Abstract
Alphavirus budding is driven by interactions between spike and nucleocapsid proteins at the plasma membrane. The binding motif, Y-X-L, on the spike protein E2 and the corresponding hydrophobic cavity on the capsid protein were described earlier. The spike-binding cavity has also been suggested to bind an internal hydrophobic motif, M113-X-I115, of the capsid protein. In this study we found that replacement of amino acids M113 and I115 with alanines, as single or double mutations, abolished formation of intracellular nucleocapsids. The mutants could still bud efficiently, but the NCs in the released virions were not stable after removal of the membrane and spike protein layer. In addition to wild-type spherical particles, elongated multicored particles were found at the plasma membrane and released from the host cell. We conclude that the internal capsid motif has a biological function in the viral life cycle, especially in assembly of nucleocapsids. We also provide further evidence that alphaviruses may assemble and bud from the plasma membrane in the absence of preformed nucleocapsids.
Semliki Forest virus (SFV) is, together with other alphaviruses, such as Sindbis virus (SINV) and Ross River virus, one of the most extensively characterized enveloped viruses (2, 17, 25). This alphavirus particle has a single positive-strand RNA genome packaged into an icosahedral nucleocapsid (NC) built up from 240 copies of the capsid protein (C) which is arranged in hexameric and pentameric capsomer rings. The NC is surrounded by a lipid bilayer derived from the host cell plasma membrane during budding. A total of 240 envelope protein heterodimers (E1-E2), arranged into 80 spikes, traverse the membrane, and the three E2 tails from one spike complex bind capsid proteins from three separate capsomers. Both the NC and the envelope layer have T = 4 symmetry (2, 7).
The structural proteins are translated as polyprotein C-p62 (precursor for E2)-6K-E1 (10). The capsid protein is autoproteolytically cleaved from the nascent chain and remains in the cytoplasm (18). The rest of the polyprotein is translocated into the endoplasmic reticulum via alternating signal and anchor sequences. The proteins are separated by host cell signal peptidases and are transported as a complex to the plasma membrane along the exocytic pathway (11, 15). The current view is that new virus particles are formed at the plasma membrane via interactions between intracellular capsid proteins and the cytoplasmic tails of the E2 spike proteins (26). In this model, an Y-X-L motif in the cytoplasmic domain of E2 interacts with a defined hydrophobic cavity of the capsid protein (19, 22, 23, 32).
Determination of a SIN capsid protein crystal structure revealed that the spike-binding cavity harbored an internal hydrophobic peptide, L-X-L, from a neighboring capsid protein molecule, mimicking the docking of the spike Y and L side chains (14). However, in the NC of the mature virion, the capsid monomers are arranged differently than in the crystal, and here the polypeptide with the motif is not long or flexible enough to reach from one capsid monomer to the other (2).
While the Y-X-L spike motif is completely conserved among alphaviruses, this is not the case for the hydrophobic cavity or for the internal motif of the capsid protein. Molecular modeling shows that the variations in the capsid motif are compensated by the alterations in the capsid cavity, suggesting that the cavity has evolved to fit the capsid motif rather than the cytoplasmic domains of the cognate spikes (13). SFV has the capsid motif M113-X-I115, while SIN has L108-X-L110. The amino acids V136 (M), G137 (E), and Y184 (Y) form the first half of the cavity, and the amino acid residues M141 (M), L168 (M), and C170 (F) form the second half (residues corresponding to SINV residues are in parentheses). A conserved residue, W251 (W), is positioned between the two halves. It should be noted that the M-X-I motif is not found to interact with the hydrophobic pocket in SFV C crystals where residues 1 to 118 have an unordered structure.
Our previous studies of point mutations in the SFV C hydrophobic pocket showed decreased viral titers, which we believed was caused by less efficient capsid-spike interactions (22). Forsell and coworkers analyzed SFV C mutants where residues 105 to 118, including the M-X-I motif, had been deleted. These mutants could not assemble intracellular nucleocapsids but did release particles at the plasma membrane (5). The released particles contained 42S RNA, showing that the region from 105 to 118 is not essential for specific RNA encapsidation (4). The capsid L-X-L motif in SINV has been replaced by charged residues (14). The mutants did not form intracellular NC and produced fewer virions than wild-type SINV. One model suggests that the capsid motif would protect the hydrophobic pocket in its hydrophilic environment prior to spike interaction. Indeed, in crystals of SINV C protein that lack the first 113 residues, both halves of the pocket harbor a dioxane molecule instead of the C motif, showing that the pockets favors ligand binding (13).
The aim of this study was to investigate whether the SFV capsid motif M-X-I plays any important role during the viral life cycle. For this purpose we mutated the M-X-I motif and analyzed the mutated capsid protein in terms of NC formation, NC stability, particle formation and release. We found that mutated particles were produced at wild-type or slightly reduced levels even though no intracellular nucleocapsids could be detected. The morphology of some of the mutated particles was altered, indicating that the internal capsid motif has a function for alphavirus assembly.
MATERIALS AND METHODS
Mutagenesis.
