Abstract
This study had 2 objectives: 1) to determine the involvement of Mycoplasma hyopneumoniae in respiratory outbreaks in herds of pigs, with the use of a nested polymerase chain reaction (nPCR) and an enzyme-linked immunosorbent assay (ELISA); and 2) to determine if the dynamics of M. hyopneumoniae infection differ between 3-site versus 1- or 2-site production systems (in which at least farrowing/gestation and nursery pigs are on the same site). Animals of different ages from 12 Spanish farms with respiratory problems were randomly sampled. Blood samples and nasal swabs were collected in a single farm visit, and ELISA and nPCR tests, respectively, were performed. All the farms demonstrated M. hyopneumoniae. According to the proportions of infected animals and the appearance of clinical signs in the different age groups, the farms were divided into 2 groups: farms in which M. hyopneumoniae probably played an important role in the observed respiratory outbreak and farms in which M. hyopneumoniae was not the main agent involved in the outbreak. Although seroconversion occurred in most herds in the finishing units, the number of seropositive pigs in the first group of farms was greater than the number in the second group. Statistically significant differences (P < 0.0001) between farms with a 1- or 2-site production system versus those with a 3-site production system were detected in nPCR results but not in rates of seroconversion. The farm effect also had a great influence on both controlled parameters: the pathogen’s DNA and antibody detection. Thus, although M. hyopneumoniae was present in all the studied farms, there were significant differences in the infection dynamics and clinical implications according to the type of production system, and M. hyopneumoniae colonization and seroconversion were greatly influenced by the effect of the individual farm.
Résumé
Les objectifs de cette étude étaient 1) déterminer l’implication de Mycoplasma hyopneumoniae lors de problèmes respiratoires dans des troupeaux de porcs à l’aide d’une épreuve nichée d’amplification en chaîne par la polymérase (nPCR) et une épreuve ELISA, et 2) determiner si la dynamique de l’infection par M. hyopneumoniae diffère entre des systèmes de production en 3 sites versus des systèmes à 1 ou 2 sites (où au moins les unités mise-bas/gestation et pouponnière sont sur un même site). Des animaux d’âges variés provenant de 12 fermes espagnoles au prise avec des problèmes respiratoires ont été échantillonnés au hasard. Des échantillons de sang et des écouvillons nasaux ont été prélevés lors d’une visite unique à la ferme, et les épreuves nPCR et ELISA effectuées. La présence de M. hyopneumoniae a été démontrée dans tous les élevages. Selon les proportions d’animaux infectés et les manifestations cliniques dans les différents groupes d’âge, les élevages ont été divisés en 2 groupes: les fermes où M. hyopneumoniae jouait un rôle important dans les problèmes respiratoires observes et les fermes où M. hyopneumoniae n’était pas l’agent principal en cause. Bien qu’une séroconversion se soit produite dans presque tous les troupeaux dans les unités de finition, le nombre de porcs positifs dans le premier groupe de ferme étaient plus grand que le nombre dans le second groupe. Des différences significatives (P < 0,0001) entre les fermes à 1 ou 2 sites de production versus celles à trois sites de production ont été trouvés en ce qui regarde les résultats de nPCR mais pas dans les taux de séroconversion. Le facteur ferme avait également une grande influence sur les deux paramètres vérifiés : la détection de l’ADN du pathogène et la détection d’anticorps. Ainsi, bien que M. hyopneumoniae soit présent dans toutes les fermes faisant partie de l’étude, il y avait des différences significatives dans les dynamiques de l’infection et les implications cliniques selon le type de système de production, de sorte que la colonisation et la séroconversion dues à M. hyopneumoniae étaient grandement influencées par les effets de chaque ferme.
(Traduit par Docteur Serge Messier)
Introduction
Mycoplasma hyopneumoniae is the primary agent of enzootic pneumonia (EP) in pigs. This chronic worldwide disease causes important economic losses. The use of just 1 diagnostic technique [culture, serology, or polymerase chain reaction (PCR)] to implement prevention programs and to perform epidemiologic studies could be inappropriate owing to limitations inherent in each technique. A definitive diagnosis should involve the combination of some of these diagnostic tools (1). The combination of serology [enzyme-linked immunosorbent assay (ELISA)] and nested PCR (nPCR) in nasal swabs has provided new and useful information about M. hyopneumoniae infection and dynamics (1). The detection of microorganisms in nasal swabs provides accurate information about the time of infection (2,3), whereas serology indicates the time of seroconversion, which is variable in mycoplasma infection in swine (1).
