Abstract
Studies of the Hepatitis C virus (HCV) replication cycle have been made possible with the development of subgenomic selectable RNAs that replicate autonomously in cultured cells. In these replicons the region encoding the HCV structural proteins was replaced by the neomycin phosphotransferase gene, allowing the selection of transfected cells that support high-level replication of these RNAs. Subsequent analyses revealed that, within selected cells, HCV RNAs had acquired adaptive mutations that increased the efficiency of colony formation by an unknown mechanism. Using a panel of replicons that differed in their degrees of cell culture adaptation, in this study we show that adaptive mutations enhance RNA replication. Transient-transfection assays that did not require selection of transfected cells demonstrated a clear correlation between the level of adaptation and RNA replication. The highest replication level was found with an adapted replicon carrying two amino acid substitutions located in NS3 and one in NS5A that acted synergistically. In contrast, the nonadapted RNA replicated only transiently and at a low level. The correlation between the efficiency of colony formation and RNA replication was corroborated with replicons in which the selectable marker gene was replaced by the gene encoding firefly luciferase. Upon transfection of naive Huh-7 cells, the levels of luciferase activity directly reflected the replication efficiencies of the various replicon RNAs. These results show that cell culture-adaptive mutations enhance HCV RNA replication.
The Hepatitis C virus (HCV) is one of the most common etiologic agents of chronic liver diseases (reviewed in reference 37). Acute infections are usually subclinical or associated with mild symptoms, but the virus persists in more than 70% of infected individuals. The long-term outcomes of these persistent infections are varied, and they can range from an apparently healthy carrier state to chronic active hepatitis, liver fibrosis, cirrhosis, and eventually hepatocellular carcinoma.
HCV was classified in the genus Hepacivirus of the family Flaviviridae, to which the genera Flavivirus (with the prototype member Yellow fever virus) and Pestivirus (with the representative member Bovine viral diarrhea virus) belong (33). These viruses have enveloped particles that harbor an RNA genome of positive polarity. The HCV genome has a length of about 9,600 nucleotides, and it carries a single long open reading frame (ORF) that is flanked at both termini by nontranslated regions (NTRs). Viral proteins are generated as a polyprotein precursor that is translated via the internal ribosome entry site (IRES) located in the 5′ NTR (45, 47). It permits the direct binding of the 40S ribosome subunit in the absence of additional translation factors (35). The polyprotein precursor is cleaved by viral and host cell enzymes into at least 10 different products (for recent reviews, see references 2 and 39). The structural proteins that are located in the amino-terminal region of the polyprotein are the core protein and the envelope glycoproteins E1 and E2 (22). The nonstructural proteins NS2, NS3, NS4A, NS4B, NS5A, and NS5B are separated from the structural proteins by the short hydrophobic polypeptide p7, which has an unknown function (27, 32). NS2 and the amino-terminal region of NS3 constitute the NS2-3 proteinase responsible for cleavage at the NS2/3 site (19, 21). Three enzymatic activities reside in NS3: a serine-type proteinase in the ∼180 amino-terminal residues and nucleoside triphosphatase- helicase activities that are located in the remainder of the protein (3, 20, 26, 41, 44). NS4A is an essential cofactor of the NS3 proteinase that forms a stable heterodimeric NS3-NS4A complex that mediates cleavages at the NS3/4A, NS4A/B, NS4B/5A, and NS5A/B sites (4, 14, 28, 42). The function of NS4B is not known. NS5A is a highly phosphorylated polypeptide that may be involved in resistance to the antiviral activity of alpha interferon (12, 13, 16, 17). Phosphorylation is mediated by an as yet unknown cellular kinase (23, 40, 43). A major phosphoacceptor site has been mapped for a genotype 1a isolate, but this site is not conserved with NS5A proteins of other HCV genotypes (38). Most HCV isolates contain two phosphoprotein variants with apparent molecular masses of 56 and 58 kDa corresponding to the basal and the hyperphosphorylated forms, respectively (25, 40, 43). It is not known whether NS5A has a direct role in RNA replication and whether phosphorylation is important for its function(s). NS5B is the RNA-dependent RNA polymerase (RdRp) (5, 31, 48).
The development of selectable subgenomic HCV replicons for the first time enabled the study of viral RNA replication in cell culture (30). These replicons are composed of the 5′ NTR, which directs translation of the gene encoding the neomycin phosphotransferase; the IRES of the Encephalomyocarditis virus (EMCV), which directs translation of the HCV NS3-NS5B region; and the 3′ NTR (Fig. 1). Upon transfection of cells of the human hepatoma cell line Huh-7 and selection with G418, cell lines that carried high levels of self-replicating HCV RNAs could be established. Subsequent analyses of the sequences of HCV RNAs isolated from selected cell lines revealed that these replicons harbored cell culture-adaptive mutations (6, 29). For instance, a single amino acid substitution in the NS5B RdRp increased the efficiency of colony formation (ECF) ∼500-fold compared with that of the unaltered replicon RNA (29). However, the mechanism by which this cell culture adaptation was achieved remained unknown. In this study we show that cell culture-adaptive mutations enhance RNA replication. Using a transient-transfection assay without subsequent selection of cells, we demonstrate a clear correlation between the level of cell culture adaptation and RNA replication. However, within selected cell lines that were obtained after transfection of replicons with different levels of adaptation, the differences in the replicon copy numbers per cell varied only slightly. These results suggest that the host cell plays an additional important role in determining the replication level.
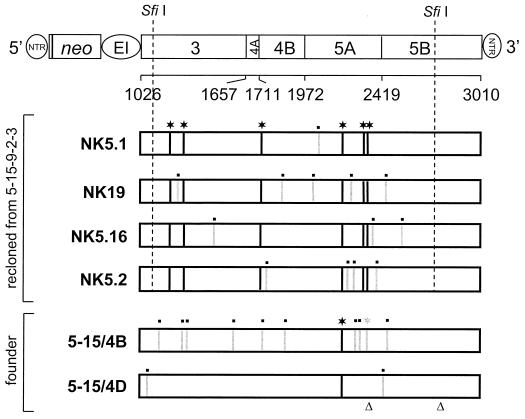
FIG. 1.
Sequence analysis of HCV replicons isolated from various cell lines. The structure of the selectable replicon construct is shown in the top, with numbers below the NS3-NS5B polyprotein referring to the P1 positions of the cleavage sites or its carboxy terminus. neo, neomycin phosphotransferase gene; EI, EMCV IRES. Since for optimal HCV IRES activity ∼36 nt of the core ORF are required, 12 amino acid residues of the core protein are fused to the amino terminus of the neomycin phosphotransferase. Coding regions of the NS3-NS5B polyprotein cloned from cell line 5-15-9-2-3 and the founder line 5-15 are drawn below. Differences in the amino acid sequences are indicated by vertical lines. A black line labeled with a star refers to an amino acid substitution that was conserved with the four sequences isolated from 5-15-9-2-3 cell lines, and gray lines labeled with a dot indicate nonconserved mutations. Note that both replicons cloned from the founder cell line 5-15 carried the proline substitution in NS5A at position 2197 but that the glutamate substitution in NS5A at position 2350 (labeled with a gray star) was found in only one of these replicons (clone 5-15/4B). The positions of single nucleotide deletions in the polyprotein coding sequences of clone 5-15/4D are indicated with triangles.
MATERIALS AND METHODS
Cell cultures.
Cell monolayers of the human hepatoma cell line Huh-7 (34) were routinely grown in Dulbecco's modified minimal essential medium (DMEM; Life Technologies, Karlsruhe, Germany) supplemented with 2 mM l-glutamine, nonessential amino acids, 100 U of penicillin, 100 μg of streptomycin, and 10% fetal calf serum. G418 (Geneticin; Life Technologies) was added at a final concentration of 500 to 1,000 μg/ml to cell lines carrying HCV replicons.
Plasmid constructions.
