Summary
Background
Biliary atresia (BA) is a devastating neonatal cholangiopathy with an unclear pathogenesis, and prompt diagnosis of BA is currently challenging.
Methods
Proteomic and immunoassay analyses were performed with serum samples from 250 patients to find potential BA biomarkers. The expression features of polymeric immunoglobulin receptor (PIGR) were investigated using human biopsy samples, three different experimental mouse models, and cultured human biliary epithelial cells (BECs). Chemically modified small interfering RNA and adenovirus expression vector were applied for in vivo silencing and overexpressing PIGR in a rotavirus-induced BA mouse model. Luminex-based multiplex cytokine assays and RNA sequencing were used to explore the molecular mechanism of PIGR involvement in the BA pathogenesis.
Findings
Serum levels of PIGR, poliovirus receptor (PVR), and aldolase B (ALDOB) were increased in BA patients and accurately distinguished BA from infantile hepatitis syndrome (IHS). Combined PIGR and PVR analysis distinguished BA from IHS with an area under the receiver operating characteristic curve of 0.968 and an accuracy of 0.935. PIGR expression was upregulated in the biliary epithelium of BA patients; Th1 cytokines TNF-α and IFN-γ induced PIGR expression in BECs via activating NF-κB pathway. Silencing PIGR alleviated symptoms, reduced IL-33 expression, and restrained hepatic Th2 inflammation in BA mouse model; while overexpressing PIGR increased liver fibrosis and IL-33 expression, and boosted hepatic Th2 inflammation in BA mouse model. PIGR expression promotes the proliferation and epithelial–mesenchymal transition, and reduced the apoptosis of BECs.
Interpretation
PIGR participated in BA pathogenesis by promoting hepatic Th2 inflammation via increasing cholangiocytes derived IL-33; PIGR has the value as a diagnostic and therapeutic biomarker of BA.
Funding
This study was financially supported by the National Natural Science Foundation of China (82170529), the National Key R&D Program (2021YFC2701003), and the National Natural Science Foundation of China (82272022).
Keywords: Biliary atresia, Biomarker, Polymeric immunoglobulin receptor, Interleukin-33, T helper cells
Research in context.
Evidence before this study
Biliary atresia (BA) is a devastating neonatal form of cholangiopathy that is currently a major indication for paediatric liver transplantation. The current diagnosis and treatment methods of BA are limited, due to the unclear pathogenesis of the disease. Traditional indicators gamma-glutamyl transpeptidase (GGT) and liver stiffness measurement (LSM) are considered helpful in diagnosing BA. However, the reliability and reproducibility of these traditional indicators offer limited accuracy in diagnosing BA. Previous evidence demonstrated the important role of immune abnormality in BA pathogenesis, but the underlying mechanisms remains elucidate.
Added value of this study
In this study, we identified the polymeric immunoglobulin receptor (PIGR) and poliovirus receptor (PVR) as superior biomarkers compared to GGT and LSM for preoperatively diagnosing BA, and the diagnostic model of PIGR combined with PVR can potentially serve as a new approach for diagnosing BA. We found PIGR expression was upregulated in the cholangiocytes of BA patients, which can be triggered by Th1-cytokines TNF-α and IFN-γ via activating NF-κB pathway. In the rotavirus-induced BA mouse model, we found silencing PIGR expression alleviated symptoms and restrained Th2 inflammation, while overexpressing of PIGR increased liver fibrosis and boosted Th2 inflammation. Moreover, we demonstrated that PIGR expression induces Th2 inflammation in the liver of BA by promoting the release of IL-33 from cholangiocytes.
Implications of all the available evidence
The results of this study validated the increase of serum PIGR in patients with BA, and provided a useful diagnostic model of combined PIGR and PVR in diagnosing BA. Our results demonstrated the important role of PIGR in the pathogenesis of BA, and uncovered a previously unrecognized mechanism of Th1/PIGR/IL-33/Th2 circuit in regulating the hepatic immune response. Evidence from current study suggest that PIGR is more than just a valuable diagnostic biomarker of BA, but also a potential therapeutic target of BA and other immuno-fibrotic biliary disorders. Our findings also add a new dimension to understanding the role of PIGR in regulating biliary immunity and have potential implications to other mucosal system, such as in the airway and gastrointestinal tract.
Introduction
Biliary atresia (BA) is a fibroinflammatory, obliterative cholangiopathy that leads to end-stage liver cirrhosis in early infancy.1 The incidence of BA varies around the world, ranging from 1 in every 5000–18,000 live birth.2 Due to the unclear pathogenesis of this disease, the current diagnosis and treatment methods of BA are limited. Timely Kasai hepato-portoenterostomy can re-establish biliary drainage and improve the prognosis, but most patients eventually require liver transplantation to survive. Despite the incidence, BA is currently the most common indication for pediatric liver transplantation.3 Early diagnosis and surgical intervention correlate with prolonged survival of the native liver.4 However, the overlapping clinical and biochemical features shared by BA and other neonatal cholestasis hinder prompt diagnosis of BA. To date, most cases of BA have been confirmed using invasive procedures, such as cholangiography, liver biopsy, and surgery.5 Hence, a simple and reliable diagnostic method for BA is urgently needed.
Blood biomarkers represent ideal, non-invasive diagnostic targets, and substantial efforts have been made to identify more accurate blood biomarkers of BA. In the past two decades, several researchers have applied proteomics analysis with blood samples to screen for biomarkers of BA, but the results varied from study to study. When studying blood samples from BA patients, Lee et al. identified apolipoprotein A-I and C-II as BA biomarkers in 2007 by applying two-dimension-electrophoresis mass spectrometry; Song et al. identified apolipoprotein C-II and C-Ⅲ in 2012 by applying surface-enhanced-desorption/ionization-time-of-flight mass spectrometry; and Chatmanee et al. identified serum matrix metalloproteinase-7 (MMP7) by applying aptamer-based multiplexed proteomics in 2017.6, 7, 8 Among the referred biomarkers, serum MMP7 has been most extensively investigated, several studies have highlighted the value of serum MMP7 in diagnosing BA.9,10 The advancement of serum MMP7 demonstrated the feasibility of researching potential blood biomarkers for diagnosing BA, encouraged further efforts to search new biomarkers.
Recently, data-independent acquisition-mass spectrometry (DIA-MS) has attracted increasing interest in screening for potential biomarkers because it overcomes the inherent irreproducibility of the traditional data-dependent acquisition scheme, provides higher quantification accuracy and data repeatability.11,12 Herein, we applied DIA-MS with serum samples from BA patients to explore possible diagnostic biomarkers. In-depth analysis of serum sample from 250 patients uncovered polymeric immunoglobulin receptor (PIGR), poliovirus receptor (PVR), and aldolase, fructose-bisphosphate B (ALDOB) as feasible diagnostic biomarkers of BA. In the research of human BA biopsy, we found PIGR expression was upregulated in the bile ducts of BA patients, and was closely related to disease progression. The pathogenesis of BA is currently unclear, but increasing evidences highlight the essential role of immune abnormalities.13 By conducting different experimental liver injury mouse models and cultivating human cholangiocytes in vitro, we investigated the mechanistic basis of the increased expression of PIGR in the biliary epithelium. By manipulating PIGR expression in both rotavirus (RRV)-induced BA mouse model and cultured human cholangiocytes, we demonstrated the important role of PIGR in BA pathogenesis and revealed a previously unknown mechanism of Th1/PIGR/IL-33/Th2 circuit in regulating hepatic immune response.
Methods
Patient cohorts and samples
This study was approved by the Ethics Committee of Shengjing Hospital of China Medical University (approval number 2018PS205K), and conducted according to the ethical guidelines of the Declaration of Helsinki. In this study, 250 consecutive infants were enrolled between January 2019 and January 2024 at Shengjing Hospital of China Medical University, including 121 BA, 90 infantile hepatitis syndrome (IHS), and 39 healthy infant peers (HC), informed consent was obtained from the guardians of all subjects. Patients with BA were identified based on operative cholangiography and liver-pathology. The following exclusion criteria were applied for BA: (a) an age of older than 90 days, (b) cholangiopathy showing a patent biliary tree, (c) genetic analysis confirming etiology other than BA, and (d) normal serum bilirubin or stool color. Serum samples from each subject were obtained during the first visit, prior to a definitive diagnosis, liver and remnant extrahepatic bile duct (EBD) biopsies of BA and control patients were obtained during surgery. Liver functional tests were taken during first visit, including: total bilirubin (TBIL), direct bilirubin (DBIL), aspartate transaminase (AST), alanine aminotransferase (ALT), and gamma-glutamyl transpeptidase (GGT). Liver stiffness measurement (LSM) was performed by the same clinician experienced in ultrasound via 2D-shear-wave elastography. Liver pathological examinations were analyzed by three experienced pathologists in a blind fashion. The liver fibrosis was staged according to the method of Scheuer et al., and hepatic inflammation was graded according to the method of Ishak and colleagues.14,15
Animal models
Animal studies were performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals, and approved by the Institutional Animal Care Committee of Shengjing Hospital of China Medical University (approval number 2021PS346K). All BALB/c mice used in this study were acquired from the Beijing Vital River Laboratory Animal Technology.
To generate RRV-induced BA mouse model, new-born BALB/c mice were intraperitoneally injected with 1.5 × 106 plaque-forming units (PFU) of rhesus rotavirus (RRV; MMU18006, American Type Culture Collection) within the first day of life; mice receiving the same volume of saline served as controls. To induce obstructive cholestasis, 6-week-old BALB/c mice underwent a bile duct ligation (BDL) and were kept feeding for 2 weeks, and control mice received a sham operation. To induce liver injury and fibrosis, 6 weeks-old BALB/c mice were intraperitoneally injected with carbon tetrachloride (CCL4; 1 μL/g of body weight, dissolved in corn oil at a 1:3 ratio, #319961, Sigma–Aldrich, USA) every 3 days for 4 weeks, and control mice were injected with same volume of corn oil. For in vivo PIGR silencing in the RRV-induced BA mouse model, 2OMe + 5Chol modified small interfering RNA (siRNA) was injected intraperitoneally at 0.5 nmol/g (dissolved in saline) every other day from day 3–11; control mice received 2OMe + 5Chol modified negative control siRNA (Ribo Bio) instead. For in vivo PIGR overexpressing in the RRV-induced BA mouse model, the “pcADV-EF1-CMV-Pigr” adenovirus expression vector (OBIO technology) was injected intraperitoneally at 1 × 108 PFU (dissolved in saline) at day 2,6, and 10; control mice received the “pcADV-EF1-CMV-MCS” control vector (OBIO) instead. To supplement mice with exogenous IL-33, mice were intraperitoneally injected with 50 ng recombinant mouse IL-33 protein (#3626-ML, R&D system) daily from day 3 to 11.
Blood and liver samples were harvested under anesthesia (induced by isoflurane inhalation) at the indicated time points. The serum levels of TBIL, DBIL, AST, ALT, and GGT were measured using colorimetric assay kits (#K760, #K761, #K235, #K236, #K126, Elabscience). Hematoxylin-Eosin (HE) staining was performed using HE-Stain-Kit (#G1120, Solarbio). Masson staining was performed using Masson's Trichrome Stain Kit (#G1340, Solarbio).
