Abstract
Rationale and Objective:
Membranoproliferative glomerulonephritis (MPGN), encompassing several distinct diseases, is a rare but significant cause of kidney failure in the US. Potential etiologies of MPGN are unclear, but prior studies have suggested dysregulation of the alternative complement pathway, and recently, autoimmunity as potential mechanisms driving MPGN pathogenesis. In this study, we examined HLA associations with end-stage kidney disease (ESKD) due to MPGN and DDD in a large racially and ethnically diverse US-based cohort.
Study design:
Case-control study.
Setting and Participants:
Using USRDS and UNOS data, we included 3570 patients with kidney failure due to MPGN and 263 due to DDD. We matched patients to kidney donor controls on designated race and ethnicity in a 1:15 ratio.
Exposures/Predictors:
58 class I and II HLA serotypes.
Outcomes:
Case/control status.
Analytic Approach:
In each disease cohort, we performed univariable and multivariable logistic regression analyses to investigate associations between the disease and 58 HLA serotypes. In subgroup analyses, we investigated HLA associations in White and Black patients. We also studied anti-GBM nephritis as a positive-control outcome. We applied a Bonferroni correction to account for multiple comparisons.
Results:
Eighteen serotypes were significantly associated with the odds of having MPGN in univariable analyses, with DR17 having the strongest association ([OR]: 1.55, 95% CI: 1.44–1.68; p-value 4.33e-28). No significant associations were found between any HLA serotype and DDD. Designated race-specific analyses showed comparable findings. We were able to recapitulate known HLA associations in anti-GBM nephritis.
Limitations:
Reliance on HLA serotypes (rather than genotype), lack of biopsy confirmed diagnoses.
Conclusion:
HLA-DR17 is associated with ESKD due to MPGN in a racially and ethnically diverse cohort. The strength of association was similar in White and Black patients, suggesting a role in the pathogenesis of MPGN. No HLA associations were observed in patients with DDD.
Title:
HLA DR-17 is associated with ESKD due to MPGN in a large racially and ethnically diverse cohort.
Prior studies have suggested dysregulation of the alternative complement pathway as a potential etiology of MPGN, but recent evidence from a British White population has implicated an autoimmune mechanism in MPGN pathogenesis. We investigated HLA associations between MPGN and DDD in a large racially and ethnically diverse cohort of patients. We find that HLA-DR17 is associated with ESKD due to MPGN in both White and Black patients. In contrast, no significant HLA associations with ESKD due to DDD were identified. These results suggest a role for autoimmunity in some cases of MPGN and highlight differences in the disease etiology of MPGN compared to DDD.
Introduction
Membranoproliferative glomerulonephritis (MPGN) accounts for up to 10% of all cases of biopsy-confirmed glomerular disease1, leading to kidney failure within 10 years in approximately 50% of patients2. MPGN, an umbrella term, describes a histologic pattern of glomerular injury that comprises several etiologically distinct disease entities. Etiologic classifications divide MPGN into immune-complex mediated MPGN (IC-MPGN) and complement-mediated MPGN or C3 glomerulopathy. C3 glomerulopathy is further subdivided into dense deposit disease (DDD) and C3 glomerulonephritis (C3GN), based on the presence or absence of characteristic dense deposits seen by electron microscopy3. IC-MPGN can be associated with infection (e.g., hepatitis B/C), autoimmune disease (e.g., cryoglobulinemia), or plasma cell dyscrasias, although in many cases the underlying etiology remains unknown. Even in patients for whom a systemic disease is identified, the underlying pathogenic mechanisms of IC-MPGN are poorly understood. In contrast, for C3 glomerulopathy (C3GN and DDD), several studies have implicated dysregulation of the alternative complement pathway in the pathogenesis of disease, either through genetic mutations or abnormalities in key complement regulators, or through autoantibodies to specific complement proteins. However, despite progress in elucidating potential mechanistic causes, the genetic mutation and/or autoantibody in C3 glomerulopathy is rarely found. Uncovering the underlying pathogenic mechanisms driving these diseases is a first step in the development of targeted, safer, and more effective treatment options.
Recently, Levine et al. performed whole genome sequencing on a cohort of 146 White British patients with MPGN (including IC-MPGN, C3GN, DDD) and, surprisingly, found no significant enrichment for rare variants in alternative complement pathway genes4. Instead, they identified increased prevalence of HLA DRB1*03:01 (serotype: HLA-DR17) in patients with MPGN compared to controls, implicating an autoimmune mechanism in MPGN pathogenesis, as opposed to dysfunction of the alternative complement pathway.
It is essential to study genetic associations in racially and ethnically diverse cohorts; validating such associations in diverse cohorts lends support for a potential causal mechanism of disease. In this study, we explore class I and II HLA serotype associations with end-stage kidney disease (ESKD) due to MPGN (including IC-MPGN and C3GN) and DDD from the United States Renal Data System (USRDS) registry5. We evaluate whether HLA antigens are associated with MPGN in a large and diverse cohort of patients and present the first comprehensive study on HLA associations in a cohort of patients with DDD.
Methods
Data source.
We extracted data from the USRDS5, which incorporates data from the United Network for Organ Sharing (UNOS). The USRDS contains records for nearly all patients who receive a kidney transplant in the United States. We assembled data regarding cause of kidney failure and patient demographics from the PATIENTS and MEDEVID files, largely derived from the Medical Evidence Report (CMS Form 2728) submitted by nephrologists within 45 days of initiation of maintenance dialysis or receipt of kidney transplantation. (ESKD due to MPGN was identified using the PDIS variable (i.e., physician-designated primary cause of kidney failure) in the PATIENTS file, with codes 5832, 58321, 5832A, 5832Z, N035 and N045; ESKD due to DDD was identified with codes 58322, 5832C, N036 and N046; ESKD due to anti-GBM nephritis was identified with codes 5834C, 44621, 5834Z, 4462Z. Unfortunately, there are no specific codes for C3GN, and as such it was not possible to separate C3GN from MPGN in our analysis. We captured data on HLA serotypes from UNOS Donor and Recipient Histocompatibility files.
Study Design.
We employed a case-control study design. The primary goal of this analysis was to identify HLA serotypes associated with kidney failure due to MPGN or DDD. Each transplant recipient with MPGN (excluding DDD) or DDD was matched to 15 kidney donor controls, with exact matching for designated race and exact or variable matching for Hispanic ethnicity. Exact matching for designated race requires cases and controls to have the same race, while variable matching for Hispanic ethnicity prefers exact matches for Hispanic ethnicity, but will allow for mismatches on Hispanic ethnicity if no exact matches can be found. Kidney donors were selected from the entire pool of available kidney donors with histocompatibility information collected by UNOS. It is not known if race was determined by self-report or designation by others. The dataset does not include information on ancestry. The exposure of interest was HLA serotype, and the outcome of interest was presence or absence of ESKD due to MPGN (or DDD). To validate the design and statistical analysis approaches used, we also studied patients with ESKD due to anti-glomerular basement membrane (anti-GBM) nephritis, as well-established HLA associations have been identified for this disease6–12.
