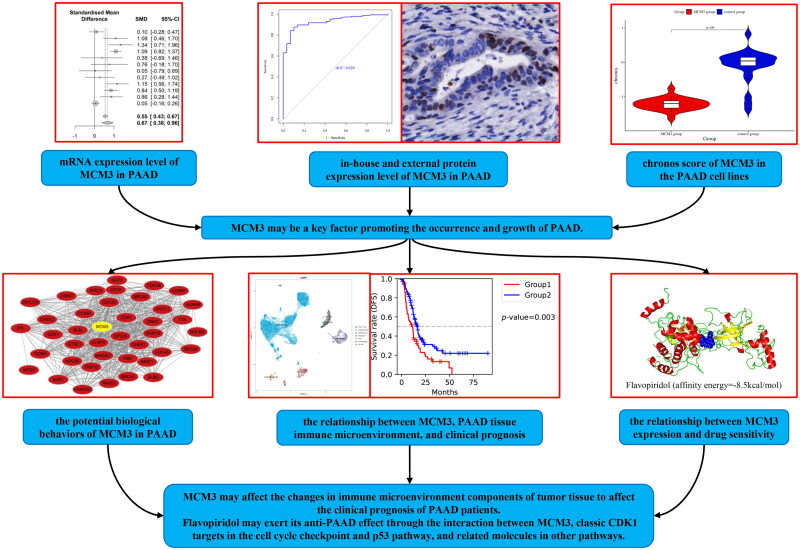
Abstract
Background
Minichromosome maintenance complex component 3 (MCM3) plays a key role in various tumours. However, it remains largely unknown what the specific role and clinical significance of MCM3 in pancreatic adenocarcinoma (PAAD) are.
Materials and Methods
We integrated high-throughput data from PAAD worldwide to analyse the expression level of MCM3 mRNA. We used immunohistochemistry to analyse MCM3 protein expression levels in 145 cases in the PAAD group and 29 cases in the non-PAAD group. We also mainly analysed the necessity of MCM3 for PAAD growth based on CRISPR screen data. In addition, we used enrichment analysis and protein–protein interaction networks to explore the molecular mechanism of MCM3 in PAAD. We also analysed the correlation between MCM3 expression, components of the immune microenvironment in PAAD tissue and clinical prognosis.
Results
In PAAD, we observed for the first time that MCM3 was significantly highly expressed at both the mRNA (SMD = 0.67, 95% CI: 0.38 ∼ 0.96) and the protein level (p < 0.05). The mRNA (AUC = 0.78, 95% CI: 0.74 ∼ 0.81; sensitivity = 0.66, 95% CI: 0.55 ∼ 0.76; specificity = 0.76, 95% CI: 0.67 ∼ 0.84) and protein (AUC = 0.929) expression levels of MCM3 had a good ability to distinguish between PAAD and non-PAAD tissue. There was heterogeneity reflected by the differential expression of MCM3 protein in PAAD cells. MCM3 played an essential role in PAAD growth, through abnormal DNA replication, p53 signalling and cell cycle checkpoints. PAAD with high MCM3 expression was sensitive to c-75, brivanib, flavopiridol and VNLG/124 drugs, with stable molecular docking models.
Conclusion
MCM3 is likely to be a critical element in promoting the initiation and growth of PAAD. Flavopiridol may exert its anti-PAAD effect through the interaction between MCM3, classic CDK1 targets in the cell cycle checkpoint and p53 pathway as well as related molecules in other pathways.
Keywords: MCM3, pancreatic adenocarcinoma, high-throughput sequencing, CRISPR screening, immunohistochemistry
KEY MESSAGES
MCM3 could potentially play a crucial role in promoting the onset and growth of PAAD.
There is heterogeneity reflected by the differential expression of MCM3 protein in PAAD cells.
The interplay between MCM3, well-established CDK1 targets in the cell cycle checkpoint and p53 pathway, along with relevant molecules in other pathways, may mediate the anti-pancreatic adenocarcinoma (PAAD) effect of flavopiridol.
Graphical abstract
1. Background
Over the last few years, some pancreatic adenocarcinoma (PAAD) patients screened out early can hope to be completely cured through surgical treatment [1]. At the same time, researchers have developed immunotherapies that induce the body’s anti-tumour immune potential to specifically resist PAAD cells [2]. However, PAAD is still a malignant neoplasm with a very pessimistic prognosis [1–5]. In 2020, there were almost as many new cases of PAAD worldwide (496,000) as there were deaths (466,000) [3]. This phenomenon is mainly due to the challenges in early detection, the aggressive and metastatic nature of PAAD and the current lack of early and effective screening methods [6–8]. Many patients with PAAD are already in the advanced stage when they first seek treatment. From this time and for the remainder of their lives, the quality of life of PAAD patients plummets due to symptoms such as diarrhoea, abnormal bowel movements, ascites, steatorrhoea, abdominal pain, back pain and weight loss [9–11]. Although several progresses were made in the treatment of pancreatic cancer, clinical trial data is still largely missing and urgently needed to verify relevant effects and for the development of more personalized treatment approaches [12–15]. Therefore, further research on the mechanisms related to PAAD is of great significance for developing more effective early screening programmes and better treatment programmes to improve the current situation of poor prognosis for PAAD [16].
The main function of minichromosome maintenance complex component 3 (MCM3) is to participate in the initiation of eukaryotic genomes [17]. It has been established that it has a crucial role in the initiation and progression of different types of tumours, and it has been explored as a potential avenue for tumour treatment. Scientific evidence has elucidated the important contribution of MCM3 in the occurrence and development of head and neck squamous cell carcinoma, highlighting its significance as a prognostic marker for this malignancy [18]. MCM3 has been implicated in the progression of the tumour microenvironment in lung squamous cell carcinoma and holds promise as a candidate for immunotherapeutic interventions targeting this particular cancer [19]. MCM3 may promote ovarian cancer progression by enhancing DNA replication and cell cycle synergy, and it is expected to serve as a therapeutic target and prognostic indicator for ovarian cancer [20]. It has also been reported that oestrogen receptor–positive breast cancer cells can resist treatment with tamoxifen and letrozole by upregulating the expression of MCM3 [21]. Multiple studies have shown that MCM3 is a reliable indicator related to early screening, predicting the prognosis and treatment of cervical cancer [17,22–24]. However, to our knowledge, there is only one report on the role of MCM3 in PAAD. Moreover, that study has a single data source and a small number of clinical samples included in the analysis, and it mainly analyses only the expression and prognosis of MCM3 without further analysing more specific mechanisms and drug treatments [25]. At the same time, the level of MCM3 protein in PAAD and the necessity of MCM3 for the growth of PAAD cells have not been verified [25]. Therefore, the specific mechanism and clinical significance of MCM3 in PAAD are still unknown.
This study is the first to integrate global high-throughput datasets, in-house protein level and CRISPR screening data to comprehensively analyse the expression of MCM3 in PAAD and its impact on the occurrence and growth of PAAD. We also investigate the potential biological behaviour of MCM3 in PAAD using gene ontology (GO) enrichment analysis, the Kyoto Encyclopedia of Genes and Genomes (KEGG) and protein-protein interaction networks (PPI). We predominantly utilise CRISPR screen data, which integrates diverse studies, to examine the association between MCM3 and drug sensitivity in PAAD cells, the components of the tissue microenvironment and the clinical significance. The insights gained from these analyses will contribute fresh understanding of the specific mechanisms and clinical implications of MCM3 in PAAD.
2. Materials and methods
2.1. Study design and collection, screening, and integration of high-throughput datasets related to PAAD worldwide
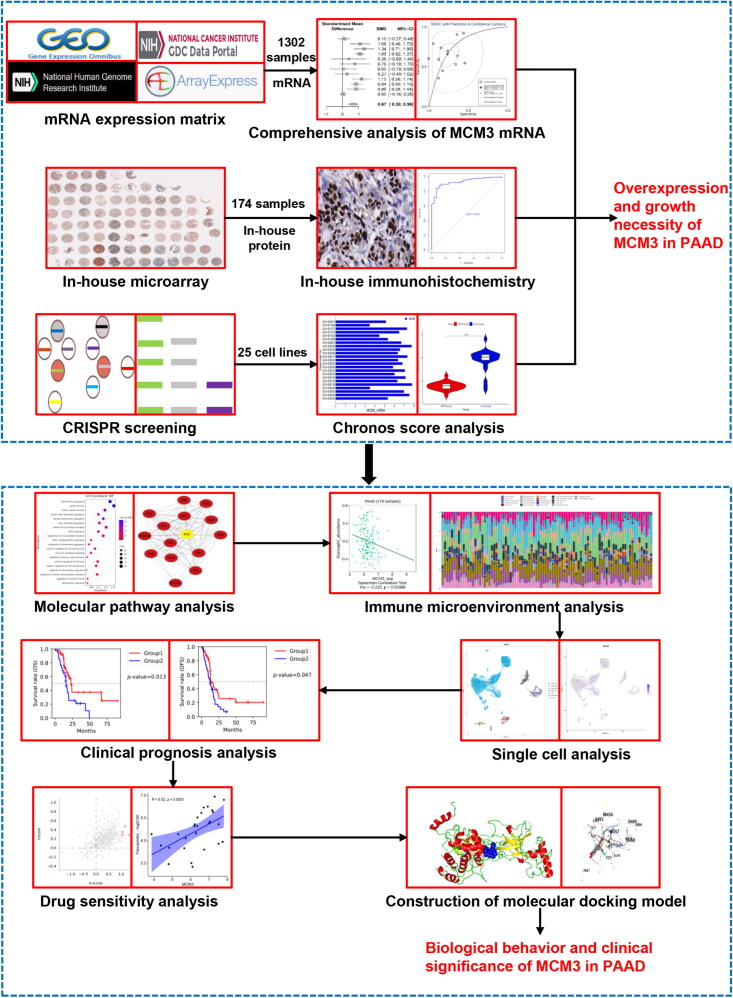
Figure 1 illustrates the overall framework of this study, demonstrating the comprehensive design. We explored the PAAD tissue expression status of MCM3 at the mRNA level by analysing relevant PAAD high-throughput datasets from the Cancer Genome Atlas (TCGA), Gene Expression Omnibus (GEO), Sequence Read Archive (SRA) and Genotype-Tissue Expression (GTEx) project. The detailed inclusion and exclusion process is shown in Figure S1. The main information of the datasets ultimately included in the RNA sequencing is detailed in Table S1-2. We used the sva and limma-voom packages of R v4.2 to eliminate batch effects and integrate expression matrices with the same platform.
