21 dpi) were different from one animal to the other (Fig. 2). This pattern of envelope variation indicated in vivo evolution of the viral envelope as opposed to a selection of minor species contained in the EIAV inoculum.
FIG. 2.
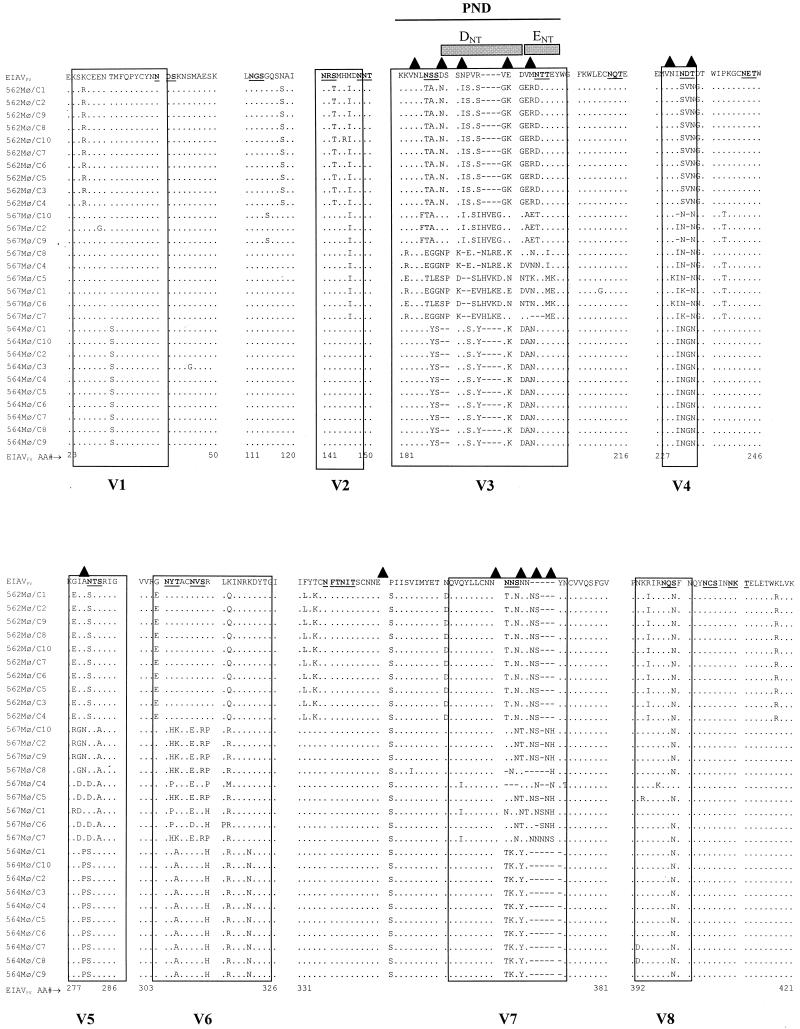
Comparison of deduced amino acid gp90 variable region sequences from EIAV isolates during sequential disease cycles in persistently infected ponies. The region of the env gene coding for the surface glycoprotein was sequenced from EIAVPV stock and from plasma viral RNA collected during febrile episodes of EIAVPV-infected infected ponies 561 (I), 562 (I), 564 (VI), and 567 (I through VI). Deduced amino acid (AA) sequences were aligned and compared to the EIAVPV inoculum sequence. Only amino acid residues different from those in EIAVPV are shown. Dots indicate residues identical to the EIAVPV sequence; dashes indicate amino acid deletions; underlined amino acids indicate potential N-glycosylation sites (NXS/T); triangles indicate newly created potential N-glycosylation sites. Previously described variable regions V1 through V8 (16) are boxed. The PND with the major neutralizing epitopes (ENT and DNT; delineated by gray boxes) localized in the V3 region (2) is indicated by a black line.
TABLE 1.
Accumulation of mutations in gp90 of the env gene
| Pony | % of envelope divergence from EIAVPV (mean ± SD)
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nucleotide
|
Amino acid
|
|||||||||||||
| Disease cycle
|
Macrophage | Disease cycle
|
Macrophage | |||||||||||
| I | II | III | IV | V | VI | I | II | III | IV | V | VI | |||
| 561 | 0.03 ± 0.05 | NAa | NA | NA | NA | NA | NA | 0.11 ± 0.14 | NA | NA | NA | NA | NA | NA |
| 562 | 0.12 ± 0.04 | NA | NA | NA | NA | NA | 2.97 ± 0.014 | 0.28 ± 0.21 | NA | NA | NA | NA | NA | 8.14 ± 0.37 |
| 564 | 0.36 ± 0.26 | 0.57 ± 0.31 | 0.57 ± 0.22 | 0.76 ± 0.14 | 2.27 ± 1.4 | 1.92 ± 0.11 | 2.15 ± 0.11 | 0.41 ± 0.34 | 1.53 ± 0.61 | 1.54 ± 0.55 | 2.24 ± 0.34 | 7.86 ± 0.96 | 5.21 ± 0.15 | 5.08 ± 0.17 |
| 567 | 0.2 ± 0.10 | 0.41 ± 0.17 | 1.63 ± 0.17 | 1.55 ± 0.11 | 2.88 ± 0.10 | 3.04 ± 0.49 | 3.22 ± 0.51 | 0.41 ± 0.13 | 1.01 ± 0.38 | 5.12 ± 0.42 | 3.22 ± 0.35 | 7.46 ± 0.69 | 7.67 ± 2.97 | 7.44 ± 0.48 |
NA, not applicable.