Mutagenesis was performed on the SFV-Helper 1 plasmid using the Quick-change mutagenesis kit (Stratagene, La Jolla, Calif.) with the primers 5′-GAGAAAGAATGTGCGCGAAGATTGAAAATGACTGTATCTTCG-3′ and 5′-CGAAGATACAGTCATTTTCAATCTTCGCGCACATTCTTTCTC-3′ for the M113A mutation, 5′-GAGAAAGAATGTGCATGAAGGCTGAAAATGACTGTATCTTCG-3′ and 5′-CGAAGATACAGTCATTTTCAGCCTTCATGCACATTCTTTCTC-3′ for the I115A mutation, and 5′-GAGAAAGAATGTGCGCGAAGGCTGAAAATGACTGTATCTTCG-3′ and 5′CGAAGATACAGTCATTTTCAGCCTTCGCGCACATTCTTTCTC-3′ for the M113A/I115A mutation. The mutations were checked by sequencing. They were also subcloned into the full-length SFV4 plasmid or the SFV-C plasmid, which express only the capsid protein.
In vitro transcription and transfection.
In vitro transcription of mRNA was carried out using SP6 polymerase as described earlier (16). BHK-21 cells were grown to subconfluent stage in 75-cm2 bottles with 10 to 15 ml of complete BHK medium (BHK21 medium [Gibco] supplemented with 5% fetal bovine serum, 10% tryptose phosphate broth, 20 mM HEPES, 2 mM glutamine, 0.1 U of penicillin/ml, and 0.1 μg of streptomycin/ml). Transfection of cells was done by electroporation as described earlier (16).
Infection of cells.
BHK-21 cell monolayers were infected with wild-type SFV (SFV-WT), SFV-M113A, SFV-I115A, or SFV-M113A/I115A particles at a multiplicity of infection of 10 in minimal essential medium (MEM) with 0.2% bovine serum albumin (Gibco) for 1 h, washed, and incubated in complete BHK medium. The virus stocks used for infection were collected in MEM from transfected cells 7 to 9 h posttransfection.
Metabolic labeling.
For short metabolic labeling of protein, cells were washed with phosphate-buffered saline, overlaid with starvation medium (methionine-free MEM [Gibco], 2 mM glutamine, 20 mM HEPES), and incubated for 30 min. The medium was then replaced by the same medium containing 100 μCi of [35S]methionine (Amersham Pharmacia Biotech)/ml and incubated for 10 to 15 min. The medium was aspirated, and the cells were washed twice with chase medium (Eagle's MEM [E-MEM] [Gibco], 2 mM glutamine, 20 mM HEPES, and 150 μg of unlabeled methionine/ml). After the chase, the cells were washed with phosphate-buffered saline before they were lysed in NP-40 buffer (1% NP-40, 50 mM Tris-HCl [pH 7.6], 150 mM NaCl, 2 mM EDTA, 1 μg of phenylmethylsulfonyl fluoride/ml, 10 mM iodoacetamide) and incubated on ice for 10 min. Alternatively, the cells were scraped off the dish and homogenized in ice-cold buffer containing 10 mM Tris-HCl [pH 7.4], 1 mM EDTA, 2.5 mg of phenylmethylsulfonyl fluoride/ml and 10% (wt/wt) sucrose by 20 strokes in a 23-gauge needle. The lysates and homogenates were cleared by centrifugation in an Eppendorf 5415C centrifuge at 6,000 rpm for 5 min. For double labeling of protein and RNA, [3H]uridine was added 3 to 4 h posttransfection, followed by [35S]methionine labeling 7 h posttransfection as described. The cells were treated with 1 mM actinomycin D prior to labeling to block de novo cellular RNA synthesis (5). For production of labeled virus, transfected cells were labeled with [35S]methionine and [3H]uridine for 15 to 20 h starting 3 h posttransfection.
Titration of virus stocks.
For titration of virus stocks, conventional plaque assay or indirect immunofluorescence was used as described earlier (20).
Gradient ultracentrifugation.
NC and other complex formations were analyzed by ultracentrifugation using either a 5-to-20% or a 15-to-30% linear sucrose gradient in TNE–NP-40 buffer (100 mM Tris-HCl [pH 7.6], 50 mM HCl, 1 mM EDTA, 0.1% NP-40). Lysate (100 to 200 μl) was incubated with 25 mM EDTA for 10 min on ice before being loaded onto the gradient and run at 40,000 rpm for 2 h at 4°C using a Kontron TST 41.14 rotor. Fractions were collected from the bottom. A 40-μl portion of each fraction was mixed with 2 ml of emulsifier-safe liquid scintillation cocktail (Packard, Groningen, The Netherlands), and counted in a beta-counter.
Electron microscopy.
At 7 h posttransfection, cells were incubated with fixation solution (2% glutaraldehyde, 0.1 M sodium cacodylate buffer [pH 7.4]) for 20 min at room temperature. Cells were scraped off and collected by pelleting. Following a wash in 0.1 M sodium cacodylate buffer, the cells were incubated in postfixation buffer (2% osmium tetroxide, 0.1 M sodium cacodylate [pH 7.4]) for 2 h at 4°C. The samples were then dehydrated for 15 min in 70% followed by 95% and finally 100% ethanol at 4°C. Uranyl acetate (2%) was added to the last dehydration step. Cells were placed in pure acetone for 15 min and embedded in LX-112 Epon resin (Ladd, Burlington, Vt.) and polymerized at 60°C. Ultrathin sections were contrasted with uranyl acetate and lead citrate and analyzed in a Philips 420 electron microscope at 80 kV.