Figure 1.
The flowchart of the main design in this study.
2.2. Comprehensive analysis of MCM3 expression levels at the mRNA level in PAAD tissue
Differences in MCM3 expression between PAAD and non-PAAD tissue worldwide were analysed using the Wilcoxon test. If p < 0.05, the difference in MCM3 expression was considered to be statistically significant. In addition, we used R v4.2 to draw a funnel plot to determine whether there was publication bias. We also used R v4.2 to analyse the number, mean and standard deviation of MCM3 expression cases in PAAD and non-PAAD tissue and drew standard the mean difference (SMD) forest plot to conduct a comprehensive analysis of the variations in MCM3 expression between PAAD and non-PAAD tissue. In addition, we used Stata v15.1 and R v4.2 to draw a summary receiver operating characteristic curve (sROC) and sensitivity and specificity forest plots.
2.3. Analysis of in-house and external MCM3 protein expression in PAAD tissue
To further validate the expression level of MCM3 in PAAD, we used 145 PAAD and 29 non-PAAD tissue samples and verified the protein expression status of MCM3 in PAAD tissue through tissue microarray and immunohistochemistry (IHC). The 29 non-PAAD tissues were pancreatic tissues from non-PAAD patients. The pathological properties of these tissues were healthy or only benign lesions. The 174 cases of pancreatic tissue were divided into PAC482 and PAC1021 tissue chips (provided by Guilin Fanpu Biotech of Guangxi, a subsidiary wholly owned by Pantomics, Inc.) for IHC, and MCM3 antibody (rabbit monoclonal antibody [EPR7080]) was purchased from Abcam Plc (Shanghai, China). These 174 pancreatic samples were collected by Guilin Fanpu Biotech of Guangxi from October 2018 to October 2021.All operational procedures adhered to the operational guidelines outlined by the manufacturers. The IHC score was independently evaluated by two senior pathologists. We selected any 10 fields of view from each pancreatic tissue sample and counted the number of positive cells out of 100 cells for each field at the highest magnification possible with the analysis tools available. The average of these 10 field-positive cells was the IHC score of this sample. This study was approved by the Ethics Committee of the First Affiliated Hospital of Guangxi Medical University (No. 2023-S382-01) and Guilin Fanpu Biotech of Guangxi (2018, Oct, 25th). All patients or their families signed informed consent forms. Our study followed the principles of the Declaration of Helsinki. We examined the protein expression of MCM3 in PAAD tissue through analysis of the Human Protein Atlas.
2.4. Exploring the effect of MCM3 on PAAD cell growth
Based on the DepMap database, we applied chronos scores to further investigate the correlation between MCM3 and the growth of PAAD cells [26, 27]. The chronos score of MCM3 in 25 PAAD cell lines was mainly knocked out by the CRISPR system to form the MCM3 group. The chronos scores of 25 randomly knocked-out genes in 25 PAAD cell lines comprised the control group. A Chronos score of 0 indicated that the growth of the cell line was not dependent on the gene, and a larger negative value indicated greater dependence [27]. The main information about these 25 PAAD cell lines was shown in Table S3.
2.5. Screening of differentially expressed genes (DEGs) in non-PAAD and PAAD tissues
We used the metafor and meta packages of R v4.2 to batch calculate the SMD of each gene in the above high-throughput datasets.If a gene was found to have p < 0.05 and the 95% confidence interval of SMD did not include 0, the gene would be considered a DEG between non-PAAD tissues and PAAD tissues. In addition, when the SMD of a DEG was greater than 0, the gene was considered to be a high-expression gene in PAAD. Otherwise, the DEG was considered to be a low-expression gene in PAAD.
2.6. Screening for MCM3 co-expression genes in PAAD
We screened the co-expressed genes of MCM3 in PAAD based on cBioPortal [28, 29]. If a certain gene in PAAD had a |Spearman’s correlation| > 0.3 and p < 0.05, the gene was classified as a co-expressed gene of MCM3 in PAAD.
2.7. Enrichment analysis and construction of the PPI network based on the intersection genes
We took the intersection of the DEGs obtained above and the co-expressed genes to obtain the intersection genes. Afterwards, we used R v4.2 to conduct GO and KEGG enrichment analyses utilising intersection genes. In addition, we used STRING v12.0 and Cytoscape v3.9.1 to build PPI networks for some pathways.
2.8. Identifying of immune microenvironment components in PAAD tissue
We determined the immune microenvironment components in PAAD tissue by using single-cell sequencing and immune infiltration analysis. In the single-cell sequencing analysis, we analysed samples of primary PAAD and untreated PAAD in the GSE205013 dataset from the GEO database [30]. We began by integrating the samples and removing batch effects. We then conducted quality control on the analysed cells to remove low-quality cells and some interfering factors, and we standardised and normalised them. Next, we conducted principal component analysis on the standardised and normalised single-cell sequencing expression matrix and selected the 20 principal components with the highest standard deviation for UMAP and TSNE dimensionality reduction. We then a conducted cluster analysis on them and used the Wilcoxon test to analyse the genes with strong features in each cell cluster. We combined the CellMarker database and the SingleR package for cell type identification [31, 32]. Additionally, we investigated the expression of MCM3 across different cell types.
To evaluate immune infiltration, we analysed the mRNA expression levels of MCM3 in 179 PAAD samples obtained from the TCGA dataset. These samples were then segregated into two groups, the MCM3 mRNA high-expression group and the MCM3 mRNA low-expression group, based on the median expression level of MCM3. Next, we used the cibersort method of the IBOR package to evaluate the MCM3_High group, and the MCM3_Low group underwent immune infiltration analysis [33].
2.9. Analysing the relationship between MCM3 expression, components of the immune microenvironment in PAAD tissue and clinical prognosis
We used Q-omics data integrating the prognosis and immune infiltration of a considerable amount of patients worldwide to analyse the components of the immune microenvironment and clinical prognosis in PAAD tissue [34]. Based on the median, we categorised the diverse cells infiltrating PAAD tissue into a group with high infiltration and another group with low infiltration. By grouping the immune cells according to their extent of infiltration, we examined the association between various infiltrating immune cells and the clinical prognosis of patients. We then plotted a Kaplan–Meier survival curve. Finally, we used TISIDB to study the correlation between the mRNA expression of MCM3 and the level of immune cell infiltration in PAAD tissue and drew a correlation scatter plot [35].
2.10. Exploring the correlation between MCM3 expression and the drug sensitivity of PAAD cells
We used Q-omics data integrating mRNA expression and drug intervention in multiple cell lines around the world to analyse the correlation between MCM3 mRNA expression and drug sensitivity in PAAD cells [34]. We began by using cross-association analysis to screen drugs that have a close relationship between MCM3 expression and the -log (IC50) of the drug. If the predictive and descriptive p-values of the analysis were both less than 0.01, the relationship was considered to be close. We divided the drugs into two groups, namely the high expression group and the low expression group, based on the median MCM3 mRNA expression level. After that, we studied the difference in -log (IC50) of the drugs between the MCM3 high expression group and the low expression group. If p < 0.05, it was determined statistically significant.
2.11. Molecular docking analysis of drugs and MCM3 protein
First, we collected the three-dimensional structure information of MCM3 macromolecule proteins and small-molecule drugs from the RCSB PDB and PubChem databases, respectively. We then used PyMOL to pre-treat the macromolecular proteins by removing irrelevant water molecules, ions and other subunits of the macromolecular proteins as well as hydrogenation. We used Open Babel to convert the format of the three-dimensional structure of small-molecule drugs and AutoDockTools to adjust its active torsion to pre-process small-molecule drugs. Next, we used AutoDock Vina to predict and computationally analyse various docking models of small-molecule drugs and MCM3 macromolecule proteins [36]. Finally, we selected the docking model with the largest |binding energy| for visual display using PyMOL. In addition, we also used CB-Dock2 to perform molecular docking on small-molecule drugs and MCM3 protein again to further verify the binding ability between MCM3 and these small-molecule drugs [37]. If the |binding energy| of the molecular docking model was less than 4 kcal/mol, it indicated that the binding effect of the drug and MCM3 was poor. If the |binding energy| was 4 ∼ 7 kcal/mol, it indicated that the binding effect of the drug and MCM3 was general. If the |binding energy| was greater than 7 kcal/mol, the combination of drugs and MCM3 was ideal.
