Abstract
The broad resistance to antibody-mediated neutralization of lentiviruses recently isolated from infected hosts is a poorly understood feature which might contribute to the ability of these viruses to persist and to the failure of experimental vaccines to protect against virulent viruses. We studied the underlying molecular mechanisms by examining the evolution of a neutralization-sensitive, tissue culture-adapted strain of feline immunodeficiency virus upon reinoculation into specific-pathogen-free cats. Reversion to broad neutralization resistance was observed in seven of seven inoculated animals and, in individual hosts, started to develop between less than 4 and more than 15 months from infection. After comparison of the envelope sequences of the inoculum virus, of an additional 4 neutralization-sensitive in vitro variants, and of 14 ex vivo-derived variants (6 neutralization sensitive, 5 resistant, and 3 with intermediate phenotype), a Lys→Asn or →Glu change at position 481 in the V4 region of the surface glycoprotein appeared as a key player in the reversion. This conclusion was confirmed by mutagenesis of molecularly cloned virus. Analysis of viral quasispecies and biological clones showed that the intermediate phenotype was due to transient coexistence of neutralization-sensitive and -resistant variants. Since the amino acid position involved was the same in four of four recent revertants, it is suggested that the number of residues that control reversion to broad neutralization resistance in FIV might be very limited. Amino acid 481 was found to be changed only in one of three putative long-term revertants. These variants shared a Ser→Asn change at position 557 in region V5, which probably collaborated with other mutations in long-term maintenance of neutralization resistance, as suggested by the study of mutagenized virus.
Feline immunodeficiency virus (FIV) is an important pathogen of domestic cats and, due to extensive analogy to human immunodeficiency virus type 1 (HIV-1), is a valuable model for AIDS studies (16, 33, 48). Antibody-mediated neutralization of FIV resembles that of HIV-1. Similarities include (i) a much greater resistance to neutralizing antibodies of viruses recently isolated from infected hosts compared to laboratory tissue culture-adapted (TCA) strains; (ii) an unpredictable sensitivity of primary isolates to inhibition by heterologous immune sera (6, 14, 27); (iii) a narrow breadth of activity of neutralizing antibodies; (iv) the presence of an important neutralization linear determinant in the variable (V) region 3 of the surface glycoprotein (SU) of TCA strains, whereas the neutralization epitopes of primary isolates appear to be mainly conformational (15, 17, 31); and (v) the dependence of neutralization on the cell substrate used (1, 39). Thus, although the functional domains of its SU and transmembrane glycoprotein (TM) are much less well characterized than those of HIV, FIV may help shed light on the mechanisms and role of antibody-mediated neutralization in lentiviral infections.
We previously reported that, following one 4-month passage in a specific-pathogen-free (SPF) cat, a highly neutralization-sensitive (NS) TCA strain of FIV had reverted to the broad neutralization resistance typical of primary isolates and that the SU of the revertant differed from the parental virus at two amino acid positions (469 and 481) within the V4 region (7). However, it remained to be determined whether reversion to the neutralization-resistant (NR) phenotype typical of wild-type virus (heretofore indicated as NS→NR reversion) was a general consequence of readaptation in vivo and, if so, whether it was associated with constant or diverse amino acid changes. Here, we show that such reversion is a general occurrence although, in individual cats, it may take variably long to develop. Moreover, analysis of numerous in vitro and ex vivo NS and NR variants as well as of biological and molecular clones has identified amino acid position 481 of SU as a major player in the reversion.
MATERIALS AND METHODS
Progenitor and variant viruses.
The progenitor TCA virus was the Petaluma strain of FIV produced by chronically infected FL4 cells (49; generous gift of Janet K. Yamamoto). In our laboratory, FL4 cells are routinely split 1:5 twice weekly. Viral stocks were obtained by harvesting supernatants at cell passages 181 (stock FL-181), 193 (FL-193), and 381 (FL-381). Female SPF cats (Iffa Credo, L'Arbresle, France) were infected intravenously with 1 ml of viral stock FL-193, corresponding to approximately 20 50% cat infectious doses, when 7 to 12 months old. Virus reisolations were performed by standard coculture of peripheral blood mononuclear cells with MBM cells. This line of feline CD3+, CD4−, and CD8− T lymphocytes is routinely cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum, 5 μg of concanavalin A, and 20 U of recombinant human interleukin-2 per ml. Reisolations varied only slightly in the time that they were first positive and the levels of reverse transcriptase produced, and three to five amplifications in MBM cells were sufficient to accumulate viral stocks of suitable titer. MBM cells were also used to produce some in vitro variants so that, with three exceptions in which supernatant of FL4 cultures was used, the virus stocks used in neutralization assays consisted of MBM cell fluids. The fluids were clarified at 350 g for 15 min, were stored in 1-ml aliquots in liquid nitrogen, and were subjected to titer determination at least twice in quadruplicate by endpoint dilution in a microtiter MBM cell assay after 1 h at 4°C, i.e., with the same incubation conditions used for the neutralization test. No differences were noted in replication kinetics between the progenitor and variant viruses. Titers ranged between 2 × 102 and 1 × 103 50% tissue culture infective doses (TCID50) per ml.
Neutralization assay.
Virus neutralization was performed as previously described (14) against 10 TCID50 of FIV and MBM cells grown in the presence of 6% pooled normal cat serum as indicator cells. Immune sera were generally diluted 1:8, 1:32, 1:128, and 1:512 (dilutions before the addition of virus and cells), and each experiment included controls receiving virus incubated with identical dilutions of normal cat serum. Neutralizing antibody titers were defined as the reciprocal of the serum dilution required to reduce by ≥50% the levels of reverse transcriptase activity produced in the presence of the corresponding dilution of normal cat serum and were calculated according to Reed and Müench (35). All experiments were repeated at least twice. In general, reproducibility was satisfactory since titers with a given virus-serum combination exhibited a maximum twofold deviation between experiments, which is within the expected error of neutralization assays with the format used.
Criteria for scoring the neutralization phenotype of virus variants.
The neutralization sensitivity of viral variants was probed against a panel of 11 heat-inactivated sera taken from eight FIV-infected cats at varying times postinfection (p.i.; see legend to Table 1). Based on the neutralization properties of primary FIV isolates (14), variants were considered NS when they were neutralized by at least nine test sera with an average titer of ≥128, NR if neutralized by no more than four sera with a mean titer of ≤32, and intermediate (NI) when they exhibited in-between sensitivity.
TABLE 1.
