Abstract
A key question in cytokinesis is how the cell division plane is positioned. Whereas microtubules of the mitotic apparatus specify the division site in animal cells, we show here that the nucleus plays this role in the fission yeast Schizosaccharomyces pombe. By centrifuging cells to move the nucleus, we find that the nucleus (or a nuclear-associated structure) actively influences the position of contractile ring assembly during early mitosis. Displacement of the nucleus during this induction period can lead to formation of multiple rings. The nucleus signals its position in a microtubule-independent manner by emitting the protein mid1p. Furthermore, movement of ring fragments together minimizes formation of multiple division sites. These dynamic mechanisms of ring positioning provide a robust coordination of nuclear and cell division.
Keywords: cytokinesis, nucleus, Schizosaccharomyces pombe
In animal cells, experiments involving physical manipulation of the mitotic apparatus demonstrate that elements of the mitotic spindle specify the position of the cleavage furrow during anaphase (1–3). For instance, Harvey (4) centrifuged sea urchin zygotes to displace the mitotic apparatus and the subsequent division plane (4). Rappaport (5) moved the mitotic apparatus repeatedly in cylindrical sand dollar eggs and observed that a single mitotic apparatus could induce the formation of multiple (up to 13) cleavage furrows; each time, a furrow formed at the new position of the mitotic apparatus, whereas the old furrow regressed. In plants and certain fungi, it has been proposed that the nucleus itself influences the positioning of the division plane (6–8). Girbardt (8) found that the site of microfilament ring formation correlated with the site of the nucleus in the hyphal fungus Trametes versicolor after displacing the nucleus by perforating the cell with a needle. Centrifugation studies also have documented a role of the nucleus in positioning of the preprophase band and future division site in higher plant cells (9). However, the molecular nature and source of the signals for division site determination in all these eukaryote organisms remain largely unknown.
Fission yeast Schizosaccharomyces pombe serves as a powerful genetic model organism for studying conserved molecular aspects of cytokinesis (10). Fission yeast cells have a closed mitosis, and both the predivisional nucleus and the ring are normally located at the cell center. In prophase, many contractile ring components, such as myosin and its light chains, accumulate first in a broad band of cortical dots overlying the nucleus in an actin-independent manner. Around metaphase, these ring components, which are associated with network of actin filaments, move together, forming a discrete ring through an actin-dependent process known as compaction (11–14). During anaphase, the mitotic spindle extends in a bidirectional manner, segregating the chromosomes to either side of the ring. At the end of anaphase, the ring marks the position of septum synthesis and contracts, leading to cell cleavage in between the daughter nuclei.
In fission yeast, it has been proposed that the position of the predivisional nucleus specifies the future division plane (6), but definitive proof and understanding of mechanisms are lacking. The primary evidence to date has been largely correlative and observational; for instance, in cells with abnormal MT organization, the nucleus often is displaced, and the cells divide asymmetrically (6). We note, however, that these correlative data also are consistent with an alternate model where, under these circumstances, the nucleus is positioned by some cortical marker located at a predetermined division site.
Molecular insights into ring positioning come from the characterization of the mid1p protein. mid1 mutants have defects in positioning the contractile ring relative to nuclear position (15, 16). Mid1p, which has some similarity to the metazoan contractile ring protein anillin (17), localizes to the nucleus and to a collection of cortical dots that overlie the nucleus during interphase (18). The position of these cortical mid1p dots correlates with the position of the nucleus, even in cells with a displaced nucleus or with multiple nuclei. During early mitosis, mid1p exits the nucleus, accumulates at the medial cortex, and functions to recruit myosin to the area near the nucleus for ring assembly (14, 18–20).
Here, to test the role of the nucleus in ring positioning in a more rigorous and systematic manner, we developed a method to experimentally manipulate nuclear position by centrifuging fission yeast cells. Our data show that the position of the nucleus dictates the site of contractile ring assembly during a period in early mitosis. Furthermore, we provide additional evidence that the nucleus may specify the division site by positioning mid1p, a protein required for ring placement (15, 16).