Different studies have shown that the clinical outcome of M. hyopneumoniae infection is dependent on environmental and management factors. The latter include high replacement rates (4), multiple-origin nursery or fattening stock (5), continuous-flow practices (6), and high animal density, all these factors having a great impact on disease outcome (4). In contrast, all-in/all-out procedures in all production stages can reduce the prevalence and severity of EP (7). Modern swine production systems include 3 sites, for the 3 stages of production (farrowing/gestation, nursery, and fattening) (8), which implies a further step in breaking transmission between different age groups (9).
The objectives of this study were: 1) to determine the involvement of M. hyopneumoniae in respiratory outbreaks in herds of pigs, with the use of nPCR and ELISA; and 2) to determine if the dynamics of M. hyopneumoniae infection differ between 3-site versus 1- or 2-site production systems (in which at least farrowing/gestation and nursery pigs are on the same site).
Materials and methods
Herds
Twelve Spanish herds with clinical respiratory symptoms (coughing) in the nursery or fattening stage, or both stages, were included in the study. Of the 12 farms, 7 had more than 2000 sows, 3 had 1000 to 2000 sows, and 2 had about 700 sows (Table I). Of the 12 farms, 5 were using M. hyopneumoniae vaccination (Stellamune; Pfizer Canada, Kirkland, Quebec), 8 were using a specific medication against the microorganism at the time of sampling, and 2 farms were not vaccinated or medicated.
Table I.
Main characteristics of the 12 farms tested for the presence of Mycoplasma hyopneumoniae
| Production (number of sows)
|
Vaccination against M. hyopneumoniae |
Medication against M. hyopneumoniae |
|||||
|---|---|---|---|---|---|---|---|
| Farm number | 1 or 2 sites | 3 sites | Yes/No | Age (wk) | Yes/No | Stage | Other respiratory pathogens detected |
| 1 | 4000 | No | Yes | F | PRRSV | ||
| 2 | 2400 + 1000 | Yes | 1–3 | Yes | N, F | SIV | |
| 3 | 6000 | No | Yes | N,F | PCV2 | ||
| 4 | 750 | No | Yes | N, F | PRRSV, App, PCV2 | ||
| 5 | 2600 | No | No | PRRSV, SIV, App | |||
| 6 | 7000 | Yes | 1–3 | No | PRRSV, PCV2, App, P multocida | ||
| 7 | 5250 | Yes | 1–3 | No | PRRSV | ||
| 8 | 1500 | Yes | 3–6 | Yes | PRRSV, App | ||
| 9 | 600 | Yes | 1–3 | No | PRRSV, ADV, SIV | ||
| 10 | 1500 | No | Yes | F | App, H. parasuis | ||
| 11 | 2400 | No | No | PRRSV, PCV2, ADV | |||
| 12 | 1064 | No | Yes | N, F | PRRSV, ADV, App, H. parasuis, P multocida | ||
N — nursery; F — finishing; PRRSV — porcine reproductive and respiratory syndrome virus; SIV — swine influenza virus; PCV2 — porcine circovirus type 2; App — Actinobacillus pleuropneumoniae; P. — Pasteurella; ADV — Aujeszky’s disease virus; H. — Haemophilus
Samples
Samples were taken from different age groups during a single visit, starting with suckling piglets and then sampling at different weekly intervals depending on the available groups in each farm. Sampled animals were selected randomly. The age range was 1 to 28 wk. Nasal swabs (Culturette; Becton Dickinson, Le Pont de Claix, France) were obtained by swabbing the mucosa of both nostrils, reaching deeply into the turbinates. Blood samples were collected from the jugular vein into tubes without anticoagulant (5-mL Venoject; Terumo Europe, Madrid, Spain). Samples were individually identified, refrigerated, and delivered by express mail to the laboratory, where they were processed.