All nucleotide numbers refer to a complete HCV genome cloned by our group (EMBL database accession number AJ238799). Construction of the vector pFK and the parental replicon construct pFK-I389neo/NS3-3′/wt (EMBL database accession number AJ242654) has been described recently (29, 30). To analyze adapted replicon variants from cell line 5-15-9-2-3 for functionality, SfiI-restricted DNA fragments (nucleotides [nt] 3622 to 8499) that were generated by long-distance reverse transcription PCR (RT-PCR) were transferred into the parental replicon pFK-I389neo/NS3-3′/wt. These constructs were designated 5.1 to 5.16 and 1 to 66. Of the four clones with the highest ECFs (5.1, 5.2, 5.16, and 19), the HCV sequence between the two SfiI sites was determined, and with clone 5.1, each mutation leading to an amino acid substitution was introduced individually into the parental replicon. Plasmids pFK2109 Asn → Asp and pFK2197 Ser → Pro were generated by transferring MluI-EcoRI (nt 6529 to 6699) and EcoRI-XhoI (nt 6699 to 7186) fragments, respectively, from pFK5.1 into pFK-I389neo/NS3-3′/wt. To obtain plasmid pFK1202 Glu → Gly/1280 Thr → Ile, which harbors both NS3 mutations, a PmeI-BstXI fragment (5′ end of the EMCV IRES up to nt 4319) from pFK5.1 was transferred into the parental replicon construct. These two mutations were separated by transferring a PmeI-EagI (nt 3991) or an EagI-EcoRI fragment that was obtained from this construct into the parental replicon, resulting in plasmid pFK1202 Glu → Gly or pFK1280 Thr → Ile, respectively. Plasmid pFK1757 Leu → Ile was generated by transferring a SalI-MluI fragment (nt 4725 to 6529) into pFK-I389neo/NS3-3′/wt. To obtain plasmids pFK2327 Pro → Ser and pFK2350 Lys → Glu, XhoI-PshAI (nt 7186 to 7338) and MluI-SpeI fragments (nt 6529 up to the 3′ end of the HCV sequence), respectively, were inserted into the parental replicon construct. Combinations of individual NS3 mutations with the major adaptive mutation in NS5A (2197 Ser → Pro) were obtained by transferring the EcoRI-XhoI fragment from pFK2197 Ser → Pro into pFK1202 Glu → Gly or pFK1280 Thr → Ile. The presence of each mutation was confirmed by sequence analysis. To generate the corresponding luciferase plasmids for the transient-replication assay, the neo gene was replaced by the gene encoding the luciferase of the firefly Photinus pyralis by using the AscI and PmeI restriction sites. These sites were introduced at the 5′ and 3′ ends of the luciferase gene by PCR.
Preparation of total RNA and quantification of HCV RNA by Northern blotting.
The methods to prepare total RNA and quantify HCV RNA by Northern blotting have been described recently (29). In brief, total RNA was prepared by a single-step isolation method (8), denatured by treatment with 5.9% glyoxal in a solution containing 50% dimethyl sulfoxide and 10 mM sodium phosphate buffer (pH 7.0), and analyzed after denaturing agarose gel electrophoresis by Northern blotting. Prior to hybridization, the membrane was stained with methylene blue and cut ∼1 cm below the 28S rRNA band and the upper strip containing the HCV replicon RNA was hybridized with a 32P-labeled negative-sense riboprobe complementary to the 3′ end of the NS5B region and part of the 3′ NTR (nt 8362 to 9408). The lower strip that was hybridized with a β-actin-specific antisense riboprobe was used to correct for total RNA amounts loaded in each lane of the gel. Specific bands were quantitated by phosphorimaging with a BAS 2500 scanner (Fuji), and the number of replicon molecules was determined by comparison with a serial dilution of in vitro transcripts loaded in parallel onto the gel.
Determination of the copy number of replicon RNAs.
Owing to the dependence of RNA replication on host cell proliferation (36), copy numbers could not be determined from single measurements. Therefore, 5 × 105 cells that had been regularly passaged three times a week at a dilution of 1:3 to 1:5 in the presence of 500 μg of G418 per ml were seeded in a 6-cm-diameter cell culture dish and harvested at daily intervals 3 to 6 days after seeding. The amounts of replicon RNA were determined by Northern blotting analysis as described above. For each replicon, at least three independent cell lines were examined.
Amplification of replicon RNA by RT-PCR and cloning of amplified DNA fragments.
Amplification of replicon RNAs was done by long-distance RT-PCR using a mixture of polymerases (7). One microgram of total RNA and 50 pmol of primer A9413 (CAG GAT GGC CTA TTG GCC TGG AG) were mixed in a total volume of 10.5 μl and denatured for 10 min at 65°C. Reverse transcription was performed with Expand-RT (Roche Biochemicals, Mannheim, Germany) in a total volume of 20 μl, and after 1 h at 42°C, different amounts of the reaction mixture were used for PCR with the Expand Long Template PCR system (Roche Biochemicals) and primers S59 (TGT CTT CAC GCA GAA AGC GTC TAG) and A9386 (TTA GCT CCC CGT TCA TCG GTT GG). Cycle conditions were two min of initial denaturation at 94°C and 40 cycles of 10 s at 94°C, 90 s at 54°C, and 540 s at 68°C. After 10 cycles, the extension time was increased 10 s for each additional cycle. After a final 10-min incubation at 68°C, PCR products were purified by preparative agarose gel electrophoresis, restricted with SfiI, and inserted into the parental construct pFK-I389neo/NS3-3′/wt.
Sequence analysis.
Sequences were verified using the Thermo Sequenase Fluorescently Labeled Primer Cycle Sequencing kit with 7-deaza-dGTP (Amersham Pharmacia Biotech, Freiburg, Germany) and IRD-41-labeled primers (MWG-Biotech, Ebersberg, Germany) according to the instructions of the manufacturer. Reaction mixtures were analyzed on a model 4000 Licor DNA sequencer (MWG-Biotech).
In vitro transcription, electroporation, and selection of G418-resistant cell lines.
In vitro transcripts were generated using the protocol described recently (29). In brief, plasmid DNA was restricted with AseI and ScaI (New England Biolabs, Bad Schwalbach/Taunus, Germany) and after extraction with phenol and chloroform and precipitation with ethanol dissolved in RNase-free water. In vitro transcription reaction mixtures contained 80 mM HEPES (pH 7.5), 12 mM MgCl2, 2 mM spermidine, 40 mM dithiothreitol (DTT), 3.125 mM each nucleoside triphosphate, 1 U of RNasin (Promega, Mannheim, Germany) per μl, 0.1 μg of restricted plasmid DNA per μl, and 0.6 U of T7 RNA polymerase (Promega) per μl. After 2 h at 37°C, an additional 0.3 U of T7 RNA polymerase per μl was added and the reaction mixture was incubated for another 2 h. Transcription was terminated by the addition of 1.2 U of RNase-free DNase (Promega) per μg of plasmid DNA and 30 min of incubation at 37°C. After one extraction with acidic phenol and chloroform, DNA was precipitated with isopropanol and dissolved in RNase-free water. The concentration was determined by measurement of the optical density at 260 nm, and RNA integrity was checked by denaturing agarose gel electrophoresis. The conditions for electroporation and selection of G418-resistant cells have been described in detail elsewhere (29). Briefly, 0.25 to 500 ng of in vitro transcripts adjusted with total RNA from naïve Huh-7 cells to a final amount of 10 μg was mixed with 400 μl of a suspension of 107 Huh-7 cells per ml. Electroporation conditions were 960 μF and 270 V using a Gene pulser system (Bio-Rad, Munich, Germany) and a cuvette with a gap width of 0.4 cm (Bio-Rad). Cells were immediately transferred to 8 ml of complete DMEM containing 1.25% dimethyl sulfoxide and seeded in a 10-cm-diameter cell culture dish. After 24 h, medium was replaced by complete DMEM supplemented with 500 μg of G418 per ml. Medium was changed weekly, and 3 to 4 weeks after electroporation, colonies were stained with Coomassie brillant blue (0.6 g/liter in 50% methanol–10% acetic acid). To determine the number of CFU of a given RNA, serial dilutions from 300 to 0.25 ng of RNA were transfected into Huh-7 cells and processed further as described above. For each replicon, 5 to 20 independent transfections were performed.