Study population.
We included all patients with kidney failure due to MPGN, DDD or anti-GBM nephritis who received a first kidney transplant in the US after 1986. Patients simultaneously missing all HLA class I and II serotype and/or race data were excluded from the analysis. As controls, we included all kidney donors with complete information on HLA serotype, race and Hispanic ethnicity. HLA haplotypes differ by ancestry because of strong linkage disequilibrium in HLA genes, and as such, it is important to attempt to distinguish between serotype associations due to strong linkage disequilibrium and serotypes that might be causally implicated in disease pathogenesis. Race is a social construct, which we use here as a proxy for ancestry. Designated race is coded in the Centers for Medicare and Medicaid Services Form 2728 according to the following 6 categories: (1) White, (2) Black or African American, (3) American Indian/Alaska Native, (4) Asian, (5) Native Hawaiian or other Pacific Islander, or (6) Other. Two categories for ethnicity are present: (1) Not Hispanic or Latino, or (2) Hispanic or Latino. Although using designated race/ethnicity as a proxy for ancestry is imperfect, it is the only available information related to ancestry available in USRDS/UNOS. Prior work has demonstrated that HLA alleles and haplotypes demonstrate clustering by race/ethnicity designation in the United States13. Of the racial groups considered, only White and Black patients had adequate sample sizes for race-stratified analyses. Because of the strong influence of ancestry on HLA inheritance, we matched cases and controls on designated race/ethnicity.
Investigated HLA Serotypes.
We considered HLA serotypes with 5% or greater prevalence in donor or disease-specific populations, resulting in the following 58 serotypes for investigation: A1, A2, A3, A11, A23, A24, A26, A29, A30, A31, A32, A68, B7, B8, B14, B18, B21, B27, B35, B39 B44, B51, B57, B60, B62, Bw4, Bw6, Cw1, Cw2, Cw4, Cw5, Cw6, Cw7, Cw8, Cw9, Cw10, Cw12, Cw16, DQ2, DQ4, DQ5, DQ6, DQ7, DQ8, DQ9, DR1, DR4, DR7, DR8, DR11, DR12, DR13, DR14, DR15, DR17, DR51, DR52, DR53.
Statistical analysis.
For each cause of kidney failure (non-DDD MPGN, DDD and anti-GBM nephritis), we first estimated the prevalence of each of the 58 serotypes among patients with ESKD and race/ethnicity-matched donor controls. We used unadjusted logistic regression models to estimate odds ratios of having a specific cause of kidney failure (i.e., MPGN, DDD, anti-GBM disease) given a specific HLA serotype. Given the multiple hypotheses tested, estimated odds ratios were considered statistically significant if associated p-values were less than 0.000862 after Bonferroni correction. In subsequent analyses, we estimated adjusted odds ratios for 57 of the 58 serotypes, adjusting for the serotype with the strongest association seen in univariate analyses for each cause of kidney failure. Finally, we repeated the aforementioned approach within subsets of White and Black patients.
HLA serotype calling and imputation.
The UNOS Recipient Histocompatibility file contains information on HLA serotypes for each transplant recipient, and the corresponding date of typing. Each patient may have multiple entries for HLA types, depending on the year of transplant listing and the year of transplantation (e.g., a patient may be retyped during the interval between waitlisting and transplantation). Additionally, HLA serotype designations have evolved over time due to improved molecular assays enabling better discrimination between specific HLA antigens. To capture these changes in HLA typing, some HLA serotypes are categorized as “broad” or “split” serotypes, where broad serotypes are subdivided into split serotypes due to evolving HLA typing methods. For example, HLA-DR3 is a broad antigen that has been subdivided into HLA-DR17 and HLA-DR18 with improved HLA typing methods. Depending on the year of HLA typing and transplantation, a patient may have only information on broad HLA antigens, without information on the corresponding split antigens.
In this analysis, we considered all antigens with 5% or greater prevalence in donor or disease-specific populations. In the majority of cases, split antigens were investigated in the final analysis in place of their broad antigens, except when the broad antigen has a prevalence of 5% or greater in either the case or control cohorts and all the corresponding splits had a <5% prevalence in both the donor and disease cohorts. Split antigens with a 5% or greater prevalence in donor or disease specific populations, and >5% missingness were imputed before use in the final analysis. Ultimately, we imputed the following split antigens: DR15, DR16, DR17, DR18, DQ5, DQ6, DQ7, DQ8, DQ9, Cw9 and Cw10. In order to impute these splits, for each broad/split group, we extracted data from donors with complete information about the broad and split HLA types. We then fit random forest models to predict the split antigens, using all antigens with a prevalence of 5% or greater as predictors. A random forest model was selected for imputation due to its strength in handling interactions among predictors, pertinent to this analysis due to the strong linkage disequilibrium among HLA serotypes. We assessed model performance on a test-set of patients with each of three causes of kidney failure with complete information on all relevant broad/split antigens. Across models, the median balanced accuracy was ~85%. Details on the imputation approach are provided in the Supplementary Methods and Table S1.
In a sensitivity analysis we report the E-value, defined as the minimum strength of association on the risk ratio scale that an unmeasured confounder would need to have with both the exposure and the outcome, conditional on the measured covariates, to fully explain away a specific exposure-outcome association14,15
We conducted all analyses in R v4.1.3 and Python v3.7.13.
Results
Patient characteristics.
Our study cohorts consisted of 3424 patients with kidney failure due to MPGN, 263 patients with kidney failure due to DDD, and 989 patients with kidney failure due to anti-GBM nephritis, who had received a kidney transplant in the US between 1986 and 2018 and had non-missing data on designated race and relevant class I/II HLA serotypes (Figure 1). Patient demographics are provided in Table 1. Patients with MPGN or DDD were predominantly White (77–83%) and non-Hispanic (85–87%), with a median age at first kidney failure therapy initiation of 29–38 years. Black patients comprised the second largest racial group (11–15%). The sex distribution was approximately balanced for the DDD cohort, while male patients slightly predominated (58%) in the MPGN cohort (Table 1).
Figure 1.