Materials and Methods
Yeast Strains, Media, and Pharmacological Inhibitors. S. pombe strains are listed in Table 1, which is published as supporting information on the PNAS web site. Standard S. pombe molecular genetics techniques and media were used as described (www-rcf.usc.edu/~forsburg/plasmids.html). Strains carrying nmt-based plasmids were grown in the presence of 5 μg/ml thiamine (Sigma). Methyl-2-benzimidazole-carbamate (MBC, Aldrich) was used at a final concentration of 25 μg/ml from a 100× stock solution made fresh in DMSO (Sigma-Aldrich). Latrunculin A (kindly provided by Phillip Cruz, University of California, Santa Cruz) was used at a final concentration of 100 μM.
Cell Centrifugation. Three different centrifugation protocols were used. First, centrifugation in ultracentrifuge tubes was commonly used in wild-type cells and for some of the experiments with cdc25 cells. Cells were grown on Edinburgh minimal media (EMM) plates for 1–2 days and diluted into liquid EMM culture for two to three generations at 30°C. One milliliter of log-phase cell culture (1–3 × 106 cells per ml) was loaded onto prewarmed, clear 14 × 89-mm ultracentrifugation tubes (Beckman) filled with 10 ml of solidified EMM plus 1% low-melt agarose (Seakem LE, FMC) (Fig. 5, which is published as supporting information on the PNAS web site). Cells were centrifuged at 25°C for 8 min at 25,000 rpm (70,000 × g) (speed reached 25,000 rpm only for 3 min) in an ultracentrifuge in an SW41 swinging bucket rotor (Beckman). Cells, which sedimented into a small indentation on the top of the agarose matrix during centrifugation, were immediately collected and prepared for microscopy (Fig. 6, which is published as supporting information on the PNAS web site). It took ≈2–3 min from the end of centrifugation to the beginning of imaging. To displace the interphase nucleus, cells were pretreated with 25 μg/ml methyl-2-benzimidazole-carbamate (MBC) for 5 min at 30°C, placed on an agarose matrix with MBC, and after centrifugation, washed twice in MBC-free EMM media by using very brief (5-sec) microfuge spins (Figs. 1 a and b, 2a, and 4 b and c). For analysis of mitotic cells, no MBC was added (Fig. 1c).
Fig. 1.
The position of the predivisional nucleus determines the position of ring assembly. Fission yeast cells expressing rlc1-GFP nup107-GFP GFP-atb2 (a and b) (RD48) (markers for the contractile ring, nuclear envelope, and MTs, respectively) and cells expressing rlc1-GFP GFP-atb2 cells (RD104) (c) were centrifuged to displace the nucleus from the middle of the cell, and then imaged by time-lapse 3D confocal microscopy (see Materials and Methods for details). Maximal projections of Z series of representative frames are shown. Shown are cells centrifuged during interphase (a and b), a cell with a displaced nucleus and ring (b), and a cell centrifuged in mid-mitosis (c). (Scale bars, 3 μm.)
Fig. 2.
Effect of nuclear displacement on contractile ring assembly. (a) Asynchronous wild-type cells were centrifuged and assayed for positions of the spindle, nucleus, and rlc1-GFP by using time-lapse microscopy. Percentages of cells forming contractile rings or ring fragments (as visualized by rlc1-GFP) at the specified positions are listed. (b) cdc25-22 cells were synchronized in mitosis by temperature shifts, centrifuged at specified cell-cycle points, and then imaged. (c) cdc25-22 cells were synchronized and centrifuged twice (see Materials and Methods).
Fig. 4.
The nucleus positions mid1p at the cortex. (a–c) Time-lapse images (Left) of cells expressing mid1-GFP. Kymographs (Center and Right) depict mid1-GFP behavior in boxed regions located at the cortex (c) and the nuclear envelope (n). (a) Interphase mid1-GFP cells (FC958) without centrifugation. (b and c) Two representative examples of interphase mid1-GFP cells in which the nucleus is recentering, after centrifugation. An arrow indicates large nuclear envelope deformation associated with nuclear movement. Asterisks mark representative mid1-GFP cortical dots that move with the nucleus. (Scale bars, 3 μm; kymographs, ×1.5.)
Second, we sometimes centrifuged elongated cdc25 cells in a microcentrifuge. This faster method also could displace the nucleus in shorter wild-type cells, but the efficiency was lower than the ultracentrifuge method. Centrifugation chambers were constructed by adding molten EMM plus 1% agarose into 1.5-ml tubes (Eppendorf) and then adding a 0.5-ml centrifuge adapter (USA Scientific, Ocala, FL) to form an indentation in the agarose (Fig. 6). In general, cdc25-22 cells were grown in EMM plates for 2–3 days at 25°C and grown in liquid EMM for one generation at 25°C and at 36°C for 4 h. One milliliter of cell culture (1–3 × 106 cells per ml) was concentrated 5- to 10-fold, loaded into the agarose well, and centrifuged for 2 min at 14,000 rpm (16,110 × g) in a microcentrifuge (model 5415C, Eppendorf). Cells were collected and imaged within 1–2 min. In some experiments, tubes were preheated at 36°C (Fig. 3f).