Neutralizability of consecutive isolates obtained from cats infected with TCA FIV, as determined with a panel of immune sera
| Immune seruma | Neutralizing antibody titer with virus isolate obtained from:b
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cat 275 at indicated mo p.i.
|
Cat 311 at indicated mo p.i.
|
Cat 583 at indicated mo p.i.
|
|||||||||||
| 1 | 3 | 6 | 10 | 15 | 10 | 15 | 1 | 3 | 6 | 10 | 15 | 20 | |
| 2581-9 | 128 | 512 | 11 | − | − | 8 | − | 128 | 181 | 181 | >512 | 45 | − |
| 857-12 | 32 | 181 | 11 | − | − | − | − | 8 | − | 8 | NDc | 45 | 11 |
| 874-12 | 512 | 512 | 152 | 32 | − | 45 | 8 | >512 | >512 | >512 | >512 | 45 | 8 |
| 902-12 | 128 | 512 | 11 | − | − | − | − | >512 | >512 | 128 | 512 | 181 | − |
| 905-12 | 128 | 512 | 11 | − | − | − | − | − | 8 | 64 | 194 | 11 | − |
| 338-32 | 181 | 128 | 8 | − | − | 38 | − | >512 | 512 | >512 | >512 | 14 | 32 |
| 2906-32 | 512 | 181 | 13 | − | − | − | − | >512 | 512 | >512 | 181 | 11 | − |
| 3368-32 | 128 | 181 | 11 | − | − | 11 | − | 512 | >512 | 512 | 181 | 45 | 45 |
| 338-41 | 181 | >512 | 11 | − | − | − | − | >512 | >512 | >512 | 512 | 181 | 157 |
| 2906-41 | 512 | 45 | 11 | − | − | − | − | 512 | >512 | >512 | 128 | 11 | ND |
| 3368-41 | >512 | 181 | 11 | − | − | − | − | >512 | 512 | >512 | 128 | 38 | 11 |
The number before the dash identifies the donor cat; the number after the dash identifies the month p.i. when the serum was obtained. Cats were infected with virus FL-181 (no. 338, 2906, and 3368) or with the same virus passaged three (no. 2581) or six (no. 857, 874, 902, and 905) times in SPF cats.
Values shown are the geometric means of the titers obtained in two independent assays. −, no neutralizing activity detected; mean titers of <8 were not considered significant and were scored as negative.
ND, not done.
DNA isolation, sequencing, and genetic analyses.
DNA was extracted from 6 × 106 infected cells with phenol-chloroform and was checked for integrity by ethidium bromide-stained 0.8% agarose gel and for amplificability by using feline tumor necrosis factor alpha primers. PCR amplification of env was performed as described previously (34). For SU sequencing, DNA was amplified by nested PCR, using flanking primers 237HS (5′-AGTAAACCCATTTAGGGTACCTG-3′; positions 6377 to 6399), 238AS (5′-CTCATCCCAGTCCACCCTTTTTTC-3′; positions 9103 to 9126), and 16AS (5′-CAGAAGAATTGATTTTGATTACA-3′; positions 8358 to 8380). Primers 237HS and 238AS were used for the first step and primers 237HS and 16AS for the second. The TM region was amplified by single-step PCR by using the primers V4S (5′-AACCTTTGCAATGAGAAGTT-3′; positions 7532 to 7551) and 238AS. Amplicons were then subjected to cycle sequencing using internal fluorescent-labeled primers overlapping the entire env. Nucleotide (nt) sequences were edited and translated by PC/Gene software (IntelliGenetics, Geel, Belgium). Selected regions were resequenced to confirm the substitutions that distinguished the variants. Predicted amino acid sequences were obtained with the TRANSL program, and glycosylation sites were determined with PROSITE of the PC/Gene package.
DNA sequences obtained from this and a previous study (7) were aligned by using the program CLUSTAL W (46). Genetic distance matrices were generated by using the DNADIST program of the PHYLIP software package, version 3.5c (University of Washington, Seattle), using Kimura's two-parameter method (20) to correct for superimposed substitutions and a transition/transversion of 2.0. Divergence per year was calculated as follows: distance between parental and isolate sequences at 15 months divided by 15 and multiplied by 12.
Quasispecies analysis.
Viral RNA from virus stocks was reverse transcribed and amplified with first-step primers pV3S (5′-TGTTATGTAGACAGAGTAGAT-3′; positions 7113 to 7133) and pV4AS (5′-TGCAAGACCAATTTCCAGCA-3′; positions 7847 to 7866), and the product of this reaction was reamplified for 25 cycles with internal primers pV4S2 (5′-TAGATGTAGATGGAATGTAG-3′; positions 7643 to 7662) and pV4AS2 (5′-CACAATAAGGTCATCTACCT-3′; positions 7789 to 7808). The 166-bp fragments of region V4 thus obtained were then subjected to single-strand conformation polymorphism (SSCP) analysis as reported previously (14) except that electrophoresis was carried out in 15% nondenaturing polyacrylamide gel. Following silver staining and washing of the gels in distilled water for at least 1 h, the SSCP bands were excised, directly amplified, and sequenced.
Biological clones.
Biological clones were obtained using a modification of a method previously described for HIV-1 (28). Briefly, 10 TCID50 of virus were incubated with 5 × 106 MBM cells at 37°C in 5% CO2 for 4 h. After thorough washing to remove any unabsorbed virus, numbers of cells ranging from 100 to 105 were seeded into microwells containing 105 MBM cells in 100 μl of complete medium (16 wells/dilution), in order to expand the virus produced by single infected cells. The supernatants were monitored for p25 twice weekly for 1 month, and the ones found positive that had been seeded with the lowest numbers of virus-exposed cells were examined for quasispecies composition as described above. The cultures that produced a single SSCP band were used as a source of biologically cloned virus. The clones were expanded, filtered, frozen in small aliquots in liquid nitrogen, subjected to titer determination, sequenced to confirm the genotype inferred from SSCP analysis, and examined for neutralization phenotype.
Molecular clones.
The entire env of the NS virus cultured from cat 275 at 1 month p.i. was amplified using primers ORFA1s (5′-GGTCGGGAGAACTATGAATGG-3′; positions 5974 to 5994) and LPCRas (5′-GCTGTCTCCCGTTGTAGAAGTCG-3′; positions 9046 to 9069) with proofreading Expand High Fidelity DNA polymerase (Hoffman-La Roche, Basel, Switzerland) and was cloned into the Topo TA cloning vector (Invitrogen, Carlsbad, Calif.). Site-specific mutagenesis was carried out on this construct by inverse PCR, using 5′ phosphorylated primers, the above polymerase, and a low number of cycles. Amplicons were ligated with T4 DNA polymerase (New England Biolabs, Beverly, Mass.) and were introduced into JM109 bacteria. Unaltered and mutagenized env fragments were then inserted into p34TF10 (44), which had been rendered able to grow in lymphocytes by removing the stop codon at position 6210 of open reading frame A. Virus stocks were produced by transfecting into CrFK cells and expanding in MBM cells. Proper insertion and the absence of unwanted mutations were checked by sequencing the entire env at each step.
RESULTS
NS→NR reversion.
In a previous study, growth in vivo for 4 months had caused FIV stock FL-181 to revert to the broad neutralization resistance typical of primary isolates (7). To assess whether this was a constant outcome of propagating TCA FIV in vivo, we tested for neutralization, by a panel of immune sera, three viruses (338:36, 2906:36, and 3368:36) reisolated from cats 3 years after infection with the same stock. All proved to be NR as primary FIV isolates (data not shown). In contrast, additional propagation of the producer FL4 cells for up to 24 months (variants FL-193 and FL-381) or mere passaging in MBM cells (variant 193/MBM in this study and variant 181/MBM of reference 7) had no effects on the neutralization sensitivity of TCA FIV.