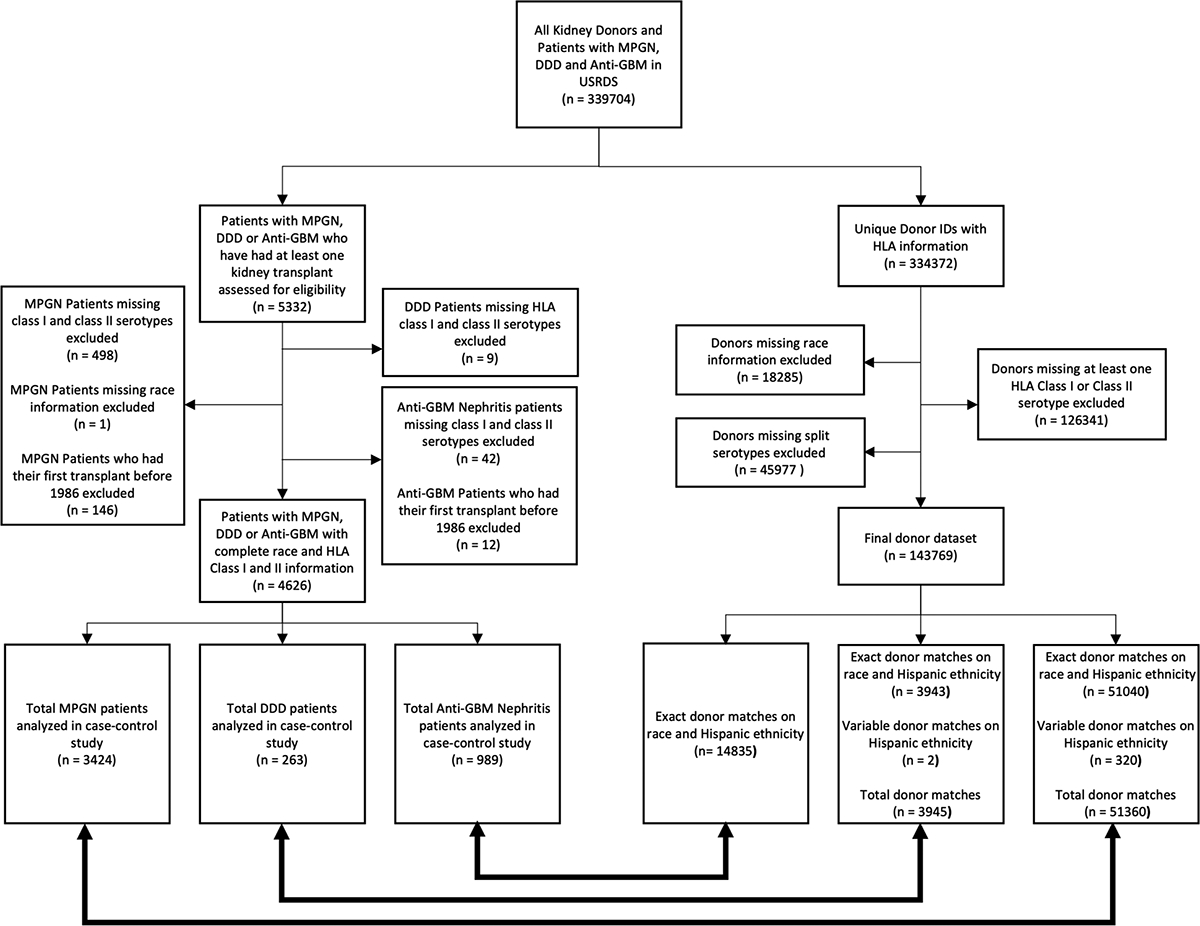
Patients who underwent transplantation within each disease group (MPGN, DDD, anti-GBM nephritis) were eligible. Patients were excluded from the final analysis if they had missing race, and/or Class I or II serotype information. Every case in the final disease cohorts is matched to 15 donors of the same ethnicity and/or race.
Table 1.
Patient characteristics and demographics among the three study cohorts: membranoproliferative glomerulonephritis (MPGN), dense deposit disease (DDD) and anti-glomerular basement membrane nephritis (anti-GBM nephritis; positive control outcome). Demographic information for the kidney donor (control) population is also provided.
| Disease | MPGN | DDD | Anti-GBM nephritis | Donors | |
|---|---|---|---|---|---|
| Number of patients meeting inclusion/exclusion criteria (%) | 3424 (100%) | 263 (100%) | 989 (100%) | 143769 (100%) | |
| Age at first ESRD service (median and IQR) | 38 (24–51) | 29 (17–45) | 41 (24–56) | - | |
| Sex | Male | 1994 (58.2%) | 128 (48.7%) | 503 (50.9%) | - |
| Female | 1430 (41.8%) | 135 (51.3%) | 486 (49.1%) | - | |
| Designated Race | White | 2647 (77.3%) | 218 (82.9%) | 904 (91.4%) | 118031 (82.1%) |
| Black | 502 (14.7%) | 30 (11.4%) | 60 (6.1%) | 20016 (13.9%) | |
| Asian | 185 (5.4%) | 8 (3.0%) | 14 (1.4%) | 4049 (2.8%) | |
| Other | 90 (2.6%) | 7 (2.7%) | 11 (1.1%) | 1673 (1.2%) | |
| Designated Hispanic Ethnicity | Yes | 463 (13.5%) | 41 (15.6%) | 70 (7.1%) | 19600 (13.6%) |
| No | 2961 (86.5%) | 222 (84.4%) | 919 (92.9%) | 124169 (86.4%) | |
| Donor Status | Living | 1394 (40.7%) | 113 (43.0%) | 463 (46.8%) | 60976 (42.4%) |
| Deceased donor | 2018 (58.9%) | 150 (57.0%) | 526 (53.2%) | 82793 (57.6%) | |
| Unspecified | 12 (0.4%) | 0 (0.0%) | 0 (0.0%) | 0 (0.0%) | |
HLA serotypes associated with MPGN.
3424 patients with kidney failure due to MPGN were matched to 51,360 donors: 99% of donors had exact matching for designated race and Hispanic ethnicity, while 1% had variable matching for Hispanic ethnicity. Figure 2 displays univariate odds ratios for each HLA serotype with associated p-values, and Table 2 highlights HLA serotypes with statistically significant associations after Bonferroni correction. Eighteen of the 58 HLA serotypes investigated were associated with kidney failure due to MPGN on univariate analysis: DR17, B8, DR52, A68, Cw16, DR7, Cw12, DQ9, B60, DR51, B7, B57, A1, Cw10, Bw6, DQ2, Cw8, and Cw9. The association with HLA-DR17 was strongest (univariate odds ratio [OR]: 1.55, 95% CI: 1.44–1.68; p-value 4.33e−28; Table 2). We estimated the strength of associations between other HLA serotypes and MPGN, adjusting for HLA-DR17 status; adjusted odds ratios are provided in Table 2. Adjusted odds ratios for 11 serotypes remained significant after conditioning on HLA-DR17 (DR52, A68, Cw16, DR7, Cw12, DQ9, B60, DR51, Cw10, DQ2, Cw9), but the strength of associations was typically attenuated (Table 2). These findings were consistent among patients with ESKD due to MPGN undergoing first transplant during the 1991–2000 (Figure S1) and 2001–2010 (Figure S2) time periods.
Figure 2.