Transient-replication assays.
Huh-7 cells were transfected by electroporation as described above using 7.5 μg of a neo replicon or 5 μg of a luciferase replicon. After addition of 10 ml of complete DMEM, 1- to 2-ml aliquots of the cell suspension were seeded in a 3-cm-diameter culture dish and harvested at time points given in Results. For Northern blot analysis, total RNA was analyzed as described above. With luciferase replicons, cells were washed three times with phosphate-buffered saline (PBS) and scraped off the plate into 350 μl of ice-cold lysis buffer (1% Triton X-100, 25 mM glycylglycine, 15 mM MgSO4, 4 mM EGTA, 1 mM DTT). One hundred microliters of lysate was mixed with 360 μl of assay buffer (25 mM glycylglycine, 15 mM MgSO4, 4 mM EGTA, 1 mM DTT, 2 mM ATP, 15 mM K2PO4 [pH 7.8]) and, after addition of 200 μl of a 200 μM luciferin stock solution, measured in a luminometer (Lumat LB9507 from Berthold, Freiburg, Germany) for 20 s. Values obtained with cells harvested 4 h after electroporation were used to determine the transfection efficiency.
Immunofluorescence.
Transfected cells (0.7 × 105) were seeded on glass coverslips and, after 4, 15.5, 24, 48, 72, and 96 h, washed three times with PBS. Cell were fixed in an ice-cold mixture of acetone and methanol (90 and 10%) for about 10 min at −20°C and washed thereafter three times with PBS, followed by a 1-h incubation in IF buffer (PBS, 3% bovine serum albumin, 0.1% Triton X-100) at 4°C. An NS3B-specific rabbit polyclonal antibody (3) was added at a dilution of 1:500 in IF buffer, and after 1 h, cells were washed three times with PBS, followed by incubation with a 1:100 dilution of an anti-rabbit antibody conjugated with fluorescein isothiocyanate (Sigma, Deisenhofen, Germany) in IF buffer. Coverslips were washed with PBS and mounted on glass slides with Permafluor (Immunotech, Marseille, France), and cells were examined under a fluorescence microscope (Zeiss, Jena, Germany).
RESULTS
Construction of a cell culture-adapted HCV replicon RNA.
To study the mechanism of cell culture adaptation, a highly efficient replicon RNA was required. Although we had recently identified a single amino acid substitution in NS5B that increased the ECF ∼500-fold (29), we attempted to develop an even more efficient replicon. On the assumption that adaptive mutations might accumulate during serial passage of replicon RNAs, total RNA was isolated from a cell line that harbored an NS3-5B replicon (cell line 5-15 [30]) and transfected into naïve Huh-7 cells. After stringent selection with 1 mg of G418 per ml, a fast-growing cell clone was isolated and total RNA was prepared and transfected into parental Huh-7 cells. After three successive passages, cell line 5-15-9-2-3 was obtained. To identify cell culture-adaptive mutations, nearly full-length replicon RNAs were amplified by long-distance RT-PCR using primers S59 and A9386 and, after restriction of the amplified DNA fragments with SfiI, almost the complete HCV ORF was inserted into the parental replicon construct (Fig. 1). In vitro transcripts from 82 different clones were prepared and transfected into naïve Huh-7 cells to determine their ECFs. As summarized in Table 1, 69 clones were replication defective, because no G418-resistant colonies were obtained. Nine of the tested clones were comparable to the wild type (20 to 50 colonies), whereas four clones were more efficient. The sequence analysis of the SfiI fragments of these clones revealed several amino acid substitutions (Fig. 1). Six of them were found in all four clones derived from the cell line 5-15-9-2-3, whereas the number of nonconserved amino acid substitutions was variable. A careful titration of in vitro transcripts derived from the two most efficient clones, which were designated 5.1 and 19, revealed that replicon 5.1 had the highest ECF (∼500,000 CFU per μg of RNA) and that replicon 19 was about sevenfold less efficient (∼70,000 CFU/μg).
TABLE 1.
ECF of HCV RNAs derived from replicon sequences recloned from cell line 5-15-9-2-3
| No. of clones tested |
CFU/μg of RNA |
|---|---|
| 69 | 0 |
| 9 | 1–100 |
| 1 | 100–1,000 |
| 1 | 1,000–10,000 |
| 2 | >10,000 |
Determination of adaptive mutations in the HCV coding sequence.
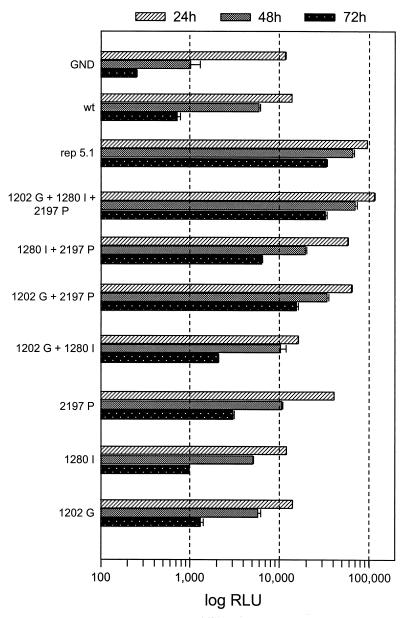
Since adaptive mutations should be generated early during selection and become fixed in all RNA progeny, we focused our further analyses on the conserved amino acid substitutions. Three of these six substitutions were located in NS5A, and two were located in NS3. Only one conserved mutation was found in NS4B, and none were found in NS4A and NS5B (see below). Interestingly, the conserved mutation in the center of NS5A at position 2197 of the HCV polyprotein was also present in two clones that were isolated from the founder cell line 5–15 (Fig. 1). In contrast, a second conserved substitution in NS5A at position 2350 was found in only one of these two clones (5–15/4B). To identify which of the six conserved mutations were responsible for cell culture adaptation, they were introduced individually into the parental replicon construct and serial dilutions of in vitro transcripts were transfected into naïve Huh-7 cells. After selection with G418, the ECF was determined. The results shown in Fig. 2 demonstrate that the most adaptive mutation was the one located in the center of NS5A at amino acid position 2197 of the polyprotein. In addition, the glycine substitution for glutamic acid and the isoleucine substitution for threonine in NS3 at positions 1202 and 1280, respectively, increased the ECF too, albeit to a much lesser extent. All other mutations did not affect the number of G418-resistant cell colonies. Interestingly, the highly adaptive NS5A substitution, but not the NS3 mutations, was already found in the replicons isolated from the founder cell line 5–15. Thus, the initial adaptive mutation was the one found in NS5A, and this mutation remained conserved during cell culture passage of the replicon RNAs. The additional adaptive mutations in NS3 must have been acquired at a later stage or were present only in a minor fraction of replicons in the founder cell line and accumulated during successive cell culture passage. Owing to the high selective pressure we used, the presence of these three mutations in the replicon RNA appeared to confer a selection advantage. In agreement with this assumption, we found that these substitutions were synergistic. When the two NS3 mutations were combined, the ECF of the replicon was ∼4-fold higher than that of the RNA harboring only the adaptive substitution at position 1280. However, when both NS3 mutations were combined with the adaptive NS5A substitution at position 2197, the ECF of the triple mutant was dramatically increased compared with the replicon that harbored only this NS5A substitution (Fig. 2). In fact, the G418 transduction efficiency of the triple mutant was as high as the one obtained with replicon 5.1. Since the analysis of sequences of the HCV ORF flanking the SfiI fragment did not reveal additional amino acid substitutions in NS3 and NS5B that were conserved between the four replicons recloned from cell line 5-15-9-2-3, we had identified the major adaptive mutations.
