Abstract
This study introduces the time-gated analysis of room-temperature phosphorescence (RTP) for the in-situ analysis of the visible and spectral information of photons. Time-gated analysis is performed using a microscopic system consisting of a spectrometer, which is advantageous for in-situ analysis since it facilitates the real-time measurement of luminescence signal changes. An RTP material hybridized with a DNA aptamer that targets a specific protein enhances the intensity and lifetime of phosphorescence after selective recognition with the target protein. In addition, time-gated analysis allows for the millisecond-scale imaging of phosphorescence signals, excluding autofluorescence, and improves the signal-to-background ratio (SBR) through the accumulation of signals. While collecting the time-gated images and spectra of RTP and autofluorescent materials simultaneously, we develop a method for obtaining phosphorescence signals by means of selective exclusion of autofluorescence signals in simulated or real cell conditions. It is confirmed that the accumulated time-gated analysis can provide ample information about luminescence signals for bioimaging and biosensing applications.
Keywords: Room-temperature phosphorescence (RTP), Time-gated microscopy, In-situ spectroscopy, Autofluorescence, Signal accumulation
Graphical abstract
Highlights
-
•
The RTP materials are hybridized with DNA aptamer for bio-recognition avoiding autofluorescence with time-gated analyses.
-
•
A spectrometer is integrated with microscopy to obtain visible and spectral data, enhancing in-situ imaging and sensing analyses.
-
•
Time-gated analyses improve image and spectra by signal accumulation and noise reduction.
1. Introduction
In the microscopic analysis of living organisms such as cells and tissues [1], autofluorescence is a drawback for overlapping photoluminescence (PL) signals [2,3]. To overcome this drawback, methods have been developed to avoid the autofluorescence of organisms, such as lifetime mapping and time-gated imaging [[4], [5], [6]]. Lifetime mapping typically uses time-correlated single-photon counting (TCSPC) for measurements, where fluorescence lifetime imaging microscopy [[7], [8], [9]] is used for fluorescent materials and phosphorescence lifetime imaging microscopy (PLIM) [10,11] for phosphorescent materials. In the case of multiple luminescence signals, a clearly distinguished mapping image can be obtained by visualizing the lifetime value of each luminescence signal [12]. However, there is a limitation that the spectral information of each signal cannot be confirmed. In addition, PLIM should be repeated to perform a new triplet population after the decay is completed and scan all x and y pixels in a detection area. This results in a long measurement time and a high cost of computational equipment [13]. Time-gated imaging is based on the combination of an optical chopper and materials with relatively longer lifetimes compared to fluorescence [[14], [15], [16]]. This method can be used to precisely control the delay time to submicroseconds using an electric signal [17,18]. A time delay exists between the excitation of luminescent materials and the detection of signals. Hence, only long-lived luminescence signals are selectively detected after autofluorescence disappears [19]. In general, signal detection becomes easier as the lifetime of a luminescent material increases [20]. For in vivo bioimaging applications, a luminescent material with a long lifetime is injected into a mouse, and the afterglow of luminescence is measured through time-gated analysis [21,22]. From a microscopic perspective, only the number of detected photons is important for visualization, even though the luminescent signal contains more information.
In this study, we have developed a time-gated microscopic system by adding a spectrometer to analyze the spectral information of photons. It is possible to simultaneously obtain the visible information from the microscope and spectral information from the spectrometer. The emission spectrum is essential for the quantitative analysis of luminescence across a broad wavelength range, and it involves parameters such as the real-time intensity decay, wavelength shift, and deconvolution of multiple peaks. A microscope coupled with a spectrometer provides a significant advantage in in-situ research because it allows for the real-time measurement of wavelength changes at desired locations in a sample. In addition, we use an organic room-temperature phosphorescence (RTP) material, which has long-lived emission over milliseconds at room temperature [[23], [24], [25], [26], [27], [28], [29], [30], [31]].as a luminescent sample that is efficient for gating time control [[32], [33], [34], [35], [36]] Organic luminescent materials can be hybridized with bioligands with high stability and biocompatibility in ambient air and aqueous conditions [[37], [38], [39], [40], [41]]. We fabricate a hybrid assembly of pure organic RTP material and DNA aptamer with specific target protein recognition. The interaction with the target protein increases the intensity of the aptamer-hybrid assembly and the lifetime of the phosphorescence signal. Furthermore, millisecond-scale images are obtained faster compared to PLIM by detecting the long lifetime of a phosphorescence signal using an electron-multiplying charge-coupled device (EMCCD). The time delay function in the shutter operation of the EMCCD is used to exclude autofluorescence, and only phosphorescence is selectively captured. The acquired signals are accumulated to reduce the signal-to-background ratio (SBR). The time-gated RTP analysis using in-situ images and spectra can provide more information for the analysis of PL signals.
2. Experimental section
2.1. Installation of time-gated microscope system with spectrometer
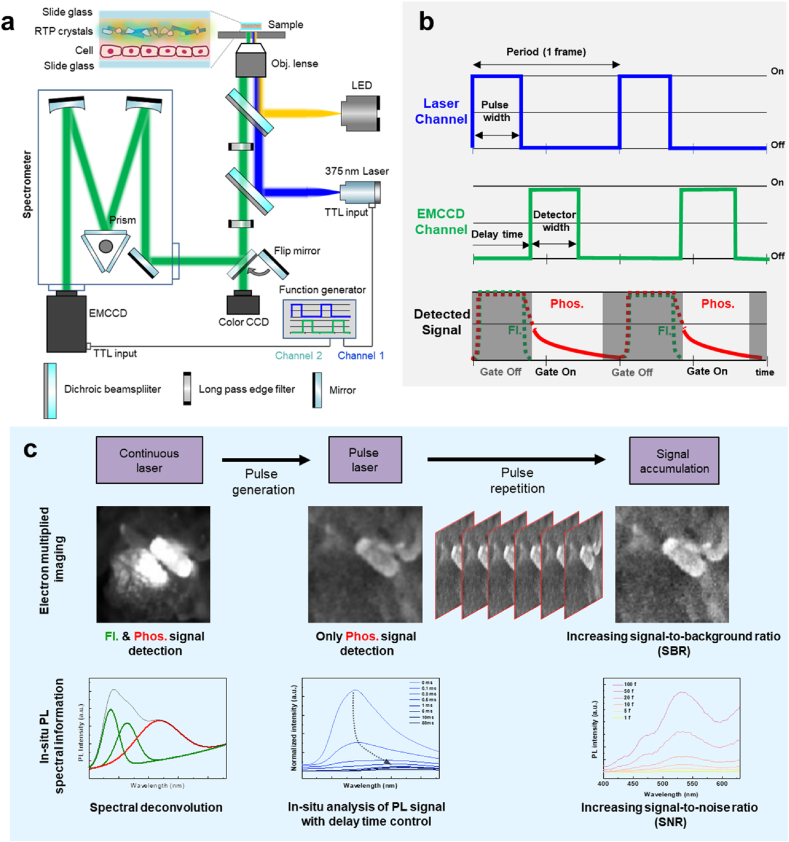
For the time-gated room-temperature phosphorescence microscopy, a 375 nm diode laser and LED lamp, each connected to the Olympus IX73 microscope body and a function generator, were used as excitation sources. A color charge-coupled device (CCD), electron-multiplying CCD (EMCCD), and spectrometer were used as the signal detectors. A two-deck noise piece was installed on the IX73 microscope to use the LED and laser light sources. The LED lamp was directly installed on the main body, and the wavelength of excitation/emission light was determined using a dichroic beam splitter and long-passage filter. A pulse function was added by connecting a function generator to the diode laser to adjust the pulse width and delay time. The pulsed laser, which started from the optical fiber, reached the body through the collimator. The collimated excitation laser was separated using a dichroic beam splitter that reflected light below 382 nm and transmitted it above 382 nm. The reflected light was collected using a 10x air gap lens (NA: 0.35) before irradiation to the sample. The sample was excited, and it emitted a photoluminescence signal that passed through a loss-passage filter with a cut-off wavelength of 405 nm. A flip mirror was added to the output port to select the output signal that passed through the color CCD or EMCCD (Pro-EM 1024) equipped with a spectrometer (HRS-300, Princeton Instruments).
2.2. Luminescent powder sampling for microscopy
The autofluorescence materials used were tryptophan (Trp), which emits blue fluorescence as a component of proteins, and riboflavin (Rf), which emits yellow fluorescence and is directly involved in cell metabolism. Isophthalic acid (IPA) has a lifetime of milliseconds or more at room temperature and an emission wavelength similar to that of the autofluorescence materials; thus, it can be differentiated only based on its lifetime. We used IPA, benzophenone (DBP), and polybenzophenone (pDBP) without further purification. The cocrystal of phenanthrene and 1,2,4,5-tetrafluoro-3,6-diiodobenzene (Phe-DITFB) was prepared according to a previous report [42]. Phenanthrene and 1,4-DITFB (1:2 M ratio) were dissolved in an ethanol–chloroform solution (1:1 vol ratio) in a 25 mL vial. The solution was kept in the dark overnight to allow for gradual solvent evaporation, and colorless crystals were obtained. All the samples were purchased from Sigma-Aldrich in powder form. Each powder (1 g) was finely ground using a grinder to remove extra-large crystals. To fix the powder for microscopic analysis, we prepared a pellet by placing 1 mg of the powder on a glass slide and pressing it with a cover glass. The mixed samples were prepared by mixing each powder in a weight ratio of 1:1.
