Abstract
This study investigated the distribution of bovine spongiform encephalopathy (BSE) in herds of cattle in Ireland over the years 1996 through 2000, prior to the introduction of widespread active surveillance. Mappings of index herds, herd density, and standardized morbidity ratios, by county, were employed to help visualize areas of potential clustering of BSE. The hypothesis of spatial clustering was tested using a spatial scan statistic applied to the location of the herd where exposure likely occurred. Both Bernoulli and Poisson spatial models indicated marked clustering of BSE herds centred on Monaghan county, with secondary clusters detected by Bernoulli approaches and some Poisson models in Wexford and Cork. The number of cases increased with time, but clear temporal-spatial clusters were rarely detected, except in the case of a cluster in Wexford. The focussed spatial scan analyses using the location of large-scale feed suppliers provided support for the hypothesis that clustering of BSE may be associated with feed source. The results of our analyses provided strong evidence in support of the hypothesis that herds, in which animals were most likely to have been exposed to the BSE agent, cluster geographically.
Résumé
La distribution de l’encéphalopathie spongiforme bovine (BSE) dans les troupeaux bovins en Irlande entre les années 1996 et 2000, avant l’introduction de méthode généralisée de surveillance active, a été investiguée. La cartographie des troupeaux index, la densité des troupeaux et les ratios standardisés de morbidité, par comté, a été employée afin d’aider à visualiser les régions de regroupement de cas de BSE. L’hypothèse de regroupement spatial a été testée en utilisant une statistique spatiale appliquée à la localisation du troupeau où l’exposition s’est probablement produite. Les modèles spatiaux de Bernoulli et Poisson indiquaient un regroupement marqué des troupeaux avec des cas de BSE centrés sur le comté de Monaghan, avec des regroupements secondaires détectés avec les approches de Bernoulli et quelques modèles de Poisson dans les comtés de Wexford et Cork. Le nombre de cas a augmenté avec le temps, mais des regroupements temporo-spatial évidents ont rarement été détectés, sauf dans le cas d’un regroupement dans le comté de Wexford. Les analyses spatiales ciblées en utilisant la localisation des distributeurs importants de nourriture ont fourni des évidences pour appuyer l’hypothèse que le regroupement des cas de BSE puisse être associer avec une source alimentaire. Les résultats de nos analyses apportent des preuves pour soutenir l’hypothèse que les troupeaux, dans lesquels des animaux sont susceptibles d’avoir été exposés à l’agent du BSE, sont regroupés géographiquement.
(Traduit par Docteur Serge Messier)
Introduction
Bovine spongiform encephalopathy (BSE), a progressive neuro-degenerative disorder of adult cattle, was first recognised in the United Kingdom (UK) in 1985 (1). Despite the continuing debate about its origin, there is clear evidence to support the hypothesis that cattle were primarily exposed to the agent through the consumption of contaminated meat and bone meal (MBM) (2–6). The age at the time of infection was estimated to be between 0 and 18 mo of age, based on simulation studies and back-calculation modelling carried out in the UK (7,8). A prominent feature of the disease in the UK was the clustering both between farms at the regional or county level and within farms (2,8–11). Authors of early epidemiological studies postulated that this geographic heterogeneity was caused by regional differences in the reprocessing of greaves and the use of solvent extraction. Later, modelling exercises provided statistical support for the conclusion that the pattern was either due to between-producer variability in feed infectivity, or between-farm variation in feed utilisation (12).
In response to the hypothesis that MBM was the source of the infectious agent, a ban on the feeding of MBM to ruminant animals was introduced in the UK, in 1988. Although the ban produced a dramatic decline in the number of new infections (11,13–15), cases of BSE continued to be diagnosed in animals born after the implementation of the ban. The spatial proximity of many cases to areas of intensive pig production led to the theory that the disease could be caused by inadvertent cross contamination of cattle rations with MBM intended for use in pig feeds (15,16).
The 1st case of BSE in Ireland was diagnosed in 1989, and early maps of the locations of case herds indicated a tendency of case herds to cluster geographically. Thus, the principal objectives of this study were to formally test whether herds with BSE in Ireland clustered geographically and, if so, to identify where and when the clusters were located and to investigate whether there was evidence that large feed-suppliers were spatially associated with any geographical clusters.
