Summary
Pancreatic cancer (PC) is one of the most lethal malignancies and new therapeutic strategies are urgently needed. β1,3-N-acetylglucosaminyltransferase V (B3GNT5) may be a potential option for cancer treatment, but its role in PC remains unknown. In this study, we first demonstrated through bioinformatics analysis that B3GNT5 was high expression in PC and predicted poor prognosis. We further constructed B3GNT5 overexpression or knockdown cell lines by employing lentivirus packaging techniques and confirmed that B3GNT5 could promote tumor cell viability and autonomous growth using cultured cells and vivo xenograft models. In addition, we found that knockdown of B3GNT5 in PC cells inhibited cell migration, invasion, and angiogenesis, as well as stemness of cancer stem cells and enhanced chemotherapy sensitivity to gemcitabine. Mechanistically, overexpression of the transcription factor STAT5B in PC cells enhanced the transcriptional activity of the B3GNT5 promoter. Our work confirmed a tumor-promotive role of B3GNT5 in PC pathogenesis.
Subject areas: Molecular biology, Cell biology, Cancer
Graphical abstract

Highlights
-
•
B3GNT5 is highly expressed in pancreatic cancer and predicts poor prognosis
-
•
B3GNT5 exacerbates malignant behaviors of pancreatic cancer cells
-
•
B3GNT5 is transcriptionally regulated by STAT5B
-
•
B3GNT5 may regulate the downstream AKT and ERK pathways
Molecular biology; Cell biology; Cancer
Introduction
Pancreatic cancer (PC) is considered to be one of the most lethal malignancies, with a very poor prognosis and it is expected to be the second leading cause of cancer-related deaths by 2030.1,2 Due to a lack of specific symptoms, the majority of patients are in the progressive stage of localized invasion or distant metastasis at the time of diagnosis.3,4 PC is also known as the “silent killer” because it is highly aggressive and has low treatment expectations.5 Despite many improvements in surgery and drug therapy, it still has a very poor diagnosis and prognosis with a median survival of <6 months and a 5-year survival rate of <5%.6,7 Since the etiology of this catastrophic tumor has not been clearly defined, it will be of great importance to explore the pathological mechanisms of PC for the development of new biomarkers as well as therapeutic targets.
Signal transducer and activator of transcription 5b (STAT5B) belongs to the STATs family of transcriptional activators and participates in diverse biological processes, such as immunomodulation, apoptosis, and lipid metabolism.8 It is also a transcription factor essential for the proliferation and survival of many solid tumors and is involved in the malignant progression of a variety of tumors including colorectal, breast, and liver cancers.9,10,11 Studies showed that STAT5B is closely related to various malignant phenotypes of PC, such as proliferation, metastasis, angiogenesis, and chemotherapy resistance, and inhibition of STAT5B can hinder the progression of PC.12,13 STAT5B regulates numerous factors, among which β1,3-N-acetylglucosaminyltransferase V (B3GNT5) has attracted our attention.
The B3GNT5 gene encodes the glycosyltransferase β-1, 3-n-acetylglucosamine transferase 5, which attaches N-acetylglucosamine (GlcNAc) from UDP-GlcNAc to Gal on the non-reducing end of the carbohydrate chain with β-1,3-linkage resulting in the precursor lactotriaosylceramide for the synthesis of lacto (type 1) and neolacto-series (type 2) glycosphingolipids.6,14,15 B3GNT5 is the fifth candidate β3GNT enzyme that has been verified to be the most viable candidate for the synthesis of lactotriaosylceramide.15 As a key glycosyltransferase in the synthesis of lacto-glycosphingolipids and neolacto series glycosphingolipids, B3GNT5 is closely related to embryonic development, cell differentiation, and some malignant diseases.15,16,17 The proliferation of cancer stem cells has an important role in malignant progression and drug resistance in PC.18 A previous study showed that B3GNT5 can affect the expression of breast cancer stem cell-related factor SSEA-1 through metabolic regulation, and thus participate in the malignant behavior of breast cancer cells.19 Zheng et al. demonstrated that B3GNT5 expression was upregulated in glioma stem cells and maintained the stemness of glioma stem cells.20 Furthermore, it is shown that enhancing B3GNT5 activity can increase neolacto-series glycosphingolipids on the surface of tumor cells, hinder the action of human leukocyte antigen class I, and then interfere with CD8+T cell activation, thereby mediating immune escape of glioma cells.21 However, the role of B3GNT5 in PC remains unknown and is rarely reported.
In this study, we evaluated the expression levels of B3GNT5 in PC and its association with prognosis. Then, we assessed the effects of B3GNT5 on PC cell proliferation, migration, invasion, and angiogenesis, as well as the stemness of cancer stem cells and chemotherapy sensitivity. Finally, we explored the potential molecular mechanisms of B3GNT5 in PC progression. Therefore, this study reveals that B3GNT5 can serve as a potential molecular biomarker and therapeutic target for PC.
Results
Identification of DEGs and their functional annotation in pancreatic cancer
The five datasets from the Gene Expression Omnibus (GEO, GSE62165, GSE63111, GSE130221, GSE15471, and GSE56560) showed that overall distribution of differentially expressed genes (DEGs) in normal and pancreatic cancer (PC) tissues (Figure 1A). The Venn diagram further revealed a total of 208 overlapping genes with upregulated expression in the five datasets (Figure 1B). To explore the potential mechanisms by which these genes may affect PC, we performed Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses on the DEGs and presented the results of top 20 enrichment analysis (Figures 1C and 1D). In the GO analysis, the top impacted cellular component (CC) terms were “collagen-containing extracellular matrix,” “protein complex involved in cell adhesion,” “cell-substrate junction,” “focal adhesion,” and “integrin complex.” The majority of DEGs were associated with the extracellular matrix (ECM) and cell adhesion, which may be because PC progression and metastasis are strongly influenced by the ECM.22,23 The top impacted biological process (BP) of GO terms were “integrin-mediated signaling pathway,” “cell adhesion mediated by integrin,” “cell-substrate adhesion,” “cell-cell adhesion via plasma-membrane adhesion molecules,” and “cell-matrix adhesion.” Since integrin-mediated cell adhesion to the extracellular matrix is essential for cell survival, its appearance at the top of the BP enrichment results is not surprising.24 In addition, the molecular function (MF) categories of “calcium-dependent protein binding,” “cell adhesion mediator activity,” “integrin binding,” “cadherin binding,” and “endopeptidase inhibitor activity” are consistent with the nature of the GO terms in CC and MF, emphasizing the significant impact of DEGs on the ECM and cell adhesion during PC. KEGG analysis showed that the most affected signal pathways were “ECM-receptor interaction,” “small cell lung cancer,” “focal adhesion,” “human papillomavirus infection,” and “mucin type O-glycan biosynthesis.” Taken together, these results suggested that up-regulated DEGs might regulate PC progression by evoking phenotypic and signaling pathways for tumor cell growth, invasion, and migration.
Figure 1.
Identification of DEGs and their functional annotation in pancreatic cancer
(A) Volcano plots of DEGs in normal and tumor samples in pancreatic cancer datasets GSE62165, GSE63111, GSE130221, GSE15471, and GSE56560. Raw data between pancreatic cancer and normal tissues were obtained from the GEO dataset (https://www.ncbi.nlm.nih.gov/). DEGs were selected by p < 0.01 and |log2FC|>1. Red for genes with significantly upregulated expression, blue for genes with significantly downregulated expression, and gray for genes with no significant difference in expression.
(B) The Venn diagram illustrates a total of 208 upregulated genes in the five datasets. GO and KEGG enrichment analysis was performed for overlapping genes in (B).
(C and D) Top 20 results of GO enrichment (C) and KEGG enrichment (D) analysis were summarized and visualized using the R package ggplot2. GEO, Gene Expression Omnibus; FC, fold change; DEGs, differentially expressed genes; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Gene and Genome.
Among the numerous enrichment analysis results, we noted the presence of biological processes and signaling pathways associated with glycosylation, such as o-glycan processes, galactosyltransferase activity, and mucin type O-glycan biosynthesis. Aberrant glycosylation has been identified in pancreatic cancer and promotes the stemness and malignant behavior of PC cells.25,26 We noted that β1,3-N-acetylglucosaminyltransferase V (B3GNT5) was enriched in the aforementioned terms (Figure S1). B3GNT5 is a common glycosyltransferase that plays an important role in a variety of solid tumors but has not been reported in PC. In summary, we selected B3GNT5 as the target gene of PC among the factors enriched in the aforementioned pathways for further study.