2.3. Characterization of luminescence signal of IPA crystals under excitation with continuous-wave laser
After a triplet population was achieved using a continuous-wave (CW) laser for a sufficiently long time, the laser was turned off to measure the decrease in the phosphorescence intensity and color changes in the IPA crystal using the color CCD in real time. The EMCCD visualized the reduction of photons at 660 ms (1.5 fps) intervals under the same excitation conditions. The grating (density of 150 groove/mm, blaze of 500 nm, and center wavelength of 500 nm) of a spectrometer was used to obtain the spectrum of IPA crystals every 0.5 s after the laser was switched off.
2.4. Characterization of luminescence signal of IPA crystals under excitation with pulsed laser
The CW laser was converted to a pulsed laser to selectively detect the phosphorescence signal of IPA crystals. A function generator generated an output pulse trigger for the CW laser in channel 1 with a period of 300 ms, pulse width of 100 ms, and no delay. Simultaneously, a second pulse trigger was generated to the EMCCD in channel 2 with a period of 300 ms, pulse width of 100 ms, and delay of 100.5 ms. The two pulses did not overlap to selectively detect the phosphorescence signal of IPA crystals. The gating time represented the time difference between the two pulses, and it was obtained by subtracting the width of the CW laser from the delay of the EMCCD (100.5 ms–100 ms = 0.5 ms). In addition, the signal-to-background ratio of the frame was reduced to improve the background of the images and obtain a clearer spectrum by accumulating signals at the same location and conditions. The time-gated images represented the brightness and shape of crystals in terms of the relative number of photons by counting nonspectral photons from the spectrometer. By contrast, spectral photons from the grating of the spectrometer appeared as a spectrum.
2.5. Fabrication of IPA–aptamer hybrid assembly and selective recognition with the target protein
The aptamer (Bioneer) had a sequence of 5′-GGT GGT GGT GGT TGT GGT GGT GGT GG-3’ (26 mer); which is a guanine-rich sequence that has a specific G-quadruplex structure. To make the specific 3D structure, 2 μM of the aptamer was dissolved in a phosphate-buffered saline (PBS) buffer (Bioneer), and 5 mM of MgCl2 (Bioneer) was added to create the G-quadruplex structure. The aptamer solution was heated at 95 °C for 10 min and then left at room temperature for 1 h. Subsequently, 20 mg/mL of commercial IPA powder was dissolved in tetrahydrofuran (THF) to form a stock solution for the reprecipitation method. 2 mL of the stock solution was injected into 20 mL of the PBS buffer containing 500 nM of the prepared aptamer solution with rapid stirring at 900 rpm for 10 min. The mixture was stored overnight at room temperature, and a visible precipitate was formed. The fabricated IPA–aptamer hybrid assembly was exposed to target nucleolin (lyophilized from 0.2 μm filtered solution in PBS, 10 % trehalose as a protectant, pH 7.4, AcroBiosystems) at concentrations of 50, 100, 200, and 500 nM at 36.5 °C for 2 h and then left at room temperature. PSMA (100 nM, lyophilized from 0.22 μm filtered solution in PBS, 0.5 % BSA, pH 7.4, AcroBiosystems) and CD30 (lyophilized from 0.22 μm filtered solution in 50 mM Tris buffer, 100 mM glycine, pH 7.5, AcroBiosystems) were used as controls. The liquid samples for microscopy were prepared by dropping 10 μL of the IPA–aptamer hybrid assembly before and after treatment with the target protein on a slide glass and covering it with cover glass.
2.6. Cell culture
Cell culture was performed according to Freshnev's method [43]. The model mouse cell line (RAW264.7 macrophages) was purchased from the American Type Culture Collection. The cells were cultured in Dulbecco's modified Eagles's medium (Gibco) containing 10 % fetal bovine serum (Gibco) and 1 % penicillin/streptomycin (Gibco) at 37 °C in a humidified incubator with 5 % CO2 atmosphere. The cell medium was replaced every 2 days, and the cells were detached for cell cryopreservation experiments until the cell density reached approximately 80 %. After the monolayer formation, the cells were detached using a 0.25 % trypsin solution with 0.02 % ethylenediaminetetraacetic acid according to the standard methodology. The cultured cell was washed with PBS buffer solution twice for sampling of time-gated microscopy.
2.7. Characterization
The surface morphology of the IPA–aptamer particles was analyzed using field-emission scanning electron microscopy (Hitachi, S-4300) at an acceleration voltage of 10 kV. A fluorescence spectrophotometer (Cary Eclipse Fluorescence Spectrophotometer, Agilent) was used to determine the phosphorescence mode. The total decay time, delay time, and gate time were 0.02 s, 0.2 ms, and 5 ms, respectively. PL data were measured 2 h after the addition of the target proteins. Powder X-ray diffraction (Rigaku, SmartLab) patterns were obtained using Cu-Kα radiation (λ = 1.5412 Å) at a voltage of 45 kV and current of 200 mA. The scanning rate was 0.017°/s, and the values of 2θ were 5°–60°. A confocal laser scanning microscope (Carl Zeiss, LSM700) was used to analyze the fluorescence images of the IPA, IPA–aptamer, and IPA–aptamer with nucleolin. The IPA molecules were excited at 405 nm and detected using a 410–550 nm filter. The cyanine 5(cy5)-labeled aptamer present on the IPA particles were excited at 555 nm and detected using a 640–800 nm filter. The phycoerythrin(PE)-labeled nucleolin proteins present on the IPA–aptamer microparticles were excited at 555 nm and detected using a 570–640 nm filter. The phosphorescence lifetimes of the IPA pellets were measured using a photomultiplier tube. Raman images were acquired with a LabRAM Soleil Raman microscope (Horiba Jobin Yvon) using a 532 nm excitation laser and a LabRAM Aramis Raman microscope (Horiba Jobin Yvon) using a 785 nm excitation laser.
3. Results and discussion
3.1. Design of an accumulated time-gated RTP analysis for avoiding autofluorescence
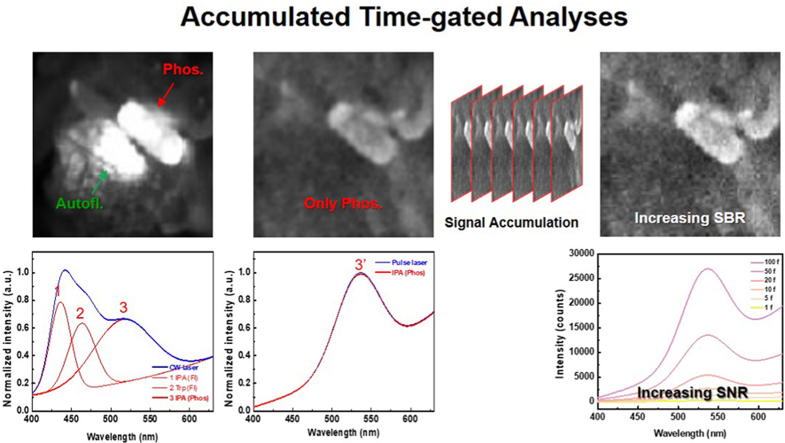
The time-gated RTP analysis equipment consisted of a light source, microscope, spectrometer, detector, and function generator (Fig. 1a). There were two light sources: an LED with an optical filter, which is commonly used in conventional optical microscopy, and a 375 nm diode laser for pulse generation. In accordance with the light source equipment, a color charge-coupled device (CCD) and EMCCD were used. Time gating was performed by connecting the function generator to the laser source and EMCCD to generate a pulse as an electrical signal. The form of the pulse signal was easily modified by controlling function parameters such as the period, pulse width, and delay time (Fig. 1b). According to the delay time between the laser and detector channels, the detected signal could be selectively obtained using a phosphorescence signal in the gate-on state. The detected signals were simultaneously expressed as images and spectra under irradiation with a continuous-wave (CW) laser or pulsed laser source (Fig. 1c). For the CW laser source, autofluorescence and phosphorescence signals could not be distinguished in an optical microscope image. In spectral information analysis, the deconvolution of the multicomponent spectrum enables the distinction between these signals after the assignment of each component. The pulsed laser source was used to selectively detect phosphorescence signals from an optical microscope image. In addition, the in-situ spectroscopy had spectral information such as change of peak intensity and peak shift over delay time. By repeating the pulse signal, the luminescence signal could be accumulated to improve the signal-to-background ratio (SBR) for optical images and the signal-to-noise ratio (SNR) for spectrum.
Fig. 1.
Accumulated in-situ time-gated room temperature phosphorescence analysis. a Schematic illustration of time-gated RTP analysis instrument. b Control parameters of the time-gated system and detected signals over time. c Detected signals expressed by images and spectra and accumulated with a reiteration of pulsed laser.