Materials and methods
The unit of concern in this study was the bovine herd, and included all of the cattle listed under the official herd number issued by the Irish Department of Agriculture and Food (DAF). Herds were eligible for this study if a herd member had been notified as a BSE suspect to DAF between January 1, 1996 and December 31, 2000. Case identification was by passive surveillance only, as this study was carried out prior to the widespread availability of rapid tests. In this paper, an index herd was the herd where the case of BSE was housed at the time of notification. Herds in which the positive animal was located at the time it was most likely to have been exposed to the infectious agent were denoted as herds of putative exposure (PE). Since the exposure could not be known with certainty, the PE herds were subdivided into categories based on the amount of time the infected animal spent in the herd: PE 1 herds were defined as herds in which the positive animal had spent its entire life; PE 2 herds included PE 1 herds, as well as herds in which a positive animal had spent its first year of life; and PE 3 herds included PE 1 and PE 2 herds, as well as herds in which a positive animal had spent its first 6 mo of life. Control herds were herds that had at least 1 BSE suspect that was subsequently deemed negative on the basis of response to treatment or post mortem histopathology and immunohistochemistry.
Source of herd data
Information pertaining to index, PE, and control herds was obtained from the National BSE Database maintained at the head-quarters of the DAF in Dublin. This database was updated using epidemiological investigation forms completed by a DAF Veterinary Inspector for each herd in which the suspect animal spent any part of its life. The 5 variables required for this analysis included: a) location of the home premises (based on the largest fragment of land attributed to the owner) in terms both of Irish National Grid (ING) map co-ordinates and District Electoral Division (DED; the smallest administrative area defined in Ireland); b) herd size (categorical variable with 4 levels based on total number of cattle present: small 1 to 20; medium 21 to 50; large 51 to 100; extra large 101+); c) herd enterprise type (categorical variable with 4 levels based on cow type: beef, suckler, dairy, mixed [both suckler and dairy cows]); d) date of clinical onset (the date of notification was used as a surrogate measure in 27 cases where this information was missing); and e) month and year of birth. Data from the Land Parcel Information System (LPIS) were converted for use with our computer software (ArcView, Windows version 3.1; Environmental Systems Research Institute, Toronto, Ontario). The location of the index and PE farm was indicated using the ING centroid of the largest farm fragment. In addition, ING coordinates were obtained for the locations of the 61 major feed suppliers in Ireland, so that these could be used in “focussed” spatial analyses.
Source of population data
The annual number of herds at risk and their herd size (the total number of cattle in the herd) were obtained from the National Tuberculosis (TB) Testing Database maintained at the Centre for Veterinary Epidemiology and Risk Assessment, University College, Dublin, Ireland. This database contains a record of all TB tests carried out in bovine herds since 1995. The population at risk in the years prior to 1996 was assumed to be the same as the 1996 population. Information regarding enterprise type in the national herd population was obtained from Teagasc, an Irish state funded organisation that provides advice on agricultural issues.
Descriptive techniques
For index herds, annual risks per county were calculated for each calendar year from 1996 to 2000, based on the assumption that the population of herds in any year represented a closed population. Annual standardized morbidity ratios (SMRs) were calculated per county by dividing the observed number of index herds, in each of the 16 herd categories (4 types times 4 sizes), by the number expected, using the overall distribution of herd type and size for all herds in Ireland as the standard population (16). The effect of index herd size, enterprise type, county, and year on risk were tested using standard analysis of variance (ANOVA) techniques (SAS, version 8; SAS Institute, Cary, North Carolina, USA). In this analysis, county risks were transformed using the natural log transformation, the hypothesis of normality was tested using the Shapiro-Wilk test, and homogeneity of variance was tested using Bartlett’s test.
Cluster analysis
Analysis of clustering was carried out using computer software (SaTScan, version 2.1; National Cancer Institute, Bethesda, Maryland, USA) (18). This software imposes circles (spatial scan statistic) or cylinders (space-time scan statistic) of varying size on the spatial data. The likelihood of a BSE case herd within and without the circle was calculated; a risk ratio created; and the statistical significance of this ratio assessed using a Monte Carlo simulation, carried out under the null hypothesis of random distribution. Two general approaches were used in our analyses and were applied to both the index herd and PE herd data. In the Bernoulli approach we compared the location of index (or PE) herds to control herds. In the Poisson approach we compared the location of index (or PE) herds relative to the population of all herds in the DED; the population at risk data were aggregated to the level of the DED and the centroid of each DED was calculated using ArcView. These methods differ in the comparative denominator used for the case herd numbers and this can lead to divergent results. In addition, the Poisson analysis allows for the explicit control of confounders and we did this using the proportional distribution of herd size and type per DED. Direct control of confounding was not available in the Bernoulli approach; however, there is an implied control in that one anticipates that, in the long run, the size and type of control herds would reflect that of the at-risk group in the source population. For each analytic method we also used focussed and unfocussed approaches. In the unfocussed analyses, SaTScan used the location of the case and control herds (Bernoulli approach) or the centroid of DEDs with aggregated count data (Poisson approach) as its background spatial grid for centring the spatial circles. In the focussed analyses, the spatial grids were the ING co-ordinates of the 61 major cattle-feed suppliers. Space-time clusters were investigated using the date of birth of the case or suspect case respectively. Analyses for spatial clustering, up to 50% of the study area and at annual and 6 monthly time scales, were carried out.