B3GNT5 is highly expressed in pancreatic cancer and predicts poor prognosis
The Cancer Genome Atlas datasets (TCGA) and seven datasets from GEO datasets (GSE15471, GSE63111, GSE130221, GSE62165, GSE56560, GSE28735, and GSE62452) showed that compared with normal tissues, B3GNT5 was overexpressed in PC tissues (Figure 2A). On this basis, we further investigated the effect of B3GNT5 on the prognosis of PC. The analysis of TCGA database showed significantly increased expression levels of B3GNT5 in stages IA, IB, IIA, IIB, III, and IV PC compared to that of stage I (Figure 2B). As shown in Figure 2C, overall survival, disease-free interval, and disease-specific survival were significantly lower in patients with high B3GNT5 expression compared to those of patients with low B3GNT5 expression. The results of the aforementioned analysis suggest that high expression of B3GNT5 may be associated with poor prognosis. Cancer stem cells (CSCs) can maintain the vitality of the tumor cell population through self-renewal and unlimited proliferation, and play a very important role in tumor development.27 Therefore, we analyzed the relationship between B3GNT5 and the expression of CSCs markers (CD44 and ALDH2) and showed that the expression of B3GNT5 showed a positive correlation with the expression of CD44 and ALDH2 (Figure 2D). We further collected five pairs of fresh PC and para-carcinoma tissues and detected the expression of B3GNT5 using quantitative real-time PCR. The results showed that compared with para-carcinoma tissues, the relative expression of B3GNT5 mRNA in pancreatic cancer tissues was significantly increased (Figure 2E). Moreover, we collected information on clinical samples from 75 PC patients and statistically analyzed the correlation between the expression of B3GNT5 and the clinicopathological features of PC. The results indicated a significant correlation between B3GNT5 expression and tumor size and pathological grading (Table 1). Figure 2F showed immunohistochemical staining maps of low and high expression of B3GNT5 and the results indicated that B3GNT5 was mainly localized in the cytoplasm. Taking the aforementioned experimental results together, we found that B3GNT5 is overexpressed in PC and that elevated B3GNT5 levels predict a poor clinical outcome.
Figure 2.
B3GNT5 is highly expressed in pancreatic cancer and predicts poor prognosis
(A) Gene expression levels of B3GNT5 in pancreatic cancer including TCGA (n = 171, T = 179), GSE15471 (n = 39, T = 39), GSE63111 (n = 4, T = 28), GSE130221 (n = 6, T = 6), GSE62165 (n = 13, T = 118), GSE56560 (n = 7, T = 28), GSE28735 (n = 45, T = 45), GSE62452 (n = 61, T = 69) databases. ∗p < 0.05, ∗∗p < 0.01.
(B) Gene expression levels of B3GNT5 in TCGA pancreatic cancer database (n = 196) based on different stages.
(C) Kaplan-Meier survival curves depicting OS, PFI, DFI, and DSS based on the B3GNT5 expression in TCGA pancreatic cancer database using the UCSC Xena online tool (https://xena.ucsc.edu/).
(D) The correlation between B3GNT5 and the expression of cancer stem cell markers ALDH2 and CD44 was analyzed by TIMER2.0 (http://timer.comp-genomics.org/timer/).
(E) The mRNA expression of B3GNT5 in para-carcinoma tissues and tumor tissues from patients with pancreatic cancer (N = 5).
(F) Immunohistochemistry staining images of high and low expression of B3GNT5. Scale bar indicates 50 μm. TCGA, The Cancer Genome Atlas; OS, overall survival; DSS, disease-specific survival; PFI, progression-free interval; DFI, disease-free interval.
Table 1.
Association between the expression of B3GNT5 and the clinicopathological features of pancreatic cancer
| Clinicopathological feature | B3GNT5 expression |
p value | |
|---|---|---|---|
| Low, |
High, |
||
| n = 28 | n = 47 | ||
| Gender | |||
| male | 17 | 27 | 0.78105 |
| female | 11 | 20 | – |
| Age (years) | |||
| ≤60 | 16 | 22 | 0.38657 |
| >60 | 12 | 25 | – |
| Tumor diameter (cm) | |||
| ≤3.5 | 20 | 20 | 0.01533 |
| >3.5 | 8 | 27 | – |
| T classification | |||
| T1 | 8 | 6 | 0.0419 |
| T2 | 14 | 18 | – |
| T3 | 6 | 23 | – |
| N stage | |||
| N0 | 19 | 37 | 0.39979 |
| N1 | 7 | 6 | – |
| N2 | 2 | 4 | – |
| M stage | |||
| M0 | 25 | 43 | 0.9259 |
| M1 | 3 | 4 | – |
| TNM stage | |||
| I + II | 24 | 40 | 0.79068 |
| III+IV | 4 | 7 | – |
Differences between groups were done by the Chi-square test.
p < 0.05 was considered significantly different.
B3GNT5 promotes malignant behaviors of pancreatic cancer cells
Considering that the high expression level of B3GNT5 was correlated with large tumor volume in clinical data, we hypothesized that B3GNT5 may influence the growth of PC cells. To investigate this hypothesis, we constructed B3GNT5 low-expressing and overexpressing human PC cell lines (CFPAC-1 and CAPAN-1). First, we performed a western blot to verify the infection efficiency. As can be seen in Figure 3A, the expression of B3GNT5 was significantly lower in B3GNT5-shRNA lentivirus-infected CFPAC-1 and CAPAN-1 cells compared to control lentivirus-infected cells, whereas the B3GNT5 overexpression group showed a significant increase in B3GNT5 protein expression compared to the mock group. Combining the aforementioned results, B3GNT5 knockdown and overexpression cell models were successfully constructed and used for subsequent experiments. Using a CCK-8 assay, we indeed found that B3GNT5 knockdown inhibited cell viability relative to the negative control cells, while B3GNT5 overexpression in CAPAN1 and CFPAN1 cells enhanced cell viability (Figure 3B). Through a colony formation assay, we additionally found that B3GNT5 knockdown inhibited the colony-forming ability of these cells compared to shNC group, while B3GNT5 overexpression promoted cell growth (Figure 3C). In addition, we also verified the effect of B3GNT5 on malignant phenotypes such as tumor cell migration and invasion, and the results showed that B3GNT5 knockdown significantly reduced the migration and invasion ability of PC cells (Figures S2A and S2B). Tube formation assay demonstrated that the tube node numbers of complete tubular structures formed by Human umbilical vein endothelial cells (HUVECs) were decreased in the conditioned medium from PC cells with B3GNT5 knockdown (Figure S2C).
Figure 3.
B3GNT5 promotes malignant behaviors of pancreatic cancer cells
The lentivirus-based systems were applied to overexpress or knockdown B3GNT5 in CFPAC-1 and CAPAN-1 cells, respectively, and screened for stably transfected cell lines.
(A) Western blot analysis of B3GNT5 expression in tumor cells with or without B3GNT5 knockdown or overexpression.
(B) Cell proliferation of respective cells was assessed with CCK8 assay. ∗∗p < 0.01, compared with the shNC or control group.
(C) Colony formation assay after plating 400 cells in each Petri dish for 2 weeks and bar graphs for numbers of colonies. Mean ± SD, N = 3.
(D) Xenograft tumors were formed by subcutaneous injection of pancreatic cancer cells into BALB/c nude mice and the tumors were stripped after 21 days. Tumor mass was photographed, measured, and weighted. B3GNT5 expression of tumors was analyzed by western blot. ∗p < 0.05, ∗∗p < 0.01, compared with the shNC group.
(E) Immunohistochemistry staining analysis of B3GNT5 in xenograft tumors. Scale bar indicates 50 μm. Mean ± SD, N = 6.
In this study, a xenograft model in nude mice was established to further verify the effect of B3GNT5 on PC. CAPAN-1 or CFPAC-1 cells successfully infected with shB3GNT5 or shNC lentivirus were subcutaneously injected into nude mice. After 21 days, the tumors were stripped, photographed, measured, and weighted. The results showed that the volume and weight of tumors caused by cells in the shB3GNT5 group were significantly reduced compared to cells infected with shNC group (Figure 3D), indicating that tumors grew more slowly in the shB3GNT5 group. The protein expression of B3GNT5 in tumor tissues was further detected using western blot, and the results showed that the protein expression of B3GNT5 in tumor tissues in the shB3GNT5 group was significantly lower compared with that in the shNC group (Figure 3D). We further demonstrated by immunohistochemical staining that the positive expression of B3GNT5 in tumor tissues of shB3GNT5 group was significantly reduced compared with that of shNC group (Figure 3E). Taken together, these data indicated that B3GNT5 promotes malignant behaviors of PC cells in vitro and in vivo.
Knockdown of B3GNT5 inhibits stemness of CSCs
Given the importance of B3GNT5 in cancer cell stemness, we hypothesize that B3GNT5 may be involved in the malignant progression of PC by affecting stemness maintenance of pancreatic CSCs. CSCs were first isolated from cells using the ALDEFLUOR kit, and then we investigated the effect of B3GNT5 expression on markers of CSCs (CD44, CD326, and CxCR4). Immunofluorescence experiments showed that B3GNT5 knockdown inhibited the expression of specific pancreatic CSCs markers when compared with control cells (Figure 4A). The western blot analysis further showed similar experimental results, which indicated that B3GNT5 knockdown reduced the number of CSCs in PC cells (Figure 4B). Finally, we tested if B3GNT5 could suppress sphere formation by pancreatic CSCs. Figure 4C showed the cell culture images the cell culture images of the first and second generations, and the experimental results showed that B3GNT5 knockdown resulted in a significant reduction in the number of CSCs forming spheres. We further determined the effect of B3GNT5 on the self-renewal capacity of CSCs by limiting dilution assay and found that decreased expression of B3GNT5 resulted in a lower spheroid initiating frequency (Figure 4D). These results suggest that B3GNT5 expression is important for the stemness maintenance of PC cells.
Figure 4.
Knock down of B3GNT5 inhibits stemness of CSCs
CSCs were isolated from CFPAC-1 and CAPAN-1 cells using the ALDEFLUOR kit.