3.2. Optical properties of isophthalic acid crystal with long-lived phosphorescence emission
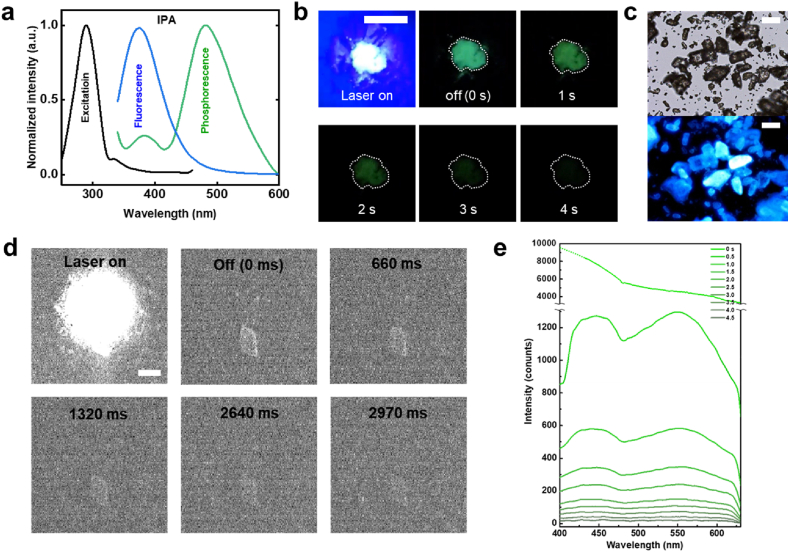
We selected isophthalic acid (IPA) as a representative RTP material because of its long lifetime of a few seconds [44,45]. First, we analyzed the visible excitation and emission spectra of IPA solution (Fig. 2a). The IPA absorbed UV light at approximately 290 nm and simultaneously emitted luminescence signals with a fluorescence peak at 375 nm and phosphorescence peak at 480 nm. White-colored pristine IPA was irradiated with a 375 nm UV lamp and observed with the naked eye (Fig. 2b). Pristine IPA emitted cyan light during UV light irradiation. After the UV light was turned off, IPA emitted only green phosphorescence for approximately 4 s. The observation of the IPA powder through optical and fluorescent microscopy revealed that the IPA powder had various sizes and morphologies (Fig. 2c). Pristine IPA also emitted cyan luminescence upon excitation with an LED lamp filtered by a 330–385 nm dichroic beam splitter. We performed in-situ analysis using time-gated microscopy (Fig. 2d and Supplementary Video 1). A single IPA crystal was irradiated with a 375 nm laser source without pulse generation. The IPA crystal was superimposed on the reflected laser signal during the irradiation. When the laser was turned off, the luminescence signal of the IPA crystal gradually decreased for approximately 3 s and then disappeared. We added a spectrometer as an additional accessory to the microscope and observed a decrease in the luminescence of IPA (Fig. 2e). After the laser was turned off, the luminescence signal of IPA decreased considerably with two peaks at 440 and 550 nm, and it faded away after a few seconds.
Fig. 2.
Long-lived phosphorescence emission spectra of IPA with CW laser excitation. a PL spectrum of IPA in THF (0.1 mg/mL). Fluorescence at 375 nm and phosphorescence at 480 nm b Photo images of IPA crystal under irradiation with 365 nm UV lamp (scale bar: 1 cm). c Optical and fluorescence microscopy images of IPA crystal (λex: 330–385 nm, λem: 420–IR nm, scale bar: 100 μm). d EMCCD images of IPA crystal in realtime observation with CW laser source (scale bar: 20 μm). e In-situ PL spectrum of IPA with CW laser source. Emission signal was filtered using 405 nm long-pass edge filter.
3.3. In situ time-gated RTP images and spectrum of IPA crystal under pulsed laser irradiation
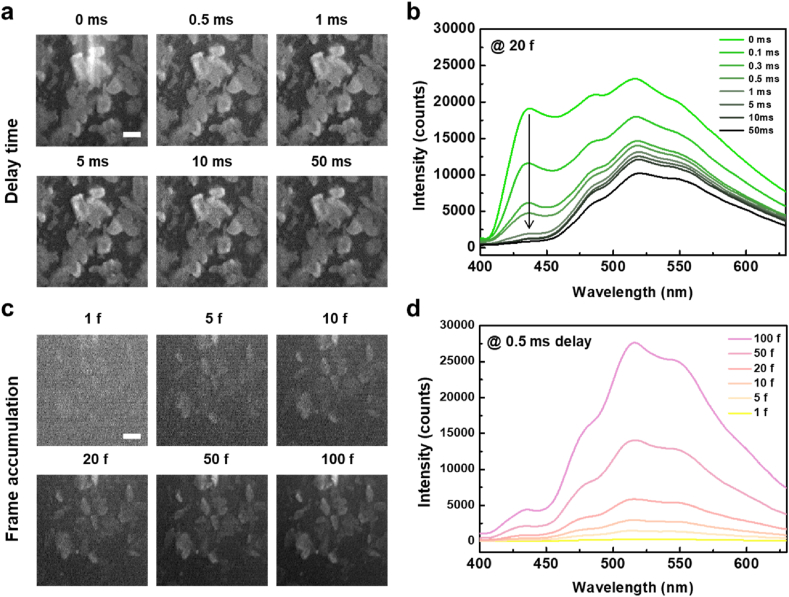
The 375 nm CW laser was converted to a pulsed laser by utilizing a function generator that imposed an electrical signal on the device. It connected the laser source and EMCCD detector through time-to-live (TTL) channels to control function parameters such as the pulse period, width, height, and delay time between them. The long-lived phosphorescence signal of the IPA crystal was obtained using a pulse period of 300 ms, laser intensity of 300 mA, laser width of 100 ms, and detector width of 100 ms. Then, we obtained the time-gated images of the IPA crystal for delay times of 0–50 ms (Fig. 3a). All EMCCD images were optimized using autoscaled contrast to generate clearer images. The time-gated images of the IPA crystal were obtained at a delay time of 0 ms, and no significant difference was observed between delay times of 0.5–50 ms. By contrast, in-situ spectrometry confirmed the differences between the fluorescence and phosphorescence signals of the IPA crystal for different delay times (Fig. 3b). At a delay time of 0 ms, the time-gated spectra of the IPA crystal showed four emission peaks in a range of 400–630 nm, with two main peaks at 435 and 516 nm and two shoulder peaks at 483 and 550 nm. The intensities of all the peaks decreased as the delay time increased; in particular, the main peak at 435 nm nearly disappeared after a delay time of 1 ms. This implied that the lifetime of the luminescence at 435 nm was relatively shorter than that of the others, representing a fluorescence signal. The others were the phosphorescence signals of the IPA crystal, which retained their intensities until a delay time of 50 ms. We accumulated the luminescence signals of IPA to improve the resolution of the time-gated images (Fig. 3c). At a delay time of 0.5 ms, it was difficult to distinguish between the IPA crystals in a single frame. The resolution of the images was significantly improved through the accumulated time-gated analysis. IPA crystals were observed by accumulating more than 20 frames of time-gated images. The calculated SBR was confirmed based on the number of accumulated frames (Supplementary Fig. 1). Similarly, in in-situ spectrometry, the SNR of the spectrum improved as the number of accumulated frames increased (Fig. 3d). The other function parameters, such as the pulse width or laser intensity, were controlled using time-gated microscopy (Supplementary Figs. 2 and 3). The time-gated images of the IPA crystal became clearer as the pulse width increased regardless of the laser or detector channel. The same trend was observed when the laser intensity was increased. The accumulated time-gated analysis of RTP enabled us to assign all the peaks of the PL spectrum and compare their lifetimes, which was not possible using microscopy. Furthermore, the SBR in the images and SNR in the spectrum increased with the number of accumulated frames at a delay time of 0.5 ms.
Fig. 3.
Time-gated RTP images and spectrum of IPA crystals under pulsed laser. a Time-gated images of IPA crystal over delay time (scale bar: 20 μm; accumulation: 20 frames). b Time-gated spectrum of IPA crystal over delay time. c Time-gated images of IPA crystal with frame accumulation (scale bar: 20 μm; delay time: 0.5 ms). d Time-gated spectrum of IPA crystal with frame accumulation. All data were obtained using 375 nm pulsed laser source. Fixed parameters: pulse period: 300 ms; laser intensity: 300 mA; laser pulse width: 100 ms; detector pulse width: 100 ms.