DNA extraction
The nasal swabs were resuspended in 1000 μL of sterile phosphate-buffered saline (PBS) and vigorously vortexed; 400 μL of the suspension was used for DNA extraction. The samples were processed with a DNA extraction kit (Nucleospin Blood; Macherey-Nagel GmbH & Co KG, Düren, Germany), according to the manufacturer’s instructions. To test for contamination during the extraction procedure, we included a negative control, using PBS as the extraction substrate, in each group of processed samples.
Polymerase chain reaction
The 4 stages of the PCR process (DNA extraction, mix preparation, DNA amplification, and electrophoresis) were conducted in 4 different rooms to minimize contamination. We performed nPCR using primers and conditions previously described (3). The specificity of this technique had previously been assessed with the use of DNA of different mycoplasmas and acholeplasmas, as well as other bacterial species found in the respiratory tract of swine, of which only M. hyopneumoniae was amplified (3). The nPCR has a sensitivity of 80 mycoplasmal cells (3).
Two sets of primers from the 16S ribosomal gene of M. hyopneumoniae were used. Briefly, 2.5 μL of the DNA preparation was used as PCR templates in the first reaction and 0.5 μL in the second reaction. Amplification was performed in a final volume of 25 μL. The reaction mixture consisted of 200 nM of each primer, 0.2 mM of each nucleotide (Amersham–Pharmacia Biotech, Barcelona, Spain), 1 × PCR buffer (Ecogen, Barcelona, Spain), 4 mM MgCl2 (Ecogen), and 1 U of Taq DNA polymerase (Ecogen).
The 2 reactions were performed in a thermocycler (GeneAmp PCR system 9700; Applied Biosystems, Madrid, Spain) under the same conditions: 30 cycles, denaturation at 94°C for 30 s, annealing at 60°C for 45 s, and extension at 72°C for 30 s. In each run, we included with every 4 test samples a negative control consisting of millipore water, as well as a positive control in the last position (DNA extracted from a pure culture of M. hyopneumoniae). If 1 negative control was PCR-positive, all positive samples were tested again.
The amplified products (300 base pairs) were run in a 2% agarose gel with 0.05 mg/mL of ethidium bromide and then visualized and pictured (Bio-1d V.96; Vilber Lourmat, Marine la Vallee, France).
Enzyme linked immunosorbent assay
Blood samples were centrifuged at 760 × g for 10 min at 4°C; the serum was used for the ELISA technique. Antibodies against M. hyopneumoniae were assayed with a monoclonal blocking ELISA (DAKO M. hyopneumoniae, ELISA kit; DAKO A/S, Glostrup, Denmark). The sensitivity of the monoclonal antibody used (mAb17, 74 kDa) has been shown to be 103 color changing unit (CCU) in the 20 M. hyopneumoniae strains tested (10). No reactions of the mAb 17 were observed with 105 CCU of M. flocculare, M. hyorhinis, M. hyosynoviae, or M. dispar.
The inhibition percentage (IP) was calculated considering the optical density (OD) of each sample as well as the negative control. Classification of individual animals on the basis of IP values was as follows: IP < 30%, negative; IP > 50%, positive; IP > 30% and < 50%, suspicious. The percentage was obtained from the following formula: % IP = (mean negative value OD − sample OD) ÷ (mean negative value OD).
On farms 4, 5, 7, and 9, where other respiratory pathogens were suspected to be present, we also performed serologic tests for antibodies against porcine respiratory and reproductive syndrome virus (PRRSV) (Porcine Respiratory and Reproductive Syndrome virus Antibody test kit; IDEXX Europe, BV, Schiphol-Rijk, The Netherlands), Aujeszky’s disease virus (Pseudorabies Virus gpI Antibody Test Kit; IDEXX Europe), swine influenza virus (SIV) (Civtest suis Influenza; Hipra, Spain), and Actinobacillus pleuropneumoniae (Civtest suis App.; Hipra).
Other techniques
Immunohistochemistry was also used to detect SIV antigen (on farm 2) and in situ hybridization to detect porcine circovirus type 2 (11) (on farms 3 and 5, where high mortality rates and poor performance in the finishing pigs were observed).