Fig. 3.
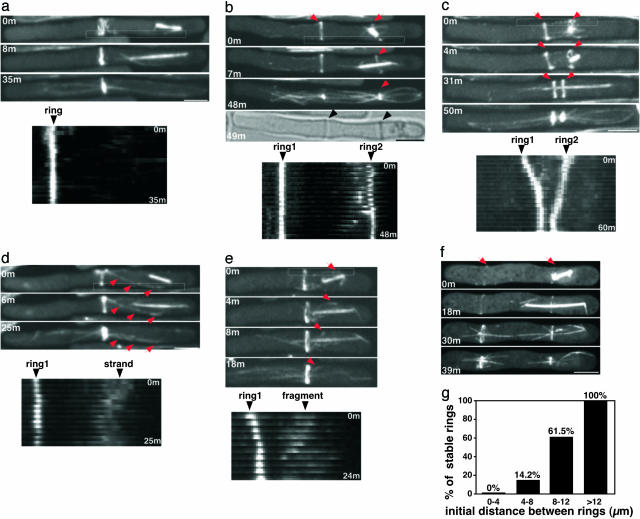
Induction of extra contractile rings and ring fragments by the nucleus. cdc25-22 rlc1-GFP GFP-atb2 cells (RD129) were synchronized in mitosis by temperature shift to 36°C for 4 h, released to 25°C, and then centrifuged to displace the nucleus. Representative images of maximum Z-series projections and kymographs of the indicated boxed regions are shown. (a) Example of a cell centrifuged during anaphase that did not form a second ring or ring fragment. (b) Example of a cell centrifuged in early mitosis that assembled a second ring at the position of the displaced nucleus (arrows). Both rings were stable and functional, because they contracted and directed septum assembly (arrow heads). (c) Example of two rings that move toward each other. (d) Example of a stable strand of rlc1-GFP that extended from the region of displaced nucleus to the central ring. (e) Example of an rlc1-GFP fragment that moved from the nucleus to the ring at the old site, fusing with it (arrow at 18 min). (f) Example of cell after a double-centrifugation procedure (see Materials and Methods) that assembled two stable rings near each cell tip. (g) In cells with two rings, the percentages of cells in which rings do not move toward each other were compared with the initial distances between the rings (n ≥ 5 each point; data from both single- and double-centrifugation protocols). (Scale bars, 5 μm; kymographs, ×1.5.)
Third, to generate rings far apart, we used a double-centrifugation protocol. cdc25-22 cells were grown at 36°C for 4 h, treated with MBC (25 μg/ml) for 2 min, centrifuged in preheated (36°C) agarose chambers with MBC at 14,000 rpm (16,110 × g) in a microcentrifuge for 2 min. Cells were incubated at 36°C for 10 min and shifted to the permissive temperature (25°C) to induce the entry into mitosis. After 20–30 min, MBC was washed out, and cells were checked for mitotic progression by using fluorescence microscopy. Cells were then centrifuged again in agarose chambers without MBC at 25°C during early mitosis to induce the formation of an extra ring (Figs. 2c and 3 f and g).
Microscopy, Image Acquisition, and Sample Preparation. For imaging, cells were mounted onto agarose pads (21).
Images were acquired by using a spinning disk confocal fluorescence upright microscope (PerkinElmer/Wallac, Wellesley, MA; Nikon) (21, 23). Typically, Z series (0.5 μm apart) were obtained every 150 sec for 20–90 min. Temperature was controlled by using an objective heater (Bioptechs, Butler, PA). open lab 3 (Improvision, Lexington, MA) and image j (http://rsb.info.nih.gov/ij) software were used for image acquisition, analysis, and kymograph construction.