(A) Immunofluorescence images of CD44, CD326, and CxCR4 expression (red) in CSCs. Cell nuclei were stained by DAPI (blue). Scale bar indicates 100 μm.
(B) Western blot analysis was performed to examine the expressions of CD44, CD326 and CxCR4.
(C) Images and the statistical results showed the effect of B3GNT5 knockdown on the spheres formation abilities of CSCs by sphere-forming assay. Scale bar indicates 100 μm.
(D) The limiting dilution assay of spheroid formation was performed in shNC and shB3GNT5 tumor cells with different gradient cell numbers as indicated and analyzed the data using ELDA software (http://bioinf.wehi.edu.au/software/elda). CSCs, cancer stem cells; DAPI, 4′6′-diamidino-2-phenylindole dihydrochloride; ELDA, extreme limiting dilution analysis.
Knockdown of B3GNT5 enhances chemosensitivity of pancreatic cancer cells to GEM
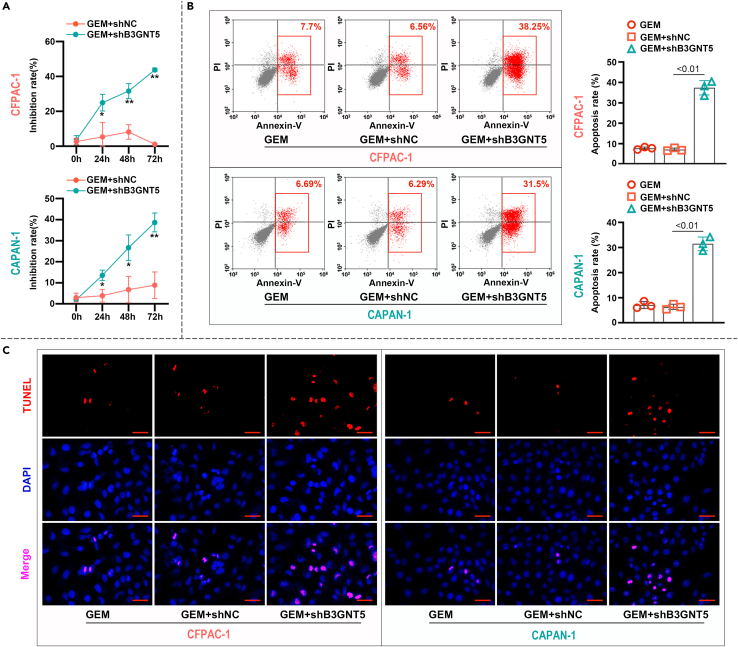
Dew to the difficulty of surgical resection, chemotherapy is still the mainstay of PC treatment, and gemcitabine (GEM) is the first-line chemotherapy for PC.28,29 To study the effect of B3GNT5 on the therapeutic efficacy of GEM, we first compared the effect of GEM on the proliferation of B3GNT5 knockdown and normal PC cells by cell proliferation assays. The results of CCK-8 experiment showed that B3GNT5 knockdown resulted in a significant increase in GEM inhibition of cells compared to shNC infection (Figure 5A). We further determined the effect of B3GNT5 knockdown on PC cells apoptosis after GEM treatment by flow cytometry (Figure 5B) and TUNEL staining (Figure 5C). Both of these two assays indicated that B3GNT5 knockdown significantly increased apoptosis of PC cells. Collectively, the aforementioned results suggested that B3GNT5 could inhibit the chemosensitivity of PC cells to GEM.
Figure 5.
Knock down of B3GNT5 enhances chemosensitivity of pancreatic cancer cells to GEM
CFPAC-1 and CAPAN-1 cells were treated with 10 nM GEM, and the cells were collected 48 h later for the following assays.
(A) Cell inhibition rates were measured by CCK-8 assay. ∗p < 0.05, ∗∗p < 0.01, compared with the GEM+shNC group.
(B) The percentage of apoptotic cells was evaluated by flow cytometry with annexin V-FITC/PI staining.
(C) Analysis of apoptosis by TUNEL staining in tumor cells. Cell nuclei (blue), TUNEL-positive cells (red). Scale bar indicates 50 μm. Mean ± SD, N = 3. GEM, Gemcitabine; TUNEL, terminal deoxynucleotidyl transferase dUTP nick end-labeling.
STAT5B promotes transcriptional activity of B3GNT5 in pancreatic cancer
The aforementioned findings have shown that B3GNT5 promotes the development of PC, but the exact mechanism of its involvement in disease progression is unknown. Previous studies have shown that STAT5B is deeply involved in the malignant progression of PC.12 The bioinformatic analysis showed that B3GNT5 can be regulated by STAT5B, so we hypothesized that the mechanism by which B3GNT5 promotes PC may be due to the regulation by STAT5B. We first transfected CFPAC-1 and CAPAN-1 cells with STAT5B overexpressing vector and its control and the overexpression of STAT5B was confirmed by western blot (Figure 6A). On this basis, we detected the expression level of B3GNT5 in STAT5B overexpressing cells using quantitative real-time PCR and western blot. The results showed that the relative expression of B3GNT5 mRNA and protein was significantly increased in cells transfected with the STAT5B overexpression vector compared with those transfected with the Mock group (Figure 6B). In addition, the knockdown of STAT5B in PC cells was accompanied by a decrease in the expression of B3GNT5 (Figure S3). Therefore, we speculated that STAT5B positively regulated the expression of B3GNT5 in PC cells. We next assessed whether STAT5B promotes B3GNT5 expression by increasing its promoter activity. The location and sequence of the predicted binding site of STAT5B to the B3GNT5 promoter are shown in Figure 6C. The luciferase reporter assays were performed 48 h after transfection with the indicated B3GNT5 promoter fragments and STAT5B overexpression vector or empty vector, and the results revealed that STAT5B overexpression significantly enhanced luciferase activity. Furthermore, we observed that STAT5B overexpression exhibited a promoting effect on the transcriptional activity of several of the predicted promoter fragments (Figure 6C). These results confirm that STAT5B promotes the activity of the B3GNT5 promoter to induce its transcription.
Figure 6.
STAT5B promotes transcriptional activity of B3GNT5 in pancreatic cancer
(A) CFPAC-1 and CAPAN-1 cells were transfected with the STAT5B overexpression vector and its controls, the transfection efficiency was verified by western blot after 48 h.
(B) Quantitative real-time PCR and western blot analysis of B3GNT5 expression in tumor cells with or without STAT5B knockdown or overexpression. ∗∗p < 0.01, compared with the Mock group.
(C) The schematic diagram of positions and sequences of the predicted binding sites on the promoter of B3GNT5. The promoter sequence of B3GNT5 was downloaded from the UCSC website (https://www.genome.ucsc.edu/) and the transcriptional regulation sites of B3GNT5 by STAT5B were predicted by the HumanTFDB database (http://bioinfo.life.hust.edu.cn/HumanTFDB). The dual-luciferase assay was performed 48 h after transfection with the indicated B3GNT5 promoter fragments and STAT5B overexpression vector or empty vector.
(D) STAT5B overexpression plasmid was transfected into B3GNT5-stabilized knockdown tumor cells, and cell proliferation of respective cells was measured by CCK8 assay. ∗p < 0.05, ∗∗p < 0.01, compared with the control group; ∗∗p < 0.01, compared with the STAT5B group.
(E) The STAT5B overexpression plasmid was transfected into B3GNT5-stabilized knockdown tumor cells, which were further treated with 10 nM GEM after 24 h, and the percentage of apoptotic cells was evaluated by flow cytometry with annexin V-FITC/PI staining. Mean ± SD, N = 3. GEM, Gemcitabine.
Finally, we co-transfected cells with STAT5B overexpression vector and B3GNT5 shRNA to assess whether inhibition of B3GNT5 reversed the tumor-promoting effects of STAT5B. The results of the CCK-8 experiment showed that compared with the control group, the cell viability of the STAT5B overexpression group was significantly increased, while compared with the STAT5B group, the cell viability of STAT5B + shB3GNT5 group was significantly decreased (Figure 6D). We further treated transfected cells with 10 nM GEM to detect whether the chemosensitivity of B3GNT5 to GEM is also regulated by STAT5B. Flow cytometry results showed that apoptosis was significantly reduced in the STAT5B overexpression group compared with the control group, while B3GNT5 knockdown partially rescued the effect of STAT5B overexpression (Figure 6E). Overall, these findings indeed suggest that B3GNT5 is regulated by STAT5B, thereby enhancing transcriptional activity and promoting malignant behavior in PC cells.
Label-free quantitative proteomic analysis in pancreatic cancer cells with B3GNT5 overexpression
The role of B3GNT5 in PC disease progression has been revealed, but whether it also promotes PC by modulating other factors or pathways also deserves continued investigation. Therefore, we collected proteins from control and B3GNT5 overexpressed cells, and used a label-free quantitative proteome method to study the proteome, followed by screened differentially expressed proteins using fold change >1.5 and <0.66, p < 0.05 as the screening criteria (Figure 7A). We further performed KEGG analysis of differentially expressed genes, as shown in Figure 7B; the heatmap showed the gene of the terms associated with tumor/pancreatic cancer progression in the KEGG enrichment results. We further subjected the genes to Venn analysis and identified three crossover proteins: MAPK1, AKT1, and MAPK3, suggesting that B3GNT5 may regulate the ERK and AKT pathways (Figure 7C). On this basis, we further examined the effect of B3GNT5 on ERK and AKT pathways in pancreatic cancer cells by western blot assay and found that B3GNT5 knockdown not only suppressed the overall ERK and AKT expression but also inhibited the activation of phosphorylation of these two pathways (Figure 7D). This provided more evidence that B3GNT5 mediated PC progression and B3GNT5 may promote a series of malignant behaviors of PC cells by regulating the abnormal activation of downstream AKT and ERK pathways.