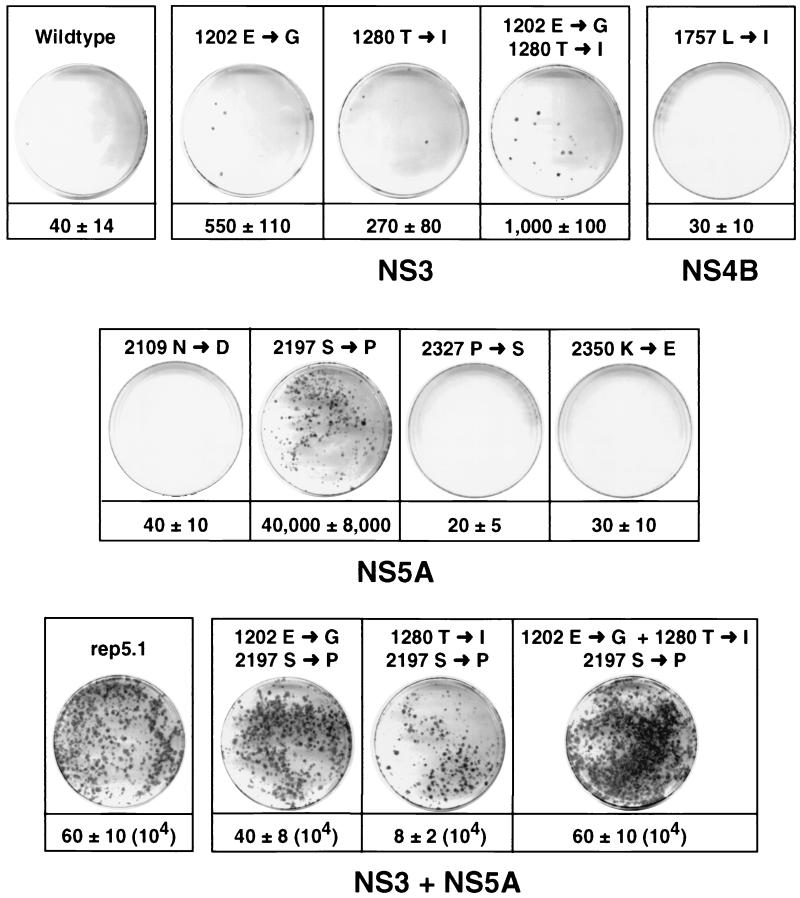
FIG. 2.
Identification of cell culture-adaptive mutations. Huh-7 cells were transfected with the parental replicon 5.1 or replicon RNAs carrying the mutations given above each plate. Numbers below the plates refer to the CFU per microgram of in vitro-transcribed replicon RNA. For comparison, representative plates that were obtained after transfection of each 10 ng of in vitro-transcribed replicon RNA or 1 ng in the cases of the replicons shown in the bottom line are shown. Note that for the determination of the CFU, serial titrations down to 0.25 ng of each RNA were transfected. With the nonadapted replicons, five independent transfections were performed, but with the adapted RNAs, 20 to 50 independent transfections were performed.
Transient replication of HCV RNAs in nonselected cells.
In all experiments performed so far we had measured RNA replication by determining the G418 transduction efficiency. Since this approach did not allow the direct determination of replication kinetics, transient-replication assays in nonselected cells were performed. For this approach we chose the most adapted RNAs, 5.1 and 19, and replicon 9-13F, which had an ECF that was ∼100-fold lower than that of RNA 5.1 (Table 2). Replicon 9-13F carried one essential adaptive mutation in NS5A at position 2163 of the polyprotein (29). A derivative of replicon 5.1 that carried a 10-amino-acid residue deletion spanning the active site of the NS5B RdRp served as a negative control (rep5.1/Δ5B) (Fig. 3A). Huh-7 cells were transfected with these neo replicons, and total RNA that was prepared after 3.5, 15, 24, 48, 72, and 96 h was analyzed by Northern blotting to determine the copy numbers of the replicons. The representative result in Fig. 3A shows that input RNA was rapidly degraded and barely detectable 15 h after transfection (lane 28). In the low-level adaptation replicon 9-13F, a much slower gradual reduction of HCV RNA could be observed, and it became undetectable at 96 h posttransfection (lanes 21 to 26). In contrast, an increase of the copy numbers of HCV RNAs was found with the two highly adapted replicons. A careful quantification of the Northern blot by phosphorimaging revealed that the copy numbers of both RNAs increased 24 h after transfection (Fig. 3B), which is the time when transfected cells started growing. In replicon 5.1, at 72 h posttransfection the level of replicating RNA reached a maximum and declined within the next 24 h, when cells became confluent. A similar kinetic was found with replicon 19, but the copy number at the time of the peak was ∼3-fold lower. As described above, the decline in the amounts of replicon RNAs in confluent cells (96 h) was most likely due to the tight linkage between RNA replication and host cell proliferation (36).
TABLE 2.
CFU of adapted replicons and copy numbers of HCV RNAs in cell lines obtained after transfection of given replicons and G418 selection
| Replicon | CFU/μg of RNAa | No. of copiesb |
|---|---|---|
| 19 | 72,000 ± 5,000 | 5 ± 1.6 × 107 |
| 5.1 | 510,000 ± 75,000 | 6.9 ± 1.4 × 107 |
| 9-13F | 4,250 ± 650c | 2.6 ± 0.5 × 107 |
| 5B2884Gly | 23,000 ± 3,000c | 5 ± 2.5 × 107 |
Mean values and standard deviations derived from 20 to 50 independent transfections of each replicon RNA.
Number of HCV replicon RNA molecules per microgram of total RNA as determined by Northern blotting of three independent cell lines of each replicon (Fig. 7). Mean values and standard deviations are from a total of three determinations for each cell line.
Data are from the work of Lohmann et al. (29).
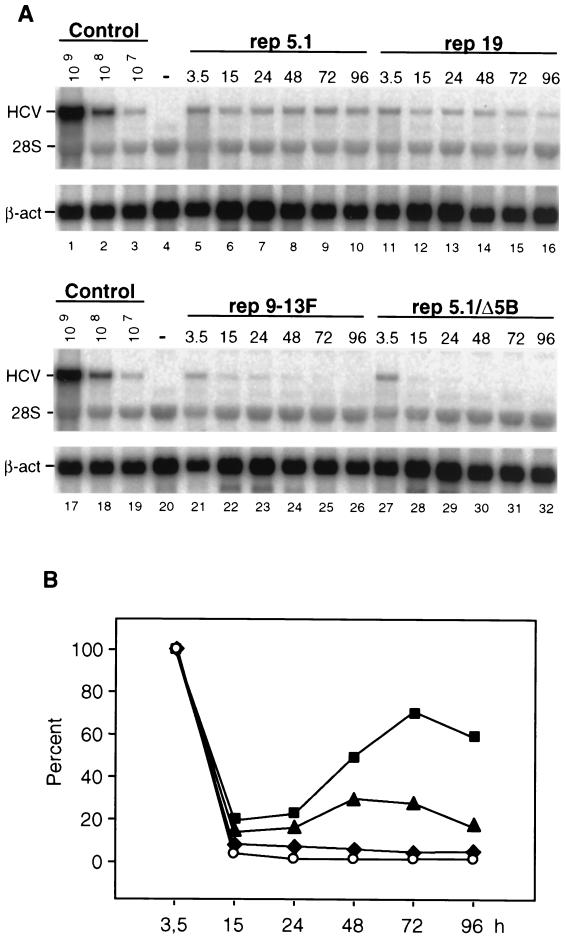
FIG. 3.
Transient replication of cell culture-adapted HCV replicons. (A) Huh-7 cells were transfected with selectable replicon RNAs and harvested at time points indicated above each lane. HCV RNAs were analyzed by Northern blotting. The number of replicon RNA molecules was determined by comparison with the serial dilution of in vitro transcripts (lanes 1 to 3). β-Actin RNA (β-act) served as a control to correct for the amount of total RNA loaded in each lane of the gel (∼2 μg). The result obtained with total RNA from naïve Huh-7 cells is shown in lanes 4 and 20. The positions of replicon RNAs, 28S RNA, and β-actin m-RNA are given in the left. The replicon carrying a deletion that spans the active site of the NS5B RdRp served as a negative control (rep5.1/Δ5B). (B) Quantification of the RNA in the Northern blots shown in panel A by phosphorimaging. Values were corrected for the RNA amounts determined 3.5 h after transfection, and they are expressed as percentages. With rep5.1, the 100% value corresponds to 1.3 × 107 replicon RNA molecules per μg of total RNA. ■, replicon 5.1; ▴, replicon 19; ⧫, replicon 9-13F; ○, replicon 5.1/Δ5B. Analogous results were obtained in three further independent experiments.