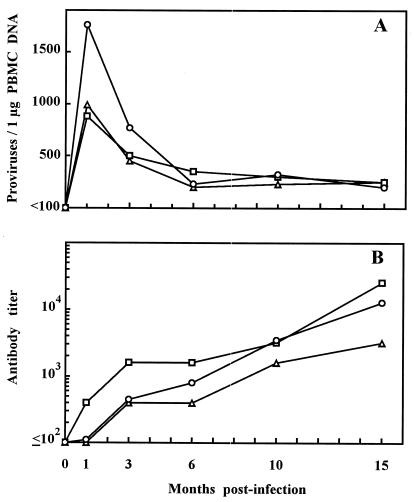
To further characterize the reversion, we studied consecutive virus samples from three SPF cats (no. 275, 311, and 583) inoculated with stock FL-193. The animals had become readily infected (Fig. 1); however, virus was cultured at all times tested only from two cats. Cat 311 was uniformly provirus positive and showed a prompt anti-FIV antibody response (Fig. 1B) but yielded the first positive culture at 10 months p.i. Failure to retrieve the virus from this cat at earlier time points was not unprecedented (2, 23) and was attributed to reduced virulence of TCA FIV for cats, as shown by relatively low provirus loads, low or negative postacute plasma viremias, and the absence of significant T-cell subset count changes (Fig. 1A and data not shown). Table 1 shows the neutralization behavior of reisolated viruses. The virus from cat 275 was as NS as the inoculum virus at 1 and 3 months, possessed an NI phenotype at 6 months, and scored NR at 10 and 15 months. The 10- and 15-month isolates from cat 311 both scored NR. In cat 583, the virus remained fully NS for at least 10 months, at 15 months was NI, and at 20 months still scored as NI although it had clearly progressed further toward global resistance. Thus, NS→NR reversion was a constant consequence of TCA FIV readaptation in vivo although it occurred after variably long time intervals in individual cats. The results also showed that the reversion could develop in a graded manner but affected in parallel the neutralizing efficiency of all panel sera, indicating that what was being observed was indeed the reacquisition of a neutralization phenotype typical of primary FIV isolates.
FIG. 1.
Follow-up of FIV infection in three cats infected with TCA FIV. (A) Numbers of proviruses in the peripheral blood mononuclear cells measured by competitive PCR as described previously (25). (B) Antibody titers to whole FIV antigen measured by enzyme-linked immunosorbent assay (25). ○, cat 275; □, cat 311; ▵, cat 583.
Timing of NS→NR reversion relative to development of resistance to autologous antibodies.
The consecutive viral samples described above were also tested in checkerboard assays for neutralization by consecutive sera derived from the same animals. Early isolates were effectively inhibited by all sera harvested 3 or more months p.i. but resisted neutralization by autologous sera collected at any time prior to virus reisolation. In contrast, late isolates resisted most sera and were inhibited exclusively, and very inefficiently, by some contemporary and subsequent autologous sera (Table 2). Thus, these findings showed that the NS→NR reversion developed considerably later than resistance to autologous antibodies. The analysis also showed that individual cats had developed at least partly distinct repertoires of neutralizing antibodies, as revealed by the partially different patterns of viral isolates neutralized (Table 2), and that this had occurred in concomitance with the marked decrease in circulating viral loads observed between month 1 and 3 p.i. (Fig. 1A), thus suggesting that neutralizing antibodies had participated in curbing the acute phase of viral replication.
TABLE 2.
Neutralizability of consecutive virus samples obtained from cats infected with TCA FIV by consecutive sera from the same cats
| Serum
|
Neutralizing antibody titer with virus isolate obtained from:a
|
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cat 275 at indicated mo p.i.
|
Cat 311 at indicated mo p.i.
|
Cat 583 at indicated mo p.i.
|
||||||||||||
| Donor cat | Mo p.i. at collection | 1 | 3 | 6 | 10 | 15 | 10 | 15 | 1 | 3 | 6 | 10 | 15 | 20 |
| 275 | 1 | − | − | − | − | − | − | − | − | 8 | − | − | − | − |
| 3 | 181 | − | − | − | − | − | − | 181 | 512 | 512 | 45 | 32 | − | |
| 6 | >512 | 45 | − | − | − | − | − | >512 | >512 | >512 | 181 | 11 | − | |
| 10 | >512 | 76 | 8 | − | − | − | − | >512 | >512 | >512 | >512 | − | − | |
| 15 | >512 | 512 | 32 | 8 | − | − | − | >512 | >512 | >512 | >512 | 181 | − | |
| 311 | 1 | − | − | − | − | − | − | − | − | − | 11 | 11 | − | − |
| 3 | >512 | 11 | 11 | − | − | − | − | >512 | >512 | >512 | >512 | − | − | |
| 6 | >512 | >512 | 8 | − | − | − | − | >512 | >512 | >512 | >512 | − | − | |
| 10 | >512 | 181 | 45 | − | − | 11 | 11 | >512 | >512 | >512 | >512 | 11 | − | |
| 15 | >512 | 512 | 57 | − | − | 20 | − | >512 | >512 | >512 | >512 | 11 | − | |
| 583 | 1 | 8 | − | − | − | − | − | − | 128 | 38 | 11 | − | − | − |
| 3 | >512 | 11 | − | − | − | − | − | 512 | 181 | 181 | 64 | 11 | − | |
| 6 | 512 | − | − | − | − | − | − | 181 | >128 | 181 | 181 | − | − | |
| 10 | >512 | 8 | − | − | − | − | − | 512 | >128 | 181 | 181 | 11 | − | |
| 15 | >512 | − | − | − | − | − | − | 512 | >128 | 181 | 181 | − | − | |
Values shown are the geometric means of the titers obtained in two independent assays. −, no neutralizing activity detected; mean titers of <8 were not considered significant and were scored as negative.
SU gene changes over time.
The SU genes (nt positions 6798 to 8097) of the above consecutive isolates and of three in vitro variants (FL-193, 193/MBM, and FL-381) were amplified from DNA of the cells used for viral stock production and were bidirectionally sequenced in their entirety. None of the nt sequences obtained had premature stop codons, deletions, or insertions, but all differed from each other and from parental virus, indicating that they were independent.
As shown in Table 3, the 15-month isolates from cats 275 and 311 diverged from parental virus by 0.85 and 0.70%, respectively, with an average divergence rate of 0.62% per year. This rate was higher than the 0.41% previously found in a long-term naturally infected cat (21) and was slightly less than that calculated for HIV-1 and simian immunodeficiency virus (5, 51). The 15-month isolate from cat 583 diverged from the inoculum by only 0.15%, and this was attributable to an especially high frequency of back mutations, as revealed by interisolate divergence rates essentially equivalent to that observed in the other cats. The latter rates were especially high during the first month, most likely as a result of exuberant virus replication in the acute phase (Fig. 1) and of the virus's need to readapt to the intrahost environment, and subsequently declined, first rapidly and later more gradually. These findings were consistent with data showing that selective pressure for change in HIV-1 is host dependent (51) and suggested that in cat 583 the selective forces driving virus evolution were weaker than in the other two animals, which might explain why in this animal NS→NR reversion had occurred later than in the others. It is unfortunate that the ratio of mutated synonymous and nonsynonymous codon sites, which might have corroborated this conclusion, was not informative due to low total numbers of mutations. It is also noteworthy that in cats 275 and 583, for which the time of reversion was precisely known, this occurred during a phase of relatively slow evolution, indicating that it had required limited mutational changes.
TABLE 3.
Genome divergence analysis of sequential isolates obtained from cats infected with TCA FIVa
| Cat | % Divergence from parental virus at 15 mo p.i.
|
% Divergence from parental virus or previous isolate per mo
|
||||||
|---|---|---|---|---|---|---|---|---|
| Total | Per year | 0–1 | 1–3 | 3–6 | 6–10 | 10–15 | 15–20 | |
| 275 | 0.85 | 0.68 | 0.54 | 0.23 | 0.10 | 0.06 | 0.14 | NDb |
| 311 | 0.70 | 0.56 | → | 0.21c | 0.03 | ND | ||
| 583 | 0.15 | 0.12 | 0.46 | 0.31 | 0.18 | 0.16 | 0.09 | 0.03 |
Kimura's two-parameter distances as calculated from pairwise comparison of the available sequential isolates.