For the progressor ponies (564 and 567), envelope mutations in the viral isolates continued to increase during the subsequent febrile episodes or chronic stage of disease, indicating an increased divergence of the viral populations from the infecting virus (Table 1 and Fig. 2). As depicted in Fig. 2, these divergent populations were not representative of accumulating mutations but signified the appearance of entirely new quasispecies. The average nucleotide and amino acid divergence increased in viral isolates of pony 567 from 0.2 and 0.4%, respectively, in disease cycle I to 3.0 and 7.7%, respectively, in disease cycle VI. Pony 564 virus species experienced similar divergence from disease cycles I to VI, with the nucleotide and amino acid percentages increasing respectively from 0.36 and 0.41% to 1.92 and 5.21%.
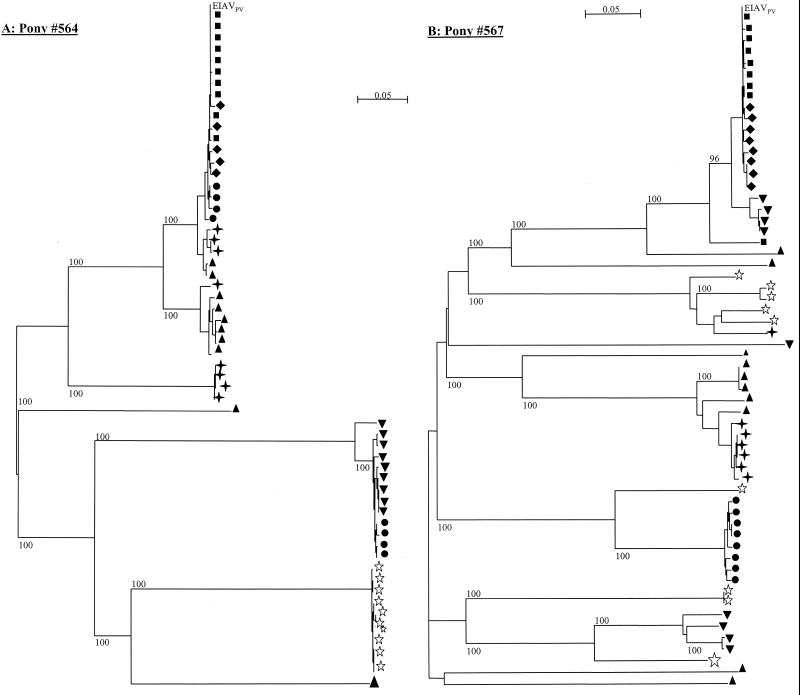
To illustrate distinctly the relationship between the viral strains associated with sequential febrile episodes in these progressor ponies, we constructed phylogenetic trees with bootstrap analysis using the nucleotide sequence encompassing variable regions V2 to V8 of the gp90 obtained from pony 564 (Fig. 3A) and pony 567 (Fig. 3B), using EIAVPV as the outgroup sequence. This analysis showed that almost all of the clones isolated from plasma RNA obtained during one febrile episode clustered together and that each febrile episode was accompanied by the emergence of novel quasispecies. Viral variants appeared to be eliminated with the cessation of a disease cycle, and a new population of viral variants was associated with the subsequent cycle of disease. This pattern of envelope variation is compatible with the model of chronic EIA in which each disease cycle is caused by a variant population of virus that is able to temporarily escape host immune surveillance.
FIG. 3.
Phylogenetic study of the gp90 nucleotide sequences of longitudinal viral isolates from progressor ponies 564 and 567. Phylogenetic trees were constructed with the neighbor joining method using V2 to V8 nucleotide sequences derived from plasma taken during fever I (■), fever II (⧫), fever III (●), fever IV (▾), fever V (▴), and fever VI (✦) and from MDM cultures obtained during asymptomatic stage at 800 dpi (⋆). Bootstrap values were determined over 1,000 iterations and are indicated at the nodes of the branches. Branch lengths are proportional to the distance existing between the sequences.
Nature of amino acid variation of gp90 during sequential febrile episodes.
Deduced amino acid data demonstrated a dynamic evolution of the viral envelope quasispecies and distinct patterns of viral envelope evolution associated with consecutive febrile episodes in parallel infections of the progressor ponies 564 and 567. Viral species evolution in the nonprogressor ponies (561 and 562) could be evaluated only through the analysis of the asymptomatic species, the ponies having experienced only one fever episode, and thus will be discussed below. During the febrile episodes observed in the progressor ponies, envelope variation was localized predominantly in the eight segments of the gp90 previously defined as variable domains (Fig. 2) and mutations were localized predominantly in V3, V5, V6, and V7.