3.4. RTP enhancement for IPA–aptamer hybrid assembly with specific recognition of target protein
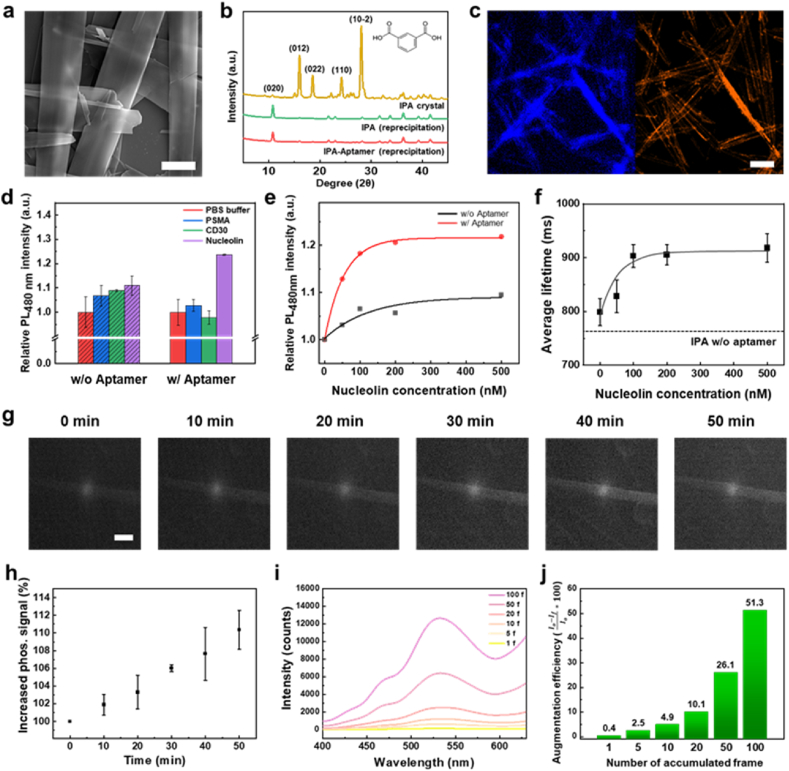
We have studied techniques for fabricating complexes between light-emitting crystals and biomaterials such as using DNA, aptamers, and peptides [[46], [47], [48], [49]]. A light-emitting crystal was recrystallized with a bioligand using the reprecipitation method, and its fluorescence increased after specific recognition with a target material. To expand the biohybrid technique to RTP materials, we successfully fabricated an IPA–aptamer hybrid assembly in which the DNA aptamer had a specific binding affinity for nucleolin, which is a multifunctional protein containing four RNA-binding domains (RBDs) [50,51]. The DNA aptamer was used as the bioligand to selectively bind with nucleolin. The guanine-rich G-quadruplex structure of the aptamer interacted with the RBDs [52]. The morphology of the IPA–aptamer hybrid assembly was a strap shape with a thickness of approximately 5 μm, as observed in a scanning electron microscopy (SEM) image (Fig. 4a). We determined the distribution of the DNA aptamer in IPA–aptamer hybrid assembly by using the Cy5-labeled aptamer for fabricating the assembly. The blue fluorescence emission in the IPA–aptamer hybrid assembly was visualized using a confocal laser scanning microscope (CLSM) under excitation with a 405 nm laser (Supplementary Fig. 4). The red-emitting Cy5 molecule was attached to the 3′ end of the aptamer. The distribution of the DNA aptamer molecules was nearly identical to that of the IPA particles. The merged image showed the coexistence of the IPA particles and aptamers. This was because the DNA aptamer was distributed entirely within the IPA–aptamer hybrid assembly. To examine the structural changes in the IPA crystals, X-ray diffraction (XRD) analysis was performed using the pristine IPA powder, recrystallized IPA, and recrystallized IPA–aptamer hybrid assembly (Fig. 4b). The XRD pattern of the pristine IPA powder showed the typical peaks of IPA at 15.9° and 27.98° corresponding to the (012) and (10-2) planes, respectively [53]. After IPA was recrystallized from the aptamer using the reprecipitation method, an XRD peak was observed at 10.66° corresponding to the (020) plane, and the intensities of the peaks at 15.9° and 27.98° decreased. As the XRD patterns of the IPA and IPA–aptamer straps were nearly identical, it could be inferred that the DNA aptamer negligibly affected the crystal structure of IPA. Then, we treated the IPA–aptamer hybrid assembly with nucleolin, which selectively interacted with the DNA aptamer of hybrid assembly. The confocal microscopy images of the IPA–aptamer hybrid assembly with nucleolin showed the blue fluorescence emission of the IPA assembly (Fig. 4c). We determined the position of nucleolin by attaching phycoerythrin (PE), which had a strong orange emission, to nucleolin. Nucleolin was mainly distributed on the surface of the IPA–aptamer hybrid assembly. To investigate the specificity of the IPA–aptamer hybrid assembly, we used different types of proteins to analyze the phosphorescence amplification of the IPA assembly with and without the aptamer (Fig. 4d). The phosphorescence intensity of the IPA particles without the aptamer was increased by approximately 10 % owing to the nonspecific binding of all proteins to IPA particles. In the case of the IPA–aptamer hybrid assembly, the phosphorescence intensity increased by 25 % when nucleolin was exposed to the IPA–aptamer microparticles. By contrast, the phosphorescence intensity of the other proteins was either maintained or decreased. These results demonstrated the selective response of the IPA–aptamer hybrid assembly to nucleolin. To analyze the enhancement of phosphorescence with the nucleolin concentration, proteins with concentrations of 0–500 nM were added to the IPA hybrid assembly with and without the DNA aptamer (Fig. 4e). In the IPA particles without the aptamer, the relative phosphorescence intensity slightly increased with the nucleolin concentration, and it became saturated at approximately 10 % at a nucleolin concentration of 500 nM. When the IPA–aptamer hybrid assembly was exposed to nucleolin, the phosphorescence intensity sharply increased to 20 % at a nucleolin concentration of 200 nM and became saturated. We utilized a photomultiplier to understand the phosphorescence lifetime of IPA pellets as a function of the nucleolin concentration (Fig. 4f). We obtained the phosphorescence intensity signal according to the decay time and calculated the average lifetime of each IPA–aptamer hybrid assembly by calculating an exponential decay and average lifetime formula. The phosphorescence lifetime of IPA particles without aptamer was 773 ms as a control, while the IPA-aptamer hybrid assembly was 798 ms which was slightly increased. When the IPA–aptamer hybrid assembly was exposed to nucleolin at concentrations of 0–500 nM, the average phosphorescence lifetime increased to 923 ms at 100 nM and became saturated without significant changes. We successfully fabricated the IPA–aptamer hybrid assembly that could selectively recognize nucleolin by simultaneously increasing the phosphorescence intensity and lifetime.
Fig. 4.
Accumulated detection of phosphorescence augmentation in IPA–aptamer hybrid assembly with specific target recognition. a SEM image of IPA–aptamer hybrid assembly (scale bar: 5 μm). b XRD spectra of IPA crystal (yellow line), IPA recrystallized using reprecipitation method (green line), and IPA–aptamer hybrid obtained using reprecipitation method (red line). c CLSM images of IPA–aptamer hybrid assembly after exposure to nucleolin. IPA was excited by a 405 nm laser, and emission was filtered in a range of 410–500 nm (left). PE-labeled nucleolin was excited with a 555 nm laser, and emission was filtered in a range of 580–640 nm (right) (scale bar: 20 μm). d Relative PL intensity of IPA without (dashed bar) or with (full filled bar) aptamer after exposure to various proteins with same concentration of 200 nM. e Relative phosphorescence max. intensity of IPA at 480 nm without or with aptamer with different concentrations of nucleolin. f Average lifetime of IPA–aptamer hybrid assembly with different concentrations of nucleolin. g Time-gated images of IPA–aptamer hybrid assembly after exposure to nucleolin (scale bar: 20 μm). h Increased phosphorescence max. intensity of IPA–aptamer hybrid assembly at 530 nm over reaction time. i Time-gated spectrum of phosphorescence augmentation with accumulated detected signal of IPA–aptamer hybrid assembly after target recognition. j Efficiency of phosphorescence augmentation with accumulated detected signal of IPA–aptamer hybrid assembly after target recognition.
3.5. Accumulation of phosphorescence signals of IPA–aptamer hybrid assembly after treatment with target protein via time-gated microscopy
We used in-situ time-gated analysis to observe the increase in the phosphorescence of the IPA–aptamer hybrid assembly after treatment with nucleolin in real time (Fig. 4g and Supplementary Fig. 5a). The function parameters of the equipment were fixed at a pulse period of 300 ms, laser intensity of 300 mA, laser width of 100 ms, detector width of 100 ms, the delay time of 0.5 ms, and accumulation of 20 frames. The time-gated images of the IPA–aptamer hybrid became clearer over time, and a strip-shaped crystal was observed. In the time-gated spectra of the IPA–aptamer hybrid assembly, the phosphorescence intensity increased to 110 % after a reaction time of 50 min after target treatment (Fig. 4h and Supplementary Fig. 5b). To amplify the phosphorescence augmentation signal, the time-gated spectra of the IPA–aptamer hybrid assembly were obtained by accumulating 1–100 frames accumulation. (Fig. 4i). The S/N ratio of the spectrum improved with accumulation by amplifying the augmented signal. To quantify this increase, we calculated the augmentation efficiency of each accumulated spectrum (Fig. 4j). The augmentation efficiency with the target treatment was only 0.4 with when 1 frame was accumulated, and it increased to 51.3 when 100 frames were accumulated. These results showed that the sensitivity and limit of detection of target recognition could be improved through frame accumulation in time-gated analysis.