Serum proteomics and enzyme-linked immunosorbent assay (ELISA) analyses
Methods of proteomics analysis are detailed in Supplementary Materials. The concentrations of biomarkers in serum samples from patients were detected using appropriate ELISA kits (Camilo Biological, China), according to the manufacturer's instructions.
Multiplex assays of cytokines and RNA-sequencing
The concentrations of 23 cytokines and chemokines in the liver homogenate samples of mice from Si-NC and Si-PIGR groups were analyzed by quantitative cytokine assays using Bio-Plex Pro Mouse Cytokine 23-plex Assay (#M60009RDPD, BIO-RAD), according to the protocols provided by the manufacturer. Data were collected and analyzed using a Luminex-X200 instrument equipped with Milli-plex Analyst software version 5.1 (Merck). The immunoassay data in mice livers were expressed in terms of per mg total protein estimated using BCA protein assay kit (#ZJ101, Epizyme).
Total RNA of cultured BECs was isolated with Steady-Pure-RNA Extraction Kit (#AG21017, Accurate-Biotechnology, China). RNA quality was assessed on an Agilent 2100 Bioanalyzer (Agilent Technologies, USA) and checked using RNase free agarose gel electrophoresis. After total RNA was extracted, eukaryotic mRNA was enriched by Oligo (dT) beads. Then the enriched mRNA was fragmented into short fragments using fragmentation buffer and transcribed into cDNA by using NEB Next Ultra RNA Library Prep Kit for Illumina (NEB #7530, New England Biolabs, Ipswich, MA, USA).The purified double-stranded cDNA fragments were end repaired, A base added, and ligated to Illumina sequencing adapters. The ligation reaction was purified with the AM Pure XP Beads (1.0×).And polymerase chain reaction (PCR) amplified. The resulting cDNA library was sequenced using Illumina Novaseq6000 by Gene Denovo Biotechnology Co (Guangzhou, China). The transcripts with the parameter of FDR (false discovery rate) < 0.05 and absolute fold change >2 were considered differentially expressed transcripts.
Reverse transcription quantitative PCR, immunoblotting, and immunostaining analyses
Total RNA was isolated using the Steady-Pure-RNA Extraction Kit (Accurate-Biotechnology). Reverse transcription quantitative PCR (RT-qPCR) was performed using the SYBR-Premix-Pro-Taq-Kit (#AG11701, Accurate-Biotechnology) on 7500 Real-Time PCR System (Applied Biosystems). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control, and the primers used in this study are listed in Supplementary Materials (Table S1).
Total protein of tissue and cell samples was extracted using a protein extraction kit (#BC3710, Solarbio). The protein extracts were separated using 10% SDS-PAGE Gel Kit (#PG112, Epizyme), transferred to polyvinylidene fluoride membranes; after incubated with primary antibodies at 4 °C for 16 h and secondary antibodies at 37 °C for 2 h, corresponding proteins were visualized using an Amersham-Imager-680 (General Electric Company). GAPDH and Beta Tubulin was used as an internal control. The following antibodies were used in immunoblotting study: anti-PIGR-antibody (1:1000 diluted in TBS, #AF7746, Beyotime), anti-E-cadherin-antibody (1:5000 diluted in TBS, #20874, Proteintech), anti-smooth-muscle-actin-antibody (1:3000 diluted in TBS, #14395, Proteintech), anti-GAPDH-antibody (1:2000 diluted in TBS, #ab9485, Abcam), anti-Beta Tubulin-antibody (1:1000 diluted in TBS, #10068, Proteintech) and goat anti-rabbit-IgG-HRP (1:2000 diluted in TBS, #ab6721, Abcam).
Immunofluorescence staining of the sections and cultured BECs were performed as following procedures: paraffin-embedded sections were dewaxed and boiled in Tris–EDTA buffer (pH 8.0) for antigen retrieval, blocked with 3% hydrogen peroxide (15 min); sections or cultured BECs were blocked with 5% bovine serum albumin (1 h); incubated with an anti-cytokeratin 19 antibody (diluted 1:200 in TBS, #60187, Proteintech), an anti-PIGR antibody (diluted 1:50 in TBS, #AF7746, Beyotime), an anti-Interleukin 33-antibody (diluted 1:200 in TBS, #66235, Proteintech) an anti-E-cadherin-antibody (diluted 1:200 in TBS, #20874, Proteintech), or an anti-smooth-muscle-actin-antibody (1:100 diluted in TBS, #14395, Proteintech) for 2 h at 21 °C. Then, sections or BECs were incubated under dark conditions for 1 h with a CoraLite594-conjugated goat anti-rabbit IgG (H + L) antibody (diluted 1:300 in TBS, #SA00013-4, Proteintech), and/or a fluorescein isothiocyanate-conjugated goat anti-mouse IgG (H + L) antibody (diluted 1:200 in TBS, #SA0003-1, Proteintech), after which they were incubated with DAPI solution (#1012, Servicebio) for 5 min. Triple fluorescent staining of PIGR, CK19 and IL-33 in the sections was performed using a TSA-Plus-Fluorescence-Triple-Label-Four-Color-Staining Kit (#G1236, Servicebio), according to the manufacturer's instructions. Antibodies used in this study were validated by knockdown and overexpression, or validated by the suppliers.
Cell culture
Human intrahepatic biliary epithelial cells (BECs) (#5100, ScienCell) were cultured in RPMI 1640 medium (#PM150110, Pricella Life Science &Technology Co., Ltd) supplemented with 10% fetal bovine serum (#164210, Pricella). The BECs were validated by immunostaining of CK19. The following sequence “GACAGACATTAGCATGTCA” was targeted using 50 nmoL/ml siRNA to inhibit PIGR expression in BECs, and negative control siRNA was used as the control. The pcSlenti-CMV-PIGR-3FLAG-PGK-Puro-WPRE expression vector (OBIO) was used to overexpress PIGR in BECs, the pcSlenti-CMV-MCS-3FLAG-PGK-Puro-WPRE vector (OBIO) served as vector control. Several groups of BECs were established for the different interventions. Briefly, the control group was cultured in complete RPMI 1640 medium, the IL-1β group was treated with 10 ng/mL recombinant human IL-1β protein (#201-LB, R&D system), the TNF-α group was treated with 10 ng/mL recombinant human TNF-α protein (#10291-TA, R&D system), the IFN-γ group was treated with 10 ng/mL recombinant human IFN-γ protein (#285-IF, R&D system), the IL-4 group was treated with 10 ng/mL recombinant human IL-4 protein (#204-IL, R&D system), the TGF-β group was treated with 10 ng/mL recombinant human TGF-β protein (#7754-BH, R&D system), groups containing PDTC were treated with 100 μmol/mL pyrrolidinedithiocarbamate ammonium (HY-18738, MedChemExpress).
Cell proliferation and apoptosis analyses
For cell counting kit-8 (CCK8)-based cell viability assays, BECs were inoculated in a 96-well-plate (5 × 103/well) and divided into groups. BECs were transinfected with siRNAs or expression vectors, cultured for 12, 24, 48 and 72 h, and then incubated with 10 μL of CCK-8 solution (#606335, Sangon Biotech) for 2 h. The apoptosis of BECs was detected with the Annexin V-FITC/PI-Apoptosis-Detection-Kit (#BL110A, Biosharp), and the apoptotic ratio was quantitatively analyzed using a FACS-Canto-Ⅱ Flow Cytometer (BD Biosciences).
Flow cytometric analysis of lymphocytes in mouse liver
To analyze the proportion of T-helper 2 cells in the liver of BA mouse model, mice livers were harvested at the time of sacrifice, dissociated, digested with enzymes and lymphocytes were isolated using the mouse-tissue-lymphocyte isolation Kit (#P8870, Solarbio). Isolated lymphocytes were incubated with CD3 Monoclonal Antibody-FITC (0.25 μg/million cells, #110032, eBioscience), CD4 Monoclonal Antibody-PE (0.125 μg/million cells, #120041, eBioscience), and IL-4 Monoclonal Antibody-APC (0.25 μg/million cells, #177041, eBioscience) in a dark condition for 25 min at 21 °C; Rat-IgG2b-kappa-Isotype-Control-FITC (0.25 μg/million cells, #114031, eBioscience), Rat-IgG2b-kappa-Isotype-Control-PE (0.125 μg/million cells, #124031, eBioscience), and Rat-IgG1-kappa-Isotype-Control-APC (0.25 μg/million cells, #174301, eBioscience) were used as isotype controls. Flow cytometric (FCM) analyses were performed using a FACS-Canto-Ⅱ Flow Cytometer (BD Biosciences), and FCM data were analyzed using Flowjo_V10.8.1 software (Tree Star). Cell populations were selected according to forward and side scatters, and gated according to isotype controls of each antibody.
Statistical analysis and data visualization
Statistical analysis was performed and visualized using the differential-enrichment test in R software (version 3.6.3), SPSS software (version 26.0, SPSS, Chicago), Med-Calc® Statistical Software (version 20.010, Med-Calc Software Ltd, Ostend, Belgium), cluster Profiler package (R software), Microsoft Office 2021, and Prism 8.0.2 (GraphPad, La Jolla, CA). The diagnostic performance of the biomarkers was assessed using the receiver operating characteristic curve (ROC) and area under the curve (AUC), and the sensitivity and specificity were determined at the optimal cut-off probability (determined by Youden index). DeLong's test was used to compare AUC values for significant differences.16 Logistic regression was used to construct combined analyses of biomarkers. A two-tailed P-value < 0.05 was considered significant.
Role of funders
The funders had no role in study design; in the collection, analysis, and interpretation of data; in the writing of the report; and in the decision to submit the manuscript for publication.
Results
Proteomics analysis identifies PIGR, PVR, and ALDOB as potential biomarkers of BA
The overall 250 subjects were divided into discovery cohort and validation cohort. We applied DIA-MS in 20 serum samples collected between January 2019 and June 2019, the 20 patients were considered as a discovery cohort. As the research progresses, we obtained interesting findings and conducted continuous research until January 2024. Considering the batch differences, technological updates, and other interfering factors, we included 230 subjects collected between July 2019 and January 2024 into the validation cohort to verify the findings. The demographic and clinical parameters of the patient cohorts are summarized in the table (Table 1). The stability and repeatability of the proteomic data were supported by quality control procedures, as presented in the Supplementary Material (Figure S1). Eventually, we identified 1403 proteins in serum samples of patients in the discovery cohort. In-depth proteomics analysis revealed distinct abnormalities in metabolism processes, KEGG pathways, and immune cell infiltrations in patients with BA, as described in the Supplementary Results (Figure S2).
Table 1.