2.12. Statistical analysis
All statistical analyses in this study were performed in Stata v15.1 and R v4.2. In this study, p < 0.05 was considered statistically significant unless otherwise specified. When analysing two sets of quantitative data, if these data satisfy the normal distribution, T-test analysis was used to analyse these data. Otherwise, the Wilcoxon test was used for analysis. The chi-square Q test and I2 statistical analysis were used to determine the calculation model to be used for calculating the SMD of the mRNA expression level. If I2 > 50%, there was a large heterogeneity, and the random effect model was used to calculate the SMD. Otherwise, the fixed effect model is used.
3. Results
3.1. Comprehensive analysis of MCM3 mRNA expression in PAAD tissue
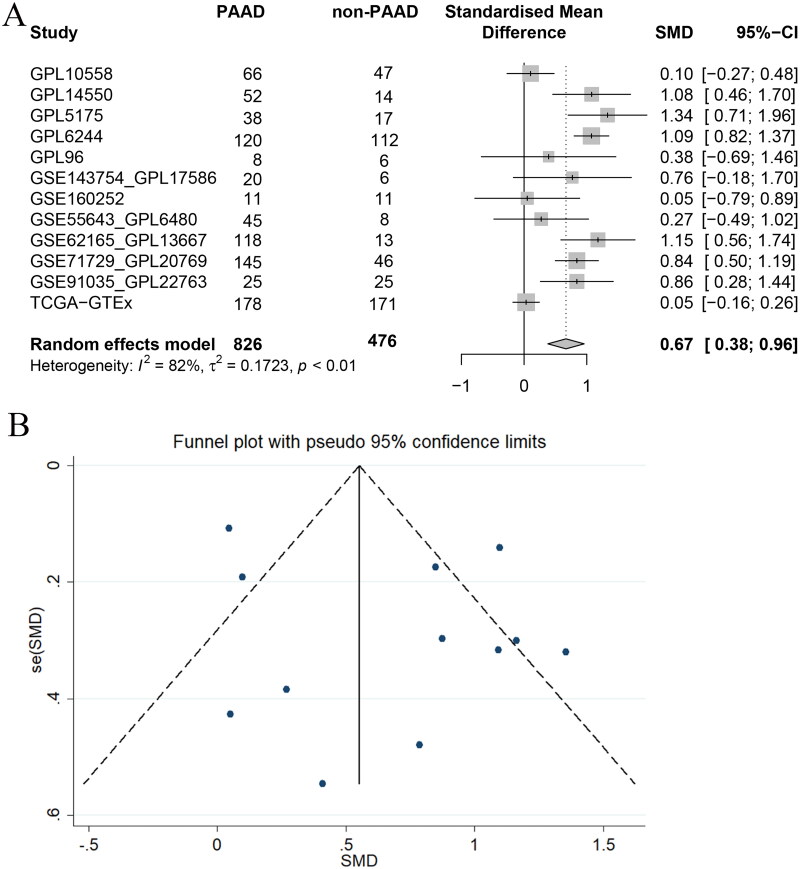
Based on the evidence-based concept, we collected and integrated 826 PAAD samples and 476 non-PAAD samples from 12 datasets from multiple datasets that met the inclusion criteria. Figure 2A shows that I2 > 50% and p < 0.05, indicating large heterogeneity, and a random effects model was employed to count SMD. Figure 2A also shows that, compared with 476 non-PAAD samples, 826 PAAD samples showed overall high expression of MCM3 at the mRNA level (SMD = 0.67, 95% CI: 0.38 ∼ 0.96). For detailed data on the SMD of MCM3 mRNA, please see Table S4. From each dataset, all the included datasets showed that MCM3 was highly expressed in PAAD tissue (SMD > 0). Figure 2B shows that our overall analysis of the various datasets did not indicate any significant publication bias.
Figure 2.
Integrated analysis of MCM3 mRNA expression in PAAD tissue worldwide.
(A) The standard mean difference forest map of MCM3 mRNA expression in PAAD tissue*;
(B) Funnel plot with pseudo 95% confidence limits;
*Analysis based on normal pancreatic tissue.
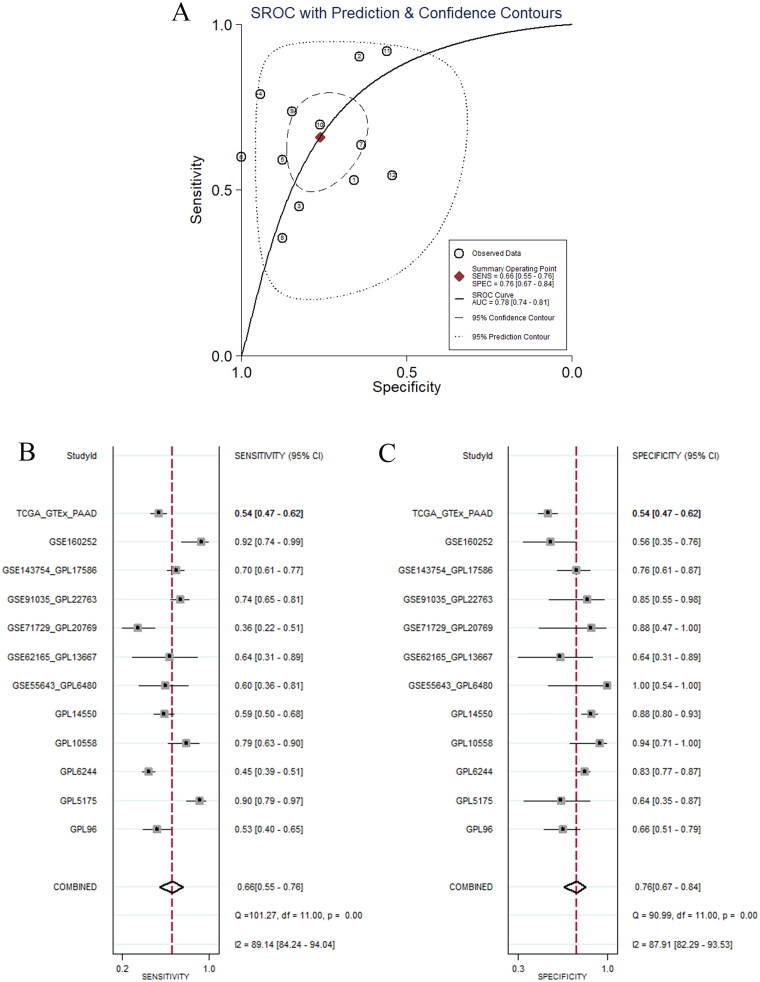
In addition, we analysed the ability of high expression of MCM3 to identify PAAD tissue by constructing sROC curve and forest plots. Figure 3A shows that the overall difference in expression levels of MCM3 between PAAD and non-PAAD tissue had strong discriminating ability (AUC = 0.78, 95% CI: 0.74 ∼ 0.81; sensitivity = 0.66, 95% CI: 0.55 ∼ 0.76; specificity = 0.76, 95% CI: 0.67 ∼ 0.84). Figures 3B–C and S2–3 also show that the expression of MCM3 within most datasets also had a strong ability to distinguish PAAD tissue. These results confirm that the expression of MCM3 has a strong ability to identify PAAD tissue and also confirm that the expression of MCM3 mRNA showed a significant increase in PAAD tissue.
Figure 3.
Discriminant analysis of PAAD tissue by highly expressed MCM3 map.
(A) Summary receiver operating characteristic curve;
(B) Sensitivity forest plots;
(C) Specificity forest plot.
3.2. Analysis of the expression status of the in-house and external MCM3 protein in PAAD tissue
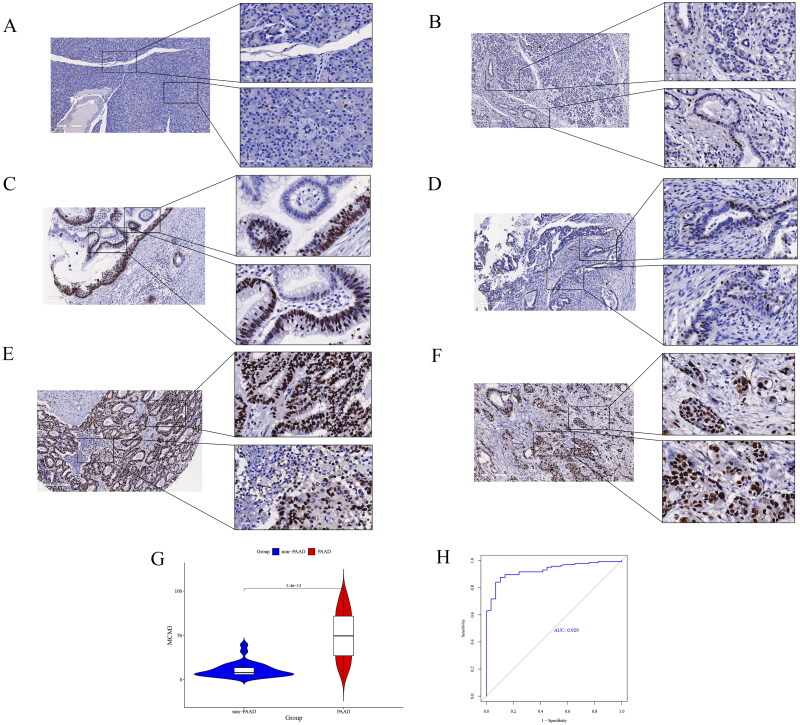
Figure 4A–F show that the positive signal of MCM3 protein was significantly more present in the PAAD tissue group than in the non-PAAD tissue group and was mainly concentrated in the nuclear region. Figure 4G–H show from a quantitative analysis perspective that the immunohistochemical score of MCM3 protein levels were significantly increased in the 145 PAAD tissue samples, compared to the 29 non-PAAD tissue samples (p < 0.05), and the immunohistochemical score of MCM3 protein had a strong ability to distinguish between the group of PAAD tissue and the group of non-PAAD tissue (AUC = 0.929). This indicates that at the protein level, MCM3 is significantly overexpressed in in-house PAAD tissue compared to non-PAAD tissue. In addition, we found that the positive signal of MCM3 protein was stronger in external PAAD tissue than in non-PAAD tissue (Figure S4). Furthermore, it is interesting to note that both in the in-house PAAD tissue and in the external PAAD tissue, we found groups of PAAD cells with similar location and morphology, some of which were positive for MCM3 protein and some negative. This may indicate the presence of different sub-populations or subtypes of tumour cells in PAAD cells, divided by differences in MCM3 protein expression. This is a manifestation of PAAD cell heterogeneity.