ND, not done; at 20 months, only the virus culture from cat 583 was performed.
% Divergence between parental virus and isolate at 10 mo.
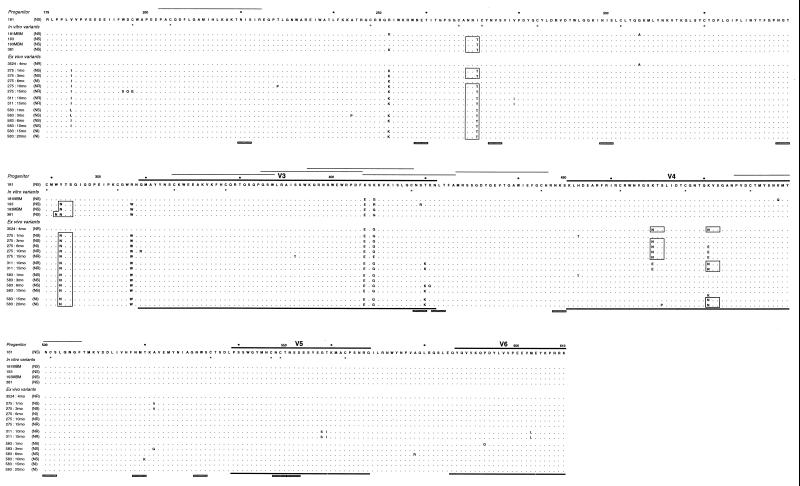
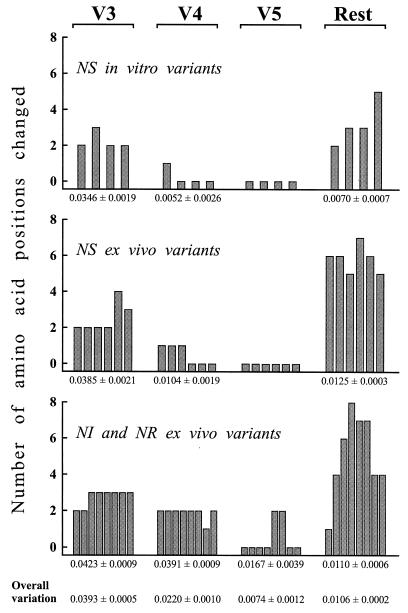
Figure 2 shows the SU amino acid alignment for all the variants described above and for three similar variants (FL-181, 181/MBM, and 3524:4mo) studied previously and described in an earlier report (7). The SU of FIV contains variable regions interspersed between more conserved segments (29). Amino acid substitutions were not randomly distributed since variation was especially high in the V3 and V4 regions. Interestingly, for V3 this was true regardless of the in vitro or ex vivo source (Fig. 3), suggesting that at least part of its variation might be independent of the in vivo environment, as observed for the corresponding region of HIV-1 (50). Furthermore, the V3 of the NR and NS variants contained similar numbers of amino acid changes, in accord with the idea that it may possibly serve as a decoy to distract the immune system from more critical regions (6, 27, 31) but is of less or no importance for determining global neutralization resistance. In contrast, the V4 of the NR and NI variants accumulated more changes than the NS counterparts, hence clearly suggesting its implication in NS→NR reversion.
FIG. 2.
Deduced amino acid sequences of the SU of in vitro and ex vivo variants derived from TCA FIV. Differences relative to the progenitor virus are shown in capital letters. The neutralization phenotype is given in parenthesis next to the variant designation. Lines over and under sequences indicate the variable regions (29). Numbers indicate amino acid positions starting with the first methionine of Env, according to the reported sequence of the 34TF10 clone of FIV Petaluma (44). Potential N-linked glycosylation sites common to all sequences are indicated by shaded bars under the alignment, and those present in only some variants are indicated by open boxes inside the alignment. Conserved cysteines are undermarked by an asterisk in the progenitor sequence. Synthetic peptides recognized by the sera of more than 50% of infected cat sera (24) are indicated by thin lines over the progenitor sequence to indicate the presence of linear B epitopes. Sequences 181, 181/MBM, and 3524:4mo have already been reported (7). In sequence 583:15mo, K and N coexisted at position 481 with an approximate ratio of 1:1.
FIG. 3.
Distribution of predicted amino acid changes relative to progenitor TCA FIV in the V3, V4, and V5 regions and in the rest of the SU of the viral variants listed in Fig. 2, grouped by source and neutralization phenotype. The figures under the bars represent the mean number of changes per amino acid position in the indicated SU regions ± the standard error for the groups of variants depicted above, while overall variation is the same parameter for all the variants. Variable regions V3 to V5 are delineated as shown in Fig. 2. Bars represent individual variants.
Table 4 summarizes the differential amino acid changes detected in the NR and NI variants and not found in any of the NS variants. Clearly, position 481 emerged as a critical residue since (i) it was substituted in each of the NR variants; (ii) it was replaced also in the three NI variants although in one (variant 583:15mo) the replacement was incomplete; (iii) timing of the replacement correlated with the onset of NS→NR reversion (Table 1); (iv) in one NR and two NI variants, it was the only position substituted, thus demonstrating that changes at this site were sufficient to confer the NR phenotype; and (v) the other positions were substituted in a maximum of two variants derived from a single cat, suggesting that they may have contributed to resistance to autologous antibodies or other immune effectors (18, 26) but played little if any role in NS→NR reversion. Regarding the residues present at position 481 of the NR variants, the Lys of the parental virus was substituted for by either Asn or Glu. In the former case, the revertants had acquired one additional potential N-linked glycosylation site in V4. Another additional predicted glycosylation site was present at position 469 in some ex vivo variants but did not correlate with neutralization resistance (Fig. 2).
TABLE 4.
Summary of differential amino acids found in NR and NI sequential isolates and not found in any of the NS variants examined
| Isolate (mo) | Phenotype | No.a | Amino acid change at position:
|
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 195 | 196 | 197 | 228 | 279 | 359 | 392 | 397 | 409 | 410 | 422 | 468 | 469 | 471 | 472 | 481 | 510 | 557 | 558 | 559 | 603 | |||
| Early revertants | |||||||||||||||||||||||
| 3524 (4) | NR | 1/5 | K→N | ||||||||||||||||||||
| 275 (6) | NI | 1/8 | K→E | ||||||||||||||||||||
| 275 (10) | NR | 3/11 | T→P | Q→R | K→E | ||||||||||||||||||
| 275 (15) | NR | 6/13 | W→S | D→Q | C→G | S→T | K→E | K→E | |||||||||||||||
| 311 (10) | NR | 6/14 | V→I | K→E | K→N | G→S | T→I | M→L | |||||||||||||||
| 311 (15) | NR | 6/14 | V→I | K→E | K→N | G→S | T→I | M→L | |||||||||||||||
| 583 (15) | NI | 1/8 | K→Nb | ||||||||||||||||||||
| 583 (20) | NI | 2/9 | S→P | K→N | |||||||||||||||||||
| Putative long-term revertants | |||||||||||||||||||||||
| 338 (36) | NR | 4/11 | R→K | K→E | V→I | S→N | |||||||||||||||||
| 3368 (36) | NR | 2/8 | N→T | S→N | |||||||||||||||||||
| 2906 (36) | NR | 7/12 | K→E | K→R | S→N | K→M | L→F | K→E | S→N | ||||||||||||||
Substitutions found in NR or NI variants only/total number of substitutions present in the variant.
