Abstract
Objectives
Serum creatinine (SCr) is the primary biomarker for assessing kidney function; however, it may lag behind true kidney function, especially in instances of acute kidney injury (AKI). The objective of the work is to develop Nephrocast, a deep-learning model to predict next-day SCr in adult patients treated in the intensive care unit (ICU).
Materials and Methods
Nephrocast was trained and validated, temporally and prospectively, using electronic health record data of adult patients admitted to the ICU in the University of California San Diego Health (UCSDH) between January 1, 2016 and June 22, 2024. The model features consisted of demographics, comorbidities, vital signs and laboratory measurements, and medications. Model performance was evaluated by mean absolute error (MAE) and root-mean-square error (RMSE) and compared against the prediction day’s SCr as a reference.
Results
A total of 28 191 encounters met the eligibility criteria, corresponding to 105 718 patient-days. The median (interquartile range [IQR]) MAE and RMSE in the internal test set were 0.09 (0.085-0.09) mg/dL and 0.15 (0.146-0.152) mg/dL, respectively. In the prospective validation, the MAE and RMSE were 0.09 mg/dL and 0.14 mg/dL, respectively. The model’s performance was superior to the reference SCr.
Discussion and Conclusion
Our model demonstrated good performance in predicting next-day SCr by leveraging clinical data routinely collected in the ICU. The model could aid clinicians in in identifying high-risk patients for AKI, predicting AKI trajectory, and informing the dosing of renally eliminated drugs.
Keywords: serum creatinine, acute kidney injury, machine learning, deep learning, critical care
Background
Acute kidney injury (AKI) is a major source of mortality and morbidity in hospitalized patients.1–4 It has been estimated that 40%-60% of patients will experience at least one AKI event during their intensive care unit (ICU) stay.5,6 The management of AKI is an ongoing challenge, especially in critically ill patients.7 Drugs cleared by the kidneys often have a narrow therapeutic window for efficacy without causing adverse reactions.8 The margin for error is even lower in the presence of nephrotoxic drugs, where the therapeutic window is frequently shifting due to AKI.9–13 Accurate assessment of glomerular filtration rate (GFR) is crucial when initiating and adjusting the dose of renally eliminated drugs in patients with AKI.14–17 Broadly, there are 2 ways to estimate kidney function: measuring the urinary clearance of solutes and estimating clearance based on observed serum levels of solutes. Because solute clearance varies based on a person’s body size, clearance can be indexed to body surface area to produce an estimated GFR as another measure of kidney function.18
A common urine-based method of estimating kidney function is creatinine clearance (CrCl), which measures the clearance using a 24-h urine collection paired with a serum creatinine (SCr) measurement.19–21 The inconvenience of collecting urine over such a long time period limits its utility in clinical settings.22–24 To mitigate this limitation, several statistical equations have been developed to estimate kidney function using a single SCr measurement, including the Cockcroft-Gault (CG),25 and Chronic Kidney Disease Epidemiology Collaboration (CKD-EPI) equations.26–28 Both the CrCl and the SCr-based estimation equations rely on the assumption of steady-state SCr. While this assumption is reasonable in ambulatory settings, it is rarely met in the ICU, where substantial fluctuations in kidney function occur on 35%-40% of patient-days in the ICU.29 The reason these equations are inaccurate during AKI is that the change in SCr lags behind the true kidney function, especially in rapidly progressing AKI.30–32 If we could anticipate the future SCr, then the SCr-based estimation equation would more accurately reflect the GFR.
While many machine learning models have been previously developed to predict the onset of AKI based on SCr change, these models typically do not estimate future SCr.33–36 This limits the utility of these models in predicting AKI trajectory, differentiating between transient and persistent AKI, and informing the dosing of renally eliminated drugs. Achieving an accurate and dynamic prediction of kidney function could address these constraints. In this study, we aim to develop and validate Nephrocast, a deep-learning model capable of accurately predicting next-day SCr levels in critically ill adult patients.
Materials and methods
Study cohort
Data were extracted from the electronic health record (EHR) system of the University of California San Diego (UCSD) Health System between January 1, 2016 and June 30, 2023 for Nephrocast development and internal validation. The UCSD Health System consists of 2 academic medical centers, including a Level I Trauma Center, that provide critical care across a wide range of specialties, including medical, cardiovascular, and surgical ICUs. Data were collected from July 1, 2023 to November 31, 2023 to perform temporal validation. Nephrocast was validated prospectively using data collected from January 1, 2024 to June 22, 2024. In this study, patients were eligible for inclusion if their age was ≥18 years old and had spent a minimum of 24 h in an ICU. Patients were excluded if they had a diagnosis of chronic kidney disease (CKD) stage 5 or end-stage kidney disease (ESKD). Patient-days were excluded if the patient received renal replacement therapy (RRT) on the prediction day or the last 7 days from the prediction day, and if the ICU stay extended beyond 14 days. The University of California San Diego Institutional Review Board (IRB) approval was obtained with the waiver of informed consent (#800257).
Outcome definition
The outcome variable, next-day SCr, was defined as the SCr value measured at 6:00 am on the next patient-day. If no measurement was available at 6:00 am, the closest SCr measurement within a 6-h range was selected. If the SCr measurement for the following day was missing within a 6-h window centered around 6:00 am, no prediction was made during model training.
Clinical features as predictors
Variables consisted of 50 vital signs and laboratory measurements, 6 demographic features, 11 Systemic Inflammatory Response Syndrome (SIRS) and Sequential Organ Failure Assessment (SOFA) criteria, 11 medication features, and 62 comorbidities. Vital signs and laboratory variables were compiled at an hourly resolution into non-overlapping bins with the median value utilized for variables with multiple measurements per hour. Old values were carried forward for up to 24 h if no new measurements were available. All remaining missing variables were imputed using the mean. For each vital sign and laboratory measurement, an additional 2 features consisting of the slope of change and mean value over the previous 72 h were calculated.
Model development and validation
The initial model of Nephrocast consisted of a feedforward neural network with 2 hidden layers of size 128 and 64 units. To enhance temporal focus and capture relevant time-based patterns, we integrated a multi-headed attention layer. This attention mechanism allowed the model to prioritize critical timestamps, improving its predictive accuracy. The final layer of the model produces a single value, representing the predicted next-day SCr. Training was conducted using L2 regularization and the Adam optimizer, with hyperparameters optimized through Bayesian hyperoptimization.37,38
To evaluate the Nephrocast’s performance, we employed 10-fold cross-validation, ensuring robust and unbiased performance assessment with a 90:10 split at the encounter level for training and testing in each fold. This approach grouped each encounter’s prediction days entirely into either the training or testing set to prevent data leakage and ensure the integrity of our evaluation. The best-performing model from cross-validation was further used for temporal validation.