Results
Data were available on a total of 460 index and 541 control herds; the distributions of index and control herds per county are shown in Table I. Cork, Cavan, Monaghan, Wexford, and Meath (in that order) had the greatest number of index case herds, while Monaghan, Cavan, Wexford, Meath, and Louth had the highest incidence risks. Meath and Cavan had the highest per population reporting of control herds followed by Kilkenny, Longford, and Carlow. The herd of PE was available for between 60% and 85% of all BSE cases, depending on the level of detail required. The PE herds were categorized into 277 PE 1 herds, 367 PE 2 herds, and 392 PE 3 herds. Thirty-four percent of the index herds (164 BSE positive cattle) had purchased the BSE positive animal, with 2/3 of the purchases occurring at an age of more than 2 y.
Table I.
Number of District Electoral Divisions (DEDs), herds, case herds, and control herds, per county, between the years 1996 and 2000, in Ireland
| County | Number of DEDs | Mean number of herds 1996 to 2000 | Number of BSE case herds | Number of control herds |
|---|---|---|---|---|
| Carlow | 50 | 1743 | 8 | 11 |
| Cavan | 91 | 6097 | 58 | 48 |
| Clare | 151 | 7369 | 24 | 27 |
| Cork | 315 | 15228 | 65 | 80 |
| Donegal | 144 | 7639 | 15 | 17 |
| Dublin | 33 | 676 | 2 | 1 |
| Galway | 214 | 14546 | 15 | 32 |
| Kerry | 163 | 9067 | 7 | 5 |
| Kildare | 86 | 2517 | 3 | 12 |
| Kilkenny | 110 | 3886 | 15 | 26 |
| Laois | 55 | 3494 | 5 | 17 |
| Leitrim | 78 | 4028 | 7 | 13 |
| Limerick | 135 | 7265 | 27 | 31 |
| Longford | 52 | 3042 | 11 | 20 |
| Louth | 37 | 1711 | 9 | 10 |
| Mayo | 155 | 12981 | 2 | 14 |
| Meath | 89 | 4800 | 35 | 47 |
| Monaghan | 65 | 4971 | 55 | 19 |
| Offaly | 87 | 3813 | 4 | 7 |
| Roscommon | 110 | 7021 | 1 | 6 |
| Sligo | 79 | 4913 | 4 | 5 |
| Tipperary | 163 | 8525 | 22 | 33 |
| Waterford | 90 | 2862 | 8 | 11 |
| Westmeath | 102 | 3743 | 12 | 22 |
| Wexford | 117 | 4172 | 35 | 19 |
| Wicklow | 73 | 2150 | 11 | 8 |
BSE — bovine spongiform encephalopathy
Overall, using ANOVA there was a significant difference in the risk of BSE among index herd enterprise types (P = 0.0001) with a cumulative herd risk of 10 per 1000 mixed herds, 7 per 1000 dairy herds, and 2 per 1000 suckler herds. There was a significant increase in risk of BSE with increasing herd size (data not shown). Counties differed significantly in the risk per 100 herds (P = 0.004) and in the SMRs (P < 0.0001) that adjusted for geographic differences in herd size and herd enterprise type. Although the number of BSE cases increased over the 5-year period, there was no significant effect of year on the risk of BSE, nor on the SMR, per county. Hence, we show maps relating to the number of index (or PE) herds over the full period only.
The population distribution indicates that the central north area, the northwest area, and Clare county have the highest density of herds, whereas the central north area, Wexford, and Cork had the highest number of index herds (Figure 1). Based on the SMRs that adjusted for enterprise type and herd size (Figure 2), it appeared that the counties surrounding Northern Ireland, especially Cavan, Monaghan, and Meath, counties on the east-coast and one county in the west (Clare) had an excess of cases.
Figure 1.