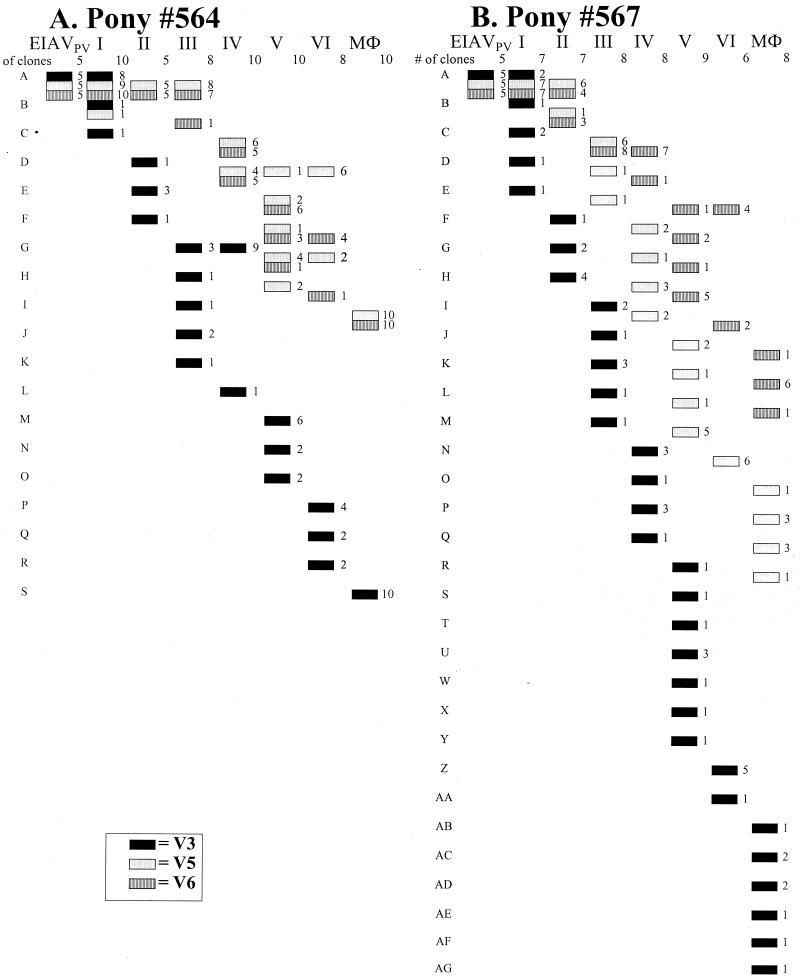
We compared the temporal generation of variant amino acid sequences of the emergent envelope species in three highly mutated regions (V3, V5, and V6) showing extensive variation in both progressor ponies 564 and 567 (Fig. 4). For pony 567, even as early as the first fever, only two of the sequenced clones had a V3 sequence identical to that of EIAVPV. By the second fever, all clones were different from EIAVPV and none of the sequences present during the first fever were maintained. The same pattern of evolution was observed for pony 564 (Fig. 4) up to the last analyzed fever, with one complex population replacing the previous quasispecies. The same pattern was observed for V5 and V6 in both ponies, but novel envelope sequences appeared more slowly and were less complex than observed for V3. In general, new envelope quasispecies that typically were present at only a single disease cycle rapidly replaced the inoculum EIAVPV species.
FIG. 4.
Temporal evolution of gp90 envelope V3, V5, and V6 domains of longitudinal viral populations in progressor ponies 564 (A) and 567 (B). The deduced amino acid sequences of gp90 variable regions V3, V5, and V6 are depicted as species A through S (pony 564) or A through AG (pony 567), where only population A (the inoculum) is common to the two ponies. The number of clones containing each genetic species is listed in the column after the species symbol, with the total number of clones analyzed indicated at the top of the same column. Viral plasma species were determined for the fever episodes (I through VI), while MDM (MΦ) viral species were characterized for long-term asymptomatic infection.
The different viral populations experienced a wide array of alterations, be they insertions, deletions, or point mutations of amino acids within the defined gp90 variable domains of the envelope. An example of the type of sequence mutations that occurred is found in the V7 region of viral isolates from pony 567. An asparagine-rich region originally containing 7 N residues was expanded to 11 residues, with the largest portion of the additional asparagines introduced as insertions rather than point mutations (Fig. 2). The frequent modification of viral envelope glycosylation sites was one of the most conspicuous changes. During the course of disease in pony 564, 33.3% (6 of 18) of the potential N-linked glycosylation sites (NXS/T) in gp90 were modified, and in pony 567 viral isolates, 38.9% (7 of 18) of potential sites were modified (Fig. 2). These glycosylation site mutations included deletions, additions, and repositioning of potential N-linked glycosylation sites in the viral gp90 sequence. In pony 564 viral isolates, the modifications consisted of five deletions, two additions, and two repositionings of glycosylation sites compared to the original EIAVPV sequence. Pony 567 viral envelope modifications resulted in three repositionings, four deletions, and four additions (Fig. 2). N-glycosylation site 5 in gp90 (V3 region) of pony 567 fever V and VI isolates experienced two changes within the same original EIAVPV glycosylation site, a shift and the creation of a new site through the process of point mutation and insertion (Fig. 2). The propensity for variations in gp90 glycosylation implies that this protein modification is an important determinant of EIAV envelope immunological properties.