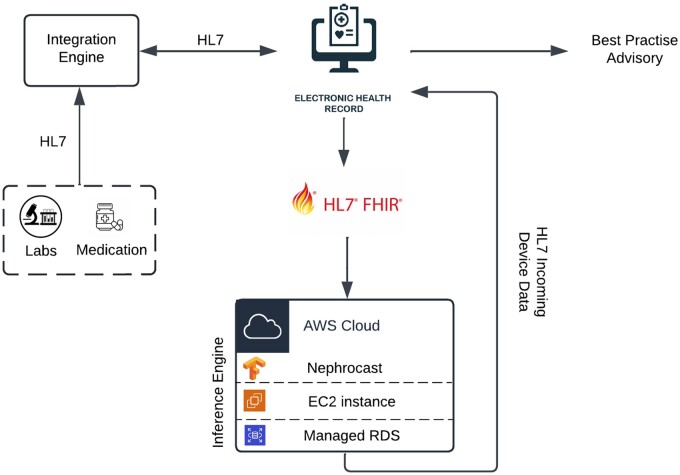
To validate Nephrocast in a production environment for real-time performance assessment, we leveraged an existing cloud-based infrastructure designed to directly access UCSDH EHR data using Fast Healthcare Interoperability Resources (FHIR) and Health Level 7 (HL7) standards with OAuth 2.0 authentication, as previously described by Boussina et al.39 The schematic diagram of this “silent mode” prospective validation environment is shown in Figure 1. The input feature set (including demographics, comorbidities, vital signs, laboratory measurements, and medications) was extracted by the platform from January 1, 2024 to June 22, 2024 and passed to Nephrocast to predict next-day SCr levels.
Figure 1.
Schematic diagram of the Nephrocast prospective validation pipeline. Abbreviations: AWS = amazon web services; EC2 = elastic compute cloud; FHIR = fast healthcare interoperability resources; HL7 = health level 7; RDS = relational database service.
To interpret the model’s predictions, we calculated feature importance scores, which quantify the relative contribution of each input feature to the model’s output. Higher importance scores indicate a greater influence of the corresponding feature on the predicted outcome. For instance, a feature with a high importance score suggests that variations in this feature have a substantial impact on the predicted next-day SCr, underscoring its clinical relevance.40
Evaluation of predicted SCr
In the context of this research, reference SCr was defined as the laboratory measured SCr on the day of making the prediction. Predicted SCr was defined as the SCr predicted by Nephrocast for the next patient-day. Measured SCr was defined as the laboratory measured SCr on the next patient-day. To further illustrate these terms, consider a hypothetical scenario involving a patient in the ICU whose SCr levels on the second and third days of their ICU stay are 1.0 mg/dL and 1.5 mg/dL, respectively. The goal is to predict SCr on the third day. In this case, the reference SCr would be 1.0 mg/dL, and the measured SCr would be 1.5 mg/dL. The predicted SCr would be Nephrocast prediction for day 3.
Error was defined as the difference between predicted SCr and measured SCr.
Errors were summarized using mean absolute error (MAE) and root mean squared error (RMSE), with the latter being more sensitive to large errors.
where is the number of analyzed SCr observations.
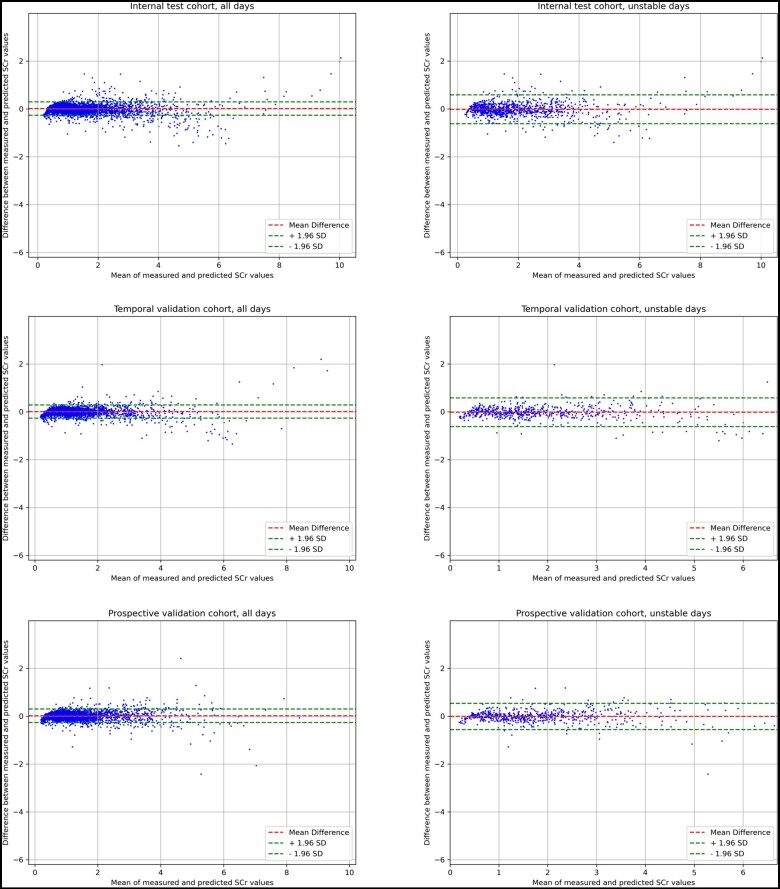
Similar to the approach of Huang et al, reference SCr served as a baseline model, and its performance was compared against that of measured SCr to assess the clinical usefulness of the model. This comparison was made under the assumption that the SCr level would remain consistent between prediction’s day and next-day, reflecting standard clinical practice.41 Additionally, we trained a multivariable linear regression model with L2 regularization using Nephrocast’s predictors, and its performance was evaluated against that of Nephrocast. To assess the performance of the Nephrocast in patients with significant fluctuating kidney function, we evaluated the Nephrocast performance on days of unstable kidney function, defined as those in which there was a change of 30% or 0.3 mg/dL or more in SCr concentration between the reference SCr and the measured SCr on the following patient day. Bland-Altman plots were used to assess the difference between predicted and measured SCr.42
AKI definition and staging
AKI was defined according to the 2012 Kidney Disease Improving Global Outcomes (KDIGO) AKI guidelines criteria using the peak-to-baseline SCr ratio.8 Baseline SCr was defined as SCr at hospital admission. The urine output criterion was not implemented due to the sparsity of data.
Descriptive analyses and software
Patient characteristics were described and summarized using descriptive statistics such as mean (SD), median (interquartile range [IQR]), or counts (%), where appropriate. Continuous variables were analyzed using the Wilcoxon rank-sum test. All hypotheses were two-sided, and significance levels were set at the 5% level. Python 3.10 was used for analysis. NumPy 1.23.5 was used for all data preprocessing. The deep learning model was implemented using TensorFlow 2.13.0.43
Results
Study population
A total of 25 243 encounters met the eligibility criteria in the training dataset, corresponding to 95 111 patient-days. In the training set, males represented 60.8% of the cohort with a median (IQR) age of 61.5 (48.4-71.5) years. The most common ethnicity was White (12 781, 50.63%). The median (IQR) days of ICU stays was 3.67 (2.26-6.54). During their hospital stay, 94.4% of the patients were admitted to the medical ICU at some point, and about 37.8% of the patients were admitted to the surgical ICU at some point during their hospital stay. The median (IQR) baseline SCr was 0.68 (0.52-0.89) mg/dL. Overall, 10.5% of patients had a diagnosis of CKD. The median (IQR) SOFA II score was 5.3–8 The mortality rate during hospital stay was 6.16%. The percentage of patients who developed AKI was 46.4%, and the percentage of patients with AKI stage I, II, and III were 25.5%, 12.8%, and 8.1%, respectively. The percentage of unstable measurements represented 11.5% of all measurements (Table 1). The demographic and clinical characteristics of the temporal and prospective validation cohorts were comparable to those of the training cohort.