Particles produced from 107 cells during 20 h were pelleted through a 20% sucrose cushion by centrifugation at 30,000 rpm for 90 min at 4°C using a Kontron TST 41.14 rotor. The medium was removed by suction, and the pellet was resuspended in TNE buffer (50 mM Tris-HCl [pH 7.4], 100 mM NaCl, 1 mM EDTA) on ice overnight. Virus suspension (3 μl) was placed on Formvar carbon-reinforced Formvar-coated 100 mesh grids for 1 min, stained with 2% uranylacetate for 30 s, and analyzed in a Philips 420 electron microscope at 80 kV.
RESULTS
To study the possible function of the internal M-X-I motif in the C protein, the amino acid residues M113 and/or I115 were replaced by alanines, giving the constructs SFV-H-M113A, SFV-H-I115A and SFV-H-M113A/I115A. To study the effects of the point mutations while eliminating the risk that revertants might spread in culture, the mutations were first introduced into a helper construct, SFV-H, which codes only for the SFV structural proteins (Fig. 1). Since the helper constructs lack the viral replicase, each mutant RNA was electroporated together with an SFV-green fluorescent protein (GFP) reporter construct (Fig. 1) into BHK cells. Production and processing of the structural proteins were analyzed by metabolic labeling followed by cell lysis after different chase times. When total cell lysates were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), we found that the C protein of all constructs was produced in wild-type amounts, it did not aggregate to any great extent (protein soluble in NP-40), and it was stable over several hours. The mutations had not affected the serine protease activity of the capsid protein, since the p62 and E1 polypeptides were produced from the nascent chain with apparently normal kinetics and in wild-type amounts. Finally, the p62 protein, precursor for E2, was cleaved in the longer chase time to E2 (and E3, which is too small to be visible on the gel in Fig. 1). This indicates that correct folding and binding to E1 had occurred followed by transport to the cell surface along the exocytic pathway (Fig. 2A) (30).
FIG. 1.
Constructs used in the study. The internal hydrophobic motif of the capsid protein as well as the capsid-binding motif of the spike is highlighted. Grey boxes represent the transmembrane domains of the proteins p62, 6K, and E1.
FIG. 2.
Analysis of SFV structural protein synthesis and processing showing autoradiographs of SDS–10% PAGE gels. (A) Total NP-40 lysates from cells cotransfected with mRNA from SFV-H and SFV-GFP constructs, labeled with [35S]methionine for 15 min, and chased for 10 min (lanes 1 to 4) or 120 min (lanes 5 to 8). Lanes 1 and 5, SFV-H; lanes 2 and 6, SFV-H-M113A; lanes 3 and 7, SFV-H-I115A; lanes 4 and 8, SFV-H-M113A/I115A. (B) Total NP-40 lysates from cells transfected with mRNA from SFV-C constructs, labeled and chased as in panel A. Lanes 1 and 5, SFV-C; lanes 2 and 6, SFV-C-M113A; lanes 3 and 7, SFV-C-I115A. Note that GFP and C are similar in size and migrate to the same position on the gel, thereby causing a thicker band. Note that the pulses were given at a time posttransfection when replicon-induced shutdown of host protein synthesis had occurred. This also eliminated the need for immunoprecipitation of the samples.
Wild-type SFV capsid protein efficiently encapsidates viral RNA into NC structures in the cytoplasm. This encapsidation is dependent on a packaging signal residing in a region of the RNA molecule that codes for the nsP2 protein of the replicase complex (31). Previous studies have shown that wild-type C folds correctly and assembles into NC when expressed as single protein in the absence of spike proteins and in the absence of C self-cleavage from the nascent chain (21, 26). Therefore, to investigate whether the mutant capsid proteins assemble into intracellular NCs in the absence of spike proteins, the mutations were subcloned into the SFV-C vector (Fig. 1). SFV-C has a stop codon immediately after the capsid gene so that the capsid protein is produced independently of its autoprotease activity. The previous experiment (Fig. 2A) had already shown that the autoprotease activity of the C protein was unaffected by the mutations. The expression of the SFV-C-M113A, SFV-C-I115A, and SFV-C-M113A/I115A mutants was assayed by pulse-chase and analyzed by SDS-PAGE as described above (Fig. 2B). No difference was found in protein expression level between the wild type and mutants. Cell lysates from the short chase time were treated with EDTA to disrupt the polysomes and loaded onto 15-to-30% sucrose gradients. The gradients were then subjected to ultracentrifugation, fractionated, and analyzed by scintillation counting (Fig. 3A). Wild-type capsid proteins assembled intracellular NCs that migrated into the sucrose gradient and gave a peak around fraction 12. In contrast, none of the mutants showed any peak in the same fractions. Instead smaller structures that migrated less far into the gradient were observed with peaks at fractions 18 and 20. Since it is possible that the detergent could disrupt mutant NCs, the experiment was also done with homogenized cells (Fig. 3B). Wild-type NCs from both lysed and homogenized cells banded in the same peak, around fraction 14, while no NC could be detected for the mutant.
FIG. 3.
Analysis of assembled intracellular NCs by sucrose gradient ultracentrifugation of lysates (lys) or homogenates (hom) of transfected cells. (A) Total 35S-labeled cell lysates from cells transfected with SFV-C, SFV-C-M113A, SFV-C-I115A or SFV-C-M113A/I115A were run on 15-to-30% (wt/wt) sucrose gradients and fractionated, and radioactivity was determined by liquid scintillation counting. The bottom of the gradient is to the left. (B) Total 35S-labeled homogenates from cells infected with SFV-WT or SFV-M113A/I115A were run on 15-to-30% (wt/wt) sucrose gradients and fractionated, and radioactivity was determined by liquid scintillation counting. The bottom of the gradient is to the left.