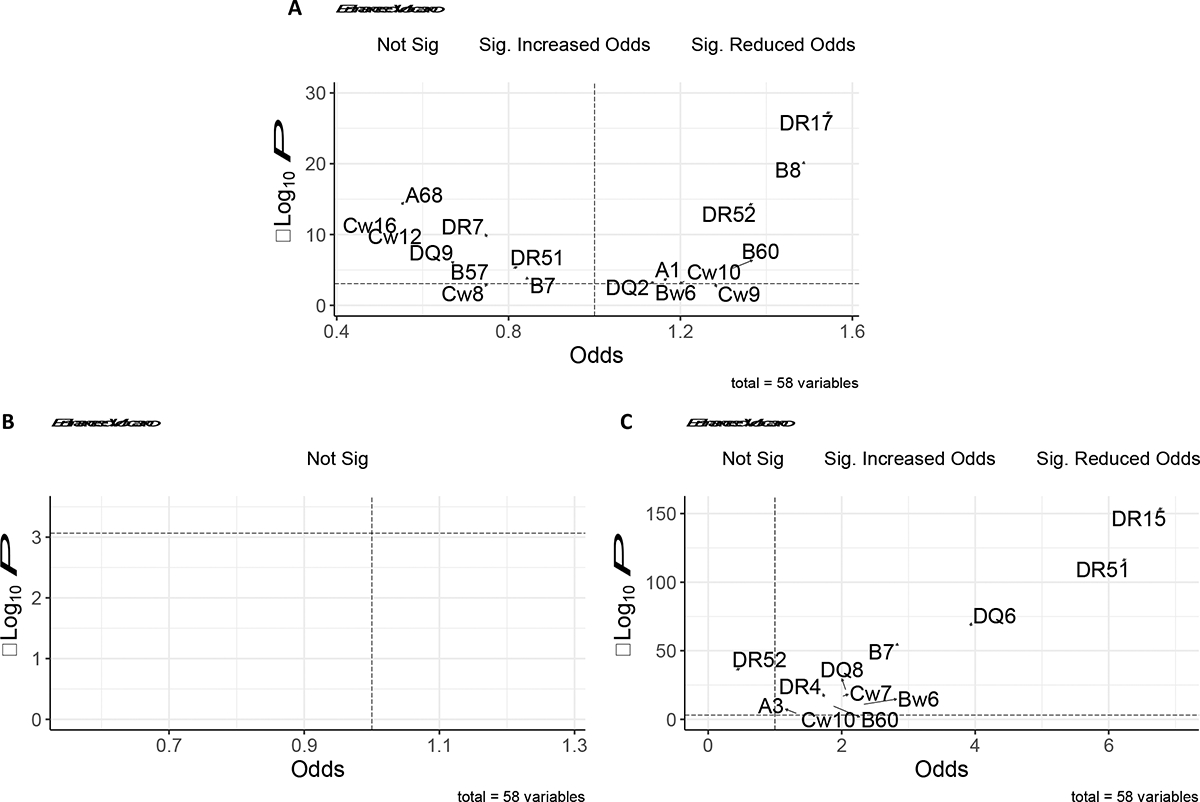
Volcano plots exploring the relationship between HLA serotype associations in (A) MPGN, (B) DDD and (C) Anti-GBM nephritis cohorts. The dashed horizontal line in each plot represents the significance threshold after Bonferroni correction (0.000862), while the dashed vertical line represents an odds ratio of 1. Serotypes with the most significant positive and negative associations are labelled. No significant associations were found between any serotype and DDD.
Table 2.
All statistically significant univariate and adjusted odds ratios associated with kidney failure due to MPGN. Odds ratios were considered statistically significant if p-values were less than 0.000862 after Bonferroni correction (58 tests). Adjusted odds ratios were estimated from a multivariable logistic regression model that included a term for DR17, as DR17 had the strongest univariate association with ESKD due to MPGN. Prevalence of each HLA serotype among cases (n=3424) and designated race/ethnicity matched controls (n=51360) is also provided. Significant results are bolded.
| Serotypes | Odds Ratio (95% CI) | P-value | Adjusted Odds Ratio* (95% CI) | P-value | Prevalence among MPGN cases (%) | Control Prevalence (%) |
|---|---|---|---|---|---|---|
| DR17 | 1.55 (1.44–1.68) | 4.33E-28 | ref | ref | 27.48 | 19.62 |
| B8 | 1.49 (1.37–1.62) | 6.32E-21 | 1.19 (1.06–1.33) | 0.00285 | 23.04 | 16.73 |
| DR52 | 1.37 (1.27–1.49) | 3.70E-15 | 1.18 (1.08–1.29) | 0.000151 | 69.61 | 62.51 |
| A68 | 0.55 (0.47–0.64) | 6.57E-15 | 0.57 (0.49–0.66) | 1.62E-13 | 5.8 | 10.04 |
| Cw16 | 0.45 (0.36–0.56) | 8.22E-13 | 0.46 (0.37–0.58) | 6.34E-12 | 3.56 | 7.58 |
| DR7 | 0.75 (0.68–0.82) | 2.57E-10 | 0.78 (0.71–0.86) | 1.19E-07 | 17.85 | 22.52 |
| Cw12 | 0.56 (0.46–0.67) | 2.39E-09 | 0.58 (0.47–0.7) | 2.67E-08 | 4.65 | 8.07 |
| DQ9 | 0.67 (0.57–0.79) | 8.38E-07 | 0.70 (0.6–0.82) | 1.45E-05 | 5.43 | 7.89 |
| B60 | 1.32 (1.17–1.5) | 6.31E-06 | 1.38 (1.22–1.56) | 2.12E-07 | 9.14 | 7.06 |
| DR51 | 0.81 (0.74–0.89) | 6.55E-06 | 0.85 (0.77–0.93) | 0.000642 | 24.47 | 28.58 |
| B7 | 0.84 (0.77–0.92) | 0.000129 | 0.87 (0.8–0.95) | 0.00261 | 18.41 | 21.18 |
| B57 | 0.75 (0.64–0.87) | 0.000235 | 0.78 (0.67–0.91) | 0.00167 | 5.27 | 6.92 |
| A1 | 1.16 (1.07–1.25) | 0.000332 | 0.99 (0.91–1.08) | 0.841 | 26.36 | 23.65 |
| Cw10 | 1.23 (1.1–1.38) | 0.00035 | 1.25 (1.12–1.41) | 9.24E-05 | 15.74 | 13.19 |
| Bw6 | 1.21 (1.09–1.35) | 0.000362 | 1.13 (1.01–1.25) | 0.0304 | 87.1 | 84.79 |
| DQ2 | 1.14 (1.06–1.23) | 0.000458 | 0.80 (0.72–0.89) | 5.45E-05 | 40.05 | 36.91 |
| Cw8 | 0.75 (0.64–0.89) | 0.000662 | 0.76 (0.65–0.9) | 0.00132 | 6.7 | 8.7 |
| Cw9 | 1.28 (1.11–1.47) | 0.000814 | 1.31 (1.14–1.52) | 0.000211 | 9.13 | 7.29 |
To gain additional insights into which HLA serotypes might plausibly be causally related to MPGN, we leveraged the racial/ethnic diversity of our patient and donor cohorts and explored HLA serotype associations among White and Black patients separately. Among the 2647 White patients, we found HLA-DR17 to have the strongest association with MPGN (OR: 1.58, 95% CI: 1.45, 1.73; p-value: 1.35e−25), and further conditioning on HLA-DR17 tended to attenuate other statistically significant associations (Figure 3, Table S2). Among HLA-serotypes with positive associations among the 502 Black patients with MPGN, HLA-DR17 also had the strongest association (OR: 1.68, 95%CI: 1.33–2.11; p-value: 1.15e−05), and conditioning on HLA-DR17 similarly attenuated other significant associations (Figure 3, Table S3).