Results
To experimentally manipulate the position of the nucleus in fission yeast cells, we developed a method of cell centrifugation. Because MT bundles normally position the interphase nucleus at the cell center (24), we first treated cells with a reversible MT inhibitor, methyl-2-benzimidazole-carbamate (MBC), and then centrifuged the cells in an agarose matrix (see Materials and Methods). Under these conditions, >80% of the cells exhibited an offset nucleus after centrifugation. In contrast with a previous centrifugation study in fission yeast (25), we used relatively low centrifugation speeds, so that >95% cells were viable and nuclear integrity and F-actin organization were unperturbed after centrifugation (data not shown). We then washed out the MBC and imaged the subsequent behavior of the nucleus, MTs, and contractile ring by using GFP fusions to nuclear pore proteins, α-tubulin, and myosin regulatory light chain rlc1p (13, 24, 26).
After being displaced by centrifugation, the nucleus in interphase cells gradually moved back to the cell center in a MT-dependent manner. Some cells entered mitosis before the nucleus recentered. All of these cells formed a ring and divided asymmetrically at the position of the displaced nucleus (100%, n = 36; Figs. 1b and 2a). In cells where the nucleus moved back to the middle, they divided in the middle (Fig. 1a), suggesting that centrifugation did not merely displace a nuclear-independent ring determinant. In all cases, cells divided at the position of the predivisional nucleus. Thus, these findings suggest that the position of the predivisional nucleus may actively specify the position of ring assembly.
We then examined the effect of moving the nucleus during mitosis. The mitotic nucleus could be moved by centrifugation even in the absence of MBC, because cytoplasmic MTs depolymerize in mitosis. Mitotic stages were assayed by spindle length and behavior over time. In analyzing asynchronous wild-type cells, we found that movement of the nucleus during metaphase did not alter the division site (Fig. 1). These results suggested that the nucleus influences ring positioning in a cell period sometime in early mitosis, before metaphase.
To examine these cell-cycle effects further, we analyzed temperature-sensitive cdc25-22 cells that were synchronized in mitosis. The cell-cycle block and release caused cdc25-22 cells to elongate, allowing for larger displacement of the nucleus (Fig. 2b) (27). All cells centrifuged in mitosis still assembled a primary ring at the central position (Fig. 2). In addition, 34% of cdc25 cells (n = 53) assembled a stable secondary ring in the vicinity of the displaced nucleus (Figs. 2b and 3b). Both the primary and secondary rings were functional, because they subsequently contracted at the same time and directed septum assembly (Fig. 3b and Movie 1, which is published as supporting information on the PNAS web site). In addition, other cells assembled incomplete rlc1-GFP rings, strands, or dots in the vicinity of the displaced nucleus (Figs. 2b and 3 d and e). Generally, these secondary structures were more stable and extensive in cells centrifuged in early mitosis and were unstable and less frequent in cells centrifuged later in mitosis (Fig. 3 a and b). Similar secondary structures, including ring-like structures, also were seen in centrifuged cdc25+ cells, showing that these effects were not dependent on the cdc25 background (Fig. 6). Thus, these results suggest that the nucleus is capable of inducing the assembly of ring structures during a discrete cell-cycle period from prophase through metaphase.
Time-lapse recordings revealed that the multiple rings or ring fragments often moved together in a “compaction” process. Ring fragments (rlc1-GFP dots or pieces) at the site of the displaced nucleus frequently moved toward the established central ring (Fig. 3e and Movie 2, which is published as supporting information on the PNAS web site). In some cases, whole secondary rings moved toward the primary ring (Fig. 3c) and often fused with it. Occasionally, both rings moved toward each other, whereas in other cells, neither ring moved (Fig. 3 b and c). The stability of the rings correlated with the distance between them (Fig. 3g). For instance, in relatively short wild-type cells, a secondary ring usually moved toward and merged with the primary ring (Fig. 6). To further test whether rings far apart may be more stable, we generated elongated cells with two rings near each cell tip (13.8 ± 2.3 μm apart) by centrifuging cells twice (see Materials and Methods). In most cases (54%, n = 11), two rings formed and contracted at the position where they were assembled (Figs. 2 c and 3 f and g). Therefore, ring compaction, which functions in normal ring assembly, also may function to bring together extra ring structures within some distance of each other (≈8 μm; wild-type mitotic cells are 14 μm long), minimizing the deleterious effects of forming multiple division planes.