Demographics and laboratory values of study subjects.
| Discovery cohort |
Validation cohort |
|||||||
|---|---|---|---|---|---|---|---|---|
| BA (n = 11) | HC (n = 9) | P | BA (n = 110) | IHS (n = 90) | HC (n = 30) | P (BA vs IHS) | P (BA vs HC) | |
| Age (days) | 41 (39–47) | 46 (40–52) | 0.064 | 53 (42–71) | 52 (39–67.5) | 46.5 (36–66) | 0.810 | 0.303 |
| Gender, n (%) | ||||||||
| Male | 5 (45.5%) | 4 (44.4%) | 0.964 | 46 (41.8%) | 40 (44.4%) | 16 (53.3%) | 0.775 | 0.302 |
| Female | 6 (54.5%) | 5 (55.6%) | 64 (58.2%) | 50 (55.6%) | 14 (46.7%) | |||
| TBIL (μm/L) | 146.3 (115.2–153.2) | 8.3 (7.7–9.8) | <0.001 | 143.65 (116.7–166.5) | 129.0 (103.7–166.9) | 7.3 (6.3–8.5) | 0.154 | <0.001 |
| DBIL (μm/L) | 93.3 (78.4–103.5) | 2.1 (1.5–2.9) | <0.001 | 96.8 (82.2–124.1) | 91.8 (70.1–122.7) | 1.9 (1.6–2.5) | 0.218 | <0.001 |
| AST (U/L) | 153.0 (103.0–175.0) | 13.0 (11.0–14.0) | <0.001 | 154.5 (104.5–194.0) | 136.5 (92.5–189.0) | 10.0 (8.0–11.0) | 0.303 | <0.001 |
| ALT (U/L) | 135.0 (85.0–189.0) | 11.0 (9.5–12.0) | <0.001 | 126.0 (80.8–181.8) | 121.5 (82.8–171.3) | 11.0 (9.0–12.0) | 0.886 | <0.001 |
| GGT (U/L) | 234.0 (198.0–275.0) | 26.0 (20.0–32.0) | <0.001 | 343.0 (235.0–545.5) | 204.5 (132.0–274.3) | 24.5 (15.8–37.3) | <0.001 | <0.001 |
| LSM (kPa) | 13.6 (11.5–15.5) | N/A | N/A | 14.0 (11.5–16.3) | 10.5 (8.5–12.0) | N/A | <0.001 | N/A |
Data were shown as Median (IQR). Gender was tested using Chi-square test, all other rows were tested using Wilcoxon Rank-sum test. Abbreviations: BA, biliary atresia; IHS, infantile hepatitis syndrome; HC, peer healthy infants; TBIL, total bilirubin; DBIL, direct bilirubin; AST, aspartate transaminase; ALT, alanine aminotransferase; TBIL, total bilirubin; DBIL, direct bilirubin; GGT, gamma-glutamyl transpeptidase; LSM, liver stiffness measurement.
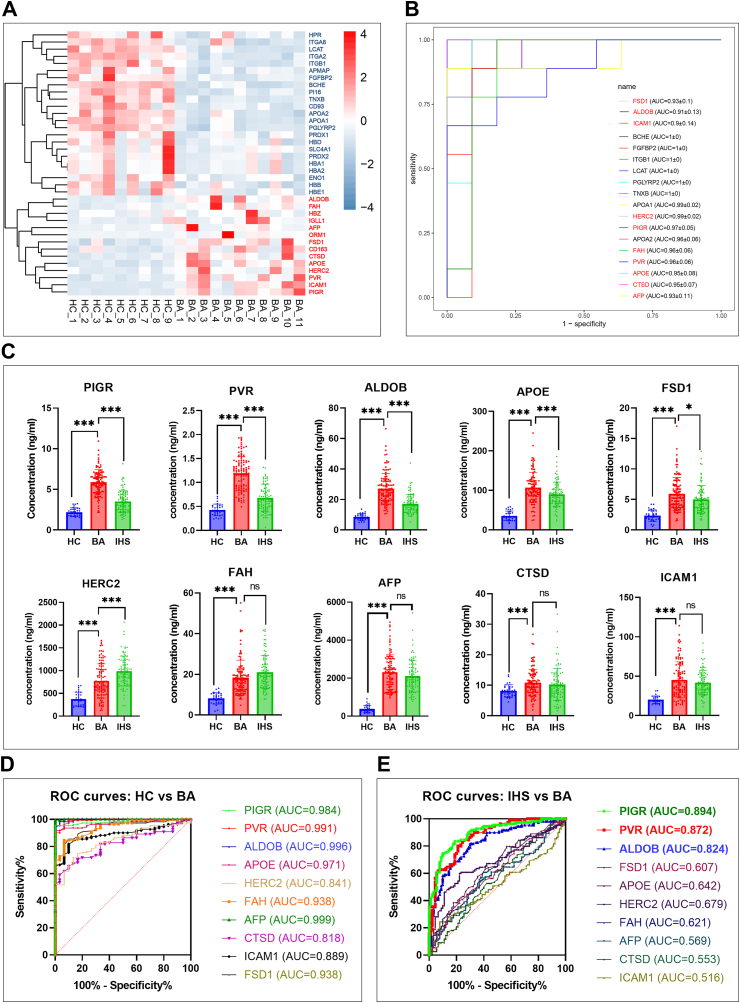
By setting FDR <0.05 and absolute fold change >2 as screening thresholds, we identified 37 differential proteins, of which 14 were upregulated and 23 were downregulated in BA (Fig. 1A). To find candidate proteins for further investigation, we made ROC curves of the 37 differential proteins, and found that 18 of them obtained an AUC higher than 0.90 (Fig. 1B). Among the 18 differential proteins, ten were significantly upregulated in BA patients, including PVR, intercellular adhesion molecule 1 (ICAM1), ALDOB, PIGR, HECT and RLD domain containing E3 ubiquitin protein ligase 2 (HERC2), apolipoprotein E (APOE), fumarylacetoacetate hydrolase (FAH), alpha fetoprotein (AFP), cathepsin D (CTSD), and fibronectin type III and SPRY domain containing 1 (FSD1). The ten elevated serum proteins were selected for further investigation.
Fig. 1.
(A) Heatmap of the 37 differentially protein proteins in the discovery cohort. Proteins upregulated in BA group are marked in red and proteins downregulated are marked in blue (B) ROC curves of the 18 differential proteins with an AUC higher than 0.9 in the discovery cohort. Ten proteins that were elevated in the BA group are marked in red (C) Serum levels of the ten candidate proteins among HC (n = 30), IHS (n = 90), and BA (n = 110) in the validation cohort; ns, P > 0.05; ∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001; two-tailed t-test (D) ROC curves of the ten candidate proteins in distinguishing HC (n = 30) and BA (n = 110) in the validation cohort (E) ROC curves of the ten candidate proteins in distinguishing IHS (n = 90) and BA (n = 110) in the validation cohort.
To validate the proteomic data, we quantified the ten candidate proteins in the serum samples from 230 subjects of the validation cohort. By comparing 110 BA with 30 HC, we found that the concentrations of the ten candidate proteins were higher in BA (Fig. 1C), and ROC analysis showed that all AUC values were higher than 0.80 (Fig. 1D). These data are consistent with the proteomics analysis. However, the challenge in prompt diagnosis of BA is differentiating it from IHS. Hence, we compared the ten candidate proteins between 110 BA and 90 IHS, and found that PIGR, PVR, ALDOB, FSD1, and APOE were significantly higher and that HERC2 was significantly lower in BA than in the IHS (Fig. 1C). According to ROC curves, AUC values of FSD1, APOE, HERC2, FAH, AFP, CTSD, and ICAM1 were less than 0.75, whereas the values of PIGR, PVR, and ALDOB were 0.894, 0.872, and 0.824, respectively (Fig. 1E). These data indicated that PIGR, PVR, and ALDOB have potential to serve as biomarkers of BA.
Serum PIGR and PVR are better diagnostic biomarkers than traditional clinical indicators
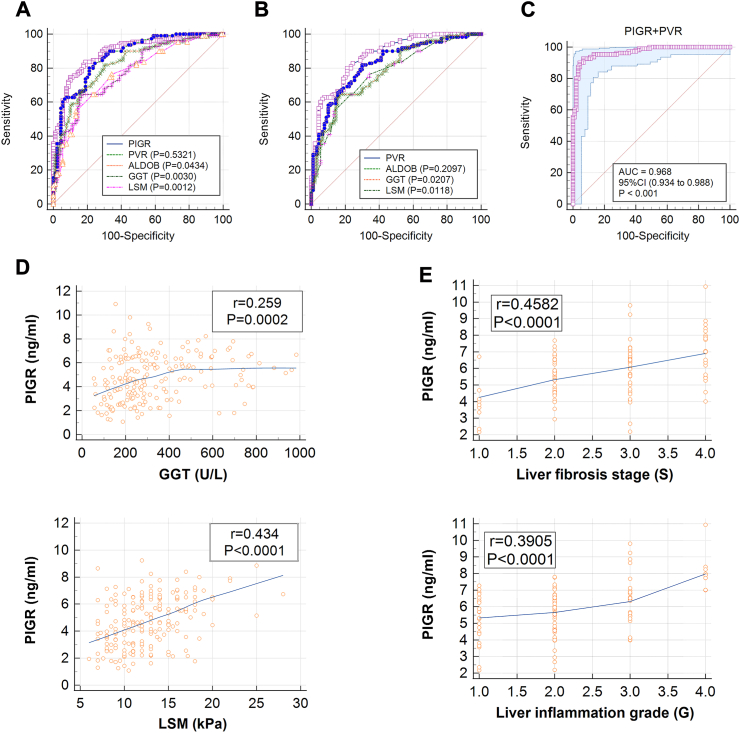
To evaluate the diagnostic value of serum levels of PIGR, PVR, and ALDOB, we compared them with six clinical indicators, including AST, ALT, TBIL, DBIL, GGT, and LSM in the validation cohort. As summarized in the table, PIGR, PVR, ALDOB, GGT, and LSM showed better properties (AUC >0.7) for distinguishing BA from IHS than AST, ALT, DBIL, and TBIL (AUC <0.7); and PIGR presented the best diagnostic performance among the nine indicators, with an AUC of 0.898, a sensitivity of 0.836, a specificity of 0.811, and an accuracy of 0.825 (Table 2). We compared the ROC curves of PIGR, PVR, ALDOB, GGT, and LSM, and found that the AUC of PIGR was 0.894, which was significantly higher than those of ALDOB, GGT, and LSM (Fig. 2A); and the AUC of PVR was 0.872, which also significantly higher than those of GGT and LSM (Fig. 2B). To further investigate the diagnostic potential of PIGR, PVR, ALDOB, GGT, and LSM, we constructed a series of combined analysis using logistic-regression and ROC curve. Among those combined analyses, we found the combination of PIGR and PVR presented the highest accuracy of 0.935, with a sensitivity of 0.927 a specificity of 0.933, and an AUC of 0.968 (Fig. 2C). These data suggest that PIGR and PVR are better diagnostic biomarkers than traditional clinical indicators GGT and LSM, and the diagnostic model of PIGR combined with PVR can potentially serve as a new approach for diagnosing BA.
Table 2.