Figure 4.
Analysis of MCM3 protein expression in in-house PAAD tissue.
(A)-(B) Expression status of MCM3 in non-PAAD tissue*;
(C)-(F) Heterogeneous expression of MCM3 in PAAD tissue*;
(G) Violin diagram for differential analysis of MCM3 protein expression levels between non-PAAD and PAAD groups;
(H) ROC curve to evaluate the ability of MCM3 protein expression levels to discriminate between non-PAAD tissue groups and PAAD tissue groups;
* Cells with positive MCM3 protein are in brown, and cells with negative MCM3 protein are in blue.
3.3. Necessity analysis of MCM3 on the growth of PAAD
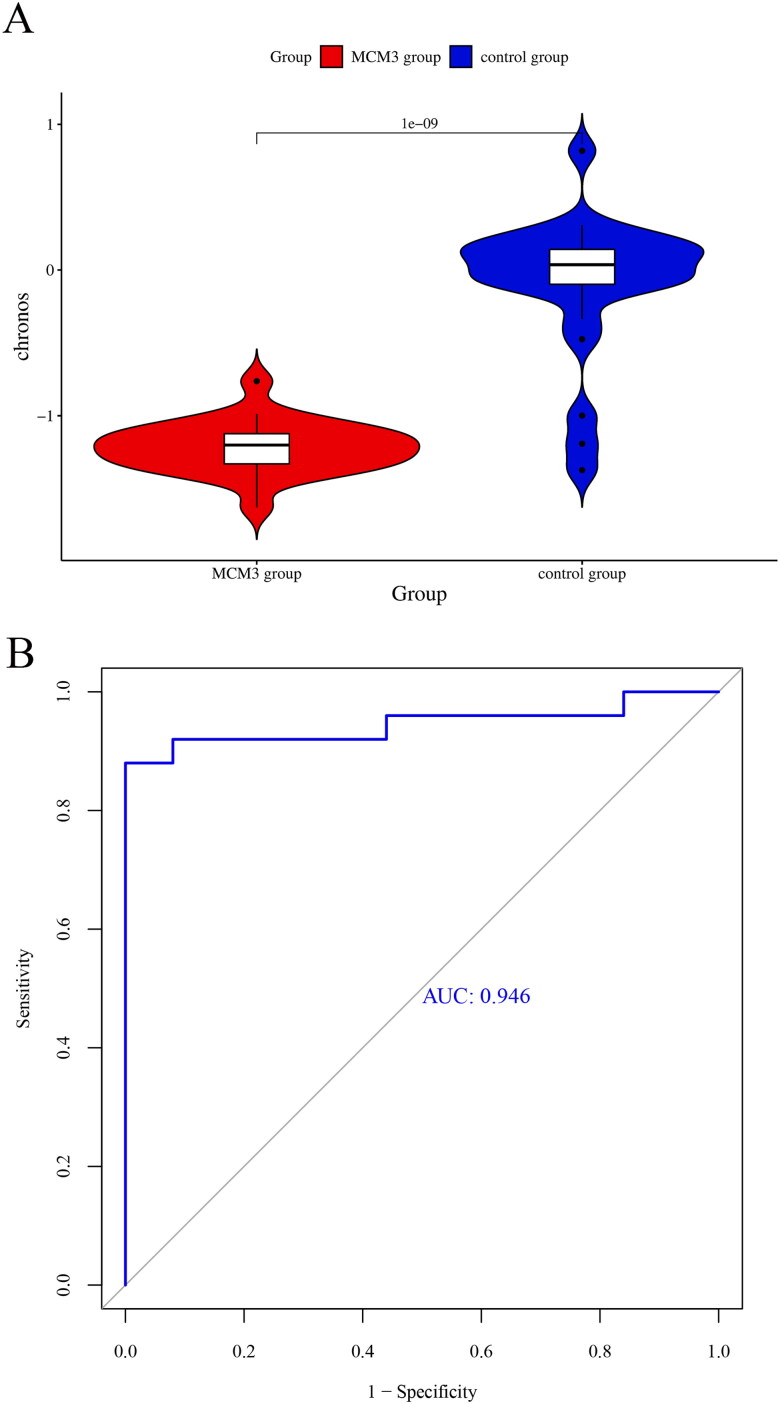
There was a significant decrease in the chronos score of the MCM3 group in contrast to the control group (p < 0.05) (Figure 5A). The score showed good discriminative ability between the MCM3 group and the control group (AUC = 0.946) (Figure 5B). There were 23 cell lines in the MCM3 group with a chronos score less than −1. These findings indicate that the survival rate of PAAD cells significantly decreased after MCM3 was knocked out. MCM3 is likely to be an essential gene for the growth of PAAD. In addition, the expression level of MCM3 mRNA in PAAD cell lines is shown in Figure S5.
Figure 5.
Comparing the effect of knocking out MCM3 in the CRISPR system on the growth of PAAD cells.
(A) Violin diagram comparing the difference in chronos scores between the MCM3 knockout group and the control group;
(B) Identification of ROC curves between MCM3 knockout group and control group using chronos score.
3.4. Exploration of the potential biological behaviour of MCM3 in PAAD
We further explored the potential molecular mechanisms of MCM3 in PAAD by constructing pathway analysis graphs and PPI networks. We obtained 10540 DEGs and 888 MCM3 co-expressed genes in PAAD, and based on this, 462 intersection genes were obtained.
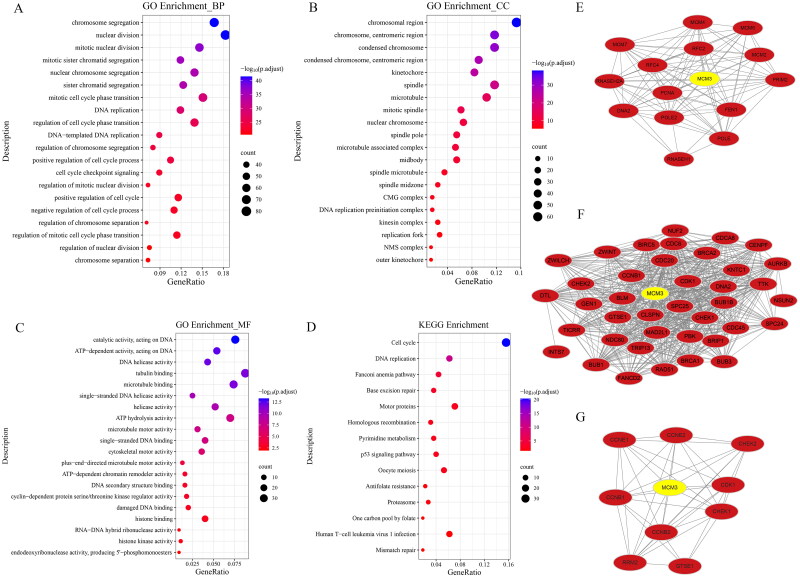
Figure 6A–D show the enrichment results of these intersecting genes. The biological process (BP) section of the GO enrichment analysis shows that MCM3 may participate in abnormal biological behaviours such as chromosomal aggregation, DNA replication and cell cycle checkpoint signalling in PAAD. The Cellular Component (CC) section displays chromosomal region, condensed chromosome, spindle and more. The Molecular Function (MF) section includes catalytic activity acting on DNA. The KEGG enrichment analysis shows that MCM3 in PAAD may participate in abnormal biological behaviour, such as cell cycle, p53 signalling pathway and, DNA replication.
Figure 6.
Mapping of potential biological behavioural analysis of MCM3 in PAAD based on intersection genes.
(A) Bubble diagram of the biological process (BP) section for GO enrichment analysis; (B) Bubble diagram of the cellular component (CC) section for GO enrichment analysis;
(C) Bubble diagram of the molecular function (MF) section for GO enrichment analysis; (D)Bubble diagram for KEGG enrichment analysis;
(E) DNA replication of protein-protein interaction networks; (F) Cell cycle checkpoint signalling of protein-protein interaction networks;
(G) P53 signalling pathway of protein-protein interaction networks.
