Abstract
PURPOSE
Validated and accurate prognostic testing is critical for precision medicine in uveal melanoma (UM). Our aims were to (1) prospectively validate an integrated prognostic classifier combining a 15-gene expression profile (15-GEP) and PRAME RNA expression and (2) identify clinical variables that enhance the prognostic accuracy of the 15-GEP/PRAME classifier.
MATERIALS AND METHODS
This study included 1,577 patients with UM of the choroid and/or ciliary body who were enrolled in the Collaborative Ocular Oncology Group Study Number 2 (COOG2) and prospectively monitored across 26 North American centers. Test results for 15-GEP (class 1 or class 2) and PRAME expression status (negative or positive) were available for all patients. The primary end point was metastasis-free survival (MFS).
RESULTS
15-GEP was class 1 in 1,082 (68.6%) and class 2 in 495 (31.4%) patients. PRAME status was negative in 1,106 (70.1%) and positive in 471 (29.9%) patients. Five-year MFS was 95.6% (95% CI, 93.9 to 97.4) for class 1/PRAME(–), 80.6% (95% CI, 73.9 to 87.9) for class 1/PRAME(+), 58.3% (95% CI, 51.1 to 66.4) for class 2/PRAME(–), and 44.8% (95% CI, 37.9 to 52.8) for class 2/PRAME(+). By multivariable Cox proportional hazards analysis, 15-GEP was the most important independent predictor of MFS (hazard ratio [HR], 5.95 [95% CI, 4.43 to 7.99]; P < .001), followed by PRAME status (HR, 1.82 [95% CI, 1.42 to 2.33]; P < .001). The only clinical variable demonstrating additional prognostic value was tumor diameter.
CONCLUSION
In the largest prospective multicenter prognostic biomarker study performed to date in UM to our knowledge, the COOG2 study validated the superior prognostic accuracy of the integrated 15-GEP/PRAME classifier over 15-GEP alone and clinical prognostic variables. Tumor diameter was found to be the only clinical variable to provide additional prognostic information. This prognostic classifier provides an advanced resource for risk-adjusted metastatic surveillance and adjuvant trial stratification in patients with UM.
INTRODUCTION
Uveal melanoma (UM) is the most common primary malignancy of the eye and has a strong propensity for metastasis.1 Despite improvements in primary tumor treatment, there has been no survival increase,2 due at least in part to early subclinical micrometastasis.3,4 Tebentafusp recently became the first drug showing a survival benefit in UM patients with metastatic disease restricted to HLA:A02-01.5-7 It is likely that such therapies will have even greater benefit in the adjuvant or early metastatic setting when disease burden is low.8-12 As such, there is a critical need for validated, standardized, and highly accurate prognostic testing to tailor surveillance to metastatic risk and to identify high-risk patients for enrollment in adjuvant clinical trials.1 Such testing can also reduce surveillance in low-risk patients, which may decrease test-related anxiety and cost to the health care system.
CONTEXT
Key Objective
The key objective of this study was to prospectively evaluate the 15-gene expression profile (15-GEP) and PRAME RNA expression status as an integrated prognostic tool in 1,577 patients with uveal melanoma (UM) enrolled in the Collaborative Ocular Oncology Study Number 2 (COOG2).
Knowledge Generated
PRAME status significantly enhanced the prognostic accuracy of 15-GEP in patients with UM. Tumor diameter made a small additional contribution to prognostic accuracy.
Relevance (G.K. Schwartz)
The addition of PRAME RNA expression to the 15-GEP offers an enhanced diagnostic tool to predict clinical outcome in patients with primary UM.*
*Relevance section written by JCO Associate Editor Gary K. Schwartz, MD, FASCO.
While there are numerous clinical, histopathologic, and molecular factors that have been proposed as prognostic variables in UM,1 gene expression profile (GEP) has been shown to provide prognostic accuracy superior to other factors.13-17 Accordingly, a standardized 15-GEP was developed using targeted cDNA amplification, microfluidics qPCR technology, and machine learning to classify primary UMs from a biopsy sample.18 The 15-GEP has undergone analytic optimization for use on fine needle and formalin-fixed samples.19 The 15-GEP was prospectively validated by the Collaborative Ocular Oncology Group Study Number 1 (COOG1) 17 and numerous subsequent studies.19-23 It is included in National Comprehensive Cancer Network guidelines and is widely used in routine clinical practice.1,20
In subsequent studies, RNA expression of the cancer-testis antigen Preferentially Expressed Antigen in Melanoma (PRAME) was found to provide additional prognostic information independent of 15-GEP, being associated with increased metastatic risk in both class 1 and class 2 tumors.24-27 Although the initial COOG1 study did not identify any clinical factors that provided prognostic information independent of 15-GEP, subsequent retrospective studies have suggested that tumor diameter may enhance the accuracy of 15-GEP.21,28-31 In this first report of the Collaborative Ocular Oncology Group Number 2 (COOG2), to our knowledge, the largest prospective biomarker study performed to date in UM, we evaluate the prognostic value of 15-GEP, PRAME, and clinical prognostic factors in developing an integrated prognostic classifier suitable for routine clinical practice and clinical trial stratification.
MATERIALS AND METHODS
Patient Enrollment
Between January 2017 and April 2020, COOG2 prospectively enrolled 1,687 patients with UM involving the choroid, ciliary body and/or iris across 26 ocular oncology centers in the United States and Canada (Appendix Table A1, online only). Informed consent was obtained from each patient. Primary treatment was performed according to the standard at each center. Federal Wide Assurance from the Office of Human Research Protections and Institutional Review Board (IRB) or Ethics Committee approval was obtained in accordance with policies at each center. Noninclusion criteria included patient age <18 years, diagnosis of a uveal tumor other than UM (eg, metastatic cancer), prior radiotherapy, and patient withdrawal from the study. Fifty-one patients who met entry criteria were not included because their tumor sample was inadequate to allow reverse transcription of RNA and/or amplification of cDNA for GEP and PRAME testing. Prior photodynamic therapy or transpupillary thermotherapy was allowed if there was evidence of tumor regrowth. No participants were excluded on the basis of sex, ethnicity, or race. A data lock was performed on March 9, 2023, and patients with primary iris melanoma (n = 101) or metastatic UM at baseline (n = 9) were excluded, resulting in 1,577 patients included in this report. Given the published distribution and metastatic rates of class 1A, class 1B, class 2, PRAME-, and PRAME+ in UM,24,25,27,32 a sample size of approximately 1,500 was estimated to yield sufficient patients with discordant PRAME versus 1A/1B results to allow detection of a relative risk of >3.0 in 5-year metastatic rates between PRAME+/1A compared with PRAME-/1B with >80% power.
Tumor Sample Analysis
All patients underwent testing of their primary UM sample with DecisionDx-UM (class 1A, class 1B or class 2) and DecisionDx-PRAME (negative or positive), as previously described.18,24 This testing was performed by the Castle Biosciences College of American Pathologists–accredited, Clinical Laboratory Improvement Amendments–certified laboratory, as per standard of care.19,33 For 369 (22%) of the patients, molecular analysis was completed on residual clinical samples collected between May 2014 and December 2016, before enrollment in COOG2, after which they continued to be monitored prospectively. The median time from sample collection to COOG2 initiation was 11 months (0.1 to 32 months).
Data Management
REDCap, a secure HIPAA compliant application,35 was used for electronic data management. Each center was given restricted access by key study personnel and were issued unique research ID numbers to assign study patients. Deidentified clinical data were entered by each center at baseline and subsequent follow-up intervals. Baseline data included date of enrollment, date and method of biopsy, cytology result (if available), date and method of primary tumor treatment, patient age at study entry, sex, self-reported race and ethnicity, iris color (blue/green, intermediate, or brown), tumor diameter, tumor thickness, ciliary body involvement, and metastatic status. The American Joint Committee on Cancer (AJCC) 8th edition36 was used for tumor staging. Follow-up data included local tumor recurrence (tumor regrowth in the eye or orbit after radiotherapy or in the orbit after enucleation), metastatic status, date and location of initial metastasis, systemic status at last follow-up, and date and cause of death. Molecular test results were entered into REDCap by Castle Biosciences, which was masked to other REDCap data. Each center was masked to data entered by other centers and by Castle Biosciences. Only the coordinating center and COOG2 Data Committee had access to all data (Data Supplement, online only).
Baseline and follow-up ophthalmic visits were performed as per standard of care at each center but typically included a comprehensive ophthalmic examination, fundus photography, optical coherence tomography, and ultrasonography performed at least every 3-4 months for the first year after treatment, every 4-6 months for the second year, and every 6-12 months thereafter. Baseline systemic imaging was typically performed with computed tomography (CT) of the chest, abdomen, and pelvis. Subsequent systemic surveillance typically included imaging of the liver with CT, magnetic resonance imaging, or ultrasound at least twice a year, along with chest CT or chest x-ray at least once a year.
Statistical Analysis
Statistical analysis was performed using SAS 9.4 (SAS Institute, Cary, NC). The Chi-square test was used to compare categorical variables, and Wilcoxon signed-rank test was used for comparing continuous variables. All statistical tests were two-sided, and statistical significance was defined as P < .05. Differences in metastasis-free survival (MFS, time from primary tumor treatment to first radiographic detection of metastatic disease) and melanoma-specific survival (MSS, time from primary tumor treatment to death due to melanoma metastasis) associated with a given factor were evaluated using Kaplan-Meier (KM) survival curves and the log-rank test. Cox regression was used to assess the contribution of multiple factors influencing metastatic risk. Univariable and multivariable Cox models were constructed to assess the impact of variables both separately and in combination. Model performance was evaluated by the concordance statistic (C-statistic), which measures the ability of a model to accurately rank individuals by their predicted risk.