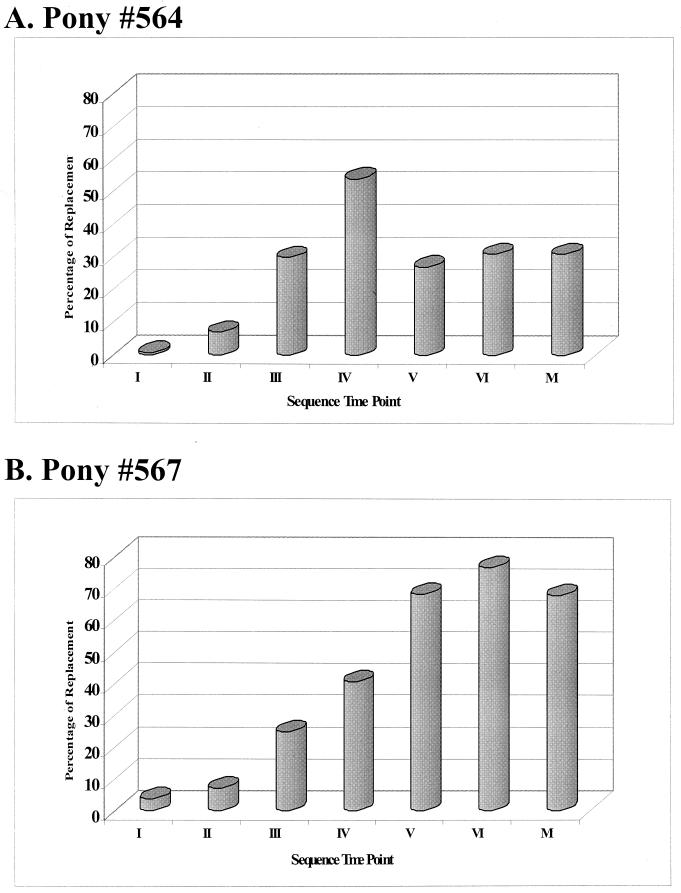
The most rapidly evolving region of gp90 throughout the multiple febrile episodes appeared to be the V3 region encompassing the EIAV principal neutralizing domain (PND) (12) (Fig. 2). The EIAVPV V3 amino acid residues of pony 564 were replaced at increasing rates starting at 0.7% in fever I to 54% by fever IV (Fig. 5). The rate of replacement of pony 567 viral isolates increased at a stunning rate, evolving from 2.6% at the first fever to 71% by fever VI (Fig. 5). The changes in the V3 domain were localized mainly to the neutralizing epitope E (ENT), in the second half of the V3 loop (Fig. 2). Evolution of V3 was the result of deletion, insertion, and/or point mutations. Virus populations evolved independently in the two progressor ponies. For example, the large deletion present in the envelope sequence during pony 564's third febrile episode was not observed in the virus sequences of pony 567. Small duplications such as the KKV motif by fever IV, VNL motif by fever V, or SSN motif by fevers V and VI (Fig. 2) characterized V3 evolution in pony 567 viral envelopes. In essence, the individual V3 mutations were not definitively fixed in the virus genomes from one fever to the next or from one pony to another.
FIG. 5.
V3 region replacement during progression of the infection in ponies 564 and 567. The percentage of V3 amino acid variation from the EIAVPV inoculum was calculated for sequential viral isolates. Percentage of variation was calculated by dividing the number of alterations within the designated V3 region by the total number of amino acids in the inoculum EIAVPV V3 region. Amino acid substitution, insertions, and deletions were all included in the assessment of V3 domain variation during sequential disease cycles (I through VI) and after long-term asymptomatic infection (MΦ).
gp90 variation during long-term asymptomatic infection.
We next sought to evaluate ongoing viral variation during asymptomatic stages of infection by characterizing prevalent EIAV species at 800 days postinoculation in the experimentally infected ponies. While the levels of EIAV plasma RNA were detectable during asymptomatic periods in our viral load short RT-PCR (11), the same levels of viral RNA were too low to be amplified by our protocol of long-range RT-PCR. We were unable to amplify the env fragment from viral RNA, even from large volumes of plasma (data not shown). To characterize the viral species in asymptomatic animals, we isolated MDM from whole blood sampled at 800 dpi and cultured the macrophage for 8 to 14 days for EIAV production. Viral RNA was repeatedly isolated from the pelleted virus of RT-positive MDM culture supernatants from ponies 562, 564, and 567. In contrast, supernatants from pony 561 MDM were routinely negative for RT activity, indicating a very low level of infection of blood monocytes that was also observed in the viral load assay (Fig. 1). Attempts to RT-PCR amplify viral RNA from 561 supernatants were also reproducibly negative. Therefore, the analysis of inapparent carrier virus population was focused on the two progressor (564 and 567) and one nonprogressor (562) pony.
Analysis of the clones derived from the MDM-derived viral RNA from each pony in the inapparent carrier state demonstrated that the viral populations continued to evolve during the asymptomatic stages of infection despite the associated low levels of virus replication (Fig. 6). For the progressor ponies (564 and 567), gp90 variation continued to increase at the nucleotide level (2.15 and 3.22%, respectively) but leveled off at the amino acid level (5.08 and 7.44%, respectively) (Table 1), suggesting multiple hits at the same nucleotide position. Although the percentage rates of variation were similar to the last disease cycle, the gp90 species were clearly different from the previous clinical episode viral isolates (Fig. 2, 3, 4, and 6). Thus, even at the low level of plasma viral replication during the inapparent stage of disease, the viral species continued to evolve. The virus species recovered from nonprogressor pony 562 also displayed a significant level of evolution regardless of the fact that the pony experienced an extremely different clinical progression with only a single fever episode. Even in the absence of multiple clinical episodes, the degree of variation for pony 562 viral isolates was 2.97% at the nucleotide and 8.14% at the amino acid level (Table 1).