Table 1.
Patient characteristics.
| Variable | Training (N = 25 243) | Temporal validation (N = 1378) | Prospective validation (N = 1570) |
|---|---|---|---|
| Patient-days, (N) | 95 111 | 5153 | 5454 |
| Age, median (IQR), years | 61.5 (48.4-71.5) | 61.1 (46.1-70.8) | 63.5 (50.6-73.2) |
| Sex, n (%) | |||
| Male | 15 348 (60.8%) | 831 (60.3%) | 937 (59.7%) |
| Female | 9905 (39.2%) | 547 (39.7%) | 633 (40.3%) |
| Ethnicity, n (%) | |||
| Black | 1913 (7.58%) | 99 (7.18%) | 99 (6.31%) |
| White | 12 781 (50.63%) | 644 (46.73%) | 782 (49.81%) |
| Asian | 1477 (5.85%) | 88 (6.39%) | 109 (6.94%) |
| Other | 9072 (35.94%) | 547 (39.64%) | 580 (36.94%) |
| SOFA score, median (IQR) | 5 (3-8) | 5 (3-8) | 5 (2-9) |
| ICU length of stay, median (IQR), days | 3.67 (2.26-6.54) | 3.83 (2.35-7.18) | 3.77 (2.49-6.80) |
| Unit type, na | |||
| MICU | 23 817 | 1454 | 1563 |
| Other | 4687 | 273 | 246 |
| ICU mortality, N (%) | 1555 (6.16%) | 61 (4.43%) | 81 (5.16%) |
| Baseline SCr, median (IQR)b | 0.68 (0.52-0.89) | 0.65 (0.49-0.87) | 0.67 (0.51-0.89) |
| AKI, stage, n (%)c | |||
| I | 6443 (25.5%) | 382 (27.7%) | 421 (26.8%) |
| II | 3230 (12.8%) | 182 (13.2%) | 234 (14.9%) |
| III | 2049 (8.1%) | 121 (8.8%) | 130 (8.3%) |
| Unstable patient-days, n (%)d | 10 948 (11.51 %) | 616 (11.95%) | 633 (9.81%) |
| Comorbidities, n (%) | |||
| Anemia | 1211 (4.8%) | 86 (6.2%) | 116 (7.4%) |
| Chronic kidney disease | 1281 (10.5%) | 97 (7.0%) | 114 (7.3%) |
| Coronary artery disease | 2034 (8.1%) | 115 (8.3%) | 178 (11.3%) |
| Diabetes | 2438 (9.7%) | 150 (10.9%) | 211 (13.4%) |
| Hypertension | 3697 (14.6%) | 236 (17.1%) | 321 (20.4%) |
| Liver disease | 116 (0.5%) | 10 (0.7%) | 21 (1.3%) |
| Severe sepsis/septic shock | 2039 (8.1%) | 103 (7.5%) | 103 (6.6%) |
| Mechanical ventilation, N (%) | 11 468 (45.43%) | 667 (48.40%) | 769 (48.98%) |
Patients may undergo transfers to various units throughout their hospitalization.
Baseline SCr was defined as the first measurement during hospital stay.
AKI was defined according to the 2012 Kidney Disease Improving Global Outcomes (KDIGO) criteria using the peak-to-baseline SCr ratio8.
Unstable patient-days were defined as those in which there was a change of 30% or 0.3 mg/dL or more in SCr concentration between the reference SCr and the measured SCr on the following patient-day.
Abbreviations: AKI = acute kidney injury; ICU = intensive care until; MICU = medical intensive care unit; SCr = serum creatinine; SICU = surgical intensive care unit; SOFA = sequential organ failure assessment.
Model performance
Nephrocast exhibited a small prediction error and outperformed reference SCr in internal testing, temporal validation, and prospective validation (Table 2). The median (IQR) MAE and RMSE across all days in the internal test dataset were 0.09 (0.085-0.09) mg/dL and 0.15 (0.146-0.152) mg/dL compared to MAE of 0.13 (0.131-0.135) mg/dL and RMSE of 0.25 (0.245-0.253) mg/dL for reference SCr. In unstable days, the median (IQR) MAE and RMSE in the internal test set were 0.20 (0.197-0.203) mg/dL and 0.31 (0.307, 0.327) mg/dL, respectively, and were superior to the MAE of 0.54 (0.532-0.548) mg/dL and RMSE of 0.67 (0.663, 0.694) mg/dL of reference SCr. The model performance in the training set is shown in Table S1.
Table 2.
Summary of model performance on all days and unstable days.
| All days |
Unstable days |
|||||
|---|---|---|---|---|---|---|
| Test set | ||||||
| Nephrocast | Reference SCr | P-value | Nephrocast | Reference SCr | P-value | |
| MAE, median (IQR), mg/dL | 0.09 (0.085, 0.09) | 0.13 (0.131, 0.135) | <.01 | 0.20 (0.197, 0.203) | 0.54 (0.532, 0.548) | <.01 |
| RMSE, median (IQR), mg/dL | 0.15 (0.146,0.152) | 0.25 (0.245, 0.253) | <.01 | 0.31 (0.307, 0.327) | 0.67 (0.663, 0.694) | <.01 |
| Temporal validation | ||||||
| MAE, mg/dL | 0.08 | 0.13 | N/A | 0.19 | 0.54 | N/A |
| RMSE, mg/dL | 0.14 | 0.25 | N/A | 0.31 | 0.66 | N/A |
| Prospective validation | ||||||
| MAE, mg/dL | 0.09 | 0.13 | N/A | 0.18 | 0.50 | N/A |
| RMSE, mg/dL | 0.14 | 0.23 | N/A | 0.28 | 0.60 | N/A |
Reference SCr was defined as the measured SCr on the day of making the prediction. Predicted SCr was defined as the predicted SCr by our model for the next patient-day. Measured SCr was defined as the laboratory measured SCr on the next patient-day. Model error was defined as the difference between predicted SCr and measured SCr, which was compared against the difference between reference SCr and measured SCr. Error was summarized using MAE and RMSE. Unstable patient-days were defined as those in which there was a change of 30% or 0.3 mg/dL or more in SCr concentration between the reference SCr and the measured SCr on the following patient-day.
Abbreviations: IQR = interquartile range; MAE = mean absolute error; RMSE = root mean squared error; SCr = serum creatinine.
The model performance was comparable between training and validation. In temporal validation, Nephrocast’s MAE and RMSE across all days were 0.08 mg/dL and 0.14 mg/dL, respectively, and were superior to reference SCr MAE of 0.13 mg/dL and RMSE of 0.25 mg/dL. In unstable days, the temporal validation MAE and RMSE were 0.19 mg/dL and 0.31 mg/dL, respectively, and were superior to reference SCr MAE of 0.54 mg/dL and RMSE of 0.66 mg/dL. In the prospective cohort, Nephrocast’s MAE and RMSE across all days were 0.09 mg/dL and 0.14 mg/dL, respectively, outperforming the reference SCr MAE of 0.13 mg/dL and RMSE of 0.23 mg/dL. In unstable days, Nephrocast’s MAE and RMSE were 0.18 mg/dL and 0.28 mg/dL, respectively, and superior to the reference SCr MAE of 0.50 mg/dL and RMSE of 0.60 mg/dL.