Replacement occurred in 50% of sequences.
V4 quasispecies during NS→NR reversion.
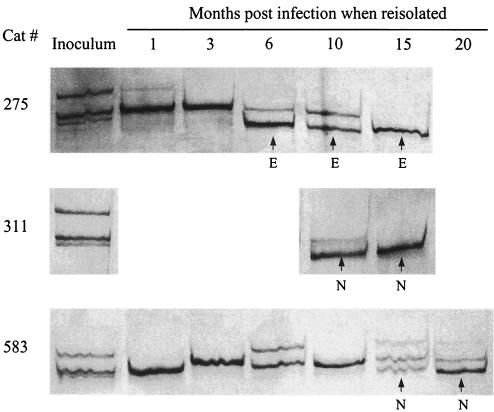
The readout of the neutralization assay used was expected to be determined by the most prominent variant in the virus stock. It was, therefore, of interest to investigate whether gradual acquisition of the NR phenotype was due to the gradual accumulation of mutations or to the time needed for the Asn-481 and Glu-481 sequences to become sufficiently abundant. We hence compared inoculum virus and the consecutive ex vivo isolates for quasispecies composition in a 166-bp segment of the V4 region encompassing position 481 (Fig. 4). The inoculum was found to be composed of three distinct sequences in the region examined, all with a Lys at position 481. The acute phase of replication in vivo was characterized by a simplification of quasispecies complexity, similar to that observed in seroconverting HIV-1 individuals (52, 53). Subsequently, the number of distinct sequences fluctuated between 1 and 3. It is of particular interest, however, that Asn-481 and Glu-481 sequences first became evident at different time points in individual hosts, initially coexisted with Lys-481 sequences for variable times, but eventually became the predominant (cat 583) sequence or the only one demonstrable. Since this gradual takeover paralleled the kinetics of NS→NR reversion (Tables 1 and 2), we concluded that the coexistence of parental and Asn-481 or Glu-481 sequences in the viral population was the most likely explanation for the NI phenotype.
FIG. 4.
Quasispecies composition in the V4 region of consecutive isolates obtained from cats infected with TCA FIV as determined by SSCP analysis and sequencing of the resulting bands. Sequences derived from individual bands differed in 1 to 6 nt. The ones with Lys-481 replaced are marked E for Glu or N for Asn.
Neutralization phenotype of biological clones.
Biological clones were derived from the NI virus cultured from cat 583 at 15 months p.i. and were studied by SSCP in the V4 region. Each clone gave a single band in a position that, in the original uncloned virus, was typical either of Lys-481 or Asn-481 sequences (data not shown). Four clones selected randomly from each of these two groups were assayed for their neutralization phenotype, and their SU was sequenced. All clones that had given a Lys-481 band proved NS while all clones with an Asn-481 band were NR (Table 5). Invariably, sequence analysis of these eight clones confirmed the genotype inferred from SSCP analysis. A similar study with the NR virus cultured from cat 275 at 10 months p.i. led to Glu-481 clones only, as determined by SSCP and sequence analysis, indicating a high prevalence of this genome, and each clone proved NR (results not shown). Thus, these findings corroborated the conclusions of quasispecies analysis.
TABLE 5.
Neutralizability of biological clones derived from the NI virus cultured from cat 583 at month 15 p.i.
| Immune seruma | Neutralizing antibody titerb
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Clone with Lys-481
|
Clone with Asn-481
|
|||||||
| B7 | C1 | G5 | H1 | B1 | C6 | C9 | H8 | |
| 902-12 | 181 | 45 | 32 | 45 | − | 8 | − | − |
| 905-12 | 11 | 8 | 8 | − | − | − | − | − |
| 338-41 | 512 | 45 | 32 | 128 | − | − | − | − |
| 2906-41 | 45 | 11 | 10 | 128 | − | − | − | − |
The number before the dash identifies the donor cat; the number after the dash identifies the month p.i. when the serum was obtained.
Values shown are the geometric means of the titers obtained in two independent assays. −, no neutralizing activity detected; mean titers of <8 were not considered significant and were scored as negative.
SU sequences of putative long-term revertants.
We also sequenced the SU of the NR variants 338:36, 2906:36, and 3368:36 (sequences available by e-mail on request), which had been obtained from animals infected with the FL-181 stock 3 years previously (see above) and were likely to have reverted to broad neutralization resistance long before sampling. Lys-481 was found replaced (by Glu) only in one of these putative long-term revertants. However, all had Ser-557 replaced by an Asn, resulting in an additional potential glycosylation site in V5. Although this suggested that such change was important for neutralization resistance, each of these variants displayed a minimum of two differential amino acids (Table 4), rendering identification of key changes problematic.
TM sequences.
In HIV-1, changes within TM have been seen to impact SU conformation and regulate susceptibility to selected neutralizing antibodies (6, 27, 31, 45). In FIV, TM changes have been seen to affect cell tropism (47), and immunization with a TM peptide conferred some protection against subsequent virus challenge (38). For investigating whether TM mutations could have contributed to the reduction of virus neutralizability, we sequenced the encoding gene of one NI and five NR variants (sequences available by e-mail on request). One NR and one NI variant showed no amino acid changes compared to the NS variant used as a reference. The others showed one to three substitutions irregularly dispersed through the extracellular, transmembrane, or intracellular domains. In particular, there was no single position which was affected in all the NR variants. These findings ruled out the possibility that the TM protein played a major role in NS→NR reversion.
Neutralization phenotype of molecularly cloned viruses.
The above data had strongly suggested that amino acid positions 481 and 557 were important in reacquisition and, possibly, maintenance of the NR phenotype by TCA FIV. To confirm the importance of these positions, we generated mutagenized molecular clones as described in Materials and Methods. When transfected into CrFK cells and expanded in MBM cells, all clones led to the production of viral stocks suitable for the neutralization assays except the ones with Glu-481, which—for reasons that have remained unclear—either gave a single cycle of replication (three clones) or rapidly reverted to Lys-481 (one clone). As shown in Table 6, Asn-481 conferred complete resistance to all the four sera tested and Asn-557 did to only two sera, thus suggesting that while the former change was sufficient for broad neutralization resistance, the latter probably required the coexistence of additional mutations.
TABLE 6.
Neutralizability of molecular clones having different amino acids at positions 481 and 557
| Immune seruma | Neutralizing antibody titerb of clone with indicated amino acids
|
||
|---|---|---|---|
| Lys-481 and Ser-557 | Asn-481 and Ser-557 | Lys-481 and Asn-557 | |
| 902-12 | 181 | − | − |
| 905-12 | 11 | − | − |
| 338-41 | >512 | − | 23 |
| 2906-41 | 128 | − | 90 |
The number before the dash identifies the donor cat; the number after the dash identifies the month p.i. when the serum was obtained.
Values shown are the geometric means of the titers obtained in two independent assays. −, no neutralizing activity detected; mean titers of <8 were not considered significant and were scored as negative.