3.6. Obtaining spectral information using RTP and autofluorescence materials under gating time control
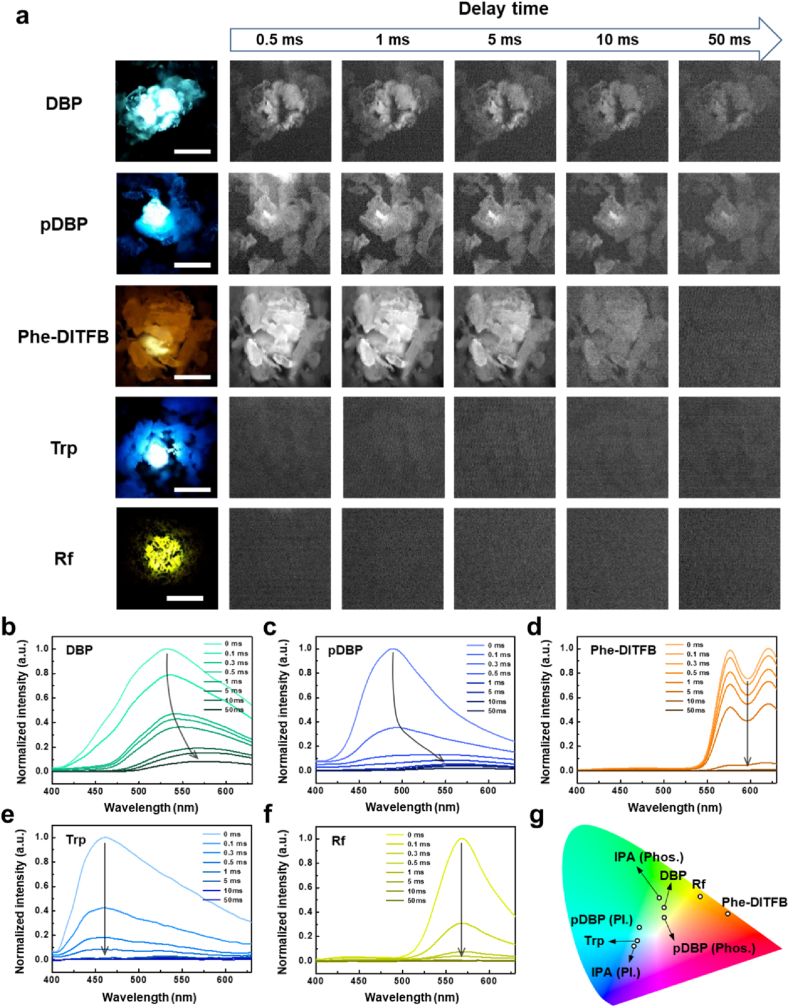
Autofluorescence materials in cells or tissues show fluorescence at each wavelengths, which causes problems in terms of the SBR in bioimaging techniques. To solve these problems using time-gated RTP analysis, we used RTP and autofluorescence materials as luminescence models to mimic the error conditions of bioimaging analysis. We selected benzophenone (DBP) [54], polybenzophenone (pDBP), and cocrystals of phenanthrene and 1,2,4,5-tetrafluoro-3,6-diiodobenzene (Phe-DITFB) [42] as representative RTP materials and tryptophan (Trp) [55] and riboflavin (Rf) [56,57] as representative autofluorescence materials. Optical and fluorescence microscopy images with various filters were obtained using conventional microscopy with LED lamp irradiation (Supplementary Fig. 6). Luminescent and time-gated images were obtained via irradiation with a pulsed laser under delay time control (Fig. 5a). DBP exhibited the longest lifetime, and it showed cyan luminescence in optical microscopy images. The luminescence signal was slightly reduced as the delay time increased from 0.5 to 50 ms in time-gated microscopy images. However, the signal remained sufficient for determining the shape of the crystals until a delay time of 50 ms because of the phosphorescence signal of DBP. Similarly, in the case of pDBP, blue luminescence was observed in the optical microscopy image, and the phosphorescence signal of pDBP crystals was maintained at a delay time of 50 ms. The orange luminescence of Phe-DITFB also decreased as the delay time increased. However, the crystals were observed up to a delay time of 10 ms but not at a delay time of 50 ms. It is necessary to adjust the gating time window according to the lifetime of the RTP materials used. For Trp and Rf, the delayed signals were not visible from a delay time of 0.5 ms onwards because of their short lifetime of autofluorescence signal. The time-gated spectra of the samples were obtained and normalized (Supplementary Fig. 7). The spectra of all the samples decreased as the delay time increased. The time-gated spectra of DBP had a main peak at 530 nm at a delay time of 0 ms, which gradually decreased, and there was a red shift to 570 nm as the delay time increased (Fig. 5b). The peaks at 530 and a red shift at 570 nm were attributed to fluorescence and phosphorescence, respectively. The time-gated spectra of pDBP were similar to those of DBP, with a main peak at 488 nm and red shift at 565 nm representing fluorescence and phosphorescence, respectively (Fig. 5c). The time-gated spectra of Phe-DITFB showed two main peaks at 575 and 620 nm, which decreased without any peak shift as the delay time increased (Fig. 5d). The peaks of the time-gated spectra of the autofluorescence materials decreased more rapidly than those of the RTP materials at 460 nm for Trp and 570 nm for Rf (Fig. 5e and f). To quantify this decrease, each time-gated spectrum was normalized based on the intensity of the main peak at a delay time of 0 ms. The normalized intensities at a delay time of 1 ms were 0.409 for DBP, 0.122 for pDBP, 0.547 for Phe-DITFB, 0.016 for Trp, and 0.007 for Rf. The decrease in the intensity of the RTP materials was less than that of the autofluorescence materials for equal delay times. The CIE color mapping of each time-gated spectrum was identical to the luminescence color obtained from the optical images (Fig. 5g).
Fig. 5.
Time-gated images and spectrum of RTP and autofluorescence materials with delay time control. a Luminescent and time-gated images of RTP and autofluorescence materials over delay time (scale bar: 50 μm). b-f Normalized timge-gated spectrum of DBP, pDBP, Phe-DITBP, Trp and Rf over delay time. g CIE color mapping of spectrum for RTP and autofluorescence materials.
3.7. Differentiation between phosphorescence and autofluorescence using time-gated microscopy and application to actual cells
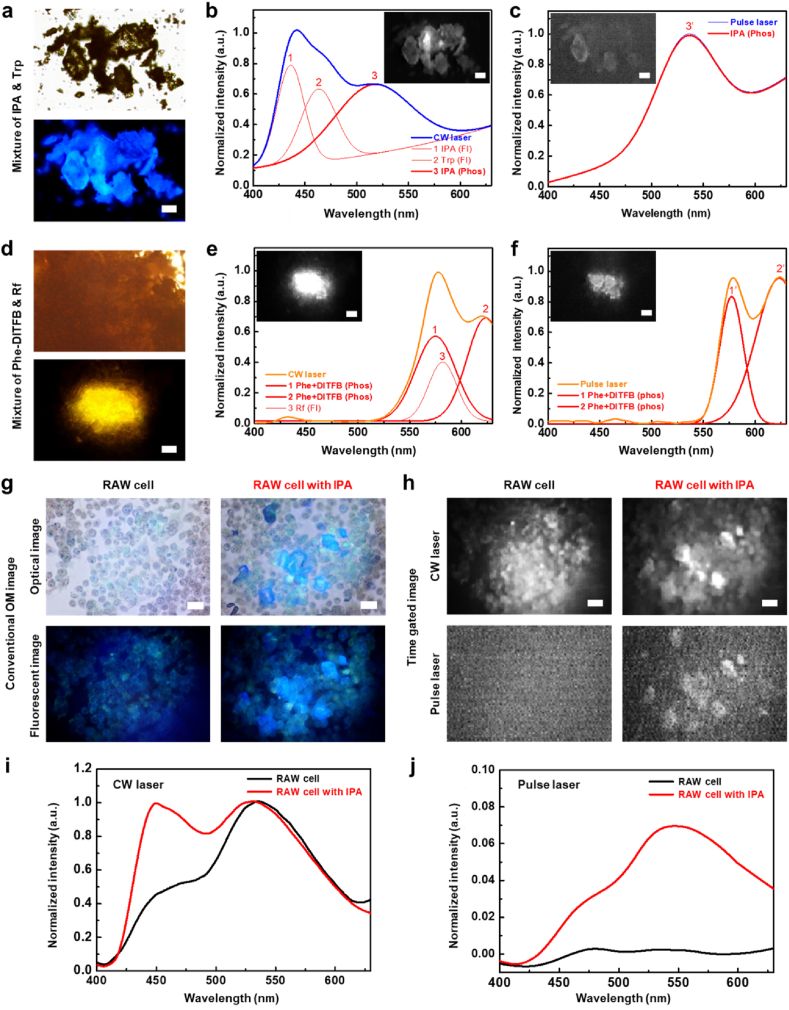
We mimicked the clinical conditions of bioimaging analysis to artificially simulate the problems caused by autofluorescence materials in bioimaging analysis. In control simulation, the RTP and autofluorescence materials had different wavelength bands and crystal shapes; thus, it was easy to differentiate between them using optical microscopy. This was represented by a mixture of IPA and Rf crystals, as observed via conventional optical microscopy (Supplementary Fig. 8). It was easy to distinguish between the blue luminescence of IPA crystals and the yellow luminescence of Rf crystals through optical microscopy. The phosphorescence signal of IPA crystals could be selectively detected using time-gated analysis with delay time control. In-situ analysis of IPA luminescence change was also detected (Supplementary Video 2). In addition, there were two simulations with errors caused by autofluorescence. In the first, the RTP and autofluorescence materials had the same wavelength bands and similar crystal shapes. In the second, the RTP and autofluorescence materials had different crystal shapes but the fluorescence was so strong that it covered the phosphorescence signals. The first simulation was represented by a mixture of IPA and Trp crystals observed using conventional optical microscopy (Fig. 6a). The IPA and Trp crystals both exhibited blue luminescence and had various morphologies; therefore, it was difficult to distinguish between them using optical and fluorescence microscopy images. With the in-situ Raman spectroscopy (Supplementary Fig. 9), we obtained the Raman spectrum of pure IPA and Trp powder, and assigned the specific Raman shift at 1635 cm−1 to IPA and 3405 cm−1 to Trp. With the Raman mapping images, the purple color represents IPA-rich region and the cyan color represents Trp-rich region. However, the resolution of the image was not enough to distinguish the crystals and compare with optical image. The IPA and Trp mixture could be differentiated using time-gated analysis via microscopy and spectroscopy. Under irradiation with a CW laser, the EMCCD image of the IPA and Trp crystal mixture was similar to the optical microscopy image (Fig. 6b). The time-gated spectrum of the mixed powder showed three peaks, which were deconvoluted for peak assignment. As we already analyzed the time-gated spectra of the pure components, we could assign the peaks at 436 nm to the fluorescence of IPA, 465 nm to the fluorescence of Trp, and 518 nm to the phosphorescence of IPA. Under the pulsed laser irradiation, only the IPA crystals were observed in the time-gated image with a phosphorescence signal (Fig. 6c). The time-gated spectrum also showed only the phosphorescence spectra of the IPA crystals at 535 nm. The second simulation was performed using a mixture of Phe-DITFB and Rf crystals, as observed via conventional optical microscopy (Fig. 6d). The Phe-DITFB and Rf crystals exhibited orange–yellow luminescence but had different crystal morphologies. However, as the quantum yield