The different efficiencies of RNA replication were reflected by the levels of HCV protein expression. As shown by immunofluorescence, 4 h after electroporation all transfected cells were positive for NS3 (Fig. 4). In contrast, at 96 h posttransfection the antigen was found only in cells transfected with the highly adapted RNAs 5.1 and 19, whereas the signal found in cells with the poorly adapted replicon 9-13F was hardly above the background. Overall, the replication kinetics determined by Northern blotting and immunofluorescence in this transient-replication assay were in good agreement with the ECF of each replicon RNA, suggesting that cell culture adaptation is mediated by a direct enhancement of RNA replication.
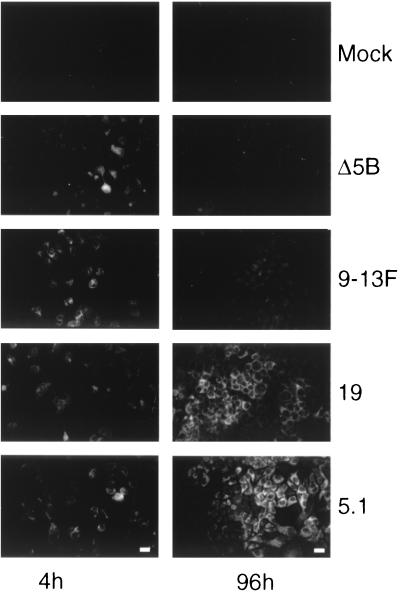
FIG. 4.
Immunofluorescence analysis of transiently replicating HCV RNAs. A portion of transfected Huh-7 cells (Fig. 3) was seeded onto glass coverslips. NS3 was detected by immunofluorescence at 4 and 96 h posttransfection (left and right sides, respectively). Mock-transfected Huh-7 cells served as a negative control. Bar = 50 μm.
Transient replication of HCV replicons carrying a luciferase reporter gene.
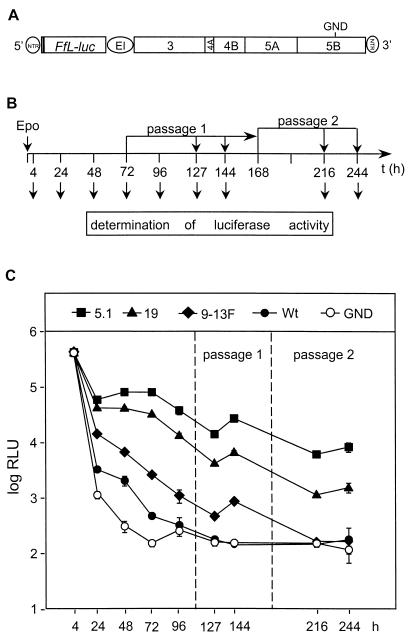
To further substantiate this assumption and to simplify the subsequent analyses, we replaced the neo genes in the various cell culture-adapted replicons by the gene encoding the luciferase of the firefly P. pyralis (Fig. 5A). Since the copy number of this reporter gene should be determined by the replication levels of the corresponding replicon RNAs, the luciferase activity in a lysate of transfected cells could be used to directly monitor the replication of an RNA. Huh-7 cells were transfected by electroporation, and each 1/10 of the cells was seeded into a cell culture dish. Cells were harvested at various time points, and luciferase activities were determined. As a negative control, a derivative of the wild-type replicon that carried a single amino acid substitution changing the GDD motif of the RdRp active site to GND was transfected in parallel. Luciferase activities measured 4 h after transfection were used to determine the transfection efficiencies. A representative result is shown in Fig. 5. In this experiment we also analyzed the replication of the HCV RNAs in cells after passage, because we wanted to avoid the reduction of replicon RNA levels when the cells reached confluence (36). Therefore, cells in one culture dish were passaged at a dilution of 1:3 at 72 h posttransfection and passaged a second time 96 h later (Fig. 5B). Cells were harvested at given time points to determine the luciferase activities. Overall, the replication kinetics observed with these luciferase replicons were very similar to the ones obtained with the neo replicons that were analyzed by Northern blot analysis up to 96 h after transfection (Fig. 5C). At every determined time point (except 4 h posttransfection), luciferase activities obtained with the most highly adapted RNA, 5.1, were consistently the highest whereas those found in 9-13F-transfected cells were much lower. Interestingly, in the cells passaged at 72 h posttransfection, an increase in luciferase activity was clearly visible with replicons 5.1, 19, and 9-13F (compare the values obtained 127 and 144 h posttransfection). When these cells were passaged a second time 96 h after the first passage, only in cells transfected with the highly adapted replicons 5.1 and 19 could a second increase of luciferase activity be observed (compare the values at 216 and 244 h posttransfection). These results show that, in unselected cells, the replicons were gradually lost, with the level of decline being determined by the level of cell culture adaptation.
FIG. 5.
Transient replication of HCV replicons carrying a reporter gene. (A) Structure of a replicon construct harboring a firefly luciferase gene (Ffl-luc). GND indicates the position of the amino acid substitution of the NS5B RdRp. For further details, see the legend to Fig. 1. (B) Flow chart of the experimental approach. Huh-7 cells were transfected by electroporation (Epo), and 1/10 of each was seeded in a culture dish. After 4, 24, 48, 72, and 96 h, cells were lysed and luciferase activities were determined. Cells that were cultured for 72 h were passaged at a dilution of 1:3. One-third of each was harvested at time points (t) corresponding to 127 and 144 h after transfection, whereas the remainder were again passaged at a time point corresponding to 168 h posttransfection. Cells of this second passage were lysed at time points corresponding to 216 and 244 h after transfection. (C) Representative results obtained with the replicons specified at the top. Values for each time point correspond to the mean and the error range of quadruplicate results. Note that, owing to low variations, error bars are in most cases not visible in the graph. Values are corrected for transfection efficiency as determined by measuring the luciferase activity 4 h after transfection. Wt, wild type.
Owing to the high sensitivity of the luciferase reporter, we also determined the replication kinetic of the parental, nonadapted replicon RNA. As shown in Fig. 5C, only at 24, 48, and 72 h posttransfection was a difference found between this replicon and the replication-defective negative control (GND). However, compared to that of even the least-adapted RNA, 9-13F, a dramatic difference in the replication level was found. Therefore, we can conclude that cell culture-adaptive mutations strongly increase RNA replication and that they were generated during a low-level replication of the nonadapted parental replicon. It should be noted that, in more than 10 independent experiments, the absolute values of luciferase activities varied significantly but that, in all cases, the relative differences between the individual replicons remained the same.
Having developed a highly sensitive and fast replication assay for HCV RNAs, we next wanted to confirm the usability of this approach for reverse-genetic studies. To this end, we generated a panel of luciferase replicon RNAs harboring the single amino acid substitutions in NS3 and NS5A we had already analyzed with respect to colony formation (Fig. 2). These replicons were transfected into naïve Huh-7 cells, and luciferase activities were measured at daily intervals up to 96 h posttransfection. The values obtained 4 h after transfection were used to correct for different transfection efficiencies (not shown). For each replicon, a clear correlation was found between the luciferase activities in the transient-transfection assay and the ECF of the corresponding neo construct (Fig. 6). For instance, the replicon harboring the adaptive mutation in NS3 at position 1202 replicated at a rather low level. In contrast, for each time point, the luciferase activities in lysates of cells transfected with the replicon with the adaptive mutation in NS5A (nt 2197, S → P) were higher. When these two mutations were combined, the resulting RNA replicated at a level that was clearly higher than the sum of the replication levels of the RNAs carrying the substitutions individually. A further increase was found with the replicon harboring in addition the substitution in NS3 at position 1280. Although this mutation, when analyzed individually, only slightly increased replication compared with that of the nonadapted replicon, it did so synergistically both in combination with the other more adaptive NS3 substitution at position 1202 and with the adaptive NS5A mutation at position 2197. In fact, the replication level of this triple mutant was as high as the one observed with replicon 5.1, confirming that these NS3 and NS5A substitutions were responsible for cell culture adaptation and enhanced replication levels synergistically. In summary, the results that were based on luciferase HCV replicons correlated well with the results obtained with the neo replicons, both in transient-replication assays and after selection of stable cell lines. Thus, HCV RNAs that harbor a reporter gene can be used to accurately determine replication of the replicon RNA.