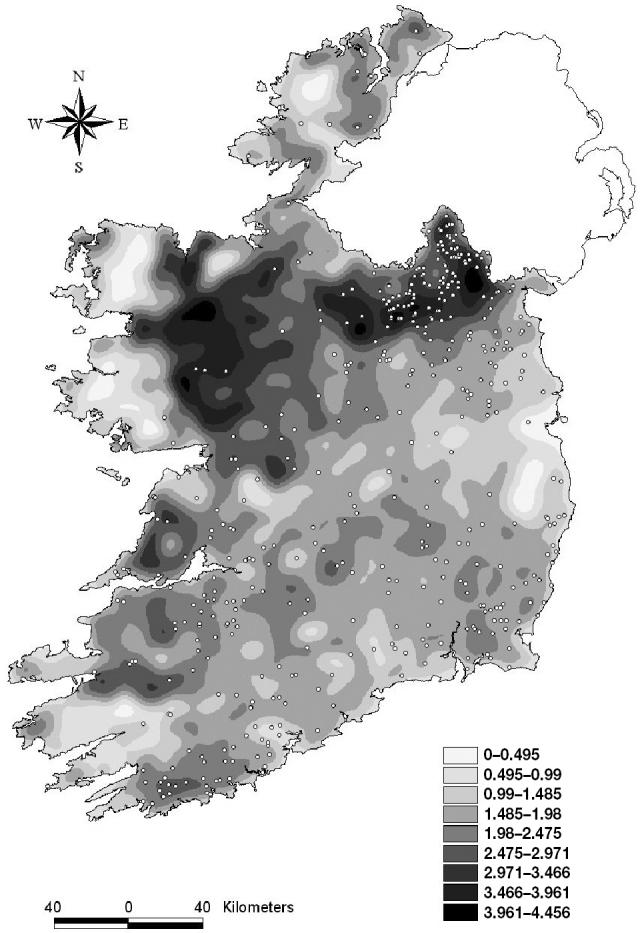
The location of bovine spongiform encephalopathy (BSE) index herds and the density per square kilometre of herds in Ireland, from 1996 to 2000.
Figure 2.
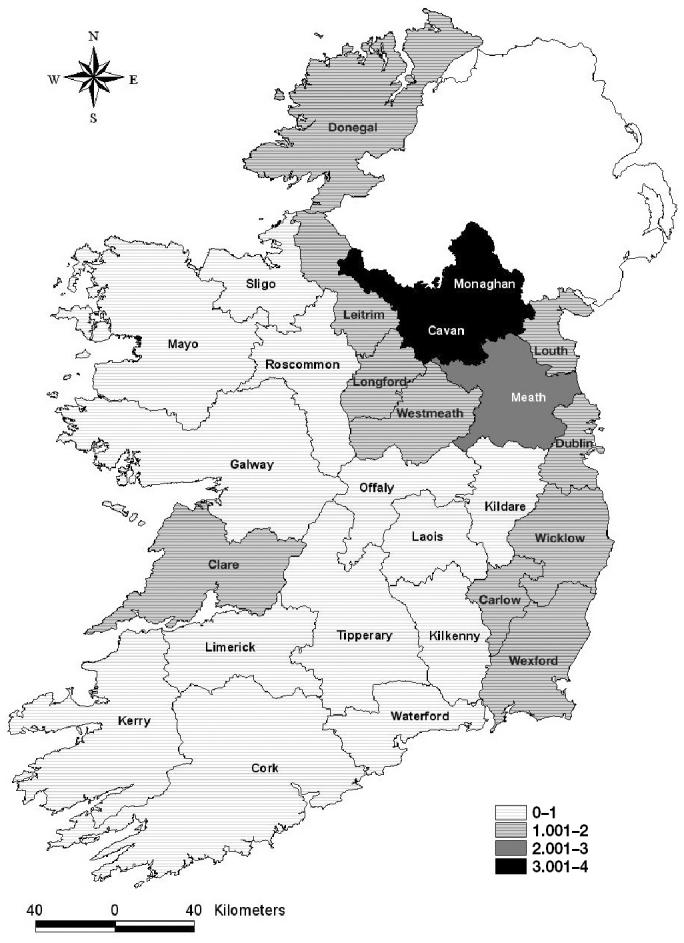
The standardized morbidity ratios per county for bovine spongiform encephalopathy (BSE) in Ireland, from 1996 to 2000.
Cluster analysis
The temporal scan component did not clearly elucidate times of increased case occurrence, except in a few instances that will be mentioned subsequently. Hence most results reported here are “spatial” only, and based on PE herd location (Table II). For PE herds, there was strong evidence of clustering, with the most likely clusters (P = 0.0001) being located in the northeast region centred most often on Monaghan county (Figure 3). These clusters were present and statistically significant, regardless of model type or whether correction for confounding effect (herd enterprise type and herd size) was used. The feed-supplier, identified as the centroid in the focussed analysis, differed between models, with Bernoulli based approaches tending to show more variability (5 different mills selected, but most often 1 of 2 very spatially close mills) in which feed-supplier was placed at the centroid of the most likely cluster. In contrast the Poisson based approach repeatedly identified the same feed-supplier as the centroid of the cluster, regardless of whether a temporal scan or correction for confounding effects was included.
Table II.