To gain a more comprehensive understanding of the more detailed mechanism of MCM3 in PAAD, our study also constructed a PPI network diagram. Figure 6E shows that MCM families such as MCM3, MCM4 and MCM7 are likely to have a crucial role in the DNA replication of PAAD abnormalities. Figure 6F shows that MCM3 interacted with numerous molecules, such as CDK1 and CCNB1, affecting abnormal cell cycle checkpoint signalling. In addition, there were also interactions between MCM3 and cell cycle–related molecules such as CDK1, CCNB1 and CCNB2 in the abnormal p53 signalling pathway (Figure 6G).
3.5. Analysis of the relationship between MCM3 expression level, PAAD tissue immune microenvironment status and clinical prognosis
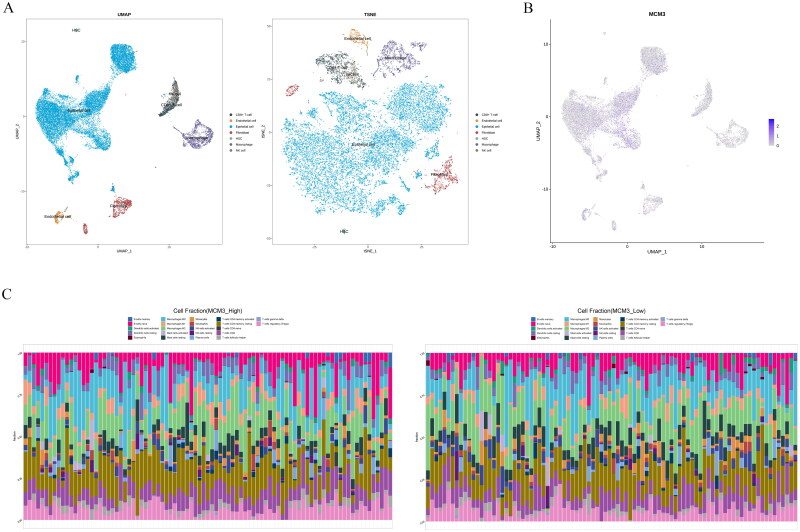
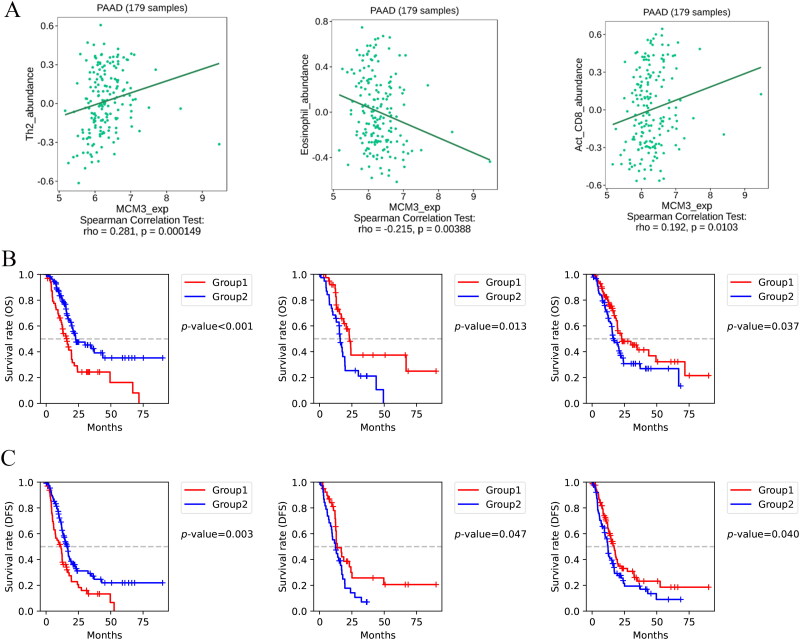
Figure 7 shows that the immune microenvironment components in PAAD tissue included CTL, eosinophils, CD4 + T cells, NK cells and macrophages. In addition, we found that MCM3 was mainly expressed in the epithelial cells of PAAD tissue. To further explore the clinical value of MCM3 expression and potential immune-related mechanisms, we analysed the connection between MCM3 expression level, PAAD tissue immune microenvironment status and clinical prognosis. The expression of MCM3 was positively correlated with the degree of Th2 cell and activated CD8 + T cell infiltration in the PAAD immune microenvironment (r > 0, p < 0.05) (Figure 8A). The expression of MCM3 was negatively correlated with eosinophils in the PAAD immune microenvironment (r < 0, p < 0.05) (Figure 8A). The overall survival (OS) (Figure 8B) and disease-free survival (DFS) (Figure 8C) of the group with high infiltration of eosinophils and activated CD8 + T cells in the immune microenvironment of PAAD tissue were better than those of the lower infiltration group, while the high infiltration of Th2 cellswas worse than the OS (Figure 8B) and the DFS (Figure 8C) of the lower infiltration group (p < 0.05).
Figure 7.
PAAD Tissue immune microenvironment status.
(A) Single cell sequencing analysis of PAAD tissue based on UMAP and TSNE dimensionality reduction;
(B) Expression of MCM3 in various cells of PAAD tissue based on UMAP dimensionality reduction;
(C) Differences in immune infiltration between the high and low expression groups of MCM3.
Figure 8.
Analysis of the relationship between MCM3 expression level, PAAD tissue immune microenvironment status, and clinical prognosis.
(A) Scatter plots of the correlation between the degree of infiltration of Th2 cells, eosinophils, and activated CD8+ T cells and the expression level of MCM3 in PAAD tissue;
(B) Kaplan-Meier survival curves comparing overall survival in PAAD tissue with Th2 cells, eosinophils, and activated CD8+ T cells in the high infiltration and low infiltration groups*;
(C) Kaplan-Meier survival curves comparing disease-free survival in PAAD tissue with Th2 cells, eosinophils, and activated CD8+ T cells in the high infiltration and low infiltration groups*;
*Divided into a high infiltration group (Group 1) and a low infiltration group (Group 2) based on the median degree of infiltration of a certain cell in PAAD tissue.
3.6. MCM3 expression and drug sensitivity analysis in PAAD cell lines
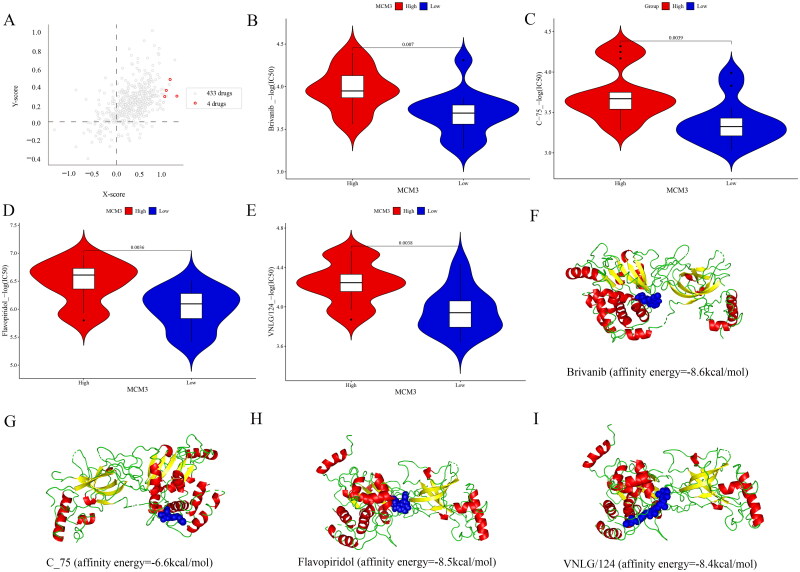
Figure 9A shows that the cross-association analysis screened out the four drugs to which the drug sensitivity of PAAD cells was most closely related to MCM3 expression, namely brivanib, c-75, flavopiridol and VNLG/124. The Wilcoxon test in Figure 9B–E shows that there was a significant difference in the sensitivity to these four drugs between the PAAD cells in the MCM3 high-expression group and the PAAD cells in the MCM3 low-expression group (p < 0.05). In addition, the drug sensitivity of these drugs shows a strong positive correlation with the expression level of MCM3 (R > 0, p < 0.05) (Figure S6). This suggests that PAAD cells with high MCM3 expression may be more sensitive to these drugs than PAAD cells with low MCM3 expression.
Figure 9.
Analysis of the relationship between drug sensitivity and MCM3 in PAAD cell lines.
(A) Cross-association analysis of MCM3 expression and drug reactivity;
(B)-(E) Differences in drug sensitivity between high and low expression groups of MCM3*;
(F)-(I) Relatively ideal models for drugs docking with the MCM3 protein;
* Divide MCM3 into high expression groups and low expression groups based on its median expression value; The -log (IC50) of these drugs is considered drug sensitivity.
We also studied the effects of these four drugs on MCM3 by analysing molecular docking models. Figure 9F–I show that except for the c-75 model with general affinity energy (-6.6 kcal/mol), the binding energy of all other models was relatively ideal (brivanib_affinity energy = −8.6 kcal/mol; flavopiridol_affinity energy = −8.5 kcal/mol; VNLG/124_affinity energy = −8.4 kcal/mol). In addition, the molecular docking model constructed using CB-Dock2 also shows that these drugs have strong binding ability with MCM3 protein (Figure S7). This further illustrates the correlation between MCM3 and the drug sensitivity of these drugs.