Figure 3.
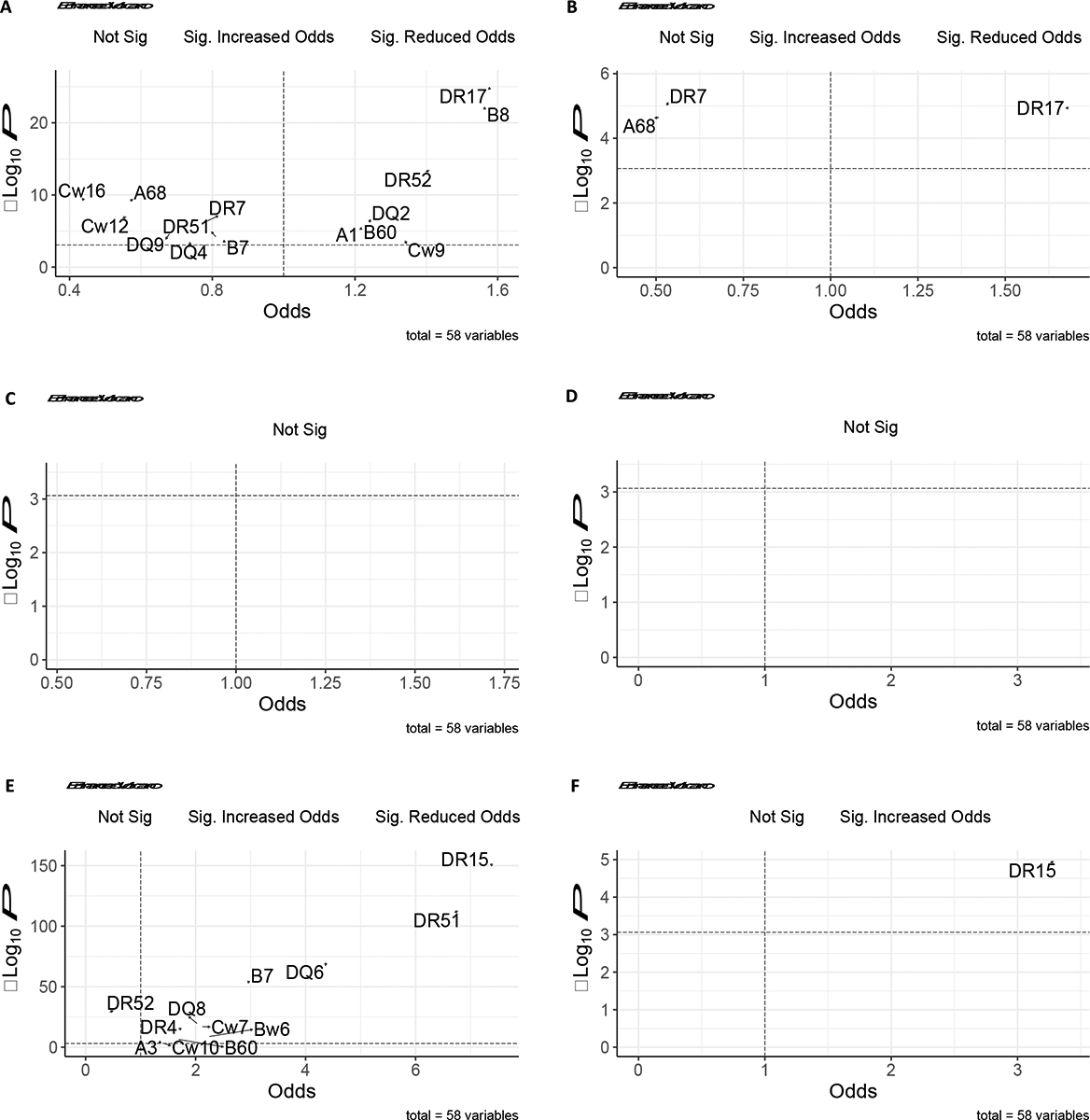
Race specific volcano plots of serotype associations for (A) White patients with MPGN, (B) Black patients with MPGN, (C) White patients with DDD, (D) Black patients with DDD, (E) White patients with anti-GBM nephritis (F) Black patients with anti-GBM nephritis. The dashed horizontal line in each plot represents the significance threshold after Bonferroni correction (0.000862), while the dashed vertical line represents an odds ratio of 1. Serotypes with the most significant positive and negative associations are labelled.
HLA serotypes associated with DDD.
Two hundred and sixty-three patients with kidney failure due to DDD were matched to 3945 donors, with 99.9% of donors matched exactly on designated race and Hispanic ethnicity, and 0.1% of donors with variable matching for Hispanic ethnicity. We found no statistically significant associations between HLA serotype and kidney failure due to DDD. We further examined whether any HLA associations could be identified when considering White (n=218) and Black patients (n=30) separately, but none were identified.
HLA serotypes associated with anti-GBM nephritis (positive outcome control).
Nine hundred and eighty nine patients with kidney failure due to anti-GBM nephritis were matched to 14835 donors on designated race and Hispanic ethnicity. Figure 2 displays univariate odds ratios for each HLA serotype with associated p-values, and Table 3 highlights those HLA serotypes with statistically significant associations after Bonferroni correction. Twenty-nine of the 58 HLA serotypes investigated were significant on univariate analysis: DR15, DR51, DQ6, B7, DR52, DQ8, DR11, DQ7, DQ5, Bw4, DR7, Cw7, DR4, DR13, DQ2, DR1, Bw6, B60, B35, Cw4, DR14, B44, Cw16, DR17, A3, DQ9, Cw2, Cw6, and Cw10. The association with HLA-DR15 was strongest (univariate odds ratio [OR]: 6.81, 95% CI: 5.91–7.85; p-value 5.26e−155; Table 3). We sought to estimate the strength of associations between other HLA serotypes and anti-GBM, controlling for HLA-DR15 status; adjusted odds ratios are provided in Table 3. Adjusted odds ratios for 17 serotypes remained significant after conditioning on HLA-DR15 (DR52, DQ8, DR11, DQ7, DQ5, Bw4, DR7, Cw7, DR4, DR13, DR1, Bw6, B60, B35, Cw4, DR14, Cw16, Cw10), but the strength of associations was typically attenuated (Table 3).