DISCUSSION
We have approached the issue of the poor sensitivity to antibody-mediated neutralization of wild-type lentiviruses by investigating the evolution of an NS TCA strain of FIV upon reinoculation into its natural host. The reversion to the NR phenotype typical of primary isolates was a uniform outcome of readaptation in vivo, thus indicating that this phenotype is an important survival factor in vivo for FIV as well as for other lentiviruses (4, 5, 10, 19). That the reversion took variable numbers of months to develop suggests that broad neutralization resistance is necessary, especially for long-term viral persistence. This, together with the fact that all the virus variants studied grew equally well in culture regardless of neutralization phenotype, also makes it unlikely that substrate-controlled epigenetic changes or improved tropism for specific target cells were important causes of NS→NR reversion (2, 3, 8, 30, 40, 43). It is therefore plausible that reversion was driven by the host's immune response. Accordingly, the transition was slowest in cat 583, which also exhibited the weakest anti-FIV enzyme-linked immunosorbent assay antibody response and the lowest rate of SU change. The present findings also indicate that TCA viruses should not be used as the challenge in vaccine experiments even if repassaged in vivo for some time, unless their complete reversal to wild-type phenotype has been verified.
Env analysis of consecutive virus samples obtained from cats infected with the TCA FIV under study identified amino acid position 481, located centrally in the V4 region, as a key determinant of NS→NR reversion since (i) substitution of either Asn or Glu for Lys-481 was common to each of the virus samples with an NI or NR phenotype but was not found in any of the NS variants; (ii) emergence and takeover of the sequences containing these substitutions coincided temporally with the development of the NR phenotype; and (iii) in one NR and two NI variants, amino acid 481 was the only differential position compared to NS variants, thus suggesting that this change alone was sufficient for acquisition of the NR phenotype. The importance of amino acid 481 was confirmed by the phenotype of biological and molecular clones. All clones with Asn-481 or Glu-481 effectively resisted neutralization, whereas the ones with Lys-481 were as NS as the TCA virus. The additional, sporadic differential amino acids observed in some consecutive virus samples were dispersed throughout Env and were temporally unrelated to the transition. Thus, they may have represented escape mutations that protected the virus from the in vivo ongoing action of specific subsets of neutralizing antibodies and other immune effectors (18, 26, 31) but appeared unimportant for global neutralization resistance.
Studies with lentiviruses and other viruses have shown that single amino acid substitutions, within critical linear or discontinuous epitopes or even located at a distance, can permit escape from specific immune sera or sets of monoclonal neutralizing antibodies. For example, substituting a Leu or Thr for Ser-483 was shown to render a molecular clone of the Amsterdam 19 strain of FIV susceptible to neutralization by an otherwise ineffective antiserum raised against a different clone of the same strain (41). In HIV-1, there is also evidence that certain amino acid positions within Env control neutralizability on a large scale (6, 27, 32). The present findings represent the first indication that a single amino acid change can bring about a general resistance of FIV to neutralization. Since the diverse polyclonal sera we used to probe neutralizability are expected to have simultaneously acted upon multiple epitopes and clusters of epitopes and most if not all the neutralization epitopes of wild-type FIV operative in the lymphoid cell-based assay used here are conformation-dependent (17, 37), they also represent evidence that distinct conformation-determining amino acids exist within SU which prevent the functionality of multiple FIV neutralization epitopes. The replacements of amino acid 481 associated with broad neutralization resistance resulted either in the introduction of a potential glycosylation site (Lys→Asn) or in a change of charge polarity (Lys→Glu), events that may have influenced tertiary and quaternary structure, such as a condensation of SU or altered intramolecular interactions within Env oligomeric spikes (10, 12, 36).
It has been calculated that lentivirus-infected hosts are confronted with every possible virus mutation on a daily basis (11). That changes at amino acid 481 were associated with NS→NR reversion in 4 of 4 hosts is, therefore, of considerable interest. Because host cats were not siblings and their sera displayed partially different patterns of neutralizing activity in checkerboard tests, it seems unlikely that this was due to a stereotyped immune response. It is, instead, more likely that changes at this position provided such a selective advantage in vivo as to represent a highly preferable pathway for virus evolution. Similar to what was suggested by recent findings with TCA chimeric simian HIV (9), this in turn may imply that the number of Env residues responsible for broad neutralization resistance of fresh isolates is very limited. It is notable that virtually all the SU sequenced from field FIV isolates possess an Asn-481 irrespective of the clade in which they are classified (34).
Finally, it is of interest that Lys-481 was found to be replaced (by an Asn) in only 1 of 3 NR variants first reisolated from cats inoculated with TCA FIV 3 years earlier. In such putative long-term revertants, precise definition of the position(s) responsible for resistance was problematic due to the presence of at least two differential SU residues relative to NS variants. Although these revertants shared a Ser→Asn mutation at position 557 in the V5 region, a molecular clone with Asn-557 proved still partially neutralizable. It is, therefore, likely that changes at this position determine the broadly resistant phenotype only when associated with other substitutions. Because no earlier samples of these variants were available, we cannot discriminate whether they developed upon viruses that already had become NR due to 481 changes, which would have demonstrated that the genetic bases of the NR phenotype evolve with the duration of infection, or instead stemmed directly from NS virus, which would have shown that alternative routes to NS→NR reversion exist. A Lys→Gln change at position 560 of V5 was previously seen to render a molecular clone of the Amsterdam 19 strain of FIV resistant to neutralization by an autologous antiserum (42). It is also noteworthy that neither position 557 nor position 481 has been implicated as a determinant of FIV tropism to different cell types (13, 22).
In conclusion, if extended to other TCA strains, the approach used here may permit identification of Env determinants that confer broad neutralization resistance to wild-type FIV, eventually providing leads for the development of vaccines effective against difficult-to-neutralize strains.
ACKNOWLEDGMENTS
This work was supported by grants from Ministero della Sanità—Istituto Superiore di Sanità, “Programma per l'AIDS,” and by the Ministero della Università e Ricerca Tecnologica, Rome, Italy. F.M. was the holder of fellowships from Ministero della Sanità and ANLAIDS, Rome, Italy.
We are indebted to Janet K. Yamamoto, University of Florida, for the generous gift of FL4 cells.