RESULTS
Patient Enrollment
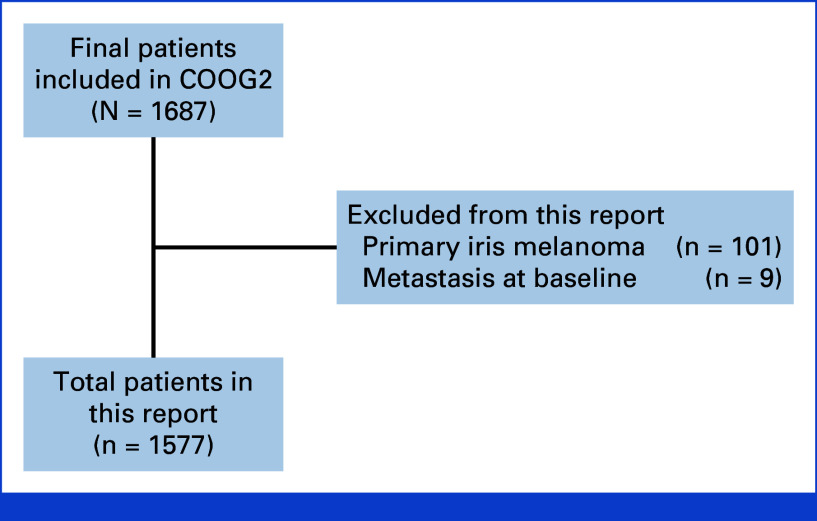
Of 1,687 total patients enrolled in COOG2, 101 patients with primary iris melanoma and nine patients with metastatic UM at baseline were excluded, resulting in 1,577 patients included in this report (Fig 1). Baseline demographic and clinical information are summarized in Table 1. The median follow-up was 43.6 months. Metastatic disease was detected in 269 (17.1%) patients, and the median time to metastasis among patients with an event was 22.6 months (range, 0.1-92.9). Local tumor recurrence was identified in 68 (4.3%) patients with a median time of 23.9 months (range, 3.5-82.2 months) after biopsy/primary enucleation, with 30 (44.1%) of these subsequently developing metastatic disease. The study included 530 (33.6%) AJCC T1 tumors, compared with 81 (17.6%) in COOG117 and 0 (0%) in The Cancer Genome Atlas (TCGA) cohort37 (Appendix Table A2). Conversely, 161 (10.2%) of patients in this study were treated with primary enucleation, compared with 92 (20.0%) in COOG1 and 77 (96.3%) in TCGA. The median follow-up time was 43.6 months for COOG2 compared with 17.4 months for COOG1 and 26.2 months for TCGA.
FIG 1.
Overview of Collaborative Ocular Oncology Group Study 2 (COOG2) and patients included in this report.
TABLE 1.
Summary of COOG2 Cohort of 1,577 Patients With Posterior Uveal Melanoma
| Characteristic | Value |
|---|---|
| Age at study entry, years | |
| Median (range) | 64 (18-99) |
| Mean (SD) | 62 (13.5) |
| Male sex, No. (%) | 809 (51.3) |
| Ethnicity, No. (%) | |
| Non-Hispanic or Latino | 1,494 (94.7) |
| Hispanic or Latino | 55 (3.5) |
| Not specified | 28 (1.8) |
| Race, No. (%) | |
| White | 1,518 (96.3) |
| Black | 12 (0.8) |
| Asian | 7 (0.4) |
| Native American/Alaskan | 3 (0.2) |
| Native Hawaiian/Pacific Islander | 2 (0.1) |
| More than one race | 5 (0.3) |
| Not specified | 30 (1.9) |
| Eye, right, No. (%) | 801 (50.8) |
| Iris color, No. (%) | |
| Blue/green | 613 (38.9) |
| Brown | 238 (15.1) |
| Intermediate | 127 (8.1) |
| Not specified | 599 (38.0) |
| Ciliary body involvement, No. (%) | 250 (15.9) |
| Melanocytosis, No. (%) | 75 (4.7) |
| Cell type, No. (%) | |
| Spindle | 252 (16.0) |
| Mixed | 234 (14.8) |
| Epithelioid | 76 (4.8) |
| Other | 309 (19.6) |
| Not performed or not specified | 706 (44.8) |
| Tumor diameter, mm | |
| Median (range) | 12 (2-32) |
| Mean (SD) | 12.1 (±4.0) |
| Tumor thickness, mm | |
| Median (range) | 4.1 (0.5-18) |
| Mean (SD) | 5.1 (±3.1) |
| AJCC T-category, No. (%) | |
| T1 | 530 (33.6) |
| T2 | 564 (35.8) |
| T3 | 360 (22.8) |
| T4 | 123 (7.8) |
| Primary tumor treatment | |
| 125I plaque brachytherapy | 1,265 (80.2) |
| Enucleation | 161 (10.2) |
| Proton beam radiotherapy | 115 (7.3) |
| External beam radiotherapy | 11 (0.7) |
| Laser therapy | 8 (0.5) |
| Other or unspecified | 17 (1.1) |
| Type of biopsy, No. (%) | |
| Transvitreal FNAB | 699 (44.3) |
| Transscleral FNAB | 638 (40.5) |
| Transcameral FNAB | 4 (0.3) |
| Vitrectomy biopsy | 67 (4.3) |
| Incisional biopsy | 6 (0.4) |
| Not specified | 163 (10.3) |
| 15-GEP test results, No. (%) | |
| Class 1 | 1,082 (68.6) |
| Class 1A | 693 (43.9) |
| Class 1B | 389 (24.7) |
| Class 2 | 495 (31.4) |
| PRAME, No. (%) | |
| Negative (–) | 1,106 (70.1) |
| Positive (+) | 471 (29.9) |
| Distant metastasis, No. (%) | 269 (17.1) |
| Time to metastasis/last follow-up, months | |
| Median (range) | 43.6 (0-104.4) |
| Mean (SD) | 45.2 (±22.4) |
| Local recurrence, No. (% of all patients) | 68 (4.3) |
| Primary tumor treatment, No. (% of 68 local recurrences) | |
| 125I plaque brachytherapy | 48 (70.6) |
| Proton beam radiotherapy | 12 (17.6) |
| Enucleation | 5 (7.4) |
| Other | 3 (4.4) |
| Time to local recurrence, months | |
| Median (range) | 23.9 (3.5-82.2) |
| Mean (SD) | 28.4 (19.6) |
Abbreviations: 15-GEP, 15-gene expression profile test; AJCC, American Joint Committee on Cancer; FNAB, fine-needle aspiration biopsy; SD, standard deviation.
15-GEP and PRAME
Molecular test results are summarized in Table 1. The 15-GEP result was class 1A in 693 patients (43.9%), class 1B in 389 (24.7%), and class 2 in 495 (31.4%). KM survival analysis and the log-rank test demonstrated no significant difference in MFS or MSS between class 1A and 1B cases (Appendix Fig A1). Therefore, these groups were combined and reported as one category (class 1) for all subsequent analyses. The development of metastatic disease was noted in 64 (5.9%) of 1,082 patients with a class 1 tumor versus 205 (41.4%) of 495 patients with a class 2 tumor (P < .001). Class 1 tumors were associated with superior actuarial survival compared with class 2 tumors, with 5-year MFS of 92.3% (95% CI, 90.2 to 94.4) for class 1 versus 52.1% (95% CI, 47.0 to 57.8) for class 2, and 5-year MSS of 97.4% (95% CI, 96.2 to 98.7) for class 1 versus 68.8% (95% CI, 63.6 to 74.5) for class 2 (Figs 2A and 2B; Appendix Table A3).
FIG 2.
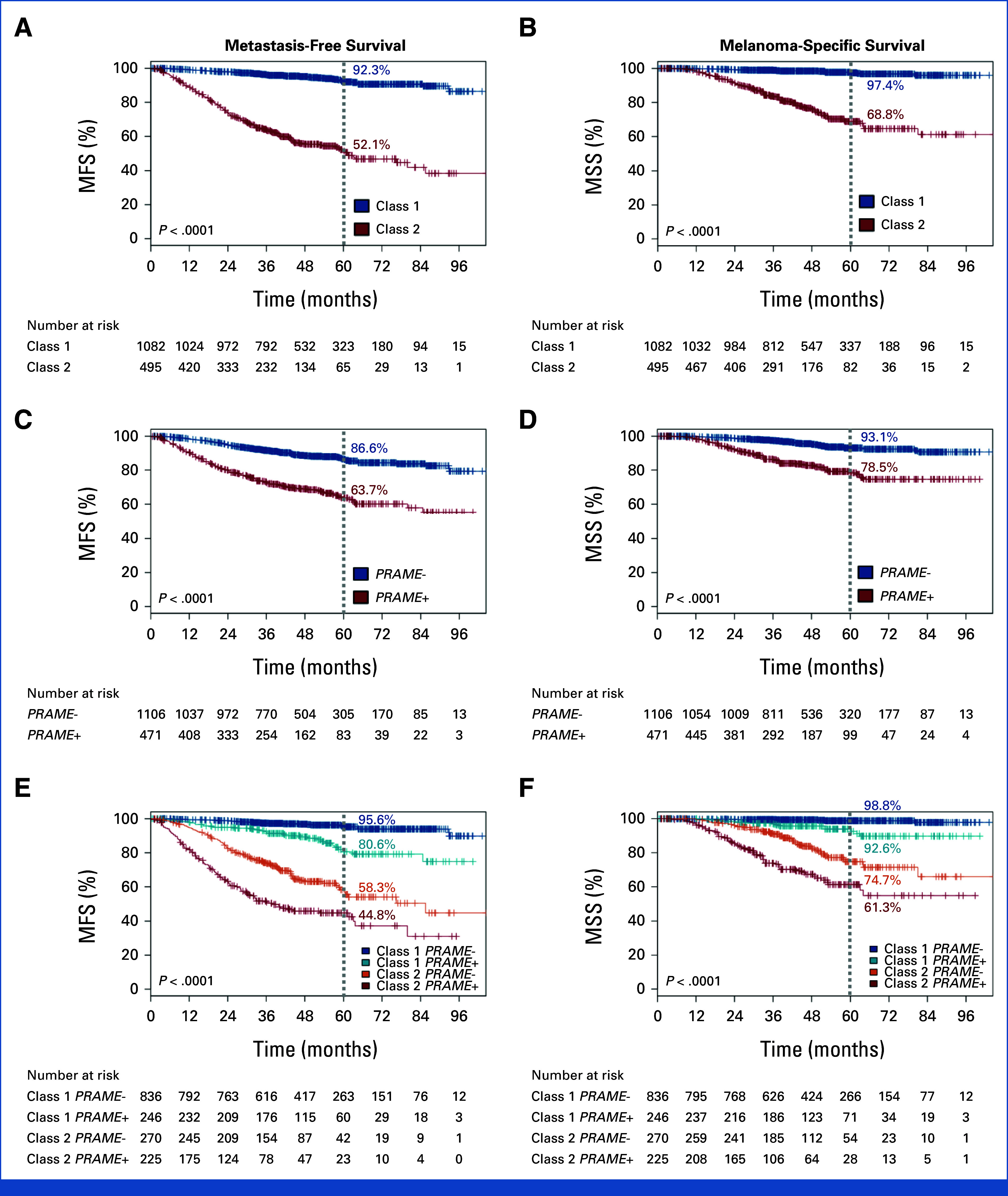
Metastasis-free survival (left panels) and melanoma-specific survival (right panels) stratified by (A) and (B) 15-gene expression profile (15-GEP) class, (C) and (D) PRAME RNA expression status, and (E) and (F) integrated 15-GEP/PRAME subclassifications. Five-year point estimates are indicated by the gray dotted line. P value, log-rank test, and number of patients at risk at each 12-month interval are shown. Censoring events are marked with a vertical hash across the survival curve (See Appendix Table A3 for complete 3-year and 5-year outcomes with 95% CIs, along with total number of metastasis/death events on study). MFS, metastasis-free survival; MSS, melanoma-specific survival.