of the luminescence with Phe-DITFB was considerably smaller than that with Rf, it was difficult to find Phe-DITFB crystals in the region where Rf was widely distributed. With the in-situ Raman spectroscopy (Supplementary Fig. 10), we obtained the Raman spectrum of pure Phe-DITFB and Rf powder, and assigned the specific Raman shift at 402 cm−1 to Phe-DITFB and 740 cm−1 to Rf. With the Raman mapping images, the purple color represents Phe-DITFB-rich region and the cyan color represents Rf-rich region. However, the resolution of the image was not enough to distinguish the crystals and compare with optical image. The time-gated analysis could provide only the phosphorescence signal of Phe-DITFB crystals using microscopy and spectroscopy. Under CW laser irradiation, the time-gated image of the Phe-DITFB and Rf crystal mixture was similar to the optical microscopy image (Fig. 6e). The time-gated spectra were deconvoluted into three peaks. We assigned the peaks at 575 and 622 nm to the phosphorescence of Phe-DITFB and the one at 580 nm to the fluorescence of Rf. Under the pulsed laser irradiation, only Phe-DITFB crystals were observed in the time-gated image with a phosphorescence signal (Fig. 6f). The time-gated spectrum also showed only the phosphorescence of Phe-DITFB crystals at 577 and 625 nm. Based on these results of the autofluorescence problems under artificial conditions, we extended it to actual cell applications. We selected the RAW264.7 macrophage (RAW) cells as the 2D cell model for autofluorescence. RAW cells exhibited green and blue autofluorescence under UV irradiation [58,59]. To confirm the cytotoxicity of RTP materials, we carried out the cell viability test with RTP materials (Supplementary Fig. 11). IPA and DBP were metal-free organic RTP materials showing high cell viability, while Ir(ppy)3 and PdOEP were heavy metal-containing RTP materials showing low cell viability. The IPA was used for the cell experiments due to the highest cell viability among them. RAW cells with or without IPA crystals were observed using optical and fluorescence microscopy (Fig. 6g). The blue and green luminescence of RAW cells was observed using optical and fluorescence microscopy images. In the mixture of RAW cells and IPA crystals, the blue luminescence of IPA crystals was surrounded by the same colored autofluorescence of RAW cells, which could not be distinguished in optical and fluorescence microscopy images. In the in-situ time-gated image with the same area, the autofluorescence signal of RAW cells was detected under excitation with the CW laser (Fig. 6h). However, no signal was detected under excitation with the pulsed laser when autofluorescence disappeared within a short lifetime. In the mixture of RAW cells and IPA crystals under CW laser excitation, the autofluorescence signal of RAW cells and the luminescence signal of IPA crystals were detected using time-gated microscopy. However, only the phosphorescence signal of IPA crystals was selectively obtained under irradiation with the pulsed laser. The selective detection of the phosphorescence signal of the RTP material within the autofluorescence cell was confirmed using the time-gated spectra of each sample. Under CW laser irradiation (Fig. 6i), the time-gated spectrum of RAW cells was obtained with a main peak at 535 nm. For RAW cells with IPA, the main peak appeared at 450 nm, which was confirmed to be the luminescence signal of IPA crystals. Under irradiation with the pulsed laser (Fig. 6j), the time-gated spectrum of RAW cells did not show any luminescence signal. By contrast, the time-gated spectrum of RAW cells with IPA had a main peak at 548 nm owing to the phosphorescence signal of IPA crystals. These results confirmed that the time-gated analysis could solve the problems caused by autofluorescence signals in bioimaging analysis.
Fig. 6.
Determination of phosphorescence signal in mixture of RTP and autofluorescence mixture and cell imaging application. a Optical (top) and fluorescent (bottom) microscopy images of IPA and Trp mixture (scale bar: 10 μm). b, c Spectral deconvolution of time-gated spectrum of IPA and Trp mixture and time-gated images (inset) under irradiation with CW laser (left) and pulsed laser (righ) (scale bar: 10 μm). d Optical (top) and fluorescence (bottom) microscopy images of Phe-DITFB cocrystal and Rf mixture (scale bar: 10 μm). e, f Spectral deconvolution of time-gated spectrum of Phe-DITFB cocrystal and Rf mixture and time-gated images (inset) under irradiation with CW laser (left) and pulsed laser (right)(scale bar: 10 μm). g Conventional optical microscopy and h time-gated images of RAW264.7 cell with or without IPA crystals (scale bar: 10 μm). i, j Time-gated spectrum of RAW264.7 cell with or without IPA crystals under irradiation with CW laser (left) and pulsed laser (right).
4. Conclusions
The autofluorescence of living organisms interferes with bioimaging analysis by competing with the luminescence signals of light-emitting materials. Lifetime mapping and time-gated imaging were presented to solve this problem. However, there was only limited information that could not provide the spectrum of signals to interpret the results. In this study, we proposed a method that combined a spectrometer with time-gated signal detection and an accumulation system with a microscope to obtain spectral information to perform quantitative spectroscopic analysis. We modified the existing time-gated microscopy and lifetime mapping to overcome the limitations of the lack of spectral information. We showed that organic RTP materials, which exhibited luminescence at room temperature and were relatively stable in the presence of moisture and oxygen, were suitable for bioimaging applications. The hybridization of RTP materials with a bioligand provided a biorecognition function that could be used in biosensing applications. When RTP material was used for RAW cell imaging, our method successfully avoided autofluorescence and obtained the accumulated phosphorescence signals, suggesting its potential applicability in the bioimaging field. These results could lead to the development in vivo or in vitro biopsy and advancements in disease diagnosis.
CRediT authorship contribution statement
Yong Ho Cho: Writing – original draft, Validation, Methodology, Investigation, Conceptualization. Seokho Kim: Writing – original draft, Validation, Methodology, Investigation, Conceptualization. Tae Kyung Won: Methodology, Investigation. Sunki Cho: Methodology, Investigation. Dong June Ahn: Writing – review & editing, Writing – original draft, Validation, Supervision, Project administration, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This work was supported by the National Research Foundation of Korea (NRF 2021R1A2C3009955) and a Korea University Grant.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mtbio.2024.101238.
Contributor Information
Yong Ho Cho, Email: cyh9312@korea.ac.kr.
Seokho Kim, Email: seokho@korea.ac.kr.
Tae Kyung Won, Email: callys@korea.ac.kr.
Sunki Cho, Email: tjsrl7823@korea.ac.kr.