Table 3.
All statistically significant univariate and adjusted odds ratios associated with kidney failure due to anti-GBM nephritis (positive outcome control). Odds ratios were considered statistically significant if p-values were less than 0.000862 after Bonferroni correction (58 tests). Adjusted odds ratios were estimated from a multivariable logistic regression model that included a term for DR15, as DR15 had the strongest association with anti-GBM nephritis. Prevalence of each HLA serotype among cases (n=989) and designated race/ethnicity matched controls (n=14835) is also provided. Significant results are bolded.
| Serotypes | Odds Ratio (95% CI) | P-value | Adjusted Odds Ratio* (95% CI) | P-value | Prevalence among Anti-GBM cases (%) | Control Prevalence (%) |
|---|---|---|---|---|---|---|
| DR15 | 6.81 (5.91–7.85) | 5.26E-155 | ref | ref | 70.59 | 26.05 |
| DR51 | 6.27 (5.36–7.32) | 8.55E-118 | 0.63 (0.39–1.02) | 0.0606 | 71.29 | 28.38 |
| DQ6 | 3.93 (3.37–4.58) | 1.74E-68 | 0.81 (0.63–1.05) | 0.113 | 73.67 | 41.57 |
| B7 | 2.85 (2.5–3.25) | 1.33E-55 | 1.27 (1.1–1.47) | 0.00113 | 46.36 | 23.26 |
| DR52 | 0.41 (0.36–0.47) | 1.91E-36 | 0.68 (0.59–0.79) | 3.98E-07 | 40.16 | 62.12 |
| DQ8 | 2.06 (1.78–2.39) | 4.26E-22 | 3.12 (2.66–3.66) | 5.88E-45 | 32.46 | 18.89 |
| DR11 | 0.26 (0.2–0.34) | 1.15E-21 | 0.37 (0.28–0.49) | 4.26E-12 | 5.49 | 18.36 |
| DQ7 | 0.44 (0.37–0.52) | 2.55E-21 | 0.67 (0.56–0.8) | 8.42E-06 | 19.18 | 35.19 |
| DQ5 | 0.40 (0.33–0.49) | 1.40E-20 | 0.52 (0.42–0.63) | 5.59E-11 | 14.07 | 29.06 |
| Bw4 | 0.54 (0.48–0.62) | 1.50E-19 | 0.63 (0.55–0.72) | 4.02E-11 | 47.47 | 62.4 |
| DR7 | 0.40 (0.32–0.48) | 3.08E-19 | 0.56 (0.45–0.68) | 2.89E-08 | 11.05 | 23.9 |
| Cw7 | 2.01 (1.71–2.36) | 1.67E-17 | 1.33 (1.13–1.58) | 0.000806 | 68.62 | 52.07 |
| DR4 | 1.74 (1.53–1.99) | 8.82E-17 | 2.76 (2.39–3.18) | 1.24E-44 | 42.39 | 29.67 |
| DR13 | 0.44 (0.35–0.54) | 3.45E-15 | 0.62 (0.5–0.77) | 9.98E-06 | 10.58 | 21.37 |
| DQ2 | 0.56 (0.48–0.65) | 1.37E-13 | 0.83 (0.71–0.98) | 0.0269 | 26.33 | 38.96 |
| DR1 | 0.48 (0.39–0.59) | 3.46E-12 | 0.65 (0.53–0.81) | 0.000102 | 10.34 | 19.5 |
| Bw6 | 2.32 (1.82–2.95) | 1.15E-11 | 1.86 (1.45–2.38) | 9.25E-07 | 92.39 | 83.97 |
| B60 | 1.87 (1.54–2.26) | 1.69E-10 | 2.19 (1.79–2.68) | 2.98E-14 | 13.6 | 7.77 |
| B35 | 0.51 (0.41–0.63) | 4.65E-10 | 0.63 (0.51–0.79) | 4.09E-05 | 9.82 | 17.68 |
| Cw4 | 0.50 (0.4–0.62) | 8.34E-10 | 0.61 (0.48–0.76) | 1.67E-05 | 12.69 | 22.63 |
| DR14 | 0.29 (0.18–0.46) | 1.45E-07 | 0.43 (0.27–0.68) | 0.000348 | 1.93 | 6.32 |
| B44 | 0.65 (0.55–0.77) | 4.07E-07 | 0.75 (0.63–0.89) | 0.000871 | 18.32 | 25.59 |
| Cw16 | 0.29 (0.17–0.48) | 2.39E-06 | 0.35 (0.21–0.6) | 9.17E-05 | 2.09 | 6.88 |
| DR17 | 0.69 (0.58–0.82) | 3.24E-05 | 1.01 (0.84–1.21) | 0.935 | 16.44 | 22.24 |
| A3 | 1.33 (1.16–1.53) | 6.01E-05 | 1.01 (0.87–1.16) | 0.943 | 31.21 | 25.42 |
| DQ9 | 0.52 (0.38–0.72) | 9.48E-05 | 0.72 (0.52–1.01) | 0.0543 | 4.43 | 8.15 |
| Cw2 | 0.55 (0.39–0.76) | 0.000346 | 0.64 (0.46–0.9) | 0.0102 | 5.3 | 9.3 |
| Cw6 | 0.68 (0.55–0.85) | 0.000708 | 0.79 (0.63–0.99) | 0.042 | 13.11 | 18.1 |
| Cw10 | 1.41 (1.16–1.73) | 0.000721 | 1.75 (1.42–2.15) | 1.54E-07 | 17.02 | 12.67 |
Among White patients, we found HLA-DR15 continued to have the strongest association with anti-GBM (OR: 7.40, 95% CI: 6.37, 8.59; p-value: 5.29e−151), and further conditioning on HLA-DR15 tended to attenuate other statistically significant associations (Figure 3, Table S4). Among Black patients, only HLA-DR15 showed a significant association with anti-GBM nephritis (OR: 3.29, 95% CI: 1.93–5.59; p-value: 1.10e−05) (Figure 3, Table S5). Taken together, these data support a strong association between HLA-DR15 and kidney failure due to anti-GBM, corroborated by several published studies.
Discussion
The HLA is a fundamental component of adaptive immunity responsible for presentation of self- and non-self-peptides to T-cell receptors. HLA is divided into class I (HLA-A, HLA-B, and HLA-C) and II alleles (HLA-DR, HLA-DQ and HLA-DP), often inherited in strong linkage disequilibrium, tending to segregate together in specific haplotypes. Polymorphisms in HLA are thought to contribute to the development of several autoimmune kidney diseases; associations between specific HLA types and diabetes, IgA nephropathy, anti-GBM disease and anti-nuclear cytoplasmic antibody (ANCA)-associated vasculitis among others, have been identified6,16–20. In this case-control study, we quantified associations between 58 class I and II HLA antigens and kidney failure due to MPGN or DDD in a large, diverse cohort of patients with kidney failure due to MPGN or DDD in the US. We determined that kidney failure due to MPGN was associated with HLA-DR17 (OR: 1.55, 95%CI: 1.44–1.68). In contrast, we found no significant associations between class I and II HLA serotypes and kidney failure due to DDD. Finally, we were able to recapitulate the direction and magnitude of known HLA associations with anti-GBM nephritis (positive-control outcome), supporting the validity of our analytic approach.