Selected statistically significant clusters of bovine spongiform encephalopathy (BSE) detected by SaTScan analysis among herds of putative exposure in Ireland, from 1996 to 2000
| PE herd category | Model type | Focussed or unfocussed | Region | Radius (kilometres) | P-value | Cluster rating |
|---|---|---|---|---|---|---|
| PE 2 | Bernoulli | Unfocussed | North east | 44.0 | < 0.001 | Most likely |
| PE 2 | Bernoulli | Focussed | North east | 43.1 | < 0.001 | Most likely |
| PE 3 | Bernoulli | Unfocussed | North east | 44.0 | < 0.001 | Most likely |
| PE 3 | Bernoulli | Focussed | North east | 43.1 | < 0.001 | Most likely |
| PE 2 | Poisson — sizea | Focussed | North east | 60.6 | < 0.001 | Most likely |
| PE 2 | Poisson — type and size | Focussed | North east | 60.6 | < 0.001 | Most likely |
| PE 2 | Poisson — no covariates | Focussed | North east | 60.6 | < 0.001 | Most likely |
| PE 1 | Bernoulli | Focussed | North east | 43.1 | < 0.001 | Most likely |
| PE 1 | Bernoulli | Unfocussed | North east | 60.6 | < 0.001 | Most likely |
| PE 2 | Poisson — type and size | Unfocussed | North east | 66.2 | < 0.001 | Most likely |
| PE 2 | Poisson — no covariates | Focussed | South east | 65.6 | < 0.001 | Secondary |
| PE 2 | Poisson — no covariates | Unfocussed | South east | 42.6 | < 0.001 | Secondary |
| PE 2 | Poisson — type and sizeb | Focussed | South east | 113.3 | < 0.001 | Secondary |
| PE 2 | Poisson — type and sizeb | Unfocussed | South east | 73.4 | < 0.001 | Secondary |
| PE 2 | Bernoulli | Focussed | South east | 42.6 | < 0.001 | Secondary |
| PE 2 | Bernoulli | Unfocussed | South east | 105.8 | < 0.001 | Secondary |
| PE 2 | Poisson — no covariates | Focussed | South | 28.2 | < 0.001 | Secondary |
| PE 2 | Poisson — no covariates | Unfocussed | South | 12.2 | 0.012 | Secondary |
| PE 2 | Bernoulli | Focussed | South | 98.9 | < 0.001 | Secondary |
| PE 2 | Bernoulli | Unfocussed | South | 100.6 | 0.012 | Secondary |
PE — putative exposure
Factors controlled in analysis
Temporal component included indicating a peak occurrence from 1992 to 1994
Figure 3.
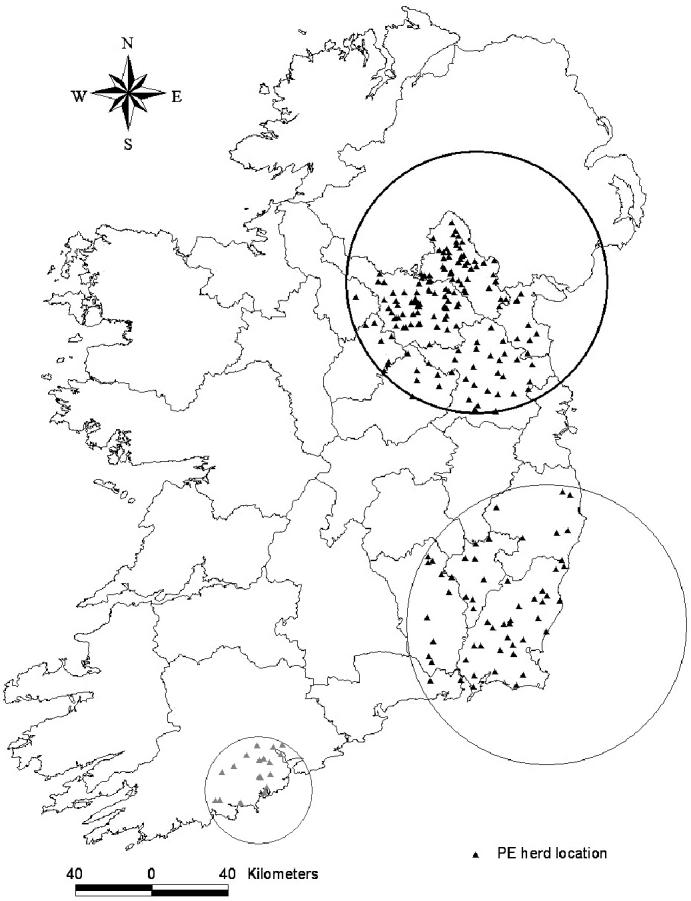
The locations of significant clusters of putative exposure bovine spongiform encephalopathy (BSE) herds, in Ireland, from 1996 to 2000, as detected by a spatial scan statistic.