Figure 7.
Label-free quantitative proteomic analysis in pancreatic cancer cells with B3GNT5 overexpression
(A) Scatterplots were used to assess DEGs between B3GNT5 overexpression samples and control samples with fold change>1.5 and <0.66, and p < 0.05. x- and y axis values are the expression mean values. The center line is y = x, the top line is y-x = 3, and the bottom line is y-x = −3.
(B) KEGG analysis was performed on the DEGs, and heatmaps showed that the DEGs were enriched in 6 terms associated with tumor/pancreatic cancer progression.
(C) The Venn diagram illustrated a total of 3 overlapping genes (AKT1, MAPK1, and MAPK3) in 6 terms.
(D) Western blot analysis of ERK and AKT phosphorylation in CFPAC-1 and CAPAN-1 cells with B3GNT5 knockdown, respectively. DEGs, differentially expressed genes; KEGG, Kyoto Encyclopedia of Gene and Genome.
Discussion
PC is one of the common malignant solid tumors of the digestive system, and with the improvement of living standards and the change of diet structure, the morbidity and mortality of PC have also been on the rise in recent years.30,31 Although immunotherapies based on therapies targeting cancer-associated fibroblasts in combination with immune checkpoint inhibitors offer clinical benefits to patients with pancreatic cancer, there has not been a significant general improvement in pancreatic cancer outcomes.32,33 Currently, a growing number of studies have focused on the aberrant glycosylation of glycoproteins and glycolipids in the progression of various types of cancer, suggesting that there is an urgent need to identify the specific glycosyltransferases responsible for tumor aggressiveness. B3GNT5 is considered to be a key glycosyltransferase in the biosynthesis of the (neo-)lacto series glycosphingolipids, and its overexpression in a variety of solid tumors has also been shown to correlate with poorer patient survival.20,34,35 While it is generally accepted that B3GNT5 is important in the formation and development of cancer, little is known about the exact role of this glycosyltransferase, particularly in the development of PC.
The stemness of CSCs has important clinical implications in driving tumor initiation, metastasis, and recurrence, as well as resistance to conventional cancer therapies.36,37 B3GNT5 was found to maintain the stemness of glioma stem cells.20 We therefore assessed whether B3GNT5 affects the maintenance of stemness in pancreatic CSCs, it was found that B3GNT5 knockdown inhibited the expression of CSCs markers (CD44, CD326, and CxCR4) in PC cells as well as sphere formation of CSCs. This suggests that B3GNT5 does have the potential ability to enhance the stemness of CSCs and thus induce tumorigenesis. Currently, CSC-associated cells are still identified by cell surface markers, and there are various markers in different solid tumors. B3GNT5 was reported to affect the expression of SSEA-1, another CSC marker in breast cancer, and there are also reports suggesting that SSEA-1 can be used as a marker for glioma stem cells, esophageal cancer stem cells, and gastric cancer stem cells.38,39,40 However, our previous detection of SSEA-1 in PC stem cells found that its expression level was extremely low, and almost no difference was observed between the two groups of shNC and shB3GNT5 (data not shown); therefore, it was hypothesized that SSEA-1 might not be a marker for PC stem cells, which is also consistent with the current literature report that the main markers of pancreatic cancer stem cells are CD44, CD326, and CxCR4, among others.27,41
GEM has been the basic regimen of chemotherapy for PC since it was put into clinical use in 1997, and it is also often used in combination with other drugs, which has achieved remarkable clinical efficacy.42,43,44 However, the clinical benefits of these regimens are progressively limited by chemotherapy resistance. Furthering understanding of the mechanisms of GEM resistance may lead to the discovery of promising targeted therapies. Protein glycosylation plays an important role in promoting chemoresistance in PC. We noted that the expression of B3GNT5 was significantly elevated in cisplatin-resistant ovarian cancer cells.35 Therefore, it is reasonable to suspect that B3GNT5 may be involved in the process of GEM resistance in PC cells. Our study demonstrated that B3GNT5 knockdown enhanced the chemical sensitivity of PC cells to GEM, thereby reducing the proliferation capacity of PC cells and promoting apoptosis. Therefore, we suggest that B3GNT5-based strategies, including modulation of B3GNT5 expression, may help overcome GEM resistance in the future.
It is well known that the PI3K/AKT and MAPK/ERK pathways are important participants in the pathology of PC, and their aberrant activation not only drives carcinogenesis but also exacerbates a series of malignant behaviors of tumor cells such as migration, invasion, maintenance of stemness and drug resistance.45,46,47 Glycosyltransferases have been widely shown to regulate tumor progression by modulating the AKT/ERK signaling pathway.48,49 Our proteomic results suggested that B3GNT5 might affect the malignant phenotype of PC cells by regulating the ERK and AKT pathways, and further proved by western blot B3GNT5 knockdown not only suppressed the overall ERK and AKT expression but also inhibited the activation of phosphorylation of these two pathways. It has been reported that interference with glycosylation leads to down-regulation of ERK and AKT pathways, further inhibiting angiogenesis.50 Glycosylation is an important post-translational modification of proteins that plays an important role in protein structure and function. When glycosylation is impaired, it may lead to misfolded proteins or abnormal function, thereby causing endoplasmic reticulum stress to reduce protein synthesis or induce the degradation of misfolded proteins.51,52,53 Therefore, we speculate that B3GNT5 deficiency may lead to abnormal glycosylation modification of ERK and AKT, resulting in protein misfolding and degradation. The aforementioned findings provided more favorable evidence to rationally explain the molecular mechanism by which B3GNT5 promotes the malignant behavior of PC cells and suggested that down-regulation of B3GNT5 inhibited the malignant phenotype of pancreatic cancer by blocking the ERK and AKT signaling pathways, and the mechanisms by which B3GNT5 regulates these two signaling pathways will be explored in a follow-up study.
STAT is a family of seven transcription factors that regulate the expression of many different genes thereby affecting core cellular processes.54 The dysregulation of this family gene is closely related to the occurrence and development of many tumors, including hematological malignancies and solid tumors.55,56 Among them, STAT5B was identified as a molecular target for PC and involved in tumor cell proliferation, invasion, angiogenesis, and chemotherapy resistance to GEM.12,13 STAT5B generally regulates the malignant phenotype of tumor cells by activating the transcription of downstream factors.57 In this study, we found that STAT5B can activate the transcription of B3GNT5 by recognizing specific sequences in its promoter region. Our results suggested that B3GNT5 was an important link in STAT5B signaling.
Taken together, B3GNT5 plays a facilitating role in PC progression and aggravates the malignant phenotype of tumor cells. Our results not only demonstrate the role of B3GNT5 in promoting stem cell maintenance and chemotherapy resistance in PC but also reveal the upstream and downstream regulatory mechanisms of B3GNT5. To our knowledge, this was the first study to reveal the underlying mechanism of B3GNT5 in PC. Due to the insidious onset of PC, B3GNT5 may be a promising biomarker for the early diagnosis of PC. In addition, the malignant degree of PC is relatively high, and commonly used chemotherapy drugs such as GEM have developed drug resistance in practical clinical applications, and our findings are important for improving the clinical efficacy of the drugs.
Limitations of the study
In this study, we demonstrated that B3GNT5 is an important driver of pancreatic cancer progression, with the roles of promoting tumor cell proliferation, migration, invasion, angiogenesis, maintaining the stemness of CSCs, and inhibiting chemosensitivity, among other malignant behaviors. Meanwhile, our research also has some limitations. Firstly, since normal pancreatic cells and pancreatic tissues are difficult to obtain, the difference in B3GNT5 expression between normal pancreatic tissues and pancreatic cancer tissues was only analyzed with the available online dataset at this time. Secondly, due to the limited clinical samples available at present, only a small amount of B3GNT5 expression in pancreatic cancer and adjacent tissues has been detected, and the sample size should be expanded for detection in future work.
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to the lead contact, Yuhe Lin (linyuhe@sj-hospital.org).
Materials availability
This study did not generate new unique reagents.
Data and code availability
-
•
The proteomic data have been deposited at PRIDE and is publicly available at the date of publication. Accession numbers are listed in the key resources table.
-
•
This paper does not report any original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Acknowledgments
The authors did not receive support from any organization for the submitted work.
Author contributions
Y.H.L. conceived and designed experiments. W.Y. conducted and analyzed the experiments. Y.H.W. collected clinical samples. X.Z. conducted a bioinformatics analysis. W.Y. and Y.H.W. performed statistical analysis. W.Y. wrote the initial draft of the manuscript. Y.H.L. revised the manuscript. All authors read and approved the final manuscript.