To our knowledge, this is the largest study exploring HLA associations with MPGN and DDD to date, and the first to include a sizeable proportion of Black patients. Recently, Levine et al. performed whole genome sequencing on 146 patients with MPGN (including DDD and C3GN) and 6442 controls, all designated as White race4. They found no significant enrichment for rare variants in alternative complement pathway genes, but did find enrichment for an HLA haplotype incorporating DQA1*05:01/DQB1*02:01, and DRB1*03:01 (corresponding to serotypes HLA-DQ2 and HLA-DR17, respectively), with estimated odds ratios of 1.5–2.5. This study was the first to suggest that HLA (and hence autoimmunity), rather than complement defects, may be contributing to the pathogenesis of MPGN, DDD and C3GN. While groundbreaking, limitations of this study include a modest sample size, a homogenous study population (all patients of northern European origin), and the aggregation of IC-MPGN, C3GN, DDD. This latter limitation is particularly noteworthy, as C3GN and DDD clearly differ in pathogenesis from IC-MPGN.
Overcoming some of these limitations, our larger study (n=3424 for MPGN and n=263 for DDD) in a more racially diverse population confirms the association between HLA-DR17 and MPGN identified by Levine et al., and the breadth of our cohort allowed us to potentially separate the role of HLA-DR17 from its associated haplotype in White patients, HLA-A1-B8-DR3-DQ2, evidenced by the fact that this haplotype was not associated with MPGN in the Black patient cohort. However, the magnitude of the association with HLA-DR17 we identified was modest, similar to that of Levine et al., highlighting that the pathogenesis of MPGN is complex and likely multifactorial. Of note, HLA-DR17 has also been found to be associated with membranous nephropathy and lupus nephritis (with larger odds ratios, >2.0), highlighting the potential importance of DR17 across several autoimmune kidney diseases19,20.
In contrast, we found no significant HLA associations with kidney failure due to DDD, suggesting that the underlying pathogenesis for MPGN and DDD are likely distinct. While other studies have highlighted an association between DDD and type I diabetes, suggesting a possible role for autoimmunity and potential HLA associations21, to our knowledge only one study specifically explored HLA associations with DDD and was limited to 16 patients22, prohibiting detailed assessment.
Several strengths of our study should be noted. First, we used real-world data, and HLA serotypes were collected and reported to UNOS/USRDS during routine clinical care. As such, due to improvements in molecular assays for HLA typing during the time period studied, several HLA serotypes were further subclassified (from “broad” antigens to “split antigens”). To account for these changes, we developed machine learning models to impute prevalent split antigens, where missing; our models had excellent performance (Supplementary Methods and Table S1). In a sensitivity analysis, we demonstrate that the findings presented are robust to the time period of transplantation selected (Figures S1–S2). In addition, we studied HLA serotypes associated with anti-GBM nephritis as a positive outcome control to verify that our statistical approach and the data used are capable of reproducing well-established associations that have been previously reported. In the anti-GBM nephritis cohort, we found strong associations with HLA-DR15 (OR: 6.81, 95% CI: 5.91–7.85; p-value 5.26e-155) and HLA-DR4 (OR: 1.74, 95% CI: 1.53–1.99; p-value 8.82e-17), both of which are similar in magnitude to reported HLA associations with anti-GBM nephritis6–8,11,12.
Several limitations must also be mentioned. By studying only patients with ESKD, one might consider the possibility that the observed associations are due to a relation between HLA and ESKD itself, rather than any disease-specific association. We believe this is unlikely because no specific HLA serotype association was identified across the three groups studied (MPGN, DDD and anti-GBM nephritis). Furthermore, an evaluation of the literature demonstrates no clear association between HLA serotypes and ESKD itself23; a recent meta-analysis also failed to identify a consistent relation between HLA and ESKD24. We relied on designated race/ethnicity as a proxy for ancestry to study HLA associations with disease while considering the strong linkage disequilibrium in HLA haplotypes. Race is a social construct. While imperfect, it is well known that the distribution of HLA antigens differs among designated racial and ethnic groups13,25–27. Our study was also limited due to the lack of information on HLA genotypes, instead relying on reported HLA serotypes. HLA serotypes can correspond to multiple genotypes, and high-dimensional genotyping of the HLA locus across diverse cohorts of patients with MPGN and DDD may be helpful in clarifying exact mechanisms by which HLA may contribute to disease pathogenesis. For example, an unmeasured genetic locus associated with both HLA-DR17 and ESKD due to MPGN at a strength of 2.26 on the risk ratio scale would explain away the observed association between HLA-DR17 and ESKD due to MPGN14,15. In addition, we relied upon glomerular disease diagnoses reported to the USRDS, which may not necessarily be biopsy confirmed. USRDS diagnoses for glomerular disease are reported to be highly specific but poorly sensitive (18). Finally, we excluded patients lacking class I and II HLA serotypes in USRDS/UNOS, thereby reducing the sample size of our study. Importantly, we do not believe exclusion of these patients introduces systematic bias in estimates of HLA associations with causes of ESKD, as evidence by our analysis of the anti-GBM nephritis cohort. Ultimately, although our study is the largest to date, MPGN and DDD are rare glomerular diseases, and the number of patients studied here was modest, especially when stratified by designated race. As such, our study may have been underpowered to detect HLA associations with small effect sizes.
In conclusion, we identified several class I and II HLA serotypes associated with kidney failure due to non-DDD MPGN, most notably HLA-DR17, but no significant HLA associations with kidney failure due to DDD. These data contribute to our understanding of the potential pathogenic mechanisms underlying MPGN, suggesting a likely role for autoimmunity in some cases, and highlight potential differences in HLA susceptibility to MGPN compared to DDD.
Supplementary Material
Footnotes
Disclaimer: The data reported here have been supplied by the United States Renal Data System (USRDS). The interpretation and reporting of these data are the responsibility of the author(s) and in no way should be seen as an official policy or interpretation of the U.S. government.