FIG. 6.
Comparison of amino sequences of gp90 derived from clones obtained during asymptomatic stages of infection. Deduced amino acid (AA) sequences recovered from viral RNA purified from supernatants of MDM taken at 800 dpi from ponies 562, 564 and 567 are compared to the EIAVPV sequence. Only residues different from those in EIAVPV are shown. Dots indicate identical residues, dashes indicate deletions of residues; underlined amino acids indicate potential N-glycosylation sites (NXS/T); triangles indicate newly created potential N-glycosylation sites. Previously described variable regions V1 through V8 (16) are boxed. The PND with the major neutralizing epitopes (ENT and DNT; delineated by gray boxes) localized in the V3 region (2) is indicated by a black line. For each pony, only clones with different sequences are represented.
In both progressor and nonprogressor ponies, the persistent evolution during long-term asymptomatic infection culminated in the creation of new quasispecies that were distinct from earlier viral populations associated with disease cycles (Fig. 3 and 6). The modifications that occurred in the long-term asymptomatic populations consisted of a wide collection of changes including insertions, deletions, and point mutations. The nature of the amino acid sequence variation observed in the isolates revealed some common patterns of evolution between the progressor and nonprogressor ponies, the majority of changes occurring in the variable regions, primarily V3 through V7, with V3 being the most variable (Fig. 4 and 6). The V7 region, containing the asparagine-rich span of amino acids, was a hot spot for insertions again in ponies 562 and 567, introducing a region with up to 12 N residues rather than the original 7 N residues. These insertions in V7 were in different locations than observed in the last febrile episode, indicating they are unique sites of evolution and not merely retained from the previous febrile episode viral population. A notable change in the amino acid sequence was the modification or creation of glycosylation sites. The percentage of modification was generally greater than that observed in sequential disease episodes, with 44% (8 of 18) of the potential N-glycosylation sites modified for pony 567 viral isolates (five repositionings, six deletions, and four additions), 33.3% (6 of 18) modified for pony 562 isolates (three repositionings, three deletions, and four additions), and 33.3% (6 of 18) modified for pony 564 isolates (two shifts, four deletions, and one new site created).
While the general location and type of mutations were comparable both between progressor and nonprogressor ponies and between chronic and asymptomatic stages of disease, each pony evolved individually distinct changes that not only distinguished one pony from another but also separated one stage of disease from another. However, interesting common mutations involving the creation and repositioning of N-linked glycosylation sites can be found in all three ponies. The first common mutation is a point mutation at amino acid 234 in the V4 region. The threonine-to-asparagine mutation shifts a predicted potential N-linked glycosylation site. It first appears in the third febrile episode in ponies 564 and 567, is carried through the rest of the disease episodes, and is found in all of the asymptomatic clones, including that from the nonprogressor pony, 562. The second common alteration is also a point mutation and is found at amino acid 345, which lies between the V6 and V7 regions (Fig. 2 and 6). This proline-to-serine mutation also appeared in the envelope sequence of the third febrile episode of ponies 564 and 567 and in all three ponies' macrophage clones (Fig. 2). This mutation may not occur in a defined variable region, however, it creates a potential N-linked glycosylation site, which could play an important role in immune evasion. These mutations are unique because they are the only two variations among numerous changes that are common to all infected ponies and retained throughout mid-disease cycle to inapparent states of infection.
The overall gp90 variation observed in progressor and nonprogressor ponies from the first fever cycle through inapparent disease can be more precisely assessed by the rate of fixation. The rate of fixation was determined from the formula R = D/2T, where R is the number of nucleotide substitutions per site per year, D is the mean pairwise nucleotide distance between EIAVPV and the pony-derived samples, and T is the length of time after infection that the samples were isolated. This was estimated as a mean of the number of nucleotide substitutions per site per year. These calculations revealed fixation rates 5.48 × 10−2 for pony 561, 2.59 (±1.68) × 10−2 for pony 562, 3.56 (±2.54) × 10−2 for pony 564 and 2.43 (±1.16) × 10−2 for pony 567. Thus, these calculated rates of fixation indicated a continuous evolution of the viruses independently of the disease progression and steady-state levels of virus replication.
Taken together, these data demonstrate for the first time an ongoing evolution of EIAV envelope quasispecies during long-term asymptomatic infections, regardless of the low level of plasma viral replication that occurred during the inapparent stage. The rates of viral variation were similar in progressor and nonprogressor ponies with dramatically different disease profiles associated with markedly different steady-state levels of apparent virus replication.
DISCUSSION
The clearly defined cycles of disease and viremia characteristic of chronic EIA provide a uniquely dynamic model for investigating the nature and role of genomic and antigenic variation in lentivirus persistence and pathogenesis. Previous studies of viral variation during persistent infection have been limited primarily to the characterization of virus populations associated with sequential clinical episodes during chronic EIA in a single experimentally infected horse or pony (16, 33). In the present study, we have for the first time followed the evolution of viral quasispecies in four persistently infected ponies inoculated with the same reference EIAVPV inoculum but displaying markedly different clinical profiles (progressor versus nonprogressor) and steady-state levels of virus replication during long-term asymptomatic infection. Thus, this study represents the first detailed analyses of EIAV envelope variation during acute, chronic, and inapparent stages of infection over a 2.5-year observation period in parallel experimental infections. The results of these studies reveal important fundamental new aspects of EIAV variation that warrant reconsideration of current models of the mechanisms that drive lentivirus variation in vivo.