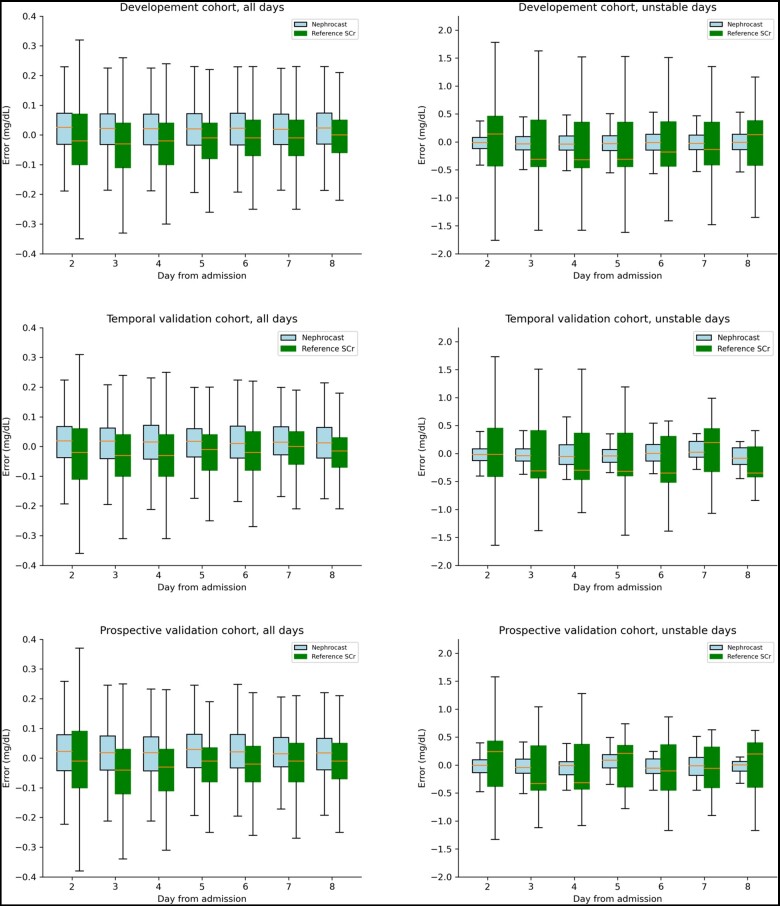
When compared to the regularized multivariable linear regression model, Nephrocast performance was statistically superior (Table S2). The difference was specifically notable in days of unstable kidney function (MAE = 0.20 versus 0.24; RMSE = 0.31 versus 0.43 mg/dL; P <.01). Additionally, Nephrocast demonstrated consistent performance throughout the first 8 days of ICU stay (Figure 2) and across a wide range of SCr value changes (Figure 3) in both stable and unstable days.
Figure 2.
Temporal trends of prediction error for all days and unstable days. Reference SCr was defined as the measured SCr on the day of making the prediction. Predicted SCr was defined as the predicted SCr by Nephrocast on the next patient-day. Measured SCr was defined as the laboratory measured SCr on the next patient-day. Model error was defined as the difference between measured SCr and predicted SCr, which was compared against the difference between measured SCr and reference SCr. Unstable days were defined as those in which there was a change of 30% or 0.3 mg/dL or more in SCr concentration between the reference SCr and the measured SCr on the following patient day. Abbreviation: SCr = serum creatinine.
Figure 3.
Bland-Altman plots of predicted and measured serum creatinine in internal test dataset, temporal validation dataset, and prospective for both all days and unstable days. Unstable days were defined as those in which there was a change of 30% or 0.3 mg/dL or more in SCr concentration between the reference SCr and the measured SCr on the following patient day. Abbreviations: SCr = serum creatinine; SD = standard deviation.
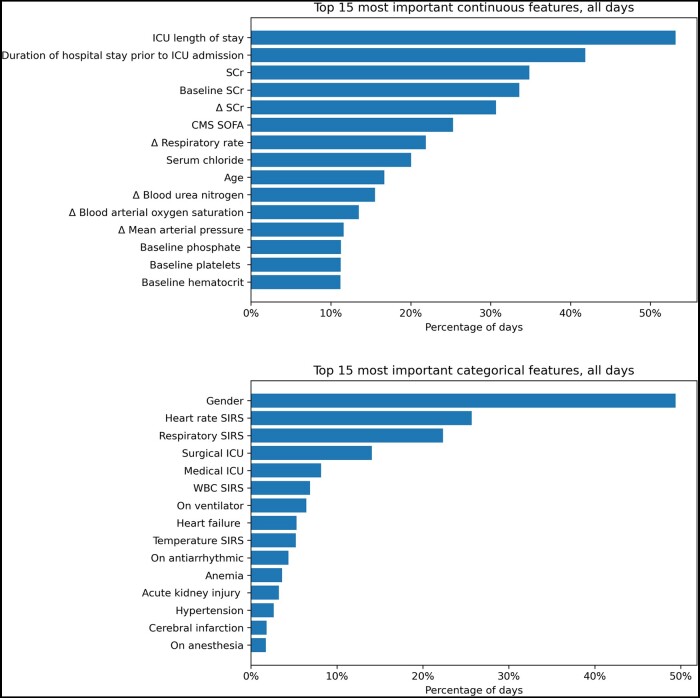
Feature importance
A total of 241 features were assessed (Table S3). The top 15 most important continuous and categorical features across all days are shown in Figure 4. The most important continuous feature was the ICU length of stay. In total, 3 features were related to SCr, 2 features were related to hospital and ICU length of stay, and the remaining features were for laboratory results and vital signs. The most important categorical feature was gender. Four features were components of the SIRS score, 2 features were for ICU unit type, and the remaining features were related to comorbidities, and medical conditions. The top 15 most important features on unstable days are shown in Figure S1. In those days, SCr-related features ranked as the most important continuous features.
Figure 4.
Top 15 features in order of importance for all days in terms of continuous and categorical features. Abbreviations: CMS = centers for medicare and medicaid services; SCr = serum creatinine; SIRS = systemic inflammatory response syndrome; SOFA = sequential organ failure assessment; WBC = white blood cell.
Discussion
In this study, we developed Nephrocast, a deep-learning model to predict next-day SCr in critically ill adult patients using EHR data. The model demonstrated good performance in internal testing, temporal validation, and prospective validation. It also exhibited consistent performance across different clinical scenarios, including stable days, unstable days, throughout the first 8 days of ICU stay, and over wide ranges of SCr concentrations. Our model outperformed reference SCr and penalized multivariable linear regression as baseline models, especially in days of unstable kidney function, demonstrating the advantage of deep learning methods. The results of prospective validation support the potential integration of Nephrocast into clinical workflows to evaluate its operational and clinical impact.