We speculate that the nucleus may specify the position of ring assembly by producing a “signal” to the cortex. Mid1p, a protein required for ring positioning, is a candidate for such a signal (15, 16). To determine how the nucleus affects mid1p, we examined mid1p behavior after moving the nucleus. In uncentrifuged cells, mid1p cortical dots were stationary and stable (Fig. 4a). Maintenance of these dots is not dependent on actin or MTs (18). Upon cell centrifugation, cortical mid1p dots relocated with the nucleus to a displaced position (Fig. 4 b and c). Time-lapse microscopy showed that mid1p dots moved toward the nucleus upon centrifugation and then moved with the nucleus as the nucleus recentered (Fig. 4 b and c and Movies 3 and 4, which are published as supporting information on the PNAS web site). Interestingly, individual dots moved with the nucleus, and groups of dots moved at different rates. By treating cells with MBC or Lat-A before centrifugation, we found that the movement of mid1p cortical dots to a new site did not depend on MTs or actin (data not shown).
We then examined mid1p during the formation of secondary rings or fragments in cdc25 cells centrifuged in early mitosis. In these cells, mid1p localized like rlc1p to the primary ring and to a secondary ring or cortical dots near the displaced nucleus. At the secondary site, the amount of mid1-GFP accumulation appeared to depend on how early in the induction period the nucleus was moved. These findings suggest that mid1p is progressively exported from the nucleus to the adjacent cortex during early mitosis.
A key question is how the nucleus may position mid1p. One current model is that nuclear shuttling concentrates mid1p in the perinuclear region. However, a conceptual difficulty of this model is that the expected rate of free diffusion of mid1p from the nucleus to the cortex is too great to account for its precise cortical distribution. We found that blocking nuclear export did not affect ring positioning. Secondary rlc1-GFP rings (Fig. 7, which is published as supporting information on the PNAS web site) and mid1p dots (data not shown) still appeared near the displaced nucleus even when the nuclear export of mid1p was blocked by leptomycin B treatment (18) before centrifugation in G2 phase. Thus, the positioning of cortical mid1p may not rely solely on nuclear export and diffusion. Rather, these findings suggest that cortical mid1p dots are somehow physically tethered to the nucleus in a manner independent of actin and MTs.
Discussion
Here, we provide evidence for two mechanisms that ensure the proper spatial coordination between cytokinesis and nuclear division in fission yeast. First, using centrifugation to move the nucleus, we show that cells divide at the position of the predivisional nucleus. These data provide the strongest evidence to date for a model that the predivisional nucleus (or another structure associated with the nucleus) induces ring assembly in its vicinity (18). Our observations are not consistent with an alternative model where cortical elements at the future division site position the nucleus. Centrifugation at specific cell-cycle periods further showed that ring positioning occurs during early mitosis; by metaphase, the position of the division plane is set. Movement of the nucleus during this inductive period often resulted in the assembly of functional rings at multiple sites, illustrating that all or most parts of the cortex are competent to respond to the putative ring-positioning signal.
Mid1p appears to be part of an inductive “signal” emitted from the nucleus. Whereas the putative “cleavage stimulus” in animal cells is thought to emanate from the mitotic spindle and travel to the cortex on MTs, S. pombe mid1p emanates from the nucleus and may be positioned by as-yet-unidentified structures that connect the nucleus and cortex. The identities of these putative connections are currently mysterious, because they are clearly independent of MTs and actin and also are not apparent in any previously characterized intracellular structures in these cells. Mid1p has some similarities to the metazoan contractile ring protein anillin, but possible roles of anillin in ring positioning are not yet evident (28).
A secondary mechanism of compaction reduces the chances of forming multiple division sites. We observed that multiple ring fragments or rings often moved together. Compaction of ring components, which is also a normal process in ring assembly, is likely to occur by myosin-driven movement of ring components together into a tight ring structure (11–14). Similar movement of multiple furrows together also has been seen in manipulated sand dollar eggs (5). The dynamic nature of these ring-placement mechanisms provides a robust process for cell division that can adapt to perturbations in cellular organization.
Supplementary Material
Acknowledgments
We thank A. Yonetani and other members of the Chang laboratory; A. Paoletti, D. Burgess, J. Moseley, B. Goode, and J. Glynn for discussion and critical reading of the paper; and V. Simanis, I. Hagan, Y. Hiraoka, and D. Ding for valuable reagents. F.C. would like to dedicate this paper to Ray Rappaport.
Author contributions: R.R.D. and F.C. designed research; R.R.D. performed research; R.R.D. analyzed data; and R.R.D. and F.C. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: MBC, methyl-2-benzimidazole-carbamate; MT, microtubule.