PRAME status was negative (PRAME–) in 1,106 patients (70.1%) and positive (PRAME+) in 471 (29.9%). PRAME– status was associated with better survival, with a 5-year MFS of 86.6% (95% CI, 84.2 to 89.1), compared with 63.7% (95% CI, 58.5 to 69.3) for PRAME+ tumors. Similarly, 5-year MSS was 93.1% (95% CI, 91.2 to 95.0) for PRAME– tumors compared with 78.5% (95% CI, 73.9 to 83.3) for PRAME+ tumors (Figs 2C and 2D; Appendix Table A3). Furthermore, PRAME+ status was associated with worse survival in both class 1 and class 2 tumors, with 5-year MFS of 95.6% (95% CI, 93.9 to 97.4) in class 1/PRAME–, 80.6% (95% CI, 73.9 to 87.9) in class 1/PRAME+, 58.3% (95% CI, 51.1 to 66.4) in class 2/PRAME–, and 44.8% (95% CI, 37.9 to 52.8) in class 2/PRAME+ (Figs 2E and 2F; Appendix Table A3). The median time to metastasis for patients with a metastatic event was 31.3 months for class 1/PRAME–, 34.9 months for class 1/PRAME+, 24.7 months for class 2/PRAME–, and 16.5 months for class 2/PRAME+ (Table 2). Overall, class 2/PRAME+ patients had a greatly increased risk of metastasis compared with class 1/PRAME– patients (hazard ratio [HR], 22.06 [95% CI, 14.86 to 32.76]; P < .001; Appendix Table A4).
TABLE 2.
Summary of Enrolled Patients by 15-GEP/PRAME Status
| Characteristic | Class 1 PRAME (–) |
Class 1 PRAME (+) |
Class 2 PRAME (–) |
Class 2 PRAME (+) |
|---|---|---|---|---|
| No. (% of 1,577 in total cohort) | 836 (53.0) | 246 (15.6) | 270 (17.1) | 225 (14.3) |
| Age at study entry, years | ||||
| Median (range) | 62 (19-94) | 61 (18-99) | 66 (32-98) | 66 (22-95) |
| Mean (SD) | 61 (13.7) | 60 (14.1) | 65 (12.2) | 66 (12.0) |
| Male sex, No. (%) | 437 (52.3) | 128 (52.0) | 136 (50.4) | 108 (48.0) |
| Eye, right, No. (%) | 434 (51.9) | 117 (47.6) | 135 (50.0) | 115 (51.1) |
| Ciliary body involvement, No. (%) | 83 (9.9) | 37 (15.0) | 55 (20.4) | 75 (33.3) |
| Tumor diameter, mm | ||||
| Median (range) | 10.2 (2-26) | 13 (4-32) | 13.6 (4-28.7) | 15 (5-28.9) |
| Mean (SD) | 10.5 (3.5) | 13.3 (3.9) | 13.4 (3.6) | 15.0 (3.8) |
| Tumor thickness, mm | ||||
| Median (range) | 3.4 (0.5-17) | 4.6 (1-14.4) | 5.3 (1-16) | 6.1 (1.6-18) |
| Mean (SD) | 4.2 (2.5) | 5.4 (3.0) | 6.1 (3.4) | 6.9 (3.5) |
| AJCC T-category, No. (%) | ||||
| T1 | 390 (46.7) | 64 (26.0) | 51 (18.9) | 25 (11.1) |
| T2 | 304 (36.4) | 89 (36.2) | 100 (37.0) | 71 (31.6) |
| T3 | 124 (14.8) | 66 (26.8) | 93 (34.4) | 77 (34.2) |
| T4 | 18 (2.2) | 27 (11.0) | 26 (9.6) | 52 (23.1) |
| Primary enucleation, No. (%) | 47 (5.6) | 28 (11.4) | 30 (11.1) | 56 (24.9) |
| 15-GEP 1A/B subclass, No. (%) | ||||
| 1A | 545 (65.2) | 148 (60.2) | NA | NA |
| 1B | 291 (34.8) | 98 (39.8) | NA | NA |
| Time to metastasis or last follow-up, median, months | 47.9 | 46.1 | 39.4 | 26.9 |
| Time to last follow-up, monthsa | ||||
| Median (range) | 49.2 (0-103.2) | 48.5 (0.5-100.3) | 45.9 (1.1-104.4) | 44.0 (1.1-95.1) |
| Mean (SD) | 51.6 (21.3) | 50.2 (20.5) | 48.1 (18.7) | 44.7 (20.3) |
| Patients with metastatic event on study, No. (%) | 32 (3.8) | 32 (13.0) | 93 (34.4) | 112 (49.8) |
| Time to metastasis, monthsb | ||||
| Median (range) | 31.3 (6.3-92.9) | 34.9 (3.6-84.7) | 24.7 (2.2-85.6) | 16.5 (0.1-79.9) |
| Mean (SD) | 35.3 (20.7) | 34.3 (20.6) | 28.5 (16.5) | 19.5 (14.4) |
| Local recurrence, No. (%) | 23 (2.8) | 12 (4.9) | 18 (6.7) | 15 (6.7) |
| Time to local recurrence, monthsb | ||||
| Median (range) | 18.7 (3.5-82.2) | 28.9 (9.2-82.0) | 21.1 (3.5-62.8) | 27.4 (6.8-58.6) |
| Mean (SD) | 26.9 (24.0) | 32.7 (19.8) | 26.6 (17.2) | 29.3 (15.4) |
Abbreviations: 15-GEP, 15-gene expression profile; AJCC, American Joint Committee on Cancer; NA, not applicable; NS, not statistically significant; SD, standard deviation.
Excludes patients with metastatic events or death from any cause.
Includes only patients with an indicated event recorded.
15-GEP/PRAME Classifier Versus AJCC Staging
To determine the performance of the integrated 15-GEP/PRAME classifier across different tumor size categories, survival outcomes were assessed on the basis of the AJCC T-category.38 The integrated 15-GEP/PRAME system robustly stratified patients by MFS in each AJCC T-category, and it revealed prognostic deficiencies in the AJCC system, which both overestimated and underestimated metastatic risk. While the 5-year MFS for T1 tumors overall was excellent at 94.4% (95% CI, 92.1 to 96.8), 76/530 (14.3%) T1 tumors were classified as class 2/PRAME– or class 2/PRAME+, and the 5-year MFS for these subgroups was only 77.6% and 69.5%, respectively. On the other hand, the 5-year MFS for T4 tumors overall was poor at 36.2% (95% CI, 26.2 to 50.0). However, 18/123 (14.6%) of patients with a T4 tumor were classified as class 1/PRAME–, of which only one patient went on to develop metastasis (Appendix Fig A2; Appendix Table A3).
Prognostic Model Optimization
Cox regression was performed to identify clinical factors that may enhance the 15-GEP/PRAME model by providing independent prognostic information. Univariate analysis revealed the following variables to be significantly associated with MFS: 15-GEP class 2 (HR, 9.77, P < .001), PRAME+ status (HR, 3.31, P < .001), ciliary body involvement (HR, 2.88, P < .001), increased tumor diameter (HR, 1.25, P < .001), increased tumor thickness (HR, 1.24, P < .001), and increased patient age (HR, 1.02, P < .001; Table 3). Sex and iris color were not significantly associated with MFS.
TABLE 3.
Univariate and Multivariate Cox Regression Analyses of Metastasis-Free Survival
| Risk Factor | Univariate HR (95% CI) | P | Multivariate HR (95% CI) | P |
|---|---|---|---|---|
| 15-GEP class 2 | 9.77 (7.36 to 12.95) | <.001 | 5.95 (4.43 to 7.99) | <.001 |
| PRAME (+) | 3.31 (2.60 to 4.20) | <.001 | 1.82 (1.42 to 2.33) | <.001 |
| Ciliary body involvement | 2.88 (2.22 to 3.74) | <.001 | ||
| Tumor diameter | 1.25 (1.21 to 1.28) | <.001 | 1.13 (1.09 to 1.17) | <.001 |
| Tumor thickness | 1.24 (1.20 to 1.27) | <.001 | 1.07 (1.03 to 1.11) | <.01 |
| Age, years | 1.02 (1.01 to 1.03) | <.001 |
NOTE. Binary variables: 15-GEP, class 2 (v class 1), PRAME+ (v PRAME–), and ciliary body involvement, yes (v no). Continuous variables: tumor diameter, tumor thickness, and patient age.
Abbreviations: 15-GEP, 15-gene expression profile; HR, hazard ratio; NS, not statistically significant.
Multivariate models were constructed on the basis of the stepwise addition of each variable according to their relative importance in the univariable analysis. When 15-GEP and PRAME were added to the model, age (HR, 1.01, P = .114) and ciliary body involvement (HR, 1.11, P = .46) were no longer significant, whereas tumor diameter (HR, 1.13, P < .001) and tumor thickness (HR, 1.07, P = .01) remained significant, albeit with diminished hazard ratios. To assess the clinical value of tumor diameter and tumor thickness, we calculated the concordance statistic (C-statistic) when these variables are included in the model (Table 4). The 15-GEP performed well on its own (C-statistic = 0.77), and the addition of PRAME status provided a substantial improvement (C-statistic = 0.81). The addition of tumor diameter to the 15-GEP + PRAME model provided a further improvement (C-statistic = 0.85). However, the addition of tumor thickness did not further increase the C-statistic. We next evaluated the performance of a model incorporating the 15-GEP + PRAME model with AJCC T-category, which includes both tumor diameter and thickness. The predictive performance of the 15-GEP + PRAME model was improved by the addition of T-category to a lesser extent (C-statistic = 0.84) than tumor diameter alone (Table 4).
TABLE 4.
Predictive Performance of Models With Stepwise Addition of Variables to the 15-GEP
| Model | C-Statistic (95% CI) |
|---|---|
| 15-GEP | 0.77 (0.75 to 0.79) |
| 15-GEP + PRAME | 0.81 (0.79 to 0.83) |
| 15-GEP + PRAME + diameter | 0.85 (0.83 to 0.87) |
| 15-GEP + PRAME + diameter + thickness | 0.85 (0.83 to 0.87) |
| 15-GEP + PRAME + AJCC T-category | 0.84 (0.82 to 0.86) |
Abbreviations: 15-GEP, 15-gene expression profile; AJCC, American Joint Committee on Cancer.