REFERENCES
- 1.Baldinotti F, Matteucci D, Mazzetti P, Giannelli C, Bandecchi P, Tozzini F, Bendinelli M. Serum neutralization of feline immunodeficiency virus is markedly dependent on passage history of the virus and host system. J Virol. 1994;68:4572–4579. doi: 10.1128/jvi.68.7.4572-4579.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barlough J E, North T W, Oxford C L, Remington K M, Dandekar S, Ellis M N, Pedersen N C. Feline immunodeficiency virus infection of cats as a model to test the effect of certain in vitro selection pressures on the infectivity and virulence of resultant lentivirus variants. Antivir Res. 1993;22:259–272. doi: 10.1016/0166-3542(93)90036-i. [DOI] [PubMed] [Google Scholar]
- 3.Bastiani L, Laal S, Kim M, Zolla-Pazner S. Host cell-dependent alterations in envelope components of human immunodeficiency virus type 1 virions. J Virol. 1997;71:3444–3450. doi: 10.1128/jvi.71.5.3444-3450.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burns D P W, Collignon C, Desrosiers R C. Simian immunodeficiency virus mutants resistant to serum neutralization arise during persistent infection of rhesus monkeys. J Virol. 1993;67:4104–4113. doi: 10.1128/jvi.67.7.4104-4113.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burns D P W, Desrosiers R C. Selection of genetic variants of simian immunodeficiency virus in persistently infected rhesus monkeys. J Virol. 1991;65:1843–1854. doi: 10.1128/jvi.65.4.1843-1854.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burton D R, Montefiori D C. The antibody response in HIV-1 infection. AIDS. 1997;11:S87–S98. [PubMed] [Google Scholar]
- 7.Cammarota G, Matteucci D, Pistello M, Nicoletti E, Giannecchini S, Bendinelli M. Reduced sensitivity to strain-specific neutralization of laboratory-adapted feline immunodeficiency virus after one passage in vivo: association with amino acid substitutions in the V4 region of the surface glycoprotein. AIDS Res Hum Retrovir. 1996;12:173–175. doi: 10.1089/aid.1996.12.173. [DOI] [PubMed] [Google Scholar]
- 8.Castilletti C, Capobianchi M R, Fais S, Abbate I, Ficociello B, Ameglio F, Cordiali Fei P, Santini S M, Dianzani F. HIV type 1 grown on interferon γ-treated U937 cells shows selective increase in virion-associated intercellular adhesion molecule 1 and HLA-DR and enhanced infectivity for CD4-negative cells. AIDS Res Hum Retrovir. 1995;11:547–553. doi: 10.1089/aid.1995.11.547. [DOI] [PubMed] [Google Scholar]
- 9.Cayabyab M, Karlsson G, Etemad-Moghadam B, Hofmann W, Steenbeke T, Halloran M, Fanton J W, Axthelm M K, Letvin N L, Sodroski J G. Changes in human immunodeficiency virus type 1 envelope glycoproteins responsible for the pathogenicity of a multiply passaged simian-human immunodeficiency virus (SHIV-HXBc2) J Virol. 1999;73:976–984. doi: 10.1128/jvi.73.2.976-984.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng-Mayer C, Brown A, Harouse J, Luciw P A, Mayer A J. Selection for neutralization resistance of the simian/human immunodeficiency virus SHIVSF33A variant in vivo by virtue of sequence changes in the extracellular envelope glycoprotein that modify N-linked glycosylation. J Virol. 1999;73:5294–5300. doi: 10.1128/jvi.73.7.5294-5300.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coffin J M. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267:483–489. doi: 10.1126/science.7824947. [DOI] [PubMed] [Google Scholar]
- 12.Davis D, Stephens D M, Willers C, Lachmann P J. Glycosylation governs the binding of antipeptide antibodies to regions of hypervariable amino acids sequence within recombinant gp120 of human immunodeficiency virus type 1. J Gen Virol. 1990;71:2889–2898. doi: 10.1099/0022-1317-71-12-2889. [DOI] [PubMed] [Google Scholar]
- 13.Dean G A, Himathongkham S, Sparger E E. Differential cell tropism of feline immunodeficiency virus molecular clones in vivo. J Virol. 1999;73:2596–2603. doi: 10.1128/jvi.73.4.2596-2603.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Del Mauro D, Matteucci D, Giannecchini S, Maggi F, Pistello M, Bendinelli M. Autologous and heterologous neutralization analyses of primary feline immunodeficiency virus isolates. J Virol. 1998;72:2199–2207. doi: 10.1128/jvi.72.3.2199-2207.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Ronde A J, Stam J G, Boers P, Langedijk H, Meloen R, Hesselink W, Keldermans L C E J M, van Vliet A, Verschoor E J, Horzinek M C, Egberink H F. Antibody response in cats to the envelope proteins of feline immunodeficiency virus: identification of an immunodominant neutralizing domain. Virology. 1994;198:257–264. doi: 10.1006/viro.1994.1028. [DOI] [PubMed] [Google Scholar]
- 16.Elder J H, Phillips T R. Feline immunodeficiency virus as a model for development of molecular approaches to intervention strategies against lentivirus infections. Adv Virus Res. 1995;45:225–247. doi: 10.1016/s0065-3527(08)60062-7. [DOI] [PubMed] [Google Scholar]
- 17.Giannecchini S, Matteucci D, Bendinelli M. Effect of enzymatic deglycosylation on feline immunodeficiency virus sensitivity to antibody-mediated neutralization. AIDS Res Hum Retrovir. 1998;14:199–204. doi: 10.1089/aid.1998.14.199. [DOI] [PubMed] [Google Scholar]
- 18.Hosie M J, Flynn J N. Feline immunodeficiency virus vaccination: characterization of the immune correlates of protection. J Virol. 1996;70:7561–7568. doi: 10.1128/jvi.70.11.7561-7568.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kimata J T, Kuller L, Anderson D B, Dailey P D, Overbaugh J. Emerging cytopathic and antigenic simian immunodeficiency virus variants influence AIDS progression. Nat Med. 1999;5:535–540. doi: 10.1038/8414. [DOI] [PubMed] [Google Scholar]
- 20.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–120. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 21.Kyaw-Tanner M T, Robinson W F. Quasispecies and naturally occurring superinfection in feline immunodeficiency virus infection. Arch Virol. 1996;141:1703–1713. doi: 10.1007/BF01718293. [DOI] [PubMed] [Google Scholar]
- 22.Lerner D L, Elder J H. Expanded host cell tropism and cytopathic properties of feline immunodeficiency virus strain PPR subsequent to passage through interleukin-2-independent T cells. J Virol. 2000;74:1854–1863. doi: 10.1128/jvi.74.4.1854-1863.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lombardi S, Garzelli C, Pistello M, Massi C, Matteucci D, Baldinotti F, Cammarota G, Da Prato L, Bandecchi P, Tozzini F, Bendinelli M. A neutralization antibody-inducing peptide of the V3 domain of feline immunodeficiency virus envelope glycoprotein does not induce protective immunity. J Virol. 1994;68:8374–8379. doi: 10.1128/jvi.68.12.8374-8379.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Massi C, Lombardi S, Indino E, Matteucci D, La Rosa C, Esposito F, Garzelli C, Bendinelli M. Most potential linear B-cell epitopes of Env glycoproteins of feline immunodeficiency virus are immunologically silent in mice. AIDS Res Hum Retrovir. 1997;13:1121–1129. doi: 10.1089/aid.1997.13.1121. [DOI] [PubMed] [Google Scholar]
- 25.Matteucci D, Pistello M, Mazzetti P, Giannecchini S, Del Mauro D, Lonetti I, Zaccaro L, Pollera C, Specter S, Bendinelli M. Studies of AIDS vaccination using an ex vivo feline immunodeficiency virus model: protection conferred by a fixed cell vaccine against cell-free and cell-associated challenge differs in duration and is not easily boosted. J Virol. 1997;71:8368–8376. doi: 10.1128/jvi.71.11.8368-8376.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McMichael A J, Phillips R E. Escape of human immunodeficiency virus from immune control. Annu Rev Immunol. 1997;15:271–296. doi: 10.1146/annurev.immunol.15.1.271. [DOI] [PubMed] [Google Scholar]