A follow-up experiment used double labeling, where the viral RNA was labeled with [3H]uridine continuously starting 3 h postinfection. The cells were treated with actinomycin D to block cellular, but not viral, RNA synthesis. To allow time for specific labeling of only viral proteins, a pulse with [35S]methionine was initiated 7 h postinfection, when host-cell protein synthesis had been shut off (Fig. 2). The cells were lysed after a short chase, and the cleared lysates were loaded onto a 5-to-20% sucrose gradient to allow better separation of the material than in the previous experiment. In the cells infected with wild-type particles, RNA was found in fractions 6, 7, 11 to 13, 17, and 18 (Fig. 4A), while capsid protein was found in fractions 5 to 8 and 15 to 17 (Fig. 4B). A sample from each fraction was run on SDS–10% PAGE to analyze the protein content in the peak fractions (Fig. 4C). The fraction 3 material represents viral NCs, while material in fraction 16 is probably what was described previously as capsid protein bound to the large ribosomal subunit (24). Fraction 13 is probably the previously identified 90S intermediate complex that is believed to be a precursor form of the NC (29). In this fraction, the RNA-to-protein ratio was clearly higher than in the NC fraction 3, consistent with previous findings (29). The mutant differed from the wild type in the total lack of NCs and in having significantly larger amounts of the 90S material.
FIG. 4.
Analysis of [3H]uridine- and [35S]methionine-labeled cell lysates by sucrose gradient ultracentrifugation. Total lysates from cells infected with SFV-WT, SFV-M113A, SFV-I115A or SFV-M113A/I115A were run on 5-to-20% sucrose gradients and fractionated. The radioactivity in each fraction was determined by liquid scintillation counting. The bottom of the gradient is to the left. (A) Labeling with [3H]uridine. The cells were treated with actinomycin D to block labeling of cellular RNAs. (B) Labeling with [35S]methionine. (C) A sample of each fraction was run on SDS–10% PAGE to see the viral proteins that band in the peak fractions 6 and 16 in panel B. Total lysates are seen in lane T.
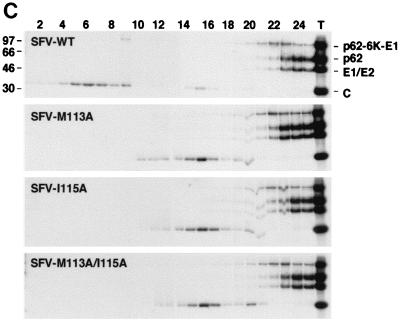
The mutants were further subcloned into the full-length infectious SFV clone, SFV-WT (Fig. 1). A pulse-chase experiment with [35S]methionine labeling of the proteins was done as described above. SDS-PAGE analysis of cleared cell lysates showed correct protein production and processing compared to the wild type (Fig. 5). Particles released into the media from cells transfected with the SFV-WT constructs were pelleted through a sucrose cushion and analyzed by SDS-PAGE. All mutants produced particles in wild-type amounts and with the same capsid-to-spike ratio. To study the budding efficiency more carefully, the medium from cells cotransfected with helper and reporter mRNAs was collected between 7 and 9 h posttransfection and used for subsequent infection of BHK cells grown on coverslips. The titers could be measured directly after 15 h by counting GFP-expressing cells. In this instance, only the reporter construct is packaged in the released particles, and new virions will not form in the absence of the helper RNA that encodes the viral structural proteins. The helper-and-reporter system was also used to minimize the risk of revertants, which may arise in alphavirus infection (1). The M113A and I115A mutations produced particles as efficiently as the wild type, while the double mutation showed about 10-times-lower production (Table 1). The same results were obtained when another reporter construct, SFV-LacZ, was used. These numbers differ from the results obtained when the full-length construct was used and released particles were analyzed by SDS-PAGE (Fig. 5). One possible explanation is that the double mutant affects the encapsidation of RNA. In the first assay, released particles were measured directly, while the second assay also involved infectivity. If the mutations disturb the specificity of RNA encapsidation, uninfectious particles will be produced when the helper RNA or subgenomic RNA is encapsidated. However, this is unlikely, since the SFV-C-105–118 deletion mutant encapsidates 42S RNA specifically (4).
FIG. 5.
Analysis of SFV structural protein synthesis, processing, and release. Total NP-40 lysates from cells transfected with SFV-WT, SFV-M113A, SFV-I115A, or SFV-M113A/I115A were run on SDS–10% PAGE. Cells were labeled with [35S]methionine 7 h posttransfection and chased for 10 min or 2 h. Medium from the longer chase time was loaded on top of a 20% sucrose cushion and centrifuged. The pelleted material was solubilized directly in loading buffer. Lanes 1 to 3, SFV (10-min chase, 2-h chase, and pellet); lanes 4 to 6, SFV-M113A; lanes 7 to 9, SFV-I115A; lanes 10 to 12, SFV-M113A/I115A.
TABLE 1.
Titers of virus stocks produced from transfected BHK cells
| Construct | Titer (IU/ml) obtained witha:
|
|
|---|---|---|
| GFP reporterb | LacZ reporterc | |
| SFV-H | 2 × 107 | 5 × 107 |
| SFV-H-M113A | 3 × 107 | 5 × 107 |
| SFV-H-I115A | 2 × 107 | 4 × 107 |
| SFV-H-M113A/I115A | 3 × 106 | 5 × 106 |
Titers of particles produced from 2.5 × 106 cells over 2 h, beginning 8 h after transfection in 2 ml of medium. Values are means of parallel dilution series in one experiment.