Declaration of interests
The authors declare no competing interests.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-B3GNT5 | Proteintech | cat# 20422-1-AP; RRID: AB_10694280 |
| Anti-STAT5B | Affinity | cat# DF6078; RRID: AB_2838046 |
| Anti-CD44 | Proteintech | cat# 60224-1-Ig; RRID: AB_11042767 |
| Anti-CD326 | Affinity | cat# DF6311; RRID: AB_2838277 |
| Anti-CxCR4 | Affinity | cat# AF5279; RRID: AB_2837765 |
| Anti-p-ERK1/2 | Affinity | cat# AF1015; RRID: AB_2834432 |
| Anti-ERK | Affinity | cat# AF0155; RRID: AB_2833336 |
| Anti-p-AKT | Affinity | cat# AF0016; RRID: AB_2810275 |
| Anti-AKT | Affinity | cat# AF6261; RRID: AB_2835121 |
| Biological samples | ||
| Fresh cancer tissues and adjacent para-carcinoma tissues | Shengjing Hospital of China Medical University | N/A |
| Pancreatic cancer tumor tissues | Shengjing Hospital of China Medical University | N/A |
| Chemicals, peptides, and recombinant proteins | ||
| Gemcitabine | Macklin | cat# G824361 |
| Fetal bovine serum | Tianhangbio | cat# 11011-8611 |
| Lipo 3000 | Invitrogen | cat# L3000015 |
| Giemsa dye | Jiancheng Bioengineering Institute | cat# D011-1-2 |
| Crystal violet | Amresco | cat# 0528 |
| Matrigel gel | Corning | cat# 356234 |
| Critical commercial assays | ||
| BCA protein assay kit | Beyotime | cat# P0011 |
| Cell counting kit-8 | KeyGen Biotech | cat# KGA317 |
| Cell apoptosis detection kit | KeyGen Biotech | cat# KGA106 |
| In situ cell death detection kit | Roche | cat# 12156792910 |
| Luciferase assay kit | KeyGen Biotech | cat# KGAF040 |
| Deposited data | ||
| The proteomic datasets generated during this study have been deposited to the Proteome Xchange Consortium via the PRIDE repository. | This paper | PXD052421 |
| Experimental models: cell lines | ||
| Human: CFPAC-1 cell | Saibaikang company | N/A |
| Human: CAPAN-1 cell | Saibaikang company | N/A |
| Experimental models: organisms/strains | ||
| BALB/c nude mice | Changzhou Cavens Laboratory Animal Co. LTD | N/A |
| Oligonucleotides | ||
| qPCR primers: B3GNT5 forward: 5′- GTTGGTTAGTGGCAGAA-3′ Reverse: 5′-AATCAAGTATTGGTAGCG-3′ |
This paper | N/A |
| Software and algorithms | ||
| Graphpad Prism 9 | GraphPad Software | https://www.graphpad.com/scientific-software/prism/ |
| Other | ||
| Microplate reader | BIOTEK | cat# 800TS |
| Flow cytometer | KeyGen Biotech | cat# KGA106 |
Experimental model and study participant details
Clinical specimens
Seventy-five cases of pancreatic cancer (PC) tumor tissues and five paired cancer tissues and adjacent para-carcinoma tissues were collected from patients who underwent surgical treatment due to PC at the Shengjing Hospital of China Medical University. The clinical and demographic information of study participants was shown in Tables S1 and S2. The study has been approved by the Ethics Committee of China Medical University (Approval No.2022PS964K), and informed consent was obtained from each patient.
Cell culture
The human pancreatic cancer cell lines (CFPAC-1 and CAPAN-1) were purchased from Saibaikang company (China). CFPAC-1 cells were cultured in DMEM (cat# G4640, Service, China) with 10% fetal bovine serum (FBS; cat# 11011-8611, Tianhangbio, China), and CAPAN-1 cells were cultured in DMEM with 20% FBS. All cells were maintained in a 37°C, 5% CO2 incubator (cat# HF-90, Heal Force, China). After the cells were in a good state of growth, they were used for experiments.
Animals
6-8 weeks healthy male BALB/c nude mice were purchased from Changzhou Cavens Laboratory Animal Co. LTD (China). The mice were provided food and water ad libitum and placed in a specific pathogen-free laboratory under 45%–55% humidity at 22 ± 1°C and a 12-h light/12-h dark cycle. All the animal experimental protocols were approved by the Ethics Committee of China Medical University (Approval No.2022PS963K).
Xenograft model
The mice were randomly divided into three groups, Control, NC shRNA, and B3GNT5 shRNA groups. 1×106 cells of each group in the logarithmic growth phase were injected subcutaneously into the right axilla of mice, and the tumor volume was measured every 3 days after growing to a tumor visible to the naked eye, then the tumor was taken after 21 days. Part of the tumor was fixed with 4% paraformaldehyde, and part of it was frozen in liquid nitrogen and then stored in an ultra-low-temperature refrigerator (cat# DW HL-668, MELING, China) at −70°C for subsequent detection.
Method details
Bioinformatics analysis
Raw data between PC and normal tissues were obtained from the GEO dataset (https://www.ncbi.nlm.nih.gov/) and 5 datasets (GSE62165, GSE63111, GSE130221, GSE15471, and GSE56560) were selected. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) functional enrichment analyses were used to predict the biological functions of differentially expressed genes and B3GNT5. Kaplan−Meier survival curves based on B3GNT5 expression and PC prognosis in the TCGA pancreatic cancer database using the UCSC Xena online tool (https://xena.ucsc.edu/). The correlation between B3GNT5 and the expression of cancer stem cell markers ALDH2 and CD44 was analyzed by TIMER2.0 (http://timer.comp-genomics.org/timer/). The promoter sequence of B3GNT5 was downloaded from the UCSC website (https://www.genome.ucsc.edu/) and the transcriptional regulation effect of B3GNT5 by Signal transducer and activator of transcription 5b (STAT5B) was predicted by the HumanTFDB database (http://bioinfo.life.hust.edu.cn/HumanTFDB).
Quantitative real-time PCR
Total RNA was isolated with the TRIzol reagent (cat# RP1001, BioTeke, China), as per the instructions of the manufacturer. The concentration of RNA in each sample was determined using a UV spectrophotometer (cat# NANO 2000, Thermo, USA). For cDNA synthesis, reverse transcription was performed with the BeyoRT II M-MLV Reverse Transcriptase (cat# D7160L, Beyotime, China). Four repeated PCR reactions were performed using 2 ×Taq PCR MasterMix (cat# PC1150, Solarbio, China), SYBR Green (cat# SY1020, Solarbio, China) in Exicycler 96 fluorescence quantifier (cat# Exicycler 96, BIONEER, Korea). β-actin was used as an internal reference for quantitative analysis of mRNA using the 2 – ΔΔ CT method (Clinical samples were analyzed by the 2 - Δ CT method). The primers were designed as follows: B3GNT5 F, 5′-GTTGGTTAGTGGCAGAA-3′, B3GNT5 R, 5′-AATCAAGTATTGGTAGCG-3’.
Immunohistochemistry
The paraffin-embedded tissue sections were removed with xylene to remove paraffin and dehydrated with a series of graded ethanol. The sections were added 0.1 M sodium citrate antigen repair solution and microwaved heating for antigen repair, followed by incubation with 3% H2O2 for 15 min to remove endogenous peroxidase, and incubation with BSA dropwise for 15 min at room temperature to block non-specific binding sites. The treated sections were further incubated overnight at 4°C with primary antibodies against B3GNT5 (1:50, cat# 20422-1-AP, Proteibtech, China), and then incubated with HRP marked goat anti-rabbit (1:50, cat# 31460, thermoFisher, USA) secondary antibody in a wet box for 1 h after washing with PBS. The sections were stained with diaminobenzidine (cat# DAB-1031, Maixinbio, China), re-stained with hematoxylin (cat# H8070, Solarbio, China), dehydrated, transparent, sealed, and photographed under a 400× microscope.
Western blot analysis
Tissue specimens and cells were lysed in a lysate consisting of Western and IP cell lysates (cat# P0013, Beyotime, China) as well as protease inhibitor (cat# ST506, Beyotime, China). Insoluble tissue fragments were removed by centrifugation at 10,000×g for 5 min at 4°C, and the resulting supernatants were collected and used for Western blot analysis. Protein concentration was determined by using the BCA Protein assay kit (cat# P0011, Beyotime, China). Proteins (20–40 μg) were separated using sodium dodecyl sulfate polyacrylamide gel electrophoresis, then transferred to polyvinylidene fluoride membranes (cat# ab133411, Abcam, China). The membrane was incubated in the containment solution (cat# P023, Beyotime, China) for 1 h at room temperature, probed with indicated primary antibodies overnight at 4°C and then incubated with secondary antibodies at 4°C for 45 min, and ECL luminol (cat# P0018, Beyotime, China) was added. The optical density values of the target bands were analyzed with Tanon Image analysis software (cat# 5200, Tanon, China). The primary antibodies including B3GNT5 antibody (1:500, cat# 20422-1-AP, Proteintech, China), STAT5B antibody (1:1000, cat# DF6078,Affinity, China), CD44 antibody (1:500, cat# DF63192, Affinity, China), CD326 antibody (1:500, cat# DF6311, Affinity, China), CxCR4 antibody (1:1000, cat# AF5279, Affinity, China), p-ERK1/2 antibody (1:5000, cat# AF1015, Affinity, China), ERK antibody (1:5000, cat# AF0155, Affinity, China), P-AKT antibody (1:5000, cat# AF0016, Affinity, China), AKT antibody (1: 5000, cat# AF6261, Affinity, China). Secondary antibodies including goat anti-rabbit (cat# A0208, Beyotime, China) or goat anti-mouse (cat# A0216, Beyotime, China).