Secondary clusters were detected in the south of the country. The location and size of secondary clusters varied, but many models detected a cluster centred in Wexford county and some detected clusters centred on Cork county (Table II). In general, the detection of secondary clusters was more frequent using the Poisson based approach. Correction for both herd enterprise type and herd size in a purely spatial Poisson analysis removed both the Cork and Wexford clusters. However, correction for confounding effects did not remove the Wexford cluster when a temporal element was included (P = 0.001), with a peak occurring from 1992 to 1994. This cluster was the largest (in area) PE cluster detected and was one of only a few instances where the focussed analyses detected a larger “cluster” area than an unfocussed approach (Table II). In relation to the Wexford cluster, Bernoulli-based approaches tended to select the same feed-supplier, whereas that selected by Poisson-based approaches differed depending on which co-variates were added. However, 5 of the 7 focussed analyses, which identified a statistically significant cluster in the Wexford area, selected the same feed mill as the centroid of the cluster.
As this study was focussed primarily on the location of exposure, the results of the index herd analysis are not reported in detail in this paper. However, no significant differences between index and PE herd analyses were detected with respect to the primary cluster, although PE herd analyses more frequently identified statistically significant secondary clusters than index herd analyses.
Discussion
This study focussed on cases of BSE identified by passive surveillance in the years 1996 to 2000. The number of cases prior to this date was too small to enable any meaningful spatial analysis, and the location of those identified after this date are not directly comparable because case ascertainment was affected by the introduction of active surveillance in 2001.
As our major objective was to identify temporal and spatial clustering, it is useful to note that there are many statistical tests available to the researcher. The majority of these tests are variations on a cross product approach, such as the Join Count Statistic, Moran’s I, and Getis’ G (19), whereas others depend on tests of adjacency (20,21). The spatial scan statistic (SaTScan) (18) was chosen because of its ability to search for both temporal-spatial and spatial clusters, to deal with confounding effects, to facilitate cluster identification independent of artificially imposed administrative boundaries (Bernoulli model approach), and to utilize a specific set of co-ordinates (feed-supplier locations) as grid-points for centroids of potential clusters.
Our initial ANOVAs indicated that both herd size and herd enterprise type were related to the risk of BSE and, therefore, they were possible confounding effects in the relationship between location and BSE case herds. Earlier UK findings revealed a similar linear trend in the relationship between risk and the herd size category (8), perhaps because of differences in feed utilisation among herd size categories; the more feed purchased by a herd owner, the greater their chance of acquiring a batch contaminated with infective material. The observation that the risk of disease in dairy and mixed herds in Ireland is substantially higher than in suckler herds is also consistent with earlier UK findings, which revealed that 46% of dairy herds had been affected by August 1993 as compared to only 10% of suckler herds (22). This difference is thought to reflect differences in exposure to the feed borne source, as replacement stock in dairy herds tend to be fed more concentrates than those in suckler herds.
Visualization is a useful first step into understanding spatial patterns. Both the plot of cases and the SMR map are useful in this regard, but they do not account for the impact of the number of case herds or the population size on the variance associated with the measure. Hence, some care is needed when interpreting the apparent pattern (Figure 2), because areas with small population sizes can have large variances. Other methods to improve the visualization of patterns, such as Bayesian smoothing, are available but were not used in this study. We have also chosen to show index herd maps, at this stage, rather than the location of the smaller subset of known location PE herds, as this reflected our progression of thinking as we analyzed the data. For the more formal assessment of clustering, we utilized the spatial scan statistic SaTScan, which includes an assessment of the impact of sampling variability using Monte Carlo techniques and locations based on PE herd distribution. This approach provided clear evidence to support the hypothesis that BSE herds cluster geographically. Furthermore, we can infer that this clustering may be related to factors affecting the herds where the animals were likely exposed, rather than in the herds where they were located at the time of diagnosis, based on research in the UK (8). Evidence from this study to support this hypothesis is provided by the detection of similar primary clusters in both index and PE herds; using PE herd location resulted in a more powerful analysis with more secondary clusters being identified than when using index herd location (data not shown). Presumably this increased power is due to the almost 25% of the index herds that had purchased the affected animal(s) at an age of 2 y or more, and the location of these herds would not reflect the likely location of exposure.