FIG. 6.
Transient replication of luciferase replicon RNAs carrying mutations in NS3 and NS5A. Huh-7 cells were transfected with the replicons specified at the left, and luciferase activities were determined in lysates of cells harvested 4, 24, 48, 72, and 96 h (not shown) after transfection. The 4-h value (not shown) was used to correct for different transfection efficiencies. Bars represent the means and the error ranges of quadruplicate results. In most cases, the variations are very small and therefore the error bars are not visible.
Lack of correlation between the ECF and replicon RNA copy number in G418-selected cell lines.
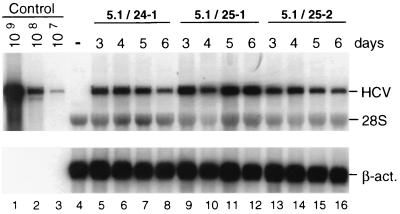
To analyze whether an increase in the ECF and the RNA replication level would directly correlate with the copy number of an HCV RNA in a selected cell line, we took advantage of several HCV replicons that differed in their levels of cell culture adaptation. Replicons 5.1, 19, and 9-13F were the ones described above. Replicon 5B2884Gly harbored a single glycine substitution in NS5B at position 2884 that led to an ∼500-fold increase in the ECF compared with that of the wild type (29) (Table 2). In vitro transcripts of these constructs were transfected into naïve Huh-7 cells, and for each replicon at least three cell clones were isolated and expanded. To determine the copy numbers of these RNAs in selected cells, we had to consider that the replication levels of the replicons depend on host cell metabolism (36). Therefore, copy numbers were determined from serial measurements using cells that had been harvested 3, 4, 5, and 6 days after seeding. A representative example of this analysis is shown in Fig. 7 for three different cell lines that were obtained after transfection with the most highly adapted replicon, 5.1, and a summary for all cell lines is given in Table 2. In agreement with the ECFs, the highest copy number was found in cell lines harboring replicon 5.1. However, in spite of the dramatic differences in the numbers of colonies obtained with a given replicon RNA, the replication levels within a selected cell line differed much less. Even between the replicons with the highest and the lowest levels of adaptation (5.1 and 9-13F, respectively), which varied in their ECFs by a factor of ∼100, the differences in the copy numbers were only ∼2-fold. With the other replicons, the replication levels were comparable. This result suggests that the ECF is primarily determined by the initial level of RNA replication and that, during selection, either particular host cells that support high-level replication of the HCV RNA are enriched or further adaptive mutations are acquired.
FIG. 7.
Determination of replicon RNA levels in selected cell lines, 24-1, 25-1, and 25-2, each harboring the adapted replicon 5.1. Cells were harvested at given times after being seeded, and HCV RNA was analyzed by Northern blotting as described in the legend to Fig. 3. A summary of the data, including analogous quantifications with cell lines harboring other replicon RNAs, is given in Table 2. β-act., β-actin.
DISCUSSION
In this study we developed a novel highly adapted HCV replicon that harbors two synergistic mutations in NS3 and one in NS5A. The ECF of this RNA was ∼5 × 105 CFU per μg of RNA, which is ∼20-fold higher than that of the best-adapted replicon we described recently (29). By analyzing this and several other HCV RNAs that differed with respect to their levels of cell culture adaptation, we found a clear correlation between the ECF and RNA replication as determined by two different transient-transfection assays. These results demonstrate that cell culture-adaptive mutations increase RNA replication. They also provide an explanation of how adaptive replicons are generated. As shown with the parental replicon carrying the luciferase gene, this nonadapted RNA replicated only transiently and at a low level in transfected cells. During this time, mutations must have been generated by the viral RNA polymerase that in a few instances were adaptive. By increasing the level of RNA replication, cells harboring such a replicon developed G418 resistance. Since most mutations did not increase but rather reduced replication or were inactivating, development of an adaptive replicon was a rare event, explaining why the numbers of G418-resistant colonies that were obtained with a nonadapted RNA were very low. Alternatively, the adaptive mutations may have been introduced by the T7 RNA polymerase used for in vitro transcription. However, given the infrequent generation of adaptive mutations, the number of replication-competent RNA molecules in the transcription reaction probably would have been too low to allow detection in the luciferase assay. The fact that we consistently observed a low level of replication with the parental replicon suggests that the adaptive mutations were generated during the initial low-level replication of this RNA.
While a clear correlation was found between replication in the transient-transfection assays and the ECF, this was not the case with respect to the copy number of replicon RNA molecules per cell after selection. For instance, the most highly adapted replicon, 5.1, had an ECF of ∼5 × 105 CFU/μg of RNA and showed the highest initial replication whereas the ECF of the least-adapted replicon was ∼100-fold lower, and this RNA replicated very inefficiently. However, in selected cell lines obtained after transfection with these replicon RNAs, the copy numbers differed only by a factor of ∼2. This result indicates that during selection of cells harboring a poorly adapted replicon, further mutations that increase the replication level are introduced into the RNA. Alternatively, during prolonged passage, we may have selected for particular host cells that support high-level replication. Currently, we cannot distinguish between the two possibilities, but in agreement with the latter assumption, we found that the copy number of replicon RNA was highest in cell line 9-13 (2.5 × 108 per μg of total RNA), which harbored a moderately adapted replicon (rep5B2884Gly with a substitution in NS5B that increased the ECF ∼500-fold over the wild-type level) (29). In contrast, in cell line 5-15-9-2-3 carrying the most highly adapted replicon (rep5.1 with an ECF that is ∼10,000-fold higher than that of the wild type), the replicon copy number was even lower (1.6 × 108 per μg of total RNA). Therefore, the level of cell culture adaptation is determined by the initial replication level, which also determines the level of G418 transduction efficiency. However, during prolonged cell passage and selection, particular host cells that support a higher level of replicon RNA replication may be selected.
By serial cell culture passage of replicon RNAs, we selected for three mutations that enhanced replication synergistically. In the founder cell line, only the highly adaptive substitution in NS5A was found, and it was the only conserved mutation in the two analyzed clones. Thus, the initial adaptation was due to this particular substitution whereas the adaptive mutations in NS3 were generated at a later stage during cell culture passage. Alternatively, these NS3 substitutions might have already been present at a low frequency in the replicons of the founder cell line but were enriched by the stringent selection procedure.
Currently, we can only speculate about the molecular mechanism of cell culture adaptation. There are certainly several ways how it can be achieved. In a previous study we had shown that a single amino acid substitution in NS5B increased the ECF ∼500-fold compared to that of the parental RNA (29). In this report, we identified two substitutions in NS3 and one in the center of NS5A. Three-dimensional structure modeling experiments suggest that the substitution at the carboxy terminus of NS3 (position 1202) is located on the surface of the molecule and probably has no effect on its various enzymatic activities (Neera Borkakoti, personal communication). The same is probably true for the mutation in domain 1 of the NS3 helicase at position 1280. We therefore favor the hypothesis that these amino acid changes affect an interaction site with a cellular or viral protein important for RNA replication. With NS5A, the substitution affected serine 2197, and this residue was shown to be important for hyperphosphorylation (43). The loss of this serine residue in the adapted replicon RNA may suggest that phosphorylation at this position is inhibitory for RNA replication. However, it is not known whether serine 2197 is a phosphoacceptor site or plays some other role in hyperphosphorylation. As deduced from the formation of pp58, the NS5A in cells harboring replicon 5.1 is still hyperphosphorylated (N. Krieger and R. Bartenschlager, unpublished results). However, the appearance of the pp56/58 double band in conventional sodium dodecyl sulfate-polyacrylamide gel electrophoresis may not be sufficient to reveal subtle differences in the phosphorylation patterns between the parental NS5A and the adaptive mutation. Therefore, further genetic and biochemical studies will be required to clarify this important point.