4. Discussion
Since the molecular mechanisms related to PAAD have not been thoroughly studied, there is great difficulty in the early diagnosis and treatment of PAAD, which in turn leads to an extremely poor prognosis for PAAD patients [38, 39]. Based on the available information, there is a paucity of comprehensive and systematic studies elucidating the potential involvement of MCM3 in PAAD. This study observed significant overexpression of MCM3 in PAAD at both the mRNA and the protein level. There is heterogeneity reflected by the differential expression of MCM3 protein in PAAD cells. MCM3 plays an essential role in the growth of PAAD. MCM3 may promote the occurrence and growth of PAAD through abnormal DNA replication, the p53 signalling pathway and the cell cycle checkpoint signalling pathway. The expression of MCM3 in PAAD may significantly affect changes in Th2 cells, eosinophils and activated CTL in the immune microenvironment, thereby affecting the clinical prognosis of PAAD patients. PAAD with high expression of MCM3 is highly sensitive to four drugs, namely c-75, bridanib, fluopiridol and VNLG/124, and the molecular docking model between MCM3 and these drugs is relatively stable. These may fill some gaps in the field of PAAD.
This study integrated for the first time 826 PAAD samples and 476 non-PAAD control samples from 12 datasets around the world. Through comprehensive analysis, it was found that MCM3 was significantly highly expressed in PAAD tissue. Although the sources and production processes of these external datasets may have many obvious differences, for example, due to different standards, the results of the above calculation and analysis show that they can be combined and analysed without significant impact. The same effect was further demonstrated through an IHC scoring analysis of the in-house protein. This indicated that the process of MCM3 from transcription to translation wasn’t interfered with by too many other factors. This is consistent with a previous study that found MCM3 mRNA to be highly expressed in PAAD and MCM3 to be highly expressed in tumours in other anatomical locations, such as head and neck squamous cell carcinoma, lung squamous cell carcinoma, ovarian cancer, breast cancer and cervical cancer [17–21, 24, 25]. This just shows that our results have broad applicability and generalisability. Furthermore, the heterogeneity of PAAD cells reflected by the MCM3 protein in PAAD tissue is similar to the differentiation of several subgroups in the epithelial cell population and the uneven expression of MCM3 in epithelial cells in the single-cell analysis mentioned above. This may hold immense importance for the prognosis of PAAD. It is necessary for us to analyse this further in the future to develop more personalised and effective treatment strategies. In view of the high expression of MCM3 mRNA and protein in PAAD and the strong necessity for MCM3 in the growth of PAAD cells, we analysed the likelihood of MCM3 being a significant contributor to the initiation and growth of PAAD.
Therefore, we explored the possible molecular mechanism of MCM3 in PAAD based on intersection genes to deepen our understanding of the process by which MCM3 promotes the occurrence and growth of PAAD. MCM3 is a member of the MCM family. The elevated expression of MCM3 contributes to sustaining the proliferative state of cancer cells [40]. Specifically, in tumour cells, the small ubiquitin-like modifier (SUMO) can act on MCM3, thereby affecting the MCM-binding protein (MCMBP) associated with MCM3 and playing a crucial role in the assembly of the MCM family, allowing cells to maintain replication permission to promote the proliferation of tumour cells [40, 41]. Studies have also reported that MCM3 can affect KAP1 by interacting with MCM6 and PCNA, thereby affecting the process of restoring heterochromatin after DNA replication. The abnormally high expression of KAP1 may enhance the metastasis ability of PAAD through the epithelial–mesenchymal transition pathway [42, 43]. MCM3 may also play a role in the occurrence and growth of PAAD through an abnormal p53 signalling pathway. Some studies have shown that mutated TP53 may upregulate the transcription factor forkhead box M1 (FoxM1) and its downstream target CCNB through abnormal interactions with the C-terminal disordered region of MCM3 [44, 45]. Research by Duo Han et al. claimed that the NF-κB/miR-488/ERBB2 axis can regulate the growth of PAAD cells through cell cycle regulatory factors such as CDK1 and CCNB [46]. p53 can promote tumour progression by activating genome-wide replication that is closely related to CCNE amplification [47]. Earlier studies confirmed that MCM3 is an important substrate of CCNB/CDK2 [48]. These are similar to our discovery that MCM3 in PAAD interacts with CDK, CCNB and CCNE in the p53 signalling pathway. In addition, the above results show that MCM3 in PAAD may play an extremely complex role in cell cycle checkpoint signalling. Firstly, some reports have shown that MCM3 is involved in the cell cycle checkpoint process of tumour cells [49, 50]. Secondly, some studies have found that abnormal expression of CDK1 not only causes abnormalities in the G2/M cell cycle checkpoint, leading to malignant proliferation that may lead to cancer, but may also promote the development of cancer stem cells (CSCs) by inducing the upregulation of proteins with stem cell properties to promote the occurrence of PAAD [51, 52]. Therefore, MCM3 may be involved in the pathogenesis of PAAD by interacting with CDKs in the cell cycle checkpoint pathway. In addition, PAAD’s MCM3 may be related to the growth, invasion, metastasis and drug resistance of tumour cells through interactions with BLM, BUB3 and CDC6 in the cell cycle checkpoint pathway [53–55].
Although there have been many reports on the immune status of PAAD tissue that clarify the important role and clinical value of the immune microenvironment in the occurrence and development of PAAD [56–58], we have not seen any research on the relationship between MCM3, the immune microenvironment and the clinical prognosis of PAAD. Even the analysis of MCM3 expression status and the tumour immune microenvironment have been rarely reported. For the first time, we found that differences in MCM3 expression levels have an impact on the clinical prognosis of patients by modulating components of the immune microenvironment in PAAD tissue (e.g. Th2 cells, eosinophils and activated CD8+ T cells). Th2 cells are significantly enriched in PAAD tissue and highly express MCM3, which may promote the occurrence of PAAD through the chronic inflammatory pathway and help PAAD cells escape from the anti-tumour process by secreting various immunosuppressive cytokines to antagonise the tumour immune microenvironment and significantly reduce the clinical prognosis of PAAD patients [59, 60]. The significantly enriched eosinophils in PAAD tissue with low expression of MCM3 may enhance anti-tumour immunity in the tumour tissue microenvironment by virtue of their ability to be chemotactic in tumour tissue, participate in inflammatory processes and tissue remodelling and have potential cytotoxic capabilities [61]. The significantly enriched activated CD8+ T cells in PAAD tissue that highly express MCM3 may induce tumour cell apoptosis through the perforin granzyme and Fas–Fas interaction and tumour cell ferroptosis to improve patient prognosis [62].
Since most patients with PAAD have already missed the best opportunity for ‘curative’ surgical treatment when they seek medical treatment, drug treatment is of great significance in the field of PAAD [63–65]. To this end, based on CRISPR screen data, we found for the first time that the relationship between MCM3 and PAAD drug (c-75, brivanib, flavopiridol and VNLG/124) sensitivity is positively correlated. This provides an in-depth study of the MCM3-related molecular mechanism of the drug and offers some insights for improving drug resistance and efficacy in the future. In the molecular docking model, from the perspective of intermolecular binding energy, except for the average docking effect between c-75 and MCM3, the docking effects between brivanib, flavopiridol, VNLG/124 and MCM3 were all ideal. However, in the molecular docking model of these four drugs and MCM3, no strong intermolecular forces were found. This may be related to the relatively strong shape complementarity of the model we constructed. Based on this, in the future, some new functional groups that can generate strong binding forces between molecules can be introduced to improve the chemical structure of existing drugs and further enhance the binding capacity between them. In addition, flavopiridol is a pan-CDK inhibitor that can inhibit the proliferation of various tumour cells by acting on targets such as CDK1, CDK2 and CDK9 [66–69]. Combining the results of our enrichment analysis and PPI network, it is not difficult to find that this drug not only directly acts on the classic CDK1 target but may also act on MCM3, classic CDK1 targets and other pathways through p53 and cell cycle checkpoints. The interaction between related molecules jointly exerts the anti-PAAD effect. However, this analysis needs more evidence in the future.
Although our research has many exciting results, there are at least the following limitations that deserve our attention. First, we used multiple methods to explore many potential molecular mechanisms of MCM3 in PAAD. However, further in vitro and in vivo experiments, such as on the specific pathway through which the highly expressed MCM3 in PAAD significantly enriches Th2 cells in the PAAD immune microenvironment, are needed to verify these results in the future. Second, in the in-house protein analysis, we only included 145 cases in the PAAD group and 29 cases in the non-PAAD group for analysis using the IHC method. In the future, we should expand the sample size and conduct further analysis and verification from multiple analytical angles to obtain more accurate results. Third, we have currently only carried out molecular docking of small-molecule drugs and MCM3. In the future, we should improve the analytical methods and study the role of drugs in MCM3-related pathways from a holistic perspective to obtain more realistic results.
5. Conclusions
In summary, we conclude that MCM3 is likely to be a critical element in promoting the initiation and growth of PAAD. MCM3 may promote the occurrence and growth of PAAD through abnormal DNA replication, the p53 signalling pathway and the cell cycle checkpoint signalling pathway. Flavopiridol may exert its anti-PAAD effect through the interaction between MCM3, classic CDK1 targets in the cell cycle checkpoint and p53 pathway as well as related molecules in other pathways.
Supplementary Material
Acknowledgements
Thank you for the technical support provided by Guangxi Zhuang Autonomous Region Clinical Medicine Research Center for Molecular Pathology and Intelligent Pathology Precision Diagnosis.