Infected cells were fixed with formaldehyde 15 h posttransfection and counted under fluorescent light.
Infected cells were fixed with methanol 6 h posttransfection and used for indirect immunofluorescence.
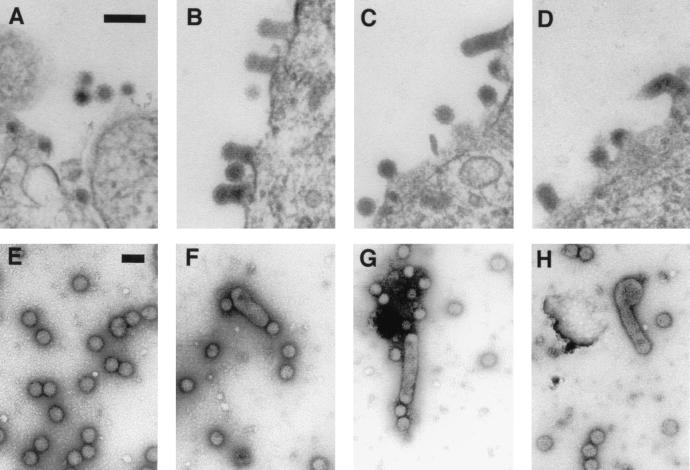
To further study virion assembly, we determined the morphology of the budding at the plasma membrane. Thin sections of cells, transfected with wild-type or mutant full-length RNA, were analyzed by electron microscopy. In addition to wild type-like particles, all mutants also showed elongated budding multicore particles (Fig. 6). Both wild-type and elongated particles could be released from the plasma membrane, as shown by electron micrographs of negatively stained particles produced from cells transfected with SFV-M113A/I115A (Fig. 6).
FIG. 6.
Analysis of particle morphology at the cell membrane and when particles are released. (A through D) Thin sections of BHK cells transfected with SFV-WT (A), SFV-M113A (B), SFV-I115A (C), and SFV-M113A/I115A (D). The cells were fixed 7 h posttransfection. (E through H) Negatively stained particles of SFV-WT (E) and three samples of SFV-M113A/I115A (F through H). Bar = 100 nm.
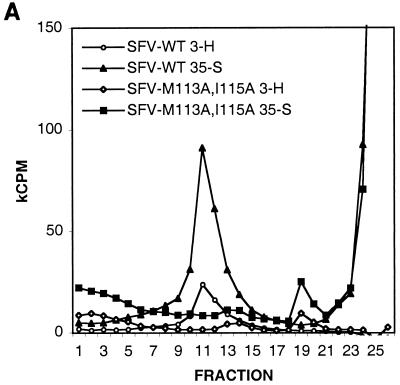
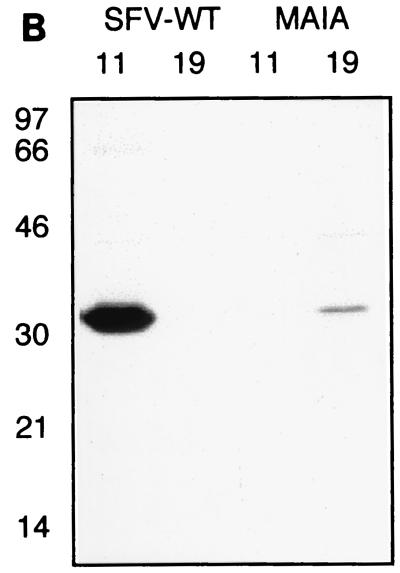
Since the mutations appeared to prohibit formation of intracellular NCs, we analyzed whether NCs of released mutant virions would be stable if the spikes were removed. Labeled virus particles were produced and purified by sedimentation through a sucrose cushion. The pellets were solubilized in NP-40 lysis buffer to disrupt the virions by removing the membrane and spike proteins from the NCs. Equal amounts (counts per minute) of wild-type and mutant material were analyzed by ultracentrifugation using a linear 15-to-30% sucrose gradient as described above (Fig. 7A). Labeled NC material of wild-type virus was found around fraction 11, similar to what was found for intracellular NCs (Fig. 4). In contrast, most of the mutant material had dissociated and was located on top of the gradient. However, a small portion of the material banded around fraction 19. The protein in fractions 11 and 19 was confirmed by SDS-PAGE to be capsid protein (Fig. 7B). Judging from the close to wild-type ratio of C to RNA, the material in fraction 19 might represent ribonucleocomplexes of unfolded NCs. These complexes were disrupted upon RNase treatment, indicating that all of them contained viral RNA (data not shown).
FIG. 7.
Stability analysis of NCs from virions. (A) NP-40 treated 3H- and 35S-labeled particles were run on a 15-to-30% sucrose gradient and fractionated, and radioactivity was determined by liquid scintillation counting. The bottom of the gradient is to the left. (B) Samples from the peak fractions, 11 and 19, were analyzed by SDS-PAGE and autoradiography.