DISCUSSION
To our knowledge, COOG2 is the largest multicenter prospective biomarker study to date in UM, with longer follow-up and more representative, real-world distribution of tumor size, ciliary body involvement, and AJCC tumor stage than COOG1 or TCGA17,37 and more similar to a large international database encompassing the full spectrum of UM36 (Appendix Table A2). This may explain, at least in part, the more favorable outcomes in COOG2. Key findings include (1) prospective validation of 15-GEP and PRAME as independent prognostic biomarkers in UM, (2) superiority of PRAME status over the 1A/1B system for class 1 tumors, (3) establishment of a new 4-group 15-GEP/PRAME system, and (4) validation of tumor diameter as the only clinical factor that improves the accuracy of 15-GEP/PRAME.
The 1A/1B system was an early attempt to subdivide class 1 UM on the basis of prognosis into low (1A) versus intermediate (1B) metastatic risk using the differential expression of two genes in the 15-GEP (CDH1 and RAB31).19 While initial reports found this system to discriminate metastatic risk among class 1 UMs,21 subsequent studies did not corroborate these findings.26,32 A transcriptome-wide search identified PRAME as the most significant biomarker for identifying class 1 tumors with increased metastatic risk.25 A subsequent study showed that PRAME may also identify class 2 tumors with shorter time to metastasis.24 The method for determining PRAME positivity was analytically validated and incorporated into the workflow of the 15-GEP platform,39 such that both are assessed from a single sample. In this study, there was no significant difference in outcome between class 1A and class 1B, whereas PRAME status demonstrated significant predictive value independent of 15-GEP. Thus, we propose that PRAME should supersede the 1A/1B system and be integrated with 15-GEP to subdivide UMs into four prognostically significant subgroups: class 1/PRAME–, class 1/PRAME+, class 2/PRAME–, and class 2/PRAME+. Interestingly, PRAME was recently shown to induce aneuploidy in UM,40 suggesting that it is not simply a biomarker but likely a driver of metastasis. Although a so-called TCGA classification system has been proposed for UM,41 TCGA performed a one-time multiomics analysis of 80 large, enucleated UMs, which was not adequate (nor intended) to form the basis for a prognostic test. Nevertheless, TCGA did confirm the fundamental 4-group molecular landscape of UM described by 15-GEP/PRAME (Fig 2E).37
In COOG1, no clinical features were found to provide prognostic information independent of the 15-GEP.17 However, subsequent retrospective studies suggested that tumor diameter could potentially improve the prognostic accuracy of the 15-GEP.21,28-31,42 Recently, it has been suggested that AJCC clinical staging needs to be combined with 15-GEP.42 Here, we investigated whether any clinical variables, including those in the AJCC system, enhance the accuracy of the 15-GEP/PRAME classifier. As expected, patient age, ciliary body involvement, tumor diameter, and thickness were significantly associated with MFS by univariate analysis (Table 3). However, when 15-GEP and PRAME were added to a multivariate model, age and ciliary body involvement were no longer significant, and the hazard ratios for tumor diameter and thickness were reduced. Using the C-statistic to assess predictive accuracy, tumor diameter provided a slight improvement over 15-GEP + PRAME, whereas tumor thickness provided no further improvement (Table 4). Furthermore, AJCC T-category, which incorporates both tumor diameter and tumor thickness, improved the performance of the 15-GEP + PRAME to a lesser extent than tumor diameter alone, consistent with previous work.43 Consequently, our findings support a parsimonious prognostic model that includes 15-GEP/PRAME plus tumor diameter, but not the AJCC T-category or other clinical variables. A method for incorporating tumor diameter into a clinically practical integrated model will be published separately.
Among the various methods that have been proposed for prognostication in UM, including chromosomal analysis, mutation profiling, and immunohistochemistry (IHC),37,44 only 15-GEP/PRAME achieves the highest level of evidence (level I) in the National Comprehensive Cancer Network Tumor Marker Utility Grading System, which requires prospective validation.45 Most of these alternative methods have only been evaluated in single institution retrospective studies using platforms that are not standardized across centers and that achieve no better than level III evidence. A direct prospective comparison showed the 15-GEP to be superior to monosomy 3,17 which is the most important chromosomal prognostic marker. While mutations in BAP1, SF3B1, and EIF1AX are the most important prognostic mutations in UM,46-48 they are not as accurate as 15-GEP for predicting metastasis.49 In the case of BAP1, whose mutational inactivation is strongly associated with class 2 GEP,46 mutations may include large deletions and other alterations that can be difficult to detect even with whole-exome sequencing.50 IHC for BAP1 and PRAME have been proposed as surrogates for 15-GEP/PRAME.44 However, aside from the inferior sensitivity, specificity, dynamic range, and analytical precision of IHC compared with quantitative PCR,45,51,52 IHC requires archival tissue from eyes that have been enucleated, which is performed in <20% of patients in modern ocular oncology practice.53,54 The 15-GEP/PRAME is covered by many third-party payers in the United States and is available internationally, thereby providing a standardized platform that can be compared across centers worldwide. A multicenter prospective study is warranted to compare the analytical performance, prognostic accuracy, cost-effectiveness, and clinical utility of 15-GEP/PRAME to the best alternative methods to establish an international standard for adjuvant trial design.
In conclusion, this report provides prospective multicenter validation, the highest level of biomarker evidence,45 for integrating 15-GEP with PRAME expression status into a 4-group prognostic classification system. This integrated prognostic tool is a uniquely valuable resource to establish standardized entry criteria for high-risk adjuvant clinical trials, and it provides a gold standard for evaluating other prognostic biomarkers.
ACKNOWLEDGMENT
The authors thank our patients who generously participated in this project. We acknowledge Katherina M. Alsina, PhD, Jason H. Rogers, MS, and Jennifer J. Siegel, PhD, from Castle Biosciences, Inc, for data management support, and the following individuals for their incredible dedication to accurate data entry: Angie Adler, Mutaz Al-Nawaflh, Corrina Azarcon, Buse Guneri Beser, Karina Bostwick, Nury Cabrera, Teja Chemudupati, Caroline Craven, Jessica Fitch, Nancy Gee, Ashley Go, Caleb Hartley, Mustafa Hashmi, Tyler Hendrickson, Gary Lamoureux, Anne Marie Lane, Kiley Lazarek, Ashton Leone, Ronan McCarthy, Audra Miller, Trece Mayhan, Monica Oxenreiter, Barbara Perez, Dayana Pineda, Nicki Plocharsky, Mary Preston, Kourtney Storey, Laurie Tavernier, Bonnie Verges, Holly Vincent, Brooke Waller, and Aaron Yeung.
APPENDIX
FIG A1.
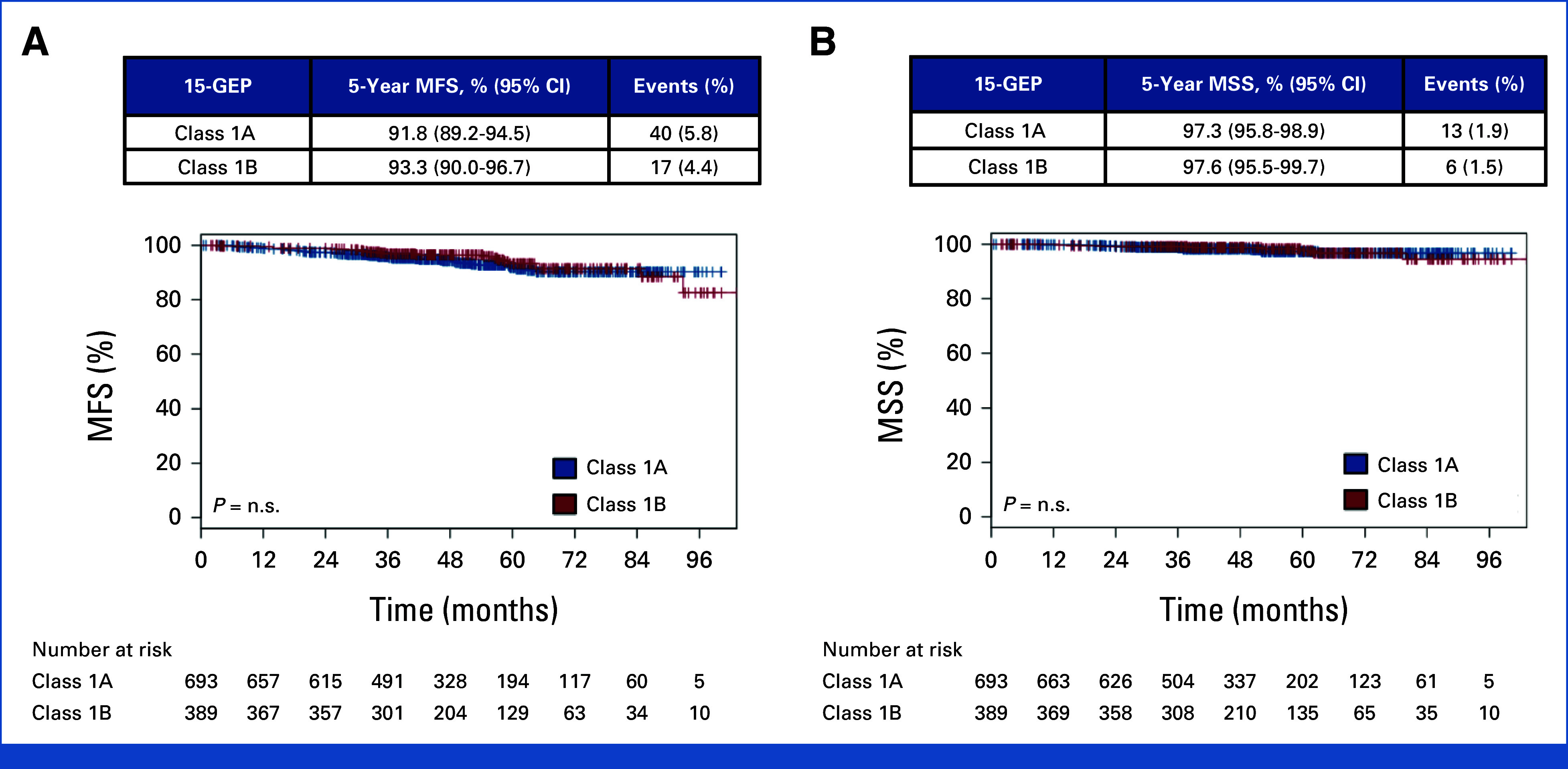
Kaplan-Meier survival curves for patients with class 1 15-gene expression profile results, separated by subclass 1A and 1B. (A) MSS and (B) MFS. Five-year point estimates and 95% CIs, along with the number of events (detection of distant metastasis or uveal melanoma-related death) are shown in the tables above each curve. P value, log-rank test. Number of patients at risk at each 12-month time point is shown below the plots. Censoring events are marked with a vertical hash across the survival curve. 15-GEP, 15-gene expression profile; MFS, metastasis-free survival; MSS, melanoma-specific survival; n.s., not statistically significant.