To date, limited work has been done to predict kidney function using machine learning techniques. Huang et al constructed a machine-learning model to predict next-day CrCl derived from measured 24-h urine collection, achieving an impressive RMSE of 18.1 (95% CI 17.9-18.3) mL/min in external validation; however, their model depends on prior CrCl measurements to make the next day prediction.41 Unlike 24-h urine collection, SCr is inexpensive to measure with a short laboratory turnaround time, making it the primary biomarker to estimate kidney function in hospital settings.44,45 In clinical practice, renal drug dose adjustments have relied on estimates derived from the CG equation, and more recently, estimated GFR based on MDRD and CKD-EPI equations.46 Although these equations assume steady-state SCr, they persist in clinical practice as the main method to guide dose adjustments in patients with AKI due to the lack of practical alternatives.47 Non-steady state equations such Jelliffe and kinetic GFR provide a more accurate assessment of kidney function for patients in AKI.48,49 These equations require 2 discrete SCr measurements at 2 different times. This situation presents an opportunity to incorporate predicted SCr from our model into these equations, thereby enhancing the ability to estimate GFR with greater accuracy.50 Regardless of the clinician’s choice of equation, a reliable prediction of SCr is required to estimate kidney function accurately in patients who are not in a steady state.
Emerging evidence suggests that AKI classification based on early SCr trajectory could provide clinically meaningful insights into AKI risk stratification. Takkavatakarn et al have recently shown that in critically ill patients with sepsis, 8 distinct SCr trajectories exist. These trajectories varied significantly in their risk for acute kidney disease (AKD), AKD or mortality by day 7, and AKD or mortality by hospital discharge.51 Similarly, Bhatraju et al evaluated the SCr trajectory in the first 72 h of ICU in critically ill patients with AKI and demonstrated that patients with non-resolving AKI trajectories had a higher risk of mortality compared to resolving AKI trajectories.52 By accurately predicting next-day SCr levels, our model could inform clinicians early about the SCr trajectory, enabling a patient-specific approach to AKI management.
There is ongoing discussion regarding when antimicrobial dose adjustments should be deferred for the first 24-48 h in hospitalized patients with acute infections with known AKI, especially for agents with a wide therapeutic index, such as β-lactams.53,54 Crass et al argued that because AKI resolves in >50% of patients within 48-72 h and SCr lags behind changes in GFR, such dosage adjustments based on equations that were derived from patients with CKD or normal renal function could result in subtherapeutic antimicrobial concentrations and potentially decreased clinical response.55 Unlike other AKI models, which only predict AKI onset, our model predicts next-day SCr; thus offering insights into the onset and recovery of AKI, and potentially informing clinicians facing decisions regarding dosage adjustments. For example, in patients with a recent onset of AKI, if our model predicts a significant improvement in kidney function in the next 24 h, indicated by a notable decline in predicted SCr level, the clinician may consider not adjusting standard doses. Conversely, if the model predicts declining kidney function, indicated by an increase in predicted SCr level, the clinician may consider intensified monitoring and perform dose adjustment if criteria are met.41 It is also worth noting that the decision for dose adjustment is drug-dependent. For example, in drugs with a narrow therapeutic index, such as vancomycin or aminoglycosides, a conservative approach may be required with more frequent monitoring. In contrast, for wide therapeutic index drugs, such as cephalosporins, more aggressive doses may be warranted for critically ill patients. Those scenarios and considerations must be considered when developing a clinical protocol to incorporate the model predictions into clinical decision-making or conducting prospective implementation studies.
The set of important predictors identified by our model aligns with previous research to predict AKI. Song et al conducted a systematic review of AKI prediction models and showed that creatinine-related variables were the most common significant predictors across machine learning models. The authors also showed that blood urea nitrogen and urine output are predictors of importance but to a lesser extent.56 Similarly, in Huang et al work to predict next-day CrCl, variables such as “CrCl of the previous day,” and “mean CrCl of all past days during ICU stay” were ranked as highly important.41 These findings are expected, given the strong correlation between repeated and longitudinal creatinine-based measurements in the same patient. Nonetheless, this issue might limit the utility of these models in patients whose SCr measurements are infrequent or far apart. Gender ranked as a feature of high importance in our model, which could be attributed to the gender-dependent difference in muscle mass and creatinine generation.57 Additionally, epidemiological studies continue to confirm a higher incidence of AKI in men compared to women.58 The relationship between gender and AKI continues to be an interesting research topic that requires further elucidation. Our model identified other clinical predictors indicative of systemic infections and organ dysfunction, underscoring the complex relationship between illness severity and SCr levels.59
We note limitations that are important to be acknowledged. First, it is well established that creatinine has several drawbacks as a biomarker to estimate kidney function in critically ill patients. Catabolic conditions may lead to an increase or decrease in the production of creatinine.60 Fluid resuscitation will increase clearance and dilute SCr concentration, resulting in a decline in SCr concentration. Medications commonly prescribed in the ICU settings (eg, cefazolin, albumin, dopamine) can interfere with the creatinine assay, resulting in biased results. Due to the increase in tubular secretion associated with the decline in GFR, a change in SCr will not be observed until 50% of GFR has been lost.60 Several biomarkers have been evaluated as potential alternatives or adjuncts to SCr, but their clinical adaptation has been limited.61 Perhaps the most notable example would be Cystatin C (CysC), a low molecular weight protein that is produced constantly by all nucleated cells and filtered at the glomerulus and not reabsorbed. Unlike SCr, CysC is less affected by sex, muscle mass, nutritional status, and frailty. In a study conducted at the Mayo Clinic hospital, CysC utilization in the ICU has been shown to increase from 4 tests/1000 patient-days in 2011 to 44 tests/1000 patient-days in 2018 and was assessed 6.4-fold more in ICU patients compared non-ICU patients.62 While the increase in CysC utilization is considerable, it is far from comparable to SCr utilization. Additionally, the Food and Drug Administration guidance on pharmacokinetic studies in kidney disease recommends using SCr and contemporary steady-state equations to estimate kidney function for drug labeling purposes. Drug manufacturers have not yet incorporated the use of CysC in their pharmacokinetic studies and drug labeling dose recommendations. Given the slow penetrance of routinely measuring CysC in clinical settings and lack of drug dosing guidance based on CysC, SCr will continue to serve as the standard biomarker to estimate kidney function and guide drug dosing, emphasizing the need for machine-learning models that can compensate for the shortcomings of SCr.
Second, although we included a prospective validation, our model has not been externally validated, limiting the generalizability of our findings and potentially the performance of our model if implemented in a different health system.63,64 Additionally, further work is required to investigate the integration of Nephrocast’s predictions into clinical practice and develop a best practice advisory in the EHR system.65,66 Third, the exclusion of patients of encounters with RRT on the predication day or the last 7 days, CKD stage 5, ESKD, and encounters after 14 days from the ICU admission date might have introduced a selection bias. While these decisions were made to ensure the reliability of our model, they further contribute to limiting the generalizability of this work. Fourth, potential predictors of AKI that were not available through the data pipeline were not included in the model. The inclusion of nephrotoxic drug count, concentration, and the drug’s risk of nephrotoxicity have been shown to be significant predictors in AKI models.67–71 Similarly, undergoing major surgical procedures is known to increase the risk of AKI.72 The inclusion of these predictors could potentially improve the performance of Nephrocast. Lastly, Previous clinical trials involving EHR alerts did not demonstrate a statistically significant benefit on AKI-related outcomes despite showing that these alerts were associated with discontinuing nephrotoxic medications, increasing fluid resuscitation, optimizing hemodynamic parameters, and timely nephrologist consultations.66,73 Subsequent research should prioritize identifying patient subgroups that would benefit from these clinical interventions.74
Conclusions
By leveraging clinical data routinely collected in the ICU, we developed a deep learning model to predict next-day SCr in critically ill adult patients. Our model demonstrated superior performance compared to the reference SCr, especially in cases of unstable kidney function. This capability holds promise in assisting clinicians in identifying high-risk patients for AKI, predicting AKI trajectory, and informing the dosing of renally eliminated drugs. Further work is needed to externally validate the model’s performance, explore its clinical applications, and integrate SCr prediction into clinical workflows and decision-making.