Funding Statement
This research project was supported by The First Affiliated Hospital of Guangxi Medical University Provincial and Ministerial Key Laboratory Cultivation Project: Guangxi Laboratory of Enhanced Recovery after Surgery for Gastrointestinal Cancer (NO: 21-220-18), Guangxi Medical High-level Key Talents Training ‘139’ Program (NO:2020), Guangxi Higher Education Undergraduate Teaching Reform Project (NO:2022JGA146), Guangxi Educational Science Planning Key Project (NO:2022ZJY2791) and Guangxi Medical University Undergraduate Education and Teaching Reform Project (NO:2023Z10).
Authors contributions
Gang Chen and Dan-Ming Wei designed the study. Rong-Quan He, Zhi-Guang Huang, Wan-Ying Huang, Jia-Yuan Luo, Yi-Wu Dang obtained, organized data, and conducted experiments. Yi Chen, Liu-Yan Li and Jian-Di Li did the analysis and interpretation of data. Yi Chen wrote the manuscript. Gang Chen and Dan-Ming Wei revised the manuscript. All authors have reviewed and endorsed the final manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Data availability statement
The public data section of this study can be obtained from the Cancer Genome Atlas (TCGA), Gene Expression Omnibus (GEO), Sequence Read Archive (SRA) and Genotype-Tissue Expression (GTEx). In-house data can be obtained from the corresponding author upon reasonable request.
References
- 1.Tonini V, Zanni M.. Pancreatic cancer in 2021: what you need to know to win. World J Gastroenterol. 2021;27(35):5851–5889. doi: 10.3748/wjg.v27.i35.5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Luo W, Wang J, Chen H, et al. Novel strategies optimize immunotherapy by improving the cytotoxic function of T cells for pancreatic cancer treatment. Cancer Lett. 2023;576:216423. doi: 10.1016/j.canlet.2023.216423. [DOI] [PubMed] [Google Scholar]
- 3.Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 4.Qian B, Liu Q, Wang C, et al. Identification of MIR600HG/hsa-miR-342-3p/ANLN network as a potential prognosis biomarker associated with lmmune infiltrates in pancreatic cancer. Sci Rep. 2023;13(1):15919. doi: 10.1038/s41598-023-43174-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou B, Zhang SR, Chen G, et al. Developments and challenges in neoadjuvant therapy for locally advanced pancreatic cancer. World J Gastroenterol. 2023;29(35):5094–5103. doi: 10.3748/wjg.v29.i35.5094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tseng Y-L, Lin Y-C, Hsu W-J, et al. Shared decision making with Oncologists and Palliative care specialists (SOP) model help advanced pancreatic cancer patients reaching goal concordant care: a prospective cohort study. Cancer Med. 2023;12(19):20119–20128. doi: 10.1002/cam4.6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu D, Wang Y, Liu X, et al. Development and clinical validation of a novel 9-gene prognostic model based on multi-omics in pancreatic adenocarcinoma. Pharmacol Res. 2021;164:105370. doi: 10.1016/j.phrs.2020.105370. [DOI] [PubMed] [Google Scholar]
- 8.Ettrich TJ, Seufferlein T.. Systemic therapy for metastatic pancreatic cancer. Curr Treat Options Oncol. 2021;22(11):106. doi: 10.1007/s11864-021-00895-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang VT, Sandifer C, Zhong F.. GI symptoms in pancreatic cancer. Clin Colorectal Cancer. 2023;22(1):24–33. doi: 10.1016/j.clcc.2022.12.002. [DOI] [PubMed] [Google Scholar]
- 10.Moffat GT, Epstein AS, O’Reilly EM.. Pancreatic cancer-A disease in need: optimizing and integrating supportive care. Cancer. 2019;125(22):3927–3935. doi: 10.1002/cncr.32423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kim CA, Lelond S, Daeninck PJ, et al. The impact of early palliative care on the quality of life of patients with advanced pancreatic cancer: the IMPERATIVE case-crossover study. Support Care Cancer. 2023;31(4):250. doi: 10.1007/s00520-023-07709-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schepis T, De Lucia SS, Pellegrino A, et al. State-of-the-art and upcoming innovations in pancreatic cancer care: a step forward to precision medicine. Cancers (Basel). 2023;15(13):3423. doi: 10.3390/cancers15133423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Petri G, Cascioferro S, El Hassouni B, et al. Biological evaluation of the antiproliferative and anti-migratory activity of a series of 3-(6-phenylimidazo[2,1-b][1,3,4]thiadiazol-2-yl)-1H-indole derivatives against pancreatic cancer cells. Anticancer Res. 2019;39(7):3615–3620. doi: 10.21873/anticanres.13509. [DOI] [PubMed] [Google Scholar]
- 14.Carbone D, De Franco M, Pecoraro C, et al. Structural manipulations of marine natural products inspire a new library of 3-amino-1,2,4-triazine PDK inhibitors endowed with antitumor activity in pancreatic ductal adenocarcinoma. Mar Drugs. 2023;21(5):288. doi: 10.3390/md21050288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carbone D, De Franco M, Pecoraro C, et al. Discovery of the 3-amino-1,2,4-triazine-based library as selective pdk1 inhibitors with therapeutic potential in highly aggressive pancreatic ductal adenocarcinoma. Int J Mol Sci. 2023;24(4):3679. doi: 10.3390/ijms24043679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng Y, Huang R, Zou W, et al. Comparing concurrent chemoradiotherapy, 125I seed implantation combined with chemotherapy, and chemotherapy alone efficacy in treating unresectable locally advanced pancreatic cancer. Prec Radiat Oncol. 2022;6(2):144–156. doi: 10.1002/pro6.1156. [DOI] [Google Scholar]
- 17.Wu B, Xi S.. Bioinformatics analysis of the transcriptional expression of minichromosome maintenance proteins as potential indicators of survival in patients with cervical cancer. BMC Cancer. 2021;21(1):928. doi: 10.1186/s12885-021-08674-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun E, Peng L, Liu Z, et al. Systematic analysis of expression and prognostic significance for MCM family in head and neck squamous cell carcinoma. Histol Histopathol. 2023;39(4):471–482. [DOI] [PubMed] [Google Scholar]
- 19.Yang X, Wang C, Nie H, et al. Minichromosome maintenance gene family: potential therapeutic targets and prognostic biomarkers for lung squamous cell carcinoma. Aging (Albany NY). 2022;14(22):9167–9185. doi: 10.18632/aging.204399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Y, Zou J, Zhang Q, et al. Systemic analysis of the DNA replication regulator MCM complex in ovarian cancer and its prognostic value. Front Oncol. 2021;11:681261. doi: 10.3389/fonc.2021.681261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Løkkegaard S, Elias D, Alves CL, et al. MCM3 upregulation confers endocrine resistance in breast cancer and is a predictive marker of diminished tamoxifen benefit. NPJ Breast Cancer. 2021;7(1):2. doi: 10.1038/s41523-020-00210-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Köse B, Laar R V D, Beekhuizen H V, et al. Quantitative proteomic analysis of MCM3 in ThinPrep samples of patients with cervical preinvasive cancer. Int J Mol Sci. 2023;24(13):10473. doi: 10.3390/ijms241310473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu J, Liu S, Yang X.. Construction of gene modules and analysis of prognostic biomarkers for cervical cancer by weighted gene co-expression network analysis. Front Oncol. 2021;11:542063. doi: 10.3389/fonc.2021.542063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ma H, Liu Z, Li H, et al. Bioinformatics analysis reveals MCM3 as an important prognostic marker in cervical cancer. Comput Math Methods Med. 2021;2021:8494260. doi: 10.1155/2021/8494260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liao X, Han C, Wang X, et al. Prognostic value of minichromosome maintenance mRNA expression in early-stage pancreatic ductal adenocarcinoma patients after pancreaticoduodenectomy. Cancer Manag Res. 2018;10:3255–3271. doi: 10.2147/CMAR.S171293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McFarland JM, Ho ZV, Kugener G, et al. Improved estimation of cancer dependencies from large-scale RNAi screens using model-based normalization and data integration. Nat Commun. 2018;9(1):4610. doi: 10.1038/s41467-018-06916-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dempster JM, Boyle I, Vazquez F, et al. Chronos: a cell population dynamics model of CRISPR experiments that improves inference of gene fitness effects. Genome Biol. 2021;22(1):343. doi: 10.1186/s13059-021-02540-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Werba G, Weissinger D, Kawaler EA, et al. Single-cell RNA sequencing reveals the effects of chemotherapy on human pancreatic adenocarcinoma and its tumor microenvironment. Nat Commun. 2023;14(1):797. doi: 10.1038/s41467-023-36296-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang X, Lan Y, Xu J, et al. CellMarker: a manually curated resource of cell markers in human and mouse. Nucleic Acids Res. 2019;47(D1):D721–D728. doi: 10.1093/nar/gky900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang Q, Liu Y, Du Y, et al. Evaluation of cell type annotation R packages on single-cell RNA-seq data. Genomics Proteomics Bioinform. 2021;19(2):267–281. doi: 10.1016/j.gpb.2020.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeng D, Ye Z, Shen R, et al. IOBR: multi-omics immuno-oncology biological research to decode tumor microenvironment and signatures. Front Immunol. 2021;12:687975. doi: 10.3389/fimmu.2021.687975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee J, Kim Y, Jin S, et al. Q-omics: smart software for assisting oncology and cancer research. Mol Cells. 2021;44(11):843–850. doi: 10.14348/molcells.2021.0169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ru B, Wong CN, Tong Y, et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics. 2019;35(20):4200–4202. doi: 10.1093/bioinformatics/btz210. [DOI] [PubMed] [Google Scholar]