Statistical analysis
To study the relationships between variables, we performed a descriptive univariate test using analysis of variance and chi-squared tests. To analyze the influence of different variables on M. hyopneumoniae infection in 1-site or 2-site versus 3-site production systems, we used 2 multivariate generalized linear models with a random effect analyzed using a generalized estimating equations (GEE) method, measuring infection by M. hyopneumoniae DNA detection in the first model and by antibody detection in the second model. The factors included in both models were age, production system (3-site versus 1- or 2-site), vaccination, medication, and farm (random effect). The farm effect included factors that were not controlled in the study, such as stocking densities, farm localization, genetics, weather, nutrition, and concomitant infections, among others. Vaccination and medication were nested within farm. All analyses were based on type III sums of squares.
To study the dynamics of M. hyopneumoniae infection in 1- or 2-site (n = 5) versus 3-site (n = 7) production systems, we tested the differences in least square means (LSMeans). To compare the prevalence of M. hyopneumoniae antibodies in the 12 tested farms, we performed a chi-squared test with the ELISA results. The level of significance was set to P values lower than 0.05.
The statistical analysis was performed with the use of the computer software (SAS System for Windows, version 8.0; SAS Institute Inc., Cary, North Carolina, USA).
Results
Respiratory outbreak by nPCR and serology results
A total of 704 nasal swabs and 706 serum samples were processed. Both techniques detected M. hyopneumoniae infection on all 12 farms, but at different percentages. Table II presents the percentages of positive pigs with each test in the 5 age groups. The 12 farms were divided into 2 groups according to the M. hyopneumoniae nPCR results and the appearance of clinical signs: A) farms in which the proportion of nPCR-positive pigs was zero or very low when coughing was observed and remained low (< 30%) throughout the older groups (farms 2, 3, 4, 5, 7, and 12); and B) farms in which the proportion of nPCR-positive pigs was high when coughing was observed and increased in the older groups (farms 1, 6, 8, 9, 10, and 11).
Table II.
Percentages of pigs positive for Mycoplasma hyopneumoniae by nested polymerase chain reaction (nPCR) or enzymelinked immunosorbent assay (ELISA) by farm and age
| % positive by nPCR; age (wk)
|
% positive by ELISA; age (wk)
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Farm number | 1–5 | 6–10 | 11–15 | 16–20 | 21–25 + | 1–5 | 6–10 | 11–15 | 16–20 | 21–25 + |
| 2 | 10 (0.0) | 10 (0.0) | 20 (5.0) | 10 (0.0) | 10 (30.0) | 10 (10.0) | 10 (60.0) | 20 (35.0) | 10 (10.0) | 10 (15.0) |
| 3 | 10 (0.0) | 30 (0.0) | 10 (0.0) | 10 (20.0) | 10 (20.0) | 30 (0.0) | 10 (0.0) | 10 (35.0) | ||
| 4 | 10 (10.0) | 30 (6.7) | 10 (30.0) | 10 (0.0) | 30 (6.7) | 10 (30.0) | 9 (33.3) | |||
| 5 | 19 (15.8) | 10 (0.0) | 10 (20.0) | 10 (30.0) | 19 (0.0) | 10 (0.0) | 10 (0.0) | 10 (35.0) | ||
| 7 | 16 (62.5) | 20 (10.0) | 20 (10.0) | 10 (30.0) | 16 (12.5) | 20 (0.0) | 10 (0.0) | 10 (0.0) | 10 (65.0) | |
| 12 | 10 (0.0) | 20 (0.0) | 10 (30.0) | 20 (10.0) | 10 (50.0) | 18 (11.1) | 8 (62.5) | 18 (27.8) | ||
| 1 | 27 (0.0) | 19 (0.0) | 8 (12.5) | 5 (40.0) | 9 (55.6) | 27 (40.7) | 19 (10.5) | 8 (12.5) | 5 (80.0) | 9 (66.7) |