DISCUSSION
Although crystal structures of the SINV C protein show interactions between the capsid cavity of one monomer and the internal motif of another monomer, this is not the case in crystals of the SFV C protein. Moreover, the final arrangement of pentameric or hexameric capsomers in NCs of both SFV and SINV places the C monomers in positions which do not allow interactions between motif and cavity, be it within a protein monomer or between monomers. Moreover, such interactions could not be maintained after spike binding and virus release, since the cavity then is occupied by the spike E2 tail motif. These considerations make it difficult to predict any role for the C protein motif in NC or virus assembly. On the other hand, substitution of the SINV L-X-L motif for amino acid residues with charged bulky side chains affected intracellular NC assembly as well as virus release (14).
One possible function of the capsid motif could be to protect the hydrophobic cavity from the water solvent prior to spike binding. Such docking could preserve the configuration of the capsid protein so that it may efficiently bind the spike complexes later during envelopment and budding. It could also serve to maintain the configuration of the C protein to allow efficient C multimerization and NC assembly. If the C motif does bind to the spike cavity, then one would predict that the C or NC structure would slightly change upon spike binding and virus maturation, since the motif would have to be displaced. Indeed, NCs from detergent-stripped particles appear to be more loosely packed than intracellular NC. They are reported to be more RNase sensitive and migrate faster in sucrose gradients when fractionated by ultracentrifugation (3).
Our present data suggest that the internal hydrophobic motif M-X-I of the SFV capsid protein is indeed involved in the assembly of nucleocapsids and virions. While the mutations did not affect the stability or protease activity of the protein, or its ability to efficiently form mature virions by budding at the plasma membrane, the effects were striking: (i) nucleocapsids could not assemble in the cytoplasm, (ii) the morphology of many budding particles as well as released virions was severely altered, and (iii) NCs from virions were not stable once the spike proteins and membrane had been removed by detergent treatment.
The budding efficiencies of the single mutants were the same as for wild-type virus. The results for the double mutant differ between assays of pelleted labeled particles (Fig. 5) and titration by infection (Table 1). Electron micrographs of released particles showed that the elongated particles were released into the medium. A higher percentage of multicored particles for the double mutant would give more labeled proteins per infectious particle, resulting in a lower titer but a protein amount similar to that of the wild type. However, multicored particles were found in all mutants, but only the double mutant showed a decreased virus titer. It is also possible that the double mutant could incorporate RNAs lacking the specific encapsidation signal. This would also decrease the titer when infectivity is assayed but not when released particles are assayed. However, it was recently shown that a mutant with a deletion of 24 amino acids including the M-X-I motif specifically and efficiently encapsidates 42S RNA (4). Therefore, while the reason for decreased virus titer for the double mutant remains unexplained, our results clearly show that efficient budding can occur in the absence of preformed NCs in the cytoplasm.
The possible interaction between the SINV C cavity and L-X-L motif has been suggested to constitute a first step in capsid assembly prior to RNA binding (14). We have earlier shown that a point mutation, Y184A, in the SFV capsid spike-binding cavity also results in an inability to form intracellular nucleocapsids, further supporting the idea that the capsid cavity plays an important role during NC assembly (22). However, when analyzing the M113A/I115A mutant, we found possible NC assembly intermediates (90S) that were disrupted by RNase treatment, suggesting that the mutants do bind RNA. This suggests that capsid-capsid interactions involving the M-X-I motif occur after RNA binding. In vitro NC assembly assays have shown that RNA is required for dimeric capsid protein complexes to be formed, but no details about the dimeric structure are known (27, 28).
The suggested 90S precursor form for NCs clearly has a higher RNA-to-protein ratio than NCs (Fig. 4, fraction 11) (29). The RNA of the complex was virus derived, since labeling was performed under actinomycin D treatment, which blocks cellular RNA synthesis. Accumulation of 90S precursors has been seen for two NC-deficient C mutants. First, the SINV temperature-sensitive mutant ts-13 (K138 replaced by I) exhibited intracellular accumulation of the 90S complex (12, 29). SINV C residue K138 corresponds to K142 in the SFV C protein and is in position +1 relative to M141, which is one component in the second half of the spike-binding cavity. Therefore, it is possible that the cavity is affected in the ts-13 mutant. Second, accumulation of a complex suggested to be the 90S precursor has also been shown for the C mutant that lacks amino acids 105 to 118, thus lacking the M-X-I motif (5).
In the present work we found particles of altered morphology that were not found in our earlier studies of mutations mapping to the spike-binding cavity. Electron micrographs revealed altered virus structures, with several nucleocapsids within the same membrane envelope in addition to spherical, wild type-like particles. The elongated structures could be caused by a slower NC assembly, so that nucleation of more than one NC starts at the site of assembly before the particle has been pinched off from the membrane. If capsid-capsid interactions are virtually nonexistent, then the observed efficient budding would be completely dependent on and driven by the capsid-spike interactions. Even if some capsid-capsid interactions are in place, they may still not be strong enough to allow formation of complete NCs in the cytoplasm. In both cases the need for achieving tight curvature of the membrane around the NC and the pulling force by the binding of the spike complex could explain the presence of both wild-type and elongated particles.
Other alphavirus mutants that also give rise to multicored particles have been described. In those cases the mutants were either in the spike (E2) cytoplasmic tail or in the small 6K protein, which catalyzes envelopment (8, 9). In the case of the 6K mutants, the defect could be caused by hampering proper binding of spikes to the NC by disturbing the structure of the spike itself or its lateral spike-spike interaction in the plane of the membrane. In the case of the E2 mutant, where a cysteine for serine mutation in the cytoplasmic tail removed a palmitate group, the presentation of the spike motif to the nucleocapsid may have been affected (8, 9, 32). Altogether, these results support a model of alphavirus assembly where correct interactions both in the spike layer and in the nucleocapsid are important for efficient and correct virion formation.