Diagnostic performance of indicators in the validation cohort.
| Indicator | AUC (95% CI) | Cut-off value | Sensitivity% | Specificity% | Accuracy |
|---|---|---|---|---|---|
| AST | 0.546 (0.475–0.617) | >94.0 (U/L) | 83.64 | 23.78 | 0.595 |
| ALT | 0.504 (0.432–0.575) | >66.0 (U/L) | 17.27 | 90.00 | 0.570 |
| TBIL | 0.565 (0.493–0.634) | >107.5 (μm/L) | 88.18 | 28.89 | 0.635 |
| DBIL | 0.561 (0.489–0.631) | >69.9 (μm/L) | 92.73 | 23.33 | 0.717 |
| GGT | 0.781 (0.718–0.837) | >290.0 (U/L) | 62.73 | 84.44 | 0.725 |
| LSM | 0.769 (0.705–0.826) | >11.0 (kPa) | 76.36 | 65.56 | 0.745 |
| PIGR | 0.894 (0.843–0.933) | >4.43 (ng/mL) | 83.64 | 81.11 | 0.825 |
| PVR | 0.872 (0.815–0.915) | >0.85 (ng/mL) | 80.00 | 78.89 | 0.790 |
| ALDOB | 0.824 (0.764–0.874) | >18.5 (ng/mL) | 81.82 | 70.00 | 0.735 |
The cut-off values were determined from the validation cohort by Youden index. Abbreviations: AUC, area under the receiver operating characteristic curve; AST, aspartate transaminase; ALT, alanine aminotransferase; TBIL, total bilirubin; DBIL, direct bilirubin; GGT, gamma-glutamyl transpeptidase; LSM, liver stiffness measurement; PIGR, polymeric immunoglobulin receptor; PVR, poliovirus receptor; ALDOB, aldolase B.
Fig. 2.
(A) Comparative ROC curves of PIGR, PVR,ALDOB, GGT, and LSM in distinguishing IHS (n = 90) and BA (n = 110) in the validation cohort, P-values within parentheses represent the P-value between each indicator and PIGR (Delong's test) (B) Comparative ROC curves of PVR, ALODB, GGT, and LSM in distinguishing IHS (n = 90) and BA (n = 110) in the validation cohort, P-values within parentheses represent the P-value between each indicator and PVR (Delong's test) (C) ROC curve of the combination of PIGR and PVR in distinguishing IHS (n = 90) and BA (n = 110) in the validation cohort (D) Correlation analyses of PIGR and clinical indicators (GGT and LSM) in the serum of BA patients (n = 110) in the validation cohort; r, Pearson correlation coefficient; P, P-value analyzed with two-tailed t-test (E) Correlation analyses of PIGR and degrees of liver pathology (inflammation grade and fibrosis stage) of BA patients (n = 110) in the validation cohort; r, Pearson correlation coefficient; P, P-value analyzed with two-tailed t-test.
Serum PIGR associated with liver inflammation and fibrosis of patients with BA
Our previous study has demonstrated the important role of PVR in BA pathogenesis, however, whether PIGR is involved in the pathogenesis of BA is currently unknown.17 To find underlying association, we made correlation analyses among 110 BA patients in the validation cohort, and found that serum PIGR was significantly and positively correlated with both GGT and LSM (Fig. 2D). GGT is a traditional BA indicator that associated with bile duct injury, and LSM is an advanced method that used to evaluate liver fibrosis. These data suggest that PIGR may associated with the pathological processes in the liver of BA patients. We then studied the relation between serum PIGR and liver pathology of BA patients, and found that the abundance of serum PIGR correlated positively with both inflammation grading and fibrotic stage (Fig. 2E). These evidences suggest that PIGR is closely associated with the progression of the disease.
Pro-inflammatory cytokines promote PIGR expression in cholangiocytes via activating NF-κB
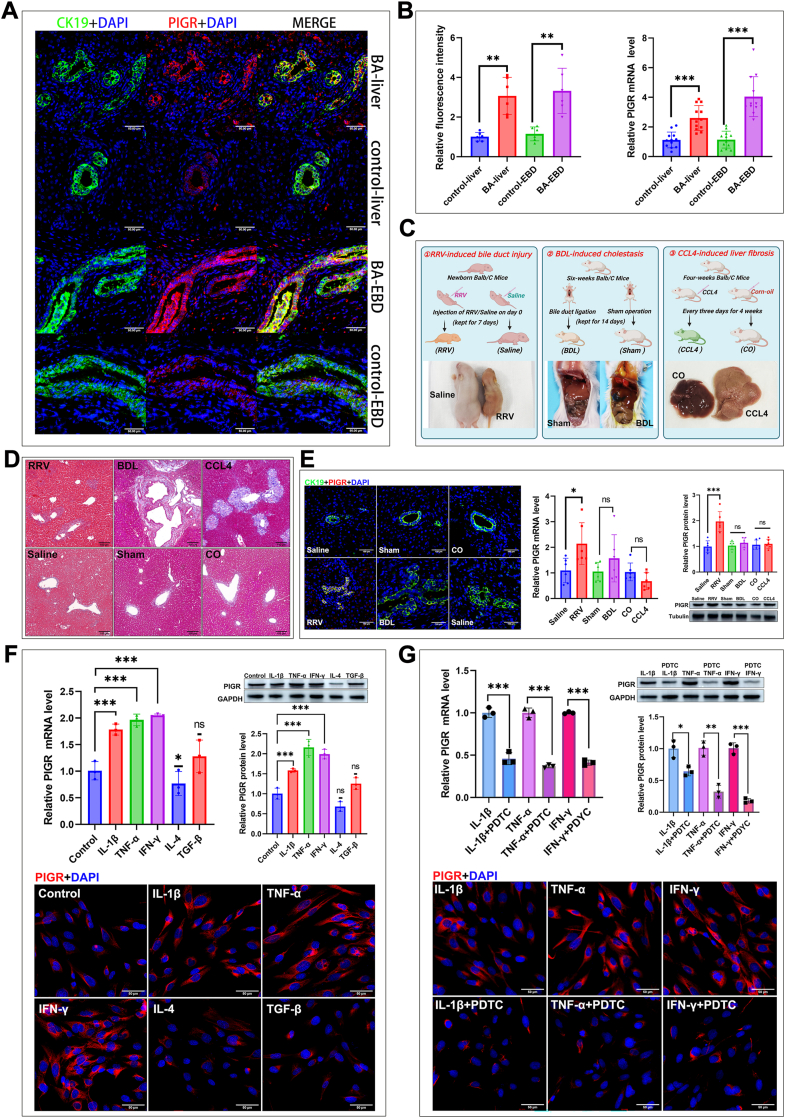
To investigate the expression feature of PIGR in the liver of BA, we detected PIGR expression in liver biopsy samples of BA and control patients. The double staining of PIGR and CK19 in the biopsy sections shows that, PIGR is mainly localized in the cholangiocytes (CK19+), and a few unidentified parenchymal cells (Fig. 3A). We calculated the fluorescence intensity of PIGR signal and found that the values in the liver and EBD sections of BA patients were significantly higher than those in the control patients (Fig. 3B). The RT-qPCR analyses also revealed that the PIGR mRNA levels were significantly elevated in the liver and EBD of BA patients compared to those in controls (Fig. 3B).
Fig. 3.
(A) Immunostaining for PIGR (Red) and CK19 (Green) in liver and EBD sections from the BA and control patients. Original magnification, × 400; Scale bars, 50 μm (B) Quantification of the fluorescence intensity of PIGR signals in the liver and EBD sections from the BA and control patients (n = 6; ∗∗, P < 0.01; Wilcoxon rank sum test); expression of PIGR mRNA in the liver and EBD of BA and control patients (n = 12; ∗∗∗, P < 0.001; Wilcoxon rank sum test) (C) Schematic illustration depicting the three experimental mouse models employed in this study (created with BioRender.com), and the representative images of each mouse model (D) Masson staining of liver sections from the experimental mouse models (E) Expression of PIGR in the liver of experimental mouse models detected by immunostaining, RT-qPCR and immunoblotting analyses (n = 6; ns, P > 0.05; ∗, P < 0.05; ∗∗∗, P < 0.001; Wilcoxon rank sum test) (F) Expression of PIGR in cultured BECs upon stimulations of different cytokines, detected by RT-qPCR and immunoblotting analyses, and immunostaining (n = 3; ns, P > 0.05; ∗, P < 0.05; ∗∗∗, P < 0.001; two-tailed t-test) (G) Expression of PIGR in cultured BECs upon stimulations of different cytokines, with or without the pretreatment of a NF-κB inhibitor PDTC; detected by RT-qPCR and immunoblotting analyses, and immunostaining (n = 3; ns, P > 0.05; ∗, P < 0.05; ∗∗, P < 0.01; ∗∗∗, P < 0.001; two-tailed t-test).
To explore the pathological basis underlying the increased expression of PIGR in the bile ducts of BA patients, we conducted three different experimental mouse models: RRV-induced bile duct injury, BDL-induced obstructive cholestasis, and CCL4-induced liver fibrosis (Fig. 3C). The HE (Figure S3) and Masson (Fig. 3D) staining revealed increased inflammation in RRV mice, dilation and proliferation of bile ducts in BDL mice, and increased periportal and septal fibrosis in CCL4 mice. The serum biochemical analyses showed significant liver dysfunction in mice received RRV injection, BDL and CCL4 injection (Figure S4). We found that the mRNA and protein levels of PIGR were significantly increased in the liver of RRV mice, but not in the BDL and CCL4 mice; and the immunostaining showed similar results that PIGR was up-regulated in the cholangiocytes of RRV-infected mice (Fig. 3E). The RRV-induced bile duct injury mouse model is mainly characterized by the immunoinflammatory damage of the bile ducts. These results suggest that the upregulation of PIGR expression in the cholangiocytes of BA may be triggered by an immunoinflammatory response, rather than a secondary reaction to cholestasis or fibrosis.
According to previous reports, inflammatory cytokines including IL-1β, TNF-α, IFN-γ, IL-4, and TGF-β are able to promote PIGR expression in certain epithelial cells, however, exposure to these factors produces divergent results.18,19 For instance, IL-4 promotes PIGR expression in Calu-3 cell line while inhibits PIGR expression in primary airway epithelial cells.20,21 How these factors affect PIGR expression in cholangiocytes is currently unknown. Hence, we cultured human primary cholangiocytes in vitro and stimulated them with the aforementioned five cytokines. We found that the three pro-inflammatory cytokines (IL-1β, TNF-α, and IFN-γ) significantly increased PIGR expression in BECs, while IL-4 significantly downregulated PIGR expression (Fig. 3F). In addition, we found that the pretreatment of BECs with PDTC, an inhibitor of nuclear factor-kappa B (NF-κB),22 significantly blocked the upregulation of PIGR expression induced by IL-1β, TNF-α, and IFN-γ (Fig. 3G). These data indicate that the pro-inflammatory immune microenvironment in the liver of BA promoted PIGR expression in the cholangiocytes via activating NF-κB.