- 27.Moore J P, Ho D D. HIV-1 neutralization: the consequences of viral adaptation to growth on transformed T cells. AIDS. 1995;9:S117–S136. [PubMed] [Google Scholar]
- 28.Orloff S L, Bandea C I, Kennedy M S, Allaway G P, Maddon P J, McDougal J S. Increase in sensitivity to soluble CD4 by primary HIV type 1 isolates after passage through C8166 cells: association with sequence differences in the first constant (C1) region of glycoprotein 120. AIDS Res Hum Retrovir. 1995;11:335–342. doi: 10.1089/aid.1995.11.335. [DOI] [PubMed] [Google Scholar]
- 29.Pancino G, Fossati I, Chappey C, Castelot S, Hurtrel B, Moraillon A, Klatzmann D, Sonigo P. Structure and variations of feline immunodeficiency virus envelope glycoproteins. Virology. 1993;192:659–662. doi: 10.1006/viro.1993.1083. [DOI] [PubMed] [Google Scholar]
- 30.Park E J, Gorny M K, Zolla-Pazner S, Quinnan G V. A global neutralization resistance phenotype of human immunodeficiency virus type 1 is determined by distinct mechanisms mediating enhanced infectivity and conformational change of the envelope complex. J Virol. 2000;74:4183–4191. doi: 10.1128/jvi.74.9.4183-4191.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Parren P W H I, Moore J P, Burton D R, Sattentau Q. The neutralizing antibody response to HIV-1: viral evasion and escape from humoral immunity. AIDS. 1999;13:S137–S162. [PubMed] [Google Scholar]
- 32.Parren P W H I, Wang M, Trkola A, Bilney J M, Purtscher M, Katinger H, Moore J P, Burton D R. Antibody neutralization-resistant primary isolates of human immunodeficiency virus type 1. J Virol. 1998;72:10270–10274. doi: 10.1128/jvi.72.12.10270-10274.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pedersen N C. Feline immunodeficiency virus infection. In: Levy J A, editor. The retroviridae. Vol. 2. New York, N.Y: Plenum Press; 1993. pp. 181–228. [Google Scholar]
- 34.Pistello M, Cammarota G, Nicoletti E, Matteucci D, Curcio M, Del Mauro D, Bendinelli M. Analysis of genetic diversity and phylogenetic relationship of Italian isolates of feline immunodeficiency virus indicates high prevalence and heterogeneity of subtype B. J Gen Virol. 1997;78:2247–2257. doi: 10.1099/0022-1317-78-9-2247. [DOI] [PubMed] [Google Scholar]
- 35.Reed L J, Müench H A. A simple method for estimating fifty percent and points. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 36.Reitter J N, Means R E, Desrosiers R. A role for carbohydrates in immune evasion in AIDS. Nat Med. 1998;6:679–684. doi: 10.1038/nm0698-679. [DOI] [PubMed] [Google Scholar]
- 37.Richardson J, Fossati I, Moraillon A, Castelot S, Sonigo P, Pancino G. Neutralization sensitivity and accessibility of continuous B cell epitopes of the feline immunodeficiency virus envelope. J Gen Virol. 1996;77:759–777. doi: 10.1099/0022-1317-77-4-759. [DOI] [PubMed] [Google Scholar]
- 38.Richardson J, Moraillon A, Crespeau F, Baud S, Sonigo P, Pancino G. Delayed infection after immunization with a peptide from the transmembrane glycoprotein of the feline immunodeficiency virus. J Virol. 1998;72:2406–2415. doi: 10.1128/jvi.72.3.2406-2415.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruppach H, Nara P, Raudonat I, Elanjikal Z, Rübsamen-Waigmann H, Dietrich U. Human immunodeficiency virus (HIV)-positive sera obtained shortly after seroconversion neutralize autologous HIV type 1 isolates on primary macrophages but not on lymphocytes. J Virol. 2000;73:5403–5411. doi: 10.1128/jvi.74.12.5403-5411.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sawyer L S W, Wrin M T, Crawford-Miksza L, Potts B, Wu Y, Weber P A, Alfonso R D, Hanson C V. Neutralization sensitivity of human immunodeficiency virus type 1 is determined in part by the cell in which the virus is propagated. J Virol. 1994;68:1342–1349. doi: 10.1128/jvi.68.3.1342-1349.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Siebelink K H J, Huisman W, Karlas J A, Rimmelzwaan G F, Bosch M L, Osterhaus A D M E. Neutralization of feline immunodeficiency virus by polyclonal feline antibody: simultaneous involvement of hypervariable regions 4 and 5 of the surface glycoprotein. J Virol. 1995;69:5124–5127. doi: 10.1128/jvi.69.8.5124-5127.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Siebelink K H J, Rimmelzwaan G F, Bosch M L, Meloen R H, Osterhaus A D M E. A single amino acid substitution in hypervariable region 5 of the envelope protein of feline immunodeficiency virus allows escape from virus neutralization. J Virol. 1993;67:2202–2208. doi: 10.1128/jvi.67.4.2202-2208.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takeuchi Y, Liong S-H, Beniasz P D, Jäger U, Porter C D, Friedmann T, McClure M O, Weiss R A. Sensitization of rhabdo-, lenti-, and spumaviruses to human serum by galactosyl(α1-3)galactosylation. J Virol. 1997;71:6174–6178. doi: 10.1128/jvi.71.8.6174-6178.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Talbott R L, Sparger E E, Lovelace K M, Fitch W M, Pedersen N C, Luciw P A, Elder J H. Nucleotide sequence and genomic organization of feline immunodeficiency virus. Proc Natl Acad Sci USA. 1989;86:5743–5747. doi: 10.1073/pnas.86.15.5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thali M, Charles M, Furman C, Cavacini L, Posner M, Robinson J, Sodroski J. Resistance to neutralization by broadly reactive antibodies to the human immunodeficiency virus type 1 gp120 glycoprotein conferred by a gp41 amino acid change. J Virol. 1994;68:674–680. doi: 10.1128/jvi.68.2.674-680.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thompson J D, Higgins D G, Gibson T J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vahlenkamp T W, Verschoor E J, Schuurman N N M P, van Vliet A L W, Horzinek M C, Egberink H F, de Ronde A. A single amino acid substitution in the transmembrane envelope glycoprotein of feline immunodeficiency virus alters cellular tropism. J Virol. 1997;71:7132–7135. doi: 10.1128/jvi.71.9.7132-7135.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Willett B J, Flynn J N, Hosie M J. FIV infection of the domestic cat: an animal model for AIDS. Immunol Today. 1997;18:182–189. doi: 10.1016/s0167-5699(97)84665-8. [DOI] [PubMed] [Google Scholar]
- 49.Yamamoto J K, Ackley C D, Zochlinski H, Louie H, Pembroke E, Torten M, Hansen H, Munn R, Okuda T. Development of IL-2-independent feline lymphoid cell lines chronically infected with feline immunodeficiency virus: importance for diagnostic reagents and vaccines. Intervirology. 1991;32:361–375. doi: 10.1159/000150220. [DOI] [PubMed] [Google Scholar]
- 50.Yoshida K, Nakamura M, Ohno T. Mutations of the HIV type 1 V3 loop under selection pressure with neutralizing monoclonal antibody NM-01. AIDS Res Hum Retrovir. 1997;13:1283–1290. doi: 10.1089/aid.1997.13.1283. [DOI] [PubMed] [Google Scholar]
- 51.Zhang L, Diaz R S, Ho D D, Mosley J W, Busch M P, Mayer A. Host-specific driving force in human immunodeficiency virus type 1 evolution in vivo. J Virol. 1997;71:2555–2561. doi: 10.1128/jvi.71.3.2555-2561.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhang L Q, MacKenzie P, Cleland A, Holmes E C, Leigh Brown A J, Simmonds P. Selection for specific sequences in the external envelope protein of human immunodeficiency virus type 1 upon primary infection. J Virol. 1993;67:3345–3356. doi: 10.1128/jvi.67.6.3345-3356.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu T, Mo H, Wang N, Nam D S, Cao Y, Koup R A, Ho D D. Genotypic and phenotypic characterization of HIV-1 in patients with primary infection. Science. 1993;261:1179–1181. doi: 10.1126/science.8356453. [DOI] [PubMed] [Google Scholar]