| 6 | 10 (10.0) | 20 (5.0) | 20 (20.0) | 10 (70.0) | 10 (20.0) | 20 (10.0) | 20 (25.0) | 10 (100.0) | ||
| 8 | 10 (20.0) | 20 (45.0) | 10 (50.0) | 10 (20.0) | 10 (60.0) | 10 (0.0) | 30 (0.0) | 10 (10.0) | 10 (20.0) | 10 (30.0) |
| 9 | 20 (15.0) | 10 (10.0) | 10 (40.0) | 8 (37.5) | 20 (15.0) | 10 (20.0) | 10 (30.0) | 8 (87.5) | ||
| 10 | 19 (15.8) | 10 (50.0) | 9 (66.7) | 9 (77.8) | 19 (42.1) | 10 (20.0) | 9 (33.3) | 9 (33.3) | ||
| 11 | 20 (20.0) | 20 (10.0) | 27 (29.6) | 9 (55.6) | 20 (0.0) | 20 (0.0) | 29 (0.0) | 10 (30.0) | 5 (60.0) | |
Note: Shading indicates occurrence of clinical signs (coughing)
In group A, the microorganism’s genome was detected in a low proportion of the tested nursery pigs (0 to 15.8%) and in growers and finishers (0 to 30%). An exception was farm 7, where 62.5 % of weaning pigs (3 wk of age) were already colonized, although the proportion decreased and remained low (10% to 30%) in the nursery and fattening pigs. Further serologic analyses indicated concurrent infections with other major respiratory pathogens when respiratory signs were observed. An outbreak of postweaning multisystemic wasting syndrome was diagnosed in farm 3, starting in late-nursery pigs. In farm 5, antibodies to a low-virulence strain of A. pleuropneumoniae appeared at 15 wk of age. In farm 12, antibodies against PRRSV appeared at 10 wk of age.
In all the farms of group B, colonization occurred several weeks earlier than the appearance of clinical signs, and the proportion of nPCR-positive animals markedly increased when the clinical signs appeared.
Seroconversion occurred in all the finishing units. However, the proportions of seropositive pigs in group A (17.4%) and group B (23.4%) were significantly different [P < 0.05, odds ratio = 0.69 (95% confidence interval, 0.47 to 1.01)].
Results for farms with 1- or 2-site versus 3-site production systems
The univariate analysis showed that the age of the tested animals had a statistically significant effect on the nPCR and ELISA results (Table III). Medication had no significant effect on nPCR or ELISA results, but vaccination had a significant effect on nPCR detection of M. hyopneumoniae (Table IV).
Table III.
Total numbers and percentages of positive pigs in the different age groups
| Number positive; age (wk)
|
||||||
|---|---|---|---|---|---|---|
| Test | 1–5 | 6–10 | 11–15 | 16–20 | 21–25 + | P value in univariate model |
| nPCR | 103 (15.53) | 217 (11.06) | 165 (16.36) | 143 (23.78) | 76 (48.68) | < 0.0001 |
| ELISA | 103 (20.39) | 215 (10.70) | 165 (13.33) | 142 (23.24) | 81 (56.79) | < 0.0001 |
Table IV.
Total numbers and percentages of positive pigs among those medicated or vaccinated
| Medicated
|
Vaccinated
|
|||||
|---|---|---|---|---|---|---|
| Test | No | Yes | P valuea | No | Yes | P valuea |
| nPCR | 223 (21.97) | 481 (18.50) | 0.2812 | 410 (16.83) | 294 (23.47) | 0.0292 |
| ELISA | 223 (21.52) | 483 (20.08) | 0.6594 | 412 (18.93) | 294 (22.79) | 0.2116 |
In univariate model
The multivariate model showed that all the variables analyzed had a significant effect on nPCR detection of M. hyopneumoniae (Table V). The farm was the variable with the greatest effect, followed by the type of production system. Vaccinated and medicated pigs reared in 3-site production systems had the lowest probability of being nPCR-positive (5%) and vaccinated and medicated pigs reared in 1- or 2-site productions systems the greatest probability (59.52%).
Table V.