Silencing PIGR expression alleviated symptoms and restrained Th2 inflammation in the RRV-induced BA mouse model
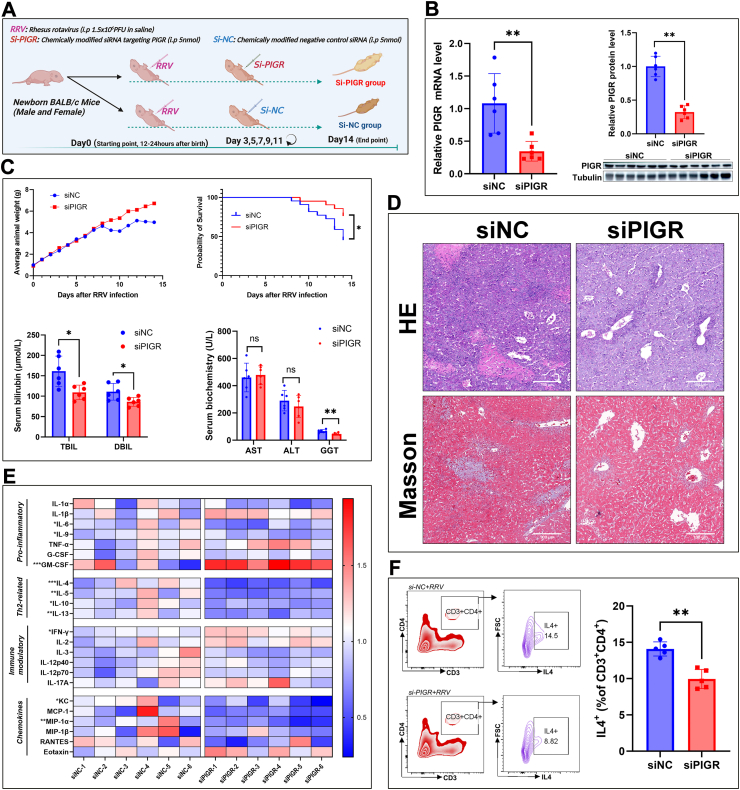
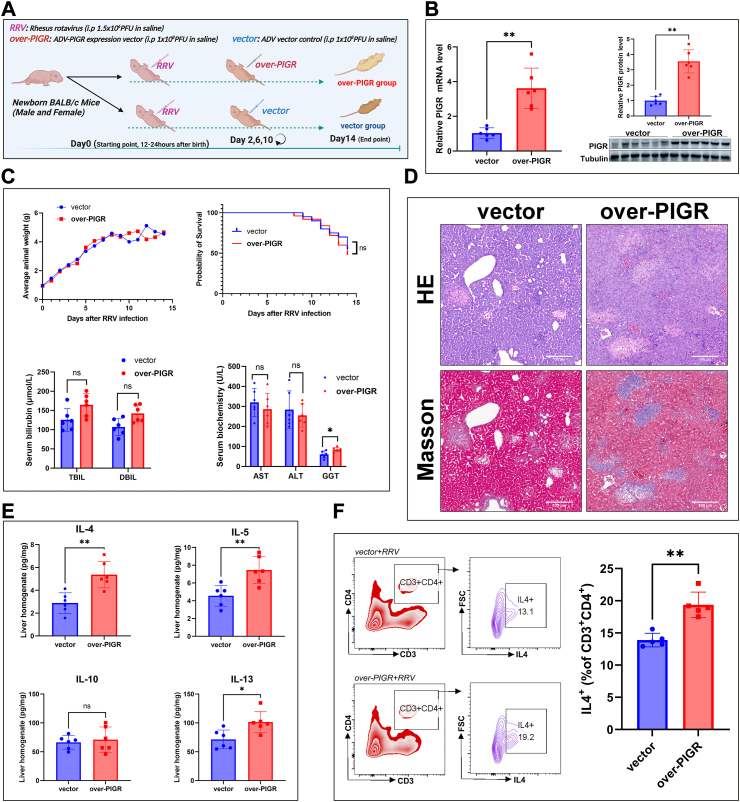
To further investigate the role of PIGR in BA pathogenesis, we silenced PIGR expression in the RRV-induced BA mouse model by injecting chemically modified siRNA (Fig. 4A). The efficiency of PIGR silencing was validated by quantitative PCR and immunoblotting analyses (Fig. 4B). We found that PIGR silencing increased body weight gain, and improved the survival of mice after RRV infection (Fig. 4C). The serum biochemical analyses showed that silencing PIGR significantly decreased levels of GGT and bilirubin on day 14 (Fig. 4C). Histologically, mice in the Si-PIGR group showed milder periportal inflammation and fibrosis than mice in the control group (Fig. 4D). These data suggest that silencing of PIGR expression alleviated the symptoms and promoted survival of the BA mouse model. To investigate the effect of PIGR silencing on the hepatic immune microenvironment of the BA mouse model, we quantified 23 cytokines and chemokines in mice liver homogenate by using multiplex assay. Interestingly, we found that Th2-cytokines, including IL-4, IL-5, IL-10, and IL-13, were significantly reduced upon PIGR silencing (Fig. 4E). We isolated hepatic lymphocytes of mice and detected Th2 (CD3+CD4+IL4+) cells with FCM, the results showed that the proportion of Th2 cells was significantly lower in the Si-PIGR group than that in the Si-NC group (Fig. 4F). These data suggest that silencing of PIGR expression alleviated symptoms and restrained Th2 inflammation in the RRV induced BA mouse model.
Fig. 4.
(A) Schematic of experimental design for PIGR silencing in the BA mouse model (created with BioRender.com) (B) PIGR mRNA and protein expression in liver tissue of mice on day 14, using RT-qPCR and immunoblotting analyses (n = 6; ∗∗, P < 0.01; Wilcoxon rank sum test) (C) Curves of the average body weight of mice from day 0–14. Kaplan-Meyer curves of the survival of mice in the Si-NC and Si-PIGR groups (Si-NC, n = 22; Si-PIGR, n = 20; ∗, P < 0.05; Gehan-Breslow-Wilcoxon test). Serum bilirubin levels, serum biochemistry analyses (AST, ALT, and GGT) of mice on day 14 (n = 6; ns, P > 0.05; ∗, P < 0.05; Wilcoxon rank sum test) (D) HE-stained images (original magnification, × 100; scale bars, 100 μm) and Masson-stained images (original magnification, × 100; scale bars, 100 μm) of mouse liver on day 14 (E) Heatmap of the multiplex assays of 23 cytokines and chemokines in the liver homogenate of mice from Si-NC and Si-PIGR groups (n = 6; ∗, P < 0.05; ∗∗, P < 0.01; Wilcoxon rank sum test) (F) FCM analyses of Th2 cell proportion in isolated hepatic lymphocytes of mice on day 14, the results represent one of five independent experiments (n = 5; ∗∗, P < 0.01; Wilcoxon rank sum test).
Overexpressing PIGR increased liver fibrosis and boosted Th2 inflammation in the RRV-induced BA mouse model
We overexpressed PIGR in the BA mouse model by using an adenovirus expression vector (Fig. 5A). The efficiency of PIGR overexpression was validated by RT-qPCR and immunoblotting analyses (Fig. 5B). Serum biochemistry analyses revealed that the overexpression of PIGR significantly increased the serum levels of GGT and bilirubin on day 14 (Fig. 5C). However, we found PIGR overexpression did not significantly alter the weight gain and survival of RRV-infected mice (Fig. 5C). Histological staining showed that the overexpression of PIGR increased periportal inflammation and fibrosis on day 14 (Fig. 5D). We quantified the concentrations of IL-4, IL-5, IL-10, and IL-13 in the liver homogenate of mice on day 14, and found that the levels of IL-4, IL-5, and IL-13 were significantly higher in the over-PIGR group than in the vector control group (Fig. 5E). FCM analyses showed that the proportion of hepatic Th2 cells was significantly increased in the over-PIGR group on day 14 (Fig. 5F). These results suggest that the overexpression of PIGR increased liver fibrosis and promoted Th2 inflammation in the RRV induced BA mouse model.
Fig. 5.
(A) Schematic of experimental design for PIGR overexpressing in the BA mouse model (created with BioRender.com) (B) PIGR mRNA and protein expression in liver tissue of mice on day 14, using RT-qPCR and immunoblotting analyses (n = 6; ∗∗, P < 0.01; Wilcoxon rank sum test) (C) Curves of the average body weight of mice from day 0–14. Kaplan-Meyer curves of the survival of mice in the vector and over-PIGR groups (vector, n = 20; over-PIGR, n = 24; ns, P > 0.05; Gehan-Breslow-Wilcoxon test). Serum bilirubin levels, serum biochemistry analyses (AST, ALT, and GGT) of mice on day 14 (n = 6; ns, P > 0.05; ∗, P < 0.05; Wilcoxon rank sum test) (D) HE-stained images (original magnification, × 100; scale bars, 100 μm) and Masson-stained images (original magnification, × 100; scale bars, 100 μm) of mouse liver on day 14 (E) Concentrations of IL-4, IL-5, IL-10, and IL-13 in the liver homogenate of mice on day14 (n = 6; ∗, P < 0.05; ∗∗, P < 0.01; Wilcoxon rank sum test) (F) FCM analyses of Th2 cell proportion in isolated hepatic lymphocytes of mice on day 14, the results represent one of five independent experiments (n = 5; ∗∗, P < 0.01; Wilcoxon rank sum test).
PIGR expression induces Th2 inflammation in the liver of BA by promoting the release of IL-33 from cholangiocytes
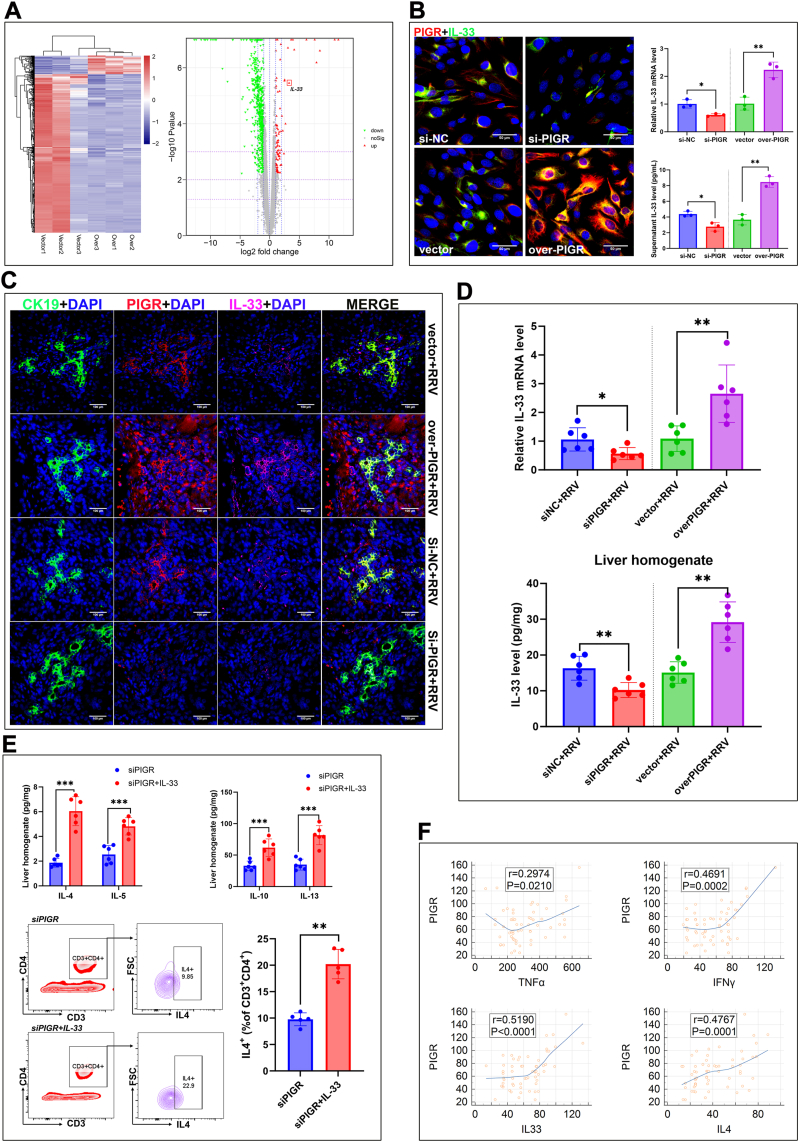
As mentioned above, we found PIGR is mainly expressed in cholangiocytes and is closely related to Th2 inflammatory response in the liver. To find underlying molecular basis, we overexpressed PIGR in cultured BECs and analyzed the gene expression alterations via RNA-sequencing. We identified 903 differential genes between the overexpression group and the vector control, among which we found that interleukin-33 (IL-33) was significantly upregulated in BECs after PIGR overexpression (Fig. 6A). To validate the result, we silenced and overexpressed PIGR expression in cultured BECs, respectively. In cultured BECs, we found PIGR silencing significantly downregulated IL-33 expression and reduced the supernatant level of IL-33; while PIGR overexpression significantly upregulated IL-33 expression and increased the supernatant level of IL-33 (Fig. 6B). These data suggest that the PIGR promotes expression and increase the release of IL-33 in the cholangiocytes.