Two other studies have recently shown that the spike proteins can contribute to the NC assembly, similar to what is found for the point mutations in this study (6, 22). They analyzed a mutant with a large deletion (amino acid 40 to 118) in the capsid protein, including the M-X-I motif. In both studies the NCs collapsed into smaller structures or monomers when mutated virus was treated with detergent to remove the spike and membrane layer. This further shows that the capsid interactions in the mutated NCs are weaker than in wild-type NCs, which are stable even after removal of the spike and membrane. The M113A/I115A mutant NC dissociated into smaller structures that were similar in size to the structures found when assaying for intracellular NC assembly. It is possible that they represent small building blocks for NC assembly. In vitro assays of SINV NC assembly suggest that the first building blocks are capsid protein dimers, rather than capsomers, in complex with RNA (28). In our study we have shown that the smaller structures are disrupted upon RNase treatment, supporting the in vitro data.
Collectively our results suggest that the internal capsid motif M-X-I is involved in the assembly of SFV nucleocapsids. The M-X-I mutants form precursors but not complete NCs, suggesting a model for NC assembly where an interaction involving the M-X-I motif and the spike-binding pocket is involved in a rather late step. The defect can be rescued by interactions with the spikes at the plasma membrane, since infectious particles are released from the cell as efficiently as the wild type. Even so, this frequently resulted in virions with altered morphology and in NCs with altered structures, which were unstable upon envelope removal. In contrast to earlier studies using NC-deficient C mutants with big deletions, including the M-X-I motif, which also decreased the efficiency of virus formation (5, 6), we have described NC-deficient mutants with unaffected virus production.
ACKNOWLEDGMENTS
We thank Kjell Hultenby for excellent technical assistance with electron microscopy assays.
This work was supported by The Swedish Medical Research Council, The Swedish Natural Science Research Council, and Swedish Research Council for Engineering Sciences.
REFERENCES
- 1.Berglund P, Sjöberg M, Garoff H, Atkins G J, Sheahan B J, Liljeström P. Semliki Forest virus expression system: production of conditionally infectious recombinant particles. Nat Biotechnol. 1993;11:916–920. doi: 10.1038/nbt0893-916. [DOI] [PubMed] [Google Scholar]
- 2.Cheng R H, Kuhn R J, Olson N H, Rossmann M G, Choi H K, Smith T J, Baker T S. Nucleocapsid and glycoprotein organization in an enveloped virus. Cell. 1995;80:621–630. doi: 10.1016/0092-8674(95)90516-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coombs K, Brown B, Brown D T. Evidence for a change in capsid morphology during Sindbis virus envelopment. Virus Res. 1984;1:297–302. doi: 10.1016/0168-1702(84)90018-2. [DOI] [PubMed] [Google Scholar]
- 4.Forsell K. The role of the capsid protein in Semliki Forest virus assembly. Ph.D. thesis. Stockholm, Sweden: Karolinska Institute; 2000. [Google Scholar]
- 5.Forsell K, Griffiths G, Garoff H. Preformed cytoplasmic nucleocapsids are not necessary for alphavirus budding. EMBO J. 1996;15:6495–6505. doi: 10.1002/j.1460-2075.1996.tb01040.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forsell K, Xing L, Kozlovska T, Cheng R H, Garoff H. Membrane proteins organize a symmetrical virus. EMBO J. 2000;19:5081–5091. doi: 10.1093/emboj/19.19.5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuller S D. The T = 4 envelope of Sindbis virus is organized by interactions with a complementary T = 3 capsid. Cell. 1987;48:923–934. doi: 10.1016/0092-8674(87)90701-x. [DOI] [PubMed] [Google Scholar]
- 8.Gaedigk-Nitschko K, Ding M X, Levy M A, Schlesinger M J. Site-directed mutations in the Sindbis virus 6K protein reveal sites for fatty acylation and the underacylated protein affects virus release and virion structure. Virology. 1990;175:282–291. doi: 10.1016/0042-6822(90)90210-i. [DOI] [PubMed] [Google Scholar]
- 9.Gaedigk-Nitschko K, Schlesinger M J. Site-directed mutations in Sindbis virus E2 glycoprotein's cytoplasmic domain and the 6K protein lead to similar defects in virus assembly and budding. Virology. 1991;183:206–214. doi: 10.1016/0042-6822(91)90133-v. [DOI] [PubMed] [Google Scholar]
- 10.Garoff H, Frischauf A-M, Simons K, Lehrach H, Delius H. Nucleotide sequence of cDNA coding for Semliki Forest virus membrane glycoproteins. Nature. 1980;288:236–241. doi: 10.1038/288236a0. [DOI] [PubMed] [Google Scholar]
- 11.Garoff H, Huylebroeck D, Robinson A, Tillman U, Liljeström P. The signal sequence of the p62 protein of Semliki Forest virus is involved in initiation but not in completing chain translocation. J Cell Biol. 1990;111:867–876. doi: 10.1083/jcb.111.3.867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hahn C S, Strauss E G, Strauss J H. Sequence analysis of three Sindbis virus mutants temperature-sensitive in the capsid protein autoprotease. Proc Natl Acad Sci USA. 1985;82:4648–4652. doi: 10.1073/pnas.82.14.4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee S, Kuhn R J, Rossman M G. Probing the potential glycoprotein binding site of Sindbis virus capsid protein with dioxane and model building. Proteins. 1998;33:311–317. [PubMed] [Google Scholar]