Type 3 generalized estimating equations (GEE) analysis of nested polymerase chain reaction (nPCR) and enzyme linked immunosorbent assay (ELISA) results
| nPCR
|
ELISA
|
|||||
|---|---|---|---|---|---|---|
| Variable | DF | Chi-squared | Pr > chi-squared | DF | Chi-squared | Pr > chi-squared |
| Age | 1 | 4.44 | 0.0351 | 1 | 8.49 | 0.0036 |
| Production system | 1 | 13838.6 | < 0.0001 | 1 | 0.22 | 0.6403 |
| Vaccination | 1 | 9.34 | 0.0022 | 1 | 1.67 | 0.1963 |
| Medication | 1 | 26.72 | < 0.0001 | 1 | 4.85 | 0.0276 |
| Farm effect | 8 | 6.06el4 | < 0.0001 | 8 | 7.18el2 | < 0.0001 |
DF — degrees of freedom
On the other hand, detection of antibodies against M. hyopneumoniae was significantly influenced by only farm, medication, and age (Table V). As in the nPCR analyses, the probability of having antibodies against M. hyopneumoniae increased with age.
Infection with M. hyopneumoniae was first detected by nPCR at different ages and in different proportions among the tested farms. Although not statistically significant, piglets were already colonized in the nurseries in higher proportions in the 1- or 2-site farms (4 out of 5) than in the 3-site farms (4 out of 7). In addition, in farms with 1- or 2-site systems, the percentage of infected pigs increased progressively with age, whereas in farms with 3-site production systems, the percentage decreased with age in the nurseries and abruptly increased in the fattening pigs (Figure 1). However, the probability of detecting the pathogen’s DNA in nasal swabs increased with age in both types of farms (P < 0.0351).
Figure 1.
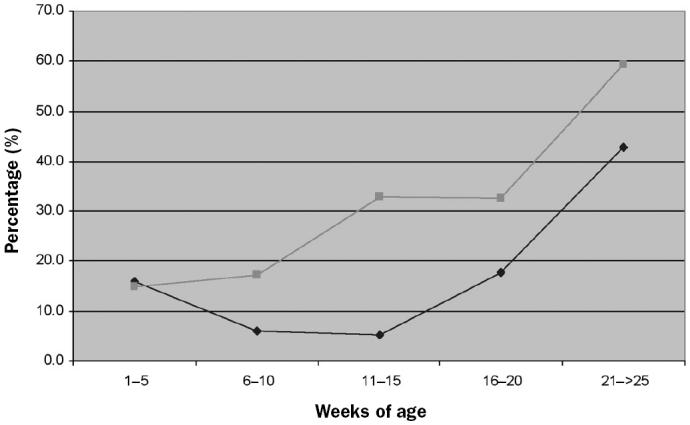
Mean percentage of animals positive for Mycoplasma hyopneumoniae infection by nested polymerase chain reaction in farms with a 1- or 2-site production system versus those with a 3-site system.
Seroconversion was observed rather late in the finishing period in both types of farm (Figure 2). In fact, no significant differences in time of seroconversion between 1- or 2-site systems and 3-site systems were found (Table V). However, in farms 1 and 3 (3-site systems) and farm 12 (1-site system), seroconversion occurred at the same time when the microorganism was first detected by nPCR.
Figure 2.
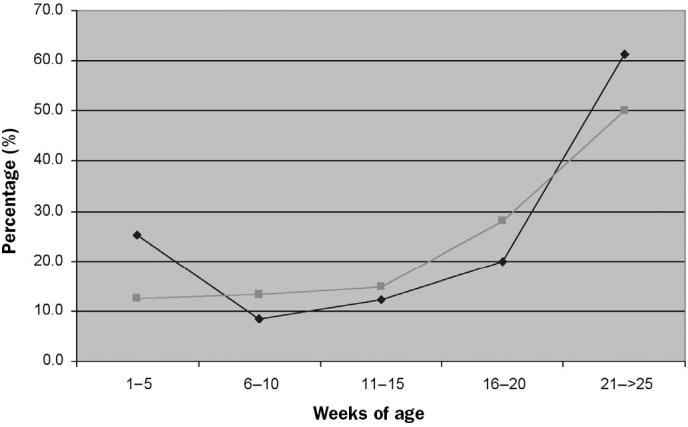
Mean percentage of positive animals by enzyme-linked immunosorbent assay in the same farms.