Fig. 6.
(A) Heatmap and volcano plot of the differential expressed genes between vector control BECs (vector) and PIGR overexpressed BECs (Over) (B) Detection of IL-33 expression in PIGR-silenced, PIGR-overexpressed, and corresponding control BECs, using immunostaining (original magnification, × 400; scale bars, 50 μm) and RT-qPCR analysis (n = 3; ∗, P < 0.05; ∗∗, P < 0.01; two-tailed t-test); quantification of supernatant levels of IL-33 of the PIGR-silenced, PIGR-overexpressed, and corresponding control BECs, using enzyme-linked immunosorbent assay (n = 3; ∗, P < 0.05; ∗∗, P < 0.01; two-tailed t-test) (C) Immunofluorescence staining of CK19 (green), PIGR (red), and IL-33 (pink) in the liver sections of PIGR-silenced, PIGR-overexpressed, and corresponding control mice on day 14 (original magnification, × 400; scale bars, 100 μm) (D) Detection of PIGR mRNA and protein levels in the liver tissue of mice on day 14 (n = 6; ∗, P < 0.05; ∗∗, P < 0.01; Wilcoxon rank sum test) (E) Detection of IL-4, IL-5, IL-10, and IL-13 concentrations in the liver homogenate of PIGR-silenced mice (with or without injection of exogenous IL-33) using enzyme-linked immunosorbent assay (n = 6; ∗∗∗, P < 0.001; Wilcoxon rank sum test); FCM analyses of Th2 cell proportion in isolated hepatic lymphocytes of mice on day 14, the results represent one of five independent experiments (n = 5; ∗∗, P < 0.01; Wilcoxon rank sum test) (F) Correlation analyses of concentrations of PIGR and four cytokines in the liver homogenate of BA patients (n = 60); r, Pearson correlation coefficient; P, P-value analyzed with two-tailed t-test.
Previous studies suggest that IL-33 is a crucial inducer of type-2 inflammation released by barrier cells (endothelial and epithelial cells).23 We hypothesized that the upregulation of PIGR in the cholangiocytes of BA promotes hepatic Th2 inflammation by enhancing the release of IL-33. To verify this hypothesis, we detected IL-33 expression in PIGR-silenced and PIGR-overexpressed BA mouse models. The triple stained (CK19, PIGR, and IL-33) mice liver sections revealed that silencing PIGR downregulated IL-33 expression while overexpressing PIGR upregulated IL-33 expression in the cholangiocytes of BA mouse model (Fig. 6C). Quantification analyses revealed that silencing PIGR significantly decreased IL-33 level, while overexpressing PIGR significantly increased IL-33 level in the liver of the BA mouse model (Fig. 6D). To find more evidence, we supplemented PIGR-silenced BA mice with exogenous IL-33 by injection of recombinant mouse IL-33 protein. We found that IL-33 administration significantly elevated IL-4, IL-5, IL-10, and IL-13 levels, and increased the proportion of Th2 cells in the liver of BA mouse model (Fig. 6E). These data suggest that PIGR expression promotes Th2 inflammation via increasing the level of cholangiocytes-derived IL-33.
To validate the association between PIGR and aforementioned 8 cytokines (IL1-β, TNF-α, IFN-γ, IL-4, IL-5, IL-10, IL-13, and IL-33) in human BA, we quantified these 9 proteins in the liver homogenate of 60 BA patients and made correlation analyses. The correlation analyses revealed that the abundance of PIGR was significantly correlated with TNF-α, IFN-γ, IL-4, and IL-33 levels (Fig. 6F). As mentioned above, TNF-alpha and IFN-γ stimulations can upregulate PIGR expression in cultured BECs, while PIGR expression increases IL-4 level in the BA mouse model by promoting the release of IL-33. These results support the hypothesis that the upregulated expression of PIGR in the cholangiocytes of BA, which triggered by Th1 cytokines TNF-α and IFN-γ, promotes Th2 inflammation in the liver by enhancing the release of IL-33.
PIGR expression regulates the proliferation, apoptosis and epithelial–mesenchymal transition of cultured BECs
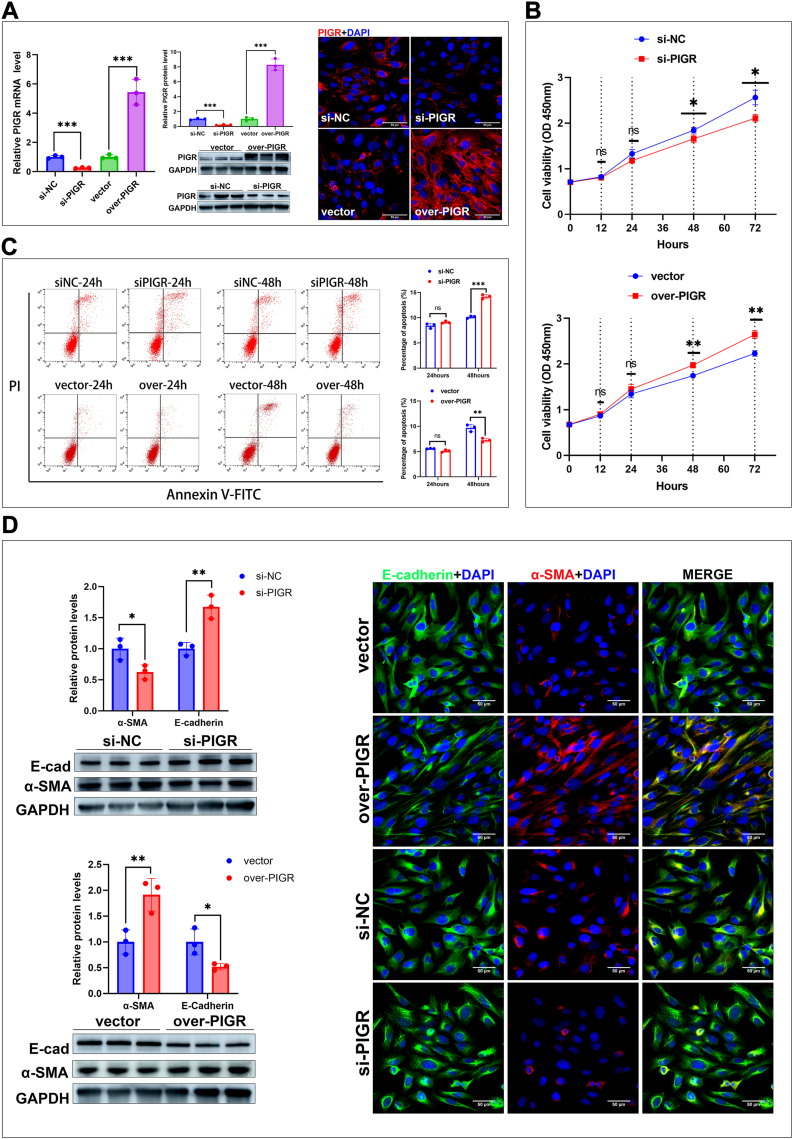
The proliferation, apoptosis, and epithelial–mesenchymal transition (EMT) of cholangiocytes are crucial for maintaining the balance between damage and repair of the biliary epithelium.24 To investigate the role of PIGR on these biological processes, we silenced and overexpressed PIGR in BECs and validated their efficiency using RT-qPCR and immunoblotting analyses (Fig. 7A).
Fig. 7.
(A) Validation of the efficiencies of silencing and overexpressing PIGR in cultured BECs by detecting PIGR expression using RT-qPCR and immunoblotting analyses (n = 3; ∗∗∗, P < 0.001; two-tailed t-test), and immunostaining (original magnification, × 400; scale bars, 50 μm) (B) Proliferation of PIGR-silenced and PIGR-overexpressed BECs from 0 to 72 h post-transfection (n = 3; ns, P > 0.05; ∗, P < 0.05; ∗∗, P < 0.01; two-tailed t-test). Data are presented as mean ± SEM of three independent experiments (C) Apoptosis of PIGR-silenced and PIGR-overexpressed BECs detected by FCM after staining with Annexin-V-FITC and PI (n = 3; ns, ∗∗, P < 0.01; ∗∗∗, P < 0.001; two-tailed t-test). Data presented are as mean ± SEM of three independent experiments (D) Detection of two EMT markers (α-SMA and E-cadherin) in PIGR-silenced and PIGR-overexpressed BECs, using RT-qPCR and immunoblotting analyses (n = 3; ∗, P < 0.05; ∗∗, P < 0.01; two-tailed t-test), and immunostaining (original magnification, × 200; scale bars, 50 μm).
We evaluated the proliferation of BECs at different post-transfection times using CCK8 assay, and found that overexpression of PIGR promoted the proliferation of BECs, whereas silencing of PIGR reduced the proliferation of BECs (Fig. 7B). The FCM analyses showed that overexpression of PIGR reduced apoptosis, whereas silencing PIGR increased apoptosis of BECs at both 24 and 48 h after transfection (Fig. 7C). We detected the expressions of two EMT markers, alpha smooth muscle actin (α-SMA) and E-cadherin, and found that silencing PIGR significantly decreased α-SMA expression and increased E-cadherin expression, while overexpressing PIGR significantly increased α-SMA expression and decreased E-cadherin expression (Fig. 7D). These data suggest that PIGR participates in regulating the balance of damage and repair of the biliary epithelium by promoting the proliferation and EMT, and inhibiting the apoptosis of cholangiocytes.