Dong June Ahn, Email: ahn@korea.ac.kr.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Frangioni J.V. In vivo near-infrared fluorescence imaging. Curr. Opin. Chem. Biol. 2003;7(5):626–634. doi: 10.1016/j.cbpa.2003.08.007. [DOI] [PubMed] [Google Scholar]
- 2.Mansfield J.R., Gossage K.W., Hoyt C.C., Levenson R.M. Autofluorescence removal, multiplexing, and automated analysis methods for in-vivo fluorescence imaging. J. Biomed. Opt. 2005;10(4):41207–41209. doi: 10.1117/1.2032458. [DOI] [PubMed] [Google Scholar]
- 3.Croce A.C., Ferrigno A., Bottiroli G., Vairetti M. Autofluorescence‐based optical biopsy: an effective diagnostic tool in hepatology. Liver Int. 2018;38(7):1160–1174. doi: 10.1111/liv.13753. [DOI] [PubMed] [Google Scholar]
- 4.Zhang K.Y., Yu Q., Wei H., Liu S., Zhao Q., Huang W. Long-lived emissive probes for time-resolved photoluminescence bioimaging and biosensing. Chem. Rev. 2018;118(4):1770–1839. doi: 10.1021/acs.chemrev.7b00425. [DOI] [PubMed] [Google Scholar]
- 5.Chang B., Chen J., Bao J., Sun T., Cheng Z. Molecularly engineered room-temperature phosphorescence for biomedical application: from the visible toward second near-infrared window. Chem. Rev. 2023;123(24):13966–14037. doi: 10.1021/acs.chemrev.3c00401. [DOI] [PubMed] [Google Scholar]
- 6.Yang W., Chen S.-L. Time-gated fluorescence imaging: advances in technology and biological applications. Journal of Innovative Optical Health Sciences. 2020;13(3) doi: 10.1142/S1793545820300062. [DOI] [Google Scholar]
- 7.Shaked N.T., Boppart S.A., Wang L.V., Popp J. Label-free biomedical optical imaging. Nat. Photonics. 2023;17:1031–1041. doi: 10.1038/s41566-023-01299-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ye S., Latham A.P., Tang Y., Hsiung C.-H., Chen J., Luo F., Liu Y., Zhang B., Zhang X. Micropolarity governs the structural organization of biomolecular condensates. Nat. Chem. Biol. 2024;20:443–451. doi: 10.1038/s41589-023-01477-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhong W., Urayama P., Mycek M.-A. Imaging fluorescence lifetime modulation of a ruthenium-based dye in living cells: the potential for oxygen sensing. J. Phys. Appl. Phys. 2003;36(14):1689. doi: 10.1088/0022-3727/36/14/306. [DOI] [Google Scholar]
- 10.Lei Z., Endo M., Ube H., Shiraogawa T., Zhao P., Nagata K., Pei X.-L., Eguchi T., Kamachi T., Ehara M. N-heterocyclic carbene-based c-centered au (i)-ag (i) clusters with intense phosphorescence and organelle-selective translocation in cells. Nat. Commun. 2022;13:4288. doi: 10.1038/s41467-022-31891-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cao X., Rao Allu S., Jiang S., Jia M., Gunn J.R., Yao C., LaRochelle E.P., Shell J.R., Bruza P., Gladstone D.J. Tissue pO2 distributions in xenograft tumors dynamically imaged by cherenkov-excited phosphorescence during fractionated radiation therapy. Nat. Commun. 2020;11:573. doi: 10.1038/s41467-020-14415-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baggaley E., Botchway S.W., Haycock J.W., Morris H., Sazanovich I.V., Williams J.G., Weinstein J.A. Long-lived metal complexes open up microsecond lifetime imaging microscopy under multiphoton excitation: from flim to plim and beyond. Chem. Sci. 2014;5(3):879–886. doi: 10.1039/C3SC51875B. [DOI] [Google Scholar]
- 13.Chelushkin P.S., Tunik S.P. Phosphorescence lifetime imaging (plim): state of the art and perspectives. Progress in Photon Science: Recent Advances. 2019;109–128 [Google Scholar]
- 14.Beverloo H., Van Schadewijk A., van Gelderen‐Boele S., Tanke H. Inorganic phosphors as new luminescent labels for immunocytochemistry and time‐resolved microscopy. Cytometry: The Journal of the International Society for Analytical Cytology. 1990;11(7):784–792. doi: 10.1002/cyto.990110704. [DOI] [PubMed] [Google Scholar]
- 15.Marriott G., Clegg R.M., Arndt-Jovin D.J., Jovin T.M. Time resolved imaging microscopy. Phosphorescence and delayed fluorescence imaging. Biophys. J. 1991;60(6):1374–1387. doi: 10.1016/S0006-3495(91)82175-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jin D., Piper J.A. Time-gated luminescence microscopy allowing direct visual inspection of lanthanide-stained microorganisms in background-free condition. Anal. Chem. 2011;83(6):2294–2300. doi: 10.1021/ac103207r. [DOI] [PubMed] [Google Scholar]
- 17.Sreenivasan V.K., Wan Razali W.A., Zhang K., Pillai R.R., Saini A., Denkova D., Santiago M., Brown H., Thompson J., Connor M. Development of bright and biocompatible nanoruby and its application to background-free time-gated imaging of g-protein-coupled receptors. ACS Appl. Mater. Interfaces. 2017;9(45):39197–39208. doi: 10.1021/acsami.7b12665. [DOI] [PubMed] [Google Scholar]
- 18.Razali W.A., Sreenivasan V.K., Bradac C., Connor M., Goldys E.M., Zvyagin A.V. Wide‐field time‐gated photoluminescence microscopy for fast ultrahigh‐sensitivity imaging of photoluminescent probes. J. Biophot. 2016;9(8):848–858. doi: 10.1002/jbio.201600050. [DOI] [PubMed] [Google Scholar]
- 19.Jin D. Demonstration of true‐color high‐contrast microorganism imaging for terbium bioprobes. Cytometry. 2011;79(5):392–397. doi: 10.1002/cyto.a.21052. [DOI] [PubMed] [Google Scholar]
- 20.Cui M., Dai P., Ding J., Li M., Sun R., Jiang X., Wu M., Pang X., Liu M., Zhao Q. Millisecond‐range time‐resolved bioimaging enabled through ultralong aqueous phosphorescence probes. Angew. Chem. Int. Ed. 2022;61(14) doi: 10.1002/anie.202200172. [DOI] [PubMed] [Google Scholar]
- 21.Fan Y., Liu S., Wu M., Xiao L., Fan Y., Han M., Chang K., Zhang Y., Zhen X., Li Q. Mobile phone flashlight‐excited red afterglow bioimaging. Adv. Mater. 2022;34(18) doi: 10.1002/adma.202201280. [DOI] [PubMed] [Google Scholar]
- 22.Chang K., Xiao L., Fan Y., Gu J., Wang Y., Yang J., Chen M., Zhang Y., Li Q., Li Z. Lighting up metastasis process before formation of secondary tumor by phosphorescence imaging. Sci. Adv. 2023;9(20) doi: 10.1126/sciadv.adf6757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao W., He Z., Tang B.Z. Room-temperature phosphorescence from organic aggregates. Nat. Rev. Mater. 2020;5(12):869–885. doi: 10.1038/s41578-020-0223-z. [DOI] [Google Scholar]
- 24.Gu L., Shi H., Bian L., Gu M., Ling K., Wang X., Ma H., Cai S., Ning W., Fu L. Colour-tunable ultra-long organic phosphorescence of a single-component molecular crystal. Nat. Photonics. 2019;13(6):406–411. doi: 10.1038/s41566-019-0408-4. [DOI] [Google Scholar]
- 25.Wang Z., Gao L., Zheng Y., Zhu Y., Zhang Y., Zheng X., Wang C., Li Y., Zhao Y., Yang C. Four-in-One stimulus-responsive long-lived luminescent systems based on pyrene-doped amorphous polymers. Angew. Chem. 2022;134(32) doi: 10.1002/ange.202203254. [DOI] [PubMed] [Google Scholar]
- 26.Gao L., Huang J., Qu L., Chen X., Zhu Y., Li C., Tian Q., Zhao Y., Yang C. Stepwise taming of triplet excitons via multiple confinements in intrinsic polymers for long-lived room-temperature phosphorescence. Nat. Commun. 2023;14(1):7252. doi: 10.1038/s41467-023-43133-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y., Gao L., Zheng X., Wang Z., Yang C., Tang H., Qu L., Li Y., Zhao Y. Ultraviolet irradiation-responsive dynamic ultralong organic phosphorescence in polymeric systems. Nat. Commun. 2021;12(1):2297. doi: 10.1038/s41467-021-22609-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang Y., Chen X., Xu J., Zhang Q., Gao L., Wang Z., Qu L., Wang K., Li Y., Cai Z., Zhao Y., Yang C. Cross-Linked Polyphosphazene Nanospheres boosting long-lived organic room-temperature phosphorescence. J. Am. Chem. Soc. 2022;144(13):6107–6117. doi: 10.1021/jacs.2c02076. [DOI] [PubMed] [Google Scholar]