FIG A2.
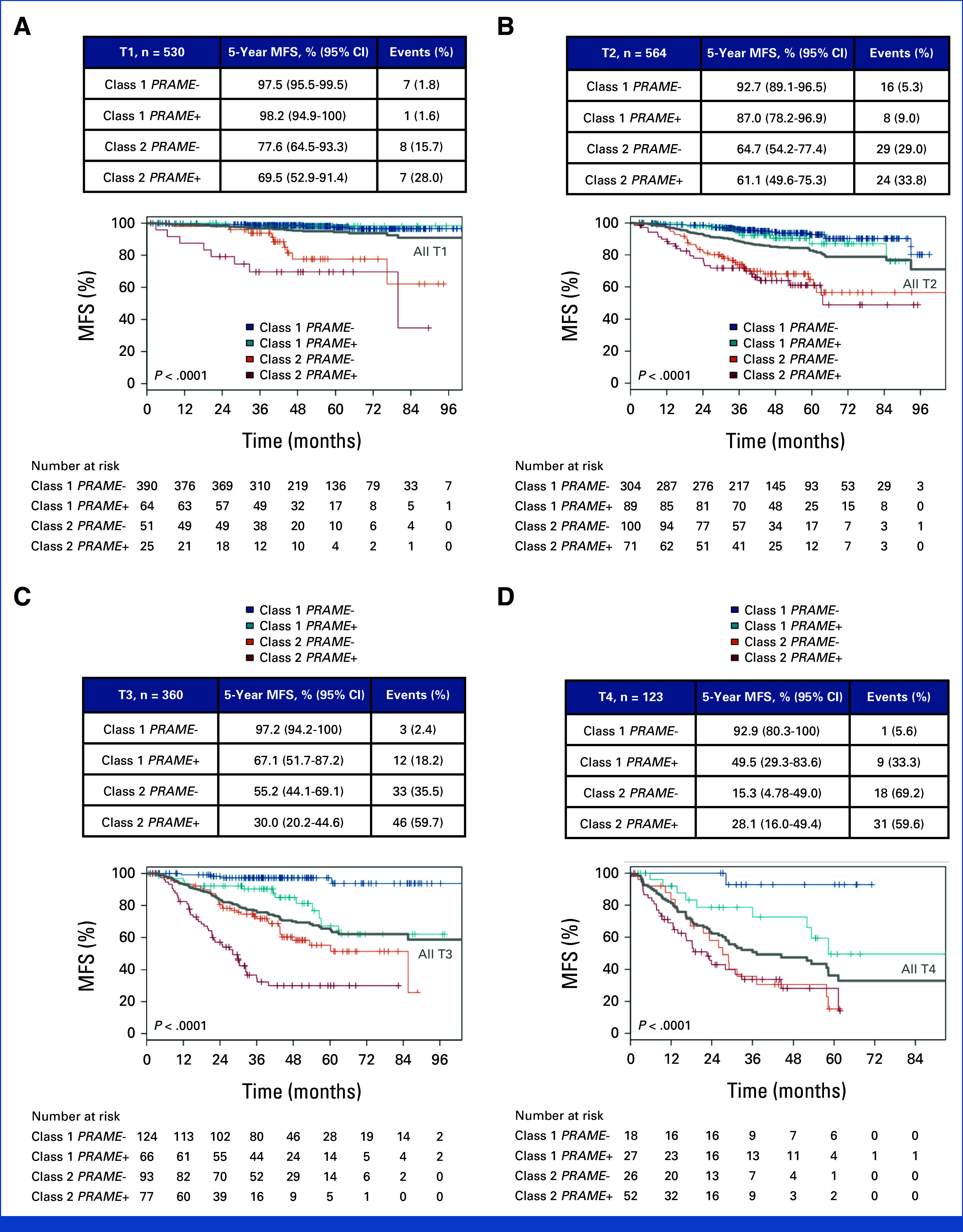
Five-year metastasis-free survival by AJCC 8th edition T stage, subclassified by GEP/PRAME groups. (A) T1, (B) T2, (C) T3, and (D) T4. P value, log-rank test. Number of patients at risk at each 12-month time point is shown below the plots. Censoring events are marked with a vertical hash across the survival curve. 15-GEP, 15-gene expression profile; AJCC, American Joint Committee on Cancer; MFS, metastasis-free survival.
TABLE A1.
COOG2 Participating Sites
| COOG2 Study Sites |
|---|
| Associated Retinal Consultants Michigan |
| Colorado Retina Associates |
| Duke University |
| Emory University |
| Hartford Hospital |
| Retina Consultants of Texas |
| Massachusetts Eye and Ear Infirmary |
| Oregon Health & Science University |
| Retina Associates of Arizona |
| Retina Consultants of Alabama |
| Retina Consultants of Sacramento |
| Retina Specialists of Michigan |
| Stanford University |
| Tennessee Retina |
| Texas Retina Associates |
| Tufts Medical Center |
| Tumori Foundation |
| University of Alberta |
| University of Cincinnati |
| University of Colorado |
| University of Miami |
| University of Michigan |
| University of Texas MD Anderson Cancer Center |
| University of Virginia |
| University of Wisconsin-Madison |
| Washington University in St Louis |
TABLE A2.
Comparison of Patient Cohorts Included in the Present Study (COOG2) Versus COOG1, AJCC, and TCGA
| Variable | Study (year) | |||
|---|---|---|---|---|
| COOG2 (2024) | COOG117 (2012) | TCGA37 (2017) | AJCC36 (2015) | |
| Patients, No. | 1,577 | 459 | 80 | 3,217 |
| Centers, No. | 26 | 12 | 6 | 10 |
| Study type | Prospective | Prospective | Retrospective | Retrospective |
| Age at study entry, years | ||||
| Median (range) | 64 (18-99) | 61 (NR) | 62 (22-86) | NR |
| Mean | 62 | 62 | 61 | |
| Male sex, No. (%) | 809 (51.3) | 235 (51.2) | 45 (56.3) | NR |
| Ciliary body involvement, No. (%) | 250 (15.9) | 139 (30.3) | 16 (20.0) | 459 (14.3) |
| Tumor diameter, mm | ||||
| Median (range) | 12 (2-32) | 12.7 (1.3-24.0) | 16.8 (10-23.6) | 11.8 (2-30) |
| Mean | 12.1 | 12.8 | 16.2 | 11.7 |
| Thickness, mm | ||||
| Median (range) | 4.1 (0.5-18) | 5.5 (1.0-17.5) | 11 (4.4-16) | 4.7 (1.1-23) |
| Mean | 5.1 | 6.3 | 10.8 | 5.4 |
| AJCC tumor stage, No. (%) | n = 1,577 | n = 425 | n = 80 | n = 3,217 |
| T1 | 530 (33.6) | 81 (19.1) | 0 | 1,115 (34.7) |
| T2 | 564 (35.8) | 170 (40.0) | 14 (17.5) | 1,128 (35.0) |
| T3 | 360 (22.8) | 140 (32.9) | 32 (40.0) | 789 (24.5) |
| T4 | 123 (7.8) | 34 (8.0) | 34 (42.5) | 185 (5.8) |
| Primary enucleation, No. (%) | 161 (10.2) | 92 (20.0) | 77 (96.3) | NR |
| 15-GEP test results, No. (%) | ||||
| Class 1 | 1,082 (68.6) | 276 (61.9) | NR | NR |
| Class 2 | 495 (31.4) | 170 (38.1) | ||
| Patients with metastatic event on study, No. (%) | 269 (17.1) | 47 (10.2) | 26 (32.5) | 325 (10.1) |
| Time to metastasis/last follow-up, months | ||||
| Median (range) | 43.6 (0-104.4) | 17.4 | 26.2 (0-85.4) | 38.4 (1.0-151.3) |
| Mean | 45.2 | 18.0 | 26.9 | NR |
Abbreviations: 15-GEP, 15-gene expression profile; AJCC, American Joint Committee on Cancer; NR, not reported; TCGA, The Cancer Genome Atlas.
TABLE A3.