A priori, it was hypothesized that BSE case herds are linked by a process of spatial diffusion from a central node (most likely feed-suppliers). Based on the results of epidemiological investigations on both case and control farms in Ireland, the vast majority of herd owners in Ireland rely on proprietary concentrates to feed both their young and adult stock and tend to source their feeds locally (Sheridan, personal communication 2003). Our results provide evidence of a spatial association between large feed-suppliers and clusters of affected herds. This hypothesis gained support from the finding that focussed analyses using ING co-ordinate locations of major feed-suppliers often produced smaller, more precisely located clusters with smaller P-values than unfocussed analyses. However, since we have no prior experience with focussed analyses, we view this finding as supportive but not conclusive. The finding that the radius of most statistically significant clusters varied between 40 and 70 kilometres (Table II) also fits with the concept of local distribution of proprietary concentrates. Thus, the spatial association between the location of feed-suppliers and BSE in Ireland is consistent with the finding of modelling processes carried out in the UK, which demonstrated that between-producer variability in the infectiousness of feed, was capable of explaining BSE case clustering in the UK (12). Between-producer variability of feed infectivity fits the pattern of cases in Ireland particularly well because the majority of cases of BSE in Ireland developed after a ban on the direct feeding of MBM to ruminant animals in Ireland was introduced in 1990. The likelihood that cattle rations manufactured by a particular feed supplier might pose a BSE risk depends on a number of factors, including the physical structure of the mill, the amount and source of MBM entering the plant for use in non-ruminant rations, and the type of feed manufactured. Thus, cross contamination of young stock feeds with contaminated MBM is likely to be a heterogeneous process and is unlikely to be occurring at the same rate throughout the country.
Although no temporal element was identified in relation to the most likely cluster among PE herds, there was evidence to suggest that clustering of exposure in the Wexford area was limited to between the years 1992 and 1994. This may reflect an increase in feed risk experienced in this area during this time.
Field studies, such as this one, have to contend with misclassification of exposure and/or outcome, as well as selection and confounding bias. We attempted to eliminate misclassification of control herds and selection bias by selecting as controls, those herds where BSE awareness had been demonstrated by the notification of a suspect animal that subsequently had been confirmed negative. Efforts to eliminate misclassification with regard to location were made by cross checking locations recorded in the BSE database with those recorded for the same herd numbers in the LPIS. However, it was a challenge to obtain reliable information relating to events that happened many years prior to the investigation, and thus we could not identify the location of all PE herds with as much certainty as the index herds. Regarding confounding bias, it is possible that variables, other than feed source, are responsible for the relationship between location and disease. Our analyses indicated that herd enterprise type and size were related to the occurrence of BSE, and thus we controlled for this effect in our spatial scan Poisson analyses. In the presence of this control using the Poisson models, the “suspected” clusters over the Cork area were deemed not to be significant. However, the Bernoulli models still identified significant clusters centred in Cork. As described above, the cluster over Wexford remained significant when the temporal-spatial Poisson model controlled for these factors, as was also the case in some of the Bernoulli based approaches. Given the apparent differences in reporting of non-confirmed suspect cases, by county, we anticipated some divergent results between the Bernoulli and Poisson models. However, the differences were only noted for suspected secondary clusters as correction for confounding effects did not affect the location or statistical significance of the most likely cluster, regardless of model. Since there could be other determinants of BSE that are geographically clustered, we consider our study as exploratory, nonetheless our findings are consistent with the hypothesis that large scale suppliers of feed and variability in feed infectivity may be responsible for the clustering of BSE cases. We believe that formal temporal-spatial analyses were helpful in meeting our objectives and should be used to supplement traditional disease investigation methods whenever they are applicable.
Acknowledgments
The authors thank the staff of Department of Food and Agriculture, in particular Mr. D. Lynch who collected and maintained the epidemiological information used in this study. The authors offer sincere thanks to the staff at the Centre for Veterinary Epidemiology and Risk Analysis, University College Dublin, who provided technical assistance. The help of Ms. Kendra Long, who proofread the paper and offered constructive advice, is also greatly acknowledged.