While this paper was in preparation, Blight and coworkers (6) identified several cell culture-adaptive mutations too. Focusing their analysis on NS5A, they identified a total of 10 different mutations that were all located in the center of the molecule. Among these was the proline substitution for serine at position 2197 that we identified here as the most adaptive one. While this mutation reduced only NS5A hyperphosphorylation, it was completely blocked with an isoleucine substitution for serine at position 2204, and this mutation conferred the highest level of adaptation (6). Thus, NS5A hyperphosphorylation appears to be nonessential for HCV RNA replication in cell culture.
Cell culture adaptation by specific mutations in the genome has been described for several other viruses, too. In many cases, adaptation was used for the attenuation of a virus. For instance, with the Sindbis virus a selection procedure was used to isolate a noncytopathic replicon (15). This RNA harbored either one of two single amino acid substitutions in nonstructural protein 2 that reduced the level of RNA replication to 1% of that of the wild-type replicon. This reduction led to a loss of cytopathogenicity concomitant with a cell type restriction (1). Mutations conferring cell culture adaptation by enhancing virus growth have been described for HAV. While the parental virus has a slow-growth property, several cell culture-adapted isolates that replicate more efficiently and to higher titers have been described (e.g., see reference 46). The mutations primarily responsible for adaptation were identified in viral proteins 2B and 2C and in the 5′ NTR (9, 18, 24), although other mutations acquired during adaptation could also contribute to efficient virus growth in cell culture (10, 11). However, the molecular basis for cell culture adaptation is not known.
The development of highly cell culture-adapted HCV replicons opens new avenues for studies of RNA replication, in particular with the help of reverse genetics. These analyses can now be performed in transient-transfection assays, circumventing the problem that, during selection of cells, the effect of a mutation to be studied is masked by second-site reversions or compensations. However, the use of selectable HCV replicons is still a valuable tool, in particular for mutants that replicate only poorly. Therefore, the combination of both assays should help to clarify the mechanism of HCV replication.
ACKNOWLEDGMENTS
We are grateful to Ulrike Herian for excellent technical assistance and to Neera Borkakoti for help with three-dimensional modelling of NS3.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (SFB490, Teilprojekt A2) and the European Community (QLK2-1999-00356).
REFERENCES
- 1.Agapov E V, Frolov I, Lindenbach B D, Pragai B M, Schlesinger S, Rice C M. Noncytopathic Sindbis virus RNA vectors for heterologous gene expression. Proc Natl Acad Sci USA. 1998;95:12989–12994. doi: 10.1073/pnas.95.22.12989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bartenschlager R, Lohmann V. Replication of hepatitis C virus. J Gen Virol. 2000;81:1631–1648. doi: 10.1099/0022-1317-81-7-1631. [DOI] [PubMed] [Google Scholar]
- 3.Bartenschlager R, Ahlborn-Laake L, Mous J, Jacobsen H. Nonstructural protein 3 of the hepatitis C virus encodes a serine-type proteinase required for cleavage at the NS3/4 and NS4/5 junctions. J Virol. 1993;67:3835–3844. doi: 10.1128/jvi.67.7.3835-3844.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartenschlager R, Ahlborn-Laake L, Mous J, Jacobsen H. Kinetic and structural analyses of hepatitis C virus polyprotein processing. J Virol. 1994;68:5045–5055. doi: 10.1128/jvi.68.8.5045-5055.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Behrens S E, Tomei L, DeFrancesco R. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 1996;15:12–22. [PMC free article] [PubMed] [Google Scholar]
- 6.Blight K J, Kolykhalov A A, Rice C M. Efficient initiation of HCV RNA replication in cell culture. Science. 2000;290:1972–1974. doi: 10.1126/science.290.5498.1972. [DOI] [PubMed] [Google Scholar]
- 7.Cheng S, Fockler C, Barnes W M, Higuchi R. Effective amplification of long targets from cloned inserts and human genomic DNA. Proc Natl Acad Sci USA. 1994;91:5695–5699. doi: 10.1073/pnas.91.12.5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 9.Cohen J I, Rosenblum B, Ticehurst J R, Daemer R J, Feinstone S M, Purcell R H. Complete nucleotide sequence of an attenuated hepatitis A virus: comparison with wild-type virus. Proc Natl Acad Sci USA. 1987;84:2497–2501. doi: 10.1073/pnas.84.8.2497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Day S P, Murphy P, Brown E A, Lemon S M. Mutations within the 5′ nontranslated region of hepatitis A virus RNA which enhance replication in BS-C-1 cells. J Virol. 1992;66:6533–6540. doi: 10.1128/jvi.66.11.6533-6540.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Emerson S U, Huang Y K, Purcell R H. 2B and 2C mutations are essential but mutations throughout the genome of HAV contribute to adaptation to cell culture. Virology. 1993;194:475–480. doi: 10.1006/viro.1993.1286. [DOI] [PubMed] [Google Scholar]
- 12.Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murakami T, Yamamoto C, Izumi N, Marumo F, Sato C. Comparison of full-length sequences of interferon-sensitive and resistant hepatitis C virus 1b. Sensitivity to interferon is conferred by amino acid substitutions in the NS5A region. J Clin Investig. 1995;96:224–230. doi: 10.1172/JCI118025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murakami T, Yamamoto C, Ogura Y, Izumi N, Marumo F, Sato C. Mutations in the nonstructural protein 5A gene and response to interferon in patients with chronic hepatitis C virus 1b infection. N Engl J Med. 1996;334:77–81. doi: 10.1056/NEJM199601113340203. [DOI] [PubMed] [Google Scholar]
- 14.Failla C, Tomei L, DeFrancesco R. Both NS3 and NS4A are required for proteolytic processing of hepatitis C virus nonstructural proteins. J Virol. 1994;68:3753–3760. doi: 10.1128/jvi.68.6.3753-3760.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frolov I, Agapov E, Hoffman T A, Pragai B M, Lippa M, Schlesinger S, Rice C M. Selection of RNA replicons capable of persistent noncytopathic replication in mammalian cells. J Virol. 1999;73:3854–3865. doi: 10.1128/jvi.73.5.3854-3865.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gale M J, Korth M J, Tang N M, Tan S L, Hopkins D A, Dever T E, Polyak S J, Gretch D R, Katze M G. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology. 1997;230:217–227. doi: 10.1006/viro.1997.8493. [DOI] [PubMed] [Google Scholar]
- 17.Gale M J, Blakely S M, Kwieciszewski B, Tan S-L, Dossett M, Tang N M, Korth M J, Polyak S J, Gretch D, Katze M G. Control of PKR protein kinase by hepatitis C virus nonstructural 5A protein: molecular mechanism of kinase regulation. Mol Cell Biol. 1998;18:5208–5218. doi: 10.1128/mcb.18.9.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graff J, Normann A, Feinstone S M, Flehmig B. Nucleotide sequence of wild-type hepatitis A virus GBM in comparison with two cell culture-adapted variants. J Virol. 1994;68:548–554. doi: 10.1128/jvi.68.1.548-554.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grakoui A, McCourt D W, Wychowski C, Feinstone S M, Rice C M. A second hepatitis C virus-encoded proteinase. Proc Natl Acad Sci USA. 1993;90:10583–10587. doi: 10.1073/pnas.90.22.10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grakoui A, McCourt D W, Wychowski C, Feinstone S M, Rice C M. Characterization of the hepatitis C virus-encoded serine proteinase: determination of proteinase-dependent polyprotein cleavage sites. J Virol. 1993;67:2832–2843. doi: 10.1128/jvi.67.5.2832-2843.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hijikata M, Mizushima H, Akagi T, Mori S, Kakiuchi N, Kato N, Tanaka T, Kimura K, Shimotohno K. Two distinct proteinase activities required for the processing of a putative nonstructural precursor protein of hepatitis C virus. J Virol. 1993;67:4665–4675. doi: 10.1128/jvi.67.8.4665-4675.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hijikata M, Kato N, Ootsuyama Y, Nakagawa M, Shimotohno K. Gene mapping of the putative structural region of the hepatitis C virus genome by in vitro processing analysis. Proc Natl Acad Sci USA. 1991;88:5547–5551. doi: 10.1073/pnas.88.13.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ide Y, Tanimoto A, Sasaguri Y, Padmanabhan R. Hepatitis C virus NS5A protein is phosphorylated in vitro by a stably bound protein kinase from HeLa cells and by cAMP-dependent protein kinase A-alpha catalytic subunit. Gene. 1997;201:151–158. doi: 10.1016/s0378-1119(97)00440-x. [DOI] [PubMed] [Google Scholar]