Supplementary Material
Contributor Information
Ghodsieh Ghanbari, Department of Biomedical Informatics, University of California San Diego (UCSD) School of Medicine, La Jolla, CA 92093, United States.
Jonathan Y Lam, Department of Biomedical Informatics, University of California San Diego (UCSD) School of Medicine, La Jolla, CA 92093, United States.
Supreeth P Shashikumar, Department of Biomedical Informatics, University of California San Diego (UCSD) School of Medicine, La Jolla, CA 92093, United States.
Linda Awdishu, Skaggs School of Pharmacy and Pharmaceutical Sciences, UCSD, La Jolla, CA 92093, United States.
Karandeep Singh, Joan and Irwin Jacobs Center for Health Innovation, UC San Diego Health, San Diego, CA 92093, United States.
Atul Malhotra, Division of Pulmonary, Critical Care, and Sleep Medicine, Department of Medicine, UCSD, La Jolla, CA 92093, United States.
Shamim Nemati, Department of Biomedical Informatics, University of California San Diego (UCSD) School of Medicine, La Jolla, CA 92093, United States.
Zaid Yousif, Skaggs School of Pharmacy and Pharmaceutical Sciences, UCSD, La Jolla, CA 92093, United States.
Author contributions
Ghodsieh Ghanbari, Jonathan Y. Lam, Supreeth P. Shashikumar, Atul Malhotra, Shamim Nemati, and Zaid Yousif conceptualized and designed the study. Ghodsieh Ghanbari and Jonathan Y. Lam performed data extraction and analysis. Ghodsieh Ghanbari, Jonathan Y. Lam, Supreeth P. Shashikumar, Linda Awdishu, Karandeep Singh, Atul Malhotra, Shamim Nemati, and Zaid Yousif were involved in data interpretation and writing the manuscript. All authors read and approved the final manuscript.
Supplementary material
Supplementary material is available at JAMIA Open online.
Funding
G.G. and J.Y.L. acknowledge grant funding from the United States National Library of Medicine (award number T15LM011271). S.N. acknowledges grant funding from the United States National Library of Medicine (award number R01LM013998) and the United States National Institute of General Medical Sciences (award number R35GM143121).
Conflicts of interest
S.N., S.P.S., and A.M. are cofounders, advisors, and hold equity in Healcisio Inc, a predictive analytics startup. The terms of this arrangement have been reviewed and approved by the UC San Diego in accordance with its conflict-of-interest policies. A.M. is funded by the National Institutes of Health. A.M. reports income related to medical education from Livanova, Eli Lilly, Zoll, and Powell Mansfield. ResMed provided a philanthropic donation to UCSD. KS’s institution receives grant funding from the National Institute for Diabetes and Digestive and Kidney Disease; K.S.’s institution previously received grant funding from Teva Pharmaceuticals; K.S. previously consulted for Flatiron Health. L.A. serves as a clinical consultant for MediBeacon. Z.Y. serves as a clinical consultant for Takeda Pharmaceuticals.
Data availability
The code supporting this research is available https://github.com/NematiLab/Serum_Creatinine_Prediction. No public repository exists for the study data, as it contains protected health information. Further inquiries can be directed to the corresponding author.
References
- 1. Andonovic M, Traynor JP, Shaw M, Sim MAB, Mark PB, Puxty KA.. Short- and long-term outcomes of intensive care patients with acute kidney disease. eClinicalMedicine. 2022;44:101291. 10.1016/j.eclinm.2022.101291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kellum JA, Romagnani P, Ashuntantang G, Ronco C, Zarbock A, Anders HJ.. Acute kidney injury. Nat Rev Dis Primers. 2021;7:52. 10.1038/s41572-021-00284-z [DOI] [PubMed] [Google Scholar]
- 3. Coca SG, Yusuf B, Shlipak MG, Garg AX, Parikh CR.. Long-term risk of mortality and other adverse outcomes after acute kidney injury: a systematic review and meta-analysis. Am J Kidney Dis. 2009;53:961-973. 10.1053/j.ajkd.2008.11.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Doyle JF, Forni LG.. Acute kidney injury: short-term and long-term effects. Crit Care. 2016;20:188. 10.1186/s13054-016-1353-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nisula S, Kaukonen KM, Vaara ST, et al. ; FINNAKI Study Group. Incidence, risk factors and 90-day mortality of patients with acute kidney injury in Finnish intensive care units: the FINNAKI study. Intensive Care Med. 2013;39:420-428. 10.1007/s00134-012-2796-5 [DOI] [PubMed] [Google Scholar]
- 6. Hoste EAJ, Bagshaw SM, Bellomo R, et al. Epidemiology of acute kidney injury in critically ill patients: the multinational AKI-EPI study. Intensive Care Med. 2015;41:1411-1423. 10.1007/s00134-015-3934-7 [DOI] [PubMed] [Google Scholar]
- 7. Chen S, Li Y, Su B.. Acute kidney failure: current challenges and new perspectives. J Clin Med. 2023;12:3363. 10.3390/jcm12103363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Khwaja A. KDIGO clinical practice guidelines for acute kidney injury. Nephron Clin Pract. 2012;120:c179-c184. 10.1159/000339789 [DOI] [PubMed] [Google Scholar]
- 9. Zhu W, Barreto EF, Li J, Lee HK, Kashani K.. Drug-drug interaction and acute kidney injury development: a correlation-based network analysis. PLoS One. 2023;18:e0279928. 10.1371/journal.pone.0279928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Awdishu L, Mehta RL.. The 6R’s of drug induced nephrotoxicity. BMC Nephrol. 2017;18:124. 10.1186/s12882-017-0536-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bartoli E. Adverse effects of drugs on the kidney. Eur J Intern Med. 2016;28:1-8. 10.1016/j.ejim.2015.12.001 [DOI] [PubMed] [Google Scholar]
- 12. Cartin-Ceba R, Kashiouris M, Plataki M, Kor DJ, Gajic O, Casey ET.. Risk factors for development of acute kidney injury in critically ill patients: a systematic review and meta-analysis of observational studies. Crit Care Res Pract. 2012;2012:691013. 10.1155/2012/691013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Schiffl H, Fischer R.. Five-year outcomes of severe acute kidney injury requiring renal replacement therapy. Nephrol Dial Transplant. 2008;23:2235-2241. 10.1093/ndt/gfn182 [DOI] [PubMed] [Google Scholar]
- 14. Blanco VE, Hernandorena CV, Scibona P, Belloso W, Musso CG.. Acute kidney injury pharmacokinetic changes and its impact on drug prescription. Healthcare (Basel). 2019;7:10. 10.3390/healthcare7010010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. DeGrado JR, Gilmore JF, Hohlfelder B, Stevens CA, Gabardi S.. Drug dosing in acute kidney injury. In: Waikar SS, Murray PT, Singh AK, eds. Core Concepts in Acute Kidney Injury. Springer US; 2018:343-361. 10.1007/978-1-4939-8628-6_23 [DOI] [Google Scholar]
- 16. Blot SI, Pea F, Lipman J.. The effect of pathophysiology on pharmacokinetics in the critically ill patient–concepts appraised by the example of antimicrobial agents. Adv Drug Deliv Rev. 2014;77:3-11. 10.1016/j.addr.2014.07.006 [DOI] [PubMed] [Google Scholar]