Lentivirus infection
The two short hairpin RNA (shRNA) targeting 5′-GCAGTTAATTATTCAGTTATT-3′ or 5′-GATCAAAGGTACAATGATATA-3′ were designed according to the sequence of B3GNT5 mRNA. The scramble shRNA targeting 5′-TTCTCCGAACGTGTCACGT-3′ was used as a control. The above interference sequences were synthesized and ligated into the viral vector pLVX-shRNA1 (cat# BR004, Fenghuibio, China) by Generalbiol Co., LTD (China), and the enzymatic cleavage site was BamHI/EcoRI. And the overexpression sequence was inserted into the viral vector pLVX-IRES-puro (cat# BR025, Fenghuibio, China) with the enzymatic cleavage site of XhoI/Xba. CFPAC-1 and CAPAN-1 cells in the logarithmic growth phase were then infected with Lentivirus at Multiplicity of Infection (MOI) 10 and 5, respectively. After mixing, cells were further cultured in an incubator at 37°C and 5% CO2.
Cell transfection
The cells were inoculated in a 6-well plate, cultured to a density of about 70%, and then transfected with Lipo 3000 (cat# L3000015, Invitrogen, USA) according to the reagent manufacturer’s instructions. The plates were further cultured in a 37°C, 5% CO2 incubator to observe whether monoclonal cell clusters visible to the naked eye were formed, and finally stable transfected cell lines were obtained and used for subsequent experiments.
Cell viability analysis
The cell viability is assessed by using cell counting kit-8 (CCK-8; cat# KGA317, KeyGen Biotech, China) according to the manufacturer’s instructions. Briefly, Cells were seeded into 96-well plates at 5×103 cells/well according to the experimental groups, and cultured in 37°C, 5% CO2 incubator. 10 μL of CCK-8 reagent was added to each well, and the CCK-8 assay was performed after 0, 24, 48, and 72 h, respectively. The cell culture medium that reached the specific time was discarded, replaced by 100 μL of normal medium with 10 μL of CCK-8, mixed well and incubated in 37°C, 5% CO2 incubator for 2 h. The absorbance at 450 nm was detected using a microplate reader (cat# 800TS, BIOTEK, USA). Independent experiments were repeated 5 times.
Colony formation assay
The cells were cultured to a density of about 90%, digested by trypsin, resuspended, counted and then seeded into petri dishes, 400 cells were inoculated in each petri dish at 37°C and 5% CO2 for 14 days to form colonies. Then the cells were fixed with paraformaldehyde (cat# C104188, Aladdin, China) for 1 min, stained with Giemsa (cat# D011-1-2, Jiancheng Bioengineering Institute, China) for 5 min, finally photographed to statistics the number of cell clones. Colony formation rate = (number of clones/number of inoculated cells) × 100%.
Immunofluorescence
Cells were fixed with paraformaldehyde (cat# 80096618, Sinopharm, China), permeabilized in 0.5% Triton X-100 for 15 min at room temperature. Nonspecific binding sites is blocked with BSA (cat# A602440-0050, Sangon Biotech, China) for 15 min at room temperature. After incubation with primary antibodies against with primary antibody against CD44 (1:500, cat# DF63192, Affinity, China), CD326 (1:50, cat# 60224-1-Ig, Proteintech, China), CxCR4 (1:500, cat# AF5279, Affinity, China) overnight at 4°C, followed by incubation with fluorescent secondary antibody Cy3 marked goat anti-rabbit (1:200, cat# A27039, Invitrogen, USA) for 60 min at room temperature. Finally, the cells were washed with PBS and the nuclei were re-stained with DAPI, the supernatant of the centrifuged cells was discarded, and 200 μL of BSA was added, which was evenly coated on a clean slide, added anti-fluorescence quencher, and photographed by observing under the 400× fluorescence microscope.
Transwell assay
Cell invasion and migration were measured using 24-well transwell chambers (cat# 14341, Labselect, China) with or without Matrigel gel (cat# 356234, Corning, USA). Briefly, the culture solution containing 10% FBS (700 μL) was seeded in the lower chamber, 300 μL of cell suspension was added to the upper chamber, and the cell number was 5×103/well. Cells were then incubated at a 37°C, 5% CO2 incubator for 24 h, fixed using 4% paraformaldehyde for 20 min, stained using 0.5% crystal violet (cat# 0528, Amresco, USA) for 5 min, photographed, and quantified by counting them in five random high-power fields.
Tube formation assay
CFPAC1 and CAPAN1 cells were cultured for 24 h and the conditioned medium was collected. Human umbilical vein endothelial cells (HUVECs) were seeded into dishes coated with Matrigel gel and cultured in the conditioned medium for 6 h. The tubule formation was observed under a microscope and the number of tube nodes was calculated.
Sphere-forming assay
Cells were resuspended in 1 mL complete medium and incubated with Trypan blue for 3 min. The total number of cells in four quadrants was counted under the microscope (n = N/4 × 104 × 10). 500 cells were seeded into a 24-well ultra-low adsorption culture plate, and 0.5 mL DMEM/F12 medium (1 × glutaMAX + 1 × B27 +1 × N2 + 20 ng/ml bFGF + 50 ng/ml EGF) was added to each well and cultured for 7 d. Cell spheres were collected by cell screen, digested, and separated into single cells by trypsin, and further cultured for 7 d.
Limiting-dilution assay
For in vitro limiting dilution assay, a 96-well ultra-low adsorption culture plate was used to seed the cells in a 200 μL DMEM/F12 medium (20 ng/ml bFGF +20 ng/ml EGF) per well. Cells were seeded at a density of 1, 20, 40, 80, 160, 320, or 640 cells/well, with 10 replicates for each density. Spheres were counted on day 7. The spheroidal synthesis efficiency was calculated by the Extreme Limiting Dilution Analysis (http://bioinf.wehi.edu.au/software/elda).
Flow cytometry
Apoptotic cells were detected using Annexin V-FITC/PI double staining according to the instructions of the apoptosis detection kit (cat# KGA106, KeyGen Biotech, China). Cells were collected by centrifugation at 150g for 5 min and washed twice with PBS, then resuspended with 500 μL Binding Buffer. 5 μL Annexin V-FITC and 5 μL of Propidium Iodide were added respectively. The mixed solution was incubated at room temperature away from light for 15 min. Apoptosis was subsequently detected by flow cytometer (cat# KGA106, KeyGen Biotech, China).
TUNEL staining
Apoptotic cells were detected using the In Situ Cell Death Detection Kit (cat# 12156792910, Roche, Switzerland). Briefly, the cells were permeabilized with 0.1% Triton X-100 (cat# ST795, Beyotime, China), and incubated with 200 μL TUNEL reaction solution (Enzyme solution and Label Solution were prepared according to 1:9, which was ready to be used and operated on ice) at 37°C for 1 h under the condition of humidification and avoiding the light. Subsequently, the cells were washed with PBS, then stained with DAPI (cat# D106471-5mg, Aladdin, China) for 5 min, and then washed with PBS again and finally sealed with anti-fluorescence quencher (cat# S2100, Solarbio, China), the cells were observed and photographed under a microscope (cat# BX53, OLYMPUS, Japan).
Dual-luciferase assay
293T cells were planted in 12-well culture plates at a cell density of approximately 70% and incubated at a 37°C and 5% CO2 incubator overnight. The B3GNT5 promoter region containing different binding sites was ligated to pGL3 vector, and cells were co-transfected with the empty vector or STAT5B overexpression vector. After 48 h, cells were washed twice with PBS and cell lysates were added. The luciferase assay kit (cat# KGAF040, KeyGen Biotech, China) was used to detect the luciferase activity of the cells in each well according to the manufacturer’s instructions. Renilla luciferase gene was used as the internal reference and the luciferase activity was expressed as the ratio of firefly luciferase activity to Renilla luciferase activity.
Label-free quantitative proteomics analysis
Cells of the Mock group and B3GNT5 overexpression group were added with lysate, and total proteins were collected and quantified with a BCA protein assay kit. Protein samples were added to 5 mM DTT, incubated at 37°C for 1 h, then added to 10 mM of iodoacetamide, incubated at room temperature for 45 min away from light. The sample was then diluted 4 times with 25 mM ammonium bicarbonate and pancreatic enzyme was added according to the ratio of protein to pancreatic enzyme 50:1, incubated at 37°C overnight and formic acid was added to adjust the pH to less than 3, and the enzyme digestion was terminated. The samples were then desalted on the C18 column. The mass spectrometry (MS) experiments were performed in the Orbitrap Exploris™ 480 mass spectrometer (Thermo Fisher Scientific), samples were loaded onto the column and separated with a linear gradient of buffer A (100% water, 0.1% formic acid) and buffer B (80% acetonitrile, 0.1% formic acid). MS data were acquired using a data-dependent method and the full scanning range of the MS is m/z 350–1500. The MS data were analyzed using Proteome Discoverer 2.4 software. MS data were searched against the Homo sapiens sp database. The proteomics data was deposited at the ProteomeXchange Consortium via the PRIDE partner repository and is accessible with the dataset identifier PXD052421. The access link is https://www.ebi.ac.uk/pride/review-dataset/acdedd21993e4b25ad633f1bf416505c, and the access code is rWYDmJYcqE1b.