- 24.Jansen R W, Newbold J E, Lemon S M. Complete nucleotide sequence of a cell culture-adapted variant of hepatitis A virus: comparison with wild-type virus with restricted capacity for in vitro replication. Virology. 1988;163:299–307. doi: 10.1016/0042-6822(88)90270-x. [DOI] [PubMed] [Google Scholar]
- 25.Kaneko T, Tanji Y, Satoh S, Hijikata M, Asabe S, Kimura K, Shimotohno K. Production of two phsosphoproteins from the NS5A region of the hepatitis C viral genome. Biochem Biophys Res Commun. 1994;205:320–326. doi: 10.1006/bbrc.1994.2667. [DOI] [PubMed] [Google Scholar]
- 26.Kim D W, Gwack Y, Han J H, Choe J. C-terminal domain of the hepatitis C virus NS3 protein contains an RNA helicase activity. Biochem Biophys Res Commun. 1995;215:160–166. doi: 10.1006/bbrc.1995.2447. [DOI] [PubMed] [Google Scholar]
- 27.Lin C, Lindenbach B D, Pragai B M, McCourt D W, Rice C M. Processing in the hepatitis C virus E2-NS2 region: identification of p7 and two distinct E2-specific products with different C termini. J Virol. 1994;68:5063–5073. doi: 10.1128/jvi.68.8.5063-5073.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin C, Pragai B M, Grakoui A, Xu J, Rice C M. Hepatitis C virus NS3 serine proteinase: trans-cleavage requirements and processing kinetics. J Virol. 1994;68:8147–8157. doi: 10.1128/jvi.68.12.8147-8157.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lohmann V, Körner F, Dobierzewska A, Bartenschlager R. Mutations in hepatitis C virus RNAs conferring cell culture adaptation. J Virol. 2001;75:1437–1449. doi: 10.1128/JVI.75.3.1437-1449.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lohmann V, Körner F, Koch J O, Herian U, Theilmann L, Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285:110–113. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- 31.Lohmann V, Körner F, Herian U, Bartenschlager R. Biochemical properties of hepatitis C virus NS5B RNA-dependent RNA polymerase and identification of amino acid sequence motifs essential for enzymatic activity. J Virol. 1997;71:8416–8428. doi: 10.1128/jvi.71.11.8416-8428.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mizushima H, Hijikata M, Asabe S, Hirota M, Kimura K, Shimotohno K. Two hepatitis C virus glycoprotein E2 products with different C termini. J Virol. 1994;68:6215–6222. doi: 10.1128/jvi.68.10.6215-6222.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Murphy F A, Fauquet C M, Bishop D H L, Ghabrial S A, Jarvis A W, Martelli G P, Mayo M A, Summers M D, editors. Classification and nomenclature of viruses: sixth report of the International Committee on Taxonomy of Viruses. Vienna, Austria: Springer Verlag; 1995. pp. 424–426. [Google Scholar]
- 34.Nakabayashi H, Taketa K, Miyano K, Yamane T, Sato J. Growth of human hepatoma cells lines with differentiated functions in chemically defined medium. Cancer Res. 1982;42:3858–3863. [PubMed] [Google Scholar]
- 35.Pestova T V, Shatsky I N, Fletcher S P, Jackson R J, Hellen C U. A prokaryotic-like mode of cytoplasmic eukaryotic ribosome binding to the initiation codon during internal translation initiation of hepatitis C and classical swine fever virus RNAs. Genes Dev. 1998;12:67–83. doi: 10.1101/gad.12.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pietschmann T, Lohmann V, Rutter G, Kurpanek K, Bartenschlager R. Characterization of cell lines carrying self-replicating hepatitis C virus RNAs. J Virol. 2001;75:1252–1264. doi: 10.1128/JVI.75.3.1252-1264.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Poynard T, Ratziu V, Benhamou Y, Opolon P, Cacoub P, Bedossa P. Natural history of HCV infection. Best Practice Res Clin Gastroenterol. 2000;14:211–228. doi: 10.1053/bega.1999.0071. [DOI] [PubMed] [Google Scholar]
- 38.Reed K E, Rice C M. Identification of the major phosphorylation site of the hepatitis C virus H strain NS5A protein as serine 2321. J Biol Chem. 1999;274:28011–28018. doi: 10.1074/jbc.274.39.28011. [DOI] [PubMed] [Google Scholar]
- 39.Reed K E, Rice C M. Overview of hepatitis C virus genome structure, polyprotein processing, and protein properties. Curr Top Microbiol Immunol. 2000;242:55–84. doi: 10.1007/978-3-642-59605-6_4. [DOI] [PubMed] [Google Scholar]
- 40.Reed K E, Xu J, Rice C M. Phosphorylation of the hepatitis C virus NS5A protein in vitro and in vivo: properties of the NS5A-associated kinase. J Virol. 1997;71:7187–7197. doi: 10.1128/jvi.71.10.7187-7197.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Suzich J A, Tamura J K, Palmer H F, Warrener P, Grakoui A, Rice C M, Feinstone S M, Collett M S. Hepatitis C virus NS3 protein polynucleotide-stimulated nucleoside triphosphatase and comparison with the related pestivirus and flavivirus enzymes. J Virol. 1993;67:6152–6158. doi: 10.1128/jvi.67.10.6152-6158.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tanji Y, Hijikata M, Satoh S, Kaneko T, Shimotohno K. Hepatitis C virus-encoded nonstructural protein NS4A has versatile functions in viral protein processing. J Virol. 1995;69:1575–1581. doi: 10.1128/jvi.69.3.1575-1581.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tanji Y, Kaneko T, Satoh S, Shimotohno K. Phosphorylation of hepatitis C virus-encoded nonstructural protein NS5A. J Virol. 1995;69:3980–3986. doi: 10.1128/jvi.69.7.3980-3986.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tomei L, Failla C, Santolini E, DeFrancesco R, LaMonica N. NS3 is a serine protease required for processing of hepatitis C virus polyprotein. J Virol. 1993;67:4017–4026. doi: 10.1128/jvi.67.7.4017-4026.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsukiyama K K, Iizuka N, Kohara M, Nomoto A. Internal ribosome entry site within hepatitis C virus RNA. J Virol. 1992;66:1476–1483. doi: 10.1128/jvi.66.3.1476-1483.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Venuti A, Di Russo C, del Grosso N, Patti A M, Ruggeri F, De Stasio P R, Martiniello M G, Pagnotti P, Degener A M, Midulla M, Pana A, Perez-Bercoff R. Isolation and molecular cloning of a fast-growing strain of human hepatitis A virus from its double-stranded replicative form. J Virol. 1985;56:579–588. doi: 10.1128/jvi.56.2.579-588.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang C, Sarnow P, Siddiqui A. Translation of human hepatitis C virus RNA in cultured cells is mediated by an internal ribosome-binding mechanism. J Virol. 1993;67:3338–3344. doi: 10.1128/jvi.67.6.3338-3344.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yamashita T, Kaneko S, Shirota Y, Qin W, Nomura T, Kobayashi K, Murakami S. RNA-dependent RNA polymerase activity of the soluble recombinant hepatitis C virus NS5B protein truncated at the C-terminal region. J Biol Chem. 1998;273:15479–15486. doi: 10.1074/jbc.273.25.15479. [DOI] [PubMed] [Google Scholar]