Discussion
By conducting proteomic and immunoassay analyses on 250 serum samples of BA and control patients, we found serum level of PIGR significantly increased in patients with BA and presented excellent diagnostic potential. The findings of upregulated PIGR expression in BA patients and its correlation with hepatic inflammation and fibrosis suggested a potential link between PIGR and disease progression. Using three different mouse models, we demonstrated the upregulation of PIGR in the biliary epithelium of BA is not caused by cholestasis or liver fibrosis, but rather by the immuno-inflammatory injury of the bile ducts. By manipulating the expression of PIGR in the RRV-induced BA mouse model with chemically modified siRNA and adenovirus expression vector, we found PIGR expression promotes the progression of hepatic inflammation and liver fibrosis. By applying Luminex-based multiplex assay and RNA-seq in the BA mouse model and cultured human cholangiocytes, we demonstrated that PIGR promotes Th2 inflammation in the liver by facilitating the release of IL-33 from cholangiocytes. We also found the upregulation of PIGR in the biliary epithelium of BA can be triggered by Th1- cytokines, especially TNF-α and IFN-γ. Collectively, these findings demonstrated the essential role of PIGR in the pathogenesis of BA, and reveal a previously unrecognized mechanism of Th1/PIGR/IL-33/Th2 circuit in regulating the hepatic immune response.
In previous reports, several traditional clinical indicators, such as GGT and LSM, were considered helpful in diagnosing BA.25 However, the diagnostic accuracy of GGT appeared to be age-dependent; and the diagnostic performance of LSM varied with an AUC between 0.787 and 0.997 in the previous studies.26, 27, 28, 29 In this study, similar results were achieved; we found GGT and LSM distinguished BA from IHS with AUC of 0.781 and 0.769. Nevertheless, comparative ROC analyses showed that serum levels of PIGR and PVR were more effective in distinguishing BA than both GGT and LSM. Interestingly, in the past year, two proteome-based studies have reported the elevation of serum PIGR in BA patients. In July 2023, Kumar et al. reported the elevation of serum PIGR in BA patients via plasma proteome profiling method; in April 2024, Fu et al. proposed a biomarker panel comprising PIGR and immunoglobulin lambda variable chain in diagnosing BA, with an accuracy of 0.89 and an AUC of 0.944.30,31 In this study, 250 patients were enrolled, which was a larger cohort than Kumar et al.‘s and Fu et al.'s study. Our study not only confirmed the significant increase of serum PIGR in BA patients, but also found the combination of PIGR and PVR was a better diagnostic tool for BA that presented a higher accuracy of 0.935 and a higher AUC of 0.968. Moreover, we found serum level of PIGR positively correlated with several important clinical indicators of BA, including GGT, LSM, and the inflammation grade and fibrosis stage of liver pathology. Most importantly, previous studies have not detected PIGR expression in the liver specimens of BA, nor discussed its role in BA pathogenesis; but in current study, we not only found the upregulation of PIGR expression in the bile ducts of BA patients, but also uncovered the underlying mechanisms of why PIGR is upregulated and how it contributes to the pathogenesis of the disease.
The upregulation of PIGR in the bile ducts of BA patients was closely related to Th1-cytokines, especially TNF-α and IFN-γ. First, we demonstrated that TNF-α and IFN-γ can directly induce PIGR expression in cultured BECs via activating NF-κB; second, in the liver of BA patients, we found significant correlations between PIGR expression and the levels of TNF-α and IFN-γ. Effectors of the Th1 inflammation have long been implicated in pathogenesis of BA.32 In human BA, tissue analyses show increased infiltration of activated CD4+ and CD8+ lymphocytes in the livers, expression of Th1 cytokines, including TNF-α and IFN-γ.33, 34, 35 Furthermore, the oligoclonal expansion of CD4+ and CD8+ lymphocytes were found in the liver and bile duct remnants of BA patients.36 The prominent Th1 microenvironment in the BA liver can trigger the upregulation of PIGR in cholangiocytes. Besides, cholangiocytes express multiple pattern recognition receptors (PRRs) that are capable of recognizing “danger signals”, including endogenous signals referred to as damage associated molecular patterns (DAMPs) and exogenous signals referred to as pathogen associated molecular patterns (PAMPs).37 When triggered by endogenous or exogenous signals, the challenged breach of the epithelium activates the biliary immune response via several proinflammatory pathways, such as NF-κB, IRF3, and MAPK pathways.38,39 Hence, exposure to DAMPs or PAMPs can directly stimulate PIGR expression by the cholangiocytes via activating NF-κB pathway, and simultaneously promotes the inflammatory microenvironment which further induces the expression of PIGR. Despite a prominent Th1 inflammatory response, studies in human and BA mouse model also reveal a Th2 commitment in disease progression. Liver and serum samples from a subgroup of BA patients have high levels of Th2 cytokines, including IL-4, IL-13, and IL-33; and study in BA mouse model also revealed Th2 signals induced bile ducts injury and are compatible with the BA phenotype.40, 41, 42 One theory proposed that the persistent immune-mediated bile ducts injury in BA liver drives the initial Th1-predominant milieu shifts toward a Th2 with the simultaneous emergence of the Th17 subset, and the mixed (Th1-2-17) immune response caused persistent liver injury and progressive fibrosis of BA patients.13,39 However, the mechanism that induce the emergence of this mixed immune response in BA livers remains elusive. The close associations between PIGR and Th2 response we found in current study suggest that PIGR may contribute to the mixed immune response.
PIGR is known as a transcytosis mediator expressed in epithelial cells that plays an important role in maintaining the mucosal immunity, which has been found to be associated with several immune disorders in the airway, biliary tract, and gastrointestinal tract.43, 44, 45 However, previous studies have mainly focused on exploring the function of PIGR in transporting IgA and IgM into the luminal space. In this study, we revealed a previously unknown mechanism of PIGR in regulating biliary immunity by promoting Th2 immune response via enhancing the release of IL-33. IL-33, one of the DAMPs that when released by cholangiocytes, engages to IL-33R (ST2) expressed on immune cells and promotes the production of profibrotic Th2 cytokines.23,46 Increased level of IL-33 has been found in serum and tissue biopsies in both human patients and BA mouse model.47,48 In BA mouse model, previous study reported that IL-33 stimulates innate helper cell (ILC2) to produce IL-13 and thereby drives the cholangiocytes proliferation.49 In this study, we manipulated PIGR expression in the BA mouse model, and found that silencing PIGR reduced the levels of IL-33 and Th2-cytokines (IL4, IL-5, IL-10, and IL-13), while overexpressing PIGR increased the levels of IL-33 and Th2-cytokines. In addition, we found silencing PIGR also increased the levels of Th1-cytokines (IL-1β, TNF-α, and IFN-γ). This may be due to the silencing of PIGR disrupting the balance between Th1 and Th2, leading to a shift of hepatic inflammation towards Th1 response. Moreover, we found administration of IL-33 significantly suppressed the Th-2 inhibitory effect caused by PIGR silencing. In human BA, previous studies reported that the IL-33 level significantly correlated with both GGT level and liver fibrosis.47,50 In current study, we also found the serum level of PIGR in the BA patients significantly correlated with Th1-cytokines (TNF-α and IFN-γ), Th2-cytokines (IL-4 and IL-33), GGT, and the progression of liver fibrosis. These results suggest that PIGR is an important modulator of IL-33 expression in the cholangiocytes, and the Th1/PIGR/IL-33/Th2 circuit contributes to the persistent hepatic inflammation and progressive liver fibrosis of BA.
In conclusion, we conducted research on serum samples of 250 patients, validated the increase of serum PIGR in patients with BA, and provided a useful diagnostic model of combined PIGR and PVR in diagnosing BA. However, this is a single-center study, a multicenter study should be conducted in the future to overcome the limited representativeness of the diagnostic model. In the liver of human and murine BA, the expression of PIGR localized mainly in the cholangiocytes and is triggered by Th1 cytokines. Experiments to investigate the role of PIGR in BA pathogenesis revealed PIGR as a potent inducer of hepatic Th2 inflammation by promoting the release of IL-33. Combined, the Th1/PIGR/IL-33/Th2 circuit may contribute to the emergence and persistent of the complicated mixed immunoinflammatory response in the BA liver. Our results suggest that PIGR is more than just a valuable diagnostic biomarker of BA, but also a potential therapeutic target of BA and other immuno-fibrotic biliary disorders. These findings also add a new dimension to understanding the role of PIGR in regulating biliary immunity and have potential implications to other mucosal system, such as in the airway and gastrointestinal tract.
Contributors
Zhi-Bo Zhang and Yuan Li conceived and designed the study. Yuan Li and Tian-Yu Li performed all the experiments and analyzed the data. Zhi-Bo Zhang, Qi Qiao, Ling-Fen Xu, Min-Ting Zhang, and Min-Ting Zhang recruited subjects, collected samples, and analyzed the clinical data. Yuan Li wrote the paper, and Zhibo Zhang reviewed and edited the manuscript. All the authors have read and approved the final manuscript.
Data sharing statement
The MS proteomics data were deposited in the ProteomeXchange Consortium via the PRIDE54 partner repository with the dataset identifier PXD029469. All data and related documents are available upon reasonable request (zhangzb@sj-hospital.org).
Declaration of interests
The authors do not have any disclosures to report.
Acknowledgements
This study was financially supported by the National Natural Science Foundation of China (82170529), the National Key R&D Program (2021YFC2701003), and the National Natural Science Foundation of China (82272022).
Footnotes
Supplementary data related to this article can be found at https://doi.org/10.1016/j.ebiom.2024.105344.