Quantification and statistical analysis
All data are presented as the mean ± standard deviation. Statistics were performed using Prism (Graph Pad Software Inc). ANOVA or Student’s t-test was used to determine the statistical rigor in all the experiments. p < 0.05 was considered to indicate a statistically significant difference.
Published: September 5, 2024
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2024.110889.
Supplemental information
References
- 1.Kamisawa T., Wood L.D., Itoi T., Takaori K. Pancreatic cancer. Lancet. 2016;388:73–85. doi: 10.1016/s0140-6736(16)00141-0. [DOI] [PubMed] [Google Scholar]
- 2.Park W., Chawla A., O'Reilly E.M. Pancreatic cancer: A review. JAMA. 2021;326:851–862. doi: 10.1001/jama.2021.13027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McGuigan A., Kelly P., Turkington R.C., Jones C., Coleman H.G., McCain R.S. Pancreatic cancer: A review of clinical diagnosis, epidemiology, treatment and outcomes. World J. Gastroenterol. 2018;24:4846–4861. doi: 10.3748/wjg.v24.i43.4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moutinho-Ribeiro P., Iglesias-Garcia J., Gaspar R., Macedo G. Early pancreatic cancer-The role of endoscopic ultrasound with or without tissue acquisition in diagnosis and staging. Dig. Liver Dis. 2019;51:4–9. doi: 10.1016/j.dld.2018.09.027. [DOI] [PubMed] [Google Scholar]
- 5.Fung S., Forte T., Rahal R., Niu J., Bryant H. Provincial rates and time trends in pancreatic cancer outcomes. Curr. Oncol. 2013;20:279–281. doi: 10.3747/co.20.1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nevala-Plagemann C., Hidalgo M., Garrido-Laguna I. From state-of-the-art treatments to novel therapies for advanced-stage pancreatic cancer. Nat. Rev. Clin. Oncol. 2020;17:108–123. doi: 10.1038/s41571-019-0281-6. [DOI] [PubMed] [Google Scholar]
- 7.Siegel R.L., Miller K.D., Jemal A. Cancer statistics, 2018. CA. Cancer J. Clin. 2018;68:7–30. doi: 10.3322/caac.21442. [DOI] [PubMed] [Google Scholar]
- 8.Smith M.R., Satter L.R.F., Vargas-Hernández A. STAT5b: A master regulator of key biological pathways. Front. Immunol. 2022;13 doi: 10.3389/fimmu.2022.1025373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laka K., Mapheto K.B.F., Mbita Z. Selective in vitro cytotoxicity effect of Drimia calcarata bulb extracts against p53 mutant HT-29 and p53 wild-type Caco-2 colorectal cancer cells through STAT5B regulation. Toxicol Rep. 2021;8:1265–1279. doi: 10.1016/j.toxrep.2021.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernaciak T.M., Zareno J., Parsons J.T., Silva C.M. A novel role for signal transducer and activator of transcription 5b (STAT5b) in β1-integrin-mediated human breast cancer cell migration. Breast Cancer Res. 2009;11 doi: 10.1186/bcr2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kang D.Y., Darvin P., Yoo Y.B., Joung Y.H., Sp N., Byun H.J., Yang Y.M. Methylsulfonylmethane inhibits HER2 expression through STAT5b in breast cancer cells. Int. J. Oncol. 2016;48:836–842. doi: 10.3892/ijo.2015.3277. [DOI] [PubMed] [Google Scholar]
- 12.Moser C., Ruemmele P., Gehmert S., Schenk H., Kreutz M.P., Mycielska M.E., Hackl C., Kroemer A., Schnitzbauer A.A., Stoeltzing O., et al. STAT5b as molecular target in pancreatic cancer-inhibition of tumor growth, angiogenesis, and metastases. Neoplasia. 2012;14:915–925. doi: 10.1593/neo.12878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sumiyoshi H., Matsushita A., Nakamura Y., Matsuda Y., Ishiwata T., Naito Z., Uchida E. Suppression of STAT5b in pancreatic cancer cells leads to attenuated gemcitabine chemoresistance, adhesion and invasion. Oncol. Rep. 2016;35:3216–3226. doi: 10.3892/or.2016.4727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Togayachi A., Kozono Y., Ikehara Y., Ito H., Suzuki N., Tsunoda Y., Abe S., Sato T., Nakamura K., Suzuki M., et al. Lack of lacto/neolacto-glycolipids enhances the formation of glycolipid-enriched microdomains, facilitating B cell activation. Proc. Natl. Acad. Sci. USA. 2010;107:11900–11905. doi: 10.1073/pnas.0914298107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alam S., Anugraham M., Huang Y.L., Kohler R.S., Hettich T., Winkelbach K., Grether Y., López M.N., Khasbiullina N., Bovin N.V., et al. Altered (neo-) lacto series glycolipid biosynthesis impairs α2-6 sialylation on N-glycoproteins in ovarian cancer cells. Sci. Rep. 2017;7 doi: 10.1038/srep45367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Biellmann F., Hülsmeier A.J., Zhou D., Cinelli P., Hennet T. The Lc3-synthase gene B3gnt5 is essential to pre-implantation development of the murine embryo. BMC Dev. Biol. 2008;8:109. doi: 10.1186/1471-213x-8-109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Daniotti J.L., Lardone R.D., Vilcaes A.A. Dysregulated expression of glycolipids in tumor cells: From negative modulator of anti-tumor immunity to promising targets for developing therapeutic agents. Front. Oncol. 2015;5:300. doi: 10.3389/fonc.2015.00300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nagaraju G.P., Farran B., Luong T., El-Rayes B.F. Understanding the molecular mechanisms that regulate pancreatic cancer stem cell formation, stemness and chemoresistance: A brief overview. Semin. Cancer Biol. 2023;88:67–80. doi: 10.1016/j.semcancer.2022.12.004. [DOI] [PubMed] [Google Scholar]
- 19.Miao Z., Cao Q., Liao R., Chen X., Li X., Bai L., Ma C., Deng X., Dai Z., Li J., Dong C. Elevated transcription and glycosylation of B3GNT5 promotes breast cancer aggressiveness. J. Exp. Clin. Cancer Res. 2022;41:169. doi: 10.1186/s13046-022-02375-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeong H.Y., Park S.Y., Kim H.J., Moon S., Lee S., Lee S.H., Kim S.H. B3GNT5 is a novel marker correlated with stem-like phenotype and poor clinical outcome in human gliomas. CNS Neurosci. Ther. 2020;26:1147–1154. doi: 10.1111/cns.13439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jongsma M.L.M., de Waard A.A., Raaben M., Zhang T., Cabukusta B., Platzer R., Blomen V.A., Xagara A., Verkerk T., Bliss S., et al. The SPPL3-defined glycosphingolipid repertoire orchestrates HLA Class I-mediated immune responses. Immunity. 2021;54:132–150.e9. doi: 10.1016/j.immuni.2020.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grzesiak J.J., Ho J.C., Moossa A.R., Bouvet M. The integrin-extracellular matrix axis in pancreatic cancer. Pancreas. 2007;35:293–301. doi: 10.1097/mpa.0b013e31811f4526. [DOI] [PubMed] [Google Scholar]
- 23.Usman O.H., Kumar S., Walker R.R., 3rd, Xie G., Sumajit H.C., Jalil A.R., Ramakrishnan S., Dooling L.J., Wang Y.J., Irianto J. Differential modulation of cellular phenotype and drug sensitivity by extracellular matrix proteins in primary and metastatic pancreatic cancer cells. Mol. Biol. Cell. 2023;34 doi: 10.1091/mbc.E23-02-0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rosenberg N., Mor-Cohen R., Sheptovitsky V.H., Romanenco O., Hess O., Lahav J. Integrin-mediated cell adhesion requires extracellular disulfide exchange regulated by protein disulfide isomerase. Exp. Cell Res. 2019;381:77–85. doi: 10.1016/j.yexcr.2019.04.017. [DOI] [PubMed] [Google Scholar]
- 25.Scott E., Munkley J. Glycans as Biomarkers in Prostate Cancer. Int. J. Mol. Sci. 2019;20 doi: 10.3390/ijms20061389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Leon F., Seshacharyulu P., Nimmakayala R.K., Chugh S., Karmakar S., Nallasamy P., Vengoji R., Rachagani S., Cox J.L., Mallya K., et al. Reduction in O-glycome induces differentially glycosylated CD44 to promote stemness and metastasis in pancreatic cancer. Oncogene. 2022;41:57–71. doi: 10.1038/s41388-021-02047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hernández-Camarero P., López-Ruiz E., Griñán-Lisón C., García M.Á., Chocarro-Wrona C., Marchal J.A., Kenyon J., Perán M. Pancreatic (pro)enzymes treatment suppresses BXPC-3 pancreatic Cancer Stem Cell subpopulation and impairs tumour engrafting. Sci. Rep. 2019;9 doi: 10.1038/s41598-019-47837-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y.J., Wu J.Y., Wang J.M., Hu X.B., Cai J.X., Xiang D.X. Gemcitabine loaded autologous exosomes for effective and safe chemotherapy of pancreatic cancer. Acta Biomater. 2020;101:519–530. doi: 10.1016/j.actbio.2019.10.022. [DOI] [PubMed] [Google Scholar]