References
- 1.Rappaport, R. (1996) in Developmental and Cell Biology Series, eds. Barlow, P. W., Bard, J. B. L., Green, P. B. & Kirk, D. L. (Cambridge Univ. Press, Cambridge, U.K.).
- 2.Rappaport, R. (1986) Int. Rev. Cytol. 105, 245-281. [DOI] [PubMed] [Google Scholar]
- 3.Burgess, D. R. & Chang, F. (2005) Trends Cell Biol. 15, 156-162. [DOI] [PubMed] [Google Scholar]
- 4.Harvey, E. B. (1935) Biol. Bull. (Woods Hole, Mass.) 69, 287-297. [Google Scholar]
- 5.Rappaport, R. (1985) J. Exp. Zool. 234, 167-171. [DOI] [PubMed] [Google Scholar]
- 6.Chang, F. & Nurse, P. (1996) Cell 84, 191-194. [DOI] [PubMed] [Google Scholar]
- 7.Verma, D. P. (2001) Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 751-784. [DOI] [PubMed] [Google Scholar]
- 8.Girbardt, M. (1979) Exp. Mycol. 3, 215-228. [Google Scholar]
- 9.Mineyuki, Y. (1999) Int. Rev. Cytol. 187, 1-49. [Google Scholar]
- 10.Guertin, D. A., Trautmann, S. & McCollum, D. (2002) Microbiol. Mol. Biol. Rev. 66, 155-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Motegi, F., Nakano, K. & Mabuchi, I. (2000) J. Cell Sci. 113, 1813-1825. [DOI] [PubMed] [Google Scholar]
- 12.Naqvi, N. I., Eng, K., Gould, K. L. & Balasubramanian, M. K. (1999) EMBO J. 18, 854-862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Le Goff, X., Motegi, F., Salimova, E., Mabuchi, I. & Simanis, V. (2000) J. Cell Sci. 113, 4157-4163. [DOI] [PubMed] [Google Scholar]
- 14.Wu, J. Q., Kuhn, J. R., Kovar, D. R. & Pollard, T. D. (2003) Dev. Cell 5, 723-734. [DOI] [PubMed] [Google Scholar]
- 15.Sohrmann, M., Fankhauser, C., Brodbeck, C. & Simanis, V. (1996) Genes Dev. 10, 2707-2719. [DOI] [PubMed] [Google Scholar]
- 16.Chang, F., Woollard, A. & Nurse, P. (1996) J. Cell Sci. 109, 131-142. [DOI] [PubMed] [Google Scholar]
- 17.Field, C. M. & Alberts, B. M. (1995) J. Cell Biol. 131, 165-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paoletti, A. & Chang, F. (2000) Mol. Biol. Cell 11, 2757-2773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bahler, J., Steever, A. B., Wheatley, S., Wang, Y., Pringle, J. R., Gould, K. L. & McCollum, D. (1998) J. Cell Biol. 143, 1603-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Motegi, F., Mishra, M., Balasubramanian, M. K. & Mabuchi, I. (2004) J. Cell Biol. 165, 685-695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tran, P. T., Paoletti, A. & Chang, F. (2004) Methods 33, 220-225. [DOI] [PubMed] [Google Scholar]
- 22.Zimmerman, S., Tran, P. T., Daga, R. R., Niwa, O. & Chang, F. (2004) Dev. Cell 6, 497-509. [DOI] [PubMed] [Google Scholar]
- 23.Pelham, R. J. & Chang, F. (2001) Nat. Cell Biol. 3, 235-244. [DOI] [PubMed] [Google Scholar]
- 24.Tran, P. T., Marsh, L., Doye, V., Inoue, S. & Chang, F. (2001) J. Cell Biol. 153, 397-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khar, A. & Mitchison, J. M. (1989) J. Cell Sci. 92, 345-348. [DOI] [PubMed] [Google Scholar]
- 26.Ding, D. Q., Chikashige, Y., Haraguchi, T. & Hiraoka, Y. (1998) J. Cell Sci. 111, 701-712. [DOI] [PubMed] [Google Scholar]
- 27.Russell, P. & Nurse, P. (1986) Cell 45, 145-153. [DOI] [PubMed] [Google Scholar]
- 28.Straight, A. F., Field, C. M. & Mitchison, T. J. (2005) Mol. Biol. Cell 16, 193-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.