References
- 1.Sethi S, Fervenza FC. Membranoproliferative glomerulonephritis--a new look at an old entity. N Engl J Med. 2012;366(12):1119–1131. doi: 10.1056/NEJMra1108178 [DOI] [PubMed] [Google Scholar]
- 2.Alchi B, Jayne D. Membranoproliferative glomerulonephritis. Pediatr Nephrol. 2010;25(8):1409–1418. doi: 10.1007/s00467-009-1322-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sethi S, Fervenza FC. Membranoproliferative glomerulonephritis: pathogenetic heterogeneity and proposal for a new classification. Semin Nephrol. 2011;31(4):341–348. doi: 10.1016/j.semnephrol.2011.06.005 [DOI] [PubMed] [Google Scholar]
- 4.Levine AP, Chan MMY, Sadeghi-Alavijeh O, et al. Large-Scale Whole-Genome Sequencing Reveals the Genetic Architecture of Primary Membranoproliferative GN and C3 Glomerulopathy. J Am Soc Nephrol. 2020;31(2):365–373. doi: 10.1681/ASN.2019040433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.U.S. Renal Data System. 2020 USRDS annual data report: Epidemiology of kidney disease in the United States. National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases, Bethesda, MD, 2020. [Google Scholar]
- 6.Phelps RG, Rees AJ. The HLA complex in Goodpasture’s disease: a model for analyzing susceptibility to autoimmunity. Kidney Int. 1999;56(5):1638–1653. doi: 10.1046/j.1523-1755.1999.00720.x [DOI] [PubMed] [Google Scholar]
- 7.Rees AJ, Peters DK, Compston DA, Batchelor JR. Strong association between HLA-DRW2 and antibody-mediated Goodpasture’s syndrome. Lancet. 1978;1(8071):966–968. doi: 10.1016/s0140-6736(78)90252-0 [DOI] [PubMed] [Google Scholar]
- 8.Rees AJ, Peters DK, Amos N, Welsh KI, Batchelor JR. The influence of HLA-linked genes on the severity of anti-GBM antibody-mediated nephritis. Kidney Int. 1984;26(4):445–450. doi: 10.1038/ki.1984.194 [DOI] [PubMed] [Google Scholar]
- 9.Huey B, McCormick K, Capper J, et al. Associations of HLA-DR and HLA-DQ types with anti-GBM nephritis by sequence-specific oligonucleotide probe hybridization. Kidney Int. 1993;44(2):307–312. doi: 10.1038/ki.1993.245 [DOI] [PubMed] [Google Scholar]
- 10.Fisher M, Pusey CD, Vaughan RW, Rees AJ. Susceptibility to anti-glomerular basement membrane disease is strongly associated with HLA-DRB1 genes. Kidney Int. 1997;51(1):222–229. doi: 10.1038/ki.1997.27 [DOI] [PubMed] [Google Scholar]
- 11.Kitagawa W, Imai H, Komatsuda A, et al. The HLA-DRB1*1501 allele is prevalent among Japanese patients with anti-glomerular basement membrane antibody-mediated disease. Nephrol Dial Transplant. 2008;23(10):3126–3129. doi: 10.1093/ndt/gfn179 [DOI] [PubMed] [Google Scholar]
- 12.Yang R, Cui Z, Zhao J, Zhao MH. The role of HLA-DRB1 alleles on susceptibility of Chinese patients with anti-GBM disease. Clin Immunol. 2009;133(2):245–250. doi: 10.1016/j.clim.2009.07.005 [DOI] [PubMed] [Google Scholar]
- 13.Cao K, Hollenbach J, Shi X, Shi W, Chopek M, Fernández-Viña MA. Analysis of the frequencies of HLA-A, B, and C alleles and haplotypes in the five major ethnic groups of the United States reveals high levels of diversity in these loci and contrasting distribution patterns in these populations. Human Immunology. 2001;62(9):1009–1030. doi: 10.1016/S0198-8859(01)00298-1 [DOI] [PubMed] [Google Scholar]
- 14.VanderWeele TJ, Ding P. Sensitivity Analysis in Observational Research: Introducing the E-Value. Ann Intern Med. 2017;167(4):268–274. doi: 10.7326/M16-2607 [DOI] [PubMed] [Google Scholar]
- 15.Mathur MB, Ding P, Riddell CA, VanderWeele TJ. Web Site and R Package for Computing E-values. Epidemiology. 2018;29(5):e45–e47. doi: 10.1097/EDE.0000000000000864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Robson KJ, Ooi JD, Holdsworth SR, Rossjohn J, Kitching AR. HLA and kidney disease: from associations to mechanisms. Nat Rev Nephrol. 2018;14(10):636–655. doi: 10.1038/s41581-018-0057-8 [DOI] [PubMed] [Google Scholar]
- 17.Valdes AM, Erlich HA, Carlson J, Varney M, Moonsamy PV, Noble JA. Use of class I and class II HLA loci for predicting age at onset of type 1 diabetes in multiple populations. Diabetologia. 2012;55(9):2394–2401. doi: 10.1007/s00125-012-2608-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Feehally J, Farrall M, Boland A, et al. HLA has strongest association with IgA nephropathy in genome-wide analysis. J Am Soc Nephrol. 2010;21(10):1791–1797. doi: 10.1681/ASN.2010010076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Xu R, Li Q, Liu R, et al. Association Analysis of the MHC in Lupus Nephritis. J Am Soc Nephrol. 2017;28(11):3383–3394. doi: 10.1681/ASN.2016121331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sekula P, Li Y, Stanescu HC, et al. Genetic risk variants for membranous nephropathy: extension of and association with other chronic kidney disease aetiologies. Nephrol Dial Transplant. 2017;32(2):325–332. doi: 10.1093/ndt/gfw001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu DF, Moon M, Lanning LD, McCarthy AM, Smith RJH. Clinical features and outcomes of 98 children and adults with dense deposit disease. Pediatr Nephrol. 2012;27(5):773–781. doi: 10.1007/s00467-011-2059-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Noël LH, Descamps B, Jungers P, et al. HLA antigen in three types of glomerulonephritis. Clin Immunol Immunopathol. 1978;10(1):19–23. doi: 10.1016/0090-1229(78)90004-1 [DOI] [PubMed] [Google Scholar]
- 23.Lowe M, Jervis S, Payton A, et al. Systematic review of associations between HLA and renal function. Int J Immunogenet. 2022;49(1):46–62. doi: 10.1111/iji.12566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noureen N, Zaidi N. Association between human leukocyte antigen (HLA) and end-stage renal disease (ESRD): a meta-analysis. PeerJ. 2023;11:e14792. doi: 10.7717/peerj.14792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buhler S, Sanchez-Mazas A. HLA DNA Sequence Variation among Human Populations: Molecular Signatures of Demographic and Selective Events. PLOS ONE. 2011;6(2):e14643. doi: 10.1371/journal.pone.0014643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Just JJ, King MC, Thomson G, Klitz W. African-American HLA class II allele and haplotype diversity. Tissue Antigens. 1996;48(6):636–644. doi: 10.1111/j.1399-0039.1996.tb02686.x [DOI] [PubMed] [Google Scholar]
- 27.Tu B, Mack SJ, Lazaro A, et al. HLA-A, -B, -C, -DRB1 allele and haplotype frequencies in an African American population. Tissue Antigens. 2007;69(1):73–85. doi: 10.1111/j.1399-0039.2006.00728.x [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