- 17. Matzke GR, Aronoff GR, Atkinson AJ, et al. Drug dosing consideration in patients with acute and chronic kidney disease—a clinical update from kidney disease: improving global outcomes (KDIGO). Kidney Int. 2011;80:1122-1137. 10.1038/ki.2011.322 [DOI] [PubMed] [Google Scholar]
- 18. Stevens LA, Levey AS.. Measurement of kidney function. Med Clin North Am. 2005;89:457-473. 10.1016/j.mcna.2004.11.009 [DOI] [PubMed] [Google Scholar]
- 19. Rowe JW, Andres R, Tobin JD, Norris AH, Shock NW.. The effect of age on creatinine clearance in men: a cross-sectional and longitudinal study. J Gerontol. 1976;31:155-163. 10.1093/geronj/31.2.155 [DOI] [PubMed] [Google Scholar]
- 20. Hahn T, Yao S, Dunford LM, et al. A comparison of measured creatinine clearance versus calculated glomerular filtration rate for assessment of renal function before autologous and allogeneic BMT. Biol Blood Marrow Transplant. 2009;15:574-579. 10.1016/J.BBMT.2009.01.015 [DOI] [PubMed] [Google Scholar]
- 21. Al-Dorzi HM, Alsadhan AA, Almozaini AS, et al. The performance of equations that estimate glomerular filtration rate against measured urinary creatinine clearance in critically ill patients. Crit Care Res Pract. 2021;2021:5520653. 10.1155/2021/5520653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mann SJ, Gerber LM.. Addressing the problem of inaccuracy of measured 24‐hour urine collections due to incomplete collection. J Clin Hypertens (Greenwich). 2019;21:1626-1634. 10.1111/jch.13696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mizéhoun‐Adissoda C, Houehanou C, Chianéa T, et al. Estimation of daily sodium and potassium excretion using spot urine and 24‐hour urine samples in a black population (Benin). J Clin Hypertens (Greenwich). 2015;18:634-640. 10.1111/jch.12722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gerber LM, Mann SJ.. Development of a model to estimate 24‐hour urinary creatinine excretion. J Clin Hypertens (Greenwich). 2014;16:367-371. 10.1111/jch.12294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cockcroft DW, Gault MH.. Prediction of creatinine clearance from serum creatinine. Nephron. 1976;16:31-41. 10.1159/000180580 [DOI] [PubMed] [Google Scholar]
- 26. Levey AS, Stevens LA, Schmid CH, et al. ; CKD-EPI (Chronic Kidney Disease Epidemiology Collaboration). A new equation to estimate glomerular filtration rate. Ann Intern Med. 2009;150:604-612. 10.7326/0003-4819-150-9-200905050-00006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Inker LA, Schmid CH, Tighiouart H, et al. ; CKD-EPI Investigators. Estimating glomerular filtration rate from serum creatinine and cystatin C. N Engl J Med. 2012;367:20-29. 10.1056/NEJMoa1114248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Inker LA, Eneanya ND, Coresh J, et al. ; Chronic Kidney Disease Epidemiology Collaboration. New creatinine- and cystatin C–based equations to estimate GFR without race. N Engl J Med. 2021;385:1737-1749. 10.1056/NEJMoa2102953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Huang CY, Güiza F, De Vlieger G, Meyfroidt G.. Daily fluctuations in kidney function in critically ill adults. Crit Care. 2022;26:347. 10.1186/s13054-022-04226-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kadivarian S, Heydarpour F, Karimpour H, Shahbazi F.. Measured versus estimated creatinine clearance in critically ill patients with acute kidney injury: an observational study. Acute Crit Care. 2022;37:185-192. 10.4266/acc.2021.01256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Bouchard J, Macedo E, Soroko S, et al. ; Program to Improve Care in Acute Renal Disease. Comparison of methods for estimating glomerular filtration rate in critically ill patients with acute kidney injury. Nephrol Dial Transplant. 2010;25:102-107. 10.1093/ndt/gfp392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Edelstein CL. Biomarkers of acute kidney injury. Adv Chronic Kidney Dis. 2008;15:222-234. 10.1053/j.ackd.2008.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Feng Y, Wang AY, Jun M, et al. Characterization of risk prediction models for acute kidney injury: a systematic review and meta-analysis. JAMA Netw Open. 2023;6:e2313359. 10.1001/JAMANETWORKOPEN.2023.13359 [DOI] [PubMed] [Google Scholar]
- 34. Haredasht FN, Vanhoutte L, Vens C, Pottel H, Viaene L, De Corte W.. Validated risk prediction models for outcomes of acute kidney injury: a systematic review. BMC Nephrol. 2023;24:133. 10.1186/s12882-023-03150-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Vagliano I, Chesnaye NC, Leopold JH, Jager KJ, Abu-Hanna A, Schut MC.. Machine learning models for predicting acute kidney injury: a systematic review and critical appraisal. Clin Kidney J. 2022;15:2266-2280. 10.1093/CKJ/SFAC181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Qian Q, Wu J, Wang J, Sun H, Yang L.. Prediction models for AKI in ICU: a comparative study. Int J Gen Med. 2021;14:623-632. 10.2147/IJGM.S289671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wu J, Chen XY, Zhang H, Xiong LD, Lei H, Deng SH.. Hyperparameter optimization for machine learning models based on Bayesian optimization. J Electron Sci Technol. 2019;17:26-40. 10.11989/JEST.1674-862X.80904120 [DOI] [Google Scholar]
- 38. Kingma DP, Ba J. Adam: A Method for Stochastic Optimization. arXiv, Published online January 29, 2017. 10.48550/arXiv.1412.6980 [DOI]
- 39. Boussina A, Shashikumar SP, Malhotra A, et al. Impact of a deep learning sepsis prediction model on quality of care and survival. NPJ Digit Med. 2024;7:153-159. 10.1038/s41746-023-00986-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Degenhardt F, Seifert S, Szymczak S.. Evaluation of variable selection methods for random forests and omics data sets. Brief Bioinform. 2017;20:492-503. 10.1093/bib/bbx124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Huang C-Y, , GüizaF, , Wouters P,. et al. Development and validation of the creatinine clearance predictor machine learning models in critically ill adults. Crit Care. 2023;27(1):272. 10.1186/s13054-023-04553-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Altman DG, Bland JM.. Measurement in medicine: the analysis of method comparison studies. J R Stat Soc Ser Stat. 1983;32:307-317. 10.2307/2987937 [DOI] [Google Scholar]
- 43. Abadi M, Agarwal A, Barham P, et al. TensorFlow: large-scale machine learning on heterogeneous distributed systems. arXiv, Published online March 16, 2016. 10.48550/arXiv.1603.04467 [DOI]
- 44. Goldstein SL. Automated/integrated real-time clinical decision support in acute kidney injury. Curr Opin Crit Care. 2015;21:485-489. 10.1097/MCC.0000000000000250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tawadrous D, Shariff SZ, Haynes RB, Iansavichus AV, Jain AK, Garg AX.. Use of clinical decision support systems for kidney-related drug prescribing: a systematic review. Am J Kidney Dis. 2011;58:903-914. 10.1053/j.ajkd.2011.07.022 [DOI] [PubMed] [Google Scholar]