- 29.Conroy T., Hammel P., Hebbar M., Ben Abdelghani M., Wei A.C., Raoul J.L., Choné L., Francois E., Artru P., Biagi J.J., et al. FOLFIRINOX or gemcitabine as adjuvant therapy for pancreatic cancer. N. Engl. J. Med. 2018;379:2395–2406. doi: 10.1056/NEJMoa1809775. [DOI] [PubMed] [Google Scholar]
- 30.Lippi G., Mattiuzzi C. The global burden of pancreatic cancer. Arch. Med. Sci. 2020;16:820–824. doi: 10.5114/aoms.2020.94845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Guo Z., Hong Y., Cheng Y. Dietary inflammatory index and pancreatic cancer risk: A systematic review and dose-response meta-analysis. Public Health Nutr. 2021;24:6427–6435. doi: 10.1017/s1368980021001579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nagaraju G.P., Malla R.R., Basha R., Motofei I.G. Contemporary clinical trials in pancreatic cancer immunotherapy targeting PD-1 and PD-L1. Semin. Cancer Biol. 2022;86:616–621. doi: 10.1016/j.semcancer.2021.11.003. [DOI] [PubMed] [Google Scholar]
- 33.Luong T., Golivi Y., Nagaraju G.P., El-Rayes B.F. Fibroblast heterogeneity in pancreatic ductal adenocarcinoma: Perspectives in immunotherapy. Cytokine Growth Factor Rev. 2022;68:107–115. doi: 10.1016/j.cytogfr.2022.09.001. [DOI] [PubMed] [Google Scholar]
- 34.Liu X.D., Xie D.F., Wang Y.L., Guan H., Huang R.X., Zhou P.K. Integrated analysis of lncRNA-mRNA co-expression networks in the α-particle induced carcinogenesis of human branchial epithelial cells. Int. J. Radiat. Biol. 2019;95:144–155. doi: 10.1080/09553002.2019.1539880. [DOI] [PubMed] [Google Scholar]
- 35.Zhao R., Qin W., Qin R., Han J., Li C., Wang Y., Xu C. Lectin array and glycogene expression analyses of ovarian cancer cell line A2780 and its cisplatin-resistant derivate cell line A2780-cp. Clin. Proteomics. 2017;14:20. doi: 10.1186/s12014-017-9155-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clevers H. The cancer stem cell: Premises, promises and challenges. Nat. Med. 2011;17:313–319. doi: 10.1038/nm.2304. [DOI] [PubMed] [Google Scholar]
- 37.Kakarala M., Wicha M.S. Implications of the cancer stem-cell hypothesis for breast cancer prevention and therapy. J. Clin. Oncol. 2008;26:2813–2820. doi: 10.1200/jco.2008.16.3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yamamuro S., Okamoto Y., Sano E., Ochiai Y., Ogino A., Ohta T., Hara H., Ueda T., Nakayama T., Yoshino A., Katayama Y. Characterization of glioma stem-like cells from human glioblastomas. Int. J. Oncol. 2015;47:91–96. doi: 10.3892/ijo.2015.2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rassouli F.B., Matin M.M., Bahrami A.R., Ghaffarzadegan K., Cheshomi H., Lari S., Memar B., Kan M.S. Evaluating stem and cancerous biomarkers in CD15+CD44+ KYSE30 cells. Tumour Biol. 2013;34:2909–2920. doi: 10.1007/s13277-013-0853-5. [DOI] [PubMed] [Google Scholar]
- 40.Barker N., Huch M., Kujala P., van de Wetering M., Snippert H.J., van Es J.H., Sato T., Stange D.E., Begthel H., van den Born M., et al. Lgr5(+ve) stem cells drive self-renewal in the stomach and build long-lived gastric units in vitro. Cell Stem Cell. 2010;6:25–36. doi: 10.1016/j.stem.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 41.Bünger S., Barow M., Thorns C., Freitag-Wolf S., Danner S., Tiede S., Pries R., Görg S., Bruch H.P., Roblick U.J., et al. Pancreatic carcinoma cell lines reflect frequency and variability of cancer stem cell markers in clinical tissue. Eur. Surg. Res. 2012;49:88–98. doi: 10.1159/000341669. [DOI] [PubMed] [Google Scholar]
- 42.Von Hoff D.D., Ervin T., Arena F.P., Chiorean E.G., Infante J., Moore M., Seay T., Tjulandin S.A., Ma W.W., Saleh M.N., et al. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N. Engl. J. Med. 2013;369:1691–1703. doi: 10.1056/NEJMoa1304369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mukherjee D., Chakraborty S., Bercz L., D'Alesio L., Wedig J., Torok M.A., Pfau T., Lathrop H., Jasani S., Guenther A., et al. Tomatidine targets ATF4-dependent signaling and induces ferroptosis to limit pancreatic cancer progression. iScience. 2023;26 doi: 10.1016/j.isci.2023.107408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Natu J., Nagaraju G.P. Gemcitabine effects on tumor microenvironment of pancreatic ductal adenocarcinoma: Special focus on resistance mechanisms and metronomic therapies. Cancer Lett. 2023;573 doi: 10.1016/j.canlet.2023.216382. [DOI] [PubMed] [Google Scholar]
- 45.Griesmann H., Mühl S., Riedel J., Theuerkorn K., Sipos B., Esposito I., Vanden Heuvel G.B., Michl P. CUX1 Enhances Pancreatic Cancer Formation by Synergizing with KRAS and Inducing MEK/ERK-Dependent Proliferation. Cancers. 2021;13:2462. doi: 10.3390/cancers13102462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang T., Liu M., Liu Q., Xiao G.G. Wogonin increases gemcitabine sensitivity in pancreatic cancer by inhibiting Akt pathway. Front. Pharmacol. 2022;13 doi: 10.3389/fphar.2022.1068855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chai X., Chu H., Yang X., Meng Y., Shi P., Gou S. Metformin Increases Sensitivity of Pancreatic Cancer Cells to Gemcitabine by Reducing CD133+ Cell Populations and Suppressing ERK/P70S6K Signaling. Sci. Rep. 2015;5 doi: 10.1038/srep14404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Khiaowichit J., Talabnin C., Dechsukhum C., Silsirivanit A., Talabnin K. Down-Regulation of C1GALT1 Enhances the Progression of Cholangiocarcinoma through Activation of AKT/ERK Signaling Pathways. Life. 2022;12 doi: 10.3390/life12020174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cheng J., Xia L., Hao X., Gan F., Bai Y., Zhang C., Mao Y., Zhu Y., Pu Q., Park D.W., et al. Targeting STT3A produces an anti-tumor effect in lung adenocarcinoma by blocking the MAPK and PI3K/AKT signaling pathway. Transl. Lung Cancer Res. 2022;11:1089–1107. doi: 10.21037/tlcr-22-396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kovács K., Decatur C., Toro M., Pham D.G., Liu H., Jing Y., Murray T.G., Lampidis T.J., Merchan J.R. 2-Deoxy-Glucose Downregulates Endothelial AKT and ERK via Interference with N-Linked Glycosylation, Induction of Endoplasmic Reticulum Stress, and GSK3β Activation. Mol. Cancer Ther. 2016;15:264–275. doi: 10.1158/1535-7163.Mct-14-0315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang Y., Maeda Y., Liu Y.S., Takada Y., Ninomiya A., Hirata T., Fujita M., Murakami Y., Kinoshita T. Cross-talks of glycosylphosphatidylinositol biosynthesis with glycosphingolipid biosynthesis and ER-associated degradation. Nat. Commun. 2020;11:860. doi: 10.1038/s41467-020-14678-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.d'Azzo A., Tessitore A., Sano R. Gangliosides as apoptotic signals in ER stress response. Cell Death Differ. 2006;13:404–414. doi: 10.1038/sj.cdd.4401834. [DOI] [PubMed] [Google Scholar]
- 53.Garcia-Ruiz C., Morales A., Fernández-Checa J.C. Glycosphingolipids and cell death: one aim, many ways. Apoptosis. 2015;20:607–620. doi: 10.1007/s10495-015-1092-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang W., Lopez McDonald M.C., Hariprasad R., Hamilton T., Frank D.A. Oncogenic STAT Transcription Factors as Targets for Cancer Therapy: Innovative Strategies and Clinical Translation. Cancers. 2024;16 doi: 10.3390/cancers16071387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Koptyra M., Gupta S., Talati P., Nevalainen M.T. Signal transducer and activator of transcription 5a/b: biomarker and therapeutic target in prostate and breast cancer. Int. J. Biochem. Cell Biol. 2011;43:1417–1421. doi: 10.1016/j.biocel.2011.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang W., Marinis J.M., Beal A.M., Savadkar S., Wu Y., Khan M., Taunk P.S., Wu N., Su W., Wu J., et al. RIP1 Kinase Drives Macrophage-Mediated Adaptive Immune Tolerance in Pancreatic Cancer. Cancer Cell. 2018;34:757–774.e7. doi: 10.1016/j.ccell.2018.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yang T., Chi Y., Wang X., Xu C., Chen X., Liu Y., Huang S., Zhu X., Zhang H., Zhuo H., Wu D. PRL-mediated STAT5B/ARRB2 pathway promotes the progression of prostate cancer through the activation of MAPK signaling. Cell Death Dis. 2024;15:128. doi: 10.1038/s41419-023-06362-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The proteomic data have been deposited at PRIDE and is publicly available at the date of publication. Accession numbers are listed in the key resources table.
-
•
This paper does not report any original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.