Summary of Survival Statistics Across Patient Subsets by 15-GEP Class, PRAME, Integrated 15-GEP/PRAME, and AJCC T-Stage in Patients With Posterior Uveal Melanoma
| Subgroup | No. (%) | MFS, % (95% CI) | Events, No. | MSS, % (95% CI) | Events, No. | ||
|---|---|---|---|---|---|---|---|
| 3-Year | 5-Year | 5-Year | 3-Year | 5-Year | 5-Year | ||
| All patients | 1,577 (100) | 86.1 (84.4 to 87.9) | 79.9 (77.5 to 82.3) | 253 | 94.1 (92.9 to 95.4) | 88.9 (86.9 to 90.9) | 126 |
| 15-GEP | |||||||
| Class 1A | 693 (43.9) | 95.9 (94.4 to 97.5) | 91.8 (89.2 to 94.5) | 40 | 98.7 (97.8 to 99.6) | 97.3 (95.8 to 98.9) | 13 |
| Class 1B | 389 (24.7) | 96.9 (95.0 to 98.7) | 93.3 (90.0 to 96.7) | 17 | 99.2 (98.3 to 100) | 97.6 (95.5 to 99.7) | 6 |
| Class 1 (A + B) | 1,082 (68.6) | 96.2 (95.1 to 97.4) | 92.3 (90.2 to 94.4) | 57 | 98.9 (98.2 to 99.5) | 97.4 (96.2 to 98.7) | 19 |
| Class 2 | 495 (31.4) | 63.9 (59.6 to 68.5) | 52.1 (47.0 to 57.8) | 196 | 83.4 (80.0 to 87.0) | 68.8 (63.6 to 74.5) | 107 |
| PRAME | |||||||
| Negative (–) | 1,106 (70.1) | 91.7 (90.0 to 93.4) | 86.6 (84.2 to 89.1) | 115 | 97.4 (96.4 to 98.4) | 93.1 (91.2 to 95.0) | 51 |
| Positive (+) | 471 (29.9) | 72.9 (68.8 to 77.3) | 63.7 (58.5 to 69.3) | 138 | 86.2 (82.9 to 89.6) | 78.5 (73.9 to 83.3) | 75 |
| 15-GEP/PRAME | |||||||
| Class 1/(–) | 836 (53.0) | 97.4 (96.2 to 98.5) | 95.6 (93.9 to 97.4) | 27 | 99.5 (99.0 to 100) | 98.8 (97.9 to 99.7) | 7 |
| Class 1/(+) | 246 (15.6) | 92.4 (89.0 to 96.0) | 80.6 (73.9 to 87.9) | 30 | 96.8 (94.5 to 99.2) | 92.6 (88.3 to 97.1) | 12 |
| Class 2/(–) | 270 (17.1) | 74.2 (68.9 to 79.8) | 58.3 (51.1 to 66.4) | 88 | 90.9 (87.3 to 94.6) | 74.7 (68.0 to 82.2) | 44 |
| Class 2/(+) | 225 (14.3) | 51.0 (44.5 to 58.5) | 44.8 (37.9 to 52.8) | 108 | 73.8 (67.8 to 80.4) | 61.3 (53.5 to 70.2) | 63 |
| AJCC T-category | |||||||
| T1 | 530 (33.6) | 96.8 (95.3 to 98.4) | 94.4 (92.1 to 96.8) | 23 | 99.6 (99.1 to 100) | 98.1 (96.5 to 99.7) | 6 |
| T2 | 564 (35.8) | 88.6 (85.9 to 91.4) | 82.8 (79.1 to 86.6) | 77 | 96.3 (94.6 to 97.9) | 90.5 (87.5 to 93.7) | 36 |
| T3 | 360 (22.8) | 76.4 (71.9 to 81.3) | 65.5 (59.4 to 72.2) | 94 | 89.1 (85.7 to 92.7) | 79.0 (73.4 to 85.1) | 50 |
| T4 | 123 (7.8) | 52.0 (43.0 to 62.7) | 36.2 (26.2 to 50.0) | 59 | 70.7 (62.0 to 80.7) | 60.7 (50.6 to 72.8) | 34 |
Abbreviations: 15-GEP, 15-gene expression profile; AJCC T-stage; American Joint Committee on Cancer tumor stage (8th edition); MFS, metastasis-free survival; MSS, melanoma-specific survival.
TABLE A4.
Cox Regression Analysis of Metastasis-Free Survival by 15-Gene Expression Profile/PRAME Status
| Risk Factor | Univariate HR (95% CI) | P |
|---|---|---|
| Class 1/PRAME– | Reference | |
| Class 1/PRAME+ | 3.66 (2.24 to 5.98) | <.001 |
| Class 2/PRAME– | 11.33 (7.57 to 16.94) | <.001 |
| Class 2/PRAME+ | 22.06 (14.86 to 32.76) | <.001 |
Abbreviation: HR, hazard ratio.
J. William Harbour
Honoraria: Castle Biosciences
Consulting or Advisory Role: Castle Biosciences
Patents, Royalties, Other Intellectual Property: I receive royalties from Washington University for IP that was licensed to Castle Biosciences regarding prognostic testing in uveal melanoma
Travel, Accommodations, Expenses: Castle Biosciences
Zelia M. Correa
Travel, Accommodations, Expenses: Castle Biosciences
Amy C. Schefler
Research Funding: Castle Biosciences
Prithvi Mruthyunjaya
Employment: Pfizer
Stock and Other Ownership Interests: Seagen, Pfizer
Consulting or Advisory Role: Castle Biosciences, Aura Biosciences, alcon, Immunogen, Amgen
Research Funding: Genentech
Miguel A. Materin
Consulting or Advisory Role: Castle Biosciences, AstraZeneca, IDEAYA Biosciences
Thomas A. Aaberg Jr
Employment: Neurotech Pharmaceuticals Inc
Leadership: Neurotech Pharmaceuticals Inc
Stock and Other Ownership Interests: Sanro, Eclipse Life Sciences
Honoraria: Castle Biosciences, Regeneron
Consulting or Advisory Role: Regeneron, Castle Biosciences
Research Funding: Genentech (Inst), aviceda (Inst), Alkeus (Inst)
Travel, Accommodations, Expenses: Neurotech Pharmaceuticals Inc
Alison H. Skalet
Honoraria: Immunocore
Consulting or Advisory Role: Castle Biosciences Inc, Immunocore
Travel, Accommodations, Expenses: Castle Biosciences, Immunocore
David A. Reichstein
Honoraria: Immunocore, Genentech
Consulting or Advisory Role: Immunocore
Speakers' Bureau: Genentech
Travel, Accommodations, Expenses: Castle Biosciences
Ezekiel Weis
Travel, Accommodations, Expenses: Castle Biosciences
Ivana K. Kim
Stock and Other Ownership Interests: KSQ Therapeutics, Rappta Therapeutics, Frontier Medicines, Riva Therapeutics, Kestrel Therapeutics, Function Oncology
Consulting or Advisory Role: Genentech, Kodiak Sciences, Thermo Fisher Scientific, KSQ Therapeutics, MPM Capital, Solasta Ventures, Calyx, Tyra Biosciences, Jubilant Therapeutics, Frontier Medicines, Rappta Therapeutics, Hexagon Bio, Function Oncology, Riva Therapeutics, Serinus Biosciences, Kestrel Therapeutics
Research Funding: Allergan
Travel, Accommodations, Expenses: Genentech, Aura Biosciences, Castle Biosciences
Timothy S. Fuller
Research Funding: Aura Biosciences
Hakan Demirci
Consulting or Advisory Role: Castle Biosciences
Research Funding: Aura Biosciences, Aura Biosciences
Kisha D. Piggott
Travel, Accommodations, Expenses: Castle Biosciences
Basil K. Williams
Stock and Other Ownership Interests: Lumata Health
Consulting or Advisory Role: Alcon, Allergan, Alimera Sciences, EyePoint Pharmaceuticals, Astellas Pharma, Immunocore, Genentech/Roche, Regeneron
Travel, Accommodations, Expenses: Castle Biosciences
Eugene Shildkrot
Employment: Roche/Genentech
Stock and Other Ownership Interests: Immunogen, Immunocore, Astellas Pharma, Astellas Pharma, Apellis Pharmaceuticals
Honoraria: Astellas Pharma, IDEAYA Biosciences
Consulting or Advisory Role: Astellas Pharma, IDEAYA Biosciences, IDEAYA Biosciences, Horizon Therapeutics
Speakers' Bureau: Frictionless solutions
Travel, Accommodations, Expenses: Castle Biosciences, Horizon Therapeutics
Antonio Capone Jr
Employment: EyeCare Partners
Leadership: EyeCare Partners
Stock and Other Ownership Interests: EyeCare Partners, Neolight, Interview Medical Sciences
Honoraria: Mianus Capital, Partners Group
Consulting or Advisory Role: Partners Group, Mianus Capital
Research Funding: Iveric Bio, Aura Biosciences, Regeneron
Patents, Royalties, Other Intellectual Property: Zone1 ROP tool, A number of patents on Wnt signaling pharmacotherapeutic
Travel, Accommodations, Expenses: EyeCare Partners
Scott C. Oliver
Research Funding: Castle Biosciences (Inst)
Travel, Accommodations, Expenses: Castle Biosciences (Inst)
Scott D. Walter
Consulting or Advisory Role: Apellis Pharmaceuticals, Bausch and Lomb, Genentech/Roche, Allergan, Alimera Sciences, Astellas Pharma, Castle Biosciences, IDEAYA Biosciences, Lupin Pharmaceuticals, Novartis, Regeneron, Regeneron
Speakers' Bureau: Apellis Pharmaceuticals, Bausch and Lomb, Genentech/Roche, Regeneron, Spark Therapeutics
John Mason III
Travel, Accommodations, Expenses: Castlebiosciences
Michael Altaweel
Consulting or Advisory Role: Oasis Therapeutics
Research Funding: Aura Biosciences (Inst)
Jill R. Wells
Travel, Accommodations, Expenses: Castle Biosciences
Jay S. Duker
Employment: EyePoint Pharmaceuticals, Aura Biosciences
Leadership: EyePoint Pharmaceuticals
Stock and Other Ownership Interests: EyePoint Pharmaceuticals
Dan S. Gombos
Honoraria: iVista
Consulting or Advisory Role: Castle Biosciences, Immunogen
Travel, Accommodations, Expenses: Castle Biosciences
Other Relationship: 3T Ophthalmics, Aura Biosciences, Castle Biosciences
Tony Tsai
Research Funding: Aura Biosciences (Inst)
Travel, Accommodations, Expenses: Castle Biosciences, Aura Biosciences
Brian P. Marr
Honoraria: Castle Biosciences
Consulting or Advisory Role: Aura Biosciences, Castle Biosciences
Travel, Accommodations, Expenses: Castle Biosciences
Christina L. Decatur
Employment: University of Miami
Honoraria: Castle Biosciences
Research Funding: University of Miami
Travel, Accommodations, Expenses: Castle Biosciences
No other potential conflicts of interest were reported.
PRIOR PRESENTATION
Presented in part at the Retina Society Annual Meeting, London, UK, September 11-15, 2019; American Academy of Ophthalmology Annual Meeting, virtual, November 13-15, 2020; Association for Research in Vision and Ophthalmology Annual Meeting, virtual, May 1-7, 2021; International Society of Ocular Oncology Meeting, Leiden, the Netherlands, June 17-22, 2022; American Society of Retina Specialists Annual Meeting, New York City, NY, July 10, 2022; and the FLORetina Meeting, Rome, Italy, November 30-December 3, 2023.
SUPPORT
Supported by NCI grant R01 CA125970 (J.W.H.); Cancer Prevention and Research Institute of Texas Recruitment of Established Investigator Award RR220010 (J.W.H.); Research to Prevent Blindness, Inc Senior Scientific Investigator Award (J.W.H.); NCI Cancer Center Support Grants P30 CA142543 to University of Texas Southwestern Simmons Comprehensive Cancer Center and P30 CA240139 to University of Miami Sylvester Comprehensive Cancer Center; NEI Center Core Grants P30 EY030413 to University of Texas Southwestern Department of Ophthalmology and P30 EY014801 to University of Miami Department of Ophthalmology; Research to Prevent Blindness, Inc Challenge Grant to University of Texas Southwestern Department of Ophthalmology; Research to Prevent Blindness, Inc Unrestricted Grant to University of Miami Department of Ophthalmology; and Castle Biosciences, Inc grant to University of Miami.
DATA SHARING STATEMENT
A data sharing statement provided by the authors is available with this article at DOI https://doi.org/10.1200/JCO.24.00447.