- 46. Ravenstijn P, Chetty M, Manchandani P.. Design and conduct considerations for studies in patients with impaired renal function. Clin Transl Sci. 2021;14:1689-1704. 10.1111/cts.13061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Awdishu L, Connor AI, Bouchard J, Macedo E, Chertow GM, Mehta RL.. Use of estimating equations for dosing antimicrobials in patients with acute kidney injury not receiving renal replacement therapy. J Clin Med. 2018;7:211. 10.3390/jcm7080211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jelliffe R. Estimation of creatinine clearance in patients with unstable renal function, without a urine specimen. Am J Nephrol. 2002;22:320-324. 10.1159/000065221 [DOI] [PubMed] [Google Scholar]
- 49. Chen S. Kinetic glomerular filtration rate in routine clinical practice—applications and possibilities. Adv Chronic Kidney Dis. 2018;25:105-114. 10.1053/j.ackd.2017.10.013 [DOI] [PubMed] [Google Scholar]
- 50. Kwong YD, Chen S, Bouajram R, et al. The value of kinetic glomerular filtration rate estimation on medication dosing in acute kidney injury. PLoS One. 2019;14:e0225601. 10.1371/journal.pone.0225601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Takkavatakarn K, Oh W, Chan L, et al. Machine learning derived serum creatinine trajectories in acute kidney injury in critically ill patients with sepsis. Crit Care. 2024;28:156. 10.1186/s13054-024-04935-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bhatraju PK, Mukherjee P, Robinson-Cohen C, et al. Acute kidney injury subphenotypes based on creatinine trajectory identifies patients at increased risk of death. Crit Care. 2016;20:372. 10.1186/s13054-016-1546-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hughes S, Heard KL, Mughal N, Moore LSP.. Optimization of antimicrobial dosing in patients with acute kidney injury: a single-centre observational study. JAC Antimicrob Resist. 2022;4:dlac080. 10.1093/jacamr/dlac080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Lew AK, Crass RL, Eschenauer G.. Evolution of equations for estimating renal function and their application to the dosing of new antimicrobials. Ann Pharmacother. 2019;54:496-503. 101177/1060028019890346 [DOI] [PubMed] [Google Scholar]
- 55. Crass RL, Rodvold KA, Mueller BA, Pai MP.. Renal dosing of antibiotics: are we jumping the gun? Clin Infect Dis. 2019;68:1596-1602. 10.1093/CID/CIY790 [DOI] [PubMed] [Google Scholar]
- 56. Song X, Liu X, Liu F, Wang C.. Comparison of machine learning and logistic regression models in predicting acute kidney injury: a systematic review and meta-analysis. Int J Med Inform. 2021;151:104484. 10.1016/J.IJMEDINF.2021.104484 [DOI] [PubMed] [Google Scholar]
- 57. Cirillo M, Anastasio P, De Santo NG.. Relationship of gender, age, and body mass index to errors in predicted kidney function. Nephrol Dial Transplant. 2005;20:1791-1798. 10.1093/ndt/gfh962 [DOI] [PubMed] [Google Scholar]
- 58. Curtis LM. Sex and gender differences in AKI. Kidney360. 2024;5:160-167. 10.34067/KID.0000000000000321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Syed M, Syed S, Sexton K, et al. Application of machine learning in intensive care unit (ICU) settings using MIMIC dataset: systematic review. Inform MDPI. 2021;8:16. 10.3390/informatics8010016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Kashani K, Rosner MH, Ostermann M.. Creatinine: from physiology to clinical application. Eur J Intern Med. 2019;72:9-14. 10.1016/j.ejim.2019.10.025 [DOI] [PubMed] [Google Scholar]
- 61. Zhang WR, Parikh CR.. Biomarkers of acute and chronic kidney disease. Annu Rev Physiol. 2019;81:309-333. 10.1146/annurev-physiol-020518-114605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Teaford HR, Rule AD, Mara KC, et al. Patterns of cystatin C uptake and utilization across and within hospitals. Mayo Clin Proc. 2020;95:1649-1659. 10.1016/J.MAYOCP.2020.03.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Ramspek CL, Jager KJ, Dekker FW, Zoccali C, van Diepen M.. External validation of prognostic models: what, why, how, when and where? Clin Kidney J. 2021;14:49-58. 10.1093/CKJ/SFAA188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Talari K, Goyal M.. Retrospective studies—utility and caveats. J R Coll Physicians Edinb. 2020;50:398-402. 10.4997/JRCPE.2020.409 [DOI] [PubMed] [Google Scholar]
- 65. Yuan N, Zhang J, Khaki R, et al. Implementation of an electronic health records–based safe contrast limit for preventing contrast-associated acute kidney injury after percutaneous coronary intervention. Circ Cardiovasc Qual Outcomes. 2023;16:e009235. 10.1161/CIRCOUTCOMES.122.009235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wilson FP, Martin M, Yamamoto Y, et al. Electronic health record alerts for acute kidney injury: multicenter, randomized clinical trial. BMJ. 2021;372:m4786. 10.1136/bmj.m4786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Gray MP, Barreto EF, Schreier DJ, et al. Consensus obtained for the nephrotoxic potential of 167 drugs in adult critically ill patients using a modified Delphi method. Drug Saf. 2022;45:389-398. 10.1007/S40264-022-01173-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wang L, McGregor TL, Jones DP, et al. Electronic health record-based predictive models for acute kidney injury screening in pediatric inpatients. Pediatr Res. 2017;82:465-473. 10.1038/pr.2017.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Moffett BS, Goldstei SL.. Acute kidney injury and increasing nephrotoxic-medication exposure in noncritically-ill children. Clin J Am Soc Nephrol. 2011;6:856-863. 10.2215/CJN.08110910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yousif ZK, Koola JD, Macedo E, et al. Clinical characteristics and outcomes of drug-induced acute kidney injury cases. Kidney Int Rep. 2023;80:2333-2344. 10.1016/j.ekir.2023.07.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Yousif Z, Awdishu L.. Drug-induced acute kidney injury risk prediction models. Nephron. 2023;147:44-47. 10.1159/000526267 [DOI] [PubMed] [Google Scholar]
- 72. Grams ME, Sang Y, Coresh J, et al. Acute kidney injury after major surgery: a retrospective analysis of veterans health administration data. Am J Kidney Dis. 2016;67:872-880. 10.1053/j.ajkd.2015.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Li T, Wu B, Li L, et al. Automated electronic alert for the care and outcomes of adults with acute kidney injury: a randomized clinical trial. JAMA Netw Open. 2024;7:e2351710. 10.1001/jamanetworkopen.2023.51710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. VanderWeele TJ, Luedtke AR, van der Laan MJ, Kessler RC.. Selecting optimal subgroups for treatment using many covariates. Epidemiology. 2019;30:334-341. 10.1097/EDE.0000000000000991 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The code supporting this research is available https://github.com/NematiLab/Serum_Creatinine_Prediction. No public repository exists for the study data, as it contains protected health information. Further inquiries can be directed to the corresponding author.