It has long been assumed that the rate of lentivirus envelope evolution under constant selective forces is directly proportional to the rate of viral replication, i.e., the more rounds of viral replication, the more opportunities for mutations by the viral RT as it copies viral genomic RNA to proviral DNA (6). This model of lentivirus variation predicts higher levels of lentivirus genomic variation in progressors than in long-term nonprogressors because of the higher steady-state levels of virus replication associated with clinical progression and chronic disease. In contrast to this prediction, however, we observed similar extents of EIAV envelope genomic and amino acid variation in viral isolates recovered at 28 months postinfection from two progressor ponies and one nonprogressor pony. The nonprogressor ponies experienced only a single acute disease episode, remained asymptomatic over the 28-month observation period, and maintained very low levels of plasma viral RNA (undetectable to <102 copies per ml). In contrast, the progressor ponies each experienced six disease episodes with accompanying viremia levels of >109 copies per ml and maintained plasma viral RNA levels of about 104 copies per ml during asymptomatic stages of infection. Taken together, these data indicate that the rate of EIAV envelope variation did not correlate with either the clinical progression or quantitative measures of viral replication as determined by EIAV viral RNA levels in plasma. These data then suggest that only relatively low levels of viral replication, perhaps in target tissues, as observed in nonprogressor ponies (12) is required to drive EIAV variation.
We have previously demonstrated low levels of virus infection and active replication in long-term asymptomatic carriers of EIAV, especially in macrophage-rich organs such as the spleen, liver, and kidney (12). We have also reported a similar development and maintenance of EIAV-specific humoral and cellular immune responses over a 3-year period in the four experimentally infected ponies, regardless of the pattern of clinical progression. This latter observation prompted us to propose that the enduring protective immunity observed in long-term inapparent carriers of EIAV may be due to the chronic low-level antigen production and immune stimulation in infected tissues (11). The present study indicates that this same low level tissue-associated virus replication is sufficient to generate viral variants and drive immune selection. According to this model of persistent EIAV infection, target tissues can be viewed as constant sources of new random viral quasispecies, even in the absence of evident disease or peripheral virus replication. As long as the immune system is able to recognize and control the evolving quasispecies, the viral replication remains suppressed and the infected animal is asymptomatic. However, when specific antigenic variants generated in tissues are able to escape established host immune surveillance, there is a rapid expansion of virus replication in tissue, high levels of plasma viremia, and disease until the immune system is able to reestablish control over the escape variants. Thus, the target tissues during asymptomatic infection can be viewed as having the potential to activate a viral infection at any time.
The individual mutations that occurred in the absence of disease in both nonprogressor and progressor ponies suggest that the observed changes are associated with viral evasion of the immune system. A majority of the alterations involved the creation, repositioning, or deletion of potential N-linked glycosylation sites. It has previously been shown that N-linked glycosylation plays a role in the escape of neutralizing antibodies and the protection of the virus from immune recognition in both simian and simian-human chimeric immunodeficiency viruses (4, 5, 25). It has also been suggested that N-linked glycosylation assists in lentivirus infection, as it has been demonstrated to facilitate interactions with the viral receptors CD4 and CCR5 in human immunodeficiency virus (18). Mutation of residues through deletion, repositioning, or creation of new N-linked glycosylation sites in the gp90 regions accessible to the immune system changes the accessibility of the envelope and therefore the residues that the immune system encounters. This alteration of the envelope could equip the virus with the ability to evade the immune system. The specific glycosylation sites that were created or repositioned in all three ponies during mid-disease cycle and carried through inapparent infection have a strong correlation with immune selection and neutralizing antibody. Preliminary studies ongoing in our lab have demonstrated it is at this point of disease that we find the concurrent appearance of neutralization resistance in the clones (unpublished data). This trend of neutralization resistance in the viral isolates appears to continue to the end of disease and therefore indicates that these could be potential important epitopes for immune evasion. The isolates and their neutralization characteristics are currently being examined.
By following the dynamic evolution of the virus populations in two chronically infected ponies, we showed that while the degrees of variation were similar in both progressor animals, the pattern of mutations from the EIAVPV strain was clearly different, with evidence of animal-specific signatures. This confirms that the mutations we observed effectively occurred in vivo and are not due to emergence of minor populations present in the inoculated strain and suggests that the phenomenon driving the changes is somehow specific to a given infected animal. These results appear to reflect the differential selection of virus populations based on tissue or cell tropism, viral fitness, and/or EIAV-specific immune response.
This extensive study of the gp90 evolution in disease progressor and nonprogressor EIAV-infected ponies in correlation with the results of our characterization of the immune response and viral replication in the same animals (11) raises important questions about protection against disease in infected animals. Mutations in gp90 are clearly associated with recurrence of the disease but alone are not sufficient to trigger clinical episodes. As we observed, the gp90 evolution rate did not correlate with disease progression, as demonstrated by the large number of mutations in the viral populations of a nonprogressor animal. The viral populations appear to continue to evolve constantly, regardless of low levels of plasma viremia. The majority of the new populations remain under immune control; however, when they are able to escape control, replication in the plasma soars and disease ensues, as in chronic EIA. As shown by the EIAV system, immune control against viral replication can be efficiently established and successfully fight against emerging virus populations. In infected ponies, host immune responses have matured to the point that virus variation occurs, but new populations are not able to escape established immune control. The cellular and humoral responses were not markedly different in progressor and nonprogressor animals. Therefore, undefined immune parameters that are able to control viral expansion in asymptomatic animals remain to be elucidated. They are the keys to elicit an effective protection against lentivirus-associated diseases.