- 36.Trott O, Olson AJ.. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Y, Yang X, Gan J, et al. CB-Dock2: improved protein-ligand blind docking by integrating cavity detection, docking and homologous template fitting. Nucleic Acids Res. 2022;50(W1):W159–W164. doi: 10.1093/nar/gkac394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kozłowska M, Śliwińska A.. The link between diabetes, pancreatic tumors, and miRNAs-new players for diagnosis and therapy? Int J Mol Sci. 2023;24(12):10252. doi: 10.3390/ijms241210252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee HS, Jung EH, Shin H, et al. Phenotypic characteristics of circulating tumor cells and predictive impact for efficacy of chemotherapy in patients with pancreatic cancer: a prospective study. Front Oncol. 2023;13:1206565. doi: 10.3389/fonc.2023.1206565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saito Y, Santosa V, Ishiguro KI, et al. MCMBP promotes the assembly of the MCM2-7 hetero-hexamer to ensure robust DNA replication in human cells. Elife. 2022;11:e77393. doi: 10.7554/eLife.77393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Quan Y, Zhang Q-Y, Zhou AL, et al. Site-specific MCM sumoylation prevents genome rearrangements by controlling origin-bound MCM. PLoS Genet. 2022;18(6):e1010275. doi: 10.1371/journal.pgen.1010275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu C, Zhan L, Jiang J, et al. KAP-1 is overexpressed and correlates with increased metastatic ability and tumorigenicity in pancreatic cancer. Med Oncol. 2014;31(7):25. doi: 10.1007/s12032-014-0025-5. [DOI] [PubMed] [Google Scholar]
- 43.Jang SM, Kauzlaric A, Quivy J-P, et al. KAP1 facilitates reinstatement of heterochromatin after DNA replication. Nucleic Acids Res. 2018;46(17):8788–8802. doi: 10.1093/nar/gky580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schaefer-Ramadan S, Aleksic J, Al-Thani NM, et al. Novel protein contact points among TP53 and minichromosome maintenance complex proteins 2, 3, and 5. Cancer Med. 2022;11(24):4989–5000. doi: 10.1002/cam4.4805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jeong JH, Ryu JH.. Broussoflavonol B from Broussonetia kazinoki siebold exerts anti-pancreatic cancer activity through downregulating FoxM1. Molecules. 2020;25(10):2328. doi: 10.3390/molecules25102328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Han D, Zhu S, Li X, et al. The NF-κB/miR-488/ERBB2 axis modulates pancreatic cancer cell malignancy and tumor growth through cell cycle signaling. Cancer Biol Ther. 2022;23(1):294–309. doi: 10.1080/15384047.2022.2054257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zeng J, Hills SA, Ozono E, et al. Cyclin E-induced replicative stress drives p53-dependent whole-genome duplication. Cell. 2023;186(3):528–542.e14. doi: 10.1016/j.cell.2022.12.036. [DOI] [PubMed] [Google Scholar]
- 48.Li J, Deng M, Wei Q, et al. Phosphorylation of MCM3 protein by cyclin E/cyclin-dependent kinase 2 (Cdk2) regulates its function in cell cycle. J Biol Chem. 2011;286(46):39776–39785. doi: 10.1074/jbc.M111.226464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gharbaran R, Sayibou Z, Atamturktur S, et al. Diminazene aceturate-induced cytotoxicity is associated with the deregulation of cell cycle signaling and downregulation of oncogenes Furin, c-MYC, and FOXM1 in human cervical carcinoma Hela cells. J Biochem Mol Toxicol. 2023;38(1):e23527. [DOI] [PubMed] [Google Scholar]
- 50.Zhou H, Xiong Y, Zhang G, et al. Elevated expression of minichromosome maintenance 3 indicates poor outcomes and promotes G1/S cell cycle progression, proliferation, migration and invasion in colorectal cancer. Biosci Rep. 2020;40(7):BSR20201503. doi: 10.1042/BSR20201503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wijnen R, Pecoraro C, Carbone D, et al. Cyclin dependent kinase-1 (CDK-1) inhibition as a novel therapeutic strategy against pancreatic ductal adenocarcinoma (PDAC). Cancers (Basel). 2021;13(17):4389. doi: 10.3390/cancers13174389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zolotovskaia MA, Modestov AA, Suntsova MV, et al. Pan-cancer antagonistic inhibition pattern of ATM-driven G2/M checkpoint pathway vs other DNA repair pathways. DNA Repair (Amst). 2023;123:103448. doi: 10.1016/j.dnarep.2023.103448. [DOI] [PubMed] [Google Scholar]
- 53.Li H, Hao C, Yang Q, et al. Identification of hub genes in chronic pancreatitis and analysis of association with pancreatic cancer via bioinformatic analysis. Gen Physiol Biophys. 2022;41(1):15–30. doi: 10.4149/gpb_2021033. [DOI] [PubMed] [Google Scholar]
- 54.Li J, Hu B, Wang T, et al. C-Src confers resistance to mitotic stress through inhibition DMAP1/Bub3 complex formation in pancreatic cancer. Mol Cancer. 2018;17(1):174. doi: 10.1186/s12943-018-0919-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lan C, Yamashita Y-I, Hayashi H, et al. High expression of bloom syndrome helicase is a key factor for poor prognosis and advanced malignancy in patients with pancreatic cancer: a retrospective study. Ann Surg Oncol. 2022;29(6):3551–3564. doi: 10.1245/s10434-022-11500-9. [DOI] [PubMed] [Google Scholar]
- 56.Zhang X, Zhou Y, Hu J, et al. Comprehensive analysis identifies cuproptosis-related gene DLAT as a potential prognostic and immunological biomarker in pancreatic adenocarcinoma. BMC Cancer. 2023;23(1):560. doi: 10.1186/s12885-023-11042-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shi Y, Wang Y, Dong H, et al. Crosstalk of ferroptosis regulators and tumor immunity in pancreatic adenocarcinoma: novel perspective to mRNA vaccines and personalized immunotherapy. Apoptosis. 2023;28(9-10):1423–1435. doi: 10.1007/s10495-023-01868-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jia Y, Guo B, Zhang W, et al. Pan-cancer analysis of the prognostic and immunological role of GJB2: a potential target for survival and immunotherapy. Front Oncol. 2023;13:1110207. doi: 10.3389/fonc.2023.1110207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhu Y, Zhang C, Zhao D, et al. BDNF acts as a prognostic factor associated with tumor-infiltrating Th2 cells in pancreatic adenocarcinoma. Dis Markers. 2021;2021:7842035–7842022. doi: 10.1155/2021/7842035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li S, Motiño O, Lambertucci F, et al. Protein regulator of cytokinesis 1: a potential oncogenic driver. Mol Cancer. 2023;22(1):128. doi: 10.1186/s12943-023-01802-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gatault S, Legrand F, Delbeke M, et al. Involvement of eosinophils in the anti-tumor response. Cancer Immunol Immunother. 2012;61(9):1527–1534. doi: 10.1007/s00262-012-1288-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang W, Green M, Choi JE, et al. CD8(+) T cells regulate tumour ferroptosis during cancer immunotherapy. Nature. 2019;569(7755):270–274. doi: 10.1038/s41586-019-1170-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pajewska M, Partyka O, Czerw A, et al. Management of metastatic pancreatic cancer-comparison of global guidelines over the last 5 years. Cancers (Basel). 2023;15(17):4400. doi: 10.3390/cancers15174400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lin S. DTX3L mediated ubiquitination of cGAS suppresses antitumor immunity in pancreatic cancer. Biochem Biophys Res Commun. 2023;681:106–110. doi: 10.1016/j.bbrc.2023.09.073. [DOI] [PubMed] [Google Scholar]
- 65.Li W, Li T, Sun C, et al. Identification and prognostic analysis of biomarkers to predict the progression of pancreatic cancer patients. Mol Med. 2022;28(1):43. doi: 10.1186/s10020-022-00467-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mounika P, Gurupadayya B, Kumar HY, et al. An overview of CDK enzyme inhibitors in cancer therapy. Curr Cancer Drug Targets. 2023;23(8):603–619. doi: 10.2174/1568009623666230320144713. [DOI] [PubMed] [Google Scholar]
- 67.Tong J, Tan X, Hao S, et al. Inhibition of multiple CDKs potentiates colon cancer chemotherapy via p73-mediated DR5 induction. Oncogene. 2023;42(12):869–880. doi: 10.1038/s41388-023-02598-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fu L, Mou J, Deng Y, et al. Design, synthesis, and activity assays of cyclin-dependent kinase 1 inhibitors with flavone scaffolds. Front Chem. 2022;10:940427. doi: 10.3389/fchem.2022.940427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sakamoto H, Ando K, Imaizumi Y, et al. Alvocidib inhibits IRF4 expression via super-enhancer suppression and adult T-cell leukemia/lymphoma cell growth. Cancer Sci. 2022;113(12):4092–4103. doi: 10.1111/cas.15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The public data section of this study can be obtained from the Cancer Genome Atlas (TCGA), Gene Expression Omnibus (GEO), Sequence Read Archive (SRA) and Genotype-Tissue Expression (GTEx). In-house data can be obtained from the corresponding author upon reasonable request.