Discussion
In this study, the presence of M. hyopneumoniae in 12 Spanish farms with respiratory problems was assessed. The results indicated the presence of the organism on all the farms, although with different infection dynamics, proportions of infected animals, and clinical implications.
In the farms in which the proportion of animals nPCR-positive for M. hyopneumoniae remained low or was absent in the age groups in which coughing was recorded, the pathogen was probably not playing a central role in the clinical respiratory problems observed. The other respiratory agents detected in the affected groups may have triggered the respiratory outbreak.
In the farms in which the proportions of infected pigs were high and increasing when clinical signs occurred, M. hyopneumoniae probably played an important role in the respiratory outbreak, although other agents were also concomitantly found.
As previously described (1,12), in most farms, colonization occurred several weeks before clinical signs appeared. It seems that a certain, unknown percentage of animals must be infected to elicit the onset of clinical signs (1). In farm 7, however, a high percentage of weaning piglets were infected without showing clinical signs. A possible explanation is the protective effect of maternal immunity (13), which is still present in 3-wk-old piglets, as indicated by the presence of antibodies. Another possible explanation is that the tested animals were still in the incubation period. Moreover, the proportion of colonized piglets decreased with age on this farm. The low stocking densities could explain the decrease in colonization observed in fattening pigs (4,14), since the fatteners sampled were selected replacement gilts.
Seroconversion occurred on all farms in the finishing stages, independent of mycoplasma involvement in the respiratory process, time of colonization, or production system. Therefore, in contrast to nPCR, the ELISA test performed was useful in determining the presence of M. hyopneumoniae but did not indicate if M. hyopneumoniae was involved in the clinical signs. These results support the idea that the time of seroconversion is highly variable; it may depend on infectious dose (15) and on the onset and severity of pneumonic lesions (16).
Although the microorganism circulated in all the farms, lower proportions of positive animals were detected than in a study conducted in the midwestern United States (1). The percentage of infected animals was lower even when M. hyopneumoniae seemed to play a central role in the respiratory process. Possible explanations for this finding include the wider use of antibiotics in feed in the Spanish herds tested compared with the US herds used in the previous study.
Significant differences were found between nPCR results from the 2 categories of production system studied. The fact that nursery colonization was greater in the farms with a 1- or 2-site system could support the idea that the use of multiple isolated sites decreases the chance of vertical transmission of disease (8). The evolution of infection observed in the 1- or 2-site systems suggests a progressive transmission of the microorganism, whereas in the 3-site systems the appearance of the microorganism was more abrupt. This difference is probably a consequence of the interruption of horizontal transmission by the physical separation of the pigs in a 3-site system. The particular infection dynamics observed in the 3-site systems may, in part, explain the new presentation of respiratory outbreaks in modern swine production systems: acute and affecting late-finishing pigs (17).
Besides the observed difference in infection dynamics in the 2 systems, overall an animal reared in a 1- or 2-site production system had a greater probability of being colonized by M. hyopneumoniae than an animal reared in a 3-site production system.
Vaccination and medication strategies significantly lowered the percentage of infected animals as determined by nPCR. However, the number of farms with 1 or 2 sites versus 3 sites with and without treatment was insufficient to properly quantify these effects.
In conclusion, this study showed that 1) M. hyopneumoniae was present in all the studied farms, although with different infection dynamics and clinical implications; 2) the dynamics of M. hyopneumoniae infection differed significantly between the farms with a 1- or 2-site system and those with a 3-site system; and 3) M. hyopneumoniae colonization and seroconversion were greatly influenced by the effect of each individual farm.
Acknowledgments
This work was financially supported by Pfizer Animal Health. The authors thank Merche Mora, Eva Huerta, and Maria de la Sierra Espinar for nPCR technical assistance. We also thank Cristina Ramírez, Unitat de Malalties Infeccioses, Facultat de Veterinària de la Universitat Autònoma de Barcelona, for conducting the ELISA tests. We are grateful to the veterinarians who helped with the collections and carefully completed the epidemiologic surveys. Special thanks to Dr. Joaquim Segalés for helping in the preparation of this manuscript.