AUTHOR CONTRIBUTIONS
Conception and design: J. William Harbour, Zelia M. Correa, Amy C. Schefler, Prithvi Mruthyunjaya, Miguel A. Materin, Thomas A. Aaberg Jr, Ezekiel Weis, Hakan Demirci, Eugene Shildkrot, Scott C. Oliver, Devron H. Char, Brian P. Marr
Financial support: J. William Harbour
Administrative support: J. William Harbour, Zelia M. Correa, Prithvi Mruthyunjaya, Thomas A. Aaberg Jr, Christina L. Decatur
Provision of study materials or patients: J. William Harbour, Zelia M. Correa, Amy C. Schefler, Prithvi Mruthyunjaya, Miguel A. Materin, Thomas A. Aaberg Jr, Alison H. Skalet, Ivana K. Kim, Hakan Demirci, Eugene Shildkrot, Antonio Capone Jr, Scott C. Oliver, Scott D. Walter, John Mason III, Devron H. Char, Michael Altaweel, Jay S. Duker, Peter G. Hovland, Tony Tsai, Brian P. Marr, Christina L. Decatur
Collection and assembly of data: J. William Harbour, Zelia M. Correa, Amy C. Schefler, Thomas A. Aaberg Jr, Alison H. Skalet, David A. Reichstein, Ezekiel Weis, Ivana K. Kim, Timothy S. Fuller, Hakan Demirci, Kisha D. Piggott, Basil K. Williams, Eugene Shildkrot, Antonio Capone Jr, Scott D. Walter, John Mason III, Devron H. Char, Michael Altaweel, Jill R. Wells, Jay S. Duker, Peter G. Hovland, Dan S. Gombos, Tony Tsai, Cameron Javid, Christina L. Decatur, James J. Dollar, Stefan Kurtenbach, Song Zhang
Data analysis and interpretation: J. William Harbour, Zelia M. Correa, Amy C. Schefler, Thomas A. Aaberg Jr, Alison H. Skalet, David A. Reichstein, Ezekiel Weis, Ivana K. Kim, Hakan Demirci, Basil K. Williams, Scott C. Oliver, Scott D. Walter, Devron H. Char, Jay S. Duker, Tony Tsai, Ang Gao, Stefan Kurtenbach Song Zhang
Manuscript writing: All authors
Final approval of manuscript: All authors
Accountable for all aspects of the work: All authors
AUTHORS' DISCLOSURES OF POTENTIAL CONFLICTS OF INTEREST
15-Gene Expression Profile and PRAME as Integrated Prognostic Test for Uveal Melanoma: First Report of Collaborative Ocular Oncology Group Study No. 2 (COOG2.1)
The following represents disclosure information provided by authors of this manuscript. All relationships are considered compensated unless otherwise noted. Relationships are self-held unless noted. I = Immediate Family Member, Inst = My Institution. Relationships may not relate to the subject matter of this manuscript. For more information about ASCO's conflict of interest policy, please refer to www.asco.org/rwc or ascopubs.org/jco/authors/author-center.
Open Payments is a public database containing information reported by companies about payments made to US-licensed physicians (Open Payments).
J. William Harbour
Honoraria: Castle Biosciences
Consulting or Advisory Role: Castle Biosciences
Patents, Royalties, Other Intellectual Property: I receive royalties from Washington University for IP that was licensed to Castle Biosciences regarding prognostic testing in uveal melanoma
Travel, Accommodations, Expenses: Castle Biosciences
Zelia M. Correa
Travel, Accommodations, Expenses: Castle Biosciences
Amy C. Schefler
Research Funding: Castle Biosciences
Prithvi Mruthyunjaya
Employment: Pfizer
Stock and Other Ownership Interests: Seagen, Pfizer
Consulting or Advisory Role: Castle Biosciences, Aura Biosciences, alcon, Immunogen, Amgen
Research Funding: Genentech
Miguel A. Materin
Consulting or Advisory Role: Castle Biosciences, AstraZeneca, IDEAYA Biosciences
Thomas A. Aaberg Jr
Employment: Neurotech Pharmaceuticals Inc
Leadership: Neurotech Pharmaceuticals Inc
Stock and Other Ownership Interests: Sanro, Eclipse Life Sciences
Honoraria: Castle Biosciences, Regeneron
Consulting or Advisory Role: Regeneron, Castle Biosciences
Research Funding: Genentech (Inst), aviceda (Inst), Alkeus (Inst)
Travel, Accommodations, Expenses: Neurotech Pharmaceuticals Inc
Alison H. Skalet
Honoraria: Immunocore
Consulting or Advisory Role: Castle Biosciences Inc, Immunocore
Travel, Accommodations, Expenses: Castle Biosciences, Immunocore
David A. Reichstein
Honoraria: Immunocore, Genentech
Consulting or Advisory Role: Immunocore
Speakers' Bureau: Genentech
Travel, Accommodations, Expenses: Castle Biosciences
Ezekiel Weis
Travel, Accommodations, Expenses: Castle Biosciences
Ivana K. Kim
Stock and Other Ownership Interests: KSQ Therapeutics, Rappta Therapeutics, Frontier Medicines, Riva Therapeutics, Kestrel Therapeutics, Function Oncology
Consulting or Advisory Role: Genentech, Kodiak Sciences, Thermo Fisher Scientific, KSQ Therapeutics, MPM Capital, Solasta Ventures, Calyx, Tyra Biosciences, Jubilant Therapeutics, Frontier Medicines, Rappta Therapeutics, Hexagon Bio, Function Oncology, Riva Therapeutics, Serinus Biosciences, Kestrel Therapeutics
Research Funding: Allergan
Travel, Accommodations, Expenses: Genentech, Aura Biosciences, Castle Biosciences
Timothy S. Fuller
Research Funding: Aura Biosciences
Hakan Demirci
Consulting or Advisory Role: Castle Biosciences
Research Funding: Aura Biosciences, Aura Biosciences
Kisha D. Piggott
Travel, Accommodations, Expenses: Castle Biosciences
Basil K. Williams
Stock and Other Ownership Interests: Lumata Health
Consulting or Advisory Role: Alcon, Allergan, Alimera Sciences, EyePoint Pharmaceuticals, Astellas Pharma, Immunocore, Genentech/Roche, Regeneron
Travel, Accommodations, Expenses: Castle Biosciences
Eugene Shildkrot
Employment: Roche/Genentech
Stock and Other Ownership Interests: Immunogen, Immunocore, Astellas Pharma, Astellas Pharma, Apellis Pharmaceuticals
Honoraria: Astellas Pharma, IDEAYA Biosciences
Consulting or Advisory Role: Astellas Pharma, IDEAYA Biosciences, IDEAYA Biosciences, Horizon Therapeutics
Speakers' Bureau: Frictionless solutions
Travel, Accommodations, Expenses: Castle Biosciences, Horizon Therapeutics
Antonio Capone Jr
Employment: EyeCare Partners
Leadership: EyeCare Partners
Stock and Other Ownership Interests: EyeCare Partners, Neolight, Interview Medical Sciences
Honoraria: Mianus Capital, Partners Group
Consulting or Advisory Role: Partners Group, Mianus Capital
Research Funding: Iveric Bio, Aura Biosciences, Regeneron
Patents, Royalties, Other Intellectual Property: Zone1 ROP tool, A number of patents on Wnt signaling pharmacotherapeutic
Travel, Accommodations, Expenses: EyeCare Partners
Scott C. Oliver
Research Funding: Castle Biosciences (Inst)
Travel, Accommodations, Expenses: Castle Biosciences (Inst)
Scott D. Walter
Consulting or Advisory Role: Apellis Pharmaceuticals, Bausch and Lomb, Genentech/Roche, Allergan, Alimera Sciences, Astellas Pharma, Castle Biosciences, IDEAYA Biosciences, Lupin Pharmaceuticals, Novartis, Regeneron, Regeneron
Speakers' Bureau: Apellis Pharmaceuticals, Bausch and Lomb, Genentech/Roche, Regeneron, Spark Therapeutics
John Mason III
Travel, Accommodations, Expenses: Castlebiosciences
Michael Altaweel
Consulting or Advisory Role: Oasis Therapeutics
Research Funding: Aura Biosciences (Inst)
Jill R. Wells
Travel, Accommodations, Expenses: Castle Biosciences
Jay S. Duker
Employment: EyePoint Pharmaceuticals, Aura Biosciences
Leadership: EyePoint Pharmaceuticals
Stock and Other Ownership Interests: EyePoint Pharmaceuticals
Dan S. Gombos
Honoraria: iVista
Consulting or Advisory Role: Castle Biosciences, Immunogen
Travel, Accommodations, Expenses: Castle Biosciences
Other Relationship: 3T Ophthalmics, Aura Biosciences, Castle Biosciences
Tony Tsai
Research Funding: Aura Biosciences (Inst)
Travel, Accommodations, Expenses: Castle Biosciences, Aura Biosciences
Brian P. Marr
Honoraria: Castle Biosciences
Consulting or Advisory Role: Aura Biosciences, Castle Biosciences
Travel, Accommodations, Expenses: Castle Biosciences
Christina L. Decatur
Employment: University of Miami
Honoraria: Castle Biosciences
Research Funding: University of Miami
Travel, Accommodations, Expenses: Castle Biosciences
No other potential conflicts of interest were reported.