- 14.Lee S, Owen K E, Choi H K, Lee H, Lu G G, Wengler G, Brown D T, Rossmann M G, Kuhn R J. Identification of a protein binding site on the surface of the alphavirus nucleocapsid and its implication in virus assembly. Structure. 1996;4:531–541. doi: 10.1016/s0969-2126(96)00059-7. [DOI] [PubMed] [Google Scholar]
- 15.Liljeström P, Garoff H. Internally located cleavable signal sequences direct the formation of Semliki Forest virus membrane proteins from a polyprotein precursor. J Virol. 1991;65:147–154. doi: 10.1128/jvi.65.1.147-154.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liljeström P, Lusa S, Huylebroeck D, Garoff H. In vitro mutagenesis of a full-length cDNA clone of Semliki Forest virus: the 6,000-molecular-weight membrane protein modulates virus release. J Virol. 1991;65:4107–4113. doi: 10.1128/jvi.65.8.4107-4113.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mancini E J, Clarke M, Gowen B E, Rutten T, Fuller S D. Cryo-electron microscopy reveals the functional organization of an enveloped virus, Semliki Forest virus. Mol Cell. 2000;5:255–266. doi: 10.1016/s1097-2765(00)80421-9. [DOI] [PubMed] [Google Scholar]
- 18.Melancon P, Garoff H. Processing of the Semliki Forest virus structural polyprotein: role of the capsid protease. J Virol. 1987;61:1301–1309. doi: 10.1128/jvi.61.5.1301-1309.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Owen K E, Kuhn R J. Alphavirus budding is dependent on the interaction between the nucleocapsid and hydrophobic amino acids on the cytoplasmic domain of the E2 envelope glycoprotein. Virology. 1997;230:187–196. doi: 10.1006/viro.1997.8480. [DOI] [PubMed] [Google Scholar]
- 20.Salminen A, Wahlberg J M, Lobigs M, Liljeström P, Garoff H. Membrane fusion process of Semliki Forest virus. II. Cleavage-dependent reorganization of the spike protein complex controls virus entry. J Cell Biol. 1992;116:349–357. doi: 10.1083/jcb.116.2.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Skoging U, Liljeström P. Role of the C-terminal tryptophan residue for the structure-function of alphavirus capsid protein. J Mol Biol. 1998;279:865–872. doi: 10.1006/jmbi.1998.1817. [DOI] [PubMed] [Google Scholar]
- 22.Skoging U, Vihinen M, Nilsson L, Liljeström P. Aromatic interactions define the binding of the alphavirus spike to its nucleocapsid. Structure. 1996;4:519–529. doi: 10.1016/s0969-2126(96)00058-5. [DOI] [PubMed] [Google Scholar]
- 23.Skoging-Nyberg U, Liljeström P. A conserved leucine in the cytoplasmic domain of Semliki Forest virus spike protein is important for budding. Arch Virol. 2000;145:1225–1230. doi: 10.1007/s007050070121. [DOI] [PubMed] [Google Scholar]
- 24.Söderlund H, Ulmanen I. Transient association of Semliki Forest virus capsid protein with ribosomes. J Virol. 1977;24:907–909. doi: 10.1128/jvi.24.3.907-909.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Strauss J H, Strauss E G. The alphaviruses: gene expression, replication, and evolution. Microbiol Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suomalainen M, Liljeström P, Garoff H. Spike protein-nucleocapsid interactions drive the budding of alphaviruses. J Virol. 1992;66:4737–4747. doi: 10.1128/jvi.66.8.4737-4747.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tellinghuisen T L, Hamburger A E, Fisher B R, Ostendorp R, Kuhn R J. In vitro assembly of alphavirus cores by using nucleocapsid protein expressed in Escherichia coli. J Virol. 1999;73:5309–5319. doi: 10.1128/jvi.73.7.5309-5319.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tellinghuisen T L, Kuhn R J. Nucleic acid-dependent cross-linking of the nucleocapsid protein of Sindbis virus. J Virol. 2000;74:4302–4309. doi: 10.1128/jvi.74.9.4302-4309.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ulmanen I. Assembly of Semliki Forest virus nucleocapsid: detection of a precursor in infected cells. J Gen Virol. 1978;41:353–365. doi: 10.1099/0022-1317-41-2-353. [DOI] [PubMed] [Google Scholar]
- 30.Wahlberg J M, Boere W A, Garoff H. The heterodimeric association between the membrane proteins of Semliki Forest virus changes its sensitivity to low pH during virus maturation. J Virol. 1989;63:4991–4997. doi: 10.1128/jvi.63.12.4991-4997.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.White C L, Thomson M, Dimmock N J. Deletion analysis of a defective interfering Semliki Forest virus RNA genome defines a region in the nsP2 sequence that is required for efficient packaging of the genome into virus particles. J Virol. 1998;72:4320–4326. doi: 10.1128/jvi.72.5.4320-4326.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao H, Lindqvist B, Garoff H, von Bonsdorff C H, Liljeström P. A tyrosine-based motif in the cytoplasmic domain of the alphavirus envelope protein is essential for budding. EMBO J. 1994;13:4204–4211. doi: 10.1002/j.1460-2075.1994.tb06740.x. [DOI] [PMC free article] [PubMed] [Google Scholar]