ACKNOWLEDGMENTS
We thank Jonathan Steckbeck for meticulous editing of the manuscript, Beth Frost and John Cardamone for excellent technical assistance in DNA sequencing, and Gary Thomas and Brian Meade for animal care.
This work was supported by National Institutes of Health grant R01 AI 25850, by funds from the Lucille P. Markey Charitable Trust and the Kentucky Agricultural Experimental Station, and by a grant from the Pittsburgh Supercomputing Center through the NIH National Center for Research Resources, resource grant 2 P41 RR06009. C.L. was a recipient of postdoctoral grants from the Fondation pour la Recherche Medicale.
REFERENCES
- 1.Bakhanashvili M, Hizi A. Fidelity of DNA synthesis exhibited in vitro by the reverse transcriptase of the lentivirus equine infectious anemia virus. Biochemistry. 1993;32:7559–7567. doi: 10.1021/bi00080a030. [DOI] [PubMed] [Google Scholar]
- 2.Ball J M, Rushlow K E, Issel C J, Montelaro R C. Detailed mapping of the antigenicity of the surface unit glycoprotein of equine infectious anemia virus by using synthetic peptide strategies. J Virol. 1992;66:732–742. doi: 10.1128/jvi.66.2.732-742.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burns D P, Desrosiers R C. Envelope sequence variation, neutralizing antibodies, and primate lentivirus persistence. Curr Top Microbiol Immunol. 1994;188:185–219. doi: 10.1007/978-3-642-78536-8_11. [DOI] [PubMed] [Google Scholar]
- 4.Chackerian B, Rudensey L M, Overbaugh J. Specific N-linked and O-linked glycosylation modifications in the envelope V1 domain of simian immunodeficiency virus variants that evolve in the host alter recognition by neutralizing antibodies. J Virol. 1997;71:7719–7727. doi: 10.1128/jvi.71.10.7719-7727.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cheng-Mayer C, Brown A, Harouse J, Luciw P A, Mayer A J. Selection for neutralization resistance of the simian/human immunodeficiency virus SHIVSF33A variant in vivo by virtue of sequence changes in the extracellular envelope glycoprotein that modify N-linked glycosylation. J Virol. 1999;73:5294–5300. doi: 10.1128/jvi.73.7.5294-5300.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coffin J M. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 7.Cunningham T P, Montelaro R C, Rushlow K E. Lentivirus envelope sequences and proviral genomes are stabilized in Escherichia coli when cloned in low-copy-number plasmid vectors. Gene. 1993;124:93–98. doi: 10.1016/0378-1119(93)90766-v. [DOI] [PubMed] [Google Scholar]
- 8.Greene W K, Meers J, del Fierro G, Carnegie P R, Robinson W F. Extensive sequence variation of feline immunodeficiency virus env genes in isolates from naturally infected cats. Arch Virol. 1993;133:51–62. doi: 10.1007/BF01309743. [DOI] [PubMed] [Google Scholar]
- 9.Group G C. Program manual for the Wisconsin Package. Madison, Wis: Genetics Computer Group; 1994. [Google Scholar]
- 10.Hammond S A, Cook S J, Lichtenstein D L, Issel C J, Montelaro R C. Maturation of the cellular and humoral immune responses to persistent infection in horses by equine infectious anemia virus is a complex and lengthy process. J Virol. 1997;71:3840–3852. doi: 10.1128/jvi.71.5.3840-3852.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hammond S A, Li F, McKeon B M, Sr, Cook S J, Issel C J, Montelaro R C. Immune responses and viral replication in inapparent carrier ponies inoculated with equine infectious anemia virus. J Virol. 2000;74:5968–5981. doi: 10.1128/jvi.74.13.5968-5981.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harrold S M, Cook S J, Cook R F, Rushlow K E, Issel C J, Montelaro R C. Tissue sites of persistent infection and active replication of equine infectious anemia virus during acute disease and asymptomatic infection in experimentally infected equids. J Virol. 2000;74:3112–3121. doi: 10.1128/jvi.74.7.3112-3121.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim C H, Casey J W. In vivo replicative status and envelope heterogeneity of equine infectious anemia virus in an inapparent carrier. J Virol. 1994;68:2777–2780. doi: 10.1128/jvi.68.4.2777-2780.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kliks S, Contag C H, Corliss H, Learn G, Rodrigo A, Wara D, Mullins J I, Levy J A. Genetic analysis of viral variants selected in transmission of human immunodeficiency viruses to newborns. AIDS Res Hum Retroviruses. 2000;16:1223–1233. doi: 10.1089/08892220050116998. [DOI] [PubMed] [Google Scholar]
- 15.Leroux C, Chastang J, Greenland T, Mornex J F. Genomic heterogeneity of small ruminant lentiviruses: existence of heterogeneous populations in sheep and of the same lentiviral genotypes in sheep and goats. Arch Virol. 1997;142:1125–1137. doi: 10.1007/s007050050147. [DOI] [PubMed] [Google Scholar]