REFERENCES
- 1.Carvajal RD, Sacco JJ, Jager MJ, et al. : Advances in the clinical management of uveal melanoma. Nat Rev Clin Oncol 20:99-115, 2023 [DOI] [PubMed] [Google Scholar]
- 2.Aronow ME, Topham AK, Singh AD: Uveal melanoma: 5-Year update on incidence, treatment, and survival (SEER 1973-2013). Ocul Oncol Pathol 4:145-151, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gill VT, Norrman E, Sabazade S, et al. : Multiorgan involvement of dormant uveal melanoma micrometastases in postmortem tissue from patients without coexisting macrometastases. Am J Clin Pathol 160:164-174, 2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eskelin S, Pyrhonen S, Summanen P, et al. : Tumor doubling times in metastatic malignant melanoma of the uvea—Tumor progression before and after treatment. Ophthalmology 107:1443-1449, 2000 [DOI] [PubMed] [Google Scholar]
- 5.Carvajal RD, Butler MO, Shoushtari AN, et al. : Clinical and molecular response to tebentafusp in previously treated patients with metastatic uveal melanoma: A phase 2 trial. Nat Med 28:2364-2373, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nathan P, Hassel JC, Rutkowski P, et al. : Overall survival benefit with tebentafusp in metastatic uveal melanoma. N Engl J Med 385:1196-1206, 2021 [DOI] [PubMed] [Google Scholar]
- 7.Hassel JC, Piperno-Neumann S, Rutkowski P, et al. : Three-year overall survival with tebentafusp in metastatic uveal melanoma. N Engl J Med 389:2256-2266, 2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gupta S, Bedikian AY, Ahrar J, et al. : Hepatic artery chemoembolization in patients with ocular melanoma metastatic to the liver: Response, survival, and prognostic factors. Am J Clin Oncol 33:474-480, 2010 [DOI] [PubMed] [Google Scholar]
- 9.Piperno-Neumann S, Servois V, Mariani P, et al. : Prospective study of surveillance testing for metastasis in 100 high-risk uveal melanoma patients. J Fr Ophtalmol 38:526-534, 2015 [DOI] [PubMed] [Google Scholar]
- 10.Marshall E, Romaniuk C, Ghaneh P, et al. : MRI in the detection of hepatic metastases from high-risk uveal melanoma: A prospective study in 188 patients. Br J Ophthalmol 97:159-163, 2013 [DOI] [PubMed] [Google Scholar]
- 11.Davanzo JM, Binkley EM, Bena JF, et al. : Risk-stratified systemic surveillance in uveal melanoma. Br J Ophthalmol 103:1868-1871, 2019 [DOI] [PubMed] [Google Scholar]
- 12.Liu AW, Wei AZ, Maniar AB, et al. : Tebentafusp in advanced uveal melanoma: Proof of principle for the efficacy of T-cell receptor therapeutics and bispecifics in solid tumors. Expert Opin Biol Ther 22:997-1004, 2022 [DOI] [PubMed] [Google Scholar]
- 13.Worley LA, Onken MD, Person E, et al. : Transcriptomic versus chromosomal prognostic markers and clinical outcome in uveal melanoma. Clin Cancer Res 13:1466-1471, 2007 [DOI] [PubMed] [Google Scholar]
- 14.Singh AD, Sisley K, Xu Y, et al. : Reduced expression of autotaxin predicts survival in uveal melanoma. Br J Ophthalmol 91:1385-1392, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van Gils W, Lodder EM, Mensink HW, et al. : Gene expression profiling in uveal melanoma: Two regions on 3p related to prognosis. Invest Ophthalmol Vis Sci 49:4254-4262, 2008 [DOI] [PubMed] [Google Scholar]
- 16.Petrausch U, Martus P, Tonnies H, et al. : Significance of gene expression analysis in uveal melanoma in comparison to standard risk factors for risk assessment of subsequent metastases. Eye 22:997-1007, 2007 [DOI] [PubMed] [Google Scholar]
- 17.Onken MD, Worley LA, Char DH, et al. : Collaborative ocular Oncology Group report number 1: Prospective validation of a multi-gene prognostic assay in uveal melanoma. Ophthalmology 119:1596-1603, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Onken MD, Worley LA, Tuscan MD, et al. : An accurate, clinically feasible multi-gene expression assay for predicting metastasis in uveal melanoma. J Mol Diagn 12:461-468, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Plasseraud KM, Wilkinson JK, Oelschlager KM, et al. : Gene expression profiling in uveal melanoma: Technical reliability and correlation of molecular class with pathologic characteristics. Diagn Pathol 12:59, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aaberg TM Jr, Cook RW, Oelschlager K, et al. : Current clinical practice: Differential management of uveal melanoma in the era of molecular tumor analyses. Clin Ophthalmol 8:2449-2460, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Demirci H, Niziol LM, Ozkurt Z, et al. : Do largest basal tumor diameter and the American Joint Commission Cancer staging influence prognostication by gene expression profiling in choroidal melanoma? Am J Ophthalmol 195:83-92, 2018 [DOI] [PubMed] [Google Scholar]
- 22.Berry DE, Schefler AC, Seider MI, et al. : Correlation of gene expression profile status and American Joint Commission on Cancer stage in uveal melanoma. Retina 40:214-224, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Correa ZM, Augsburger JJ: Sufficiency of FNAB aspirates of posterior uveal melanoma for cytologic versus GEP classification in 159 patients, and relative prognostic significance of these classifications. Graefes Arch Clin Exp Ophthalmol 252:131-135, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Field MG, Durante MA, Decatur CL, et al. : Epigenetic reprogramming and aberrant expression of PRAME are associated with increased metastatic risk in Class 1 and Class 2 uveal melanomas. Oncotarget 7:59209-59219, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Field MG, Decatur CL, Kurtenbach S, et al. : PRAME as an independent biomarker for metastasis in uveal melanoma. Clin Cancer Res 22:1234-1242, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schefler AC, Koca E, Bernicker EH, et al. : Relationship between clinical features, GEP class, and PRAME expression in uveal melanoma. Graefes Arch Clin Exp Ophthalmol 257:1541-1545, 2019 [DOI] [PubMed] [Google Scholar]
- 27.Cai L, Paez-Escamilla M, Walter SD, et al. : Gene expression profiling and PRAME status versus tumor-node-metastasis staging for prognostication in uveal melanoma. Am J Ophthalmol 195:154-160, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Correa ZM, Augsburger JJ: Independent prognostic significance of gene expression profile class and largest basal diameter of posterior uveal melanomas. Am J Ophthalmol 162:20-27.e1, 2016 [DOI] [PubMed] [Google Scholar]
- 29.Walter SD, Chao DL, Feuer W, et al. : Prognostic implications of tumor diameter in association with gene expression profile for uveal melanoma. JAMA Ophthalmol 134:734-740, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Binkley EM, Bena JF, Davanzo JM, et al. : Gene expression profiling prognostication of posterior uveal melanoma: Does size matter? Ophthalmol Retina 4:620-629, 2020 [DOI] [PubMed] [Google Scholar]
- 31.Roelofsen CDM, Wierenga APA, van Duinen S, et al. : Five decades of enucleations for uveal melanoma in one center: More tumors with high risk factors, No improvement in survival over time. Ocul Oncol Pathol 7:133-141, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Augsburger JJ, Skinner CC, Correa ZM: Comparative metastatic rates in GEP class 1A versus 1B posterior uveal melanoma. Results contrary to expectations. Ocul Oncol Pathol 8:242-249, 2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plasseraud KM, Cook RW, Tsai T, et al. : Clinical performance and management outcomes with the DecisionDx-UM gene expression profile test in a prospective multicenter study. J Oncol 2016:5325762, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Reference deleted.
- 35.Harris PA, Taylor R, Thielke R, et al. : Research electronic data capture (REDCap)—A metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform 42:377-381, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.AJCC Ophthalmic Oncology Task Force : International validation of the American Joint Committee on Cancer's 7th Edition Classification of Uveal Melanoma. JAMA Ophthalmol 133:376-383, 2015 [DOI] [PubMed] [Google Scholar]
- 37.Robertson AG, Shih J, Yau C, et al. : Integrative analysis identifies four molecular and clinical subsets in uveal melanoma. Cancer Cell 32:204-220 e15, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kivelä T, Simpson ER, Grossniklaus HE: Uveal Melanoma, American Joint Committee on Cancer (AJCC) Staging Manual (ed 8). New York, NY, Springer, 2017, pp 805-817 [Google Scholar]
- 39.Plasseraud KM, Field MG, Qin T, et al. : Analytical validation of a clinical test for PRAME gene expression status in primary uveal melanomas. Pigment Cell Melanoma Res 30:131, 2017 [Google Scholar]
- 40.Kurtenbach S, Sanchez MI, Kuznetsoff J, et al. : PRAME induces genomic instability in uveal melanoma. Oncogene 43:555-565, 2023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vichitvejpaisal P, Dalvin LA, Mazloumi M, et al. : Genetic analysis of uveal melanoma in 658 patients using the cancer Genome Atlas classification of uveal melanoma as A, B, C, and D. Ophthalmology 126:1445-1453, 2019 [DOI] [PubMed] [Google Scholar]
- 42.Stacey AW, Dedania VS, Materin M, et al. : Improved prognostic precision in uveal melanoma through a combined score of clinical stage and molecular prognostication. Ocul Oncol Pathol 8:35-41, 2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Skinner CC, Augsburger JJ, Augsburger BD, et al. : Comparison of alternative tumor size classifications for posterior uveal melanomas. Invest Ophthalmol Vis Sci 58:3335-3342, 2017 [DOI] [PubMed] [Google Scholar]
- 44.Kumar N, Singh MK, Singh L, et al. : Diagnostic utility of immunohistochemistry in concordance with mRNA analysis of PRAME in the stratification of high-risk uveal melanoma patients. Hum Cell 36:342-352, 2023 [DOI] [PubMed] [Google Scholar]
- 45.Febbo PG, Ladanyi M, Aldape KD, et al. : NCCN Task Force report: Evaluating the clinical utility of tumor markers in oncology. J Natl Compr Canc Netw 9:S1-S32, 2011. (suppl 5); quiz S33 [DOI] [PubMed] [Google Scholar]
- 46.Harbour JW, Onken MD, Roberson ED, et al. : Frequent mutation of BAP1 in metastasizing uveal melanomas. Science 330:1410-1413, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harbour JW, Roberson ED, Anbunathan H, et al. : Recurrent mutations at codon 625 of the splicing factor SF3B1 in uveal melanoma. Nat Genet 45:133-135, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Martin M, Masshofer L, Temming P, et al. : Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3. Nat Genet 45:933-936, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Decatur CL, Ong E, Garg N, et al. : Driver mutations in uveal melanoma: Associations with gene expression profile and patient outcomes. JAMA Ophthalmol 134:728-733, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Field MG, Durante MA, Anbunathan H, et al. : Punctuated evolution of canonical genomic aberrations in uveal melanoma. Nat Commun 9:116, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Govindan S, Siraganahalli Eswaraiah M, Basavaraj C, et al. : Androgen Receptor mRNA levels determine the prognosis in triple-negative breast cancer patients. BMC Cancer 20:745, 2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sinn HP, Schneeweiss A, Keller M, et al. : Comparison of immunohistochemistry with PCR for assessment of ER, PR, and Ki-67 and prediction of pathological complete response in breast cancer. BMC Cancer 17:124, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rao YJ, Sein J, Badiyan S, et al. : Patterns of care and survival outcomes after treatment for uveal melanoma in the post-coms era (2004-2013): A surveillance, epidemiology, and end results analysis. J Contemp Brachytherapy 9:453-465, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chevli N, Zuhour RJ, Messer JA, et al. : Contemporary trends in management of uveal melanoma. J Contemp Brachytherapy 14:123-129, 2022 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
A data sharing statement provided by the authors is available with this article at DOI https://doi.org/10.1200/JCO.24.00447.