- 29.Zhang Y., Su Y., Wu H., Wang Z., Wang C., Zheng Y., Zheng X., Gao L., Zhou Q., Yang Y., Chen X., Yang C., Zhao Y. Large-area, flexible, transparent, and long-lived polymer-based phosphorescence Films. J. Am. Chem. Soc. 2021;143(34):13675–13685. doi: 10.1021/jacs.1c05213. [DOI] [PubMed] [Google Scholar]
- 30.Yang X., Yan D. Strongly enhanced long-lived persistent room temperature phosphorescence based on the Formation of metal–organic hybrids. Adv. Opt. Mater. 2016;4(6):897–905. doi: 10.1002/adom.201500666. [DOI] [Google Scholar]
- 31.Xing C., Zhou B., Yan D., Fang W.-H. Dynamic photoresponsive ultralong phosphorescence from one-dimensional Halide Microrods toward Multilevel information storage. CCS Chem. 2023;5(12):2866–2876. doi: 10.31635/ccschem.023.202202605. [DOI] [Google Scholar]
- 32.Xing C., Qi Z., Zhou B., Yan D., Fang W.-H. Solid-state photochemical cascade process boosting Smart ultralong room-temperature phosphorescence in Bismuth Halides. Angew. Chem. 2024;136(21) doi: 10.1002/ange.202402634. [DOI] [PubMed] [Google Scholar]
- 33.Xing C., Zhou B., Yan D., Fang W.-H. Integrating full-color 2D optical waveguide and heterojunction engineering in Halide Microsheets for Multichannel photonic logical gates. Adv. Sci. 2024;11(17) doi: 10.1002/advs.202310262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Q., Qu L., Hou H., Huang J., Li C., Zhu Y., Wang Y., Chen X., Zhou Q., Yang Y., Yang C. Long lifetimes white afterglow in slightly crosslinked polymer systems. Nat. Commun. 2024;15(1):2947. doi: 10.1038/s41467-024-47378-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang X., Yan D. Long-afterglow metal–organic frameworks: reversible guest-induced phosphorescence tunability. Chem. Sci. 2016;7(7):4519–4526. doi: 10.1039/C6SC00563B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fang X., Tang Y., Ma Y.-J., Xiao G., Li P., Yan D. Ultralong-lived triplet excitons of room-temperature phosphorescent carbon dots located on g-C3N4 to boost photocatalysis. Sci. China Mater. 2023;66(2):664–671. doi: 10.1007/s40843-022-2163-9. [DOI] [Google Scholar]
- 37.Cui C., Park D.H., Ahn D.J. Organic semiconductor–dna hybrid assemblies. Adv. Mater. 2020;32(51) doi: 10.1002/adma.202002213. [DOI] [PubMed] [Google Scholar]
- 38.Kim S., Cui C., Huang J., Noh H., Park D.H., Ahn D.J. Bio‐photonic waveguide of a dna‐hybrid semiconductor prismatic hexagon. Adv. Mater. 2020;32(45) doi: 10.1002/adma.202005238. [DOI] [PubMed] [Google Scholar]
- 39.Jung W.H., Park J.H., Kim S., Cui C., Ahn D.J. Molecular doping of nucleic acids into light emitting crystals driven by multisite-intermolecular interaction. Nat. Commun. 2022;13(1):6193. doi: 10.1038/s41467-022-33999-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cui C., Park D.H., Choi H., Joo J., Ahn D.J. Protein recognition by phase transition of aptamer-linked polythiophene single nanowire. Small. 2015;12(9):1154–1158. doi: 10.1002/smll.201501908. [DOI] [PubMed] [Google Scholar]
- 41.Kang M.J., Cho Y.H., Kim S., Ahn D.J. Simultaneous enhancement in phosphorescence and its lifetime of PtOEP-peptide assembly triggered by protein interaction. Int. J. Biol. Macromol. 2024;266 doi: 10.1016/j.ijbiomac.2024.131195. 2024. [DOI] [PubMed] [Google Scholar]
- 42.Shen Q.J., Pang X., Zhao X.R., Gao H.Y., Sun H.-L., Jin W.J. Phosphorescent cocrystals constructed by 1, 4-diiodotetrafluorobenzene and polyaromatic hydrocarbons based on c–i⋯ π halogen bonding and other assisting weak interactions. CrystEngComm. 2012;14(15):5027–5034. doi: 10.1039/c2ce25338k. [DOI] [Google Scholar]
- 43.Ian Freshney R. John Wiley & Sons; USA: 2005. Culture of Animal Cells: A Manual of Basic Technique; p. 2005. 642. [Google Scholar]
- 44.Kuno S., Akeno H., Ohtani H., Yuasa H. Visible room-temperature phosphorescence of pure organic crystals via a radical-ion-pair mechanism. Phys. Chem. Chem. Phys. 2015;17(24):15989–15995. doi: 10.1039/C5CP01203A. [DOI] [PubMed] [Google Scholar]
- 45.Kuno S., Kanamori T., Yijing Z., Ohtani H., Yuasa H. Long persistent phosphorescence of crystalline phenylboronic acid derivatives: photophysics and a mechanistic study. ChemPhotoChem. 2017;1(3):102–106. doi: 10.1002/cptc.201600031. [DOI] [Google Scholar]
- 46.Back S.H., Park J.H., Cui C., Ahn D.J. Bio-recognitive photonics of a dna-guided organic semiconductor. Nat. Commun. 2016;7(1) doi: 10.1038/ncomms10234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang J., Park J.H., Back S.H., Feng Y., Cui C., Jin L.Y., Ahn D.J. Mercury ion–dna specificity triggers a distinctive photoluminescence depression in organic semiconductor probes guided with a thymine-rich oligonucleotide sequence. Nanoscale. 2018;10(37):17540–17545. doi: 10.1039/C8NR03879A. [DOI] [PubMed] [Google Scholar]
- 48.Shin S., Cho Y.H., Park J.H., Ahn D.J. Light-emitting crystals of aptamer-hybrid organic semiconductor signaling on human cells expressing epcam. J. Ind. Eng. Chem. 2022;116:268–275. doi: 10.1016/j.jiec.2022.09.017. [DOI] [Google Scholar]
- 49.Seo J.S., Liu H., Cho Y.H., Jung W.H., Kim S., Ahn D.J. Triple-peak photoluminescence of dna-hybrid alq3 crystals emitting a depressed single peak upon bio-recognition. ACS Appl. Mater. Interfaces. 2023 doi: 10.1021/acsami.2c21946. [DOI] [PubMed] [Google Scholar]
- 50.Bates P.J., Laber D.A., Miller D.M., Thomas S.D., Trent J.O. Discovery and development of the g-rich oligonucleotide as1411 as a novel treatment for cancer. Exp. Mol. Pathol. 2009;86(3):151–164. doi: 10.1016/j.yexmp.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cong R., Das S., Bouvet P. vols. 185–212. 2011. The multiple properties and functions of nucleolin. (The Nucleolus). [Google Scholar]
- 52.Santos T., Salgado G.F., Cabrita E.J., Nucleolin C. Cruz. A binding partner of g-quadruplex structures. Trends Cell Biol. 2022 doi: 10.1016/j.tcb.2022.03.003. [DOI] [PubMed] [Google Scholar]
- 53.Liu Y., Jing X., Hu X., Wu G. Morphology of strontium carbonate particle adjusted by phthalic acid and isophthalic acid. IOP Conf. Ser. Mater. Sci. Eng. 2018;389(1) doi: 10.1088/1757-899X/389/1/012020. [DOI] [Google Scholar]
- 54.Zhao W., He Z., Lam J.W., Peng Q., Ma H., Shuai Z., Bai G., Hao J., Tang B.Z. Rational molecular design for achieving persistent and efficient pure organic room-temperature phosphorescence. Chem. 2016;1(4):592–602. doi: 10.1016/j.chempr.2016.08.010. [DOI] [Google Scholar]
- 55.Brancaleon L., Lin G., Kollias N. The in vivo fluorescence of tryptophan moieties in human skin increases with uv exposure and is a marker for epidermal proliferation. J. Invest. Dermatol. 1999;113(6):977–982. doi: 10.1046/j.1523-1747.1999.00799.x. [DOI] [PubMed] [Google Scholar]
- 56.Benson R., Meyer R., Zaruba M., McKhann G. Cellular autofluorescence--is it due to flavins? J. Histochem. Cytochem. 1979;27(1):44–48. doi: 10.1177/27.1.438504. [DOI] [PubMed] [Google Scholar]
- 57.König K. Clinical multiphoton tomography. J. Biophot. 2008;1(1):13–23. doi: 10.1002/jbio.200710022. [DOI] [PubMed] [Google Scholar]
- 58.Kozlova A.A., Verkhovskii R.A., Ermakov A.V., Bratashov D.N. Changes in autofluorescence level of live and dead cells for mouse cell lines. J. Fluoresc. 2020;30(6):1483–1489. doi: 10.1007/s10895-020-02611-1. [DOI] [PubMed] [Google Scholar]
- 59.Urasaki Y., Johlfs M.G., Fiscus R.R., Le T.T. Imaging immune and metabolic cells of visceral adipose tissues with multimodal nonlinear optical microscopy. PLoS One. 2012;7(6) doi: 10.1371/journal.pone.0038418. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.