References
- 1.Calsamiglia M, Pijoan C, Bosch GJ. Profiling Mycoplasma hyopneumoniae in farms using serology and a nested PCR technique. Swine Health Prod. 1999;7:263–268. [Google Scholar]
- 2.Mattson JG, Bergstrom K, Wallgren P, Johansson KE. Detection of Mycoplasma hyopneumoniae in nose swabs from pigs by in vitro amplification of the 16S rRNA gene. J Clin Microbiol. 1995;33:893–897. doi: 10.1128/jcm.33.4.893-897.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Calsamiglia M, Pijoan C, Trigo A. Application of a nested polymerase chain reaction assay to detect Mycoplasma hyopneumoniae from nasal swabs. J Vet Diagn Invest. 1999;11:246–251. doi: 10.1177/104063879901100307. [DOI] [PubMed] [Google Scholar]
- 4.Done SH. Enzootic pneumonia (mycoplasmosis) revisited. J Pig Vet Soc. 1996;38:40–61. [Google Scholar]
- 5.Castryk F, Devriese LA, Hormmez J, Cassimon P, Miry CL. Bacterial agents associated with respiratory disease in young feeder pigs. Proc 12th Int. Pig Vet Soc Congr, Lausanne, Switzerland 1990:112.
- 6.Lindqvist JO. The relationship between environmental factors and pathological lesions recorded at slaughter. Acta Vet Scand. 1974;51:17–54. [Google Scholar]
- 7.Clark LK, Scheidt AB, Armstrong CH. The effect of all-in/all-out management on pigs from a herd with enzootic pneumonia. Vet Med. 1991;86:946–951. [Google Scholar]
- 8.Harris LD, Alexander TJ. Methods of disease control. In: Straw BE, D’Allaire S, Mengeling WL, Taylor DJ, eds. Disease of Swine, 8th ed. Ames, Iowa: Iowa State University Press, 1999:1077–1078.
- 9.Harris LD. Alternative approaches to eliminating endemic diseases and improving performance of pigs. Vet Rec. 1988;123:422–423. doi: 10.1136/vr.123.16.422. [DOI] [PubMed] [Google Scholar]
- 10.Feld NC, Qvist P, Ahrens P, Friis NF, Meyling A. A monoclonal blocking ELISA detecting serum antibodies to Mycoplasma hyopneumoniae. Vet Microbiol. 1992;30:35–46. doi: 10.1016/0378-1135(92)90092-8. [DOI] [PubMed] [Google Scholar]
- 11.Rosell C, Segalés J, Plana-Durán J, et al. Pathological, immunohistochemical, and in-situ hybridazation studies of natural cases of postweaning multisystemic wasting syndrome (PMWS) in pigs. J Comp Pathol. 1999;120:59–78. doi: 10.1053/jcpa.1998.0258. [DOI] [PubMed] [Google Scholar]
- 12.Sørensen V, Ahrens P, Barford K, et al. Mycoplasma hyopneumoniae infection in pigs: duration of the disease and evaluation of four diagnostic assays. Vet Microbiol. 1997;54:23–34. doi: 10.1016/s0378-1135(96)01266-7. [DOI] [PubMed] [Google Scholar]
- 13.Lam KM, Switzer WP. Mycoplasmal pneumonia of swine: active and passive immunizations. Am J Vet Res. 1971;32:1737–1741. [PubMed] [Google Scholar]
- 14.Maes D, Verdonck M, Deluyker H. Enzootic pneumonia in pigs. Vet Q. 1996;12:104–109. doi: 10.1080/01652176.1996.9694628. [DOI] [PubMed] [Google Scholar]
- 15.Piffer IA, Young TF, Petenate A, Ross RF. Comparison of complement fixation test and enzyme-linked immunosorbent assay for detection of early infection with Mycoplasma hyopneumoniae. Am J Vet Res. 1984;45:1122–1126. [PubMed] [Google Scholar]
- 16.Sitjar M, Noyes EP, Simon X, Pijoan C. Relationship among seroconversion to Mycoplasma hyopneumoniae, lung lesions, and production parameters in pigs. Swine Health Prod. 1996;4:273–277. [Google Scholar]
- 17.Dee S. The porcine respiratory disease complex: Are subpopulations important? Swine Health Prod. 1996;4:147–149. [Google Scholar]