- 16.Leroux C, Issel C J, Montelaro R C. Novel and dynamic evolution of equine infectious anemia virus genomic quasispecies associated with sequential disease cycles in an experimentally infected pony. J Virol. 1997;71:9627–9639. doi: 10.1128/jvi.71.12.9627-9639.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lichtenstein D L, Issel C J, Montelaro R C. Genomic quasispecies associated with the initiation of infection and disease in ponies experimentally infected with equine infectious anemia virus. J Virol. 1996;70:3346–3354. doi: 10.1128/jvi.70.6.3346-3354.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ly A, Stamatatos L. V2 loop glycosylation of the human immunodeficiency virus type 1 SF162 envelope facilitates interaction of this protein with CD4 and CCR5 receptors and protects the virus from neutralization by anti-V3 loop and anti-CD4 binding site antibodies. J Virol. 2000;74:6769–6776. doi: 10.1128/jvi.74.15.6769-6776.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Montelaro R C, Ball J M, Rushlow K E. Equine retroviruses. In: Levy J A, editor. The retroviridae. Vol. 2. New York, N.Y: Plenum Press; 1993. pp. 257–360. [Google Scholar]
- 20.Oaks J L, Ulibarri C, Crawford T B. Endothelial cell infection in vivo by equine infectious anaemia virus. J Gen Virol. 1999;80:2393–2397. doi: 10.1099/0022-1317-80-9-2393. [DOI] [PubMed] [Google Scholar]
- 21.Payne S L, Fang F D, Liu C P, Dhruva B R, Rwambo P, Issel C J, Montelaro R C. Antigenic variation and lentivirus persistence: variations in envelope gene sequences during EIAV infection resemble changes reported for sequential isolates of HIV. Virology. 1987;161:321–331. doi: 10.1016/0042-6822(87)90124-3. [DOI] [PubMed] [Google Scholar]
- 22.Payne S L, Salinovich O, Nauman S M, Issel C J, Montelaro R C. Course and extent of variation of equine infectious anemia virus during parallel persistent infections. J Virol. 1987;61:1266–1270. doi: 10.1128/jvi.61.4.1266-1270.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Preston B D, Poiesz B J, Loeb L A. Fidelity of HIV-1 reverse transcriptase. Science. 1988;242:1168–1171. doi: 10.1126/science.2460924. [DOI] [PubMed] [Google Scholar]
- 24.Raabe M R, Issel C J, Montelaro R C. Equine monocyte-derived macrophage cultures and their applications for infectivity and neutralization studies of equine infectious anemia virus. J Virol Methods. 1998;71:87–104. doi: 10.1016/s0166-0934(97)00204-8. [DOI] [PubMed] [Google Scholar]
- 25.Reitter J N, Means R E, Desrosiers R C. A role for carbohydrates in immune evasion in AIDS. Nat Med. 1998;4:679–684. doi: 10.1038/nm0698-679. [DOI] [PubMed] [Google Scholar]
- 26.Roberts J D, Bebenek K, Kunkel T A. The accuracy of reverse transcriptase from HIV-1. Science. 1988;242:1171–1173. doi: 10.1126/science.2460925. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook J E, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 28.Simmonds P, Balfe P, Ludlam C A, Bishop J O, Brown A J. Analysis of sequence diversity in hypervariable regions of the external glycoprotein of human immunodeficiency virus type 1. J Virol. 1990;64:5840–5850. doi: 10.1128/jvi.64.12.5840-5850.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Starcich B R, Hahn B H, Shaw G M, McNeely P D, Modrow S, Wolf H, Parks E S, Parks W P, Josephs S F, Gallo R C, et al. Identification and characterization of conserved and variable regions in the envelope gene of HTLV-III/LAV, the retrovirus of AIDS. Cell. 1986;45:637–648. doi: 10.1016/0092-8674(86)90778-6. [DOI] [PubMed] [Google Scholar]
- 30.Suarez D L, Whetstone C A. Identification of hypervariable and conserved regions in the surface envelope gene in the bovine lentivirus. Virology. 1995;212:728–733. doi: 10.1006/viro.1995.1532. [DOI] [PubMed] [Google Scholar]
- 31.Wang S Z, Rushlow K E, Issel C J, Cook R F, Cook S J, Raabe M L, Chong Y H, Costa L, Montelaro R C. Enhancement of EIAV replication and disease by immunization with a baculovirus-expressed recombinant envelope surface glycoprotein. Virology. 1994;199:247–251. doi: 10.1006/viro.1994.1120. [DOI] [PubMed] [Google Scholar]
- 32.Yang Z, Nielsen R, Goldman N, Pedersen A M. Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics. 2000;155:431–449. doi: 10.1093/genetics/155.1.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng Y H, Nakaya T, Sentsui H, Kameoka M, Kishi M, Hagiwara K, Takahashi H, Kono Y, Ikuta K. Insertions, duplications and substitutions in restricted gp90 regions of equine infectious anaemia virus during febrile episodes in an experimentally infected horse. J Gen Virol. 1997;78:807–820. doi: 10.1099/0022-1317-78-4-807. [DOI] [PubMed] [Google Scholar]
- 34.Zheng Y H, Sentsui H, Nakaya T, Kono Y, Ikuta K. In vivo dynamics of equine infectious anemia viruses emerging during febrile episodes: insertions/duplications at the principal neutralizing domain. J Virol. 1997;71:5031–5039. doi: 10.1128/jvi.71.7.5031-5039.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]