References
- 1.Wells GAH, Scott AC, Johnson CT, et al. A novel progressive spongiform encephalopathy in cattle. Vet Rec. 1987;121:419–420. doi: 10.1136/vr.121.18.419. [DOI] [PubMed] [Google Scholar]
- 2.Wilesmith JW, Wells GAH, Cranwell MP, Ryan JBM. Bovine Spongiform Encephalopathy: Epidemiological Studies. Vet Rec. 1988;123:638–644. [PubMed] [Google Scholar]
- 3.Wilesmith JW, Ryan JBM, Atkinson MJ. Bovine spongiform encephalopathy; epidemiological studies on the origin. Vet Rec. 1991;128:199–203. doi: 10.1136/vr.128.9.199. [DOI] [PubMed] [Google Scholar]
- 4.Wilesmith JW, Ryan JBM, Hueston WD. Bovine spongiform encephalopathy: Case-control studies of calf feeding practices and meat and bonemeal inclusion in proprietary concentrates. Res Vet Med. 1992;52:325–331. doi: 10.1016/0034-5288(92)90032-w. [DOI] [PubMed] [Google Scholar]
- 5.Taylor DM, Woodgate SL, Atkinson MJ. Inactivation of the bovine spongiform encephalopathy agent by rendering processes. Vet Rec. 1995;137:605–610. [PubMed] [Google Scholar]
- 6.Schreuder BEC, Geertsma RE, Van Keulen LJM, et al. Studies on the efficacy of hyperbaric rendering procedures in inactivating bovine spongiform encephalopathy (BSE) and scrapie agents. Vet Rec. 1998;142:474–480. doi: 10.1136/vr.142.18.474. [DOI] [PubMed] [Google Scholar]
- 7.Wilesmith JW, Wells GA, Cranwell MP, Ryan JB. Bovine spongiform encephalopathy: Epidemiological studies. Vet Rec. 1988;123:638–44. [PubMed] [Google Scholar]
- 8.Donnelly CA, Ferguson MA, Ghani AC, Woolhouse MEJ, Watt CJ, Anderson RM. The epidemiology of BSE in cattle herds in Great Britain I. Epidemiological processes, demography of cattle and approaches to control by culling. Phil Trans Royal Soc London-Series B: Biological Sciences. 1997;352:781–801. doi: 10.1098/rstb.1997.0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilesmith JW, Ryan JBM, Hueston, WD, Hoinville LJ. Bovine spongiform encephalopathy; epidemiological features 1985 to 1990. Vet Rec. 1992;130:90–94. doi: 10.1136/vr.130.5.90. [DOI] [PubMed] [Google Scholar]
- 10.Stevenson MA, Wilesmith JW, Ryan JBM, et al. Descriptive spatial analysis of the epidemic of bovine spongiform encephalopathy in Great Britain to June 1997. Vet Rec. 2000;147:379–384. doi: 10.1136/vr.147.14.379. [DOI] [PubMed] [Google Scholar]
- 11.Stevenson MA, Wilesmith JW, Ryan JBM, et al. Temporal aspects of the epidemic of bovine spongiform encephalopathy in Great Britain: Individual animal-associated risk factors for the disease. Vet Rec. 2000;147:349–354. doi: 10.1136/vr.147.13.349. [DOI] [PubMed] [Google Scholar]
- 12.Hagenaars TJ, Ferguson NM, Donnelly CA, Ghani AC, Anderson RM. Feed-borne transmission and case clustering of BSE. Proc Royal Soc London-Series B: Biological Sciences. 2000;267:205–215. doi: 10.1098/rspb.2000.0989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilesmith JW, Ryan JBM. Bovine spongiform encephalopathy: Recent observations on age-specific incidences. Vet Rec. 1992;130:491–492. doi: 10.1136/vr.130.22.491. [DOI] [PubMed] [Google Scholar]
- 14.Wilesmith JW, Ryan JBM. Bovine spongiform encephalopathy: Observations on the incidence during 1992. Vet Rec. 1993;132:300–301. doi: 10.1136/vr.132.12.300. [DOI] [PubMed] [Google Scholar]
- 15.Hoinville LJ, Wilesmith JW, Richards MS. An investigation of risk factors for cases of bovine spongiform encephalopathy born after the introduction of the ‘feed ban’. Vet Rec. 1995;136:312–331. doi: 10.1136/vr.136.13.312. [DOI] [PubMed] [Google Scholar]
- 16.Hornlimann B, Audige L, Somaini B, Guidon G, Griot C. Case study of BSE in animals born after the feed ban (BAB) in Switzerland. Epidemiol Santé Anim. 1999;11:31–32. [Google Scholar]
- 17.Fleiss JL. Statistical Methods for Rates and Proportions. New York: J Wiley & Sons, 1973.
- 18.Kulldorff M, Gherman G, Williams G, Defrancesco D. SaTScan Version 2.1: Software for the spatial and space-time scan statistics. Bethesda, Maryland: National Cancer Institute, 1998.
- 19.Getis A. Spatial interaction and spatial autocorrelation: a cross product approach. Enviro Plan A. 1991;23:1269–1277. [Google Scholar]
- 20.Ohno Y, Aoki K. Cancer deaths by city and county in Japan (1969–1971): A test of significance for geographic clusters of disease. Soc Sci Med. 1981;15:251–255. doi: 10.1016/0160-8002(81)90035-6. [DOI] [PubMed] [Google Scholar]
- 21.Grimson RC, Wang KC, Johnson PWC. Searching for hierarchical clusters of disease: Spatial patterns of sudden infant death syndrome. Soc Sci Med. 1981;15D:287–292. [Google Scholar]
- 22.Wilesmith JW. An epidemiologist’s view of bovine spongiform encephalopathy. Phil Trans Royal Soc London-Series B: Biological Sciences. 1994;343:357–361. doi: 10.1098/rstb.1994.0029. [DOI] [PubMed] [Google Scholar]