Appendix A. Supplementary data
References
- 1.Hartley J.L., Davenport M., Kelly D.A. Biliary atresia. Lancet. 2009;374(9702):1704–1713. doi: 10.1016/S0140-6736(09)60946-6. [DOI] [PubMed] [Google Scholar]
- 2.Chung P.H.Y., Zheng S., Tam P.K.H. Biliary atresia: east versus west. Semin Pediatr Surg. 2020;29(4) doi: 10.1016/j.sempedsurg.2020.150950. [DOI] [PubMed] [Google Scholar]
- 3.Kasahara M., Umeshita K., Sakamoto S., et al. Liver transplantation for biliary atresia: a systematic review. Pediatr Surg Int. 2017;33(12):1289–1295. doi: 10.1007/s00383-017-4173-5. [DOI] [PubMed] [Google Scholar]
- 4.Serinet M.O., Wildhaber B.E., Broué P., et al. Impact of age at Kasai operation on its results in late childhood and adolescence: a rational basis for biliary atresia screening. Pediatrics. 2009;123(5):1280–1286. doi: 10.1542/peds.2008-1949. [DOI] [PubMed] [Google Scholar]
- 5.Lendahl U., Lui V.C.H., Chung P.H.Y., et al. Biliary Atresia - emerging diagnostic and therapy opportunities. eBioMedicine. 2021;74 doi: 10.1016/j.ebiom.2021.103689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee C.W., Lin M.Y., Lee W.C., et al. Characterization of plasma proteome in biliary atresia. Clin Chim Acta. 2007 Jan;375(1-2):104–109. doi: 10.1016/j.cca.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 7.Song Z., Dong R., Fan Y., et al. Identification of serum protein biomarkers in biliary atresia by mass spectrometry and enzyme-linked immunosorbent assay. J Pediatr Gastroenterol Nutr. 2012;55(4):370–375. doi: 10.1097/MPG.0b013e31825bb01a. [DOI] [PubMed] [Google Scholar]
- 8.Lertudomphonwanit C., Mourya R., Fei L., et al. Large-scale proteomics identifies MMP-7 as a sentinel of epithelial injury and of biliary atresia. Sci Transl Med. 2017;9(417) doi: 10.1126/scitranslmed.aan8462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang L., Zhou Y., Xu P.P., et al. Diagnostic accuracy of serum matrix metalloproteinase-7 for biliary atresia. Hepatology. 2018;68(6):2069–2077. doi: 10.1002/hep.30234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chi S., Xu P., Yu P., et al. Dynamic analysis of serum MMP-7 and its relationship with disease progression in biliary atresia: a multicenter prospective study. Hepatol Int. 2022;16(4):954–963. doi: 10.1007/s12072-022-10322-x. [DOI] [PubMed] [Google Scholar]
- 11.Müller F., Kolbowski L., Bernhardt O.M., et al. Data-independent acquisition improves quantitative cross-linking mass spectrometry. Mol Cell Proteomics. 2019;18(4):786–795. doi: 10.1074/mcp.TIR118.001276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang H., Zhang G.Y., Su W.C., et al. High throughput isolation and data independent acquisition mass spectrometry (DIA-MS) of urinary extracellular vesicles to improve prostate cancer diagnosis. Molecules. 2022;27(23):8155. doi: 10.3390/molecules27238155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Asai A., Miethke A., Bezerra J.A. Pathogenesis of biliary atresia: defining biology to understand clinical phenotypes. Nat Rev Gastroenterol Hepatol. 2015;12(6):342–352. doi: 10.1038/nrgastro.2015.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Desmet V.J., Gerber M., Hoofnagle J.H., et al. Classification of chronic hepatitis: diagnosis, grading and staging. Hepatology. 1994;19(6):1513–1520. [PubMed] [Google Scholar]
- 15.Ishak K., Baptista A., Bianchi L., et al. Histological grading and staging of chronic hepatitis. J Hepatol. 1995;22(6):696–699. doi: 10.1016/0168-8278(95)80226-6. [DOI] [PubMed] [Google Scholar]
- 16.DeLong E.R., DeLong D.M., Clarke-Pearson D.L. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44(3):837–845. [PubMed] [Google Scholar]
- 17.Li Y., Li T.Y., Qi Q., et al. Human poliovirus receptor contributes to biliary atresia pathogenesis by exacerbating natural-killer-cell-mediated bile duct injury. Liver Int. 2022;42(12):2724–2742. doi: 10.1111/liv.15457. [DOI] [PubMed] [Google Scholar]
- 18.Gohy S.T., Detry B.R., Lecocq M., et al. Polymeric immunoglobulin receptor down-regulation in chronic obstructive pulmonary disease. Persistence in the cultured epithelium and role of transforming growth factor-β. Am J Respir Crit Care Med. 2014;190(5):509–521. doi: 10.1164/rccm.201311-1971OC. [DOI] [PubMed] [Google Scholar]
- 19.de Fays C., Carlier F.M., Gohy S., et al. Secretory immunoglobulin A immunity in chronic obstructive respiratory diseases. Cells. 2022;11(8):1324. doi: 10.3390/cells11081324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ladjemi M.Z., Gras D., Dupasquier S., et al. Bronchial epithelial IgA secretion is impaired in asthma. Role of IL-4/IL-13. Am J Respir Crit Care Med. 2018;197(11):1396–1409. doi: 10.1164/rccm.201703-0561OC. [DOI] [PubMed] [Google Scholar]
- 21.Loman S., Jansen H.M., Out T.A., et al. Interleukin-4 and interferon-gamma synergistically increase secretory component gene expression, but are additive in stimulating secretory immunoglobulin A release by Calu-3 airway epithelial cells. Immunology. 1999;96(4):537–543. doi: 10.1046/j.1365-2567.1999.00731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Németh Z.H., Deitch E.A., Szabó C., et al. Pyrrolidinedithiocarbamate inhibits NF-kappaB activation and IL-8 production in intestinal epithelial cells. Immunol Lett. 2003;85(1):41–46. doi: 10.1016/s0165-2478(02)00208-0. [DOI] [PubMed] [Google Scholar]
- 23.Liew F.Y., Girard J.P., Turnquist H.R. Interleukin-33 in health and disease. Nat Rev Immunol. 2016;16(11):676–689. doi: 10.1038/nri.2016.95. [DOI] [PubMed] [Google Scholar]
- 24.Banales J.M., Huebert R.C., Karlsen T., et al. Cholangiocyte pathobiology. Nat Rev Gastroenterol Hepatol. 2019;16(5):269–281. doi: 10.1038/s41575-019-0125-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lendahl U., Lui V.C.H., Chung P.H.Y., Tam P.K.H. Biliary Atresia - emerging diagnostic and therapy opportunities. eBioMedicine. 2021;74 doi: 10.1016/j.ebiom.2021.103689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Noorulla F., Dedon R., Maisels M.J. Association of early direct bilirubin levels and biliary atresia among neonates. JAMA Netw Open. 2019;2(10) doi: 10.1001/jamanetworkopen.2019.13321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dong R., Jiang J., Zhang S., et al. Development and validation of novel diagnostic models for biliary atresia in a large cohort of Chinese patients. eBioMedicine. 2018;34:223–230. doi: 10.1016/j.ebiom.2018.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leschied J.R., Dillman J.R., Bilhartz J., et al. Shear wave elastography helps differentiate biliary atresia from other neonatal/infantile liver diseases. Pediatr Radiol. 2015;45(3):366–375. doi: 10.1007/s00247-014-3149-z. [DOI] [PubMed] [Google Scholar]
- 29.Wang X., Qian L., Jia L., et al. Utility of shear wave elastography for differentiating biliary atresia from infantile hepatitis syndrome. J Ultrasound Med. 2016;35(7):1475–1479. doi: 10.7863/ultra.15.08031. [DOI] [PubMed] [Google Scholar]
- 30.Ram A.K., Kanojia R.P., Bhatia A., et al. Polymeric immunoglobulin receptor and galectin-3-binding protein are raised in biliary atresia: reveals a proteomic-based study. J Proteomics. 2023;287 doi: 10.1016/j.jprot.2023.104974. [DOI] [PubMed] [Google Scholar]
- 31.Fu M., Guo Z., Chen Y., et al. Proteomics defines plasma biomarkers for the early diagnosis of biliary atresia. J Proteome Res. 2024;23(5):1744–1756. doi: 10.1021/acs.jproteome.3c00873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alvarez F. Is biliary atresia an immune mediated disease? J Hepatol. 2013;59(4):648–650. doi: 10.1016/j.jhep.2013.06.006. [DOI] [PubMed] [Google Scholar]
- 33.Mack C.L., Tucker R.M., Sokol R.J., Kotzin B.L. Armed CD4+ Th1 effector cells and activated macrophages participate in bile duct injury in murine biliary atresia. Clin Immunol. 2005;115(2):200–209. doi: 10.1016/j.clim.2005.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mack C.L., Tucker R.M., Sokol R.J., et al. Biliary atresia is associated with CD4+ Th1 cell-mediated portal tract inflammation. Pediatr Res. 2004;56(1):79–87. doi: 10.1203/01.PDR.0000130480.51066.FB. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang Y., Jin Z., Dong R., et al. MicroRNA-29b/142-5p contribute to the pathogenesis of biliary atresia by regulating the IFN-γ gene. Cell Death Dis. 2018;9(5):545. doi: 10.1038/s41419-018-0605-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mack C.L., Falta M.T., Sullivan A.K., et al. Oligoclonal expansions of CD4+ and CD8+ T-cells in the target organ of patients with biliary atresia. Gastroenterology. 2007;133(1):278–287. doi: 10.1053/j.gastro.2007.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chuang Y.H., Lan R.Y., Gershwin M.E. The immunopathology of human biliary cell epithelium. Semin Immunopathol. 2009;31(3):323–331. doi: 10.1007/s00281-009-0172-5. [DOI] [PubMed] [Google Scholar]
- 38.Harada K., Nakanuma Y. Biliary innate immunity: function and modulation. Mediators Inflamm. 2010;2010 doi: 10.1155/2010/373878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ortiz-Perez A., Donnelly B., Temple H., Tiao G., Bansal R., Mohanty S.K. Innate immunity and pathogenesis of biliary atresia. Front Immunol. 2020;11:329. doi: 10.3389/fimmu.2020.00329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ganschow R., Broering D.C., Nolkemper D., et al. Th2 cytokine profile in infants predisposes to improved graft acceptance after liver transplantation. Transplantation. 2001;72(5):929–934. doi: 10.1097/00007890-200109150-00031. [DOI] [PubMed] [Google Scholar]
- 41.Li J., Bessho K., Shivakumar P., et al. Th2 signals induce epithelial injury in mice and are compatible with the biliary atresia phenotype. J Clin Invest. 2011;121(11):4244–4256. doi: 10.1172/JCI57728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jian Z.H., Wang L.C., Lin C.C., Wang J.D. The correlation between plasma cytokine levels in jaundice-free children with biliary atresia. World J Pediatr. 2015;11(4):352–357. doi: 10.1007/s12519-015-0023-5. [DOI] [PubMed] [Google Scholar]
- 43.Richmond B.W., Brucker R.M., Han W., et al. Airway bacteria drive a progressive COPD-like phenotype in mice with polymeric immunoglobulin receptor deficiency. Nat Commun. 2016;7 doi: 10.1038/ncomms11240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li X., Li Y., Xiao J., et al. Unique DUOX2+ACE2+ small cholangiocytes are pathogenic targets for primary biliary cholangitis. Nat Commun. 2023;14(1):29. doi: 10.1038/s41467-022-34606-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lin H., Lin J., Pan T., et al. Polymeric immunoglobulin receptor deficiency exacerbates autoimmune hepatitis by inducing intestinal dysbiosis and barrier dysfunction. Cell Death Dis. 2023;14(1):68. doi: 10.1038/s41419-023-05589-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kotsiou O.S., Gourgoulianis K.I., Zarogiannis S.G. IL-33/ST2 Axis in organ fibrosis. Front Immunol. 2018;9:2432. doi: 10.3389/fimmu.2018.02432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Narayanaswamy B., Gonde C., Tredger J.M., Hussain M., Vergani D., Davenport M. Serial circulating markers of inflammation in biliary atresia--evolution of the post-operative inflammatory process. Hepatology. 2007;46(1):180–187. doi: 10.1002/hep.21701. [DOI] [PubMed] [Google Scholar]
- 48.Dong R., Dong K., Wang X., Chen G., Shen C., Zheng S. Interleukin-33 overexpression is associated with gamma-glutamyl transferase in biliary atresia. Cytokine. 2013;61(2):433–437. doi: 10.1016/j.cyto.2012.10.035. [DOI] [PubMed] [Google Scholar]
- 49.Li J., Razumilava N., Gores G.J., et al. Biliary repair and carcinogenesis are mediated by IL-33-dependent cholangiocyte proliferation. J Clin Invest. 2014;124(7):3241–3251. doi: 10.1172/JCI73742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu J., Yang Y., Zheng C., et al. Correlation of interleukin-33/ST2 receptor and liver fibrosis progression in biliary atresia patients. Front Pediatr. 2019;7:403. doi: 10.3389/fped.2019.00403. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.